Abstract
S-nitrosoglutathione reductase 1 (GSNOR1) is the key enzyme that regulates cellular homeostasis of S-nitrosylation. Although extensively studied in Arabidopsis, the roles of GSNOR1 in tetraploid Nicotiana species have not been investigated previously. To study the function of NtGSNOR1, we knocked out two NtGSNOR1 genes simultaneously in Nicotiana tabacum using clustered regularly interspaced short palindromic repeats (CRISPR)/caspase 9 (Cas9) technology. To our surprise, spontaneous cell death occurred on the leaves of the CRISPR/Cas9 lines but not on those of the wild-type (WT) plants, suggesting that NtGSNOR1 negatively regulates cell death. The natural cell death on the CRISPR/Cas9 lines could be a result from interactions between overaccumulated nitric oxide (NO) and hydrogen peroxide (H2O2). This spontaneous cell death phenotype was not affected by knocking out two Enhanced disease susceptibility 1 genes (NtEDS11a/1b) and thus was independent of the salicylic acid (SA) pathway. Unexpectedly, we found that the NtGSNOR1a/1b knockout plants displayed a significantly (p < 0.001) enhanced resistance to paraquat-induced cell death compared to WT plants, suggesting that NtGSNOR1 functions as a positive regulator of the paraquat-induced cell death. The increased resistance to the paraquat-induced cell death of the NtGSNOR1a/1b knockout plants was correlated with the reduced level of H2O2 accumulation. Interestingly, whereas the N gene-mediated resistance to Tobacco mosaic virus (TMV) was significantly enhanced (p < 0.001), the resistance to Pseudomonas syringae pv. tomato DC3000 was significantly reduced (p < 0.01) in the NtGSNOR1a/1b knockout lines. In summary, our results indicate that NtGSNOR1 functions as both positive and negative regulator of cell death under different conditions and displays distinct effects on resistance against viral and bacterial pathogens.
Keywords: cell death, disease resistance, nitric oxide, hypersensitive responses, reactive oxygen species, S-nitrosoglutathione reductase
Introduction
Nitric oxide (NO) is a reactive free radical gas molecule with a plethora of functions in both animals and plants (Wendehenne et al., 2014). In plants, NO is involved in diverse biological processes such as stomatal closure (Desikan et al., 2002; Neill et al., 2008), cell death and disease resistance (Durner et al., 1998; Klessig et al., 2000; Wendehenne et al., 2004; Zeidler et al., 2004; Lin et al., 2012; Wang et al., 2013; Wang and Chu, 2020), abiotic stress (Lee et al., 2008; Xuan et al., 2010), flowering (He et al., 2004), growth/development (Yun et al., 2016), and many other processes (Lamattina et al., 2003; Wendehenne et al., 2014). Among all these processes, the most significant function of NO is to potentiate the induction of hypersensitive cell death by reactive oxygen species (ROS) (Durner et al., 1998; Delledonne et al., 2001).
S-nitrosylation, attachment of NO moiety to a target protein, is a newly emerged mechanism by which NO regulates the function of various target proteins and thus various biological processes (Hess and Stamler, 2012; Wendehenne et al., 2014). This reversible posttranslational modification is analogous to protein phosphorylation (Stamler et al., 1992; Hess et al., 2005). Hundreds of proteins have been identified as targets of S-nitrosylation whose functions are regulated by this posttranslational modification (Lindermayr et al., 2005; Forrester et al., 2009; Hess and Stamler, 2012; Yang et al., 2015). In plants, the S-nitrosylated cysteine residues of some target proteins have been identified, and the functions of this posttranslational modifications are revealed (Lindermayr et al., 2006, 2010; Romero-Puertas et al., 2007; Tada et al., 2008; Yun et al., 2011; Skelly et al., 2019; Gupta et al., 2020).
S-nitrosoglutathione (GSNO), S-nitrosylated form of glutathione (GSH), functions as a major mediator of protein S-nitrosylation by a process known as transnitrosylation (Hess et al., 2005). S-nitrosoglutathione reductase (GSNOR) is the key enzyme controlling GSNO levels by reducing GSNO to oxidized GSH and NH3 and thus indirectly controlling the levels of protein S-nitrosylation (Liu et al., 2001, 2004; Feechan et al., 2005). In mice, loss of GSNOR1 function results in substantial increases in whole-cell S-nitrosylation, tissue damage, and mortality following endotoxic or bacterial challenge (Liu et al., 2004). In Arabidopsis, the hypersensitive response (HR) cell death triggered by avirulent Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) carrying the AvrB or AvrRPS4 is accelerated in loss of function mutant, gsnor1-3, but is delayed in overexpressing mutant gsnor1-1 (Yun et al., 2011), suggesting that the function of GSNOR1 is conserved between mice and Arabidopsis in negatively regulating HR cell death in response to biotic stress (Liu et al., 2004; Yun et al., 2011). The cell death in response to incompatible pathogen infections in gsnor1-3 mutant is due to high SNO concentrations and is SA- and hydrogen peroxide (H2O2)-independent (Yun et al., 2011). Interestingly, it has also been revealed that GSNOR1 is a key positive regulator of cell death induced by herbicides (Chen et al., 2009), suggesting that depending on cellular conditions, GSNOR1 could regulate cell death either positively or negatively. NO has been demonstrated to have both proapoptotic and antiapoptotic roles, depending on a variety of factors, including the type of cells involved, redox state of the cell, and the flux and dose of NO (Wang et al., 2013; Wendehenne et al., 2014; Huang et al., 2019). In general, S-nitrosylation resulting from basal low-level NO production in cells has antiapoptotic effects, whereas S-nitrosylation resulting from higher-level stimulated NO production has either proapoptotic effects or serves as a negative feedback mechanism to downregulate apoptotic responses (Mannick, 2007).
NO exerts its effects on cell death mostly through modulating H2O2 production or detoxification. S-nitrosylation of an Arabidopsis cytosolic ascorbate peroxidase (cAPX), a key enzyme controlling H2O2 levels in plants, inhibits its enzyme activity, leading to Programmed cell death (PCD) (de Pinto et al., 2013). Loss of H2O2 detoxifying enzyme, catalase in rice, results in both NO and H2O2 overaccumulation and leaf cell death (Lin et al., 2012). Overexpression of rice GSNOR1 alleviates the leaf cell death in noe1 (nitric oxide excess 1) mutant (Lin et al., 2012), suggesting a conserved role of S-nitrosylation in cell death in plants. S-nitrosylation not only can induce cell death but also can function as a negative feedback loop to limit or prevent excessive cell death. Instead of inhibiting the function of a cAPX1, S-nitrosylation of the Arabidopsis cytosolic APX1 at Cys32 enhances its enzymatic activity of scavenging H2O2, resulting in an increased resistance to oxidative stress (Yang et al., 2015). NADPH oxidase is the major source of H2O2 production under biotic stress. Arabidopsis AtRBOHD subunit of NADPH oxidase complex is S-nitrosylated at Cys890, and this modification abolishes its ability to synthesize ROS and thus avoids excessive cell death (Yun et al., 2011).
In addition to cell death, GSNOR1 plays a critical role in disease resistance. Loss of GSNOR1 function in Arabidopsis results in compromised basal resistance, R gene-mediated resistance and non-host resistance (NHR) (Feechan et al., 2005). Silencing SlGSNOR1 in tomato also compromises basal resistance against Pst DC3000 (Hussain et al., 2019). On the contrary, Arabidopsis and tomato lines that overexpress GSNOR1 lead to enhanced resistance to virulent pathogens (Feechan et al., 2005; Hussain et al., 2019). However, silencing GSNOR1 by antisense approach in Arabidopsis enhances Systemic acquired resistance (SAR) and basal resistance (Rustérucci et al., 2007). Recently, it has been shown that the accelerated cell death observed in gsnor1-3 in response to pathogen infection confers RPP4-mediated resistance to Hyaloperonospora arabidopsidis isolate Emwa1 in an SA-independent manner (Yun et al., 2011). These seemingly contradictory results suggest that the roles of GSNOR1 in plant disease resistance are more complicated than we thought. Besides cell death and disease resistance, GSNOR1 is also required for growth/development (Feechan et al., 2005; Lee et al., 2008; Chen et al., 2009; Albertos et al., 2015; Shi et al., 2015; Yun et al., 2016; Kawabe et al., 2018; Gong et al., 2019; Hussain et al., 2019), thermotolerance (Lee et al., 2008), abiotic stress responses (Zhang and Liao, 2019), hypoxia responses (Zhan et al., 2018), cytokinin signaling (Feng et al., 2013), auxin signaling (Terrile et al., 2012; Shi et al., 2015; Ni et al., 2017; Iglesias et al., 2018) and transport (Shi et al., 2015), abscisic acid (ABA)-mediated stomatal closure (Albertos et al., 2015; Wang et al., 2015), nitrogen assimilation (Frungillo et al., 2014), and nodule development (Matamoros et al., 2020).
To investigate the roles of GSNOR1 in tetraploid N. tabacum, the NtGSNOR1a and NtGSNOR1b knockout plants were generated via clustered regularly interspaced short palindromic repeats (CRISPR)/caspase 9 (Cas9) technology. Unexpectedly, spontaneous cell death was observed in the NtGSNOR1a/1b knockout plants under natural growth conditions, which has not been reported for the Arabidopsis gsnor1 mutant previously. This spontaneous cell death phenotype was not rescued by knocking out NtEDS1a/1b and thus was SA-independent. To our surprise, the NtGSNOR1a/1b knockout plants displayed increased resistance to paraquat (1,1′-dimethyl-4,4′-bipyridinium dichloride)-induced cell death compared to wild-type (WT) plants, indicating that GSNOR1 in Nicotiana species functions as both positive and negative regulator of cell death under different cellular conditions. Interestingly, whereas the resistance to Tobacco mosaic virus (TMV) was enhanced in the NtGSNOR1a/1b knockout plants, the resistance to Pst DC3000 was significantly reduced (p < 0.001), indicating that NtGSNOR1a/1b have opposite effects on viral and bacterial pathogen infections in N. tabacum.
Results
Generation of the CRISPR/Cas9 Lines That Simultaneously Knock Out Two Alleles of NtGSNOR1a and Two Alleles of NtGSNOR1b in Tetraploid N. tabacum
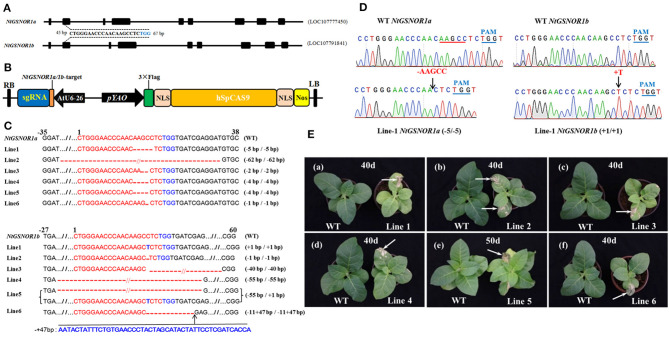
It has been known that NO promotes an increase in ROS-induced cell death (Delledonne et al., 1998; Durner et al., 1998). However, the mechanism behind this NO-promoted cell death is not understood. To investigate the roles of GSNOR1 in cell death and disease resistance in tetraploid N. tabacum, we designed to simultaneously knockout four alleles of two highly related orthologs of Arabidopsis GSNOR1 gene, namely, NtGSNOR1a (LOC107777450) and NtGSNOR1b (LOC107791841) and created NtGSNOR1a/NtGSNOR1b double knockout lines in N. tabacum cv. Samsun, which carries TMV-resistant gene, N, using CRISPR/Cas9 technology. The NtGSNOR1a and NtGSNOR1b share overall 98% identity at the nucleotide level. A consensus 20-bp sequence shared between NtGSNOR1a and NtGSNOR1b within the second exon of NtGSNOR1a/1b was used as a guide to target both genes (Figures 1A,B). The blast searching using the target sequence against the tobacco genome revealed that the target sequence only specifically matched to the NtGSNOR1a/1b target sites but not the other sites within the tobacco genome, with the closest match displayed 4-nt mismatches, which potentially could rule out the possibility of off-target effect. Multiple T0 transgenic lines were generated through Agrobacterium-mediated transformation, and six independent CRISPR/Cas9 lines, in which two alleles of NtGSNOR1a and two alleles of NtGSNOR1b, respectively, were simultaneously knocked out, were identified in the T0, T1, or T2 populations. The types of mutations for each of the four alleles in these six CRISPR/Cas9 lines were summarized in Figure 1C. The sequencing chromatograms showing the insertions and deletions (InDels) in the sequences of NtGSNOR1a and NtGSNOR1b of the CRISPR/Cas9 lines 1 were presented in Figure 1D. The sequencing chromatograms of the CRISPR/Cas9 line 2 to line 6 were presented in Supplementary Figures 1–5. Translational analysis indicated that the mutations (InDels) within all four alleles of NtGSNOR1a/1b in these six CRISPR/Cas9 lines could result in premature stop codons and would generate truncated proteins. Thus, these CRISPR/Cas9 lines can be considered independent knockout lines of both NtGSNOR1a and NtGSNOR1b (NtGSNOR1a/1b).
Figure 1.
Knocking out NtGSNOR1a and NtGSNOR1b simultaneously results in spontaneous cell death on the leaves of N. tabacum. (A) Gene model of NtGSNOR1a (LOC107777450) and NtGSNOR1b (LOC107791841). A consensus sequence between NtGSNOR1a and NtGSNOR1b at the second exon (45–65 bp downstream of start codon ATG of the open reading frame) was used as a guide to target both genes simultaneously. The TGG in blue represents PAM sequence. (B) The diagram of the CRISPR/Cas9 construct used for generating transgenic plants. (C) The sequence alignment of the six identified CRISPR/Cas9 knockout lines. The short horizontal lines in red represent the deletions; the TGG in blue represents PAM sequence; and the red letters represent the insertions; // represent some bases were omitted for the purpose of saving space. The mutation types of these six CRISPR/Cas9 lines were shown by the labeling on the right. (D) Comparison of the sequencing chromatograms between the wild type (WT) and the CRISPR/Cas9 knockout line 1 at the edited region. Arrows pointed to the sites of deletions and insertions. + represents insertion, and - represents deletion. The PAMs are marked in blue letters and lines. (E) The spontaneous cell death phenotype is displayed on the leaves of all these six CRISPR/Cas9 lines. Arrows pointed to the regions with cell death.
The Spontaneous Cell Death Occurs on the Leaves of the NtGSNOR1a/1b Knockout Plants
These six CRISPR/Cas9 knockout lines shared a similar morphological phenotype with a smaller overall stature compared to the WT plants through the entire life cycle (Figures 1E, 2). To our surprise, a severe spontaneous cell death phenotype was observed on the leaves of these NtGSNOR1a/1b knockout lines (Figure 1E), which was not observed in the WT plants throughout the entire growth season. The spontaneous cell death was not synchronous even for the same batch of the knockout plants. The cell death showed up earlier on some plants than on the others. The spontaneous cell death was mainly on the fully expanded true leaves independent of developmental stage. It could occur as early as the two-leaf stage (Figure 2A). It usually appeared on the lower leaves and spread progressively to the upper systemic leaves (Figures 1B–E). The cell death was similar to HR cell death, which initially displayed as water-soaked spots and eventually dried out. These results indicate that NtGSNOR1 is a negative regulator of cell death.
Figure 2.
The spontaneous cell death phenotype of NtGSNOR1a/1b knockout plants at different developmental stages. (A) The spontaneous cell death phenotype could occur as early as 25 days post germination. (B–F) The cell death was aggravated with the development and progressed to upper systemic leaves. (D–F) Almost all the leaves of the knockout lines eventually died at 90 days post germination. (E,F) The enlarged image in the rectangle area in (E) was shown in (F). Arrows pointed to the dead cells.
Both NO and H2O2 Were Overaccumulated in the NtGSNOR1a/1b Knockout Plants
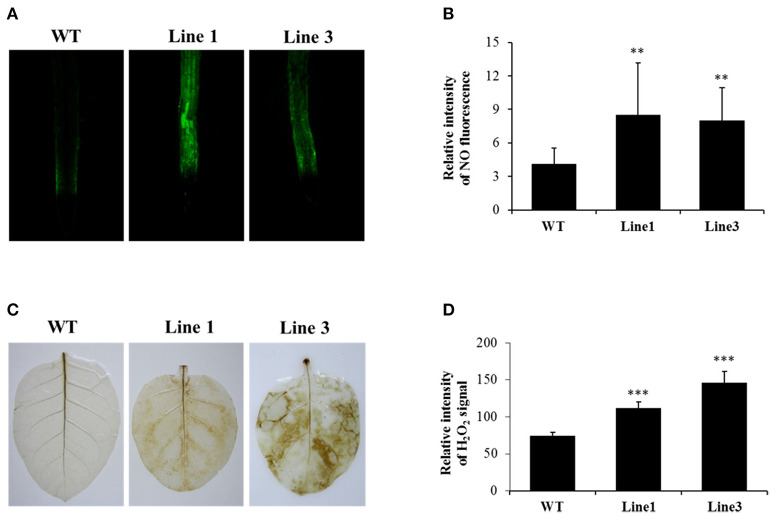
To investigate the effect of NtGSNOR1a/1b knockout on NO and H2O2 accumulation in tobacco, we compared the NO and H2O2 levels between WT and the CRISPR/Cas9 lines using 4,5-diaminoflorescein diacetate and 3,3′-diaminobenzidine (DAB) staining, respectively. Consistent with the results observed for Arabidopsis hot5-2/gsnor1-3/par2-1 mutants, NO level was significantly higher (p < 0.001) in the roots of the NtGSNOR1a/1b knockout plants of two independent lines compared with that of the WT plants (Figures 3A,B). Contrary to the results observed for gsnor1-3 mutant in Arabidopsis, H2O2 level on the leaves of the NtGSNOR1a/1b knockout lines showing the sign of cell death was also higher than those of WT plants (Figures 3C,D), although little or no H2O2 was detectable on the leaves of the NtGSNOR1a/1b knockout lines without showing any cell death. Together, these results indicate that NO and H2O2 were overaccumulated in the NtGSNOR1a/1b knockout plants and the spontaneous cell death that occurred on the NtGSNOR1a/1b knockout plants could result from synergistic interactions between the overaccumulated NO and H2O2.
Figure 3.
Knocking out NtGSNOR1a/1b leads to enhanced accumulation of both NO and H2O2. (A) NO level in the roots of 15-day-old plants was visualized by staining using 4,5-diaminoflorescein diacetate according to He et al. (2004). (B) The fluorescence intensity shown in (A) was quantified by ImageJ. (C) H2O2 was detected by staining the leaves of 40-day-old plants with 3,3'-diaminobenzidine (DAB) as described in section Materials and Methods. Oxidized DAB formed a reddish-brown deposit (examples of these deposits are indicated by the white arrows). (D) The intensity of the reddish-brown deposit shown in (C) was quantified by ImageJ. ** and *** indicate significant differences at 0.01 and 0.001 levels, respectively, by the Student's t-test.
The Spontaneous Cell Death on the Leaves of the NtGSNOR1a/1b Knockout Plants Is Not Dependent on the SA Pathway
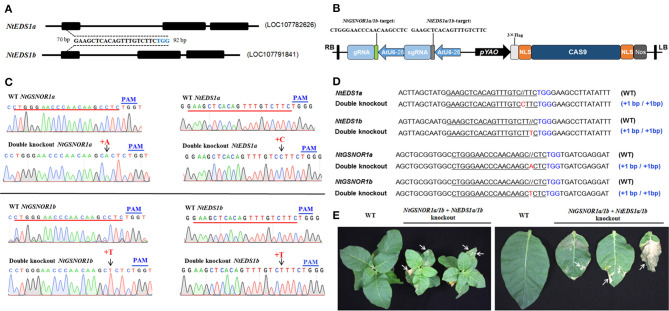
EDS1 is required for SA accumulation (Rustérucci et al., 2001), and the constitutive cell death of many autoimmune mutants is dependent on EDS1 (Zhou and Zhang, 2020). To examine whether the spontaneous cell death on the NtGSNOR1a/1b knockout plants is SA-dependent, we knocked out four alleles of NtEDS1a/1b and four alleles of NtGSNOR1a/1b simultaneously in N. Samsun (NN) using the CRISPR/Cas9 construct carrying two guides (Figure 4A). The same 20-bp guide was used for targeting NtGSNOR1a/1b (Figure 1A), while a 20-bp identical sequence shared between NtEDS1a and NtEDS1b was used for targeting NtEDS1a/1b (Figure 4B). In the T1 or T2 populations, a CRISPR/Cas9 line that simultaneously knocked out four alleles of NtGSNOR1a/1b as well as four alleles of NtEDS1a/1b was identified. A single-base homozygous insertion was presented in both NtGSNOR1a/1b and NtEDS1a/1b alleles of this double knockout line (Figures 4C,D). A single “A” homozygous insertion was presented 4 bp upstream of PAM (TGG) for the two alleles of NtGSNOR1a. A single “T” homozygous insertion was presented 4 bp upstream of PAM for the two alleles of NtGSNOR1b. A single “C” homozygous insertion was presented 4 bp upstream of PAM for the two alleles of NtEDS1a. A single “T” homozygous insertion was presented 4 bp upstream of PAM for the two alleles of NtEDS1b (Figures 4C,D). Again, the single-base insertion in all these alleles of both the NtGSNOR1a/1b and the NtEDS1a/1b could lead to premature stop codons and would generate truncated proteins. Thus, both NtGSNOR1a/1b (four alleles) and NtEDS1a/1b (four alleles) were simultaneously knocked out in this CRISPR/Cas9 line. These results reveal the power of CRISPR/Cas9 technology in resolving gene redundancy in polyploidy plant species.
Figure 4.
Knocking out NtEDS1a/1b cannot reverse the spontaneous cell death phenotype observed in NtGSNOR1a/1b knockout lines. (A) Gene model of NtEDS1a (LOC107782626) and NtEDS1b (LOC107791841). A consensus sequence between NtEDS1a and NtEDS1b at the first exon (70–92 bp downstream of start codon ATG of the open reading frame) was used as a guide to target both genes simultaneously. The TGG in blue represents PAM sequence. (B) The diagram of the CRISPR/Cas9 construct used for generating transgenic plants that knocks out both NtEDS1a/1b and NtGSNOR1a/1b. (C) Comparison of the sequencing chromatograms near the edited regions between the wild type (WT) and the CRISPR/Cas9 line, in which both NtEDS1a/1b and NtGSNOR1a/1b are knocked out simultaneously. Arrows pointed to the sites of insertions. + represents insertion. The PAM sequences are marked in blue. (D) The sequence alignments of the identified CRISPR/Cas9 double knockout line at both the NtEDS1a/1b and NtGSNOR1a/1b loci. The short horizontal lines in red represent the deletions; the TGG in blue represents PAM sequence; and the red letters represent the insertions; // represents extra space added for the purpose of the perfect sequence alignment. The mutation types of the CRISPR/Cas9 line were presented by the labeling on the right. (E) The spontaneous cell death phenotype is displayed on the leaves of the CRISPR/Cas9 double knockout line, in which both NtEDS1a/1b and NtGSNOR1a/1b are knocked out simultaneously. Arrows pointed to the regions with cell death.
To further confirm that the NtEDS1a/1b are indeed knocked out in the NtGSNOR1a/1b/NtEDS1a/1b double knockout lines, we transiently overexpressed GmMEKK1 via agro-infiltration on the leaves of the double knockout line, which can trigger SA-dependent cell death (Xu et al., 2018). As expected, the HR cell death induced by overexpressing GmMEKK1 was observed on the leaves of the NtGSNOR1a/1b knockout plants but not on the leaves of the NtGSNOR1a/1b/NtEDS1a/1b double knockout plants (Supplementary Figure 6), indicating that the NtEDS1a/1b is indeed knocked out in the double knockout lines. The fact that the spontaneous cell death was still observed on the NtGSNOR1a/1b/NtEDS1a/1b knockout plants (Figure 4E) indicated that the spontaneous cell death of NtGSNOR1a/1b knockout plants is independent of the SA pathway.
The NtGSNOR1a/1b Knockout Plants Are More Resistant to Paraquat-Induced Cell Death
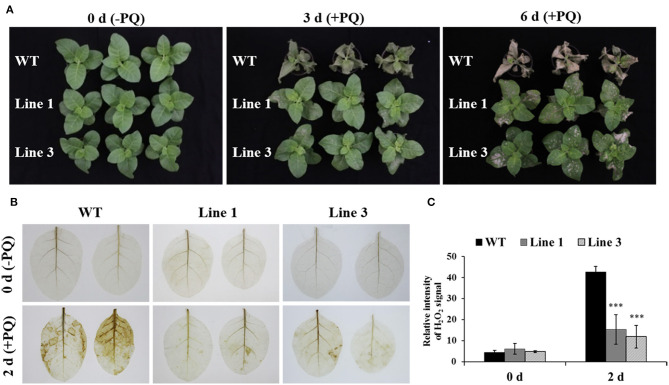
Paraquat, a non-selective herbicide, can induce cell death in plants (Dodge, 1971; Suntres, 2002). To examine whether knocking out NtGSNOR1a/1b in N. tabacum has a similar paraquat-resistant phenotype observed in Arabidopsis (Chen et al., 2009), 0.25% paraquat was sprayed onto the leaves of both the WT and the NtGSNOR1a/1b knockout plants before showing spontaneous cell death. While the WT plants almost died at 3 days post spraying (dps), two different NtGSNOR1a/1b knockout lines were still alive and relatively healthy (Figure 5A), indicating that the function of GNSOR1 on paraquat-induced cell death is conserved across plant species.
Figure 5.
The NtGSNOR1a/1b knockout plants exhibited strong resistance to paraquat (PQ)-induced cell death through reducing PQ-induced H2O2 accumulation. (A) Here, 0.25% PQ was sprayed on the leaves of 40-day-old wild type (WT) and the CRISPR/Cas9 knockout lines 1 and 3. The images were taken at 0, 3, and 6 days post spraying. (B) The H2O2 accumulation on the leaves of WT and NtGSNOR1a/1b knockout plants at 0 and 2 days post-paraquat spraying visualized by DAB staining. (C) The intensity of the reddish-brown deposit shown in (B) was quantified by ImageJ. ** and *** indicate significant differences of p-value at 0.01 and 0.001, respectively, by the Student's t-test.
To investigate the resistant mechanism of the NtGSNOR1a/1b knockout plants to the paraquat-induced cell death, we visualized the H2O2 accumulation on the leaves of both WT and the NtGSNOR1a/1b knockout plants after paraquat spraying. Consistent with the result in Arabidopsis (Chen et al., 2009), we found that the level of H2O2 accumulation was much lower on the leaves of the NtGSNOR1a/1b knockout plants than that of the WT plants (Figures 5B,C), indicating that reduced level of paraquat-induced H2O2 production or accumulation could be at least one of the molecular mechanisms behind the resistance of the NtGSNOR1a/1b knockout plants to paraquat-induced cell death.
Knocking Out NtGSNOR1a/1b Has Opposite Effects on Tobacco mosaic virus and P. syringae pv. tomato DC3000 Infections
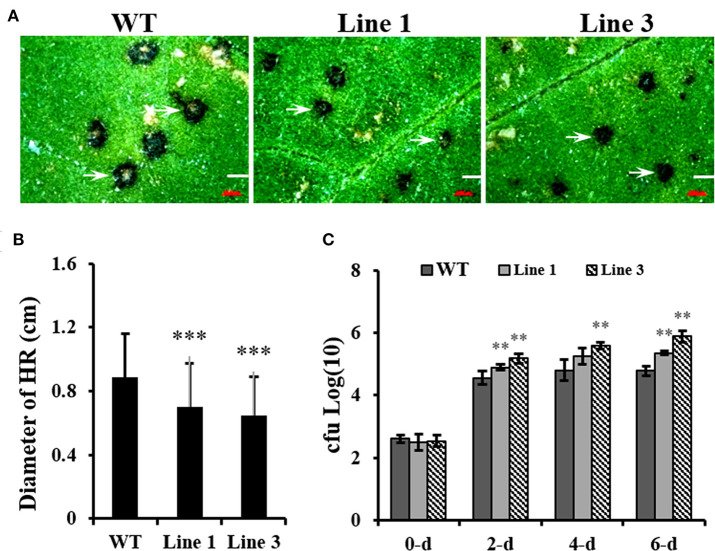
Contradictory results were obtained regarding the role of Arabidopsis GSNOR1 in disease resistance using gsnor1-3 null mutant (Feechan et al., 2005) and GSNOR1 antisense lines (Rustérucci et al., 2007). However, the role of GSNOR1 in other plant species has not been investigated. To test the effect of NtGSNOR1a/1b knockout on disease resistance in Nicotiana species, we inoculated TMV on both WT N. tabacum Samsun (NN) and the NtGSNOR1a/1b knockout plants. It was found that the sizes of HR lesions triggered by TMV infection on the leaves of the NtGSNOR1a/1b knockout plants were significantly smaller than those on the leaves of N. tabacum cv. Samsun (NN) at 5 days post inoculation (dpi) (Figures 6A,B). Appearance of local HR lesions reflects both replication and cell-to-cell movement of the virus (Meshi et al., 1982). Consistent with this statement, we showed that the transcript level of TMV capsid protein (CP) was much lower on the knockout leaves than on the WT leaves (Supplementary Figure 7). However, we cannot exclude a possibility that the smaller sizes of the HR on the leaves of the knockout plants could also reflect a decreased ability for N protein to trigger the HR upon TMV infection.
Figure 6.
Knocking out NtGSNOR1a/1b in N. tabacum results in the enhanced N gene mediated resistance against TMV but reduced resistance against Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). (A) The hypersensitive response (HR) lesions formed on the leaves of the 40-day-old NtGSNOR1a/1b knockout lines (Lines 1 and 3) are reduced in comparison to on that of wild-type (WT) Samsun (NN) plants in response to Tobacco mosaic virus (TMV) infection. (B) The sizes of HR lesions shown in (A) were measured under a dissecting microscopy. The lesions on the leaves of three independent plants of each line were measured. Over 30 lesions were measured on each infected leaf. *** represents significant difference at 0.001 level by the Student's t-test. (C) NtGSNOR1a/1b knockout plants support greater growth of Pst DC3000 compared to WT Samsun (NN) plants. The leaves of 40-day-old plants were infiltrated with Pst DC3000 (OD600 = 0.00001). The numbers of Pst DC3000 were counted at 0-, 2-, 4-, and 6-day post-inoculation and expressed as log10. ** represents significance at p-value 0.01 by the Student's t-test.
To examine the effect of knocking out NtGSNOR1a/1b on bacterial infection, the upper leaves of both WT and NtGSNOR1a/1b knockout plants without spontaneous cell death were infiltrated with Pst DC3000 as the cell death on the lower leaves interferes with the bacterial growth assay. As shown in Figure 6C, Pst DC3000 multiply better on the leaves of the NtGSNOR1a/1b knockout plants than on the leaves of the WT plants (Figure 6C), indicating that NtGSNOR1a/1b positively regulates the basal resistance to a bacterial pathogen. Together, these results indicate that knocking out NtGSNOR1a/1b has opposite effects on resistance to different types of pathogens with different infecting strategies.
Discussion
In this report, we found that both anti- and pro-cell death effects were observed in the NtGSNOR1a/1b knockout plants, reinforcing the notion that NO can play either anti- or pro-death roles depending on cellular conditions (Wang et al., 2013; Wendehenne et al., 2014) and the altered NO homeostasis plays key roles in switching its functions during the HR. In addition, we found that knocking out NtGSNOR1a/1b has opposite effects on viral and bacterial resistance.
The cell death phenotype of the NtGSNOR1a/1b knockout plants under natural conditions was not reported for the gsnor1-3/hot5/par2-1 mutants (Feechan et al., 2005; Lee et al., 2008; Chen et al., 2009) or antisense lines of GSNOR1 in Arabidopsis (Rustérucci et al., 2007), although spontaneous cell death was observed on the leaves of GSNOR1-silenced tomato plants by virus-induced gene silencing (VIGS) (Liu et al., 2017) and an accelerated cell death was observed on the leaves of gsnor1-3 mutant upon infections by avirulent bacterial strains or by an avirulent oomycete isolate (Rustérucci et al., 2007; Yun et al., 2011). The generation of both NO and H2O2 is simultaneously induced under many different stress conditions (Delledonne et al., 1998; Clarke et al., 2000; Lin et al., 2012; Scheler et al., 2013), indicating that they coexist in most cellular conditions. Both NO and H2O2 was significantly higher in the NtGSNOR1a/1b knockout plants compared to WT plants (Figure 3). The increased H2O2 level in the NtGSNOR1a/1b knockout plants could be induced by the overaccumulation of NO in these plants, since it has been reported previously that NO is essential for generating elicitor- or pathogen-inducible ROS and that ROS likely functions downstream of NO (Rasul et al., 2012; Wang L. et al., 2014; Kulik et al., 2015). It has been reported that efficient activation of hypersensitive cell death required a balance between NO and ROS production (Delledonne et al., 1998, 2001). NO/H2O2 ratio between 0.25 and 2 was effective in inducing cell death. NO/H2O2 ratio during pathogen induction of the HR or during induction of cell death by exogenous NO and artificial oxidative burst falls within this range (Delledonne et al., 2001). Based on this ratio model, we propose the following explanations for the naturally occurring cell death (Figures 1E, 2) and the reduced paraquat-induced cell death observed in the NtGSNOR1a/1b knockout plants (Figure 5A). Under normal growth conditions, NO level is high in the NtGSNOR1a/1b knockout plants, but H2O2 level is relatively low compared to the NO level. As a result, the NO/H2O2 ratio falls above the high end of the optimal ratio range so that cell death is not triggered. At certain development stages or under mild stress conditions, H2O2 is induced gradually. Once the H2O2 level is accumulated to a certain threshold level, the NO/H2O2 ratio falls within the optimal range, natural cell death occurs (Figures 1E, 2). In paraquat-sprayed leaves, tremendous amount of H2O2 is induced, causing the NO/H2O2 ratio to fall below the low end of the optimal ratio range and thus the cell death is prevented.
However, given the promiscuous natures of both NO and H2O2, the mechanism of NO- and H2O2-induced cell death could be multifaceted. Firstly, in plants, H2O2 and NO can chemically react to produce singlet oxygen or hydroxyl radicals, both of which have effects on causing cell death (Zaninotto et al., 2006). However, hydroxyl radical ONOO−, which can cause cell death in animals, is not toxic to plants (Delledonne et al., 2001). Rather, it is likely that the formation of ONOO− can serve as a protective mechanism by scavenging from reactions causing cellular damage (Delledonne et al., 2001). Consistent with this statement, we found that the H2O2 level in the NtGSNOR1a/1b knockout plants was much lower than that in WT plants sprayed with paraquat (Figure 5B), suggesting that increased RNS accumulation can neutralize and/or detoxify the effects of ROS.
Given the role of GSNOR1 in maintaining homeostasis of cellular S-nitrosylation (Liu et al., 2001; Feechan et al., 2005), it is also possible that the naturally occurring cell death (Figure 1) and resistance to paraquat-induced cell death (Figure 5) in the NtGSNOR1a/1b knockout plants could be mediated by S-nitrosylation through modulating different target proteins. S-nitrosylation of proapoptotic and antiapoptotic regulators is a common mechanism to regulate apoptosis in animal systems (Mannick, 2007; Iyer et al., 2014). Stimulation or stabilization of antiapoptotic factors by S-nitrosylation inhibits cell death (Mannick, 2007; Iyer et al., 2014), whereas stimulation of proapoptotic factors by S-nitrosylation promotes or initiates apoptosis (Mannick, 2007; Iyer et al., 2014). In plants, it has been shown that NO increases H2O2 levels by inhibiting catalase and ascorbate peroxidase, two major ROS-scavenging enzymes, through S-nitrosylation (Lin et al., 2012; Ortega-Galisteo et al., 2012; de Pinto et al., 2013). It is possible that the increased accumulation of H2O2 (Figure 3C) and naturally occurring cell death observed in the NtGSNOR1a/1b knockout plants (Figure 1) are at least partially caused by inhibiting ROS detoxification through S-nitrosylation of both catalase and ascorbate peroxidase. On the other hand, S-nitrosylation of the Arabidopsis cytosolic APX1 at Cys32 enhances its enzymatic activity of scavenging H2O2, resulting in an increased resistance to oxidative stress (Yang et al., 2015). It also has been recently shown that S-nitrosylation of the NADPH oxidase, AtRBOHD, at Cys890, abolishing its ability to synthesize ROS and thus functioning as a negative feedback loop limiting the HR (Yun et al., 2011), raises the possibility that the resistance to paraquat-induced cell death observed in the NtGSNOR1a/1b knockout plants is at least partially attributed to the feedback inhibition of H2O2 production by AtRBOHD and APX1. We recently provide evidence that NO may trigger cell death in tomato (Solanum lycopersicum) by inhibiting the activity of phosphoinositide-dependent kinase 1 (SlPDK1), a conserved negative regulator of cell death in yeasts, mammals, and plants, via S-nitrosylation (Liu et al., 2017). SlPDK1 is primarily S-nitrosylated on Cys128 and substitution of Cys128 with serine completely abolished SlPDK1 kinase activity, suggesting that S-nitrosylation of Cys128 is responsible for SlPDK1 inhibition (Liu et al., 2017). These results establish a potential link between NO-triggered cell death and inhibition of the kinase activity of tomato PDK1. It is possible that the spontaneous cell death observed on the leaves of NtGSNOR1a/1b knockout plants is a consequence of inhibition of the NtPDK1 activity.
Even though the naturally occurring spontaneous cell death is not observed in Arabidopsis gsno1-3 plants, HR was accelerated in both GSNOR1 antisense plants and in atgsnor1-3 mutant plants upon infection by avirulent bacterial strains or by an avirulent oomycete isolate (Rustérucci et al., 2007; Yun et al., 2011), indicating a conserved negative role of GSNOR1 from different plant species in triggering cell death. Interestingly, accelerated cell death is independent of SA and H2O2, as the cell death in gsnor1-3/sid2 and gsnor1-3/atrbohD, gsnor1-3/atrboF double mutants, and gsnor1-3/atrbohD/atrbohF triple mutants displays the same degree of cell death as in gsnor1-3 plants, and H2O2 level is less in gsnor1-3 plants than that in Col-0 plants (Yun et al., 2011). Consistent with this result, we found that the spontaneous cell death occurring on the NtGSNOR1a/1b knockout plants was not suppressed by knocking out the NtEDS1a/1b (Figure 4), strongly indicating that the spontaneous cell death is SA-independent.
The NtGSNOR1a/1b knockout plants displayed increased resistance to TMV but reduced resistance to Pst DC3000 (Figure 6). The opposite effects of silencing or knocking out GSNOR1 on the resistance to different types of pathogens were also observed in Arabidopsis (Feechan et al., 2005; Rustérucci et al., 2007; Yun et al., 2011). Whereas, the basal resistance to Pst DC3000 and Noco2 race of the H. parasitica, RPM1- or RPS4-mediated R resistance to Pst DC3000 (avrB) and Pst DC3000 (avrRps4), and non-host resistance to wheat powdery mildew pathogen Bgt are all compromised in Arabidopsis gsnor1-3 mutant (Feechan et al., 2005), an enhanced basal resistance to oomycete Pp Noco2 and an increased SAR were observed in Arabidopsis GSNOR1 antisense plants (Rustérucci et al., 2007). Espunya et al. (2012) attributes these opposing phenotypes to the partial reduction (antisense line) vs. the complete loss of function (knockout mutant) in GSNOR1 activity. Interestingly, contrary to their own report that various types of resistance are compromised in gsnor1-3 mutant plant (Feechan et al., 2005), Yun et al. (2011) recently showed that an enhanced resistance to Hyaloperonospora arabidopsidis isolate Emwa1, which is recognized by RPP4, was observed in gsnor1-3 mutant in an SA- and H2O2-independent manner (Yun et al., 2011). This resistance seemed to be attributed sorely to increased SNO levels in gsnor1-3 mutant (Yun et al., 2011). Thus, the opposing results in resistance to different types of pathogens observed in gsnor1-3 mutant cannot be explained sorely by the degree of reduction in GSNOR1 activity. Rather, it reflects the complexity and hierarchy of NO signaling events (Wendehenne et al., 2014).
Multiple proteins involved in defense responses could be differentially or simultaneously S-nitrosylated in response to pathogen infections. Depending on the severity of nitrosative stress, the subcellular compartmentalization of both GSNO and its target proteins, GSNO could either promote or inhibit defense responses. As the functions of the positive regulators of SA-dependent pathway, NPR1, TGA1, and SBP3, and the function of a suppressor of negative regulator of defense responses, SRG1, are negatively regulated by S-nitrosylation (Tada et al., 2008; Wang et al., 2009; Lindermayr et al., 2010; Cui et al., 2018), it is reasonable to assume that the compromised resistance of the NtGSNOR1a/1b knockout plants to Pst DC3000 (Figure 6C) could also be combined results of compromised SA-mediated resistance and de-repression of immunity inhibition. However, Yun et al. (2011) reported that the cell death triggered by incompatible pathogens in gsnor1-3 is SA- and H2O2-independent and the cell death triggered in gsnor1-3 is enough to confer the RPP4-mediated resistance to Hyaloperonospora arabidopsidis isolate Emwa1, suggesting that the resistance is SA-independent (Yun et al., 2011). However, our results indicated that the spontaneous cell death on the NtGSNOR1a/1b knockout plants (Figures 1, 2) does not confer bacterial resistance (Figure 6B). Intriguingly, contrary to the response to bacterial infection, an enhanced resistance against TMV was observed in the NtGSNOR1a/1b knockout plants (Figure 6A). The increased resistance to TMV could be due to inhibitory effects of the overaccumulated Reactive nitrogen species (RNS) and ROS (Figure 3) on virus replication or movement or both. It is also possible that the viral proteins involved in replication or movement is inhibited by S-nitrosylation. Alternatively, the enhanced resistance of the NtGNSOR1a/1b knockout plant to TMV could also be due to the inhibition of negative regulators of defense responses by S-nitrosylation. For example, Arabidopsis SUMO E2 enzyme, SCE1, plays a negative role in immune responses. S-nitrosylation SCE1 at Cys139 inhibits its SUMO-conjugating activities, leading to immune activation by relieving SUMO1/2-mediated suppression (Skelly et al., 2019). Depending on the cell type, subcellular localization, developmental stages, and plant–pathogen interactions, as well as its diffuse and promiscuous nature, NO can exert differential or even opposite effects on cell death and immunity through both S-nitrosylation-dependent and -independent mechanisms, which might explain the opposite effects of NtGSNOR1a/1b knocking out on Pst DC3000 and TMV resistance.
The CRISPR/Cas9-mediated genome editing tool has been extensively applied in plant biotechnology since its development (Xie and Yang, 2013; Wang Y. et al., 2014; Wang et al., 2015; Char et al., 2017, 2020; Chen et al., 2019; Oliva et al., 2019). Our results strongly indicated that CRISPR/Cas9 is a powerful technology not only in resolving gene redundancy in tetraploid tobacco (Figures 1, 4) but also in studying the functional connections and/or interactions of different genes (Figure 4).
Materials and Methods
Plant Materials
Nicotiana tabacum cv. Samsun, carrying a TMV-resistant gene, N, was used in this study. The tobacco plants were maintained in the growth room at 22°C with a photoperiod of 16-h light/8-h dark.
Agro-Infiltration
Agrobacterium infiltration was performed as described (Liu et al., 2005). Agrobacterium GV3101 strain carrying the 35S::GmMEKK1a construct (Xu et al., 2018), which is known to induce HR, was infiltrated into the leaves of the 30–40-day-old N. tabacum plants as indicated. The 35S::GFP construct was included as a negative control. The leaf was photographed at 2 days post infiltration. The experiment was repeated three times (at least three plants each time) with similar results.
CRISPR/Cas9 Constructs and Plant Transformation
Oligos of 20 nucleotides were chosen from the second exon of NtGSNOR1a/1b (CTGGGAACCCAACAAGCCTC, 45–65 bp downstream of start codon ATG of the open reading frame, was used as a guide to target both genes simultaneously) and first exon of NtEDS1a/1b (GAAGCTCACAGTTTGTCTTC, 70–92 bp downstream of the start codon ATG) (Figures 1A, 4B). Blast analysis indicated these two guides only perfectly matched the target sequences of NtGSNOR1a/1b and NtEDS1a/1b, respectively, but not any other sequences in the N. tabacum genome. The closest sequences in the genome to both guides displayed at least 4-nt mismatches. Thus, the off-target effects could potentially be ruled out with great probability. Both the sense and antisense strands of the chosen oligos were synthesized with ATTG and AAAC that are compatible to Bsa I sticky ends attached to the sense and antisense strands, respectively. The sense and antisense strands of the synthesized oligos were mixed in TE buffer and annealed to each other by heating at 98°C for 5 min followed by cooling to room temperature. The annealed double-strand oligos were subsequently ligated into the intermediate AtU6-26-sgRNA-SK vector (Yan et al., 2015) predigested with Bsa I. The constructed plasmid was double digested with Nhe I and Spe I (Nhe I and Spe I are isocaudomers), and the fragment of 642 bp containing the guide sequence was ligated into the destination vector pCAMBIA-1300-pYAO:Cas9 predigested with Spe I and treated with alkaline phosphatase (Yan et al., 2015). The authenticity of the final construct was confirmed by sequencing.
For double targets (NtGSNOR1a/1b + NtEDS1a/1b) construct, the Nhe I and Spe I digested fragment from the intermediate vector containing NtEDS1a/1b guide was ligated into the destination vector containing NtGSNOR1a/1b guide digested with Spe I. The authenticity of the final construct was confirmed by sequencing.
The oligos used for target guides are:
NtGSNOR1a/1b-F: 5′-ATTGCTGGGAACCCAACAAGCCTC-3′;
NtGSNOR1a/1b-R: 5′-AAACGAGGCTTGTTGGGTTCCCAG-3′;
NtEDS1a/1b-F: 5′-ATTGGAAGCTCACAGTTTGTCTTC-3′;
NtEDS1a/1b-R: 5′-AAACGAAGACAAACTGTGAGCTTC-3′.
The CRISPR/Cas9 constructs were transformed into N. tabacum cv. Samsun (NN) as described previously (Horsch et al., 1985). Briefly, fully expanded leaves from 3-week-old tobacco plants were surface-sterilized with 70% ethanol and 0.5–2% NaClO3, respectively. Leaf discs of 0.5 cm in diameter were precultivated in MS medium for 3 days followed by incubating in Agrobacterium (GV3101) suspension solution for 20–30 min. Agrobacterium-infected leaf discs were cultivated in MS medium containing 1 mg/L Indole-3-acetic acid (IAA), 1 mg/L 6-BA in the growth room for 2 days, and then transferred to MS medium supplemented with 1 mg/L IAA, 1 mg/L 6-BA, 50 mg/L Hygromycin, and 25 mg/L rifampicin. Shoots were transferred to rooting medium containing IAA (1 mg/L), 50 mg/L Hygromycin, and 25 mg/L Timentin. The rooted transformants were finally transferred to soil and grown in a growth room.
Screening of the Homozygous CRISPR/Cas9 Lines
The transgenic CRISPR/Cas9 plants generated were characterized by genomic PCR with specific primers listed below and followed by DNA sequencing. DNA was extracted from leaf tissue with the DNA extraction kit (Invitrogen). Sequence results were aligned against the wild type sequence to identify insertions and deletions (InDels). The sequencing chromatograms contained no double peaks but contained InDels indicating homozygous mutations. The double peaks in the chromatograms indicate a heterozygous insertion/deletion or WT/InDels. In this case, the genomic PCR products were cloned into the PMD19-T vector (Takara), and multiple clones were subjected to sequencing to determine the exact nature of InDels. The primers used for the genomic PCR or for screening positive clones by colony PCR are:
NtGSNOR1a/1b-F: 5′-CACTACTAGTATACCACTTA-3′
NtGSNOR1a-R: 5′-GTGGTTAACTTTATGAATAATTTGC-3′
NtGSNOR1b-R: 5′-TTAGAAACTCACATAGCCGG-3′
NtEDS1a-F: 5′-CCATCTTTGAGCTGCTAGCA-3′
NtEDS1a-R: 5′-GGGAACATATTCTTGGACATG-3′
NtEDS1b-F: 5′-CCATCTTTGAGCTGCTAAGT-3′
NtEDS1b-R: 5′-GGAATTAAGCTGATCAAACTA-3′
Tobacco mosaic virus Infection and P. syringae pv. Tomato DC3000 Growth Assay
The properly diluted sap extracted from the TMV (U1 strain) infected leaves was rub-inoculated on the upper surface of the N. tabacum cv. Samsun (NN) leaves, as described previously (Whitham et al., 1994). The HR normally is visible at 2 dpi, and the photos were taken at 4 dpi. The primers used for qRT-PCR quantification of the TMV CP are:
CP-F: GCTCTCGAAAGAGCTCCGAT
CP-R: TTTATCGCGCTCCTTATGGC
The primers for endogenous reference gene NtActin are:
NtActin-F: TGGCATCACACTTTCTACAA
NtActin-R: CCACTGAGCACAATGTT
The Pst DC3000 of OD600 = 0.00001 was infiltrated into the leaves of respective tobacco lines, as described previously (Qi et al., 2018). The leaf discs of 0.5 cm in diameter were punched from the infiltrated areas and grounded in the 10 mM MgSO4 buffer. The saps were diluted 10, 100, and 1,000 times and spread on KB agar plates. Colony-forming unit (cfu) was obtained by counting the number of the colonies on a serial of plates.
H2O2 Detection by 3,3′-Diaminobenzidine Staining
H2O2 was detected by an endogenous peroxidase-dependent in situ histochemical staining procedure using DAB (Sigma-Aldrich; Ren et al., 2002). Leaves of 30–40-day-old plants were detached and placed in a solution containing 1 mg/ml DAB (pH 5.5) for 2 h. The leaves were cleared by boiling in ethanol (96%) for 10 min and then stored in 96% ethanol. H2O2 production was visualized as a reddish-brown precipitate in cleared leaves (Thordal-Christensen et al., 1997; Ren et al., 2002).
NO Detection by DAF-2DA Staining
NO detection was performed by DAF-FM DA staining as described (He et al., 2004). Briefly, leaf discs and 15-day-old seedlings were preincubated for 2 h at room temperature in a solution of 0.1 mM CaCl2, 10 mM KCl, 10 mM MES-Tris, pH 5, and then stained with 10 μM DAF-FM DA for 45 min. After rinsing with water, the sample was observed and photographed with an inverted Axiophot microscope (Zeiss) equipped with a digital camera (Diagnostic Instruments).
Accession Numbers
The accession numbers for NtGSNOR1a, NtGSNOR1b, NtEDS1a, and NtEDS1b are LOC107777450, LOC107791841, LOC107782626, and LOC107791841, respectively.
Statistics
Statistics were performed using the Student's t-test. ** and *** represent the significant differences between the WT control and the knockout plants at levels of 0.01 and 0.001, respectively.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
J-ZL designed the experiments. Z-CL, Q-WR, JR, YG, X-TR, N-NW, H-YX, and XL performed the experiments. J-ZL wrote the manuscript with Z-CL's input. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Qi Xie for the pCAMBIA-1300-pYAO:Cas9 vector. We also thank Dr. Kewei Zhang for sharing materials and MS Jun-Mei Li for technique support.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (31571423 and 31371401 to J-ZL) and the Department of Education of Zhejiang Province (Y201840726 to XL).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.596234/full#supplementary-material
References
- Albertos P., Romero-Puertas M. C., Tatematsu K., Mateos I., Sánchez-Vicente I., Nambara E., et al. (2015). S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 6:8669. 10.1038/ncomms9669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Char S. N., Neelakandan A. K., Nahampun H., Frame B., Main M., Spalding M. H., et al. (2017). An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 15, 257–268. 10.1111/pbi.12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Char S. N., Wei J., Mu Q., Li X., Zhang Z. J., Yu J., et al. (2020). An Agrobacterium-delivered CRISPR/Cas9 system for targeted mutagenesis in sorghum. Plant Biotechnol. J. 18, 319–321. 10.1111/pbi.13229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wang Y., Zhang R., Zhang H., Gao C. (2019). CRISPR/cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. 10.1146/annurev-arplant-050718-100049 [DOI] [PubMed] [Google Scholar]
- Chen R., Sun S., Wang C., Li Y., Liang Y., An F., et al. (2009). The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 19, 1377–1387. 10.1038/cr.2009.117 [DOI] [PubMed] [Google Scholar]
- Clarke A., Desikan R., Hurst R. D., Hancock J. T., Neill S. J. (2000). NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J. 24, 667–677. 10.1046/j.1365-313x.2000.00911.x [DOI] [PubMed] [Google Scholar]
- Cui B., Pan Q., Clarke D., Villarreal M. O., Umbreen S., Yuan B., et al. (2018). S-nitrosylation of the zinc finger protein SRG1 regulates plant immunity. Nat. Commun. 9:4226. 10.1038/s41467-018-06578-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pinto M. C., Locato V., Sgobba A., Romero-Puertas M. D. C., Gadaleta C., Delledonne M., et al. (2013). S-nitrosylation of ascorbate peroxidase is part of programmed cell death signaling in tobacco bright yellow-2 cells. Plant Physiol. 163, 1766–1775. 10.1104/pp.113.222703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M., Xia Y., Dixon R. A., Lamb C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. 10.1038/29087 [DOI] [PubMed] [Google Scholar]
- Delledonne M., Zeier J., Marocco A., Lamb C. (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. U.S.A. 98, 13454–13459. 10.1073/pnas.231178298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R., Griffiths R., Hancock J., Neill S. (2002). A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 16314–16318. 10.1073/pnas.252461999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge A. D. (1971). The mode of action of the bipyridylium herbicides, paraquat, and diquat. Endeavour 30, 130–135. 10.1016/0160-9327(71)90039-1 [DOI] [PubMed] [Google Scholar]
- Durner J., Wendehenne D., Klessig D. F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADPribose. Proc. Natl. Acad. Sci. U.S.A. 95, 10328–10333. 10.1073/pnas.95.17.10328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espunya M. C., De Michele R., Gómez-Cadenas A., Martínez M. C. (2012). S-Nitrosoglutathione is a component of wound- and salicylic acid-induced systemic responses in Arabidopsis thaliana. J. Exp. Bot. 63, 3219–3227. 10.1093/jxb/ers043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A., Kwon E., Yun B. W., Wang Y., Pallas J. A., Loake G. J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U.S.A. 102, 8054–8059. 10.1073/pnas.0501456102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Wang C., Chen Q., Chen H., Ren B., Li X., et al. (2013). S-Nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat. Commun. 4:1529. 10.1038/ncomms2541 [DOI] [PubMed] [Google Scholar]
- Forrester M. T., Thompson J. W., Foster M. W., Nogueira L., Moseley M., Stamler J. S. (2009). Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat. Biotechnol. 27, 557–559. 10.1038/nbt.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frungillo L., Skelly M. J., Loake G. J., Spoel S. H., Salgado I. (2014). S-nitrosothiols regulate nitric oxide production and storage in plants through the nitrogen assimilation pathway. Nat. Commun. 5:5401. 10.1038/ncomms6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B., Yan Y., Zhang L., Cheng F., Liu Z., Shi Q. (2019). Unravelling GSNOR-mediated S-nitrosylation and multiple developmental programs in tomato plants. Plant Cell Physiol. 60, 2523–2537. 10.1093/pcp/pcz143 [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Kolbert Z., Durner J., Lindermayr C., Corpas F. J., Brouquisse R., et al. (2020). Regulating the regulator: nitric oxide control of post-translational modifications. New Phytol. 227, 1319–1325. 10.1111/nph.16622 [DOI] [PubMed] [Google Scholar]
- He Y., Tang R. H., Hao Y., Stevens R. D., Cook C. W., Ahn S. M., et al. (2004). Nitric oxide represses the Arabidopsis floral transition. Science 305, 1968–1971. 10.1126/science.1098837 [DOI] [PubMed] [Google Scholar]
- Hess D. T., Matsumoto A., Kim S. O., Marshall H. E. (2005). Stamler JS. protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166. 10.1038/nrm1569 [DOI] [PubMed] [Google Scholar]
- Hess D. T., Stamler J. S. (2012). Regulation by S-nitrosylation of protein post-translational modification. J. Biol. Chem. 287, 4411–4418. 10.1074/jbc.R111.285742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R. B., Fry J. E., Hofmann N. L., Eichholtz D., Rogers S. G., Fraley R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. 10.1126/science.227.4691.1229 [DOI] [PubMed] [Google Scholar]
- Huang D., Huo J., Zhang J., Wang C., Wang B., Fang H., et al. (2019). Protein S-nitrosylation in programmed cell death in plants. Cell. Mol. Life Sci. 76, 1877–1887. 10.1007/s00018-019-03045-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A., Yun B. W., Kim J. H., Gupta K. J., Hyung N. I., Loake G. J. (2019). Novel and conserved functions of S-nitrosoglutathione reductase in tomato. J. Exp. Bot. 70, 4877–4886. 10.1093/jxb/erz234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias M. J., Terrile M. C., Correa-Aragunde N., Colman S. L., Izquierdo-Álvarez A., Fiol D. F., et al. (2018). Regulation of SCF(TIR1/AFBs) E3 ligase assembly by S-nitrosylation of Arabidopsis SKP1-like1 impacts on auxin signaling. Redox Biol. 18, 200–210. 10.1016/j.redox.2018.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A. K., Rojanasakul Y., Azad N. (2014). Nitrosothiol signaling and protein nitrosation in cell death. Nitric Oxide 42, 9–18. 10.1016/j.niox.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe H., Ohtani M., Kurata T., Sakamoto T., Demura T. (2018). Protein S-nitrosylation regulates xylem vessel cell differentiation in Arabidopsis. Plant Cell Physiol. 59, 17–29. 10.1093/pcp/pcx151 [DOI] [PubMed] [Google Scholar]
- Klessig D. F., Durner J., Noad R., Navarre D. A., Wendehenne D., Kumar D., et al. (2000). Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. U.S.A. 97, 8849–8855. 10.1073/pnas.97.16.8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik A., Noirot E., Grandperret V., Bourque S., Fromentin J., Salloignon P., et al. (2015). Interplays between nitric oxide and reactive oxygen species in cryptogein signalling. Plant Cell Environ. 38, 331–348. 10.1111/pce.12295 [DOI] [PubMed] [Google Scholar]
- Lamattina L., Garcia-Mata C., Graziano M., Pagnussat G. (2003). Nitric oxide: the versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 54, 109–136. 10.1146/annurev.arplant.54.031902.134752 [DOI] [PubMed] [Google Scholar]
- Lee U., Wie C., Fernandez B. O., Feelisch M., Vierling E. (2008). Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20, 786–802. 10.1105/tpc.107.052647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., Wang Y., Tang J., Xue P., Li C., Liu L., et al. (2012). Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 158, 451–464. 10.1104/pp.111.184531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Saalbach G., Bahnweg G., Durner J. (2006). Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J. Biol. Chem. 81, 4285–4291. 10.1074/jbc.M511635200 [DOI] [PubMed] [Google Scholar]
- Lindermayr C., Saalbach G., Durner J. (2005). Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930. 10.1104/pp.104.058719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Sell S., Mu ller B., Leister D., Durner J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907. 10.1105/tpc.109.066464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Z., Blancaflor E. B., Nelson R. S. (2005). The tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone or within the complex aligns with and traffics along microfilaments. Plant Physiol. 138, 1853–1865. 10.1104/pp.105.065722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Z., Duan J., Whitham S. A., Qian W. J. (2017). S-nitrosylation inhibits the kinase activity of tomato phosphoinositide-dependent kinase 1 (PDK1). J. Biol. Chem. 292, 19743–19751. 10.1074/jbc.M117.803882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Hausladen A., Zeng M., Que L., Heitman J., Stamler J. S. (2001). A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490–494. 10.1038/35068596 [DOI] [PubMed] [Google Scholar]
- Liu L., Yan Y., Zeng M., Zhang J., Hanes M. A., Ahearn G., et al. (2004). Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell 116, 617–628. 10.1016/S0092-8674(04)00131-X [DOI] [PubMed] [Google Scholar]
- Mannick J. B. (2007). Regulation of apoptosis by protein S-nitrosylation. Amino Acids 32, 523–526. 10.1007/s00726-006-0427-6 [DOI] [PubMed] [Google Scholar]
- Matamoros M. A., Cutrona M. C., Wienkoop S., Begara-Morales J. C., Sandal N., Orera I., et al. (2020). Altered plant and nodule development and protein S-nitrosylation in Lotus japonicus mutants deficient in S-nitrosoglutathione reductases. Plant Cell Physiol. 61, 105–117. 10.1093/pcp/pcz182 [DOI] [PubMed] [Google Scholar]
- Meshi T., Takamatsu N., Ohno T., Okada Y. (1982). Molecular cloning of the complementary DNA copies of the common and cowpea strains of tobacco mosaic virus RNA. Virology 118, 4–75. 10.1016/0042-6822(82)90320-8 [DOI] [PubMed] [Google Scholar]
- Neill S., Barros R., Bright J., et al. (2008). Nitric oxide, stomatal closure, and abiotic stress. J. Exp. Bot. 59, 165–176. 10.1093/jxb/erm293 [DOI] [PubMed] [Google Scholar]
- Ni M., Zhang L., Shi Y. F., Wang C., Lu Y., Pan J., et al. (2017). Excessive cellular S-nitrosothiol impairs endocytosis of auxin efflux transporter PIN2. Front. Plant Sci. 8:1988. 10.3389/fpls.2017.01988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R., Ji C., Atienza-Grande G., Huguet-Tapia J. C., Perez-Quintero A., Li T., et al. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. 10.1038/s41587-019-0267-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Galisteo A. P., Rodríguez-Serrano M., Pazmiño D. M., Gupta D. K., Sandalio L. M., Romero-Puertas M. C. (2012). S-Nitrosylated proteins in Pea (Pisum Sativum L.) leaf peroxisomes: changes under abiotic stress. J. Exp. Bot. 63, 2089–2103. 10.1093/jxb/err414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Seong K., Thomazella D. P. T., Kim J. R., Pham J., Seo E., et al. (2018). NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. U.S.A. 15, E10979–E10987. 10.1073/pnas.1814856115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasul S., Wendehenne D., Jeandroz S. (2012). Study of oligogalacturonides-triggered nitric oxide (NO) production provokes new questioning about the origin of NO biosynthesis in plants. Plant Signal. Behav. 7, 1031–1033. 10.4161/psb.20658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Yang H., Zhang S. (2002). Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277, 559–565. 10.1074/jbc.M109495200 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas M. C., Laxa M., Matté A., Zaninotto F., Finkemeier I., Jones A. M., et al. (2007). S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell 19, 4120–4130. 10.1105/tpc.107.055061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C., Espunya M. C., Diáz M., Chabannes M., Martínez M. C. (2007). S-nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiol. 143, 1282–1292. 10.1104/pp.106.091686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci D. H., Aviv B. F., Holt J. L., Dangl J. E., Parker. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13, 2211–2224. 10.1105/tpc.010085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheler C., Durner J., Astier J. (2013). Nitric oxide and reactive oxygen species in plant biotic interactions. Curr. Opin. Plant Biol. 16, 534–539. 10.1016/j.pbi.2013.06.020 [DOI] [PubMed] [Google Scholar]
- Shi Y. F., Wang D. L., Wang C., Culler A. H., Kreiser M. A., Suresh J., et al. (2015). Loss of GSNOR1 function leads to compromised auxin signaling and polar auxin transport. Mol. Plant 8, 1350–1365. 10.1016/j.molp.2015.04.008 [DOI] [PubMed] [Google Scholar]
- Skelly M. J., Malik S. I., Le B. T., Bo Y., Jiang J., Spoel S. H., et al. (2019). A role for S-nitrosylation of the SUMO-conjugating enzyme SCE1 in plant immunity. Proc. Natl. Acad. Sci. U.S.A. 116, 17090–17095. 10.1073/pnas.1900052116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler J. S., Simon D. I., Osborne J. A., Mullins M. E., Jaraki O., Michel T., et al. (1992). S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc. Natl. Acad. Sci. U.S.A. 89, 444–448. 10.1073/pnas.89.1.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntres Z. E. (2002). Role of antioxidants in paraquat toxicity. Toxicology 180, 65–77. 10.1016/S0300-483X(02)00382-7 [DOI] [PubMed] [Google Scholar]
- Tada Y., Spoel S. H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., et al. (2008). Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. 10.1126/science.1156970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrile M. C., Pari's R., Caldero' n-Villalobos L. I., Iglesias M. J., Lamattina L., Estelle M., et al. (2012). Nitric oxide inflfluences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 70, 492–500. 10.1111/j.1365-313X.2011.04885.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z. G., Wei Y. D., Collinge D. B. (1997). Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. 10.1046/j.1365-313X.1997.11061187.x [DOI] [Google Scholar]
- Wang L., Guo Y, Jia L., Chu H., Zhou S., Chen K., et al. (2014). Hydrogen peroxide acts upstream of nitric oxide in the heat shock pathway in Arabidopsis seedlings. Plant Physiol. 164, 2184–2196. 10.1104/pp.113.229369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Du Y., Hou Y. J., Zhao Y., Hsu C. C., Yuan F., et al. (2015). Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. U.S.A. 112, 613–618. 10.1073/pnas.1423481112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. 10.1038/nbt.2969 [DOI] [PubMed] [Google Scholar]
- Wang Y., Chu C. (2020). S-Nitrosylation control of ROS and RNS homeostasis in plants: the switching function of catalase. Mol. Plant 13, 946–948. 10.1016/j.molp.2020.05.013 [DOI] [PubMed] [Google Scholar]
- Wang Y., Loake G. J., Chu C. (2013). Cross-talk of nitric oxide and reactive oxygen species in plant programed cell death. Front. Plant Sci. 4:314. 10.3389/fpls.2013.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Q., Feechan A., Yun B. W., Shafifiei R., Hofmann A., Taylor P., et al. (2009). S-Nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 284, 2131–2137. 10.1074/jbc.M806782200 [DOI] [PubMed] [Google Scholar]
- Wendehenne D., Durner J., Klessig D. F. (2004). Nitric oxide: a new player in plant signalling and defence responses. Curr. Opin. Plant Biol. 7, 449–455. 10.1016/j.pbi.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Wendehenne D., Gao Q. M., Kachroo A., Kachroo P. (2014). Free radical-mediated systemic immunity in plants. Curr. Opin. Plant Biol. 20C, 127–134. 10.1016/j.pbi.2014.05.012 [DOI] [PubMed] [Google Scholar]
- Whitham S., Dinesh-Kumar S. P., Choi D., Hehl R., Corr C., Baker B. (1994). The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78, 1101–1115. 10.1016/0092-8674(94)90283-6 [DOI] [PubMed] [Google Scholar]
- Xie K., Yang Y. (2013). RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant 6, 975–1983. 10.1093/mp/sst119 [DOI] [PubMed] [Google Scholar]
- Xu H. Y., Zhang C., Li Z. C., Wang Z. R., Jiang X. X., Shi Y. F., et al. (2018). The MAPK kinase kinase GmMEKK1 regulates cell death and defense responses. Plant Physiol. 178, 907–922. 10.1104/pp.18.00903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan Y., Zhou S., Wang L., Cheng Y., Zhao L. (2010). Nitric oxide functions as a signal and acts upstream of AtCaM3 in thermotolerance in Arabidopsis seedlings. Plant Physiol. 153, 1895–1906. 10.1104/pp.110.160424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., et al. (2015). High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8, 1820–1823. 10.1016/j.molp.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Yang H., Mu J., Chen L., Feng J., Hu J., Li L., et al. (2015). S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 167, 1604–1615. 10.1104/pp.114.255216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B. W., Feechan A., Yin M., Saidi N. B., Le Bihan T., Yu M., et al. (2011). S-Nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268. 10.1038/nature10427 [DOI] [PubMed] [Google Scholar]
- Yun B. W., Skelly M. J., Yin M., Yu M., Mun B. G., Lee S. U., et al. (2016). Nitric oxide and S-nitrosoglutathione function additively during plant immunity. New Phytol. 211, 516–526. 10.1111/nph.13903 [DOI] [PubMed] [Google Scholar]
- Zaninotto F., Camera S. L., Polverari A., Delledonne M. (2006). Cross talk between reactive nitrogen and oxygen species during the hypersensitive disease resistance response. Plant Physiol. 141, 379–383. 10.1104/pp.106.078857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler D., Za hringer U., Gerber I., Dubery I., Hartung T., Bors W., et al. (2004). Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. U.S.A. 101, 15811–15816. 10.1073/pnas.0404536101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan N., Wang C., Chen L., Yang H., Feng J., Gong X., et al. (2018). S-nitrosylation targets GSNOR reductase for selective autophagy during hypoxia responses in plants. Molecular. Cell. 71, 142–154. 10.1016/j.molcel.2018.05.024 [DOI] [PubMed] [Google Scholar]
- Zhang J., Liao W. (2019). Protein S-nitrosylation in plant abiotic stresses. Funct. Plant Biol. 47, 1–10. 10.1071/FP19071 [DOI] [PubMed] [Google Scholar]
- Zhou J., Zhang Y. (2020). Plant immunity: danger perception and signaling. Cell. 18, 978–989. 10.1016/j.cell.2020.04.028 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.