Summary
This protocol describes stable in vivo recordings of neuronal membrane potential in awake behaving, head-fixed mice. Previous protocols often highlight the need to minimize animal movements by anesthesia or restraint. This protocol is optimized to minimize brain movements during animal motion and has been used to record neurons in the olfactory bulb and visual cortex during active licking and locomotion behaviors. Under optimal conditions, success rates lie between 30% and 50% (recordings per microelectrode), with durations of up to 30 min.
For complete details on the use and execution of this protocol, please refer to Jordan et al. (2018) and Jordan and Keller (2020).
Subject areas: Single cell, Model organisms, Neuroscience
Graphical Abstract

Highlights
-
•
Protocol for stable head plate and recording chamber implantation
-
•
Instructions for stable and clean craniotomy and durectomy
-
•
Step-by-step guide for blind whole-cell recordings in mouse dorsal brain structures
-
•
Additional instructions for pipette retraction after neuronal filling
This protocol describes stable in vivo recordings of neuronal membrane potential in awake behaving, head-fixed mice. Previous protocols often highlight the need to minimize animal movements by anesthesia or restraint. This protocol is optimized to minimize brain movements during animal motion and has been used to record neurons in the olfactory bulb and visual cortex during active licking and locomotion behaviors. Under optimal conditions, success rates lie between 30% and 50% (recordings per microelectrode), with durations of up to 30 min.
Before you begin
Before attempting whole-cell recordings in behaving animals, a recording rig must be prepared with the following essential elements (example products in Materials and equipment):
-
1.
A typical denoized whole-cell recording system, consisting of a patch-clamp amplifier, head stage, microelectrode holder, 4-axis micromanipulators, oscilloscope, and a 20 kHz (or more) data acquisition device and software.
-
2.
A rigid head-fixation system, with the parts connecting to the head plate ideally electrically isolated from the grounding point.
-
3.
A stereoscope for visual targeting of the micropipette to the craniotomy.
-
4.
A custom micropipette pressure system with digital pressure sensors for rapid optimization and reliable repetition of the protocol (e.g., design in Figure 1).
-
5.
A treadmill, running wheel, or air-supported ball to allow smooth locomotion and comfortable animal posture. Some “give” in the locomotion apparatus is essential, so that the animal cannot exert strong forces against the headplate implant.
-
6.
A puller program must also be optimized to generate 5–7 MΩ micropipettes. An example of an optimized micropipette shape is shown in Figure 2.
Figure 1.
Electronic micropipette pressure system design
(A) Front panel of the valve box, with digital pressure sensors for both high-pressure and low-pressure lines. Valves and connections are housed within this box. Left shows the remote for controlling for the valves. The two metal switches control each two-way valve to lock pressure into each line independently, while the red switch controls the three-way valve to switch between high- and low-pressure lines connecting to the micropipette tubing.
(B) Schematic of the system, with the low-pressure line in blue and high-pressure line in pink. Arrows show connections via 4–5 mm diameter tubing. Note that the pipette tip connected to the low-pressure line is for mouth control of the pressure. Pressure range shown in sensors indicates the expected range of values experienced for each line.
Figure 2.
Example micropipette shape
Example functional in vivo micropipette tip under 40× (top) and 100× magnification (bottom), made from filamented, fire-polished 1.5 mm diameter glass capillaries on a Narishige PC-10 puller. Ideal microelectrode tip diameter should be roughly 1 μm, corresponding to 5–7 MΩ resistance. A relatively long shank and rounded tip are ideal for patching in vivo.
Test the setup
Timing: up to 3 weeks
-
7.Test the pressure control system.
-
a.Assemble the pressure control system as shown in Figure 1.
-
b.Assess the low- and high-pressure lines. Load an empty micropipette onto the head stage, add 1 bar pressure (100 mbar for testing low-pressure line) and lock the pressure in. After 5 min, assess how much pressure is lost. In an adequately air-tight system, no more than 5% should be lost.
-
a.
-
8.Ensure the system enables whole-cell recordings in anesthetized mice.
-
a.Perform steps 1–9 (Implantation of head plate) and steps 15–22 (Craniotomy and durectomy) of the main protocol consecutively, under general injection anesthesia (e.g., ketamine/xylazine).
-
b.Transfer the anesthetized mouse to the electrophysiology rig and maintain body heat there using a heat-pad.
-
c.Attempt whole-cell recordings as outlined in steps 24–40 (Whole-cell recordings) of the main protocol, while monitoring and maintaining anesthesia.
-
d.Via successive rounds of improvement (see Troubleshooting), increase success rates of obtaining a stable (> 10 min duration) whole-cell recording up to 30%–50% (recordings per micropipette descended).
-
a.
Note: Unless stable recordings are possible in anesthetized mice, it will be extremely difficult to obtain recordings in awake mice.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| NaCl | Sigma-Aldrich | S9625 |
| MgSO4 | Sigma-Aldrich | M7506 |
| CaCl2 | Sigma-Aldrich | 499609 |
| Glucose | Sigma-Aldrich | G8270 |
| NaOH | Sigma-Aldrich | S8045 |
| KMeSO3 | Sigma-Aldrich | 83000 |
| KCl | Sigma-Aldrich | P9333 |
| EGTA | Sigma-Aldrich | 03777 |
| HEPES | Sigma-Aldrich | H3375 |
| Mg-ATP | Sigma-Aldrich | A9187 |
| Na2-GTP | Sigma-Aldrich | G8877 |
| Na2-phosphocreatine | Sigma-Aldrich | P7936 |
| KOH | Sigma-Aldrich | 757551 |
| Agar | Sigma-Aldrich | A9793 |
| Biocytin | Sigma-Aldrich | B4261 |
| Lidocaine | Bichsel AG | N/A |
| Ropivacain | Presenius Kabi AG | N/A |
| Metacam | Boehringer Ingelheim | N/A |
| Buprenorphine | Reckitt Benckiser Healthcare | N/A |
| Isoflurane | Piramal | N/A |
| Experimental models: organisms/strains | ||
| Mus musculus: C57BL/6 | Charles River Laboratories | IMSR_JAX:000664 |
| Software and algorithms | ||
| Spike2 Software | Cambridge Electronic design | RRID:SCR_000903 |
| Other | ||
| Multiclamp 700B | Molecular Devices | https://www.moleculardevices.com/ |
| Headstage CV-7B | Molecular Devices | https://www.moleculardevices.com/ |
| Mini 25 micromanipulators | Luigs Neumann | https://www.luigs-neumann.org/micromanipulators |
| Microelectrode holder, ISO-S-1.5G | G23 Instruments | https://g23instruments.com/ |
| AgCl 2 × 1 mm pellet ground electrode | World Precision Instruments | Order code: EP1 |
| AC to DC converter, Micro1401 | Cambridge Electronic Design | http://ced.co.uk/products/mic4in |
| Mouse treadmill or air-floated ball | Custom made | N/A |
| Head-fixation apparatus | Custom made | N/A |
| Pipette pressure control unit | Custom made | N/A |
| Oscilloscope TPS2000 | Tektronix | https://uk.tek.com/oscilloscope/tps2000 |
| Digital pressure sensors, DP-102-M-P and DP-101-M-P | Panasonic | N/A |
| 2-way solenoid valve, | SMC | Part #: VDW22PAA |
| 3-way solenoid valve | SMC | Part #: VDW250-5G-2-01F-Q |
| PC-10 microelectrode puller | Narishige | http://products.narishige-group.com/ |
| Borosilicate glass capillaries (outer diameter: 1.5 mm, inner diameter: 0.86 mm, length: 100 mm, filamented, fire-polished) | Sutter Instrument | Item #: BF150-86-10 |
| 0.22 μm syringe filter | Merck Millipore | N/A |
| Histoacryl tissue adhesive | B. Braun | https://www.bbraun.com/en.html |
| Paladur dental cement | Kulzer | https://www.kulzer.com |
| 30-gauge needle tips | BD Micro-Fine | N/A |
| 20-gauge needle tips | BD Micro-Fine | N/A |
| Mouse temperature controller, homeothermic blanket system | Stoelting | Item #: 50300 |
| Betadine surgical skin disinfectant | MundiPharma | N/A |
| Kwik-cast silicone sealant | World Precision Instruments | Order code: KWIK-CAST |
| Mouse stereotax | World Precision Instruments | Order code: 505313 |
| Isoflurane anesthesia system | Vet-tech | N/A |
| Humigel ophthalmic gel | Virbac Schweiz, AG | N/A |
| Digital compact dry bath/heating block | Thermo Scientific | N/A |
| Surgical tools: fine scissors, scalpel, fine forceps, coarse forceps | Fine Science Tools | https://www.finescience.com/en-US/ |
| Dental drill, with 0.5 mm tip drill bits | Meisinger | N/A |
| Standard light microscope (Axioskop 50) with 40× and 100× air objectives (Epiplan 40×/0.6 and 100×/0.8) | Zeiss | https://www.micro-shop.zeiss.com/ |
| TTL generator | Custom made | N/A |
| Photonic F3000 LED light source | Warner Instruments | Model #: F3000 |
| Standard surgical stereoscope, M50 | Leica | https://www.leica-microsystems.com/ |
Materials and equipment
Electrophysiology rig:
-
•
Patch-clamp amplifier (e.g., Multiclamp 700B, Molecular Devices).
-
•
Head stage, with micromanipulator mounting adapter (e.g., CV-7B for Multiclamp 700B, Molecular Devices).
-
•
Four-axis micromanipulators (e.g., Mini 25, Luigs Neumann), with digital control box and remote (e.g., SM-5, Luigs Neumann).
-
•
Microelectrode holder (e.g., ISO-S-1.5G, G23 Instruments).
-
•
1 mm AgCl ground electrode pellet (EP1, World Precision Instruments).
-
•
AC to DC converter for 20–30 kHz data acquisition (e.g., Micro1401, Cambridge Electronic Design).
-
•
Data acquisition software (e.g., Spike2, Cambridge Electronic Design).
-
•
Head-fixation apparatus, and corresponding head plates.
-
•
Treadmill, wheel, or air-supported ball to allow head-fixed locomotion.
-
•
2-channel oscilloscope.
-
•
Custom pipette pressure control unit (Figure 1). Components: digital pressure sensors for high (DP-102-M-P, Panasonic) and low-pressure ranges (DP-101-M-P, Panasonic), two 2-way 24 V solenoid valves (VDW22PAA, SMC), one 3-way 24 V solenoid valve (VDW250-5G-2-01F-Q, SMC), 3 switches, housing for front panel and remote, 24 V power source, 5 mm tubing, 50 mL syringe, and pipette tips for control by mouth.
-
•
5× magnification stereoscope for visual guidance of microelectrode descent.
-
•
Illumination source for the craniotomy.
-
•
Optional: If not built into the patch-clamp amplifier, a device for 20 Hz TTL generation to generate test pulse.
-
•
Optional: Relevant behavioral monitoring equipment (movement sensors, lick sensors, flow sensors, etc.)
For micropipette fabrication and filling:
-
•
Micropipette puller (e.g., PC-10, Narishige).
-
•
1.5 mm, fire-polished and filamented glass capillaries for microelectrode fabrication (e.g., BF150-86-10, Sutter Instrument).
-
•
Light microscope with 40× and 100× magnification air objectives for inspecting micropipette tip shape.
-
•
Intracellular solution (example recipe below).
For surgery:
-
•
Local and general analgesics (dissolved in sterile saline, 0.9% NaCl). Local: e.g., lidocaine, 10 mg/kg (<1 h duration of action), Ropivacain 3 mg/kg (4–8 h duration of action), injected subcutaneously around site of implant. General: e.g., Metacam, 5 mg/kg, subcutaneous (24 h duration of action), and/or buprenorphine, 0.1 mg/kg, subcutaneous (4–6 h duration of action).
-
•
Isoflurane anesthesia system.
-
•
Ophthalmic gel.
-
•
60°C water bath.
-
•
Surgical tools: fine scissors, scalpel, fine forceps, coarse forceps.
-
•
Dental drill, with 0.5 mm tip drill bits.
-
•
Cold sterile saline (0.9% NaCl).
-
•
30-gauge needle tips.
-
•
20-gauge needle tips.
-
•
Heat-pad and controller.
-
•
Surgical skin disinfectant.
-
•
Kwik-cast silicone sealant (World Precision Instruments).
-
•
Mouse stereotax.
-
•
Stereoscope (with up to 5× magnification).
-
•
Bath recording solution (recipe below).
-
•
Dental cement (Paladur, Kulzer).
-
•
Tissue adhesive (Histoacryl, B. Braun).
-
•
4% low melting point agar dissolved in bath recording solution (A9793, Sigma-Aldrich).
Alternatives: Note that the specific products listed above are examples that have been used successfully. Other similar products may suffice.
Bath recording solution (Cortex Buffer)
| Reagent | Final concentration (mM) | Amount to add (g) for a 1× 500 mL stock |
|---|---|---|
| NaCl | 126 | 3.68 |
| KCl | 5 | 0.19 |
| HEPES | 10 | 1.19 |
| MgSO4 | 2 | 0.12 |
| CaCl2 | 2 | 0.11 |
| Glucose | 12 | 1.08 |
| NaOH | add as needed to adjust pH to 7.3 | n/a |
| ddH2O | - | Up to 500 mL |
| Final osmolarity | 290 mOsm |
Note: Make with deionized water. Adjust to pH 7.3 with NaOH and to an osmolality of 290 mOsm, before storing at 4°C for up to a week. For longer periods of storage up to a year, store in 50 mL aliquots at −20°C).
Intracellular solution
| Reagent | Final concentration (mM) | Amount to add (mg) for a 1× 50 mL stock |
|---|---|---|
| KMeSO3 | 135 | 905.85 |
| KCl | 5 | 18.64 |
| EGTA | 0.1 | 1.90 |
| HEPES | 10 | 119.15 |
| Mg-ATP | 4 | 101.44 |
| Na2-GTP | 0.5 | 13.08 |
| Na2-Phosphocreatine | 4 | 51.00 |
| KOH | add as needed to adjust pH to 7.3 | n/a |
| ddH2O | - | Up to 50 mL |
| Final osmolarity | 280 – 290 mOsm |
Note: make the intracellular solution on ice with ultrapure deionized water, and add the ATP, GTP, and phosphocreatine last, after roughly adjusting the osmolarity to just above the final desired osmolarity, since these molecules are unstable. Due to their acidity, pH should be adjusted to 7.3 with KOH solution after addition of these molecules. Filter the solution with an 0.22 μm pore filter and store the final solution in 0.5–1 mL aliquots at −20°C for up to 6 months.
Alternatives: Many variants of intracellular solution have been made and used successfully for whole-cell recordings, both in vitro and in vivo. For example, potassium ions are often provided by K-Gluconate, instead of KMeSO3. 10 mM biocytin can also be added for filling neurons and recovering morphologies with post-mortem histology.
Step-by-step method details
Implantation of head plate
Timing: 1 h
The aim of this step is to implant a headplate for head fixation in a manner which minimizes brain movement. Note that strict sterile procedures must be adhered to in order to prevent infections. Mice should ideally be 5–6 weeks old at the start of the protocol for optimal recording success. Both male and female mice have been used successfully. The age of the mouse at the time of patching should be no greater than 12 weeks. In older animals, the chances of obtaining a recording drop, and access resistances of successful recordings tend to be higher, likely due to changes in the properties of the tissue and increases in bone thickness making the craniotomy more difficult to achieve cleanly.
See Figure 3 for a graphical demonstration of this stage.
-
1.Prepare mouse for surgery.
-
a.Anesthetize the mouse with isoflurane (3% for induction, 1.5% for maintenance).
-
b.Ensure the mouse is adequately anesthetized by monitoring breathing rate and assessing the toe pinch reflex.
-
c.Apply ophthalmic gel to the eyes.
-
d.Place the mouse on a heated pad to ensure body temperature is maintained.
-
e.Fix the mouse’s head securely in a stereotax.
-
a.
-
2.Prepare the surgical area.
-
a.Shave the fur off from the entire dorsal surface of the head.
-
b.Disinfect the shaved area.
-
c.Inject a local analgesic (e.g., lidocaine and ropivacain) subcutaneously around the site of the implant, prior to incision.
-
a.
-
3.
Remove a circular piece of skin overlying the skull using surgical scissors.
-
4.
Remove all periosteal soft tissue overlying the parietal and interparietal bones by scraping it away using a scalpel.
CRITICAL: Pay careful attention to remove all soft tissue overlying the sutures of the skull. Failure to remove all soft tissue can lead to destabilization of the implant.
Note: Moistening the skull with sterile saline allows clearer visualization of soft tissue.
-
5.
Seal the edges of the skin with tissue adhesive.
-
6.
When the bone is dry, use the dental drill to lightly roughen the bone surface, omitting the area where you want to record.
-
7.Adhere the skull bones together to optimize stability of the brain:
-
a.Apply tissue adhesive along the sutures between parietal and interparietal bones to adhere the bones together.
-
b.Once the glue is dry, repeat until 3–4 layers of glue have been applied, and no relative movement of the parietal and interparietal bones occurs when gentle forces are applied.
-
c.Apply a thin layer of tissue adhesive to the remaining skull surface.
-
a.
-
8.Affix the headplate to the skull.
-
a.Glue the headplate to the bone with tissue adhesive.
-
b.Apply ample dental cement to affix the headplate to the skull. Ensure all exposed skull is covered in cement, except the area overlying the brain region of interest.
-
a.
-
9.
Build 1–2 mm high recording chamber walls around the craniotomy site out of dental cement. (This is to allow space for later immersion of a 1 × 3 mm AgCl ground electrode pellet, alongside space for descent of the microelectrode).
CRITICAL: If the headplate is made of metal, ensure that all parts within the recording chamber are covered by dental cement to ensure electrical isolation of the recording chamber.
-
10.
Cover the craniotomy area with Kwik-cast silicone sealant.
-
11.
Inject the animal with a general analgesic (e.g., Metacam, buprenorphine).
-
12.
Provide general analgesia for 48 h following surgery, as well as wet food on the cage floor.
Figure 3.
Headplate implantation surgery
Left diagram shows the surgical area in which skin and periosteum is removed (dotted line represents the edge of surgical area), as well as relevant landmarks and bones of the skull. Below shows an example headplate design, which can be stabilized via two screws. Right boxes (1–5) show diagrams for key steps of the surgery. Blue shading indicates areas with tissue adhesive application, and pink shows areas with dental cement application (darker indicates a deeper layer).
Behavioral training and habituation to head fixation
Timing: 3–7 days
This step allows the animal to acclimatize to head fixation and perform smooth locomotion by the time of recording, such that disruptive movements do not occur during the recording. This time can also be used to train the animal on any specific behavioral paradigms being studied. Make sure to handle the mice regularly prior to this step, to habituate them to the experimenter.
-
13.
Habituate the mouse to head fixation: Head fix the mouse at the setup for up to 1 h each day, until the mouse shows regular locomotion, and shows a comfortable posture (e.g., a lowered tail, and no contortions of the body).
-
14.
Optional: train the mouse on the relevant head-fixed task.
Craniotomy and durectomy
Timing: 20 min
The goal of this step is to make a small clean craniotomy with part of the dura removed to ensure clean insertion of the micropipette. See Figure 4 for a graphical demonstration of this stage.
-
15.
Prepare enough micropipettes for the recording session (around 10–12), as well as a means of filling them (e.g., a syringe with a thin tapered tip filled with 250 μL intracellular solution, kept on ice).
-
16.
Prepare 4% low melting point agar (dissolved in bath recording solution) and maintain in a heat bath at 60°C.
-
17.
Anesthetize the mouse with isoflurane, as before (3% for induction, 1.5% for maintenance), and head fix the mouse at the surgery station on a heated pad to maintain body temperature.
-
18.
Inject a general analgesic (e.g., Metacam, buprenorphine).
-
19.Perform a small circular craniotomy, around 1 mm in diameter over the site of interest.
-
a.Remove the silicone covering the bone.
-
b.Drill the edge of the circle slowly, regularly pausing to irrigate the bone with cold bath recording solution. Dry the bone thoroughly before resuming drilling, and clear away bone particles to prevent contamination of the craniotomy.
-
c.Once the bone is thin enough, you will see the bone circle move when lightly pressed with forceps (check this while the bone is damp).
-
d.Use a 20-gauge needle, or fine forceps, to lift and remove the bone circle from the skull, while immersed in recording solution. Make sure that at no point the brain is touched by your instrument, and that the brain does not contact air (this can cause sticking of the dura to the brain surface, making it hard to remove cleanly).
-
e.Use a syringe to lightly wash the brain with recording solution until any small bleeds have stopped.
-
a.
-
20.Perform the durectomy:
-
a.Abruptly tap a 30-gauge needle tip perpendicularly downwards onto a hard, clean surface. This should bend the needle tip to create a very small hook.
-
b.With the hook side facing down toward the brain, carefully attempt to catch onto the dura using a shallow angle (<30°).
-
c.Once attached carefully tear and pull the dura to the side of the craniotomy with the hook, without touching the underlying brain surface directly. Remove as much dura as possible from the center of the craniotomy to provide ample space for electrode descent.
-
d.Use a Pasteur pipette to lightly wash the brain with bath recording solution until any small bleeds have stopped.
-
a.
Note: A light source at a shallow angle (<30°) is helpful for visualizing the dura.
-
21.
Remove the recording solution and place a drop of warm melted 4% agar onto the craniotomy.
CRITICAL: You are aiming for an 0.5–1 mm agar layer overlying the brain and surrounding bone, allowing clear visualization of the underlying brain surface for later electrode targeting. If this is too thin, brain movement will still occur, while if it is too thick, microelectrode clogging is more likely, and the brain will be less easy to target.
Note: Make sure this is not too hot by first allowing it to cool in the pipette tip for as long as possible without it solidifying (gelling point of the suggested agar is 36°C). Applying hot agar, exceeding 40°C, could damage the brain surface, however cooling/gelling will be very rapid (on the order of seconds) given the small volume of agar and the thinness of the layer.
-
22.
Once the agar solidifies, re-immerse in bath recording solution.
-
23.
Transfer the mouse to head fixation on the recording setup to recover from anesthesia for at least 20 min, and until the mouse shows regular locomotion, or (if performing a task) licking for reward.
Figure 4.
Craniotomy and durectomy
Boxes 1–6 show key steps of the procedure. Note that all stages take place submerged in bath recording solution (Cortex buffer), aside from the initial drilling in box 1.
Whole-cell recordings
Timing: up to 2 h
See Figure 5 for a graphical demonstration of this stage.
-
24.
Place the AgCl ground electrode into the recording chamber. Make sure this cannot move and is immersed when the chamber is full.
-
25.
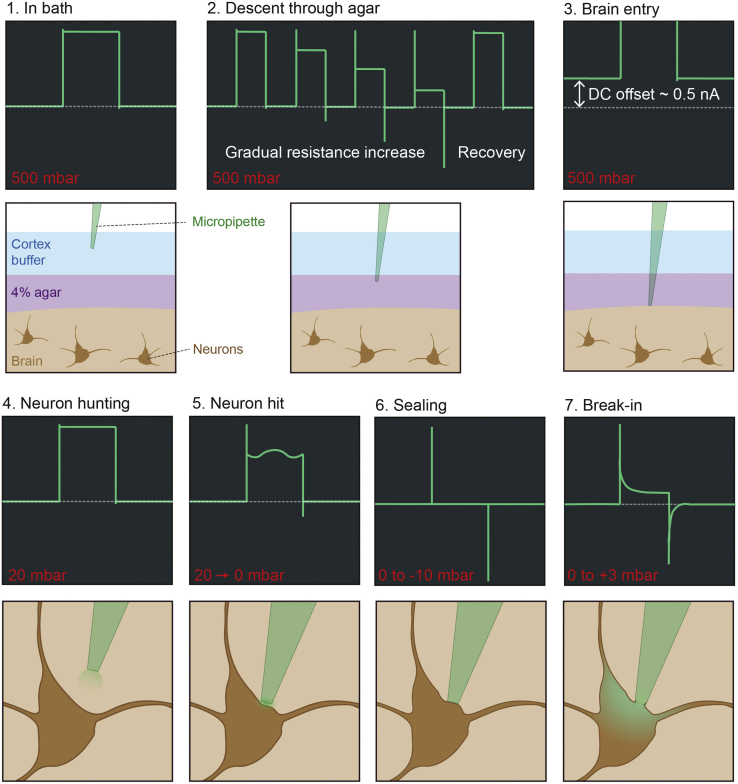
Backfill a micropipette with intracellular solution (make sure there are no trapped bubbles), load into the micropipette holder, and apply 500 mbar pressure using the high-pressure line. Apply a 10 mV voltage step at 20 Hz in voltage clamp for monitoring electrode resistance (display step-triggered electrode current on an oscilloscope).
-
26.
Descend the electrode into the recording solution. Note down the electrode resistance, which should be between 5 and 7 MΩ (Box 1, Figure 5).
CRITICAL: Do not allow the micropipette pressure to drop to 0 mbar or below while in the bath or tissue, until attempting to seal. This creates an opportunity for contamination of the electrode tip.
-
27.
Remove the bath recording solution, and visually guide the micropipette down until it touches the surface of the agar overlying the craniotomy (identified visually or through electrode current). Re-immerse the recording chamber.
-
28.
Slowly descend the micropipette into the agar at a slow speed (e.g., 30 μm/s) targeted at the craniotomy center (Box 2, Figure 5). As you move through the agar, you should see gradual increases in resistance of the micropipette which periodically decreases back to the starting resistance (i.e., a sawtooth profile of electrode resistance).
Note: If the resistance does not reduce periodically, the pipette is getting clogged on the way down. Retract the electrode to unclog the tip and descend again if this occurs (if this recurs, it may be due to too low microelectrode resistance, too low pressure, or too low agar concentration).
-
29.
Identify brain tissue entry: Watch out for a sudden change in current offset coinciding with a transient increase in resistance (Box 3, Figure 5). Zero the manipulator descent axis at this point.
Note: Do not move the micropipette laterally while in the tissue.
-
30.
Drive the micropipette down to around 100 μm above the depth of interest, switch the micropipette pressure line from high to low pressure, and apply 20 mbar pressure.
-
31.
Wait for 60 s, to allow the ejected internal solution to clear, and to allow the pressure and pipette offset to stabilize.
CRITICAL: Make sure electrode resistance is comparable to its initial resistance (when recorded in the bath). If this has increased or decreased, this indicates a clogged or broken pipette tip, respectively.
-
32.
Neuron hunting (Box 4, Figure 5): with 20 mbar pressure applied, start descending the micropipette in 2 μm steps.
Note: If possible, only perform neuron hunting while the animal is stationary. Small brain movements during locomotion can make hitting a neuron harder to detect.
-
33.
When electrode resistance increases consistently and substantially (a doubling) across 2–3 steps, this indicates collision with a neuron (Box 5, Figure 5). A mild oscillation on top of the current trace, caused by heart rate driven brain motion, is another good indicator of proximity to a neuron.
-
34.
Abruptly reduce pressure to 0 mbar (i.e., open the low-pressure line) to seal onto the cell membrane (Box 6, Figure 5). This is indicated by a rapid increase in electrode resistance toward the GΩ range. Applying gentle negative pressure (up to −10 mbar) can aid this.
-
35.
Slowly bring the command voltage down to −70 mV. This aids in sealing and prevents depolarization when breaking in.
-
36.
Carefully retract the micropipette 2–4 μm after GΩ resistance seal formation. If attempting cell filling for morphological reconstruction, see optional steps 41–45 below.
-
37.
Breaking in (Box 7, Figure 5): Using the low-pressure line, apply a brief and gentle suction pulse (up to −50 mbar, for 1 s) to break into whole-cell mode.
Note: Applying a small positive pressure (+3 mbar) in whole-cell mode can help prevent resealing.
-
38.
Switch to current clamp and record membrane potential as required by the experiment.
-
39.
Once finished, slowly retract the micropipette until outside of the tissue/agar.
-
40.
Between every 1–2 micropipette descents, very carefully remove the agar layer with fine forceps, and replace it with a fresh layer. This is to prevent agar layer break down and subsequent brain movement.
Figure 5.
Electrode resistance monitoring and obtaining of whole-cell recording
Boxes 1–7 show the stages of gaining a whole-cell recording via monitoring electrode resistance with a test pulse in voltage clamp. Green traces in the dark boxes are diagrams of the current trace as would be seen on an oscilloscope at various stages of the process. Dashed gray line shows 0 nA. Note that electrode offset is re-zeroed between “brain entry” and “neuron hunting.” The pressure range applied to the micropipette is shown in red for each stage (note: arrows between pressure values indicate a rapid transition between pressures). Diagrams below each oscilloscope diagram depict what is happening at the micropipette tip for each stage (see also Margrie et al., 2002; Rancz et al., 2011).
Neuronal filling and morphology recovery (optional)
Timing: 1 h
If neuronal morphology is desired, the following steps should also be followed during whole-cell recordings. Note that this will further limit the number of recordings obtained to one per mouse, to be sure of matching the morphology to the recording. In my experience, morphological recovery has particularly low success rates in the awake mouse, with only partial morphological recoveries most likely. See Figure 6 for a graphical depiction of this stage.
-
41.
Perform recordings as outlined in steps 24–38 (Whole-cell recordings), with an intracellular solution additionally containing 10 mM biocytin or neurobiotin. For a good neuronal fill, recording duration should be between 5 and 10 min, with an electrode resistance on the lower end of the workable range (5–6 MΩ), and an access resistance below 80 MΩ.
-
42.
Retract the microelectrode slowly after 5–10 min of recording, while the recording quality is still high, and monitor the electrode current with a 10-mV test pulse in voltage clamp, as before. For a successful fill, microelectrode resistance should gradually increase as the microelectrode is retracted (at 5–10 μm/s, until resealing), reflecting the formation of an outside-out patch.
-
43.
Once outside of the tissue and in the bath, applying a strong positive pressure to the micropipette should return the electrode resistance to close to its original value. This is an indicator of a good retraction that has retained minimal cellular material in/on the micropipette.
-
44.
Fix the brain with 4% paraformaldehyde (PFA) within an hour of recording via transcardial perfusion and store the brain for 24 h in 4% PFA.
-
45.
For a detailed procedure for biocytin/neurobiotin staining for morphological reconstruction, see Marx et al., 2012.
Figure 6.
Retraction after neuronal filling for morphological reconstruction
Boxes 1–3 show electrode current signatures for a good chance of morphological recovery. After filling the neuron in whole-cell mode for 5–10 min, the pipette is slowly retracted before access resistance deteriorates. This evokes membrane resealing and formation of an outside-out patch on the electrode tip that is indicated by a high electrode resistance. Green coloration of the neuron in the diagram indicates intracellular solution that has diffused into the cytoplasm from the micropipette tip.
Expected outcomes
In optimal conditions, this protocol allows whole-cell recording success rates between 30% and 50% of descended micropipettes. Of the successful recordings, usable recording time typically is between 3 and 30 min (mean ± standard deviation: 14 ± 4 min), with minimal changes in access resistances during a variety of mouse movements (see Table 1 and Figure 7).
Table 1.
Recording parameters from neurons recorded in behaving animals from the olfactory bulb (Jordan et al., 2018) and V1 (Jordan and Keller, 2020)
| Quantification | Putative Mitral/tufted cells | Putative pyramidal neurons |
|---|---|---|
| Number of included recordings in dataset | 57 | 46 |
| Input resistance, mean ± SD (included recordings) | 58 ± 32 MΩ | 42 ± 20 MΩ |
| Input resistance range (included recordings) | 17–138 MΩ | 14–86 MΩ |
| Access resistance, mean ± SD (included recordings) | 41 ± 18 MΩ | 56 ± 21 MΩ |
| Access resistance range (included recordings) | 10–91 MΩ | 25–140 MΩ |
| Duration, mean ± SD (included recordings) | 14 ± 5 min | 14 ± 4 min |
| Duration range (included recordings) | 5–26 min | 3–32 min |
| Number of rejected recordings (due to duration < 3 min or access resistance > 150 MΩ) | 46 | 25 |
| Number of included recordings per craniotomy, mode (range) | 1 (1–3) | 1 (1–3) |
Figure 7.
Example traces of stable recordings during different mouse behaviors
Example membrane voltage traces (black) recorded from a putative mitral neuron in the olfactory bulb (top trace, adapted from Jordan et al., 2018) and a putative pyramidal neuron in layer 2/3 of primary visual cortex (bottom trace, adapted from Jordan and Keller, 2020), during different behaviors: licking in a task and locomotion in virtual reality, respectively.
At the start of the recording, access resistances are around 41 ± 18 MΩ (mean ± standard deviation) in putative principal neurons of the olfactory bulb (Jordan et al., 2018) and 56 ± 21 MΩ in putative pyramidal neurons in V1 (Jordan and Keller, 2020). Access resistance does tend to gradually increase and therefore should be carefully monitored throughout the recording. If access resistance increases beyond acceptable levels, the experimenter may attempt to “re-open” the patch the same way one would enter whole-cell mode initially. However, in many cases this leads to loss of the recording.
In my experience, up to around 12 micropipettes can be descended into a 1 mm craniotomy, because success rates tend to drop with consecutive descents and are very low beyond 12. This is likely due to damage to the tissue with consecutive descents. If the targeted area is not too small, and the stereotactic coordinates of the descent are not too critical (e.g., in the olfactory bulb, which is anatomically rather isolated from other regions), one can attempt to visually target each electrode to different regions of the craniotomy. It is generally a good idea to vary the X-Y coordinates by around 50–100 μm with each descent, to avoid descending in the very same area. One must also keep in mind the length of time that mice can be kept comfortably head fixed and show the relevant behavior – for rewarded tasks this may be lower due to eventual satiation, while for spontaneous behaviors like locomotion, this will depend on the individual mouse, and the length of training time. Typically, one can record between one and three neurons per animal, however in a subset of mice conditions may be too poor to make whole-cell recordings that are stable during movement (5%–10% of mice).
Limitations
While brain motion caused by animal movement will be minimized using the steps outlined in this protocol, it is often not possible to eliminate motion entirely. This means that there will often be slow changes in access resistance which should be carefully monitored so that data can be excluded if this gets too high or variable. This also means that recording durations will be limited, with usable traces ranging between 3 and 30 min in duration.
If the experimental question necessitates quantifying changes over time (e.g., learning-related changes), control animals should also be used to assess the effects of changes due to intracellular solution dialysis or increases in access resistance.
Due to the need for a clean brain surface with minimal regrowth and bleeding, recordings are performed within 2 h of the craniotomy and durectomy surgery. Due to the invasiveness of this surgery, anesthesia and analgesia must be applied to the animal. While isoflurane has been chosen due to its short half-life and rapid behavioral recovery, it is possible that the anesthesia or analgesic drugs could affect some aspects of neuronal or network physiology, which should be kept in mind when interpreting the results.
It must also be noted that in the awake animal (compared to anesthetized and in vitro recordings), access resistances will typically be higher and more variable, while input resistances are typically very low (putative mitral and tufted neurons: 58 ± 32 MΩ (mean ± standard deviation), putative pyramidal neurons: 42 ± 20 MΩ). This creates highly suboptimal conditions for voltage clamp due to space clamp errors (Williams and Mitchell, 2008).
Troubleshooting
Problem 1: Unstable electrical signals during descent through the bath solution (step 26)
If you see large changes in the electrode offset as you move the micropipette through the recording solution, the recording chamber is unlikely to be electrically isolated.
Potential solution
Look for gaps in the dental cement where the recording solution may be contacting the metal headplate and creating a path to ground. Electrically isolating the entire head fixation apparatus, e.g., using PEEK instead of metal headplates or headplate holders, can also help prevent this.
Problem 2: Micropipette resistance increases after descent into tissue (step 31)
If the resistance of the micropipette increases between the recording chamber and the entry into the tissue, detecting a hit with a neuron and obtaining a recording will be very difficult. This is likely due to a lack of clean entry into the tissue.
Potential solution
This could be due to a lack of high enough pressure during the initial descent. Try increasing the descent pressure by 100 mbar at a time. Make sure the pipette pressure system is not leaking.
Make sure the dura is well cleared from the center of the craniotomy prior to attempting recording. Visually guide the micropipette descent to avoid superficial blood vessels. Check for bleeds on the brain surface. If there are no large clean areas to target the micropipette, obtaining a recording will be extremely difficult – at this point it is best to terminate the experiment.
Problem 3: Inability to detect neuronal hits (step 33)
Sometimes micropipette resistance is unchanged after descent, but collisions with neurons are hard or impossible to detect.
Potential solution
(A) This could be due either to too low or too high search pressure during “neuron hunting”: if the micropipette resistance changes often during stepping, but neuron hits are not obvious, try increasing the search pressure by 5 mbar each time. If micropipette resistance barely ever shows a change during stepping, try reducing the search pressure by 5 mbar each time.
(B) Depending on the structure being targeted, altering the descent angle may aid in increasing the chances of a clean somatic hit.
Problem 4: Inability to obtain a GΩ seal (step 34)
In some cases, you can detect a neuronal hit, but it is difficult to obtain a GΩ resistance seal.
Potential solution
Common causes for this problem are:
First, the micropipette tip is getting contaminated during descent (a clean glass surface is crucial to sealing). For this, see potential solutions to Problem 2.
Second, the pressure system is not performing well. Make sure that there are no leaks or blockages in the pressure system, and that fast step changes in pressure can be achieved. This can be checked by loading an empty micropipette, breaking the tip and applying changes in pressure while immersed in solution – you should be able to control the rate of bubbles coming out of the pipette tip on a fast timescale.
Third, the micropipette resistance is too low (< 5 MΩ). Make micropipettes with resistances of 5–7 MΩ.
Finally, the micropipette tip shape is suboptimal. A somewhat rounded tip, as shown in Figure 2, is ideal.
Problem 5: Inability to break in after GΩ seal formation (step 37)
Sometimes it will be possible to get a GΩ resistance seal onto a cell, but it will not be possible to break in despite repeated attempts.
Potential solution
This is usually due to the micropipette resistance being too high (i.e., the tip diameter is too narrow). Make lower resistance micropipettes (5–7 MΩ).
Sometimes you will seal onto something other than a neuron, for example a blood vessel. Watch the current trace for characteristic features of hitting a neuron, such as a mild oscillation on top of the current step. Hitting blood vessels (or other non-neural structures) often looks subtly different.
Problem 6: Sudden increase of access resistance or loss of recording (step 38)
Slow increases in access resistance are common in awake recordings and cannot be fully prevented. However, sudden increases, or sudden losses of the recording altogether (e.g., Figure 8), especially if frequent, usually indicate significant mechanical instability.
Figure 8.
Example loss of high-quality olfactory bulb recording during mouse movement
Dotted line indicates 0 mV.
Potential solution
Make sure to carefully replace the 4% agar layer every 1–2 pipette descents, as gradual break down of this layer will allow the brain to move.
Check that all parts of the head-fixation apparatus are rigid and there are no loose screws.
If this occurs more than once in the same animal, and the head-fixation apparatus appears rigid, this is unlikely to be fixable and the experiment should be terminated. After euthanizing the animal, check for any instabilities in the headplate (for example, gaps between cement and bone, which are often caused by incomplete removal and regrowth of the periosteum).
Repeated, sudden increases in access resistance can also be caused if there are clogging particles in the intracellular solution. This can often be determined by applying suction to the micropipette – if the electrode resistance drops when negative pressure is applied, and increases when the positive pressure is applied, this is due to a blockage within the micropipette tip. Check the failed micropipette under a light microscope to look for particulates. Potential culprits include dusty glass capillaries or improperly filtered intracellular solution.
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Rebecca Jordan (rebecca.jordan@fmi.ch).
Materials availability
No new mouse lines or materials were generated for this protocol.
Data and code availability
No new data or code was generated for this protocol.
Acknowledgments
I would like to thank Andreas Schaefer, Izumi Fukunaga, Georg Keller, Mihaly Kollo, Ede Rancz, and members of Troy Margrie’s lab for their advice and support in fine-tuning this protocol over the years. R.J. is funded by a Human Frontiers Science Program Long Term Fellowship (LT000077/2019-L). This protocol was written by R.J. while working in the lab of Georg Keller at the Friedrich Miescher Institute, who fully supports this work.
Author contributions
R.J. wrote the protocol.
Declaration of interests
The author declares no competing interests.
References
- Jordan R., Keller G.B. Opposing influence of top-down and bottom-up input on excitatory layer 2/3 neurons in mouse primary visual cortex. Neuron. 2020;108:1194–1206.e5. doi: 10.1016/j.neuron.2020.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan R., Fukunaga I., Kollo M., Schaefer A.T. Active sampling state dynamically enhances olfactory bulb odor representation. Neuron. 2018;98:1214–1228.e5. doi: 10.1016/j.neuron.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margrie T.W., Brecht M., Sakmann B. In vivo, low-resistance, whole-cell recordings from neurons in the anaesthetized and awake mammalian brain. Pflugers Arch. 2002;444:491–498. doi: 10.1007/s00424-002-0831-z. [DOI] [PubMed] [Google Scholar]
- Marx M., Günter R.H., Hucko W., Radnikow G., Feldmeyer D. Improved biocytin labeling and neuronal 3D reconstruction. Nat. Protoc. 2012;7:394–407. doi: 10.1038/nprot.2011.449. [DOI] [PubMed] [Google Scholar]
- Rancz E.A., Franks K.M., Schwarz M.K., Pichler B., Schaefer A.T., Margrie T.W. Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics. Nat. Neurosci. 2011;14:527–532. doi: 10.1038/nn.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.R., Mitchell S.J. Direct measurement of somatic voltage clamp errors in central neurons. Nat. Neurosci. 2008;11:790–798. doi: 10.1038/nn.2137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data or code was generated for this protocol.