Abstract
Objective
Brown adipose tissue (BAT) thermogenesis offers the potential to improve metabolic health in mice and humans. However, humans predominantly live under thermoneutral conditions, leading to BAT whitening, a reduction in BAT mitochondrial content and metabolic activity. Recent studies have established mitophagy as a major driver of mitochondrial degradation in the whitening of thermogenic brite/beige adipocytes, yet the pathways mediating mitochondrial breakdown in whitening of classical BAT remain largely elusive. The transcription factor EB (TFEB), a master regulator of lysosomal biogenesis and autophagy belonging to the MiT family of transcription factors, is the only member of this family that is upregulated during whitening, pointing toward a role of TFEB in whitening-associated mitochondrial breakdown.
Methods
We generated brown adipocyte-specific TFEB knockout mice, and induced BAT whitening by thermoneutral housing. We characterized gene and protein expression patterns, BAT metabolic activity, systemic metabolism, and mitochondrial localization using in vivo and in vitro approaches.
Results
Under low thermogenic activation conditions, deletion of TFEB preserves mitochondrial mass independently of mitochondriogenesis in BAT and primary brown adipocytes. However, this does not translate into elevated thermogenic capacity or protection from diet-induced obesity. Autophagosomal/lysosomal marker levels are altered in TFEB-deficient BAT and primary adipocytes, and lysosomal markers co-localize and co-purify with mitochondria in TFEB-deficient BAT, indicating trapping of mitochondria in late stages of mitophagy.
Conclusion
We identify TFEB as a driver of BAT whitening, mediating mitochondrial degradation via the autophagosomal and lysosomal machinery.
This study provides proof of concept that interfering with the mitochondrial degradation machinery can increase mitochondrial mass in classical BAT under human-relevant conditions. However, it must be considered that interfering with autophagy may result in accumulation of non-functional mitochondria. Future studies targeting earlier steps of mitophagy or target recognition are therefore warranted.
Keywords: TFEB, Brown adipose tissue, Whitening, Mitophagy, Thermogenesis, UCP1
Graphical abstract
Highlights
-
•
BAT whitening is associated with TFEB-dependent mitochondrial degradation.
-
•
TFEB-KO BAT retains higher mitochondrial levels upon thermoneutral adaptation.
-
•
This does not translate into higher thermogenic capacity or protection from obesity.
-
•
TFEB is dispensable for thermogenic adipose tissue function in the cold.
-
•
Mitochondria in thermoneutral TFEB-KO BAT are presumably trapped in lysosomes.
1. Introduction
The re-discovery of brown adipose tissue (BAT) in adult humans [[1], [2], [3], [4], [5]] as well as the negative correlation of detectable BAT with age and obesity in humans [2,3,6] have brought this tissue to the attention of metabolic research.
Cold exposure leads to the activation of brown adipose tissue via the sympathetic nervous system, triggering an adrenergic signaling cascade within brown adipocytes, ultimately resulting in the activation of non-shivering thermogenesis via the inner mitochondrial membrane protein-uncoupling protein 1 (UCP1) [7]. To meet the high metabolic demand of this process, BAT takes up substantial amounts of nutrients, mainly in the form of lipoprotein-derived triglycerides [[8], [9], [10]] but also fatty acids [9,[11], [12], [13]] and glucose [9,[14], [15], [16]]. This can consequently lead to weight loss and improvements in lipid and glucose metabolism in mice and humans [6,8,10,[17], [18], [19], [20], [21]].
However, while cold exposure undoubtedly exerts various beneficial metabolic effects, humans spend most of their time in thermoneutral or near-thermoneutral environments and rarely experience prolonged cold exposure due to well-controlled temperatures in homes and workplaces [22,23]. For clothed humans, the thermoneutral zone is approximately 22 °C [[23], [24], [25]], while mice display a thermoneutral zone at around 30 °C [[26], [27], [28], [29], [30]]. Thermoneutral housing of mice has been shown to have a tremendous impact on the outcome of metabolic experiments [[31], [32], [33], [34], [35]] and directly affects the recruitment state of BAT [7,[36], [37], [38]]. Under thermoneutral conditions, BAT oxidative capacity is strongly reduced, while lipid accumulation is increased, leading to a “white-like” appearance of the tissue [39]. Thermoneutral housing (especially in conjunction with high-calorie diets) can be used to thermally humanize mice [40,41], and BAT of mice housed under such conditions remarkably resembles human BAT [41,42].
During thermoneutral adaptation and aging, BAT and brite/beige adipocytes lose their metabolic capacity and mitochondrial content [[43], [44], [45]]. Thus, understanding the mechanism driving BAT whitening might help identify novel pharmacological targets to preserve or regain BAT metabolic capacity in humans. The temperature-induced whitening process in brite/beige adipose tissue has been shown to depend on mitophagy [46,47], while direct evidence of a role of mitophagy in BAT whitening at thermoneutrality remains lacking.
The lysosome- and autophagy-regulating transcription factor TFEB is a member of the MiT family consisting of microphthalmia-associated transcription factor (MITF), transcription factor E3 (TFE3), transcription factor EB (TFEB), and transcription factor EC (TFEC), which regulate the expression of genes within the so-called CLEAR network of lysosomal proteins [[48], [49], [50]]. TFEB and TFE3 not only regulate the expression of lysosomal genes but also the autophagosomal machinery [51], thereby controlling mitochondrial degradation [52] and whole-body metabolism by affecting mitochondrial dynamics in the liver [53]. Autophagy has also been shown to directly influence the differentiation of adipocytes [54,55]. Interestingly, TFEB and TFE3 interact with the master regulators of mitochondrial biogenesis, PGC1-alpha and PPAR-gamma [56,57], regulating mitochondrial mass and exercise endurance in skeletal muscle [58,59]. TFEB overexpression in livers also resulted in enhanced hepatic lipid metabolism, positively influencing whole-body metabolism [60]. Given the overwhelming evidence of a fundamental role of PGC1/PPARG-mediated mitochondrial biogenesis for cold-induced brown and brite/beige adipose tissue thermogenic function [61,62] it has been hypothesized that TFEB might affect these processes via a direct interplay with the PGC1A pathway. Indeed, overexpression of TFEB in adipose tissue has been shown to result in enhanced PGC1A-dependent mitochondrial biogenesis and browning of white adipose tissue, resulting in improved metabolic health in diet-induced obesity, independent of TFEB's effects on lysosomal homeostasis [63].
In the present study, we show that genetic deletion of TFEB in BAT led to preserved protein levels of UCP1 and respiratory chain complexes under thermoneutral conditions. Notably, this represents a cell autonomous process that was not linked to increased mitochondrial biogenesis but rather to decreased mitochondrial degradation via the autophagosomal machinery. However, gene expression of known TFEB target genes was only modestly affected by TFEB deficiency, indicating that the homeostatic expression of lysosomal genes in BAT is most likely mediated by other transcription factors. Surprisingly, despite increased UCP1 levels, TFEB deficiency did not protect from diet-induced obesity and associated metabolic abnormalities. This was accompanied by a lower metabolic response to injection of the adrenergic agonist CL316,243, a well-known activator of BAT thermogenesis. This indicates that higher levels of UCP1 were not readily activatable, presumably due to trapping of mitochondria within lysosomes/autophagosomes or an accumulation of damaged mitochondria. Overall, this shows that TFEB-dependent mitophagy/autophagy modulates BAT whitening, while other transcription factors seem to be responsible for regulating lysosomal biogenesis in BAT under homeostatic conditions.
2. Materials and methods
2.1. Mice and standard experimental procedures
All mouse experiments were approved by the Animal Welfare Officers of University Medical Center Hamburg-Eppendorf (UKE) and Behörde für Gesundheit und Verbraucherschutz Hamburg. To generate brown adipocyte-specific TFEB-deficient mice, TFEBfl/fl mice [60] were crossed with mice expressing Cre-recombinase under the control of the UCP1 promotor (Jackson Laboratories stock #024670 B6.FVB-Tg(Ucp1-Cre)1Evdr). These mice will hereafter be referred to as TFEBBAT KO. Littermates without Cre transgene expression were used as controls (TFEBBAT WT). The mice were housed at 30 °C, 22 °C, or 6 °C in a temperature-controlled cabinet (Memmert) in a 12-h light–dark rhythm with ad libitum access to water and a standard laboratory chow diet (P1324, Altromin) unless otherwise indicated. Obesity was induced by feeding a high-fat diet (HFD) (EF Bio-Serv no. F3282 mod., 60% calories from fat, Sniff) starting at 6 weeks of age for 16 weeks at thermoneutrality. For all thermoneutral adaptation studies, the mice were previously housed at room temperature and transferred to thermoneutrality for 1 or 4 weeks as indicated in the figure legends and results section.
The 4-h fasted mice were euthanized with CO2 followed by cervical dislocation or decapitation. Organs were harvested, and samples for RNA and protein isolation were snap-frozen in liquid nitrogen and stored at −80 °C for further analysis. Samples for histology were fixed in 4% v/v formaldehyde in PBS at 4 °C. Detailed procedures are described in the following sections.
2.2. Indirect calorimetry and body temperature measurements
For body temperature measurements in response to cold ambient temperatures, rectal temperature was measured using a BAT-12 Microprobe thermometer (Physitemp).
One week prior to indirect calorimetry and continuous body temperature measurements, the mice were implanted with telemetric transponders (G2 E-Mitter, Respironics) in the peritoneal cavity under isoflurane anesthesia as previously described [27]. Indirect calorimetric measurements were performed using a TSE PhenoMaster system. The mice were transferred to the measurement chamber in new cages without additional nesting material one day before the measurements started to reduce measurement artifacts caused by stress. The mice were maintained at a 12-h light–dark rhythm with free access to a standard chow diet and water. Oxygen and carbon dioxide levels in each cage were measured for 2.5 min in each 15 min measurement interval. The respiratory quotient (RQ) was calculated as the ratio of carbon dioxide produced to oxygen consumed. Energy expenditure (EE) was calculated using a modified Weir equation [64,65].
For gradual cold exposure studies, the TFEBBAT WT and TFEBBAT KO mice were acclimated at 22 °C and gradually exposed to 30 °C, 22 °C, and 6 °C for 24 h starting at 7:00 am. To investigate the capacity for non-shivering thermogenesis, the TFEBBAT WT and TFEBBAT KO mice were acclimated to 30 °C for one week and then transferred to metabolic chambers. The next day, non-shivering thermogenesis was induced at 30 °C by subcutaneous injection of 1 mg/kg body weight CL316,243. Baseline metabolic rates, RQ, and body temperature were defined as the levels over a time of 4 h starting 5 h before injection. The response to CL was quantified as the metabolic rates over 4.5 h starting 30 min after CL injection. ΔEE was calculated by normalization of the CL response values to baseline.
2.3. Metabolic tracer studies
To investigate the uptake of glucose and lipids into metabolically active tissues, 4-h fasted mice received an oral gavage containing 50 mg/kg body weight triglycerides from intralipid and 2 g/kg body weight glucose with 3H-triolein (0.72 MBq/kg) and 2-deoxy-D[14C]-glucose (0.15 MBq/kg) or with 2-deoxy-D[3H]-glucose (0.72 MBq/kg) in a total volume of 300 μL. Two h after oral gavage, the mice were anesthetized with ketamine (300 mg/kg) and xylazine (30 mg/kg) (i.p.) and transcardially perfused with PBS heparin (10 U/mL). The samples were homogenized in 10-fold (v/w) Solvable (PerkinElmer) and counted using a Tri-Carb scintillation counter (PerkinElmer).
2.4. Oral glucose tolerance tests and plasma parameters
Oral glucose tolerance tests were performed in 4-h fasted TFEBBAT WT and KO mice on the HFD using an oral gavage of glucose (2 g/kg body weight). Blood samples were obtained via a small cut on the tip of the tail. Blood glucose concentrations were measured using commercially available AccuCheck Aviva sticks (Roche). Plasma insulin was measured using a rat/mouse insulin assay kit (Chrystal Chem). Plasma cholesterol, triglycerides, and non-esterified free fatty acids (NEFAs) were measured using a Cholesterol CHOD-PAP kit (Roche), a Triglycerides GOP-PAP kit (Roche), and a NEFA-HR(2) kit (FUJIFILM Wako Diagnostics), respectively, according to the manufacturers' protocol.
2.5. Cell cultures
Three- to six-week-old mice were euthanized with CO2 followed by cervical dislocation. Their iBAT was harvested under sterile conditions and digested in isolation buffer (125 mM of NaCl, 5 mM of KCl, 1.3 mM of CaCl2, 5 mM of glucose, and 100 mM of HEPES at a pH of 7.4) containing collagenase II (600 U/mL, Merck) and 1.5% w/v BSA for 40 min at 37 °C. The stromal vascular fraction was collected after filtration through a 100 μm nylon filter and 30 min of incubation on ice. The samples containing pre-adipocytes were cultured and differentiated to mature brown adipocytes in DMEM Glutamax-I 4.5 g/L of glucose containing 10% NCS (Sigma, lot number 15D230), 1% penicillin and streptomycin, 1% antibiotic-antimycotic, 2.4 nM of insulin (Sigma), and 1 μM of rosiglitazone (Sigma).
For differentiation studies, cells were harvested after 4, 6, and 8 days of differentiation in TriFast (Peqlab) for mRNA isolation. Adipocytes were starved in starvation medium (DMEM Glutamax-I 1 g/L glucose containing 0.1% NCS (Sigma, lot number 15D230)). To analyze the autophagic flux in vitro, mature brown adipocytes were treated with 50 μM of chloroquine (Sigma) for 2 h to prevent lysosomal acidification. The cells were subsequently harvested in RIPA buffer and proteins were isolated as described below.
2.6. Isolation of mitochondria from brown adipose tissue
To isolate mitochondria from brown adipose tissue [38,66], the mice were exposed to 30 °C for 2 days and either fasted or fasted and subsequently refed. iBAT and scBAT of 5 mice were pooled in ice-cold 0.25 M sucrose. Tissues were minced, further homogenized in a glass homogenizer, filtered through a 250 μm nylon filter, and centrifuged at 8500 g (JA-25.50, Beckman Coulter) for 10 min. The supernatant was discarded by inverting the tube, the pellet was resuspended in 0.25 M of sucrose, homogenized in a glass homogenizer, and centrifuged at 800×g for 10 min. The supernatant containing mitochondria was centrifuged at 8500×g for 10 min and the mitochondrial pellet was resuspended in TES buffer (100 mM of KCl, 20 mM of TES, 0.6% BSA, and pH of 7.2) to induce mitochondrial swelling. After centrifugation at 8500 g for 10 min, the supernatant was discarded, and the mitochondria were resuspended in the remaining TES buffer. The solution was transferred to a small glass homogenizer, homogenized, and pelleted. The weight of the mitochondrial pellet was determined, and the mitochondria were stored at −80 °C. For immunoblotting analysis, the mitochondria were resuspended in 10 times v/w RIPA buffer and sonicated twice for 30 s at 20% amplitude, with a 30 s incubation on ice in between. Protein concentration measurement and immunoblotting was performed as described below.
2.7. RNA isolation and gene expression analysis
RNA was extracted from BAT or WAT with TriFast (Peqlab) and a NucleoSpin RNAII kit (Macherey and Nagel) according to the manufacturer's protocol using standard procedures [67]. cDNA was synthesized from 400 ng of RNA using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer's protocol. Quantitative real-time PCR was performed with a 7900HT Sequence Detection System (Thermo Fisher Scientific) using assays-on-demand primer (Applied Biosystems, assay IDs: mAcaca, Mm01304285_m1; mAdrb3, Mm00442669_m1; mAtp5a1, Mm00431960_m1; mAtp5b, Mm01160389_g1; mClcn7, Mm00442400_m1; mCox2, Mm03294838_g1; mCox4i1, Mm00438289_g1; mCox7a1, Mm00438297_g1; mCox8b, Mm00432648_m1; mCtsa, Mm00447197_m1; mCtsc, Mm00515580_m1; mCtsk, Mm00484039_m1; mDio2, Mm00515664_m1; mElovl3, Mm00468164_m1; mFasn, Mm00662319_m1; mGlut4, Mm01245502_m1; mLipa, Mm00498820_m1; mMap1lc3a, Mm01249999_g1; mMitf, Mm00434954_m1; mNd2, Mm04225288_s1; mNrf1, Mm00447996_m1; mPparg, Mm00440945_m1; mPpargc1a, Mm00447183_m1; mTbp, Mm00446973_m1; mTfam, Mm00447485_m1; mTfe3, Mm01341186_m1; mTfeb, Mm00448964_m1; mTmem55b, Mm01319582_m1; and mUcp1, Mm00494069_m1) or SYBR green primers (Table 1) that were premixed with TaqMan Univeral MasterMix II (Applied Biosystems) or SYBR Green PCR Master Mix (Applied Biosystems), respectively. Relative mRNA levels were calculated using the ΔΔCT method and normalized to expression levels of the housekeeper genes Tbp or TfIIb.
Table 1.
SYBR Green Primer.
| Name | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Atg4 | CCCTCACACAACCCAGACTT | CCCCTGTGGTTGTCACTTCT |
| Atg5 | GGAGAGAAGAGGAGCCAGGT | TGTTGCCTCCACTGAACTTG |
| Atg7 | CATGGGAACCCTGCACAAC | GGAGCTTCTTGTCTGCTGTCTTAA |
| Atg10 | CAAAACACAGTTTCGAATAA | TCATGTTTAATCACTTCTGC |
| Atg12 | GGAGACACTCCTATAATGAAA | ATAAATAAACAACTGTTCCGA |
| Atp6v0a1 | CCGAGGACGAAGTGTTTGACT | ATCAGCAGGATAGCCACGGT |
| Atp6v0a2 | TGGTGCAGTTCCGAGACCT | GCAGGGGAATATCAGCTCTGG |
| Atp6v0c | ACTTATCGCTAACTCCCTGACT | ACACCAGCATCTCCGACGA |
| Atp6v0d2 | TGCGGCAGGCTCTATCCAGAGG | CCACTGCCACCGACAGCGTC |
| Atp6v0e | GCATACCACGGCCTTACTGT | TGATAACTCCCCGGTTAGGAC |
| Atp6v1a | ACAGAGGAAGCGTGACTTACA | CACTTGGACCATGCTGAACTT |
| Atp6v1b2 | ATGCGGGGAATCGTGAACG | AGGCTGGGATAGGTAGTTCCG |
| Atp6v1c1 | ACTGAGTTCTGGCTCATATCTGC | TGGAAGAGACGGCAAGATTATTG |
| Atp6v1e1 | GAATCAAGCAAGGCTCAAAGTCC | CGGGTCGTATCTTTTACCACC |
| Atp6v1f | GCGGGCAGAGGTAAGCTAATC | TTAGGGTGGCGGTTCTTGTTT |
| Atp6v1g1 | CCCAGGCTGAAATTGAACAGT | TTCTGGAGGACGGTCATCTTC |
| Atp6v1h | GGATGCTGCTGTCCCAACTAA | TCTCTTGCTTGTCCTCGGAAC |
| Lamp1 | CAGCACTCTTTGAGGTGAAAAAC | ACGATCTGAGAACCATTCGCA |
| Lamp2 | TGTATTTGGCTAATGGCTCAGC | TATGGGCACAAGGAAGTTGTC |
| p62 | AGGATGGGGACTTGGTTGC | TCACAGATCACATTGGGGTGC |
| RagD | CTGTTTGACGTGGTCAGTAAGAT | GTTGAGTCCTTGTCATACGGG |
| Rheb | AAGTCCCGGAAGATCGCCA | GGTTGGATCGTAGGAATCAACAA |
| Tf2b | TGGAGATTTGTCCACCATGA | GAATTGCCAAACTCATCAAAACT |
2.8. Histology
To analyze the morphology of the brown adipose tissue, subscapular brown adipose tissue depots were dissected and fixed in 4% v/v formaldehyde in PBS for 24 h at 4 °C. After automated dehydration of the tissues using an Autotechnicon (Leica) and embedding in paraffin, 4 μm sections were cut using a semi-motorized rotary microtome (Leica RM2245). The sections were then mounted on glass slides (Histobond, Paul Marienfeld), deparaffinized, rehydrated, and stained with hematoxylin and eosin using standard procedures [64]. Images were taken using a Nikon eclipse Ti microscope equipped with a DS-Fi2-U3 camera for brightfield imaging and a 10× air objective. Lipid droplet sizes were quantified using the ImageJ plugin Adiposoft [68].
2.9. Immunohistochemistry
Immunostaining was principally performed as previously described [64]. Sections of BAT were prepared as previously described. For antigen retrieval, slides were boiled in 10 mM of citric acid with a pH of 6 for 1 h, cooled to room temperature, and subsequently washed in PBS. To reduce background signals, the sections were stained with 1% w/v Sudan Black in 80% v/v ethanol for 10 min and then washed. After blocking unspecific antibody-binding sites with 3% w/v BSA for 1 h at room temperature, the slides were subsequently incubated with first antibodies (Table 2) overnight at 4 °C in a humid incubation chamber. After washing with PBS, the sections were incubated with secondary antibodies (Table 3) for 1 h at room temperature in the dark. The sections were then washed with PBS and stained with 0.05% DAPI v/v in PBS for 10 min before mounting with ProLong Gold antifade reagent (Invitrogen). Fluorescence microscopy was performed using a Nikon eclipse Ti confocal laser scanning microscope with 405 nm, 488 nm, and 561 nm lasers and the following detectors: 425–475 nm; 500–550 nm, and 570–620 nm. Total cytosolic and mitochondria-associated LC3B puncta were counted independently by three people in a blinded fashion.
Table 2.
Primary antibodies.
| Name | Supplier | Reference number | Host | Dilution | |
|---|---|---|---|---|---|
| AKT | Cell Signaling | 9272 | Rabbit | WB 1:1000 | Polyclonal |
| Cathepsin D | Abcam | 75852 | Rabbit | WB 1:2000 | Monoclonal |
| DIO2 | Abcam | ab77779 | Rabbit | WB 1:1000 | Polyclonal |
| γ-Tubulin | Abcam | ab179503 | Rabbit | WB 1:2000 | Monoclonal |
| HSL | Cell Signaling | 4107 | Rabbit | WB 1:1000 | Polyclonal |
| LAMP1 | DSHB | 1D4B | Rat | WB 1:250 | Monoclonal |
| LC3B | Cell Signaling | 2775 | Rabbit | WB 1:1000 IHC 1:200 |
Polyclonal |
| OXPHOS cocktail | Abcam | ab110413 | Mouse | WB 1:25000 IHC 1:1000 |
Monoclonal |
| p62/sqstm1 | Cell Signaling | 5114 | Rabbit | WB 1:1000 | Polyclonal |
| Parkin | Abcam | ab77924 | Mouse | WB 1:1000 | Monoclonal |
| p-HSL (Ser563) | Cell Signaling | 4139 | Rabbit | WB 1:1000 | Polyclonal |
| PINK1 | Abcam | ab23707 | Rabbit | WB 1:1000 | Polyclonal |
| Phospho-PKA substrate (RRXS∗/T∗) | Cell Signaling | 9621 | Rabbit | WB 1:1000 | Polyclonal |
| Tyrosine hydroxylase (TH) | Abcam | ab137869 | Rabbit | WB 1:5000 | Monoclonal |
| Ubiquitin | Cell Signaling | 3933 | Rabbit | WB 1:1000 | Polyclonal |
| UCP1 | Cannon and Nedergaard Lab | N.A. | Rabbit | WB BAT: 1:125000 Cells: 1:5000 |
Polyclonal |
| VDAC | Cell Signaling | 4661 | Rabbit | WB 1:1000 | Monoclonal |
Table 3.
Secondary antibodies.
| Name | Supplier | Reference Number | Dilution |
|---|---|---|---|
| Peroxidase-conjugated AffiniPure goat anti-rabbit IgG (H + L) | Jackson ImmunoResearch | 111-035-144 | 1:5000 |
| Peroxidase-conjugated AffiniPure goat anti-mouse IgG (H + L) | Jackson ImmunoResearch | 115-035-146 | 1:5000 |
| Peroxidase-conjugated AffiniPure F(ab’)2 fragment donkey anti-rat IgG (H + L) | Jackson ImmunoResearch | 712-036-153 | 1:5000 |
| Cy3-AffiniPure F(ab')2 fragment donkey anti-mouse IgG (H + L) | Jackson ImmunoResearch | 715-166-150 | 1:500 |
| Alexa Fluor 488-conjugated AffiniPure F(ab’)2 fragment donkey anti-rabbit IgG (HL) | Jackson ImmunoResearch | 711-546-152 | 1:250 |
2.10. Electron microscopy
Mice were transcardially perfused with a mixture of 4% paraformaldehyde and 1% glutaraldehyde in 0.1 M of PB buffer at a pH of 7.4. The brown fats were removed, and small pieces were prepared with a razor blade. The samples were rinsed three times in 0.1 M of sodium cacodylate buffer (pH 7.2–7.4) and osmicated using 1% osmium tetroxide in cacodylate buffer. Following osmication, the samples were dehydrated using ascending ethyl alcohol concentration steps, followed by two rinses in propylene oxide. Infiltration of the embedding medium was performed by immersing the pieces in a 1:1 mixture of propylene oxide and Epon and finally in neat Epon and hardened at 60 °C. Semi-thin sections (0.5 μm) were prepared for light microscopy. They were mounted on glass slides and stained for 1 min with 1% toluidine blue. Ultra-thin sections (60 nm) were cut and mounted on copper grids. The sections were stained using uranyl acetate and lead citrate. Thin sections were examined and photographed using an EM902 (Zeiss) electron microscope.
2.11. Protein isolation and immunoblotting analysis
Protein analysis was performed as previously described [16]. Briefly, one iBAT lobe was lysed in 10 × (v/w) RIPA buffer using a TissueLyser (Qiagen). Cells were lysed in 150 μL of RIPA buffer. Protein concentrations were determined using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific). The samples were diluted to a final protein concentration of 1 μg/μL in reducing sample buffer (Invitrogen) and denatured at 60 °C for 10 min. For western blotting, 20 μg of total protein was separated on 10% or 12.5% Tris-glycine gels and proteins were subsequently transferred to 0.45 μm nitrocellulose blotting membranes (GE Healthcare Life Sciences). The membranes were blocked for 1 h at room temperature in 5% (w/v) milk in TBST and incubated overnight at 4 °C in primary antibodies (Table 2) diluted in 5% (w/v) BSA with 0.1% (v/v) Na azide in TBST. After washing in TBST, the membranes were incubated in corresponding HRP secondary antibodies (Table 3) in 5% (w/v) milk TBST for 1 h at room temperature. The membranes were washed in TBST and the detection was performed with enhanced chemiluminescence substrate (SuperSignal West Femto, Thermo Fisher Scientific) using an Amersham Imager 600 (GE Healthcare) and analyzed with Image Studio Lite (Licor). AKT was chosen as a loading control due to low variability and temperature dependence of this marker in BAT.
2.12. Isolation and measurement of TGs from cells and tissue
For triglyceride quantification in brown adipose tissue, the samples were lysed in 20× (v/w) PBS using a TissueLyser (Qiagen). For measurement in cells, the cells were harvested in 150 μl PBS/6 per well. Then 50 μl (BAT) or 40 μl (cells) were filled to 100 μl with PBS, 375 μl of chloroform:methanol (2:1) was added, vortexed, and kept in a shaker at RT for 3 h. Then, 125 μl of chloroform was added and the samples were mixed well, followed by the addition of 125 μl of dH2O and additional mixing. The samples were centrifuged at 10000 g for 5 min, the organic lower phase was harvested, dried under air, and resuspended in 50 μl of 0.1% TritonX100 in PBS. Then, 1 μl of the resuspended lipids was used for TG quantification using a Triglyceride FS 5′ kit (DiaSys GmbH 1-5760∗).
2.13. Statistical analyses and data processing
Two-tailed unpaired Student's t tests, one-way ANOVA, or two-way ANOVA followed by Tukey's multiple comparison test were performed to calculate the statistical significance. P < 0.05 was considered statistically significant. GraphPad Prism 8 and Microsoft Excel were used for calculations.
3. Results
3.1. Thermoneutrality diminished the expression of mitochondrial biogenesis markers and increased the expression of mitochondrial degradation markers
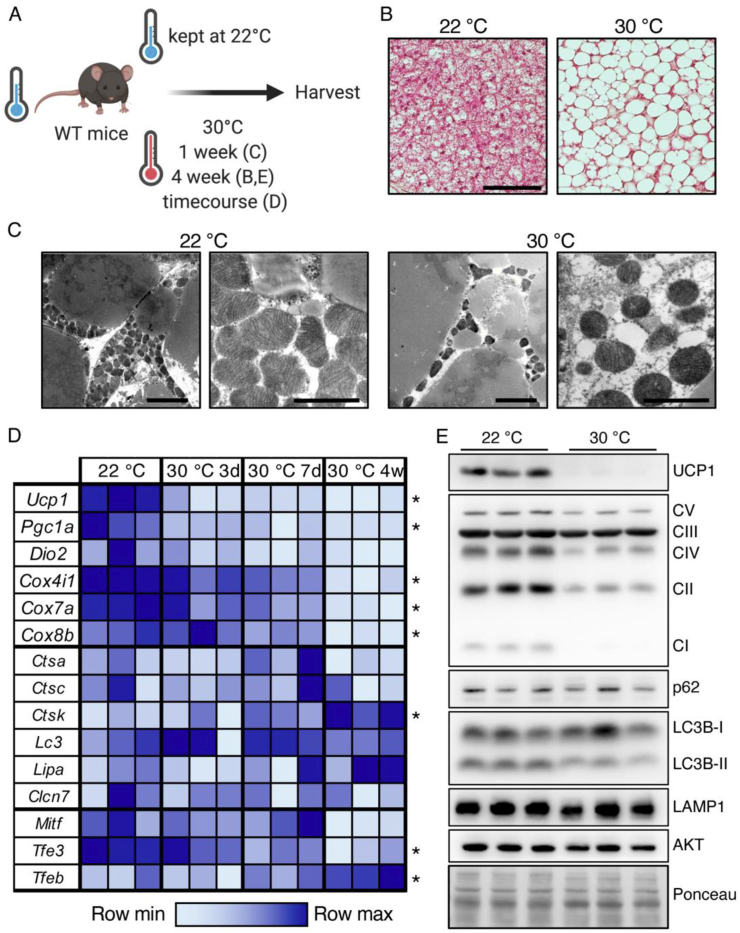
Histological and electron microscopic analysis of BAT under conditions of BAT activation or BAT whitening (Figure 1A) showed vast lipid accumulation after one week (Figure 1C) or four weeks (Figure 1B) of thermoneutral adaptation, transforming the multilocular brown adipocytes into “white-like” unilocular adipocytes with reduced mitochondrial density (Figure 1B,C). Mitochondrial mass was controlled by the opposing actions of mitochondrial biogenesis and mitochondrial degradation via the autophagic system [69,70]. To investigate the effects of BAT whitening on these pathways' activity, the gene expression of mitochondrial and thermogenesis markers as well as lysosome and autophagy markers was analyzed (Figure 1D). Thermoneutral housing led to reduced expression of the thermogenic marker genes uncoupling protein 1 (Ucp1), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (Pgc1a), cytochrome c oxidase subunit 4 isoform 1 (Cox4i1), cytochrome c oxidase subunit 7A (Cox7a), and cytochrome c oxidase subunit 8B (Cox8b), while mRNA levels of the lysosomal proteases cathepsin a and c (Ctsa and Ctsc), chloride voltage-gated channel 7 (Clcn7), autophagophore-marker microtubule-associated proteins 1A/1B light-chain 3A (Lc3a), and lysosomal acid lipase (Lipa) were unaffected by the environmental temperature, and the levels of cathepsin k (Ctsk) increased with exposure to thermoneutrality. Interestingly, the expression levels of lysosome- and autophagy-regulating transcription factors Mitf and Tfe3 were unaffected or decreased respectively with prolonged exposure at thermoneutrality, whereas the expression levels of Tfeb continuously increased with exposure to thermoneutrality (Figure 1D). When comparing Tfeb gene expression levels in BAT and iWAT of the WT mice acclimated to thermoneutrality for one week, we observed significantly higher Tfeb levels in BAT (Figure S1A). This could be indicative of higher autophagic activity in BAT compared to WAT under whitening conditions. Western blotting analysis of BAT from wild-type mice kept under standard housing conditions or adapted to thermoneutrality for 4 weeks confirmed the thermoneutrality-induced loss of mitochondrial proteins such as UCP1 and respiratory chain complexes, while protein levels of autophagy and lysosome-related proteins sequestosome1/p62 (p62), microtubule-associated proteins 1A/1B light-chain 3B (LC3B), and lysosome-associated membrane glycoprotein 1 (LAMP1) were largely unaffected by the environmental temperature (Figure 1E). Of note, the levels of lipidated LC3-II decreased in the thermoneutral BAT, indicating that after four weeks of thermoneutral adaptation, mitochondrial turnover might have decreased. Collectively, these data indicated that under thermoneutral conditions, the ratio of mitochondrial breakdown and biogenesis shifted. While the expression of genes involved in mitochondrial biogenesis was downregulated in response to thermoneutral conditions, the expression of lysosome and autophagy-related genes and proteins was largely unaffected, consequently favoring reductions in mitochondrial mass.
Figure 1.
Thermoneutrality shifted the levels of genes of mitochondrial breakdown and biogenesis toward reducing mitochondrial mass. (A) Schematic model of the experimental design. (B) Representative H&E staining of BAT of WT mice maintained at 22 °C or adapted to 30 °C for four weeks, scale bar 50 μm. (C) Representative EM images of WT mice maintained at 22 °C or acclimated to 30 °C for one week, scale bar 5 μm (left) and 2 μm (right). (D) Relative mRNA levels of genes encoding BAT markers, lysosomal-, and autophagy-related proteins, and MiT family transcription factors in BAT of WT mice acclimated to thermoneutrality for 3 days, 7 days, or 4 weeks, or maintained at room temperature (n = 3 for each time point). (E) Representative western blotting of UCP1, OXPHOS complexes and autophagy, and lysosomal marker proteins in BAT of the WT mice maintained at 22 °C or acclimated to 30 °C for four weeks. ∗P ≤ 0.05 by one-way ANOVA describing the effect of the environmental temperature.
The lysosomal gene network is under tight control by members of the MiT transcription factor family. While the Mitf and Tfe3 expression levels generally decreased at thermoneutrality, the expression of Tfeb was induced by high ambient temperatures (Figure 1D), suggesting that this transcription factor might be involved in the coordination of the autophagy-dependent mitochondrial degradation during BAT whitening.
3.2. Attenuated whitening in TFEB-deficient BAT
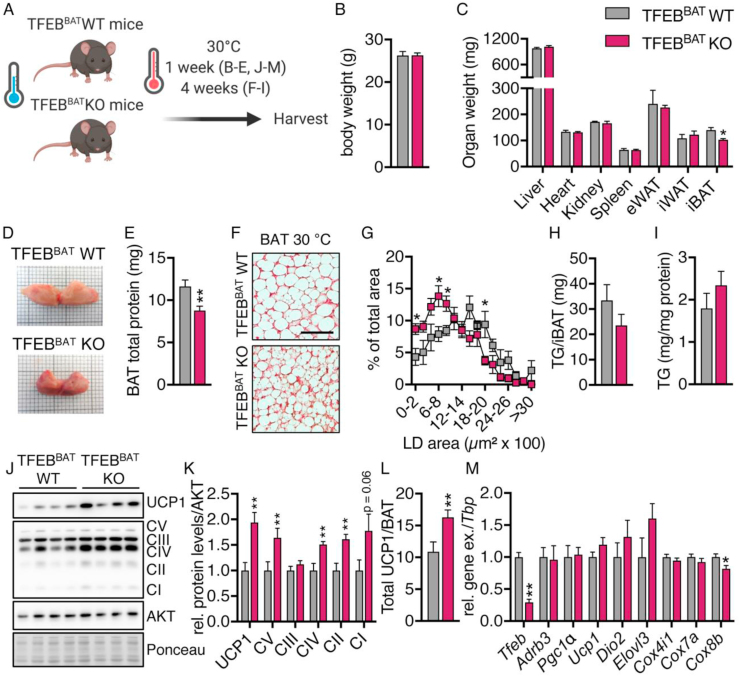
To investigate the role of TFEB for BAT plasticity during whitening, we generated brown adipocyte-specific TFEB knockout animals by crossing TFEBfl/fl mice with mice expressing Cre-recombinase under control of the Ucp1-promoter, resulting in BAT-specific TFEB knockout mice (TFEBBAT KO) and their respective Cre-negative TFEBfl/fl littermates (TFEBBAT WT). The TFEBBAT WT and TFEBBAT KO mice were exposed to thermoneutrality for one week to induce BAT whitening (Figure 2A). BAT TFEB deficiency did not affect the body weight or weights of metabolically active organs such as the liver, heart, kidney, spleen, or epididymal and inguinal white adipose tissues (eWAT and iWAT), but significantly reduced iBAT tissue weights (Figure 2B,C). Macroscopically, TFEB-deficient BAT appeared more brownish in color compared to the pale-looking BAT of the control mice and contained a reduced total protein content (Figure 2C,D). In line with the gross BAT morphology, TFEB-deficient BAT presented smaller lipid droplets following 4 weeks of adaptation to thermoneutrality (Figure 2A,F, and G) while the total amount of triglycerides and triglycerides per mg protein was unaltered (Figure 2H,I). TFEB-deficient BAT maintained higher UCP1 and OXPHOS complex protein levels compared to the whitened BAT of the WT littermates after one week of thermoneutral adaptation (Figure 2J,K). Total UCP1 levels per BAT, a readout of the total thermogenic capacity of BAT [27,38,[71], [72], [73], [74]], were significantly increased in the TFEBBAT KO mice (Figure 2L), indicating a potentially higher BAT thermogenic capacity.
Figure 2.
TFEB-deficient BAT retained signs of activity under thermoneutrality. TFEBBAT WT and KO mice maintained at room temperature were transferred to thermoneutrality for one week (B-E and J-M) or four weeks (F-I). The mice were fasted for 4 h before organ harvest. (A) Schematic model of the experimental design. (B) Body weights, (C) organ weights (n = 5 each genotype and representative for at least 2 independent experiments), and (D) representative macroscopic morphology of iBAT and (E) total iBAT protein content (n = 9 each genotype) of the TFEBBAT WT and KO mice acclimated to thermoneutrality for 1 week. (F) Representative H&E staining of BAT from the TFEBBAT WT and KO mice acclimated to thermoneutrality for 4 weeks, scale bar 50 μm. (G) Lipid droplet size distribution from BAT (n = 3 TFEBBAT WT and 4 TFEBBAT KO, 3 images per mouse), (H) total triglycerides (TG) per iBAT, and (I) triglycerides per mg protein (n = 5 each genotype). (J) Representative western blotting of BAT markers in BAT, (K) quantification of protein levels, and (L) total UCP1 levels per iBAT of the TFEBBAT WT and KO mice acclimated to thermoneutrality for 1 week (n = 9 each genotype, representative for at least 2 independent experiments). (M) Relative mRNA levels of brown adipocyte identity genes in BAT of the TFEBBAT WT and KO mice acclimated to thermoneutrality for 1 week (n = 5 each genotype). Values are presented as mean ± SEM. ∗P ≤ 0.05 and ∗∗P ≤ 0.01 by two-tailed unpaired Student's t test.
Gene expression analysis in BAT of the mice acclimated to thermoneutrality for one week confirmed the TFEB deletion (Figure 2M). The remaining expression was likely caused by Tfeb expression by other cell types within the BAT, such as macrophages, endothelial cells, and immune cells. Expression levels of genes encoding established markers of BAT activity such as adrenoceptor beta 3 (Adrb3), Pgc1a, Dio2, and elongation of very long-chain fatty acid 3 (Elovl3) were largely unchanged between genotypes, as were Ucp1 mRNA levels and expression of genes encoding cytochrome c oxidase subunits (Cox4i1, Cox7a, and Cox8b), indicating that the increased OXPHOS and UCP1 protein levels could not be explained by enhanced mitochondrial biogenesis (Figure 2M). Moreover, these data showed that the transcriptional alterations occurring in BAT upon thermoneutral adaptation were intact in TFEB-deficient BAT.
The thermogenic capacity and recruitment state of BAT are predominantly regulated by the sympathetic nervous system [7,64]. Although our brown adipocyte-specific TFEB knockout model should not directly affect the neuronal circuits regulating sympathetic nerve activity, direct effects of adipocyte-derived factors on sympathetic nerve function have been described [75]. To identify potential effects of brown adipocyte TFEB deficiency on sympathetic innervation, we measured protein levels of tyrosine hydroxylase (TH), the rate-limiting enzyme of norepinephrine biosynthesis, and its downstream effects on protein kinase A (PKA)-mediated phosphorylation of hormone-sensitive lipase (HSL) and general PKA substrates. Levels of TH, PKA-substrate phosphorylation, and HSL phosphorylation were similar between the TFEBBAT KO and WT mice, indicating that adrenergic signaling was largely unaffected by BAT TFEB deficiency and therefore cannot explain the higher UCP1 protein levels in TFEB-deficient BAT (Figure S1B and C). In summary, these data indicated that BAT TFEB deficiency attenuates BAT whitening under thermoneutral conditions in a biogenesis-independent manner.
3.3. TFEB controlled UCP1 levels in a cell-autonomous manner
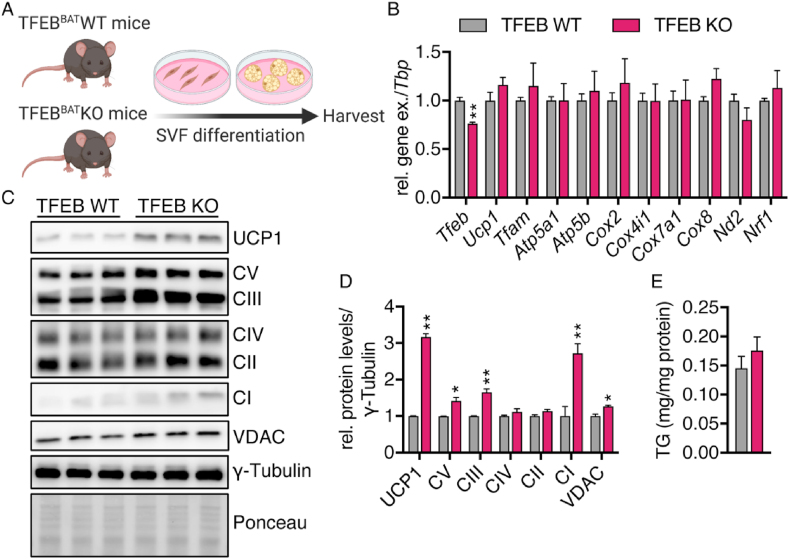
To elucidate whether TFEB modulates UCP1 protein levels cell autonomously, we isolated and differentiated primary brown adipocytes from the TFEBBAT WT and KO mice (Figure 3A). The TFEB KO cells presented with slightly but significantly reduced Tfeb mRNA levels (Figure 3B). The Tfeb mRNA detected could have originated from remaining preadipocytes and fibroblasts. Similar to in vivo, there was no difference in the gene expression of both nucleus- and mitochondria-encoded mitochondrial biogenesis markers such as Ucp1, Tfam, Cox2,4,7, and 8, ATP-synthase subunits (Atp5a1 and Atp5b), or Nd2 between the WT and KO cells (Figure 3B), but we detected significantly higher UCP1, VDAC, and OXPHOS complex protein levels (Figure 3C,D), indicating that TFEB indeed modulates UCP1 protein levels/mitochondrial content in a cell-autonomous manner. The modulation in mitochondrial mass was not accompanied by altered triglyceride deposition (Figure 3E). To determine whether the mitochondrial changes were due to an effect of TFEB deficiency on brown adipocyte differentiation, we evaluated the gene expression of brown adipocyte markers established during differentiation. As expected, the expression of adipocyte differentiation and identity markers increased during adipogenesis in both genotypes and the expression of most markers peaked on day 6 of differentiation (Figure S2A-H). Elovl3, Dio2, and Acaca levels were slightly higher in TFEB-deficient brown adipocytes on days 4–6, suggesting a somewhat faster differentiation that was normalized on day 8. The expression of other markers of brown adipocyte identity was unaffected by reduced Tfeb expression (Figure S2A–H), indicating that increased differentiation could not explain the differences in mitochondrial protein levels observed in vitro and in vivo.
Figure 3.
TFEB controlled UCP1 protein levels in a cell-autonomous manner. Primary brown adipocytes were differentiated from stroma vascular fraction (SVF) isolated from BAT of 3- to 6-week-old TFEBBAT WT and KO mice. (A) Schematic model of the experimental procedures. (B) Relative Tfeb and mitochondrial marker mRNA levels in differentiated primary brown adipocytes (n = 6 each genotype) (C) Representative western blotting of UCP1, OXPHOS, and VDAC in primary brown adipocytes and (D) quantification of protein levels (n = 3 each genotype, representative for at least 2 independent experiments). (E) Triglycerides (TG) per mg protein. Values are presented as mean ± SEM. ∗P ≤ 0.05 and ∗∗P ≤ 0.01 by two-tailed unpaired Student's t test.
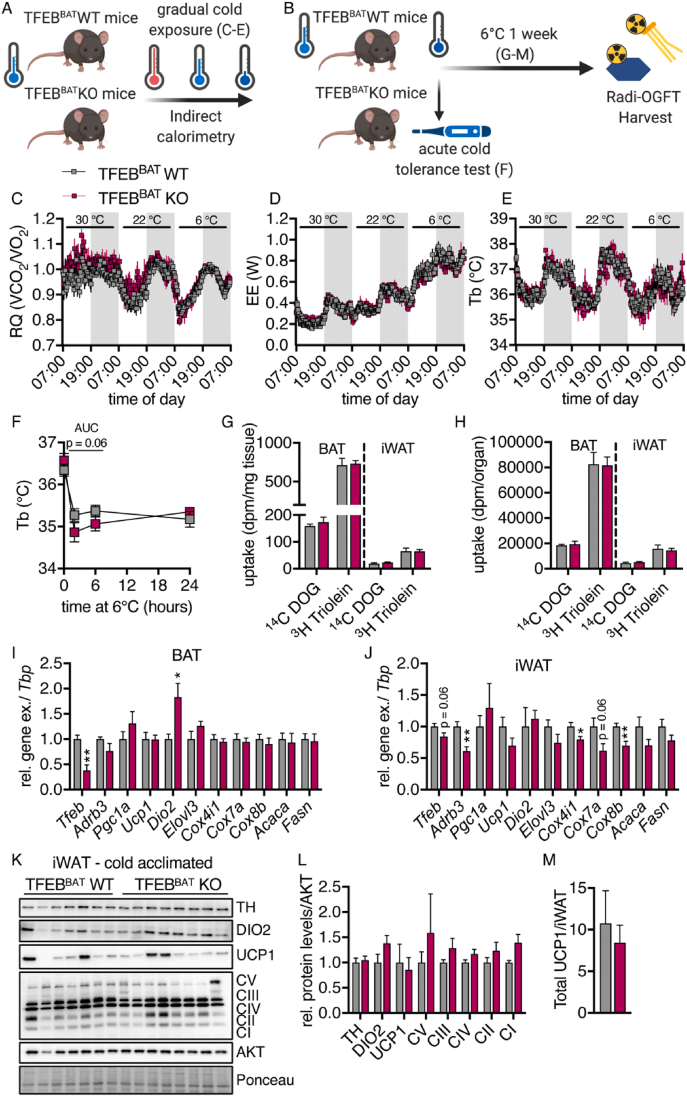
3.4. Unaltered response to the cold in TFEB-deficient BAT
TFEB overexpression has recently been shown to drive browning of white adipose tissue in a PGC1A-dependent manner [63]. Additionally, autophagy is thought to be an important regulator of adipose tissue browning and adipocyte differentiation [54,55,76] and mitochondrial homeostasis in BAT during cold acclimation [77]. To investigate the role of BAT TFEB in the adaptation and response to cold stress, we monitored metabolic parameters of the TFEBBAT WT and KO mice that had previously been housed under standard conditions in response to different ambient temperatures (Figure 4A). Neither the respiratory quotient, energy expenditure, nor body temperature differed between genotypes at thermoneutrality (30 °C), room temperature (22 °C), or under conditions of severe cold stress (6 °C) (Figure 4C–E). To test the general ability of these mice to defend their body temperature against acute cold stress by induction of shivering thermogenesis, the mice were directly transferred to 6 °C from room temperature, and their rectal temperature was measured (Figure 4B,F). The TFEBBAT KO mice experienced a more pronounced initial decrease in body temperature during the acute cold challenge but defended their body temperature similar to the WT controls after 24 h (Figure 4F).
Figure 4.
Unaltered response to cold in TFEB-deficient brown/beige adipocytes. (A) Schematic model of the experimental conditions for graphs C-D. (B) Schematic model of the experimental conditions for graphs F-M. (C) Respiratory quotient (RQ), (D) energy expenditure (EE), and (E) body temperature (Tb) of the TFEBBAT WT and KO mice in response to different environmental temperatures (30 °C, 22 °C, and 6 °C, n = 6 TFEBBAT WT and 4 TFEBBAT KO). The mice had previously been maintained at room temperature. (F) Rectal temperature in response to acute cold exposure after transfer from room temperature (n = 7 TFEBBAT WT and 8 TFEBBAT KO). (G–M) The 1-week cold-acclimated TFEBBAT WT and KO mice were fasted for 4 h, followed by gavage of a glucose/lipid emulsion containing tracer doses of 14C-deoxyglucose (DOG) and 3H-triolein. Organs were harvested 2 h after gavage following transcardial perfusion. (G) Uptake of 14C-deoxyglucose (DOG) and 3H-triolein per mg tissue and (H) per total organ (n = 7 TFEBBAT WT and 8 TFEBBAT KO). (I and J) Relative mRNA levels of brown adipocyte identity genes in (I) BAT and (J) iWAT of the 1-week cold-acclimated TFEBBAT WT and KO mice (n = 5 each genotype). (K) Representative western blotting of browning markers in iWAT of the 1-week cold acclimated TFEBBAT WT and KO mice, (L) quantifications of respective protein levels, and (M) total UCP1 protein levels per iWAT (n = 7 TFEBBAT WT and 8 TFEBBAT KO). Values are presented as mean ± SEM. ∗P ≤ 0.05 and ∗∗P ≤ 0.01 by two-tailed unpaired Student's t test.
The one-week cold-acclimated TFEBBAT WT and KO mice (Figure 4B) displayed similar body weights (Figure S3A), fasting plasma triglyceride, total cholesterol, non-esterified fatty acid levels, and decreased fasting glucose levels (Figure S3B–E). Organ weights were also comparable (Figure S3F).
To measure the metabolic activity of adipose tissues under these conditions, we performed a combined oral fat and glucose tolerance test [13] and observed no genotype effect on the uptake of radiolabeled glucose or lipids into BAT or iWAT (Figure 4G,H). When analyzing the gene expression in BAT of the one-week cold-acclimated TFEBBAT WT and KO mice, brown adipocyte markers were largely unchanged while Dio2 levels increased (Figure 4I). Conversely and in line with the described effect of TFEB overexpression on enhancing browning [63], we found reduced expression of some thermogenic and mitochondrial biogenesis markers in brite/beige iWAT (Figure 4J). Notably, the reduction in Tfeb gene expression was only marginal, presumably due to the low penetrance of the Ucp1-Cre in brite/beige WAT [78,79]. Despite the reductions in thermogenic gene expression, we did not observe alterations in thermogenic protein levels per mg of protein (Figure 4K and L) or UCP1 levels per total iWAT (Figure 4M).
Overall, these data indicated that while the TFEB-PGC1a axis seems dispensable in BAT, it appears to be involved in the transcriptional regulation of white adipose tissue browning. The unaltered thermogenic protein levels and metabolic response to cold and unaltered nutrient uptake, however, argue against profound consequences of this browning effect for whole-body physiology.
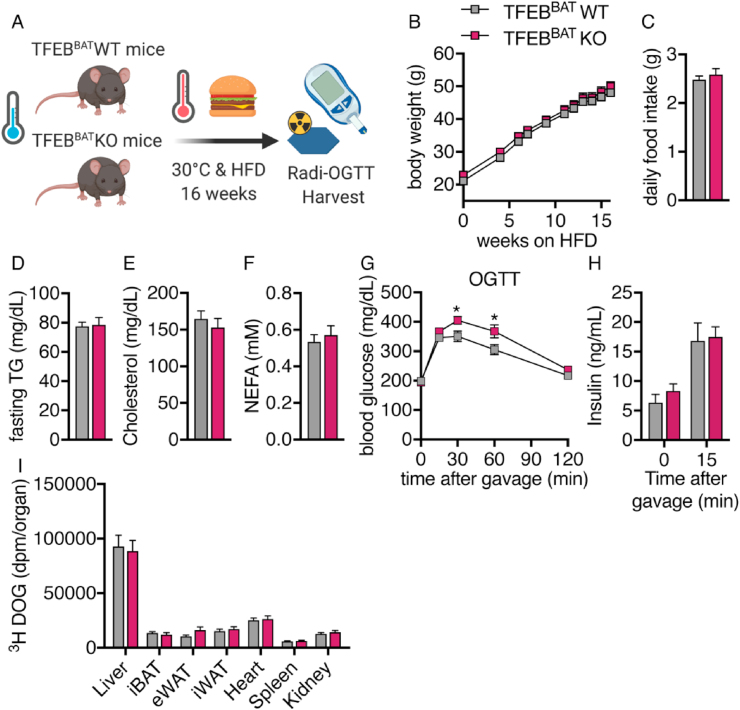
3.5. BAT TFEB deficiency did not protect from diet-induced obesity
Stimulation of BAT thermogenesis has been suggested to be a novel treatment strategy to combat obesity [[80], [81], [82]]. This assumption is mainly based on observations that obese individuals display lower levels of detectable BAT and that cold exposure can induce weight loss in humans as well as the fact that UCP1-deficient animals can become more obese [35,83]. To examine whether TFEB deficiency in BAT at thermoneutrality can protect mice from diet-induced obesity, we exposed mice to a HFD at 30 °C for 16 weeks (Figure 5A). Both genotypes displayed a similar weight gain and daily food intake throughout the feeding study (Figure 5B,C). Plasma fasting triglyceride, total cholesterol, and non-esterified fatty acid levels were unchanged between genotypes after 16 weeks of the HFD, suggesting an unaltered lipid homeostasis in the fasting state (Figure 5D–F). We next investigated whether BAT TFEB deficiency modulates systemic glucose homeostasis in obese mice by performing an oral glucose tolerance test (OGTT). Surprisingly, the TFEBBAT KO mice showed a modestly delayed blood glucose clearance after glucose gavage (Figure 5G), while fasting and glucose-induced insulin levels were unchanged between genotypes (Figure 5H). The uptake of glucose into metabolically active organs was similar in the control and TFEBBAT KO mice (Figure 5I), suggesting an overall comparable postprandial glucose handling and unaltered insulin sensitivity in the obese TFEBBAT KO mice.
Figure 5.
BAT-specific TFEB deletion did not protect against diet-induced obesity. TFEBBAT WT and TFEBBAT KO mice were transferred to thermoneutrality at 6 weeks of age and fed a high-fat diet (60% calories from fat) for 16 weeks. The mice were fasted for 4 h and received an oral gavage of 2 g/kg of glucose solution containing a tracer dose of 3H-deoxyglucose (DOG). Organs were harvested 2 h after gavage following transcardial perfusion. (A) Schematic model of the experimental design. (B) Body weight, (C) daily food intake, and (D–E) fasting levels of (D) plasma triglyceride (TG), (E) plasma cholesterol, and (F) plasma non-esterified fatty acids (NEFA). (G) Oral glucose tolerance test (OGTT) with (H) insulin levels before and 15 min after glucose administration (n = 14 TFEBBAT WT and 10 TFEBBAT KO). (I) Uptake of 3H-deoxyglucose (DOG) per total organ (n = 14 TFEBBAT WT and 10 TFEBBAT KO). Values are presented as mean ± SEM. ∗P ≤ 0.05 by two-tailed unpaired Student's t test.
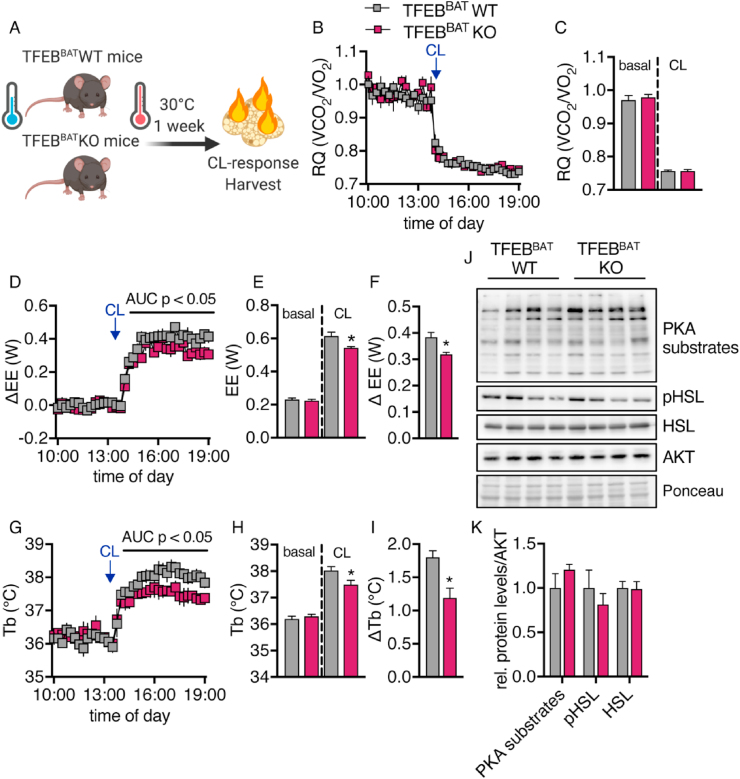
3.6. TFEB deletion reduced thermogenic capacity despite increased UCP1 levels
To understand the discrepancy between higher UCP1 levels and lack of protection against diet-induced obesity, we quantified BAT thermogenic capacity by measuring the increase in energy expenditure following injection of the β3-adrenoceptor agonist CL 316,243 (CL) in a thermoneutral environment where facultative thermogenic pathways are inactive [71,72]. The mice had previously been housed at thermoneutrality for one week (Figure 6A).
Figure 6.
TFEB deficiency reduced thermogenic capacity in response to adrenergic stimulation. TFEBBAT WT and KO mice bred and maintained at room temperature were adapted to thermoneutrality for one week. Following acclimation, they were transferred to an indirect calorimetry system and maintained at 30 °C. Following a 1-day acclimation to the system, they were injected with 1 mg/kg of CL316,243 to induce maximum BAT thermogenic activity. Five h after injection, their organs were harvested. (A) Schematic model of the experimental conditions. (B) Respiratory quotient (RQ) and (C) quantification of basal- and CL-stimulated RQ. (D) Δ energy expenditure (ΔEE), (E) quantification of basal- and CL-stimulated EE, and (F) change over baseline of EE. (G) Body temperature (Tb), (H) quantification of basal- and CL-stimulated Tb, and (I) change over baseline of Tb (n = 5 TFEBBAT WT and 4 TFEBBAT KO, representative of 2 independent experiments). (J) Western blotting analysis of PKA substrate and HSL phosphorylation in BAT of the TFEBBAT WT and TFEBBAT KO mice after CL treatment and (K) quantification. Values are presented as mean ± SEM. ∗P ≤ 0.05 by two-tailed unpaired Student's t test.
The respiratory quotient decreased similarly in the TFEBBAT WT and KO mice after injection of CL, indicating a switch from glucose to lipid metabolism (Figure 6B,C). Surprisingly, the TFEBBAT KO mice had significantly lower CL-induced increases in energy expenditure and body temperature (Figure 6D–I) despite displaying significantly higher UCP1 protein levels per mg protein and total UCP1 levels per BAT depot (Figure 2J–L). Of note, the activity of the adrenergic signaling cascade, general PKA-mediated and HSL phosphorylation, was unaltered by TFEB deficiency under these conditions (Figure 6J,K). This highlighted that this model showed a dissociation of the total UCP1 protein levels and thermogenic capacity of the tissue despite unaltered general adrenergic responsiveness. In summary, our results demonstrated that the increased UCP1 protein levels in TFEB-deficient BAT did not correlate with an increased thermogenic capacity, which suggested that TFEB might modulate the function of BAT mitochondria or accessibility to adrenergic stimulation.
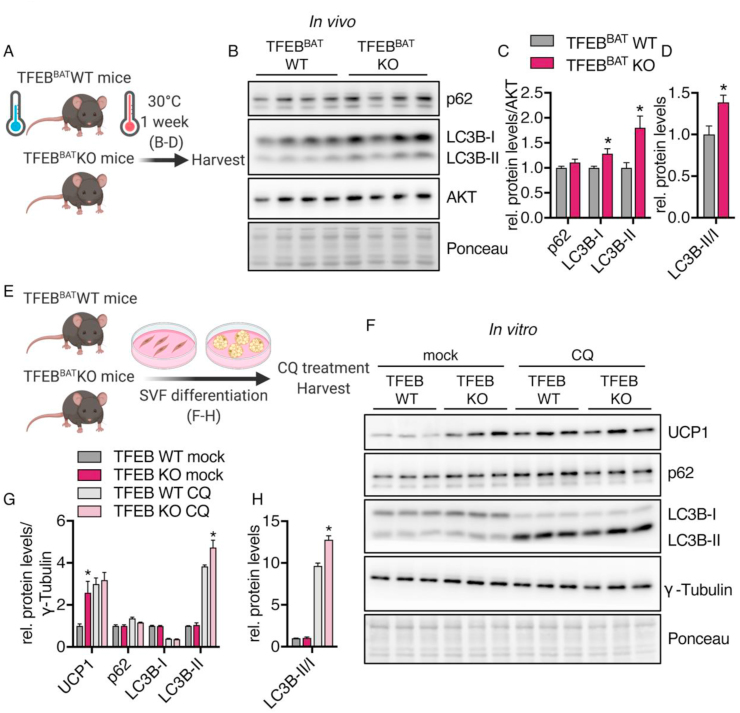
3.7. Brown adipocyte-specific TFEB deficiency modulated autophagy in vivo and in vitro
As TFEB is known to be an important transcriptional regulator of lysosomal and autophagy-related gene networks, we next sought to identify whether BAT-specific TFEB deficiency resulted in perturbations of the autophagy machinery that might explain the discrepancy between the total UCP1 levels and thermogenic capacity.
To evaluate whether TFEB deletion in BAT causes alterations in autophagy during BAT whitening, we measured the levels of autophagy-related proteins after one week of adaptation to thermoneutrality (Figure 7A) and detected increased LC3B-I and II levels and an increased LC3B-II/I ratio in TFEB-deficient BAT while the protein levels of autophagy receptor p62 were unchanged between genotypes (Figure 7B–D). LC3B-II levels directly correlate with the number of autophagosomes [84], and the accumulation of LC3B-II in TFEB-deficient BAT may indicate a dysregulated autophagic pathway. The ratio of LC3-II to LC3-I can indicate changes in the conversion of LC3-I to LC3-II and describe autophagic activity. In addition to the observed alterations in vivo, differentiated primary brown adipocytes lacking TFEB also showed alterations in autophagy-related protein levels upon serum starvation. Both genotypes demonstrated a starvation-induced increase in LC3B-II levels, whereas we observed a significant additional accumulation of LC3B-II in cells lacking TFEB and thus an increased LC3-II/I ratio (Figure S4A–C).
Figure 7.
TFEB deficiency modulated autophagy-related protein levels. TFEBBAT WT and KO mice were adapted to thermoneutrality for one week. (A) Schematic model of the experimental design for graphs B-D. (B) Representative western blotting of autophagy markers in BAT of the fasted TFEBBAT WT and KO mice. (C) Quantification of protein levels and (D) LC3II/I ratio (n = 4 each genotype). Primary brown adipocytes were differentiated from SVF cells isolated from the TFEBBAT WT and KO mice. (E) Schematic model of experimental procedures for graphs F-H. (F) Representative western blotting of UCP1, autophagy, and lysosomal markers in differentiated primary brown adipocytes treated with or without chloroquine (CQ). (G) Quantification of protein levels and (H) LC3II/I ratios (n = 3 each genotype and treatment, representative of 2 independent experiments). Values are presented as mean ± SEM. ∗P ≤ 0.05 by two-tailed unpaired Student's t test (C and D) or by two-way ANOVA with Tukey's multiple comparison test (G and H).
Autophagy is a highly dynamic process driven by the production of autophagosomal and lysosomal vesicles and the degradation of engulfed cargo. A static view on this dynamic process only reveals limited information on whether and how autophagic flux is altered. As TFEB-deficient primary brown adipocytes showed both increased UCP1 levels and alterations in autophagy-related proteins, we used these cells as a tool to evaluate whether TFEB deficiency caused alterations in autophagic flux.
Using chloroquine as an inhibitor of lysosomal acidification and fusion of autophagosomes and lysosomes [85,86], we investigated the impact of TFEB on the production of autophagic vesicles by quantifying autophagy-related protein levels (Figure 7E). The effects of chloroquine became apparent in the reduction of LC3B-I due to the continuous conversion to LC3B-II and the accumulation of LC3B-II. LC3B-II is normally either degraded or recycled and thus accumulated when lysosomal degradation was impaired by chloroquine treatment (Figure 7F,G). Consistent with our previous findings, TFEB deficiency led to an accumulation of UCP1 (Figure 7F,G). Upon inhibition of lysosomal degradation, UCP1 levels in the WT cells increased, suggesting a role of lysosomal degradation in the turnover of UCP1. Interestingly, chloroquine treatment did not affect UCP1 levels in the TFEB KO cells. In fact, the WT and KO cells displayed the same UCP1 levels upon inhibition of lysosomal and autophagic degradation. However, autophagic flux under chloroquine treatment seemed to be largely unaltered, as autophagy-related protein levels were similar between genotypes with the exception of LC3B-II, which accumulated at higher levels in the TFEB KO cells and increased the LC3II/I ratio (Figure 7F–H). This suggested that autophagic flux, more specifically the production of autophagic vesicles, was enhanced under conditions of TFEB deficiency. However, the regulation of UCP1 levels by lysosomal degradation might have been impaired in TFEB-deficient adipocytes, which could have been caused by dysregulated lysosomal acidification or fusion with autophagosomes since chloroquine treatment did not change UCP1 levels.
When measuring TFEB target gene expression in vivo in the mice acclimated to thermoneutrality for one week under starvation conditions that should induce nuclear translocation of TFEB, we did not observe any major alterations in the expression of genes encoding for lysosomal proteases (Ctsa and Ctsc), chloride channels (Clcn7), and the membrane protein Tmem55b (Figure S4D). The gene expression of the vATPase subunit Atp6v0d2 was reduced by trends in BAT of the TFEBBAT KO mice (Figure S4E), while other TFEB target genes and their related family members were unchanged between genotypes (Figure S4F).
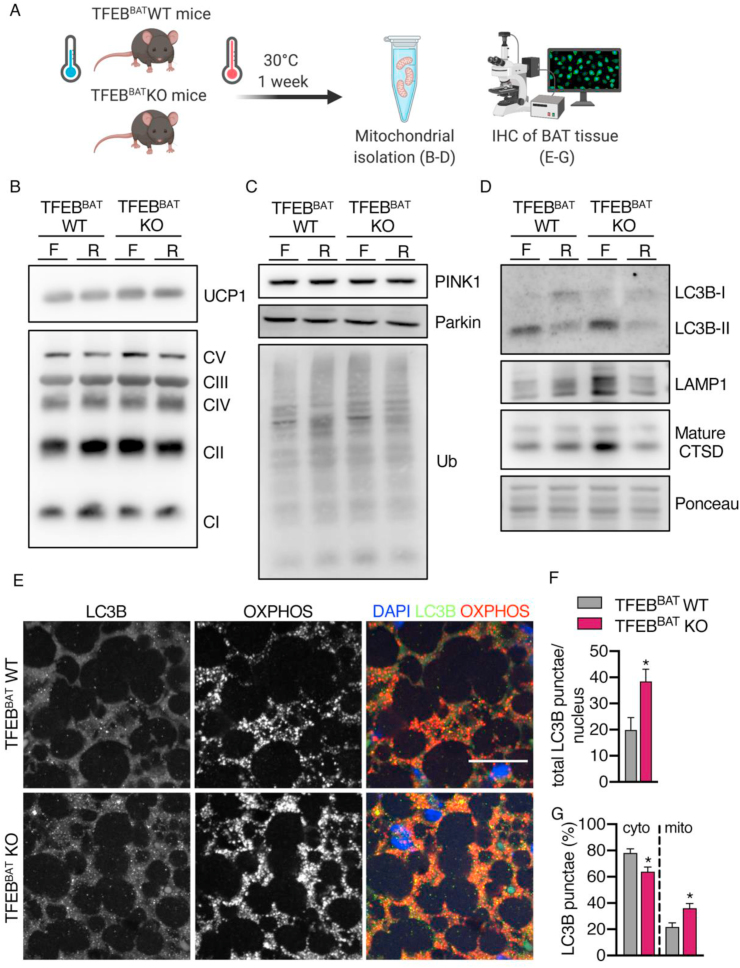
To further investigate the possible underlying mechanisms leading to higher UCP1 levels paired with a reduced thermogenic capacity in TFEB-deficient BAT, we isolated BAT mitochondria from the fasted and refed TFEBBAT WT and KO mice and analyzed the mitochondrial and autophagy-related protein levels (Figure 8A). Mitochondrial markers such as UCP1 and OXPHOS complex proteins were similar between genotypes (Figure 8B), suggesting no alterations in UCP1 protein content per mitochondria but rather more UCP1-positive mitochondria per BAT in the TFEB-deficient mice.
Figure 8.
Mitochondria were associated with autophagy and lysosome markers in TFEB-deficient BAT. TFEBBAT WT and KO mice were acclimated to thermoneutrality and either fasted (F) or fasted followed by 2 h of refeeding (R). (A) Schematic model of the experimental procedures. (B–D) Representative western blotting of (B) UCP1 and OXPHOS complex proteins, (C) PINK1, Parkin and ubiquitin (Ub), and (D) LC3B-I and II, LAMP1 and mature CTSD of isolated mitochondria from BAT of the fasted or fasted and refed TFEBBAT WT and KO mice (n = 1 each genotype and treatment). (E) Immunofluorescence analysis of OXPHOS (red) and LC3B (green) in whitened BAT from the fasted TFEBBAT WT and KO mice, scale bar 20 μm, with (F) total LC3B puncta count per nucleus, (G) relative distribution of cytosolic (cyto), and mitochondria-associated (mito) LC3B puncta (n = 5 each genotype). Values are presented as mean ± SEM. ∗P ≤ 0.05 by two-tailed unpaired Student's t test.
The recruitment of PINK1 and Parkin to the mitochondria and the mitochondrial ubiquitination status was similar between genotypes (Figure 8C). Robust recruitment of LC3B was observed in both genotypes and decreased with refeeding. Interestingly, we found an accumulation of LC3B-II associated with mitochondria from the fasted TFEBBAT KO mice compared to the WT controls (Figure 8D). The lysosomal membrane protein LAMP1 and the mature form of the lysosomal protease cathepsin D were detected in every sample, indicating that lysosomes were attached to mitochondria or some mitochondria were engulfed by autolysosomes (Figure 8D). Interestingly, mitochondria from the fasted TFEBBAT KO mice displayed increased levels of LAMP1 and mature CTSD.
To further investigate whether mitochondria in TFEB-deficient BAT might be associated with autophagic vesicles during BAT whitening, we performed immunofluorescence staining on BAT sections from the fasted TFEBBAT WT and KO mice acclimated to 30 °C for one week (Figure 8A). As expected, whitened TFEB-deficient BAT displayed a higher OXPHOS complex abundance (Figure 8E). Additionally, we detected an increased total number of LC3B puncta per nucleus (per cell) in TFEB-deficient BAT (Figure 8E,F). Interestingly and in line with the enhanced co-purification of autophagy and lysosomal proteins with mitochondria from TFEB-deficient BAT (Figure 8D), LC3B and OXPHOS complexes co-localized to a greater extent in TFEB-deficient BAT compared to BAT from the control mice (Figure 8E–G).
Together, these results suggested that BAT-specific TFEB deletion did not affect mitophagy initiation or the recruitment of the phagophore membrane but changed lysosomal protein levels associated with mitochondria of brown adipocytes.
4. Discussion
4.1. Mitochondrial homeostasis shifted toward degradation in thermoneutral BAT
An induction of mitochondrial non-shivering thermogenic capacity in brown or brite/beige adipose tissue can be achieved by physiological methods such as cold exposure or high-fat diet feeding [7,87]. Most recent studies therefore aimed at identifying processes that recruit or activate BAT or brite/beige WAT to ameliorate obesity and associated comorbidities.
This activation is necessary as humans spend most of their life under thermoneutral conditions (∼22 °C), resulting in whitening of BAT, which is characterized by diminished thermogenic capacity and activity [41,88]. We and others have observed a rapid decrease in mitochondrial mass/proteins, reduced mRNA levels of mitochondrial biogenesis markers, and continuous lipid accumulation upon exposure to thermoneutrality [36,37,41] or aging [44,89].
Accordingly, in contrast to activating brown or brite/beige mitochondrial biogenesis as a potential therapeutic strategy, mitochondrial mass may also be increased by lowering mitochondrial degradation during BAT whitening, thereby shifting the balance toward increased mitochondrial mass [70]. The major pathway of mitochondrial degradation, mitophagy, is downregulated upon cold activation of brown tissue [90,91] and recruitment of brite/beige adipose tissue [92]. It should however be noted that others reported elevated mitophagy under conditions of cold-induced BAT recruitment [77]. It may be speculated that this mainly serves as a quality control mechanism to eliminate damaged mitochondria [76,93], especially since severe blockage of autophagy in adipose tissues results in the loss of mitochondrial capacity, inflammation, and induction of stress pathways [94].
4.2. TFEB altered mitochondrial dynamics in BAT independently of biogenesis
It has been reported that deletion of the gene essential for autophagosome assembly, Atg7, in brown adipose tissue results in elevated levels of UCP1 [95,96] and improved metabolic health, as does deletion of the mitophagy-initiating protein Parkin [91]. Similarly, the whitening of brite/beige adipocytes is an autophagy-dependent process [46,47,70], and deletion or inhibition of key components of the autophagy pathway such as Atg5, Atg12, or Park2 results in elevated mitochondrial content and UCP1 levels [46,47,70,92,[96], [97], [98]]. Targeting early steps of mitophagy, such as Parkin, has been shown to result in retained mitochondrial mass in brite/beige adipose tissue during whitening following the withdrawal of browning stimuli [47]. Therefore, the function of autophagy in regulating white adipose tissue browning or the maintenance of brite/beige adipocytes has been posited to represent a novel target to enhance thermogenic capacity [55]. In addition to regulating mitochondrial turnover, autophagy also affects adipocyte differentiation and adipogenesis in vivo and in vitro [54,96,98], but we did not find strong effects of TFEB deletion on adipocyte differentiation.
Effects of TFEB overexpression on UCP1 levels have previously been observed only in white, but not brown adipose tissue [63]. However, under standard housing (22 °C) or thermoneutral conditions (30 °C), the expression levels of thermogenic markers in white adipose tissue are neglectable [64,[99], [100], [101]]. We therefore acclimated the wild-type and TFEBBAT KO mice to the cold (6 °C) to induce browning of white adipose tissue. We did not observe an altered metabolic response to the cold in the BAT TFEB-deficient mice and no alterations in the expression levels of thermogenic markers and nutrient uptake into BAT, indicating that the TFEB-PGC1a axis is dispensable or non-existent in classical brown adipose tissue (principally in line with [63]). However, we observed significant reductions in gene expression of markers of thermogenesis and mitochondrial biogenesis in white adipose tissue of the TFEB-deficient mice. This is in line with the observation that TFEB can activate mitochondrial biogenesis in white but not brown fat in a PGC1a-dependent manner [63]. However, these alterations did not translate into reductions in OXPHOS complex and UCP1 protein levels or nutrient uptake. Therefore, the physiological significance of this finding is questionable, especially in light of the generally low levels of UCP1 in brite/beige WAT compared to BAT [100].
4.3. Increased UCP1 levels in TFEB-deficient BAT were not thermogenically active
Several groups have reported increased mitochondrial mass upon genetic or pharmacological inhibition of autophagy and mitophagy in brite/beige or brown adipocytes [46,47,91,95,102]. While these models result in beneficial metabolic effects such as increased oxygen consumption, protection from diet-induced obesity, and insulin resistance, we did not observe advantageous effects on systemic metabolism caused by higher UCP1 levels and mitochondrial mass in BAT of the TFEBBAT KO mice. The TFEBBAT KO and wild-type mice exhibited comparable diet-induced body weight gain and showed a slightly impaired glucose clearance. The TFEBBAT KO mice also displayed a significantly lower increase in energy expenditure and body temperature in response to beta-adrenergic stimulation despite higher UCP1 levels. This dissociation of the thermogenic potential (as judged based on the total UCP1 levels) and actual thermogenic capacity (as judged by the response to CL injection) seemed surprising at first. One explanation would be insufficient activatability of the adrenergic pathway. However, both genotypes exhibited comparable stimulation of lipid oxidation (RQ), PKA activation, and HSL phosphorylation in BAT following CL injection. Another setting in which UCP1 levels do not correlate with the thermogenic response to adrenergic stimulation are conditions of UCP1 inhibition or lack of activation [38,103]. Indeed, BAT mitochondria isolated from the TFEBBAT KO mice displayed increased co-purification with lysosomal and autophagosomal markers compared to wild-type mitochondria. Mitochondria in TFEB-deficient BAT also showed enhanced co-localization with autophagic vesicles. It therefore seems possible that the increased UCP1 protein levels were within mitochondria that were trapped at later stages of the autophagosomal pathway, and thus rendered inaccessible to activation. This is clearly different from the results obtained in Parkin-deficient adipocytes [47,91], where earlier target recognition is blocked, presumably retaining mitochondrial function.
4.4. TFEB modulated stimulated but not basal autophagy protein levels
TFEB has been shown to regulate the expression of lysosomal and autophagy-related genes in a variety of cell types and cellular states [[48], [49], [50], [51]]. It therefore seems surprising that we could not find a strong effect of TFEB deletion on gene expression levels of several known TFEB target genes or basal autophagic protein levels. It should however be noted that TFEB-binding sites are found in the promoter regions of more than 400 genes [104] and in this study, we only measured a fraction of these genes. Interestingly, the overexpression or deletion of TFEB in muscle, which shares the same development origin as brown adipose tissue [[105], [106], [107]], only led to minor changes in the levels of autophagy and lysosome-related genes [59], as did TFEB overexpression in white adipose tissue [63]. Under conditions of TFEB activation due to starvation, we observed altered protein levels of LC3, a marker of the autophagosomal machinery, indicating somewhat altered activity of the autophagosomal/lysosomal pathway that could ultimately result in reduced mitochondrial degradation in TFEB-deficient BAT.
5. Conclusion
Our findings highlight that targeting mitophagy, in this case the lysosomal pathway, can increase UCP1 levels under conditions of BAT whitening. Targeting the whitening process might be beneficial for metabolic health since increased BAT thermogenesis has been shown to be associated with improved glucose and lipid metabolism and weight loss in mice and humans. The increased UCP1 levels in the TFEBBAT KO mice were not thermogenically active and did not provide protection against high-fat diet-induced glucose intolerance and weight gain. This was presumably due to inaccessibility of the mitochondria to thermogenic stimuli. Nevertheless, our model did not present the severe side effects of autophagy and lysosomal degradation deficiency observed in other models, highlighting that TFEB was not responsible for homeostatic expression of lysosomal genes in BAT. Targeting the lysosomal/autophagosomal pathway at an earlier stage at the level of target recognition and delivery to autophagosomes therefore appears superior over targeting the lysosomal pathway in general, yet the risk of inducing lysosomal storage defects must be carefully considered and monitored.
Author contributions
F.S. conducted the experiments, analyzed the data, prepared the figures, and drafted and commented on the manuscript. C.S., M.Y.J., P.P., and M.S. performed experiments and commented on the manuscript. T.S. and A.B. provided the reagents, designed the experiments, and commented on the manuscript. L.S. and J.H. designed the experiments and edited the manuscript. A.W.F. designed and conducted the experiments, analyzed the data, and edited the manuscript.
Funding
J.H. and L.S. were supported by the German Research Foundation DFG (Heisenberg Professorship HE3645/10-1 to J.H. and Research Grant SCHE 522/4-1 to L.S.). C.S. was supported by a fellowship from Gertraud und Heinz Rose Stiftung and the UKE MD/PhD program fellowship. A.B. was supported by grants from the Italian Telethon Foundation (TGM16CB6), MIUR FIRB RBAP11Z3YA, and the European Research Council Advanced Investigator no. 694282 (LYSOSOMICS). A.W.F. was supported by the DFG research training group GRK1459, the DACH Society for Lipidology, a DFG Research Grant (FI 2476/1-1), and a Human Frontier Science Program (HFSP) fellowship.
Acknowledgments
The authors thank Sandra Ehret, Walter Tauscher, Eva-Marie Azizi, and Birgit Henkel for excellent technical assistance. Schematic drawings were created using BioRender (biorender.com).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101173.
Conflict of interest
A.B. is co-founder of CASMA Therapeutics, Inc., Cambridge, MA, USA 02139. All of the other authors have nothing to declare.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Nedergaard J., Bengtsson T., Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. American Journal of Physiology - Endocrinology And Metabolism. 2007;293(2):E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 2.Saito M., Okamatsu-Ogura Y., Matsushita M., Watanabe K., Yoneshiro T., Nio-Kobayashi J. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58(7):1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M., Kemerink G.J., Bouvy N.D. Cold-activated brown adipose tissue in healthy men. New England Journal of Medicine. 2009;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 4.Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B. Identification and importance of brown adipose tissue in adult humans. New England Journal of Medicine. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Virtanen K.A., Lidell M.E., Orava J., Heglind M., Westergren R., Niemi T. Functional brown adipose tissue in healthy adults. New England Journal of Medicine. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 6.Yoneshiro T., Aita S., Matsushita M., Kayahara T., Kameya T., Kawai Y. Recruited brown adipose tissue as an antiobesity agent in humans. Journal of Clinical Investigation. 2013;123(8):3404–3408. doi: 10.1172/JCI67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 8.Bartelt A., Bruns O.T., Reimer R., Hohenberg H., Ittrich H., Peldschus K. Brown adipose tissue activity controls triglyceride clearance. Nature Medicine. 2011;17(2):200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- 9.Heine M., Fischer A.W., Schlein C., Jung C., Straub L.G., Gottschling K. Lipolysis triggers a systemic insulin response essential for efficient energy replenishment of activated Brown adipose tissue in mice. Cell Metabolism. 2018 doi: 10.1016/j.cmet.2018.06.020. [DOI] [PubMed] [Google Scholar]
- 10.Berbee J.F., Boon M.R., Khedoe P.P., Bartelt A., Schlein C., Worthmann A. Brown fat activation reduces hypercholesterolaemia and protects from atherosclerosis development. Nature Communications. 2015;6:6356. doi: 10.1038/ncomms7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furler S.M., Cooney G.J., Hegarty B.D., Lim-Fraser M.Y., Kraegen E.W., Oakes N.D. Local factors modulate tissue-specific NEFA utilization: assessment in rats using 3H-(R)-2-bromopalmitate. Diabetes. 2000;49(9):1427–1433. doi: 10.2337/diabetes.49.9.1427. [DOI] [PubMed] [Google Scholar]
- 12.Henkin A.H., Cohen A.S., Dubikovskaya E.A., Park H.M., Nikitin G.F., Auzias M.G. Real-time noninvasive imaging of fatty acid uptake in vivo. ACS Chemical Biology. 2012;7(11):1884–1891. doi: 10.1021/cb300194b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlein C., Talukdar S., Heine M., Fischer A.W., Krott L.M., Nilsson S.K. FGF21 lowers plasma triglycerides by accelerating lipoprotein catabolism in white and Brown adipose tissues. Cell Metabolism. 2016;23(3):441–453. doi: 10.1016/j.cmet.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Liu X., Perusse F., Bukowiecki L.J. Chronic norepinephrine infusion stimulates glucose uptake in white and brown adipose tissues. American Journal of Physiology. 1994;266(3 Pt 2):R914–R920. doi: 10.1152/ajpregu.1994.266.3.R914. [DOI] [PubMed] [Google Scholar]
- 15.Ma S.W., Foster D.O. Uptake of glucose and release of fatty acids and glycerol by rat brown adipose tissue in vivo. Canadian Journal of Physiology and Pharmacology. 1986;64(5):609–614. doi: 10.1139/y86-101. [DOI] [PubMed] [Google Scholar]
- 16.Fischer A.W., Albers K., Schlein C., Sass F., Krott L.M., Schmale H. PID1 regulates insulin-dependent glucose uptake by controlling intracellular sorting of GLUT4-storage vesicles. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2019;1865(6):1592–1603. doi: 10.1016/j.bbadis.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nedergaard J., Bengtsson T., Cannon B. New powers of brown fat: fighting the metabolic syndrome. Cell Metabolism. 2011;13(3):238–240. doi: 10.1016/j.cmet.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Bartelt A., Heeren J. Adipose tissue browning and metabolic health. Nature Reviews Endocrinology. 2014;10(1):24–36. doi: 10.1038/nrendo.2013.204. [DOI] [PubMed] [Google Scholar]
- 19.Bartelt A., John C., Schaltenberg N., Berbee J.F.P., Worthmann A., Cherradi M.L. Thermogenic adipocytes promote HDL turnover and reverse cholesterol transport. Nature Communications. 2017;8:15010. doi: 10.1038/ncomms15010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanford K.I., Middelbeek R.J., Townsend K.L., An D., Nygaard E.B., Hitchcox K.M. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. Journal of Clinical Investigation. 2013;123(1):215–223. doi: 10.1172/JCI62308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwen K.A., Backhaus J., Cassens M., Waltl M., Hedesan O.C., Merkel M. Cold-induced Brown adipose tissue activity alters plasma fatty acids and improves glucose metabolism in men. Journal of Clinical Endocrinology & Metabolism. 2017;102(11):4226–4234. doi: 10.1210/jc.2017-01250. [DOI] [PubMed] [Google Scholar]
- 22.Maloney S.K., Fuller A., Mitchell D., Gordon C., Overton J.M. Translating animal model research: does it matter that our rodents are cold? Physiology. 2014;29(6):413. doi: 10.1152/physiol.00029.2014. [DOI] [PubMed] [Google Scholar]
- 23.Johnson F., Mavrogianni A., Ucci M., Vidal-Puig A., Wardle J. Could increased time spent in a thermal comfort zone contribute to population increases in obesity? Obesity Reviews. 2011;12(7):543–551. doi: 10.1111/j.1467-789X.2010.00851.x. [DOI] [PubMed] [Google Scholar]
- 24.Fischer A.W., Cannon B., Nedergaard J. Optimal housing temperatures for mice to mimic the thermal environment of humans: an experimental study. Molecular Metabolism. 2018;7:161–170. doi: 10.1016/j.molmet.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speakman J.R., Keijer J. Not so hot: optimal housing temperatures for mice to mimic the thermal environment of humans. Molecular Metabolism. 2012;2(1):5–9. doi: 10.1016/j.molmet.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scholander P.F., Hock R., Walters V., Johnson F., Irving L. Heat regulation in some arctic and tropical mammals and birds. Biological Bulletin. 1950;99(2):237–258. doi: 10.2307/1538741. [DOI] [PubMed] [Google Scholar]
- 27.Fischer A.W., Hoefig C.S., Abreu-Vieira G., de Jong J.M., Petrovic N., Mittag J. Leptin raises defended body temperature without activating thermogenesis. Cell Reports. 2016;14(7):1621–1631. doi: 10.1016/j.celrep.2016.01.041. [DOI] [PubMed] [Google Scholar]
- 28.Fischer A.W., Csikasz R.I., von Essen G., Cannon B., Nedergaard J. No insulating effect of obesity. American Journal of Physiology - Endocrinology And Metabolism. 2016;311(1):202–213. doi: 10.1152/ajpendo.00093.2016. [DOI] [PubMed] [Google Scholar]
- 29.Fischer A.W., Cannon B., Nedergaard J. The answer to the question "What is the best housing temperature to translate mouse experiments to humans?" is: thermoneutrality. Molecular Metabolism. 2019 doi: 10.1016/j.molmet.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nedergaard J., Cannon B. The browning of white adipose tissue: some burning issues. Cell Metabolism. 2014;20(3):396–407. doi: 10.1016/j.cmet.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Tian Xiao Y., Ganeshan K., Hong C., Nguyen Khoa D., Qiu Y., Kim J. Thermoneutral housing accelerates metabolic inflammation to potentiate atherosclerosis but not insulin resistance. Cell Metabolism. 2016;23(1):165–178. doi: 10.1016/j.cmet.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giles D.A., Moreno-Fernandez M.E., Stankiewicz T.E., Graspeuntner S., Cappelletti M., Wu D. Thermoneutral housing exacerbates nonalcoholic fatty liver disease in mice and allows for sex-independent disease modeling. Nature Medicine. 2017;23(7):829–838. doi: 10.1038/nm.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganeshan K., Chawla A. Warming the mouse to model human diseases. Nature Reviews Endocrinology. 2017;13(8):458–465. doi: 10.1038/nrendo.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudele A., Rasmussen G.M., Mayntz D., Malte H., Lund S., Wang T. Effects of ambient temperature on glucose tolerance and insulin sensitivity test outcomes in normal and obese C57 male mice. Physiological Reports. 2015;3(5) doi: 10.14814/phy2.12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feldmann H.M., Golozoubova V., Cannon B., Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metabolism. 2009;9(2):203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 36.Cui X., Nguyen N.L., Zarebidaki E., Cao Q., Li F., Zha L. Thermoneutrality decreases thermogenic program and promotes adiposity in high-fat diet-fed mice. Physiological Reports. 2016;4(10) doi: 10.14814/phy2.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez-Gurmaches J., Tang Y., Jespersen N.Z., Wallace M., Martinez Calejman C., Gujja S. Brown fat AKT2 is a cold-induced kinase that stimulates ChREBP-mediated De Novo lipogenesis to optimize fuel storage and thermogenesis. Cell Metabolism. 2018;27(1):195–209. doi: 10.1016/j.cmet.2017.10.008. e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer A.W., Shabalina I.G., Mattsson C.L., Abreu-Vieira G., Cannon B., Nedergaard J. UCP1 inhibition in Cidea-overexpressing mice is physiologically counteracted by brown adipose tissue hyperrecruitment. American Journal of Physiology - Endocrinology And Metabolism. 2017;312(1):E72–E87. doi: 10.1152/ajpendo.00284.2016. [DOI] [PubMed] [Google Scholar]
- 39.Kotzbeck P., Giordano A., Mondini E., Murano I., Severi I., Venema W. Brown adipose tissue whitening leads to brown adipocyte death and adipose tissue inflammation. Journal of Lipid Research. 2018;59(5):784–794. doi: 10.1194/jlr.M079665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abreu-Vieira G., Fischer A.W., Mattsson C., de Jong J.M.A., Shabalina I.G., Rydén M. Cidea improves the metabolic profile through expansion of adipose tissue. Nature Communications. 2015;6:7433. doi: 10.1038/ncomms8433. [DOI] [PubMed] [Google Scholar]
- 41.de Jong J.M.A., Sun W., Pires N.D., Frontini A., Balaz M., Petrovic K. Human brown adipose tissue is phenocopied by classical brown (but not beige) adipose tissue in physiologically humanized mice. Nature Metabolism. 2019;1(8):830–843. doi: 10.1038/s42255-019-0101-4. [DOI] [PubMed] [Google Scholar]
- 42.Cannon B., de Jong J.M.A., Fischer A.W., Nedergaard J., Petrovic N. Human brown adipose tissue: classical brown rather than brite/beige? Experimental Physiology. 2020 doi: 10.1113/EP087875. [DOI] [PubMed] [Google Scholar]
- 43.Graja A., Gohlke S., Schulz T.J. Aging of Brown and beige/brite adipose tissue. Handbook of Experimental Pharmacology. 2019;251:55–72. doi: 10.1007/164_2018_151. [DOI] [PubMed] [Google Scholar]
- 44.Goncalves L.F., Machado T.Q., Castro-Pinheiro C., de Souza N.G., Oliveira K.J., Fernandes-Santos C. Ageing is associated with brown adipose tissue remodelling and loss of white fat browning in female C57BL/6 mice. International Journal of Experimental Pathology. 2017;98(2):100–108. doi: 10.1111/iep.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cinti S. Adipose organ development and remodeling. Comprehensive Physiology. 2018;8(4):1357–1431. doi: 10.1002/cphy.c170042. [DOI] [PubMed] [Google Scholar]
- 46.Altshuler-Keylin S., Shinoda K., Hasegawa Y., Ikeda K., Hong H., Kang Q. Beige adipocyte maintenance is regulated by autophagy-induced mitochondrial clearance. Cell Metabolism. 2016;24(3):402–419. doi: 10.1016/j.cmet.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu X., Altshuler-Keylin S., Wang Q., Chen Y., Henrique Sponton C., Ikeda K. Mitophagy controls beige adipocyte maintenance through a Parkin-dependent and UCP1-independent mechanism. Science Signaling. 2018;11(527) doi: 10.1126/scisignal.aap8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 49.Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Human Molecular Genetics. 2011;20(19):3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 50.Napolitano G., Ballabio A. TFEB at a glance. Journal of Cell Science. 2016;129(13):2475–2481. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nezich C.L., Wang C., Fogel A.I., Youle R.J. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. Journal of Cell Biology. 2015;210(3):435–450. doi: 10.1083/jcb.201501002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pastore N., Vainshtein A., Klisch T.J., Armani A., Huynh T., Herz N.J. TFE3 regulates whole-body energy metabolism in cooperation with TFEB. EMBO Molecular Medicine. 2017;9(5):605–621. doi: 10.15252/emmm.201607204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh R., Xiang Y., Wang Y., Baikati K., Cuervo A.M., Luu Y.K. Autophagy regulates adipose mass and differentiation in mice. Journal of Clinical Investigation. 2009;119(11):3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ro S.H., Jang Y., Bae J., Kim I.M., Schaecher C., Shomo Z.D. Autophagy in adipocyte browning: emerging drug target for intervention in obesity. Frontiers in Physiology. 2019;10:22. doi: 10.3389/fphys.2019.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salma N., Song J.S., Arany Z., Fisher D.E. Transcription factor Tfe3 directly regulates Pgc-1alpha in muscle. Journal of Cellular Physiology. 2015;230(10):2330–2336. doi: 10.1002/jcp.24978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salma N., Song J.S., Kawakami A., Devi S.P., Khaled M., Cacicedo J.M. Tfe3 and Tfeb transcriptionally regulate peroxisome proliferator-activated receptor gamma2 expression in adipocytes and mediate adiponectin and glucose levels in mice. Molecular and Cellular Biology. 2017;37(15) doi: 10.1128/MCB.00608-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erlich A.T., Brownlee D.M., Beyfuss K., Hood D.A. Exercise induces TFEB expression and activity in skeletal muscle in a PGC-1alpha-dependent manner. American Journal of Physiology - Cell Physiology. 2018;314(1):C62–C72. doi: 10.1152/ajpcell.00162.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mansueto G., Armani A., Viscomi C., D'Orsi L., De Cegli R., Polishchuk E.V. Transcription factor EB controls metabolic flexibility during exercise. Cell Metabolism. 2017;25(1):182–196. doi: 10.1016/j.cmet.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Settembre C., De Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nature Cell Biology. 2013;15(6):647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inagaki T., Sakai J., Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nature Reviews Molecular Cell Biology. 2016;17(8):480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spiegelman B.M., Puigserver P., Wu Z. Regulation of adipogenesis and energy balance by PPARgamma and PGC-1. International Journal of Obesity and Related Metabolic Disorders. 2000;24(Suppl 4):S8–S10. doi: 10.1038/sj.ijo.0801492. [DOI] [PubMed] [Google Scholar]
- 63.Evans T.D., Zhang X., Jeong S.J., He A., Song E., Bhattacharya S. TFEB drives PGC-1alpha expression in adipocytes to protect against diet-induced metabolic dysfunction. Science Signaling. 2019;12(606) doi: 10.1126/scisignal.aau2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischer A.W., Schlein C., Cannon B., Heeren J., Nedergaard J. Intact innervation is essential for diet-induced recruitment of brown adipose tissue. American Journal of Physiology: Endocrinology and Metabolism. 2018 doi: 10.1152/ajpendo.00443.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weir J.B. New methods for calculating metabolic rate with special reference to protein metabolism. Journal of Physiology. 1949;109(1–2):1–9. doi: 10.1113/jphysiol.1949.sp004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shabalina I.G., Petrovic N., de Jong J.M., Kalinovich A.V., Cannon B., Nedergaard J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Reports. 2013;5(5):1196–1203. doi: 10.1016/j.celrep.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 67.Fischer A.W., Albers K., Krott L.M., Hoffzimmer B., Heine M., Schmale H. The adaptor protein PID1 regulates receptor-dependent endocytosis of postprandial triglyceride-rich lipoproteins. Molecular Metabolism. 2018 doi: 10.1016/j.molmet.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galarraga M., Campion J., Munoz-Barrutia A., Boque N., Moreno H., Martinez J.A. Adiposoft: automated software for the analysis of white adipose tissue cellularity in histological sections. Journal of Lipid Research. 2012;53(12):2791–2796. doi: 10.1194/jlr.D023788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palikaras K., Tavernarakis N. Mitochondrial homeostasis: the interplay between mitophagy and mitochondrial biogenesis. Experimental Gerontology. 2014;56:182–188. doi: 10.1016/j.exger.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 70.Altshuler-Keylin S., Kajimura S. Mitochondrial homeostasis in adipose tissue remodeling. Science Signaling. 2017;10(468) doi: 10.1126/scisignal.aai9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cannon B., Nedergaard J. Nonshivering thermogenesis and its adequate measurement in metabolic studies. Journal of Experimental Biology. 2011;214(Pt 2):242–253. doi: 10.1242/jeb.050989. [DOI] [PubMed] [Google Scholar]
- 72.Virtue S., Vidal-Puig A. Assessment of brown adipose tissue function. Frontiers in Physiology. 2013;4:128. doi: 10.3389/fphys.2013.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trayhurn P., Milner R.E. A commentary on the interpretation of in vitro biochemical measures of brown adipose tissue thermogenesis. Canadian Journal of Physiology and Pharmacology. 1989;67(8):811–819. doi: 10.1139/y89-128. [DOI] [PubMed] [Google Scholar]
- 74.Fischer A.W., Cannon B., Nedergaard J. Leptin - is it thermogenic? Endocrine Reviews. 2019 doi: 10.1210/endrev/bnz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeng X., Ye M., Resch J.M., Jedrychowski M.P., Hu B., Lowell B.B. Innervation of thermogenic adipose tissue via a calsyntenin 3beta-S100b axis. Nature. 2019;569(7755):229–235. doi: 10.1038/s41586-019-1156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y., Zeng X., Jin S. Autophagy in adipose tissue biology. Pharmacological Research. 2012;66(6):505–512. doi: 10.1016/j.phrs.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 77.Lu Y., Fujioka H., Joshi D., Li Q., Sangwung P., Hsieh P. Mitophagy is required for brown adipose tissue mitochondrial homeostasis during cold challenge. Scientific Reports. 2018;8(1):8251. doi: 10.1038/s41598-018-26394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee Y.H., Kim S.N., Kwon H.J., Granneman J.G. Metabolic heterogeneity of activated beige/brite adipocytes in inguinal adipose tissue. Scientific Reports. 2017;7:39794. doi: 10.1038/srep39794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berry D.C., Jiang Y., Graff J.M. Mouse strains to study cold-inducible beige progenitors and beige adipocyte formation and function. Nature Communications. 2016;7:10184. doi: 10.1038/ncomms10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loh R.K.C., Kingwell B.A., Carey A.L. Human brown adipose tissue as a target for obesity management; beyond cold-induced thermogenesis. Obesity Reviews. 2017;18(11):1227–1242. doi: 10.1111/obr.12584. [DOI] [PubMed] [Google Scholar]
- 81.Chechi K., Nedergaard J., Richard D. Brown adipose tissue as an anti-obesity tissue in humans. Obesity Reviews. 2014;15(2):92–106. doi: 10.1111/obr.12116. [DOI] [PubMed] [Google Scholar]
- 82.Lidell M.E., Betz M.J., Enerback S. Brown adipose tissue and its therapeutic potential. Journal of Internal Medicine. 2014;276(4):364–377. doi: 10.1111/joim.12255. [DOI] [PubMed] [Google Scholar]
- 83.Luijten I.H.N., Feldmann H.M., von Essen G., Cannon B., Nedergaard J. In the absence of UCP1-mediated diet-induced thermogenesis, obesity is augmented even in the obesity-resistant 129S mouse strain. American Journal of Physiology – Endocrinology And Metabolism. 2019;316(5):E729–E740. doi: 10.1152/ajpendo.00020.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoshii S.R., Mizushima N. Monitoring and measuring autophagy. International Journal of Molecular Sciences. 2017;18(9) doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.J. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14(8):1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seglen P.O., Grinde B., Solheim A.E. Inhibition of the lysosomal pathway of protein degradation in isolated rat hepatocytes by ammonia, methylamine, chloroquine and leupeptin. European Journal of Biochemistry. 1979;95(2):215–225. doi: 10.1111/j.1432-1033.1979.tb12956.x. [DOI] [PubMed] [Google Scholar]
- 87.Heeren J., Scheja L. Brown adipose tissue and lipid metabolism. Current Opinion in Lipidology. 2018;29(3):180–185. doi: 10.1097/MOL.0000000000000504. [DOI] [PubMed] [Google Scholar]
- 88.Moonen M.P.B., Nascimento E.B.M., van Marken Lichtenbelt W.D. Human brown adipose tissue: underestimated target in metabolic disease? Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2019;1864(1):104–112. doi: 10.1016/j.bbalip.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 89.Cui X., Xiao W., You L., Zhang F., Cao X., Feng J. Age-induced oxidative stress impairs adipogenesis and thermogenesis in brown fat. The FEBS Journal. 2019;286(14):2753–2768. doi: 10.1111/febs.14838. [DOI] [PubMed] [Google Scholar]
- 90.Cairo M., Villarroya J., Cereijo R., Campderros L., Giralt M., Villarroya F. Thermogenic activation represses autophagy in brown adipose tissue. International Journal of Obesity (London) 2016;40(10):1591–1599. doi: 10.1038/ijo.2016.115. [DOI] [PubMed] [Google Scholar]
- 91.Cairó M., Campderrós L., Gavaldà-Navarro A., Cereijo R., Delgado-Anglés A., Quesada-López T. Parkin controls brown adipose tissue plasticity in response to adaptive thermogenesis. EMBO Reports. 2019;20(5) doi: 10.15252/embr.201846832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taylor D., Gottlieb R.A. Parkin-mediated mitophagy is downregulated in browning of white adipose tissue. Obesity (Silver Spring) 2017;25(4):704–712. doi: 10.1002/oby.21786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ashrafi G., Schwarz T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death & Differentiation. 2013;20(1):31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cai J., Pires K.M., Ferhat M., Chaurasia B., Buffolo M.A., Smalling R. Autophagy ablation in adipocytes induces insulin resistance and reveals roles for lipid Peroxide and Nrf2 signaling in adipose-liver crosstalk. Cell Reports. 2018;25(7):1708–1717. doi: 10.1016/j.celrep.2018.10.040. e1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim D., Kim J.H., Kang Y.H., Kim J.S., Yun S.C., Kang S.W. Suppression of Brown adipocyte autophagy improves energy metabolism by regulating mitochondrial turnover. International Journal of Molecular Sciences. 2019;20(14) doi: 10.3390/ijms20143520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Y., Goldman S., Baerga R., Zhao Y., Komatsu M., Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(47):19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singh R., Cuervo A.M. Lipophagy: connecting autophagy and lipid metabolism. International Journal of Cell Biology. 2012;2012:282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Parray H.A., Yun J.W. Combined inhibition of autophagy protein 5 and galectin-1 by thiodigalactoside reduces diet-induced obesity through induction of white fat browning. IUBMB Life. 2017;69(7):510–521. doi: 10.1002/iub.1634. [DOI] [PubMed] [Google Scholar]
- 99.de Jong J.M., Larsson O., Cannon B., Nedergaard J. A stringent validation of mouse adipose tissue identity markers. American Journal of Physiology - Endocrinology and Metabolism. 2015;308(12):E1085–E1105. doi: 10.1152/ajpendo.00023.2015. [DOI] [PubMed] [Google Scholar]
- 100.Kalinovich A.V., de Jong J.M., Cannon B., Nedergaard J. UCP1 in adipose tissues: two steps to full browning. Biochimie. 2017;134:127–137. doi: 10.1016/j.biochi.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 101.Nedergaard J., Cannon B. UCP1 mRNA does not produce heat. Biochimica et Biophysica Acta. 2013;1831(5):943–949. doi: 10.1016/j.bbalip.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 102.Deng J., Guo Y., Yuan F., Chen S., Yin H., Jiang X. Autophagy inhibition prevents glucocorticoid-increased adiposity via suppressing BAT whitening. Autophagy. 2019:1–15. doi: 10.1080/15548627.2019.1628537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Johann K., Cremer A.L., Fischer A.W., Heine M., Pensado E.R., Resch J. Thyroid-hormone-induced browning of white adipose tissue does not contribute to thermogenesis and glucose consumption. Cell Reports. 2019;27(11):3385–3400. doi: 10.1016/j.celrep.2019.05.054. e3383. [DOI] [PubMed] [Google Scholar]
- 104.Sardiello M. Transcription factor EB: from master coordinator of lysosomal pathways to candidate therapeutic target in degenerative storage diseases. Annals of the New York Academy of Sciences. 2016;1371(1):3–14. doi: 10.1111/nyas.13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Walden T.B., Timmons J.A., Keller P., Nedergaard J., Cannon B. Distinct expression of muscle-specific MicroRNAs (myomirs) in brown adipocytes. Journal of Cellular Physiology. 2009;218(2):444–449. doi: 10.1002/jcp.21621. [DOI] [PubMed] [Google Scholar]
- 106.Timmons J.A., Wennmalm K., Larsson O., Walden T.B., Lassmann T., Petrovic N. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proceedings of the National Academy of Sciences. 2007;104(11):4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sanchez-Gurmaches J., Guertin D.A. Adipocyte lineages: tracing back the origins of fat. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2014;1842(3):340–351. doi: 10.1016/j.bbadis.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.