Abstract
This study aimed to identifying the presence of SARS-CoV-2 RNA in raw and treated wastewater during the COVID-19 outbreak in Tehran, Qom and Anzali cities (Iran). From three wastewater treatment plants (WWTPs), 28 treated and untreated wastewater composite samples were collected from April 4 to May 2, 2020. In this study, polyethylene glycol 6000 (PEG 6000) was used through one-step real-time quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) for identification of RNA viruses. SARS-CoV-2 RNA was elicited from wastewater composite samples in all inlet samples taken from the three above mentioned cities. The results of outlet samples were as follows: 1) Results from Qom and East Anzali outlets showed no trace of SARS-CoV-2 RNA despite the difference in treatment disinfection method used (chlorine vs. ultraviolet (UV) disinfection). 2. In Tehran, SARS-CoV-2 RNA was not detected in any of the outlet samples taken from the modules disinfected by UV. Out of the four samples taken from the modules disinfected by chlorine, two were positive for the SARS-CoV-2 RNA which could have been caused by deficiencies in operation and maintenance. It can be concluded that meeting the standards of operation and maintenance (O&M) in WWTPs can considerably ensure that wastewater does not act as one of the roots of transmission for the disease.
Keywords: Wastewater treatment, COVID-19, SARS-CoV-2, Disinfection, Pandemic
Introduction
The rapid outbreak of coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that is spread via airborne, droplet, contact, and fecal-oral transmission modes constitutes a worldwide and disruptive challenge to societies [1, 2]. Both symptomatic and asymptomatic carriers have been recognized to transmit the SARS-CoV-2 virus via various modes of transmission [3–6]. Causing severe respiratory infections among patients receiving medical care in hospitals [7–12]. Hence, the COVID-19 outbreak has been declared a global public health emergency of international concern by the World Health Organization (WHO) [13–15]. The original source and the way of transmission to humans is the subject of current studies [6, 14, 16–18].
Usual human coronaviruses, containing kinds alpha coronavirus and beta coronavirus, commonly create mild to moderate and upper respiratory tract infections (URTI), similar to the common cold disease [19–22], while coronaviruses, containing the SARS-CoV-2 (a new coronavirus) has created severe respiratory illness and even death in some patients [16–18, 20, 23–25]. In addition, it should be noted that SARS-CoV-2 belongs to beta-CoVs and its size is between 60 and 140 nm [26].
In addition, the monitoring of viruses and bioaerosols in wastewater and water have been reported many years ago [27, 28]. For example, former studies have confirmed the presence of rotavirus, measles virus and poliovirus in water and wastewater [29, 30]. Besides, previous studies have shown that coronaviruses can exist and maintain viable in sewages originated from the fecal discharge of infected patients [31, 32]. In the case of SARS-CoV epidemic in March 2003, there had been a potential link to water and sewage, and more than 300 people in Hong Kong had been infected with the virus through a faulty sewage system [33], due to SARS-CoV which could multiply the intestinal tract with possibility of gastrointestinal transmission of the virus [34].
In the case of SARS, Chan et al. (2004) reported that the symptom of diarrhea was observed between 8 and 73% in patients, and they declared the raising concerns about the potential of environmental transmission of the virus [34]. Furthermore, Liu et al. (2004) and reported that the SARS-CoV virus was also cultured from the feces of SARS-CoV patients up to 3 weeks after infection [35]. In addition, the recent works declared that coronaviruses can also remain infectious for a few days in sewage [36, 37].
The transmission of SARS-CoV-2 is still unknown, while according to previous works, the main way of transmission of SARS-CoV-2 is human-to-human [14, 38]. Furthermore, it is not yet known whether the SARS-CoV-2 was first transmitted to humans through animals or through virus-infected surfaces; however, transmission of the virus from surfaces is also another main way of transmitting the virus [39]. Besides, presence of SARS-CoV-2 RNA in the feces of COVID-19, demonstrates that transmission can potentially create by fecal- oral routes [40–42].
Hence, although direct transmission through respiratory aerosols, where the persons are in close contact and contact with contaminated surfaces, are considered as the most important routes of transmission [43–46], there is still generally limited information about the potential for transmission through the environment like water and wastewaters. However, some researchers reported that sewage/domestic wastewater surveillance could be a sensitive tool to monitor the circulation of the virus in the population [30, 47–49]; For example, Ahmed et al. (2020) studied on the wastewater in a catchment in Australia for identification of SARS-CoV-2 RNA [50]. The results of their work described that RNA copies were counted in two positive wastewater samples in a six-day period by reverse transcriptase quantitative polymerase chain reaction (RT- qPCR) [50].
In addition, whereas the new virus has already begun to spread worldwide, it is important for water engineers and professionals to understand the nature and fate of coronavirus after discharging into aquatic environments and the effective measures to protect public health. Due to the outbreak of coronavirus disease 2019 (COVID-19) in Iran and lack of sufficient knowledge and information about the environmental persistence of this the RNA viruses in wastewater in this country, it is important to conduct research on the identification of the RNA viruses in raw and treated wastewater in main epicenters of the country. Moreover, the use of raw and treated wastewater for agriculture in Iran is a common practice in many cities especially the COVID-19 epicenters cities, including Tehran, Anzali and Qom. Therefore the presence of this the RNA viruses in produced wastewater of such cities may be associated with the possibility of its presence in agricultural produces and make it possible for the virus to be transmitted to humans.
The basic novelty of this research is that of the presence of SARS-CoV-2 RNA in raw and treated municipal wastewater in the most densely populated cities of Iran (Tehran, Anzali and Qom) with high outbreak of COVID-19. Hence, the goal of this work is multi-fold: 1) study the presence of SARS-CoV-2 RNA in inlets (raw municipal wastewater) and outlets (treated municipal wastewater) of three wastewater treatment plants (WWTPs) in the three cities of Iran: Tehran, Qom and Anzali; 2) investigate the impact on the composition of wastewater on SARS-CoV-2 RNA; 3) study the impact on disinfection (UV and chlorine) on the SARS-CoV-2 RNA in treated municipal wastewater (outlets of wastewater treatment plants (WWTPs)). The results of this work can have broad implications for other regions owing to the pervasiveness of COVID-19.
Materials and methods
Chemicals
Sodium chloride (NaCl) (Merck: 7647-14-5), sodium hydroxide (NaOH) anhydrous pellets (CAS Number: 1310-73-2), dextran sulfate sodium salt (Sigma-Aldrich: Lot Number, 8C8R4068V), polyethylene glycol (PEG 6000) (Sigma-Aldrich: CAS Number: 81255) were purchased and used for identification of SARS-CoV-2 RNA from wastewater samples. In addition, SPL Centrifuge Tubes (falcon tubes 50 mL- SPL Life Sciences Co-50,050) was applied for sample preparation.
Study area
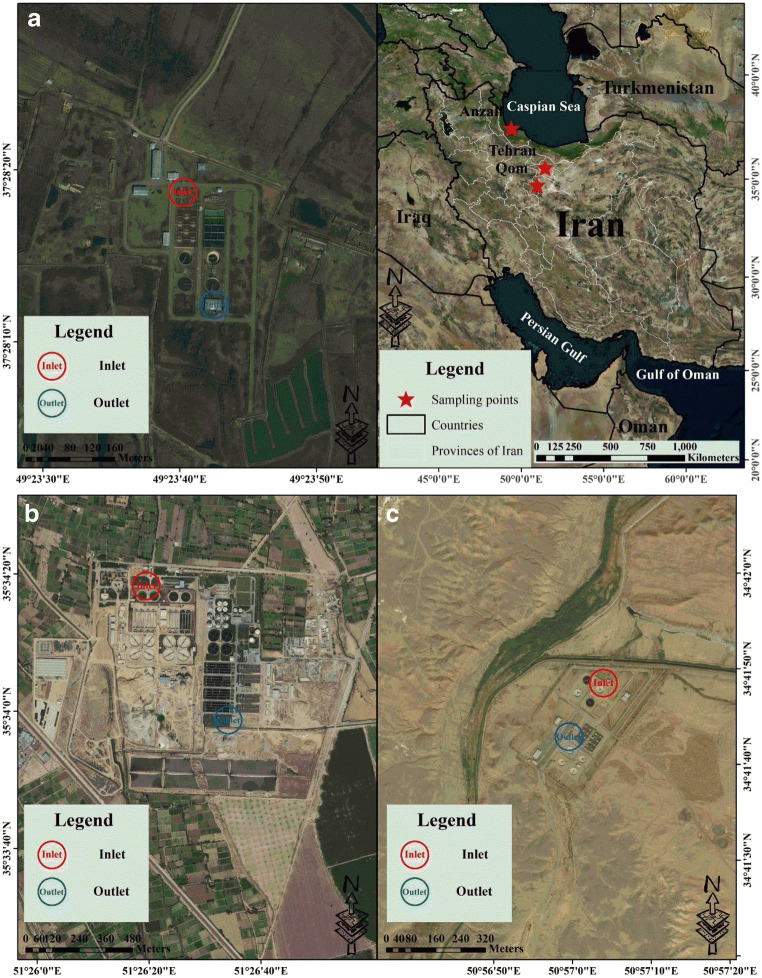
In this study, sampling were performed in three WWTPs in the three cities of Iran: Tehran, Qom and Anzali (Fig. 1).
Fig. 1.
The locations and sampling sites of each WWTP: A: East Anzali WWTP, B: Southern Tehran WWTP and C: Qom WWTP No. 3
The southern Tehran wastewater treatment plant (southern Tehran WWTP)
Tehran is the capital city of Iran with 22 distinct geographic regions and it is the largest city in this country. The population of Tehran according to Statistical Centre of Iran (SCI) is over 13 million [51]. This city is located in latitude of 35.6892° N and longitude of 51.3890° E (Fig. 1) [52–54]. Sampling was performed in inlets (untreated/raw municipal wastewater) and outlets (treated municipal wastewater) of Southern Tehran WWTP. The Southern Tehran WWTP is placed in the southeast of Tehran, Iran (in latitude of 35.5724° N and longitude of 51,5539° E) (Fig. 1) [51, 55]. The Southern Tehran WWTP is planned for three phases for treatment of wastewater with a capacity of 4.2 × 106 people. The first phase of this WWTP (i.e., from module one to four) the extended aeration activated sludge (EAAS) system. The second phase (i.e., from module five to six) is planned using the oxidation ditch method. Another phase (i.e., from module seven to eight) will be established in the future [55]. In addition, it should be noted that sampling were performed in the first phase (i.e., one to four modules) and the second phase (i.e., from module five to six). The major specifications and design parameters of this WWTP are shown in Table 1.
Table 1.
The major specifications and design parameters of Southern Tehran WWTP, East Anzali WWTP and Qom WWTP No. 3
| Southern Tehran WWTP | ||
|---|---|---|
| Parameters | Numbers of samples | Unit |
| Number of inhabitant served | – | 4.2 million people |
| Maximum flow | – | 450,000 m3/day |
| Mean flow | – | 18,750 m3/h |
| A capacity of each module | – | 525 thousand people |
| Wastewater treatment process (1–4 modules) | – | Extended aeration activated sludge (EAAS) system |
| Wastewater treatment process (5–6 modules) | – | Oxidation ditch method |
| Wastewater treatment process (7–8 modules) | – | Will be established |
| Average inlet Biochemical Oxygen Demand (Inlet-BOD5) | 31 | 215 mg/L |
| Average outlet Biochemical Oxygen Demand (Outlet-BOD5) | 31 | 24 mg/L |
| Average inlet Chemical Oxygen Demand (Inlet-COD) | 31 | 385 mg/L |
| Average outlet Chemical Oxygen Demand (Outlet-COD) | 31 | 58 mg/L |
| Average inlet Suspended Solids (Inlet-SS) | 31 | 179 mg/L |
| Average outlet Suspended Solids (Outlet-SS) | 31 | 30 mg/L |
| *Disinfection unit for 1–4 modules | – | Chlorine disinfection |
| *Disinfection unit for 5–6 modules | – | Ultraviolet (UV) disinfection |
| Wastewater treatment process (7–8 modules) | – | Will be established |
| Application of the wastewater effluent | – | Irrigate the Varamin plain and aquifer recharge |
| East Anzali WWTP | ||
| Number of inhabitant served | – | 40 thousand people |
| Mean daily inlet flow | – | 7800 m3/day |
| Mean monthly flow | – | 241,800 m3/month |
| Wastewater treatment process | – | Extended aeration activated sludge (EAAS) system |
| Average inlet Biochemical Oxygen Demand (Inlet-BOD5) | 30 | 180 mg/L |
| Average outlet Biochemical Oxygen Demand (Outlet-BOD5) | 30 | 10 mg/L |
| Average inlet Chemical Oxygen Demand (Inlet-COD) | 30 | 331 mg/L |
| Average outlet Chemical Oxygen Demand (Outlet-COD) | 30 | 19 mg/L |
| Average inlet Suspended Solids (Inlet-SS) | 30 | 170 mg/L |
| Average outlet Suspended Solids (Outlet-SS) | 30 | 22 mg/L |
| **Disinfection unit | – | Chlorine disinfection |
| Application of the wastewater effluent | – | Discharged into Anzali lagoon |
| Qom WWTP No. 3 | ||
| Number of inhabitant served | – | 249 thousand people |
| Mean daily inlet flow | – | 51,756 m3/day |
| Mean monthly flow | – | 1,542,354 m3/month |
| Wastewater treatment process | – | Conventional Activated Sludge (CAS) system with diffuser aeration |
| Average inlet Biochemical Oxygen Demand (Inlet-BOD5) | 26 | 235 mg/L |
| Average outlet Biochemical Oxygen Demand (Outlet-BOD5) | 26 | 10 mg/L |
| Average inlet Chemical Oxygen Demand (Inlet-COD) | 31 | 401 mg/L |
| Average outlet Chemical Oxygen Demand (Outlet-COD) | 31 | 36 mg/L |
| Average inlet Suspended Solids (Inlet-SS) | 31 | 254 mg/L |
| Average outlet Suspended Solids (Outlet-SS) | 31 | 10 mg/L |
| **Disinfection unit | – | Chlorine disinfection |
| Application of the wastewater effluent | – | Agriculture (mainly irrigation) |
*It should be noted that sampling were performed in the first phase (i.e., one to four modules) and the second phase (i.e., from module five to six).
**It should be noted that sampling were performed in this unit.
East Anzali wastewater treatment plant (east Anzali WWTP)
Anzali city, also known as Bandar-e Anzali, is located in the north of Iran, in Gilan Province. The population of Anzali based on Statistical Centre of Iran (SCI) in 2016 was over 118,564. This city is located in latitude of 37.4639° N and longitude of 49.4799° E (Fig. 1). Sampling was performed in inlets (untreated/raw municipal wastewater) and outlets (treated municipal wastewater) of East Anzali WWTP. This WWTP is located in the east of Anzali city, (in latitude of 35.8055° N and longitude of 41.4831° E) (Fig. 1). The major specifications and design parameters of East Anzali WWTP are shown in Table 1, an activated sludge process in used. Activated sludge of this WWTP is the biological wastewater treatment process, that comprises two grit and grease removal, two circular primary sedimentation tanks, a wastewater pumping station, two biological units, two secondary sedimentation tanks and two coagulation unit tanks, four gravity sand filter units, and a UV disinfection unit.
Qom wastewater treatment plant no. 3 (Qom WWTP no. 3)
Qom city (is) situated 140 km to the south of Tehran (34.6416° N, 50.8746° E). Qom is the seventh largest city of Iran (Fig. 1). The population of Qom according to Statistical Centre of Iran (SCI) in 2016 was 1,201,158. Sampling was performed in inlet and outlet of Qom WWTP No. 3, like both previous WWTPs. Target WWTP is placed in latitude of 38.3940° N and longitude of 49, 5787° E) (Fig. 1). It is one of the most advanced wastewater treatment plants in Iran, which contains a screen bar unit, four primary settling tanks, two aerated tanks, four secondary settling tanks, and a chlorination disinfection unit. The major specifications and design parameters of this WWTP are provided in Table 1.
Table 1. The major specifications and design parameters of Southern Tehran WWTP, East Anzali WWTP and Qom WWTP No. 3.
Untreated and treated wastewater sampling in three WWTPs
In this study, sampling points were selected in inlets (untreated/raw municipal wastewater) and outlets (treated municipal wastewater) of three WWTPs in the three cities of Iran: Tehran, Qom and Anzali. Due to the COVID-19 outbreak in these cities the first cases were found in Qom and regarding the RNA viruses entrance into, sampling in inlets (raw municipal wastewater) and outlets (treated municipal wastewater) of three WWTPs can be a representative of the covered population according to pervious works [50, 56]. For these reasons, taking composite samples of inlet and outlet of each WWTPs could be considered to be sufficient to estimate the probability of the presence or absence of the RNA viruses in the municipal wastewater. Sampling was performed in spring between April 4, 2020 and April 17, 2020 (a two-week period) and between April 20, 2020 and May 2, 2020 (a two-week period). Twelve composite untreated wastewater samples with a frequency of one composite sample each week were collected from each WWTPs during a two-week period (from April 4, 2020 to April 17, 2020). Hence, we collected a total of 12 untreated/raw wastewater samples during a two-week period owing to three composite sample at the 3 sites ((3 sites +3 inlets) × a two-week period = 12) (Table 2).
Table 2.
Detection of SARS-CoV-2 RNA in untreated (raw) and treated wastewater samples in Southern Tehran WWTP, East Anzali WWTP and Qom WWTP No. 3
| Presence (+) or absence (−) of SARS-CoV-2 RNA | ||||||
|---|---|---|---|---|---|---|
| Sampling (Inlet) | Sampling date | Types of samples | Southern Tehran WWTP: | Qom WWTP No. 3 | East Anzali WWTP | |
| Untreated wastewater | April 4, 2020 | Composite sample | + | + | + | |
| Untreated wastewater | April 17, 2020 | Composite sample | + | + | + | |
| Untreated wastewater | April 20, 2020 | Composite sample | + | + | + | |
| Untreated wastewater | May 2, 2020 | Composite sample | + | + | + | |
| Sampling (Outlet) | Sampling date | Types of samples | 1–4 modules: Chlorine | 5–6 modules: Ultraviolet | Chlorine | Chlorine |
| Treated wastewater | April 4, 2020 | Composite sample | – | – | – | – |
| Treated wastewater | April 17, 2020 | Composite sample | – | – | – | – |
| Treated wastewater | April 20, 2020 | Composite sample | + | – | – | – |
| Treated wastewater | May 2, 2020 | Composite sample | + | – | – | – |
The same exact mode of operation used to the treated wastewater sampling, yielding 16 total treated wastewater samples (Note: the inlet Southern Tehran WWTP is divided into two outlets (Tables 1 and 2)). As there were three WWTPs, we collected 28 total treated and untreated wastewater samples altogether.
Wastewater samples were moved on ice to the laboratory and kept at 4 °C until further analysis. It should be emphasized that authors have used standard personal protective equipment (PPE) for wastewater sampling and analyzing samples in laboratory, like N95 mask, steel capped boots, face shields, hard hats, long pants safety glasses/goggles and gloves. A summary of the locations and sampling sites in each of the WWTPs is shown in Fig. 1.
Preparation of sewage samples using the phase-separation method (preparation of half a liter of sewage sample)
The concentration of sewage samples for the isolation of viral RNA were performed based on WHO guidelines [27, 57, 58]. The preparation of the sewage samples and its inoculation into cell culture should be done in two consecutive days [27, 57]. The first day includes three steps, while the second day includes two steps.
The first day, which includes three steps:
Step 1: Sample preparation
At the first, sewage sample was allowed to sediment for five minutes for removing large solid materials. Then, about 500 mL of the sample was divided into two 250 mL centrifuge bottles within a biosafety cabinet (class II). Those bottles were centrifuged at 1500 g for 20 min at +4 °C. After that, the supernatant was poured in a 1 l beaker. In addition, for further using, the closed centrifuge bottles containing the solid pellet were kept in the refrigerator at 4 °C [27, 57].
Step 2: Adjusting the pH of supernatant
At first, the pH solution within a 1 l beaker were adjusted by 1 N HCl and 1 N NaOH (pH = 7.0–7.5). Then, a 500 mL of adjusted supernatant by HCl and NaOH was transferred to a 1 l beaker for further study. The rest of the supernatant and the raw sewage samples were stored as a backup sample at 4 °C until the cells inoculated with the concentrate under the microscope showed no toxicity. Finally, after ensuring that the samples were not toxic, they were stored at 20 °C until the full test result is known [27, 57].
Step 3: Glass separatory funnel with different separation phases
At first, a 39.5 mL of 22% dextran, 287 mL of 29% polyethylene glycol (PEG 6000), and 35 mL 5 M NaCl were added to 500 mL of the supernatant obtained from the previous step, respectively. Then, the mixture obtained at this stage was stirred with sufficient speed using a magnetic stirrer (vortex formation) for one hour at +4 °C. After that, a sterile one liter conical glass separatory funnel were prepared for each samples. The funnel stood on the foundation. A very thin layer of the stopcock grease was prepared on the sliding glass surface of the valves but did not clog the holes (note: Teflon valves did not need grease). Finally, the stirred mixture was slowly poured into a 1 L separatory funnel and then kept at +4 °C overnight for separating mixture to three phases (upper phase, interphase and lower phase). Polyethylene glycol created the more hydrophobic, less dense and upper phase, whereas dextran created the more hydrophilic, denser and lower phase (Fig. 2) [27, 57, 58].
Fig. 2.
Glass separatory funnel with different separation phases [27, 57]
The second day, which includes two steps:
Step 1: Interphase separation
In the second day, the bottom of the glass separatory funnel was carefully checked. The lower and intermediate phase (interphase) were seen with solids. The entire lower phase and the interphase were poured firstly dropwise into a sterile 50 mL centrifuge tube. Then, the combined volume (lower phase and the interphase) was measured and recorded. In this step, the volume of the solid precipitate was recorded in the lower layer and/or in the interphase, whether it was more than half a mL. It should also be pointed out that when the interphase was too thick, it may be collected separately, because it will be hard to gather. In addition, it should be noted that once the interphase layer was collected, the glass separatory funnel and upper phase were disinfected using appropriate disinfectants (such as sodium hypochlorite (NaOCl)) for one hour [27, 57].
Step 2: Adding pellet to the interphase solution
In this step, 5 mL of the interphase solution under a biosafety cabinet (class II) was added to a sterile centrifuge tube which contained pellets and dissolved well. Then, this dissolved solution (containing the pellet) was returned to the interphase solution, prepared in step one in the second day. Hence, the final prepared solution of interphase was prepared and used for RNA extraction [27, 57]. In addition, it should be noted that in case which the sewage samples are very clean, it helps to add a small amount of solid from pallet to facilitate the identification of the interfaces in the glass separatory funnel.
One-step real-time PCR analysis
Total RNA was extracted from 200 μL of the concentrated samples using FastPure Viral RNA Mini Kit (Vazyme-China) according to the manufacturers’ procedures. Briefly, the sample was mixed with lysis buffer and then loaded onto the filter membrane column. Then, the filter column was washed twice with washing solution. Finally, the RNA was eluted in 50 μL of RNase free water and stored at −80 °C until required.
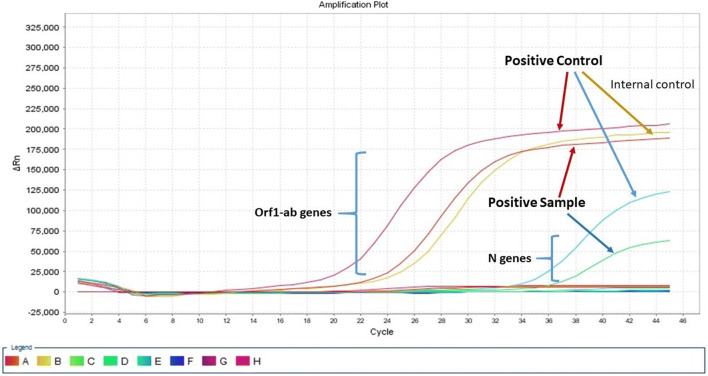
For qualitative detection of SARS-CoV-2, One-Step Taq-man Real-Time PCR was used to amplify the ORF1ab and N gene according to Sansure SARS-CoV-2 Nucleic Acid Test Kits (Sansure Biotech- China). To confirm extraction of samples, the RNase P gene was selected as Endogenous control and positive and negative samples were included in each PCR reaction (Fig. 2).
The final PCR volume reactions were performed in 50 μL reaction mixtures containing 4 μL of enzyme mix, 26 μL PCR Master mix contain primers and probe and 20 μL RNA template. Amplification was carried out under the following temperature program: cDNA synthesis at 500 C for 30 min, preheat at 95 °C for 1 min, followed by 45 two-step cycles of 95 °C for 15 s and 60 °C for 30s on Step One Plus™ Real-Time PCR System (Applied Biosystems, California, USA) instrument. The cycle threshold (Ct value) was calculated automatically.
Results and discussions
In this study, Taq-man Multiplex Real-Time PCR was used to detect SARS-CoV-2 RNA. The Kit targets ORF1ab and N genes (Sansure Biotech- China) are used for detection of SARS-CoV-2 RNA. The N gene encoded nucleocapsid that surrounded the genomic RNA and the orf1ab gene encodes RNA-dependent RNA polymerase required for viral RNA replication. This kit provides positive and negative controls to ensure correct kit performance. In this kit, human RNAse P was used as an endogenous control gene. The use of endogenous control indicates the validation of RNA extraction and presence of inhibitors in the RNA sample. Plus human RNase P gene was taken as internal and extraction control (Fig. 3). Furthermore, amplification plot for one positive sample with positive and negative controls are shown in Fig. 3. Accordingly, a positive sample is a sample that both orf1ab and N genes are positive. In addition, it should be noted that newly reported Taq-man Multiplex Real-Time PCR has been utilized for finding of human coronaviruses such as SARS-CoV-2 RNA and human respiratory syncytial virus (HRSV) in samples of wastewater and hospitalized children [59–61].
Fig. 3.
Amplification plot for one positive sample with positive and negative controls
Detection of SARS-CoV-2 RNA in wastewater samples
East Anzali WWTP
In the case of East Anzali WWTP, among two wastewater samples collected in raw wastewater (inlets) during a two-week period, two samples (100%) were positive for SARS-CoV-2 RNA using Taq-man Real-Time PCR method (Table 2). Besides, the results of four collected outlet (treated) samples from chlorine disinfection of East Anzali WWTP on April 20, 2020 and on May 2, 2020 showed that none of these samples were positive for SARS-CoV-2 RNA. Recent work has described that 71% gathered untreated wastewater samples (5 from 7 samples) from seven treatment plants in Istanbul (Turkey) was positive for SARS-CoV-2 RNA using polyethylene glycol 6000 (PEG 6000) adsorption SARS-CoV-2 concentration method, which the amount of SARS-CoV-2 in seven untreated wastewater samples ranged from 1.80 × 104 (for Pasakoy WWTP) to 8.26 × 103 (for Ambarli WWTP) [62]. Moreover, a study of Gonzalez et al. (2020), a three RT-ddPCR assays (N1, N2, N3) were applied to identify SARS-CoV-2 RNA in weekly samples from 9 WWTPs in Virginia (a southeastern U.S. state) [63]. They found that during the twenty-one week study, SARS-CoV-2 concentrations ranged from 101 to 104 copies 100 mL−1 in samples where viral RNA was measured [63].
Qom WWTP no. 3
The results of raw wastewater samples gathered from the Qom WWTP No. 3 were positive for SARS-CoV-2 RNA. However, four collected outlet (treated) samples from chlorine disinfection of Qom WWTP No. 3 during a two-week period showed that these samples were negative (Table 2). Likewise, Kocamemi et al. (2020) collected two composite samples from sewage manholes placed close to the pandemic hospitals in in Istanbul (Turkey), which both samples were examined positive (100%). In addition, the amount of SARS-CoV-2 in sewage manholes ranged from 4.49 × 104 to 9.33 × 104 [62]. In addition, Cascella et al., (2020) declared that SARS-CoV-2 virus can be efficaciously inactivated using chlorine, ultraviolet rays, heat, and ethanol, which is consistent with our results [26]. Furthermore, Wurtzer et al. (2020) collected twenty three raw and eight treated wastewater samples from 3 main WWTP of the Parisian area and they found all raw wastewater samples scored positive for SARS-CoV2 [64]. In addition, they reported that six out of eight samples from treated wastewater scored positive by RT-qPCR [64]. Moreover, treated wastewater effluents displayed a 100 times reduction in the viral load compared to the corresponding raw wastewater samples [64]. Additionally, previous work studied the presence of SARS-CoV-2 RNA in untreated and treated wastewater samples in southern Louisiana (USA). SARS-CoV-2 RNA was identified in two out of fifteen wastewater samples using two reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assays (CDC N1 and N2) [65]. Trottier et al. (2020) identified the amount of SARS-CoV-2 RNA at the inflow point of the major WWTP of Montpellier, France [66]. They found that amounts of SARS-CoV-2 RNA at the WWTP from mid-June on started increasing [66]. Generally, results of past works showed that the SARS-CoV-2 RNA existed in raw and treated wastewaters [67, 68].
Southern Tehran WWTP
Among two wastewater samples collected in inlets of Southern Tehran WWTP during a two-week period, two samples (100%) was positive for SARS-CoV-2 RNA by the Taq-man Real-Time PCR (Table 2). Furthermore, we collected two outlet samples from chlorine disinfection (module outlet 1 to 4) and two samples from ultraviolet (UV) disinfection (module outlet 5 to 6) of Southern Tehran WWTP during a two-week period and the results showed that only two samples collected from chlorine disinfection unit were positive (Table 2).
For comparison, previous investigation reported that 22% collected samples (two from nine samples) from WWTP in Southeast Queensland (Australia) were positive for SARS-CoV-2 RNA by N_Sarbeco method [50]. In addition, Wurtzer et al. (2020) collected untreated wastewater samples in WWTP of Parisian area in Paris (France) that the results of their study showed the concentration of SARS-CoV-2 RNA in untreated wastewater samples was about 5.104 genome units/L [69] and the results of the same work concluded that the increase of genome units in untreated wastewater identically pursued the increase of human SARS-CoV-2 cases showed in the study area, which is consistent with the results of our study [69]. Hence, the outbreak of COVID-19 created by SARS-CoV-2 virus has been dispersed swiftly around the world [70, 71]. Previous works reported that the COVID-19 patients can excrete SARS-CoV-2 virus into wastewater via gastrointestinal tract (feces) and those can survive from hours to days in untreated wastewater [30, 50, 69]. In addition, Bogler et al. (2020) reported that SARS-CoVs were present in wastewater for several days, leading to potential health risks via waterborne and aerosolized wastewater pathways [72]. In addition, whereas SARS-CoV-2 virus can be identified in feces samples, Wurtzer et al. (2020) has recently suggested that SARS-CoV-2 RNA in wastewater can be used as a supplementary tool to inspect novel coronavirus circulation in human populations [69]. Hence, the identification of SARS-CoV-2 RNA in wastewater makes it possible to monitor the spread of infections among the population through wastewater-based epidemiology [50, 56].
In addition to the aforementioned, according to results of outlet samples of Southern Tehran WWTP described that the UV disinfection was more effective than chlorine disinfection and for effective disinfection, WWTP operator should increase the concentration of free residual chlorine to amounts greater than or equal to 0.5 mg/L at retention time (RT) thirty minutes at pH lower than eight, which is in agreement with technical brief reported by WHO on March 03, 2020 [73].
Furthermore, based on the recent survey, the UV disinfection has numerous benefits such as formation of no disinfection byproducts, short retention time, effective on a wide range of resilient viruses, and no residues compared with chlorine disinfection [74].
Regarding that in the three WWTPs, activated sludge (AS) processes (biological wastewater treatment process) are used, hence these systems have the ability to remove SARS-CoV-2 RNA, especially WWTP that have suitable disinfection systems (as can be seen in Table 2 and Fig. 4) and the removal efficiency of SARS-CoV-2 RNA in Southern Tehran WWTP, East Anzali WWTP and Qom WWTP No. 3 were 50%, 100% and 100%, respectively.
Fig. 4.
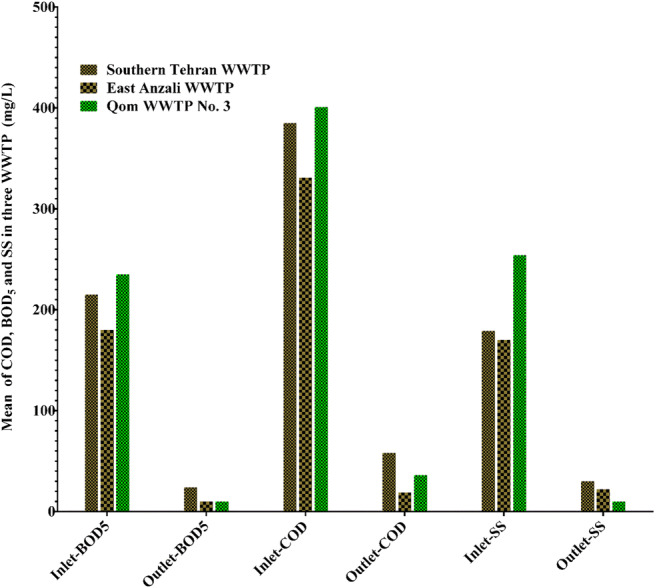
The composition of wastewater in inlets and outlets of Southern Tehran WWTP, East Anzali WWTP and Qom WWTP No. 3
On the other side, we should not forget that proper sludge management of those WWTPs are essential due to the sludge of these treatment plants can contain active viruses [62, 75–77]. For example, past work described that nine Human coronavirus 229E and one Human coronavirus HKU1 were identified from class B bio-solids in an anonymous US wastewater treatment facility [78]. In addition, during the COVID-19 outbreak, using of untreated wastewater and even treated wastewater in agricultural irrigation poses a serious threat to all members of society, especially in the cities of developing countries.
As a result, it can threaten the health of workers at WWTPs during sludge production, transportation and disposal, and the residents living near the treatment plants and even entire communities. Finally, according to the findings of this research work, it should be emphasized that workers at WWTPs should wear standard personal protective equipment (PPE), like N95 mask, steel capped boots, face shields, hard hats, long pants safety glasses/goggles and gloves.
Table. 2. Detection of SARS-CoV-2 RNA in untreated (raw) and treated wastewater samples in Southern Tehran WWTP, East Anzali WWTP and Qom WWTP No. 3.
Impact on the composition of wastewater on SARS-CoV-2 RNA
As reported in the previous work, it is essential to evaluate the effects of the composition of wastewater and meteorological parameters on SARS-CoV-2 RNA during the outbreak [50]. Hence, for this reason we measured important factors such as BOD5, COD and SS in inlet and outlets of three WWTPs. The composition of wastewater in inlets and outlets of Southern Tehran WWTP, East Anzali WWTP and Qom WWTP No. 3 are described in Fig. 4, and it can be seen in this figure that, the high outlet BOD5, COD and SS values in Southern Tehran WWTP compared to East Anzali WWTP and Qom WWTP No. 3 may explain the reason of SARS-CoV-2 RNA founding in the outlet of Southern Tehran WWTP.
In other words, the removal efficiency percentage of parameters such as BOD5, COD and SS in East Anzali WWTP and Qom WWTP No. 3 were increased. So the question arises: is there a direct relationship between the efficiencies of virus removal and those parameters according to Fig. 4 and Table 1?
Generally, it is essential to investigate the physical and chemical properties of wastewater (e.g., pH, temperature, dissolved oxygen (DO) concentration, total nitrogen (TN) concentration, salinity), composition in waste activated sludge (WAS), and even population of society for providing precise data about transmission of SARS-CoV-2.
Conclusions
This work was concentrated on the presence of SARS-CoV-2 RNA in untreated and treated wastewater during the outbreak of coronavirus disease 2019 (COVID-19) in Southern Tehran WWTP, East Anzali WWTP and Qom WWTP No. 3. The results of this work showed that all samples taken from inlets of Southern Tehran WWTP, East Anzali WWTP and Qom WWTP No. 3 were positive (four positive samples for each of the WWTPs), while the SARS-CoV-2 RNA in outlet samples were two positive samples for Southern Tehran WWTP, negative for East Anzali WWTP and negative for Qom WWTP No. 3. The results of outlet samples of Southern Tehran WWTP described that the UV disinfection was more effective than chlorine disinfection and for effective disinfection; hence, WWTP operator should increase the concentration of free residual chlorine to amounts ≥0.5 mg/L, based on WHO on March 03, 2020. In addition, the high outlet BOD5, COD and SS values in Southern Tehran WWTP compared to East Anzali WWTP and Qom WWTP No. 3 may explain the reason of SARS-CoV-2 RNA founding in the outlet of Southern Tehran WWTP. As a result, the WWTP facilities could pose a threat on the health of workers during sludge production, transportation and disposal, residents living near to the treatment plants. According to the findings of this work, it should be emphasized that workers at WWTPs should wear standard personal protective equipment (PPE), like N95 mask, steel capped boots, face shields, hard hats, long pants safety glasses/goggles and gloves. In addition, we should not forget that proper sludge management of those WWTPs are essential due to the fact that the sludge from these treatment plants may contain active viruses. Finally, it is essential to investigate the physical and chemical properties of wastewater, composition in WAS, and even population of society for providing precise data about transmission of SARS-CoV-2 via water environmental and communities.
Acknowledgements
The authors would like to thank the financial grant supported by the Deputy of Research, Tehran University of Medical Sciences, for this research as grant number 47164-99-1-99. We thank the staff of Departments of environmental health engineering and Virology, School of Public Health, Tehran University of Medical Sciences.
Authors’ contributions
MA, SN and RN supervised of this study, and participated in its design and coordination and participated in writing - review & editing. JY, TMA, AN, and NZSJ participated in methodology, validation, and visualization of this study and detection of SARS-CoV-2 in untreated and treated wastewater using Real-Time PCR analysis. MH, BV, SKAV, and MB collected samples, performed the statistical analysis and participated in the design of the study. ANB participated in the design of the study, prepared to draft the manuscript (writing - original draft) and revised the manuscript (writing - review & editing). ANB, SY and SN participated in the design of the study and in the sequence alignment. All authors read and approved the final manuscript.
Declarations
Consent for publication
Not applicable
Ethics approval and consent to participate
Not applicable
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guzman MI. An overview of the effect of bioaerosol size in coronavirus disease 2019 transmission. Int J Health Plann Manag. 2020:1–10. [DOI] [PMC free article] [PubMed]
- 2.Daniela DA, Gola M, Letizia A, Marco D, Fara GM, Rebecchi A, et al. COVID-19 and living spaces challenge. In: Well-being and public health recommendations for a healthy, safe, and sustainable housing; 2020. p. 1–15. [DOI] [PMC free article] [PubMed]
- 3.Li H, Wang Y, Ji M, Pei F, Zhao Q, Zhou Y, Hong Y, Han S, Wang J, Wang Q, Li Q, Wang Y. Transmission routes analysis of SARS-CoV-2: a systematic review and case report. Front. Cell Dev. Biol. 2020;8:618. doi: 10.3389/fcell.2020.00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abkarian M, Mendez S, Xue N, Yang F, Stone HA. Speech can produce jet-like transport relevant to asymptomatic spreading of virus. Proc. Natl. Acad. Sci. 2020;117(41):25237–25245. doi: 10.1073/pnas.2012156117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chong KL, Ng CS, Hori N, Yang R, Verzicco R, Lohse D. Extended lifetime of respiratory droplets in a turbulent vapour puff and its implications on airborne disease transmission. arXiv preprint arXiv:200801841. 2020;126(3):034502. [DOI] [PubMed]
- 6.Delikhoon M, Guzman MI, Nabizadeh R, Norouzian BA. Modes of transmission of severe acute respiratory syndrome-Coronavirus-2 (SARS-CoV-2) and factors influencing on the airborne transmission: a review. Int. J. Environ. Res. Public Health. 2021;18(2):395. doi: 10.3390/ijerph18020395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S, Yang J, Yang W, Wang C, Bärnighausen T. COVID-19 control in China during mass population movements at new year. Lancet. 2020;395:764–766. doi: 10.1016/S0140-6736(20)30421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ping K. Epidemiologic characteristics of COVID-19 in Guizhou, China. medRxiv. 2020:1–16.
- 9.WHO WHO. Health workers exposure risk assessment and management in the context of COVID-19 virus: interim guidance, 4 March 2020: World Health Organization2020.
- 10.Xu C, Luo X, Yu C, Cao S-J. The 2019-nCoV epidemic control strategies and future challenges of building healthy smart cities. London: SAGE Publications Sage UK; 2020. [Google Scholar]
- 11.Wu H, Huang J, Zhang CJ, He Z, Ming W-K. Facemask shortage and the coronavirus disease (COVID-19) outbreak: reflection on public health measures. medRxiv. 2020;21(1):100329. [DOI] [PMC free article] [PubMed]
- 12.Rodriguez-Morales AJ, Sánchez-Duque JA, Botero SH, Pérez-Díaz CE, Villamil-Gómez WE, Méndez CA, et al. Preparación y control de la enfermedad por coronavirus 2019 (COVID-19) en América Latina. Acta Med. Peruana. 2020;37(1):3–7. doi: 10.35663/amp.2020.371.909. [DOI] [Google Scholar]
- 13.Xiao Y, Torok ME. Taking the right measures to control COVID-19. Lancet Infect. Dis. 2020;20:523–524. doi: 10.1016/S1473-3099(20)30152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C, Wang J. A mathematical model for the novel coronavirus epidemic in Wuhan, China. Math. Biosci. Eng. 2020;17(3):2708–2724. doi: 10.3934/mbe.2020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO WHO. Considerations for quarantine of individuals in the context of containment for coronavirus disease ( COVID-19): interim guidance, 29 February 2020: World Health Organization2020.
- 16.CDC CfDCaP. Centers for Disease Control and Prevention. 2019 Novel Coronavirus (2019-nCoV) Situation Summary. Available from: https://www.cdc.gov/coronavirus/2019-nCoV/summary.html. Accessed 3 February 2020. 2020.
- 17.RCP RCP. Novel Coronavirus 2019 | Rubbermaid Commercial Products- ©2020 Rubbermaid Commercial Products LLC 8900 Northpointe Executive Park Drive, Huntersville, NC 28078. 2020. 2020.
- 18.Bonilla-Aldana DK, Cardona-Trujillo MC, García-Barco A, Holguin-Rivera Y, Cortes-Bonilla I, Bedoya-Arias HA et al. MERS-CoV and SARS-CoV infections in animals: a systematic review and meta-analysis of prevalence studies. 2020. [PubMed]
- 19.CDC CfDCaP. Common Human Coronaviruses- Available from: https://www.cdc.gov/coronavirus/types.html Accessed February 13, 2020. 2020.
- 20.Millán-Oñate J, Rodriguez-Morales AJ, Camacho-Moreno G, Mendoza-Ramírez H, Rodríguez-Sabogal IA, Álvarez-Moreno C. A new emerging zoonotic virus of concern: the 2019 novel coronavirus (COVID-19). Infectio. 2020;24(3):187–92.
- 21.Bonilla-Aldana DK, Villamil-Gómez WE, Rabaan AA, Rodríguez-Morales AJ. Una nueva zoonosis viral de preocupación global: COVID-19, enfermedad por coronavirus 2019. Iatreia. 2018.
- 22.Ghobadi H, Ebrahimi Kalan M, Mohammad-Shahi J, Ben Taleb Z, Ebrahimi Kalan A, Fazlzadeh M. COVID-19 and acute kidney injury; a case report. J Renal Inj Prev. 2020;9(3):e26-e. doi: 10.34172/jrip.2020.26. [DOI] [Google Scholar]
- 23.Far WWKT. What is coronavirus? 2020.
- 24.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.WHO WHO. The COVID-19 risk communication package for healthcare facilities. 2020.
- 26.Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, evaluation and treatment coronavirus (COVID-19). Statpearls [internet]. Treasure Island, FL, USA: StatPearls Publishing; 2020. Available online: http://www.ncbi.nlm.nih.gov/books/NBK554776/. Accessed 31 Mar 2020
- 27.WHO WHO. Guidelines for environmental surveillance of poliovirus circulation: Geneva: World Health Organization2003.
- 28.Dehghani M, Sorooshian A, Ghorbani M, Fazlzadeh M, Miri M, Badiee P, Parvizi A, Ansari M, Baghani AN, Delikhoon M. Seasonal variation in Culturable bioaerosols in a wastewater treatment plant. Aerosol Air Qual. Res. 2018;18(11):2826–2839. doi: 10.4209/aaqr.2017.11.0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shamsollahi HR, Ghoochani M, Sadeghi K, Jaafari J, Masinaei M, Sillanpää M, Yousefi M, Mirtalb ST, Alimohammadi M. Evaluation of the physical and chemical characteristics of water on the removal efficiency of rotavirus in drinking water treatment plants and change in induced health risk. Process. Saf. Environ. Prot. 2019;130:6–13. doi: 10.1016/j.psep.2019.07.014. [DOI] [Google Scholar]
- 30.Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-Coronavirus-2 in sewage. medRxiv. 2020; 03.29.20045880. 10.1101/2020.03.29.20045880. [DOI] [PubMed]
- 31.Leung WK, To K-F, Chan PK, Chan HL, Wu AK, Lee N, et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125(4):1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung LS. The SARS epidemic in Hong Kong: what lessons have we learned? J. R. Soc. Med. 2003;96(8):374–378. doi: 10.1177/014107680309600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peiris JSM, Chu C-M, Cheng VC-C, Chan K, Hung I, Poon LL, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan KH, Poon LL, Cheng V, Guan Y, Hung I, Kong J, et al. Detection of SARS coronavirus in patients with suspected SARS. Emerg. Infect. Dis. 2004;10(2):294–299. doi: 10.3201/eid1002.030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu W, Tang F, Fontanet A, Zhan L, Zhao Q-M, Zhang P-H, Wu XM, Zuo SQ, Baril L, Vabret A, Xin ZT, Shao YM, Yang H, Cao WC. Long-term SARS coronavirus excretion from patient cohort, China. Emerg. Infect. Dis. 2004;10(10):1841–1843. doi: 10.3201/eid1010.040297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gundy PM, Gerba CP, Pepper IL. Survival of coronaviruses in water and wastewater. Food. Environ. Virol. 2009;1(1):10. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- 37.Casanova L, Rutala WA, Weber DJ, Sobsey MD. Survival of surrogate coronaviruses in water. Water Res. 2009;43(7):1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimmond T. Trustworthy facts on 2019-nCoV and COVID-19 waste handling–an update. Accessed Feb 13, 2020. 2020:2–5.
- 39.Chan JF-W, Yuan S, Kok K-H, To KK-W. Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guzman M. Bioaerosol size effect in COVID-19 transmission. 2020. https://www.preprints.org/manuscript/202004.0093/v2. (Accessed June 17 2020)
- 41.Kazi Abdul M, Khandaker Mursheda F. Knowledge and perception towards novel coronavirus (COVID 19) in Bangladesh. 2020.
- 42.Prates ET, Garvin MR, Pavicic M, Jones P, Shah M, Alvarez C et al. Confronting the COVID-19 Pandemic with Systems Biology. bioRxiv. 2020.
- 43.Z-y G, Yang L-m, J-j X, X-h F, Y-z Z. Possible aerosol transmission of COVID-19 and special precautions in dentistry. J. Zhejiang Univ. Sci. B. 2020:1–8. [DOI] [PMC free article] [PubMed]
- 44.Liu Y, Ning Z, Chen Y, Guo M, Liu Y, Gali NK et al. Aerodynamic characteristics and RNA concentration of SARS-CoV-2 aerosol in Wuhan hospitals during COVID-19 outbreak. bioRxiv. 2020.
- 45.WHO. Coronavirus disease 2019 (COVID-19): situation report, 72. 2020.
- 46.Kalan ME, Ghobadi H, Taleb ZB, Adham D, Cobb CO, Ward KD, Behaleh R, Fazlzadeh M COVID-19 and beliefs about tobacco use: an online cross-sectional study in Iran. Environ. Sci. Pollut. Res. 2020. doi:10.1007/s11356-020-11038-x. [DOI] [PMC free article] [PubMed]
- 47.Zhang H, Kang Z, Gong H, Xu D, Wang J, Li Z et al. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. BioRxiv. 2020.
- 48.Zhang C-M, Xu L-M, Xu P-C, Wang XC. Elimination of viruses from domestic wastewater: requirements and technologies. World J. Microbiol. Biotechnol. 2016;32(4):69. doi: 10.1007/s11274-016-2018-3. [DOI] [PubMed] [Google Scholar]
- 49.Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett.\ 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 50.Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O'Brien JW et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020:138764. [DOI] [PMC free article] [PubMed]
- 51.Golkhorshidi F, Sorooshian A, Jafari AJ, Baghani AN, Kermani M, Kalantary RR, Ashournejad Q, Delikhoon M. On the nature and health impacts of BTEX in a populated middle eastern city: Tehran. Iran Atmos. Pollut. Res. 2019;10(3):921–930. doi: 10.1016/j.apr.2018.12.020. [DOI] [Google Scholar]
- 52.Norouzian Baghani A, Sorooshian A, Delikhoon M, Nabizadeh R, Nazmara S, Bakhtiari R. Pollution characteristics and noncarcinogenic risk assessment of fungal bioaerosol in different processing units of waste paper and cardboard recycling factory. Toxin Rev. 2020:1–12. 10.1080/15569543.2020.1769135.
- 53.Nabizadeh R, Sorooshian A, Delikhoon M, Baghani AN, Golbaz S, Aghaei M et al. Characteristics and health effects of volatile organic compound emissions during paper and cardboard recycling. Sustainable Cities and Society. 2020;56:102005. doi: 10.1016/j.scs.2019.102005.
- 54.Norouzian Baghani A, Bahmani Z, Sorooshian A, Farzadkia M, Nabizadeh R, Delikhoon M, et al. Characterization of polycyclic aromatic hydrocarbons associated with PM10 emitted from the largest composting facility in the Middle East. Toxin Rev. 2020:1–15. 10.1080/15569543.2020.1737823.
- 55.Momenia S, Alimohammadia M, Naddafia K, Nabizadeha R, Changania F, Zareid A, et al. Study of sludge from the largest wastewater treatment plant in the Middle East (southern Tehran, Iran) based on chemical and microbiological parameters for use in agriculture. Desalin. Water Treat. 2019;160:153–160. doi: 10.5004/dwt.2019.24369. [DOI] [Google Scholar]
- 56.Lorenzo M, Picó Y. Wastewater-based epidemiology: current status and future prospects. Curr. Opin. Environ. Sci. Health. 2019;9:77–84. doi: 10.1016/j.coesh.2019.05.007. [DOI] [Google Scholar]
- 57.WHO WHO. Polio laboratory manual: Geneva: World Health Organization2004.
- 58.Shuval HI, Cymbalista BFS, Goldblum N. The phase-separation method for the concentration and detection of viruses in water. Water Res. 1969;3(4):225–240. doi: 10.1016/0043-1354(69)90019-0. [DOI] [Google Scholar]
- 59.Fajardo A, Pereira-Gomez M, Echeverria N, Lopez-Tort F, Perbolianachis P, Aldunate F et al. Evaluation Of SYBR Green Real Time PCR For Detecting SARS-CoV-2 From Clinical Samples. bioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 60.Aghamirmohammadali FS, Sadeghi K, Shafiei-Jandaghi NZ, Khoban Z, Mokhtari-Azad T, Yavarian J. Survey of WU and KI polyomaviruses, coronaviruses, respiratory syncytial virus and parechovirus in children under 5 years of age in Tehran. Iranian Journal of Microbiology: Iran; 2020. [PMC free article] [PubMed] [Google Scholar]
- 61.Shirato K, Nao N, Katano H, Takayama I, Saito S, Kato F et al. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Japanese Journal of Infectious Diseases. 2020:JJID. 2020.061. [DOI] [PubMed]
- 62.Kocamemi BA, Kurt H, Hacioglu S, Yarali C, Saatci AM, Pakdemirli B. First Data-Set on SARS-CoV-2 Detection for Istanbul Wastewaters in Turkey. medRxiv. 2020.
- 63.Gonzalez R, Curtis K, Bivins A, Bibby K, Weir MH, Yetka K, Thompson H, Keeling D, Mitchell J, Gonzalez D. COVID-19 surveillance in southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186:116296. doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wurtzer S, Marechal V, Mouchel J, Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. medRxiv. 2020:2020.04.12.20062679. doi:10.1101/2020.04.12.20062679.
- 65.Sherchan SP, Shahin S, Ward LM, Tandukar S, Aw TG, Schmitz B, Ahmed W, Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana. USA Sci. Total Environ. 2020;743:140621. doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trottier J, Darques R, Mouheb NA, Partiot E, Bakhache W, Deffieu MS, et al. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier. Fr. One Health. 2020;10:100157. doi: 10.1016/j.onehlt.2020.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Widera M, Greve C, Janke A, Hollert H, Wintgens T, Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany–suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751:141750. doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tran HN, Le GT, Nguyen DT, Juang R-S, Rinklebe J, Bhatnagar A, et al. SARS-CoV-2 coronavirus in water and wastewater: a critical review about presence and concern. Environ. Res. 2020;110265. [DOI] [PMC free article] [PubMed]
- 69.Wurtzer S, Marechal V, Mouchel J-M, Maday Y, Teyssou R, Richard E et al. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 70.Mao K, Zhang H, Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? : ACS Publications; 2020. [DOI] [PubMed]
- 71.Ivanov D. Predicting the impacts of epidemic outbreaks on global supply chains: a simulation-based analysis on the coronavirus outbreak (COVID-19/SARS-CoV-2) case. Transport Res E-Log. 2020;136:101922. doi: 10.1016/j.tre.2020.101922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bogler A, Packman A, Furman A, Gross A, Kushmaro A, Ronen A, Dagot C, Hill C, Vaizel-Ohayon D, Morgenroth E, Bertuzzo E, Wells G, Kiperwas HR, Horn H, Negev I, Zucker I, Bar-Or I, Moran-Gilad J, Balcazar JL, Bibby K, Elimelech M, Weisbrod N, Nir O, Sued O, Gillor O, Alvarez PJ, Crameri S, Arnon S, Walker S, Yaron S, Nguyen TH, Berchenko Y, Hu Y, Ronen Z, Bar-Zeev E. Rethinking wastewater risks and monitoring in light of the COVID-19 pandemic. Nat. Sustain. 2020;3(12):981–990. doi: 10.1038/s41893-020-00605-2. [DOI] [Google Scholar]
- 73.WHO WHO. Water, sanitation, hygiene and waste management for COVID-19: technical brief, 03 March 2020: World Health Organization2020.
- 74.Bourouache M, Mimouni R, Alla AA, Hamadi F, El Boulani A, Bihadassen B. Bacteriological and physicochemical quality of treated wastewater of the Mzar treatment plant. Appl Water Sci. 2019;9(4):86. doi: 10.1007/s13201-019-0958-0. [DOI] [Google Scholar]
- 75.Lund E, Rønne V. On the isolation of virus from sewage treatment plant sludges. Water Res. 1973;7(6):863–871. doi: 10.1016/0043-1354(73)90102-4. [DOI] [Google Scholar]
- 76.Núñez-Delgado A. What do we know about the SARS-CoV-2 coronavirus in the environment? Sci. Total Environ. 2020:138647. [DOI] [PMC free article] [PubMed]
- 77.Barcelo D. An environmental and health perspective for COVID-19 outbreak: meteorology and air quality influence, sewage epidemiology Indicator, hospitals disinfection, drug therapies and recommendations. J. Environ. Chem. Eng. 2020:104006. [DOI] [PMC free article] [PubMed]
- 78.Bibby K, Viau E, Peccia J. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett. Appl. Microbiol. 2011;52(4):386–392. doi: 10.1111/j.1472-765X.2011.03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]