Junyent et al. describe how the cell communication through cytonemes that leads to synthetic embryogenesis is altered upon pluripotency state transition in stem cells. They show that in more developmentally advanced stem cells, Wnt-iGluR crosstalk in the cytonemes is impaired, resulting in reduced formation of synthetic embryo structures.
Abstract
To coordinate cell fate with changes in spatial organization, stem cells (SCs) require specific and adaptable systems of signal exchange and cell-to-cell communication. Pluripotent embryonic stem cells (ESCs) use cytonemes to pair with trophoblast stem cells (TSCs) and form synthetic embryonic structures in a Wnt-dependent manner. How these interactions vary with pluripotency states remains elusive. Here we show that ESC transition to an early primed ESC (pESC) state reduces their pairing with TSCs and impairs synthetic embryogenesis. pESCs can activate the Wnt/β-catenin pathway in response to soluble Wnt ligands, but their cytonemes form unspecific and unstable interactions with localized Wnt sources. This is due to an impaired crosstalk between Wnt and glutamate receptor activity and reduced generation of Ca2+ transients on the cytonemes upon Wnt source contact. Induced iGluR activation can partially restore cytoneme function in pESCs, while transient overexpression of E-cadherin improves pESC–TSC pairing. Our results illustrate how changes in pluripotency state alter the mechanisms SCs use to self-organize.
Introduction
Pluripotent stem cells (PSCs) have an unlimited capacity to self-renew and can give rise to the three germ layers that make all adult tissues. In vitro, PSCs can exist in at least two defined pluripotent states (naive and primed) that likely recapitulate different developmental stages of the early embryo (Nichols and Smith, 2009). Naive embryonic stem cells (ESCs) are derived from the inner cell mass of the blastocyst before implantation and display robust self-renewal and differentiation potential (Ying et al., 2008; Bradley et al., 1984). Conversely, primed ESCs (pESCs) encompass a range of pluripotent states that resemble the more developmentally advanced post-implantation epiblast (Wu and Izpisua Belmonte, 2015) and have biases toward lineage-specific differentiation (Tsakiridis et al., 2014; Brons et al., 2007). Importantly, naive ESCs can colonize the blastocyst and contribute extensively to all lineages, resulting in chimeric animals (Bradley et al., 1984). Early pESCs retain a reduced capacity to contribute to blastocyst chimaeras (Kinoshita et al., 2020 Preprint), while later pESC populations are only able to engraft in the post-implantation embryo (Ohtsuka et al., 2012; Huang et al., 2012). While much is known about the transcriptional and epigenetic changes in these pluripotent states (Neagu et al., 2020), the signaling mechanisms driving these differences remain poorly understood. By comparing naive and early pESCs, we can investigate how transitions in pluripotency state change the signals and mechanisms cells use to communicate.
In vitro, both intrinsic and extrinsic cues regulate the state of PSCs. Addition of soluble Wnt ligands or small molecules that activate the Wnt/β-catenin pathway promote the self-renewal of naive ESCs (ten Berge et al., 2011; Ying et al., 2008; Augustin et al., 2017; Merrill, 2012). Blocking Wnt signaling leads to their transition to an early pESC stage (ten Berge et al., 2011; Neagu et al., 2020). In these conditions, pESCs grow in flattened colonies that down-regulate NANOG and alkaline phosphatase expression, up-regulate epiblast markers (e.g., Otx2 and Fgf5), and undergo X chromosome inactivation in female cell lines (ten Berge et al., 2011).
Wnt ligands undergo post-translational acylation by the O-acyltransferase Porcupine (Kadowaki et al., 1996; Takada et al., 2006), which makes them hydrophobic (Langton et al., 2016; Willert et al., 2003). Therefore, Wnts are often secreted locally in vivo and presented in a restricted manner to responsive stem cells (Mills et al., 2017). To activate the Wnt/β-catenin pathway, Wnt proteins bind to the Frizzled receptor and the coreceptors low-density lipoprotein receptor-related protein 5 and 6 (LRP5/6) on the recipient cell. This binding induces receptor clustering and phosphorylation (Bilic et al., 2007), leading to the inactivation of the destruction complex that targets β-catenin for degradation. Consequently, β-catenin is stabilized and translocated to the nucleus to initiate the Wnt-mediated transcription program (Garcin and Habib, 2017).
We have previously explored how Wnt-responsive stem cells interact with localized sources of Wnt (Junyent et al., 2020). We have shown that ESCs generate specialized cytonemes that selectively react to Wnt ligands required for self-renewal. When co-cultured with trophoblast stem cells (TSCs), ESC cytonemes respond to locally TSC-produced Wnts and establish cell–cell pairing. This is achieved through a crosstalk between LRP6, localized Ca2+ transients on the cytonemes, and members of the ionotropic glutamate receptor family (iGluRs). As a result, stable ESC–TSC pairing activates the Wnt/β-catenin pathway in ESCs and initiates synthetic embryogenesis (Junyent et al., 2020). In this study, we induced the transition of naive ESCs to an early pESC state by the inhibition of Wnt/β-catenin signaling. We examined how this transition affects the interaction of pESCs with Wnt signals, their pairing with TSCs, and subsequently the formation of synthetic embryo structures.
We found that, similar to naive ESCs, pESCs contact TSCs through cytonemes. However, the frequency of pESC–TSC pairing, and consequently the formation of synthetic embryo-like structures, was significantly reduced compared with ESCs. Wnt ligands regulate ESC–TSC pairing, and ESCs can selectively recruit localized Wnt ligands that are covalently immobilized to a microbead. However, while pESCs activate the Wnt/β-catenin pathway upon exogenous addition of soluble Wnt3a proteins (a cytoneme-independent mechanism), their cytonemes are nonselective and cannot form a stable interaction with a Wnt source. To explore the mechanisms behind these differences, we investigated the components that underpin cytoneme functionality in ESCs: Wnt-iGluR crosstalk and stable cell adhesion with TSCs. ESCs and pESCs have functional iGluRs, but in pESCs, interactions between cytonemes and Wnt beads fail to generate localized Ca2+ transients at the contact site. This correlates with a significantly reduced capacity of pESCs to polarize components of the Wnt/β-catenin pathway upon Wnt bead contact. Up-regulation of iGluR activity, but not overexpression of the Wnt coreceptor LRP6, ameliorates cytoneme-mediated communication and cell polarization of pESCs. Furthermore, transient overexpression of E-cadherin in pESCs facilitates their pairing with TSCs but does not rescue synthetic embryogenesis. Altogether, our results show that changes in developmental potential alter the mechanisms that stem cells use to self-organize, and that a complex protein network, rather than a single factor, orchestrates this process.
Results
pESCs have reduced ability to form synthetic embryo-like structures
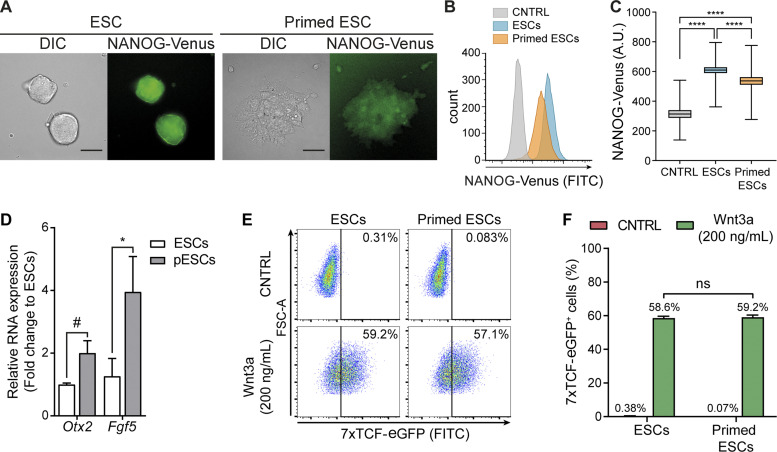
The activation of the Wnt/β-catenin pathway supports the self-renewal of naive ESCs, while its inhibition leads ESCs to progress to a more developmentally restricted “primed” pluripotent population (ten Berge et al., 2011; Neagu et al., 2020). ESCs cultured with soluble Wnt3a grew round, dome-shaped colonies that express high levels of NANOG and β-catenin (Fig. 1 A and Fig. S1 A). Inhibition of the O-acyltransferase Porcupine with IWP2 blocks the secretion of Wnts (Chen et al., 2009). Treatment of ESCs with IWP2 for 3 d led to flattened pESC colonies that express lower levels of NANOG and β-catenin and up-regulate epiblast markers Otx2 and Fgf5 (Fig. 1, A–D; and Fig. S1 A). The ESC-to-pESC transition (mediated by Wnt pathway inhibition) enabled us to compare two stem cell populations that represent different pluripotency states (ten Berge et al., 2011) but remain closely related.
Figure 1.
ESCs and pESCs represent progressive developmental stages of Wnt responsive pluripotent stem cells. (A) Representative images of DIC and NANOG-Venus levels in colonies of ESCs or pESCs. pESCs were generated by treating ESCs with 2 µM IWP2 for 3 d. Scale bars, 50 µm. (B) Representative flow cytometry histogram of the NANOG-Venus intensity in ESCs or pESCs, compared with control (CNTRL) cells. CNTRL are ESCs without the NANOG-Venus transgene. NANOG-Venus intensity for >10,000 cells/condition, expressed in log10. (C) Quantification of NANOG-Venus intensity by flow cytometry. Box and whiskers plots represent pooled data from n = 3 experiments, >10,000 cells analyzed/n. Error bars are range, and middle line is median. Statistical significance calculated by one-way ANOVA with Tukey’s multiple comparison tests: ****, P < 0.0001. A.U., arbitrary units. (D) Fold-change expression of Otx2 and Fgf5 in pESCs relative to ESCs. Bars are mean of n = 3. Error bars are SEM. Statistical significance calculated by two-way ANOVA with Šídák’s multiple comparison test: #, P = 0.061; *, P < 0.05. (E) Flow cytometry plot of 7xTCF-eGFP–expressing ESCs and pESCs upon addition of 200 ng/ml soluble Wnt3a protein or CNTRL solution. >10,000 cells/condition. Vertical line indicates threshold of 7xTCF-eGFP+ cells. (F) Percentage of 7xTCF-EGFP+ cells as shown in E for n = 3. Bars are mean, error bars are SEM, and ns are nonsignificant differences, calculated by one-way ANOVA with Tukey’s multiple comparisons test.
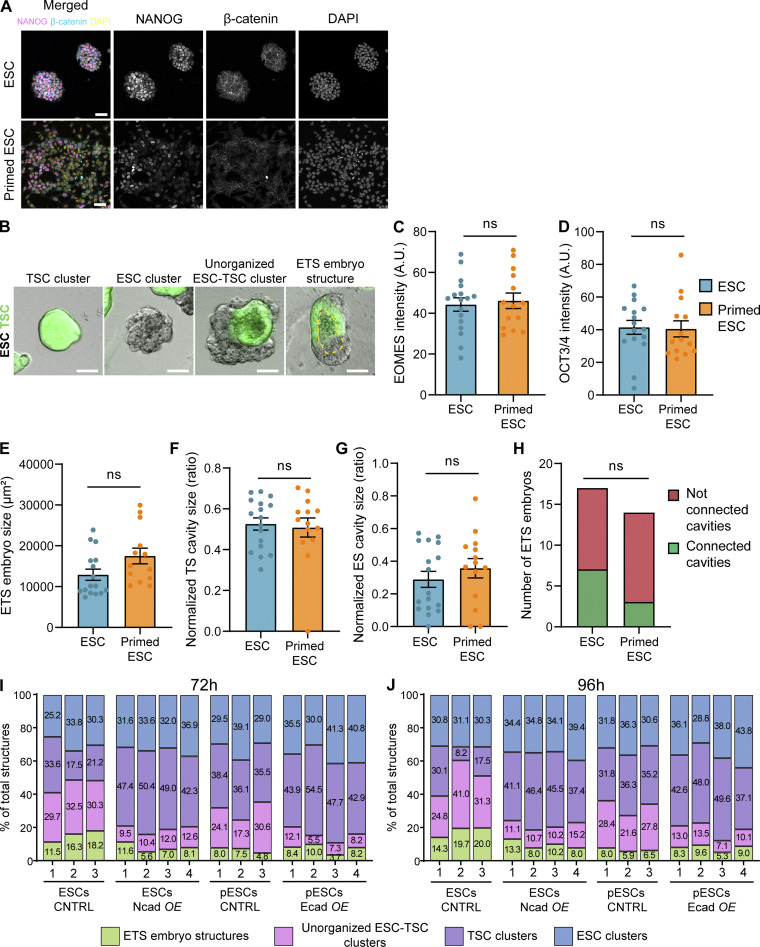
Figure S1.
Immunofluorescence of NANOG and β-catenin in pESCs and quantification of ETS structures. (A) Representative images of ESC or pESC colonies stained with antibodies against NANOG or β-catenin or with DAPI. pESCs are obtained by culturing ESCs for 3 d in media supplemented with 2 µM IWP2. Intensity range displayed is equal between the two conditions. Scale bar, 50 µm. (B) Representative images of cell clusters/structures formed at 96 h of ETS induction. From left to right, TSC-only cell cluster, ESC-only cell cluster, unorganized ESC–TSC cluster, and ETS embryo structure. Images are merged of brightfield and GFP (TSCs). Yellow dashed line highlights cavity. Scale bar, 50 µm. For all quantifications, total structures are the sum of the quantified structures for all cluster/structure types. (C and D) Quantification of marker expression in ETS structures formed by ESCs (blue) or pESCs (orange) at 96 h of co-culture with TSCs. (C) EOMES intensity on the TSC compartment, normalized to background intensity. (D) OCT3/4 intensity on the ESC compartment, normalized to the background intensity. For C and D, ns indicates nonsignificant differences, calculated by unpaired two-sided t tests. n = 17 ETS structures for ESCs and 14 for pESCs, pooled from three independent experiments. Bars are mean, and error bars are SEM. (E–G) Quantification of the size of ETS embryo structures formed by ESCs (blue) or pESCs (orange) at 96 h of co-culture with TSCs. (E) ETS embryo structure size at maximum width, in square micrometers. (F) TSC compartment cavity size normalized to TSC compartment size, expressed as a ratio. (G) ESC compartment cavity size normalized to ESC compartment size, expressed as a ratio. n = 17 ETS structures for ESCs and 14 for pESCs, pooled from three independent experiments. For E–G, ns indicates nonsignificant differences, calculated by unpaired two-sided t tests. Bars are mean, and error bars are SEM. (H) Number of ETS embryos formed by ESCs or pESCs at 96 h of co-culture with TSCs with connected or nonconnected cavities. ns indicates nonsignificant differences, calculated by Fisher’s exact two-sided test. n = 17 ETS structures for ESCs and 14 for pESCs, pooled from three independent experiments. (I and J) Breakdown of the quantification of ETS embryo structures, unorganized ESC–TSC clusters, TSC clusters, or ESC clusters at 72 h (I) and 96 h (J) of co-culture. Numbers within bars indicate percentage. Number of total structures counted is at least 80 per independent experiment, and a minimum of three independent experiments per condition. CNTRL, control; Ncad OE, N-cadherin overexpression; Ecad OE, E-cadherin overexpression.
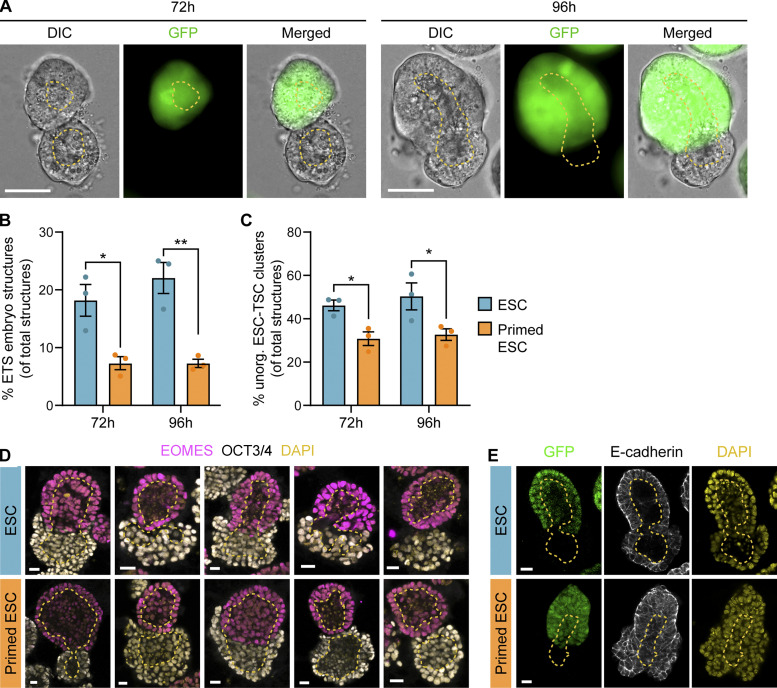
Synthetic embryos, resembling aspects of naturally developing embryos (Shahbazi and Zernicka-Goetz, 2018), are tractable and easy-to-observe experimental systems to explore how mechanisms of cell communication direct self-organization. We investigated how the ESC-to-pESC transition affects synthetic embryo formation by co-culturing ESCs or pESCs with GFP-expressing TSCs in 3D conditions that promote ESC-TSC synthetic (ETS) embryo structure formation (Harrison et al., 2018, 2017). As well as ETS embryo structures, cells in these conditions form unorganized structures containing both ESCs and TSCs, or clusters of only one cell type (Fig. 2 A and Fig. S1 B). Overall, a higher proportion of mixed-cell structures was observed with ESCs than with pESCs when mixed with TSCs (ETS embryo structures and unorganized ESC–TSC structures; Fig. 2, B and C). By 72 h, ETS embryo structures form with internal cavitation. At 96 h, some larger ETS embryo structures had a connected cavity, indicating structural maturation (Fig. 2 A; Harrison et al., 2017, 2018). pESCs formed a significantly lower proportion of ETS embryo structures than ESCs did (7.27% and 22.04% of total structures at 96 h, respectively; total structures are the sum of the structures/clusters of all types; Fig. 2 B; and Fig. S1, B, I, and J). Immunostaining of the ETS embryo structures at 96 h indicated that expression of OCT3/4 and EOMES (Eomesodermin), and localization of E-cadherin, were similar in ETS embryo structures formed by ESCs or pESCs (Fig. 2, D and E; and Fig. S1, C and D). Moreover, the overall ETS embryo structure size, the cavity size within the ESC and TSC compartments, and the proportion of ETS embryo structures with a connected cavity were comparable in both ESC–TSC and pESC–TSC cultures (Fig. S1, E–H).
Figure 2.
pESCs have a reduced ability to form synthetic embryo-like structures. (A) Representative DIC, GFP, and merged images of ETS embryo structures after 72 h and 96 h of co-culture. TSCs constitutively express GFP. At 72 h, opposing TSC and ESC clusters with internal cavitation are observed. At 96 h, larger structures with merged cavities appear. Dashed yellow line outlines cavities. Scale bars, 50 µm. (B and C) Proportion of structures formed by ESCs (blue) or pESCs (orange) at 72 h and 96 h of co-culture with TSCs. (B) Proportion of ETS embryo structures. Values are percentage of total structures (i.e., ETS embryo structures plus unorganized ESC–TSC clusters plus TSC clusters plus ESC clusters). For detailed quantification, see Fig. S1, B, I, and J. (C) Proportion of unorganized ESC–TSC clusters. Values are percentage of total structures (same as B). For extended quantification, see Fig. S1, B, I, and J. For B and C, n = 3, ≥80 total structures per n. Bars are mean, and error bars are SEM. Statistical significance calculated by two-way ANOVA with Šídák’s multiple comparison test: *, P < 0.05; **, P < 0.01. (D and E) Representative images of ETS embryo structures formed by ESCs (blue, top) and pESCs (orange, bottom) after 96 h of co-culture with TSCs. (D) ETS structures are labeled with antibodies against EOMES (magenta) and OCT3/4 (white) plus DAPI (yellow), presented as merged. (E) ETS structures labeled with antibodies against E-cadherin (white) and DAPI (yellow). TSCs express GFP. For D and E, yellow dashed lines indicate internal cavities. Scale bars, 20 µm.
Our findings indicate that pESCs formed a significantly lower proportion of organized (ETS embryos) and unorganized ESC–TSC clusters than ESCs did. This suggests that the pluripotency state transition, induced by Wnt inhibition, affects the interaction of pESCs with TSCs, which is required to initiate synthetic embryogenesis.
pESCs have an impaired capacity to pair with TSCs
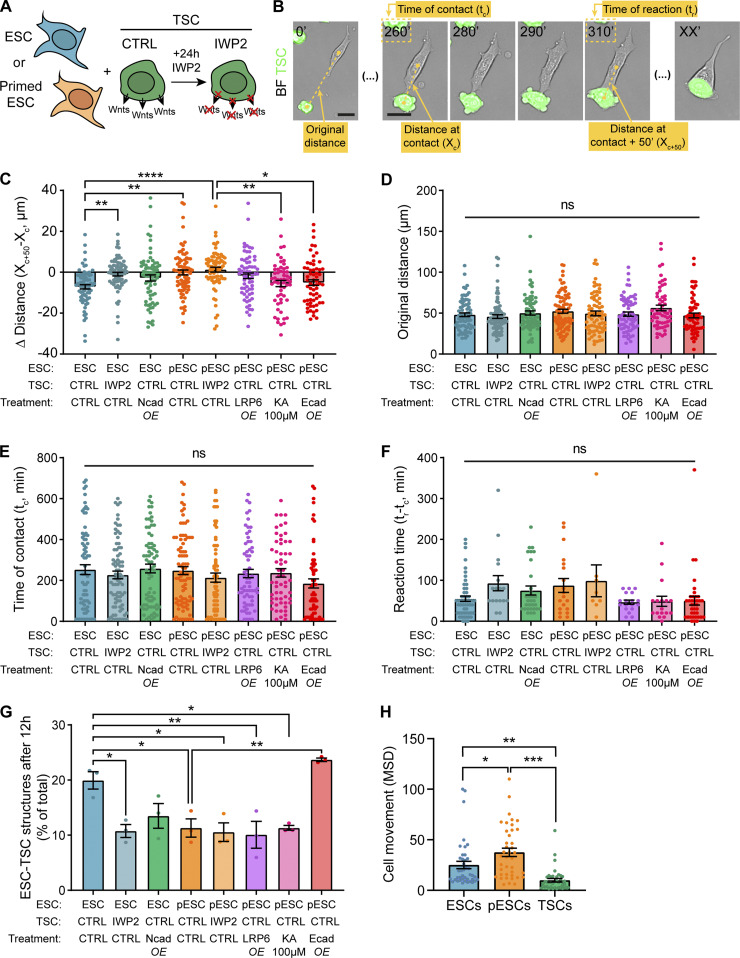
We have previously shown that ESCs generate specialized protrusions, termed cytonemes, that interact with TSCs and initiate a stable contact and ESC–TSC pairing, an essential step in synthetic embryogenesis (Fig. 3 A; Junyent et al., 2020). We examined whether pESCs use a similar mechanism. We co-cultured TSCs with ESCs or pESCs and followed their interaction by time-lapse imaging. As observed in ESCs (Junyent et al., 2020), pESCs use cytonemes to contact TSCs (Fig. 3 A). pESCs have significantly higher motility compared with ESCs, whereas TSCs have significantly restricted motility (Fig. S2 H). We measured the distance between cells after initial contact with a TSC through a cytoneme. On average, ESC-to-TSC distance was reduced after initial cell contact. However, pESC to TSC distance remained unchanged (Fig. S2, A–C). Detailed analysis revealed that an ESC cytoneme–mediated interaction with a TSC often resulted in a stable ESC–TSC pairing through reactive interactions (RIs; 74% of total interactions), whereas pESC–TSC interactions resulted in significantly reduced pESC–TSC pairing (25% RIs; Fig. 3 B). To investigate if reduced pESC–TSC interaction contributes to impaired ETS structure formation, we measured the number of mixed cell clusters (containing TSCs and either ESCs or pESCs) after short-term (12 h) co-culture. Indeed, cluster formation was significantly compromised in pESCs when compared with ESCs (Fig. S2 G).
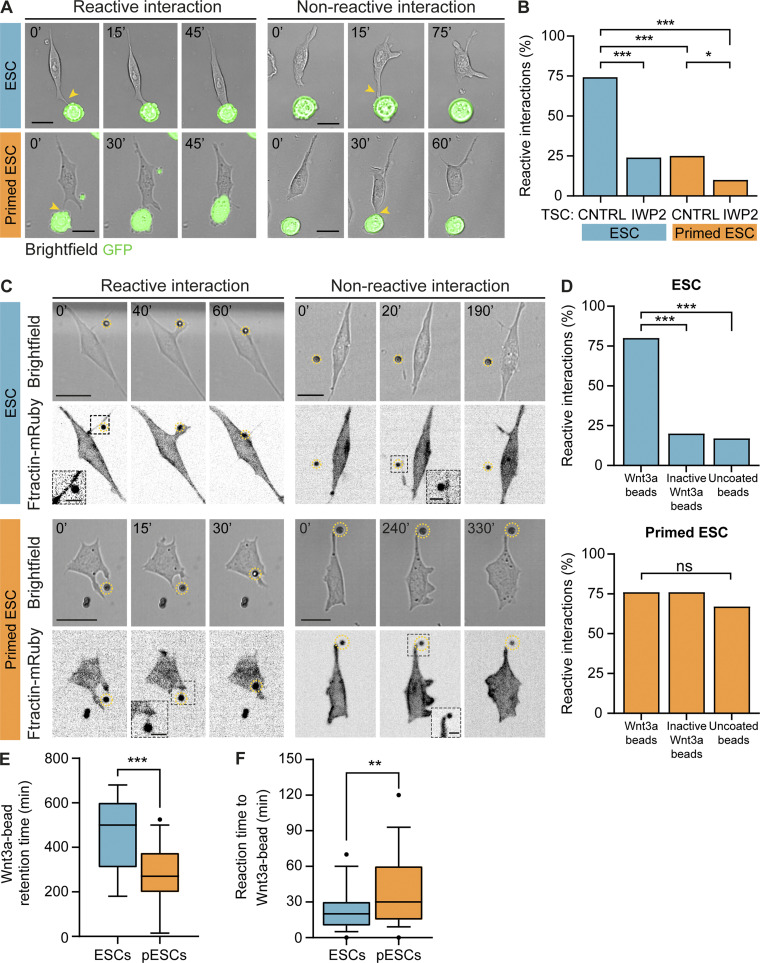
Figure 3.
pESCs cytonemes are non-selective and cannot facilitate stable interactions with TSCs. (A) Frames from time-lapse imaging of ESCs (blue, top) or pESCs (orange, bottom) interacting with TSCs that express GFP. Examples of reactive interactions (left) or non-reactive interactions (right) are shown. Time in minutes. Arrowheads (yellow) indicate initial contact through cytonemes. Scale bars, 20 µm. (B) Percentage of reactive interactions between ESCs (blue) or pESCs (orange) and TSCs in different conditions. Where indicated, TSCs were pretreated with 2 µM IWP2 for 24 h. n ≥ 71 cells pooled from three experiments. Statistical significance calculated by multiple Fisher’s exact two-sided tests: *, P < 0.05; ***, P < 0.001. CNTRL, control. (C) Frames from time-lapse imaging of ESCs (blue, top) or pESCs (orange, bottom) expressing the F-actin reporter Ftractin-mRuby3 (presented as inverted grayscale), interacting with Wnt beads. Reactive (left) and non-reactive (right) interactions are shown. Time in minutes. Wnt beads are highlighted by yellow dashed circle. Inserts are magnified and contrast-enhanced for clarity. Scale bars, 20 µm for larger images or 5 µm for inserts. (D) Percentage of reactive interactions between ESCs (blue, top) or pESCs (orange, bottom) and different types of beads. n ≥ 40 cells pooled from ≥3 experiments. Statistical significance calculated by multiple Fisher’s exact two-sided tests: ns, nonsignificant, P > 0.05; ***, P < 0.001. (E and F) Box and whiskers plots describing the distribution of the Wnt3a bead retention time (E) and time between initial Wnt3a bead contact and recruitment (F) for ESCs (blue) or pESCs (orange). For details on the measurement, see Fig. S3 B. Whiskers are 5–95% of data, middle line is median, and dots are data outside range. n ≥ 40 cells pooled from ≥3 experiments. Statistical significance calculated by unpaired two-sided t tests: **, P < 0.01; ***, P < 0.001.
Figure S2.
Quantification of ESC or pESC interaction with TSCs. (A) Schematic depicting the experimental conditions. (B) Representative annotated frames of a time-lapse live-cell imaging showing an ESC contacting a TSC with a cytoneme and pairing with it. TSCs express GFP and are labeled in green. Annotations refer to measurements in C–F, as follows: “Original distance” between the cells at t = 0’; distance between the cells at the time of cytoneme-mediated contact (“Distance at contact”; Xc); time at initial cytoneme-mediated contact (“Time at contact”; tc); distance between cells 50 min after contact (“Distance at contact + 50 minutes”; Xc+50; found empirically to be enough to capture the behavior of the cells after initial contact); time at which cell–cell pairing is established (“Time of reaction”; tr; only quantified for reactive interactions). Time is in minutes. Scale bars, 20 µm. BF, brightfield. (C) Quantification of the differential between the distance at contact (Xc) and distance at contact + 50 min (Xc+50) expressed in micrometers, for all conditions. n ≥ 58 cells pooled from ≥3 independent experiments. (D) Quantification of the original ESC–TSC distance (in micrometers) for all conditions. n ≥ 58 cells pooled from ≥3 independent experiments. (E) Quantification of the time of ESC–TSC contact through a cytoneme (in minutes) for all conditions. n ≥ 58 cells pooled from ≥3 independent experiments. (F) Quantification of reaction time between ESCs and TSCs, calculated as the differential between time at contact (tc) and time at reaction (tr) in minutes, for all conditions. Reaction time is only calculated for cells that react (according to Fig. 3 A). n ≥ 8 cells from ≥3 independent experiments. (G) Quantification of the number of mixed ESC–TSC structures after 12 h co-culture, presented as percentage of total TSC clusters. n = 3, ≥66 total structures per n. (H) Quantification of cell movement for ESCs (blue), pESCs (orange), and TSCs (green), presented as mean squared displacement (MSD). n = 40 cells pooled from ≥3 experiments. For C–H, bars indicate mean, and error bars are SEM. Symbols indicate statistical significance calculated by one-way ANOVA with Tukey’s multiple comparison tests: ns, nonsignificant, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. CTRL, control; Ecad, E-cadherin; KA, kainate; Ncad, N-cadherin; OE, overexpression.
ESC–TSC interaction requires the secretion of Wnt by the TSCs (Junyent et al., 2020). TSCs secrete Wnts locally that can be recognized by ESC cytonemes, resulting in ESCs pairing with TSCs and activating the Wnt/β-catenin pathway (Junyent et al., 2020). To examine if pESCs respond to Wnt ligands, we generated ESC and pESC lines that harbor the Wnt reporter 7xTCF-eGFP (enhanced GFP; Fuerer and Nusse, 2010). We incubated them with exogenous soluble Wnt3a ligands and monitored eGFP expression after 24 h by FACS. Our results indicate that pESCs can activate the Wnt/β-catenin pathway similarly to ESCs (Fig. 1, E and F).
Next, we investigated locally presented Wnts on the pESCs–TSCs interaction. Culturing TSCs with IWP2 for 24 h blocks the secretion of Wnt ligands (Chen et al., 2009; Junyent et al., 2020). Pretreatment of TSCs with IWP2 significantly reduced ESC–TSC pairing (24% RIs; Fig. 3 B; and Fig. S2, A–C), as previously reported (Junyent et al., 2020). Similarly, the low number of pESC–TSC pairs was further reduced by this treatment to only 10% RIs (Fig. 3 B and Fig. S2 C). The initial distance between ESCs or pESCs and TSCs, the time of initial cell contact, and the time between contact and pairing (reaction time) remained similar between conditions (Fig. S2, B and D–F).
Together, this indicates that while pESCs respond to soluble Wnt3a added globally to the media, pESC cytonemes had an impaired ability to form stable interactions with TSCs upon initial contact, resulting in fewer pESC–TSC structures.
Soluble Wnt ligands added to the media can reach the cell membrane by diffusion, a cytoneme-independent mechanism (Lippert et al., 2017). In contrast, ESCs extend cytonemes to recruit and respond to locally secreted Wnts by TSCs. We previously used a reductionist approach that allows the examination of cell–ligand interaction. We covalently immobilized Wnt3a proteins (and controls) onto microbeads and presented them to single ESCs. Our results demonstrated that Wnt3a bead recruitment by the ESC requires a directional, active, and selective process, which is cytoneme mediated (Junyent et al., 2020).
To further explore the differences between ESCs and pESCs, we compared their selectivity and response to immobilized Wnt3a ligands covalently tethered to microbeads.
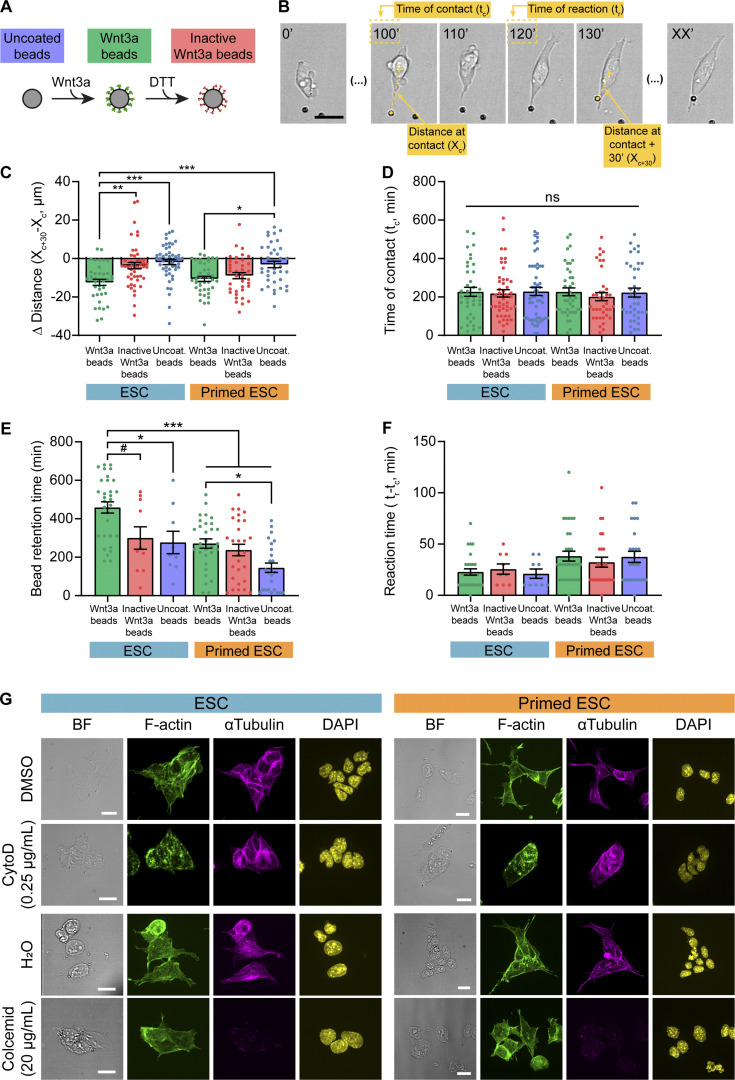
pESC cytonemes are nonselective and have unstable interactions with localized Wnt3a ligands
We incubated single ESCs or pESCs near Wnt3a beads or control beads: inactive Wnt3a beads (DTT-treated to break the disulfide bridges within Wnt ligands, disrupting the tertiary structure of the protein and rendering it inactive) or uncoated beads (Fig. S3 A). To improve the visualization of the cell–bead contact, we used cells that express the F-actin reporter Ftractin-mRuby (Hayer et al., 2016), and monitored the initial cell–bead contact by live cell imaging. Similar to the interaction with TSCs, both cell lines use cytonemes to contact the bead and recruit it to the plasma membrane (reactive interaction; Fig. 3 C). We also observed non-reactive interactions, where the contact does not lead to recruitment (non-reactive interaction; Fig. 3 C).
Figure S3.
Quantification of ESC or pESC interaction with Wnt3a or control beads, and representative images of cytochalasin D or colcemid treatment. (A) Schematic depicting the three types of beads used in the experiments. (B) Representative annotated frames of a time-lapse live imaging of an ESC contacting and recruiting a Wnt3a bead though a cytoneme. Annotations refer to measurements in C, D and F, as follows: distance between the cell and the bead at the time of cytoneme-mediated contact (“Distance at contact”; Xc); time at initial cytoneme-mediated contact (“Time at contact”; tc); distance between cell and bead 30 min after contact (“distance at contact + 30 minutes”; Xc+30; found empirically to be enough to capture the behavior of the cells after initial bead contact); time at which the bead is recruited by the cell (“Time of reaction”; tr; only quantified for reactive interactions). Scale bar, 20 µm. (C) Quantification of the differential between distance at contact (Xc) and distance at contact + 30 min (Xc+30) expressed in micrometers, for all conditions. n ≥ 40 cells pooled from ≥3 independent experiments. (D) Quantification of the time of ESC bead initial contact with a cytoneme (tc, in minutes). n ≥ 40 cells pooled from ≥3 independent experiments. (E) Quantification of the time of bead retention after reaction (in minutes) for all conditions. Bead retention time is only calculated for cells that react (according to Fig. 3 C). Data for ESC or pESC with Wnt3a beads are reused from Fig. 3 E. n ≥ 9 cells pooled from three experiments. F. Quantification of the reaction time, calculated as the difference between time at contact (tc) and time at reaction (tr) in minutes, for all conditions. Reaction time is only calculated for cells that react (according to Fig. 3 C). Data for ESC or pESC with Wnt3a beads are reused from Fig. 3 E. n ≥ 9 cells pooled from three experiments. For C–F, bars indicate mean, and error bars are SEM. Symbols indicate statistical significance calculated by one-way ANOVA with Tukey’s multiple comparison tests: ns, nonsignificant, P > 0.05; #, P = 0.051; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (G) Representative images of ESCs (blue, left) or pESCs (orange, right) treated with DMSO, 0.25 µg/ml cytochalasin D (CytoD), H2O, or 20 µg/ml colcemid (top to bottom) for 4 h, and stained with antibodies against α-tubulin (magenta) or phalloidin (F-actin, green) and DAPI (yellow). BF is brightfield. Scale bars, 20 µm.
ESCs had a significantly higher proportion of reactive interactions when cells contacted Wnt3a beads (80% RIs) than inactive Wnt3a beads or uncoated beads (20% and 17% RIs, respectively), as previously reported (Fig. 3 D; Junyent et al., 2020). In comparison, pESCs were efficient in the recruitment of both Wnt3a and inactive Wnt3a beads (76% and 76% RIs, respectively) as well as uncoated beads, but to a lower extent (67% RIs; Fig. 3 D). Measurement of the cell–bead distance after initial cytoneme-mediated contact reinforced these results (Fig. S3, B and C). However, the bead retention time on pESCs was significantly shorter for all bead types, compared with ESCs, with uncoated bead retention time being the shortest (Fig. 3 E and Fig. S3 E). The reaction time (time between initial Wnt3a bead contact and recruitment) was longer in pESCs (Fig. 3 F; and Fig. S3, B and F). Importantly, the time of initial cell–bead contact was similar between cell lines (Fig. S3, B and D), indicating that the starting conditions were comparable between experiments.
In summary, our results suggest that the transition of ESCs to pESCs, mediated by inhibition of Wnt signaling, alters cytoneme function. As a result, the cytonemes of pESCs are not selective and do not form stable contacts with the Wnt source, subsequently reducing the efficiency of synthetic embryogenesis.
To investigate underlying changes that may alter the function of pESC cytonemes, we analyzed their composition and dynamics.
F-actin and tubulin are required for pESCs cytoneme formation
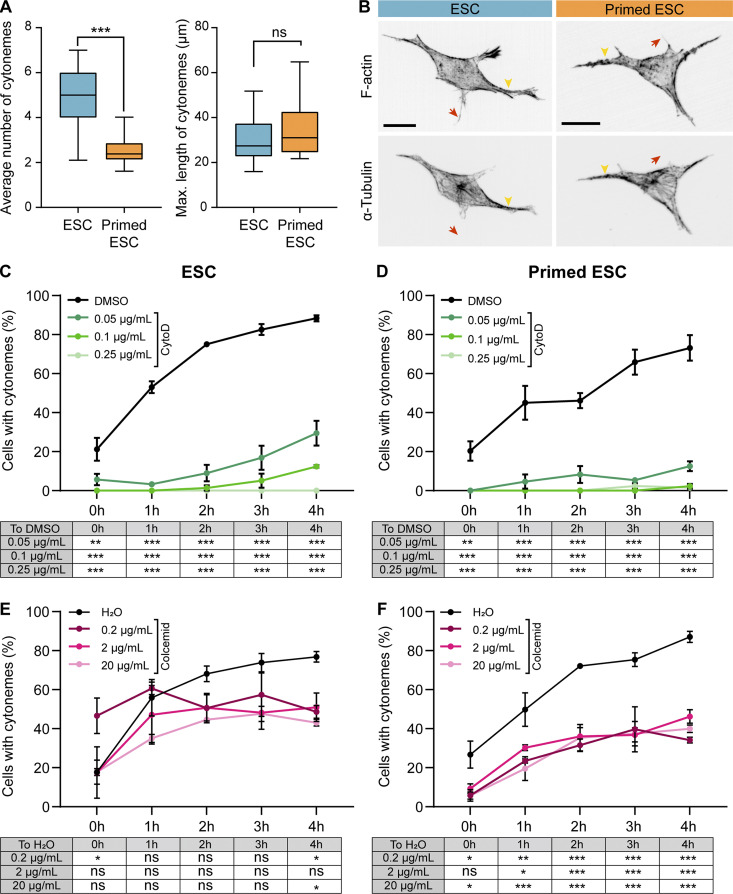
ESCs generate a median of five cytonemes per cell, while pESCs form two or three cytonemes per cell (Fig. 4 A). The maximum cytoneme length in both cell types is comparable, with a mean of ∼30 µm for ESCs and ∼35 µm for pESCs (Fig. 4 A). Next, we analyzed the cytoskeleton composition of the cytonemes. Immunostaining revealed that all cytonemes in ESCs and pESCs contain F-actin, with tubulin restricted to larger cytonemes (Fig. 4 B). Inhibition of F-actin polymerization by cytochalasin D treatment blocked cytoneme formation in both cell types (Fig. 4, C and D; and Fig. S3 G), while inhibition of tubulin polymerization by colcemid affected cytoneme formation only in pESCs (Fig. 4, E and F; and Fig. S3 G). This indicates that pESCs rely on both F-actin and tubulin polymerization for cytoneme generation, pointing to a change in the composition of the cytonemes that might contribute to their function.
Figure 4.
F-actin and tubulin are required for pESC cytoneme formation. (A) Box and whiskers plots describing the average number of cytonemes (left) or maximum cytoneme length (right) in ESCs (blue) and pESCs (orange). Whiskers are 5–95% of data, and middle line is median. n ≥ 24 cells. Statistical significance calculated by unpaired two-sided t tests: ns, nonsignificant, P > 0.05; ***, P < 0.001. (B) Representative images of ESCs (blue) and pESCs (orange) stained with phalloidin (F-actin) or antibodies against α-tubulin, presented as inverted grayscale. Arrowheads (yellow) indicate larger cytonemes, and arrows (red) indicate thin cytonemes. Scale bars, 20 µm. (C and D) The percentage of cells with cytonemes in ESCs (C) or pESCs (D) treated with a range of cytochalasin D (CytoD) concentrations or DMSO. x axis is time in hours. Points are mean of n = 3 independent experiments, ≥25 cells per n. Error bars are SEM. Table below indicates statistical significance against DMSO, calculated by two-way ANOVA with Tukey’s multiple comparison tests: **, P < 0.01; ***, P < 0.001. (E and F) The percentage of cells with cytonemes in ESCs (E) or pESCs (F) treated with a range of colcemid concentrations or H2O. x axis is time, in hours. Points are mean of n = 3 independent experiments, ≥23 cells per n. Error bars are SEM. Table below indicates statistical significance against H2O control calculated by two-way ANOVA with Tukey’s multiple comparison tests: ns, nonsignificant, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, we characterized the signaling properties of the cytonemes.
pESCs have reduced Wnt-mediated Ca2+ response at the cytonemes upon contact with a localized Wnt source
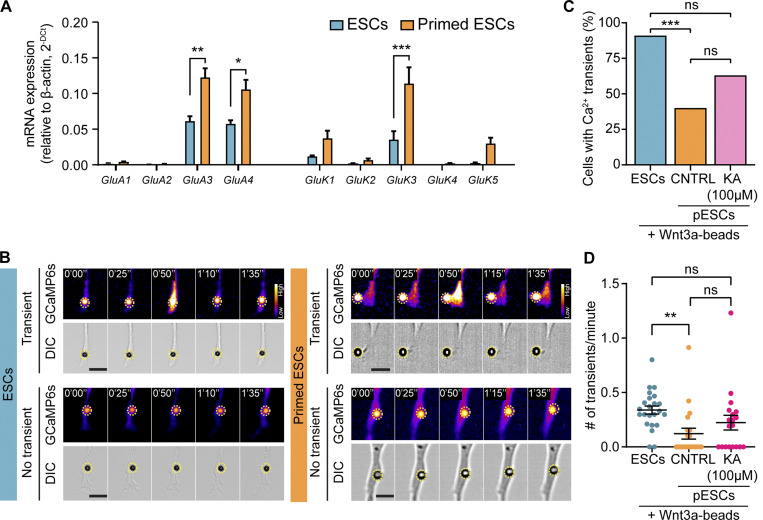
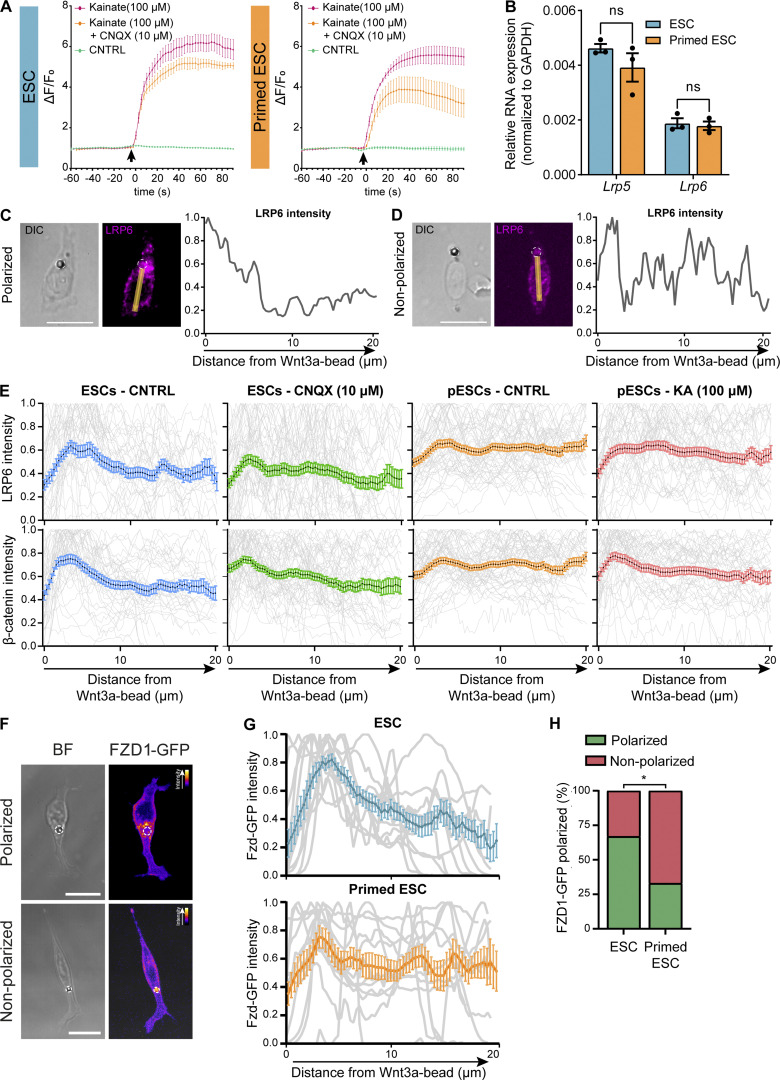
ESCs express subunits of the iGluRs (Fig. 5 A; Junyent et al., 2020; Gundry et al., 2010; Nagano et al., 2005). Using an ESC line stably expressing the free-cytoplasmic Ca2+ sensor GCaMP6s (Chen et al., 2013), we demonstrated that a contact with a TSC or a Wnt3a bead induces localized Ca2+ transients in the ESC cytoneme (Fig. 5, B–D). Pharmacological inhibition of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)/kainate iGluRs impairs the generation of Wnt-induced Ca2+ transients and ESC–TSC pairing and reduces the formation of ETS embryo structures (Junyent et al., 2020).
Figure 5.
pESCs have reduced glutamate receptor activity at the cytonemes. (A) Transcription levels of ionotropic glutamate receptor subunits in ESCs (blue) and pESCs (orange). RNA levels are expressed relative to β-actin expression. Bars are mean of n = 3, and error bars are SEM. Statistical significance calculated by one-way ANOVA with Šídák’s multiple comparison test: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) Frames from time-lapse imaging of an ESC (blue, left) or a pESC (orange, right) expressing GCaMP6s, where a cytoneme contacts a Wnt3a bead. Top: Generation of Ca2+ transients upon cytoneme-Wnt3a–bead contact. Bottom: Absence of Ca2+ transients. Bead is highlighted with a yellow dashed circle. Time is expressed in minutes and seconds. GCaMP6s intensity is presented using the Fire LUT (Fiji), and the calibration bar is shown in the figure. Scale bars, 10 µm. (C and D) Percentage (%) of cells with Ca2+ transients in the cytonemes (C), and number (#) of transients per minute, per cell (D) in ESCs (blue, n = 20), control pESCs (CNTRL, orange, n = 24) or pESCs treated with 100 µM kainate (KA, pink, n = 19). Cells for each condition are pooled from multiple independent experiments. In D, bar indicates mean, and error bars indicate SEM. Statistical significance calculated by multiple Fisher’s exact two-sided tests (C) or one-way ANOVA with Tukey’s multiple comparison test (D): ns, nonsignificant, P > 0.05; **, P < 0.01; ***, P < 0.001.
We investigated whether the cytonemes of pESCs have similar functionality. pESCs express subunits of the AMPA and kainate receptors at similar or higher levels than ESCs (Fig. 5 A). To analyze the activity of the iGluRs on pESCs, we used GCaMP6s-expressing cells and recorded whole-cell Ca2+ levels by fast time course live imaging (Fig. S4 A). Addition of 100 µM kainate (an agonist of AMPA/kainate iGluRs) induced a sustained Ca2+ increase in pESC, comparable to that observed in ESCs (Fig. S4 A). Importantly, pretreatment of the cells with 10 µM cyanquixaline (CNQX, a competitive inhibitor of the AMPA and kainate receptors) reduced the Ca2+ response to kainate in both pESCs and ESCs (Fig. S4 A). Our results indicated that pESCs expressed functional iGluRs.
Figure S4.
pESCs present functional iGluR receptors and similar Lrp5/6 levels to ESCs but exhibit impaired polarization of Wnt pathway components upon contact with a Wnt source; pESC polarization is recovered by kainate addition. (A) Whole-cell time course Ca2+ measurements of ESCs (blue, left) or pESCs (orange, right) expressing GCaMP6s. Lines indicate Ca2+ response to the addition of control (CNTRL) solution (green), 100 µM kainate (pink), or 100 µM kainate to cells pretreated with 10 µM CNQX (orange). GCaMP6s intensity is expressed as fold-change to basal intensity before addition (ΔF/F0). Points are mean of n ≥ 4, and error bars are SEM. Black arrow indicates time of addition. (B) Lrp5 and Lrp6 RNA expression levels in ESCs (blue) or pESCs (orange), presented as normalized expression to GAPDH. Bars are mean of n = 3, and error bars are SEM. ns is not significant, calculated by two-way ANOVA with Šídák’s multiple comparison test. (C and D) Examples of the quantification in E. Left: Representative image of a cell contacting a Wnt3a bead and exhibiting a polarized (C) or nonpolarized (D) distribution of Lrp6. Yellow line and arrow represent a 10-pixel-wide, 20-µm-long line used to measure the intensity profile. Wnt3a bead is highlighted with white dashed line. Scale bar, 20 µm. Right: Normalized quantification of the Lrp6 intensity profile, expressed as fold-change to maximum value across the distance from the Wnt3a bead. (E) Intensity profile of LRP6 (top) and β-catenin (bottom) in ESCs (CNTRL, blue), 10 µM CNQX-treated ESCs (green), control pESCs (CNTRL, orange) or 100 µM kainate (KA)-treated pESCs (pink) contacting Wnt3a beads. Background-normalized intensity is plotted relative to the distance from Wnt3a bead and reported as fold-change to maximum intensity value (expressed as ratio). Gray lines represent individual intensity measurements, colored lines represent mean, and error bars are SEM. n ≥ 41 cells. (F) Representative images of ESCs expressing FZD1-GFP and contacting a Wnt3a bead. Wnt3a bead is black sphere in brightfield (BF) panel, highlighted by white dashed circle. FZD1-GFP intensity is presented using the Fire LUT (ImageJ), and the calibration bar is shown in the figure. Scale bars, 20 µm. (G) Intensity profile of FZD1-GFP in ESCs (top) or pESCs (bottom) contacting a Wnt3a bead. Background-normalized intensity is plotted relative to the distance from Wnt3a bead and reported as fold-change to maximum intensity value (expressed as ratio). Gray lines represent individual intensity measurements, colored lines represent mean, and error bars are SEM. n ≥ 12 cells. (H) Percentage of ESCs or pESCs contacting Wnt3a beads that show polarized FZD1-GFP. n ≥ 12 cells. Statistical significance calculated by Fisher’s exact two-sided test: *, P < 0.05.
We examined the generation of Ca2+ transients in the cytonemes of pESCs upon contact with a Wnt3a bead. Only 40% (n = 20 cells) of pESCs generated localized Ca2+ transients at the area of Wnt3a bead contact, compared with 91% of ESCs (n = 24 cells; Fig. 5, B and C, first two conditions). The mean rate of Ca2+ transients per minute was 0.12 transients/min in pESCs in comparison to 0.33 transients/min in ESCs (Fig. 5 D, first two conditions).
Our results demonstrate that, although pESCs express functional iGluRs, the prevalence and rate of Wnt-mediated Ca2+ transients in pESC cytonemes were significantly reduced. We speculated that this impairment could be due to a compromised crosstalk between Wnt receptors and iGluRs.
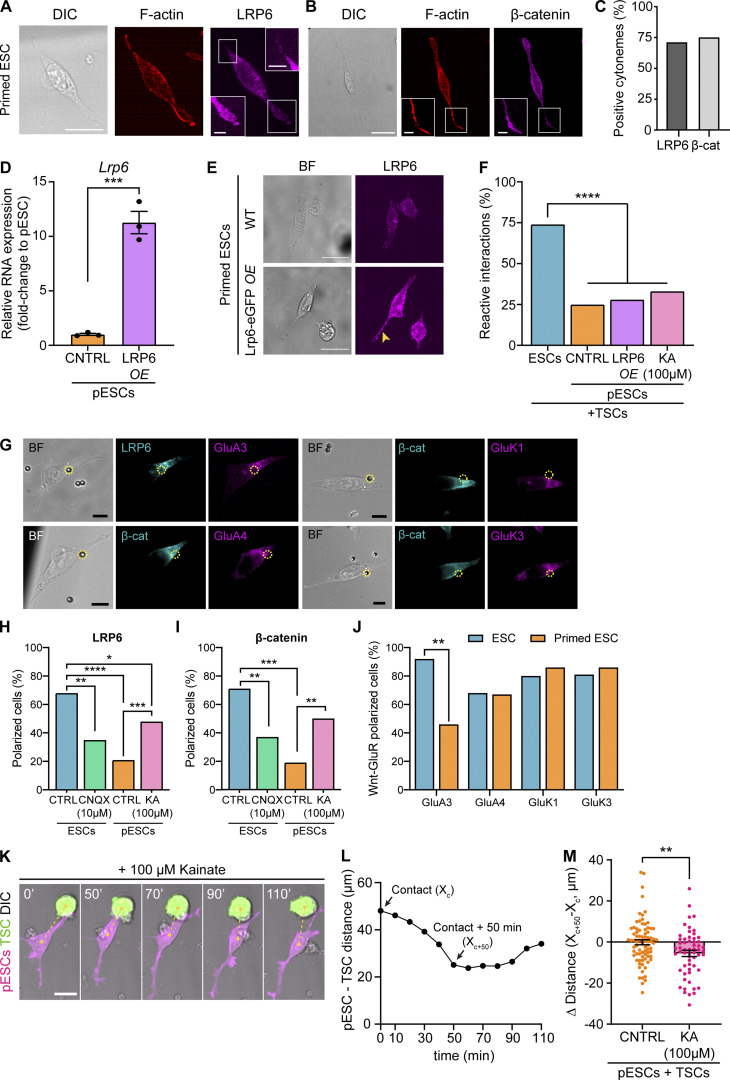
LRP6 overexpression cannot recover pESC–TSC pairing
LRP6 is key for the function of ESC cytonemes, and knockout of LRP5/6 reduces the generation of localized Ca2+ transients and impairs cytoneme-mediated ESC–TSC pairing (Junyent et al., 2020). Thus, we asked whether differences in LRP6 expression in pESCs may underlie the loss of Wnt-iGluR crosstalk in these cells. Transcriptionally, ESCs and pESCs express similar levels of Lrp5 and Lrp6 (Fig. S4 B). The prevalence of LRP6 and β-catenin–positive cytonemes is similar between ESCs (LRP6+, 58.9%; β-catenin+, all cytonemes; Junyent et al., 2020) and pESCs (∼71.5% LRP6+and β-catenin+ cytonemes; Fig. 6, A–C). Nevertheless, we tested whether LRP6 overexpression recovers pESC cytoneme activity. Transfection of pESCs with LRP6-eGFP followed by cell sorting (Fig. S5, A and B) led to increased levels of Lrp6 mRNA (Fig. 6 D) and LRP6 protein (Fig. 6 E), with LRP6 distributed in the cytonemes of sorted pESCs. However, this did not improve their cytoneme-mediated pairing with TSCs (Fig. 6 F, first three conditions; and Fig. S2, B–G), suggesting that reduced LRP6 expression does not drive the impaired Wnt-iGluR crosstalk in pESCs.
Figure 6.
iGluR activity, but not LRP6 overexpression, control cell polarization to Wnt3a beads, and pESC–TSC approximation. (A and B) Representative images of pESCs stained with antibodies against LRP6 (A) and β-catenin (B), and with phalloidin (F-actin). Inserts are magnification of boxes, contrast enhanced for clarity. Scale bars, 20 µm for larger images, 5 µm for inserts. (C) The percentage of cytonemes positive for LRP6 or β-catenin in pESCs. n = 62 cells. (D) Lrp6 RNA expression levels in control (CNTRL) pESCs or pESCs transiently overexpressing LRP6-eGFP (LRP6 OE), presented as fold-change to CNTRL pESCs. Bars are mean of n = 3 experiments. Error bars are SEM. Statistical significance calculated by unpaired two-sided t test: ***, P < 0.001. (E) Representative images of CNTRL or LRP6-eGFP overexpressing pESCs, stained with antibodies against LRP6. BF is brightfield. Yellow arrowhead indicates high levels of LRP6 in the cytoneme. Images are maximum-intensity projections presented at equal intensity range to allow comparison between panels. Scale bars, 20 µm. (F) The percentage of reactive interactions (defined in Fig. 3, A and B) between ESCs, CNTRL pESCs, pESCs overexpressing LRP6 (LRP6 OE), or pESCs treated with 100 µM kainate and TSCs. n ≥ 58 cells pooled from ≥3 independent experiments. Statistical significance calculated by multiple Fisher’s exact two-sided tests: ****, P < 0.0001. (G) Representative images of ESCs contacting a Wnt3a bead at the base of the cytonemes, stained with antibodies against LRP6 or β-catenin (cyan) and GluA3, GluA4, GluK1, and GluK3. Bead is black sphere in brightfield (BF) panel and is highlighted with a dashed yellow circle. Scale bars, 10 µm. (H and I) The percentage of control ESCs (CTRL, blue), 10 µM CNQX-treated ESCs (green), pESCs (CTRL, orange), or pESCs treated with 100 µM kainate (pink) with polarized presentation of LRP6 (H) or β-catenin (I) upon Wnt3a bead contact. n ≥ 41 cells. Further quantification is shown in Fig. S4, C–E. Statistical significance calculated by multiple Fisher’s exact two-sided tests: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (J) The percentage of ESCs (blue) or pESCs (orange) with polarized distribution of both Wnt/β-catenin pathway components and GluA3, GluA4, GluK1, or GluK3. n ≥ 26 cells. Statistical significance calculated by multiple Fisher’s exact two-sided tests: **, P < 0.01. (K and L) Representative frames of a time course live-cell imaging experiment showing a pESC (magenta) treated with 100 µM kainate contacting a TSC (green) through a cytoneme, approaching it, and then separating (K). Time is minutes, and yellow dashed line indicates distance between cells. Scare bar, 20 µm. Plot on L indicates pESC-TSC distance over time (cell in K only). Arrows point to distance at initial contact (Xc) and distance at 50 min after initial cytoneme-mediated contact (Xc+50). (M) The difference in distance between CNTRL pESCs (orange) or 100 µM kainate–treated pESCs (KA, pink) and TSCs at the initial cytoneme-mediated contact (Xc) or 50 min after contact (Xc+50, Δ Distance = Xc+50 – Xc). Bars are mean of n ≥ 58 cells pooled from ≥3 experiments. Error bars are SEM. Asterisks indicate statistical significance calculated by unpaired two-sided t test: **, P < 0.01.
Figure S5.
Gating strategy for FACS of ESCs or pESCs overexpressing tagged proteins. (A, C, and D) Schematic representation of the overexpression strategy. (B and E) Representative example of the FACS sorting of pESCs overexpressing LRP6-eGFP (B), ESCs overexpressing N-cadherin–eGFP, or pESCs overexpressing E-cadherin–mCherry (E). Gates used were SSC-A versus FSC-A for cells, SSC-A versus SSC-W for single cells, FSC-A, DAPI− for alive cells, followed by sorting by eGFP (FITC) or mCherry (PE-CF549). Control populations were used to set DAPI+/DAPI−, eGFP+/eGFP−, and mCherry+/mCherry− gates. eGFP+ cells were sorted for LRP6-eGFP (pESC) and N-cadherin–eGFP (ESC) conditions. mCherry+ cells were sorted for E-cadherin–mCherry condition. CMV, cytomegalovirus promoter; CNTRL, control; Ecad, E-cadherin; mCh, mCherry; Ncad, N-cadherin.
Up-regulated iGluR activity improves cell polarization to Wnt3a but cannot rescue pESC–TSC pairing
Contact between an ESC cytoneme and a localized Wnt source (Wnt bead or TSC) results in the polarization of LRP6 and β-catenin at the base of the cytoneme, toward the Wnt3a bead (Fig. 6, G–I; Junyent et al., 2020). Cell polarization (including Wnt pathway components) is an evolutionarily conserved feature in the response to localized Wnt signals (Garcin and Habib, 2017). Therefore, we measured cell polarization in pESCs. We observed that, while ∼68% of ESCs exhibited an accumulation of LRP6 and β-catenin near the area of Wnt bead contact, only ∼20% of pESCs were polarized (Fig. 6, H and I; and Fig. S4, C–E). We also expressed FZD1-GFP in ESCs and pESCs and performed live cell imaging. Of the cells contacting a Wnt3a bead, 67% of ESCs, but only 33% of pESCs, showed polarization of FZD1-GFP in the bead contact area (Fig. S4, F–H). Together, these results indicate that the ESC-to-pESC transition impairs the polarization of Wnt pathway components toward a localized Wnt signal.
Cell polarization and the generation of iGluR-mediated Ca2+ transients are impaired in pESC cytonemes. Therefore, we investigated the distribution of AMPA/kainate iGluRs in response to localized Wnt3a. We found that iGluR subunits GluA4, GluK1, and GluK3 appear to colocalize with polarized LRP6 and β-catenin at the Wnt bead contact area in both ESCs and pESCs. GluA3 colocalization was reduced in pESCs (Fig. 6, G and J).
We next tested whether regulation of iGluR activity modified the observed phenotype. First, we treated ESCs with CNQX, which reduces the generation of localized Ca2+ transients in response to Wnt-cytoneme contact (Junyent et al., 2020). Only ∼36% of CNQX-treated ESCs displayed polarized LRP6 and β-catenin at the area of Wnt3a bead contact (Fig. 6, H and I). Second, we stimulated iGluR activity in pESCs using kainate. 100 µM kainate increased the proportion of pESCs generating localized Ca2+ transients in the cytonemes in response to Wnt, from 40% in control cells to 63.2% in treated cells (n = 19; Fig. 5 C). The mean transient rate was increased from 0.12 transients/min in control pESCs to 0.22 transients/min in kainate-treated cells (Fig. 5 D). Moreover, kainate treatment improved pESC polarization, with ∼49% of kainate-treated pESCs with polarized LRP6 and β-catenin near the Wnt bead (Fig. 6, H and I).
Finally, we assessed pESC–TSC pairing upon treatment with 100 µM kainate. In these conditions, most cytoneme-mediated interactions between the pESC and TSC did not lead to stable cell pairing (only 33% RIs; Fig. 6 F; and Fig. S2, B–G). However, cell–cell contact through a cytoneme did lead to cell approximation, as the distance between kainate-treated pESCs and TSCs was reduced after initial contact, in contrast with untreated cells (Fig. 6, K–M; and Fig. S2 C).
Altogether, our results show that the ESC-to-pESC transition is associated with impaired Wnt-iGluR crosstalk at the cytonemes by down-regulation of Wnt-mediated iGluR activity. Up-regulation of iGluR activity with kainate partially restores pESC cytoneme function. However, stable pESC–TSC pairing, the process involving adhesion between the two cells, remained compromised.
E-cadherin overexpression improves pESCs–TSC pairing but does not result in increased ETS embryo structure formation
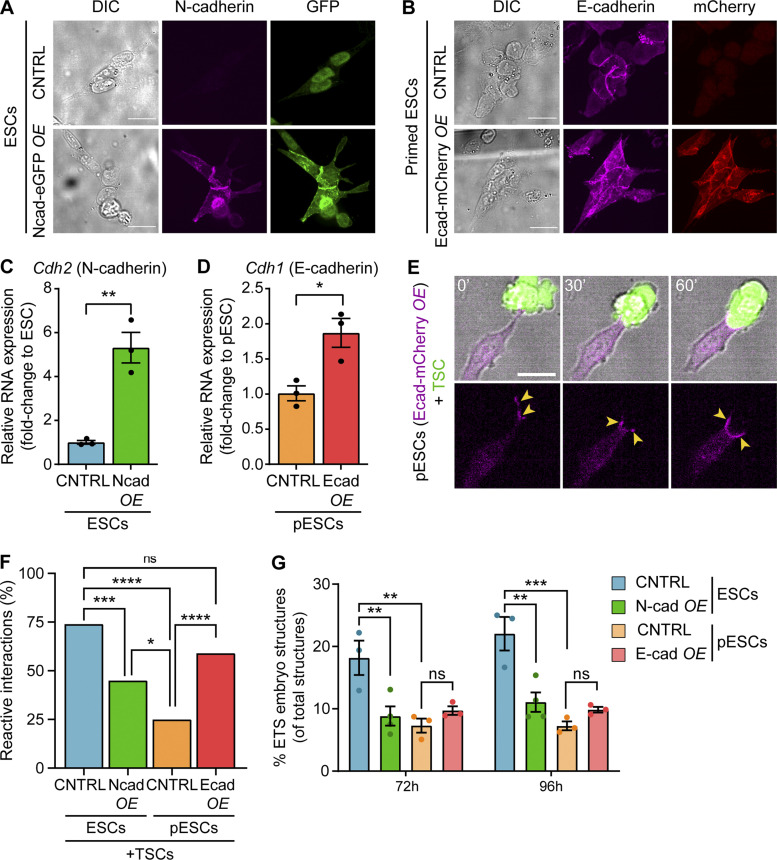
Selective cell adhesion is mediated by adherens junctions, involving cadherins (Takeichi, 2011). TSCs (Ishiuchi et al., 2019) and ESCs express high levels of E-cadherin (Takeichi et al., 1981), which are decreased in pESCs (Tesar et al., 2007; Brons et al., 2007). To investigate the requirements of cell adhesion in the cell pairing of ESCs/pESCs and TSCs, we modified the expression of cadherins. In E-cadherin–expressing cells, N-cadherin up-regulation leads to cell separation in many systems (Niessen et al., 2011; Thiery, 2002). Hence, we overexpressed N-cadherin in ESCs to disrupt their interaction with TSCs, and we overexpressed E-cadherin in pESCs to promote pESC–TSC pairing.
We transfected ESCs or pESCs with N-cadherin–eGFP or E-cadherin–mCherry, respectively, sorted them to pure populations, and validated overexpression at both protein and RNA levels (Fig. 7, A–D). Then, we analyzed the interaction of these cells with TSCs. N-cadherin overexpression in ESCs led to a reduction in ESC–TSC pairing following initial cytoneme contact (45% RIs versus 74% RIs in control ESCs; Fig. 7 F). Consequently, the percentage of ETS embryo structures formed by these cells was also reduced (Fig. 7 G; and Fig. S1, I and J). However, approximation in these cells still occurs, although to a lesser extent (Fig. S2 C). Meanwhile, TSC contact by an E-cadherin–overexpressing pESC triggered the clustering of E-cadherin complexes at the contact site (Fig. 7 E). As a result, a majority of cytoneme-mediated interactions in this condition resulted in pESC–TSC pairing (59% RIs versus 25% RIs in control pESCs; Fig. 7, E and F; and Fig. S2, B–G). However, transient E-cadherin overexpression in pESCs did not significantly improve ETS embryo structure formation (Fig. 7 G; and Fig. S1, I and J). These results show that although cadherins play a role in the pairing of ESCs/pESCs and TSCs, their transient actions alone are not sufficient to allow synthetic embryogenesis.
Figure 7.
E-cadherin overexpression in pESCs improves their pairing with TSCs but cannot sustain synthetic embryogenesis. (A) Representative images of control (CNTRL) or N-cadherin–eGFP—expressing pESCs stained with antibodies against N-cadherin (magenta) and GFP (green). For each staining, images are shown at equal intensity ranges to allow comparison between panels. Scale bars, 20 µm. (B) Representative images of CNTRL or E-cadherin–mCherry-expressing pESCs stained with antibodies against E-cadherin (magenta) and mCherry (red). For each staining, images are shown at equal intensity ranges to allow comparison between panels. Scale bars, 20 µm. (C and D) Cdh2 (N-cadherin) and Cdh1 (E-cadherin) RNA expression levels in CNTRL or overexpressing ESCs (C) or pESCs (D), presented as fold-change to the control population. Bars are mean of n = 3 experiments. Error bars are SEM. Statistical significance calculated by unpaired two-sided t test: *, P < 0.05; **, P < 0.01. (E) Representative frames of a time course live imaging showing a pESC overexpressing E-cadherin–mCherry (magenta) contacting and pairing with a TSC (green). Yellow arrowheads indicate high levels of E-cadherin at the cell–cell contact zone. Time in minutes. Scale bar, 20 µm. (F) The percentage of reactive interactions (defined in Fig. 3, A and B) between CNTRL ESCs (blue), ESCs overexpressing N-cadherin (Ncad OE, green), CNTRL pESCs (orange), or pESCs overexpressing E-cadherin (Ecad OE, red) and TSCs. n ≥ 63 cells pooled from ≥3 independent experiments. Statistical significance calculated by multiple Fisher’s exact two-sided tests: ns, nonsignificant, P > 0.05; *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (G) The percentage of ETS embryo structures over the number of total structures (i.e., the sum of all quantified structure types, according to Fig. S1 B) in CNTRL ESCs (blue), ESCs overexpressing N-cadherin (green), CNTRL pESCs (orange), and pESCs overexpressing E-cadherin (red). Bars are mean of n ≥ 3, ≥80 total structures/n. Error bars are SEM. Statistical significance calculated by two-way ANOVA with Tukey’s multiple comparisons test: ns, nonsignificant, P > 0.05; **, P < 0.01; ***, P < 0.001. A complete breakdown of the quantification can be found in Fig. S1, I and J.
Discussion
During embryogenesis and adult tissue homeostasis, transitions in cell fate coincide with changes in spatial organization (Shahbazi et al., 2017; Jones and Wagers, 2008). This coordination can be achieved through specific and adaptable mechanisms of cell-to-cell communication. In vivo tissues are complex, which can make studying such mechanisms and their dynamics throughout development challenging. On the other hand, reductionist systems offer advantageous platforms to scrutinize how cells communicate and organize at the single-cell level.
Here, we explored how differences in the pluripotency state between two stem cell populations affected the mechanisms they use to self-organize in vitro. We have previously described that ESCs use cytonemes to interact with TSCs and initiate synthetic embryogenesis (Junyent et al., 2020). Synthetic embryos represent powerful tools to understand the mechanisms of cell communication that lead to the self-organization of structures, in a technically auspicious system. In pluripotent stem cells, inhibition of autocrine Wnt signaling in naive ESCs leads to their progression to a more developmentally restricted early pESC population (ten Berge et al., 2011; Neagu et al., 2020). Recent reports indicate that early primed populations retain the capacity to form blastocyst chimaeras in a reduced manner (Kinoshita et al., 2020 Preprint). To interrogate how this transition affects self-organization, we cultured ESCs or pESCs with TSCs in conditions that allow the formation of ETS embryos (Harrison et al., 2018, 2017). The culture with pESCs formed significantly fewer synthetic embryos in comparison with the ESC-containing culture, suggesting that pESCs may be defective in the initiation step of embryo structure formation. Indeed, detailed examination revealed that the ESC-to-pESC transition significantly reduces the stable pairing with TSCs after initial contact through a cytoneme, a crucial first step for synthetic embryogenesis.
Developmental progression has been shown to impair the self-organization of stem cells. In many tissues (Sato et al., 2009; Rock et al., 2009; Zhang et al., 2017; Karthaus et al., 2014; Kale et al., 2000; Jamieson et al., 2017), multipotent tissue-specific stem cells can form organoids when cultured in vitro. However, when organoids are initiated from more developmentally restricted tissue progenitors, the efficiency of organoid formation is reduced. Using pluripotent stem cells, some studies have shown that forms of pESCs cannot generate blastoids when cultured together with TSCs (Rivron et al., 2018), or gastruloids when cultured alone in suspension (Cermola et al., 2019 Preprint). However, the mechanisms behind these intriguing results remained unstudied.
Wnt signaling regulates a wide range of cellular functions. Throughout development and adulthood, Wnt ligands function as patterning cues that control tissue formation and organization, in coordination with other developmental signals (Garcin and Habib, 2017). All mammalian Wnts undergo post-translational acylation, which makes them hydrophobic (Willert et al., 2003; Boutros and Niehrs, 2016; Langton et al., 2016). Consequently, patterning can be achieved through the localized production of Wnt from specialized cells in the stem cell niche (Farin et al., 2016; Clevers et al., 2014; Mills et al., 2017; Alexandre et al., 2014). Long-range diffusion of Wnts has also been observed in various developmental systems (Pani and Goldstein, 2018; Neumann and Cohen, 1997; Tian et al., 2019; Mulligan et al., 2012; Zecca et al., 1996; Ching et al., 2008). We have previously demonstrated that ESC–TSC pairing is dependent on locally produced Wnt by the TSCs, and that inhibition of Wnt exchange impairs this process (Junyent et al., 2020). Now we show that, while pESCs activate the Wnt/β-catenin pathway when presented with diffusible solubilized Wnt ligands, pESC cytonemes fail to form stable and selective interactions with sources of locally presented Wnts, impacting their ability to form synthetic embryos. Previously, we also explored how the difference in Wnt ligand presentation can affect cellular response. Soluble Wnt3a induces self-renewal of ESCs and human skeletal stem cells through symmetric divisions, but local presentation of Wnts to one side of the cell promotes asymmetric cell division in ESCs and a 3D cascade of osteogenic differentiation in human skeletal stem cells (Habib et al., 2013; Lowndes et al., 2016, 2017; Okuchi et al., 2021). Here we further emphasize that two cell populations with similar ability to activate the Wnt/β-catenin reporter can react differently to localized Wnts. This difference coincides with a developmental stage transition and impacts the capacity of the cells to form tissue structures.
At the mechanistic level, the composition and functionality of the cytonemes change with the ESC-to-pESC transition. In contrast with ESCs, pESCs require both F-actin and tubulin polymerization to form cytonemes, suggesting structural and dynamic differences. Although both ESCs and pESCs express functional iGluRs, developmental-stage transition to pESCs uncouples the crosstalk between Wnt and the iGluRs exhibited by ESCs. This is driven by the loss of iGluR activity, as LRP6 levels (the main Wnt coreceptor involved in the Wnt-iGluR crosstalk) are similar between ESCs and pESCs, and LRP6 overexpression does not affect pESC–TSC interaction. Meanwhile up-regulation of iGluR activity partially rescues pESC cytoneme functionality.
Upon Wnt source contact, pESCs or CNQX-treated ESCs show reduced polarization of Wnt/β-catenin machinery and lower prevalence and rate of Ca2+ transients on their cytonemes. In contrast, polarization and Ca2+ transient generation are improved in pESCs treated with kainate. We have previously shown that Wnt-iGluR crosstalk is important for ESC–TSC pairing, and alteration of these processes in pESCs might contribute to the differences observed. Indeed, iGluR activation in pESCs led to cell approximation after initial cytoneme-mediated interaction with TSCs. Interestingly, glutamatergic signaling has evolutionarily conserved roles in chemotaxis and spatial cell–cell communication (Ortiz-Ramírez et al., 2017).
Stable cell pairing was not achieved solely with iGluRs activation. Transient overexpression of the cell adhesion molecule E-cadherin on pESCs improved their stable pairing with TSCs, and overexpression of N-cadherin interfered with ESC–TSC interaction. This suggests that pluripotency state transition from ESCs to pESCs alters the mechanisms of cytoneme-mediated communication in a multifactorial manner. Loss of Wnt-mediated iGluR activity in the cytonemes of pESCs disrupts the Wnt-iGluR crosstalk driving cytoneme-mediated self-organization in ESCs. While cell adhesion mediates cell–cell pairing, it is not sufficient to allow synthetic embryogenesis alone. Notably, E-cadherin overexpression in pESCs incompletely restores the capacity of these cells to form blastocyst chimaeras (Ohtsuka et al., 2012). It will be important to investigate how cell adhesion and iGluR-mediated cell polarization are functionally connected to localized Wnt presentation in different contexts.
In summary, we use a reductionist approach that allows studying the dynamics of cell–cell communication in stem cells. Our data illustrate how changes in developmental potential impact the mechanisms that stem cells use to self-organize, leading to tissue formation. By comparing the response of ESCs and pESCs to TSCs, as well as to soluble or immobilized Wnt ligands, we have gained unique insights into the modes of ligand recognition at different developmental stages. The mechanisms identified in this study may also prove relevant to Wnt-responsive and iGluR-expressing adult stem cells and their progeny.
Materials and methods
Cell culture and differentiation of pESCs
W4 (129S6/SvEvTac) mouse ESCs were maintained in ESC basal media containing Advanced DMEM/F-12 (cat. no. 12634028; Life Technologies), 10% ESC-qualified FBS (eFBS; cat. no. ES-009-B; Millipore), 1% penicillin-streptomycin (P-S; cat. no. P4333; Sigma-Aldrich), 2 mM Glutamax (cat. no. 35050061; Life Technologies), 50 µM β-mercaptoethanol (2-ME, cat. no. 21985–023; Gibco) and 1,000 U/ml recombinant leukemia inhibitory factor (cat. no. 130–095-775; Miltenyi Biotec), supplemented with 100 ng/ml soluble Wnt3a (purified in-house). Media were changed daily, and cells were grown at low density (∼103 cells/cm2) until formation of mid-sized (100–200 µm) colonies before passaging (every 3–4 d). To passage ESCs, colonies were washed with PBS, trypsinized, neutralized, and centrifuged at 1.2 × 103 g for 4 min. Pelleted cells were resuspended in ESC basal media and counted to obtain 7,000 cells/well and transferred to a clean tissue culture–treated six-well plate. Cells were grown at 37°C, 5% CO2. ESC lines with knock-in NANOG-Venus (Habib et al., 2013) or stably expressing Ftractin-mRuby3 (Addgene plasmid #85146), GCaMP6s (Addgene plasmid #40753; Junyent et al., 2020), and 7xTCF-eGFP//SV40-mCherry (Fuerer and Nusse, 2010) were used in some experiments.
To induce ESC-to-pESC transition, ESCs were passaged as described above, but pelleted cells were resuspended in ESC basal media (without Wnt3a) supplemented with 2 µM inhibitor of Wnt production-2 (IWP2; cat. no. 72122; StemCell Technologies). Cells were cultured for 3 d in ESC basal media plus 2 µM IWP2, changing media daily, before using them for experiments.
TSCs expressing GFP (derived by the Rossant Laboratory [Hospital for Sick Children and the Department of Molecular Genetics, University of Toronto, Toronto, Canada]; Tanaka et al., 1998) were cultured on a layer of mitotically inactivated mouse embryonic fibroblasts (MEFs; Tanaka, 2006; Junyent et al., 2020). Briefly, irradiated MEFs (cat. no. PSC001; R&D Systems) were thawed in a six-well plate at 3 × 105 cells/well in MEF media containing DMEM (Life Technologies), 10% FBS, 100 µM 2-ME, 2 mM Glutamax, and 1% P-S. MEFs were cultured for at least 24 h before thawing TSCs. TSCs were cultured on MEFs in TSC media containing RPMI 1640 (cat. no. 11875093; Life Technologies), 20% eFBS, 100 µM 2-ME, 2 mM Glutamax, 1% P-S, 2 mM sodium pyruvate (Life Technologies), 25 ng/ml FGF4 (cat. no. 5846-F4; R&D Technologies), and 1 µg/ml heparin (cat. no. H3393; Sigma-Aldrich). Media were changed daily, and colonies were split as required. 24 h before the experiment, TSCs were weaned from MEFs; cells were trypsinized (0.05% trypsin-EDTA), centrifuged (4 min, 1,000 g), and resuspended in TSC media. To remove MEFs, cells were twice transferred to a clean tissue culture–treated plate, and MEFs were allowed to attach for 15 min at 37°C, 5% CO2. TSCs in the supernatant were then transferred to a clean culture plate and incubated for 24 h in TSC-conditioned media (70% TSC-MEF conditioned media for 3 d plus 30% fresh TSC media, with a final concentration of 25 ng/ml FGF4 and 1 µg/ml heparin).
In some experiments, 2 µM IWP2, 10 µM CNQX (cat. no. C127; Sigma-Aldrich), or 100 µM kainate (cat. no. 15467999; Thermo Fischer Scientific) were added to the media (indicated in the text).
All cell lines were maintained at 37°C, 5% CO2, and were routinely tested for mycoplasma infection.
ETS embryo structure formation
ETS embryo structures were generated following published protocols (Harrison et al., 2018). Briefly, ESCs, pESCs, and TSCs cultured as described above were dissociated to single cells (ESCs and pESCs) or small cell clusters (2–4 cells, TSCs). After three washes with PBS, 4,000 cells or clusters of ESC–TSC or pESC–TSC were seeded together on a Matrigel (cat. no. 354330; Corning)-coated IBIDI μ-well glass slide (cat. no. 80827; IBIDI). After 10 min incubation at 37°C, 5% CO2, to allow cell attachment, wells were filled with ETS culture media composed of 40% RPMI 1640, 25% Advanced DMEM/F-12, 25% Neurobasal A (cat. no. 10888022; Gibco), 10% eFBS, 2 mM Glutamax, 0.1 mM 2-ME, 0.5 mM sodium pyruvate, 0.25× N2 supplement (cat. no. 17502048; Life Technologies), 0.5× B27 supplement (cat. no. A3582801; Life Technologies), 12.5 ng/ml FGF4, and 500 ng/ml heparin, plus 10% Matrigel. ETS embryo structures were cultured for 4 d at 37°C, 5% CO2, changing the media daily. For analysis of structure formation, wells were imaged daily for Brightfield and GFP channels on a Zeiss inverted Axio Imager (equipped with a CoolSNAP HQ2 camera) using a Plan-Neofluar 10×/0.3 dry objective at 37°C, 5% CO2, using the Zen software (Blue edition; Zeiss). An average of seven representative positions were chosen per condition, replicate, and day. Images were analyzed in Fiji (ImageJ) by counting the number of ETS embryo structures, unorganized ESC–TSC structures, and ESC-only or TSC-only structures in each n. Results are reported as percentage of total structures, which is the sum of all the quantified structures in each n. Details and a breakdown of the quantification can be found in Fig. S1.
Preparation of Wnt3a microbeads
Recombinant Wnt3a proteins were produced in Drosophila S2 cells grown in suspension in Schneider’s Drosophila Medium (cat. no. 21720024; Life Technologies). Conditioned media were collected, filtered, and passed through a Blue Sepharose Column at constant flow rate to recover the majority of the Wnt3a proteins. Following loading of the conditioned media, the column was washed with binding buffer (1% [wt/vol] CHAPS, 150 mM KCl, and 20 mM Tris-HCl, pH 7.5, sterile-filtered). Protein was eluted with elution buffer (1% [wt/vol] CHAPS, 1.5 M KCl, and 20 mM Tris-HCl, pH 7.5, sterile-filtered) and collected as fractions, which were all tested for the presence of Wnt proteins via Western blotting. Wnt3a activity was tested via LS/L assay: L cells stably transfected with the SuperTOPFlash Wnt/β-catenin pathway reporter (Fuerer and Nusse, 2010; LS/L cells) cultured in DMEM containing 10% FBS and 1% P-S were exposed to soluble Wnt3a proteins or control conditions for >14 h before cell lysates were collected. Wnt-induced luciferase activity was determined via the Dual-Light System (cat. no. T2176; Applied Biosystems), and Luciferase readings were taken on a Glomax-Multi detection system (Promega). Alternatively, recombinant Wnt3a proteins were purchased (cat. no. 1324-WN; R&D Systems).
Wnt3a proteins were immobilized to 2.8 µm carboxylic acid–coated Dynabeads (cat. no. 14305D; Thermo Fisher Scientific) following published protocols (Junyent et al., 2020; Habib et al., 2013; Lowndes et al., 2017). Briefly, the carboxylic acid groups on the Dynabeads were activated by 30-min incubation with N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (cat. no. E7750-1G; Sigma-Aldrich) and N-hydroxysuccinimide (cat.no. 56480-25G; Sigma-Aldrich), 50 mg/ml each, dissolved in 25 mM cold 2-(N-morpholino)-ethanesulfonic acid buffer (MES; cat. no. M3671-50G; Sigma-Aldrich), pH 5, with constant rotation. Following activation, beads were retained by using a magnet and washed three times with 25 mM MES buffer, pH 5. Soluble Wnt3a protein (500 ng) was diluted 1:5 in MES buffer, pH 5, and incubated with the beads for 1 h with constant agitation, at room temperature. Beads were washed again three times with PBS, pH 7.4, before storage in media containing 10% FBS at 4°C. Inactivation of Wnt3a beads was achieved through incubation with 20 mM DTT (cat. no. P2325; Life Technologies) for 30 min at 37°C. Following incubation with DTT, beads were washed three times in PBS before storage in media containing 10% FBS at 4°C (up to 10 d). Bead activity was validated by LS/L assay (described above).
Cell transfection
For transient transfection, 8 × 104 ESCs or pESCs were seeded into one well of a 12-well plate and incubated overnight at 37°C, 5% CO2. 2 µg DNA was transfected using JetPEI (cat. no. 101–10; Polyplus-transfection). Cells were incubated overnight, and construct expression was verified prior to use in the experiments.
Plasmids used in this manuscript include Frizzled-1-GFP-CS2P+ (#16817; Addgene), pEGFP-N3-LRP6 (Habib et al., 2013), N-cadherin–eGFP (#18870; Addgene), and E-cadherin–mCherry (#71366; Addgene).
FACS
FACS was employed to detect Wnt3a response of ESCs or pESCs stably transduced with 7xTCF-eGFP//SV40-mCherry (Junyent et al., 2020). Transduced cells were sorted to gain a pure mCherry+ population. To do so, cells were trypsinized and centrifuged as described above. Pelleted cells were resuspended in FACS buffer (3% FBS in PBS, with or without 0.1 µg/ml DAPI), passed through a 35-µm nylon cell strainer, and stored on ice until analysis. Cell sorting was performed using a FACSAria system (BD Biosciences). The gating strategy included gating for side scatter (SSC)–forward scatter (FSC), SSC-area (A)–SSC-width (W), FSC-A–DAPI (alive cells), and SSC-Phycoerythrin (PE)–Texas Red (mCherry+ cells), with the necessary controls. Sorted cells were collected in ESC basal media and transferred to a tissue culture plate for expansion. To assess Wnt responsiveness, cells were cultured in standard conditions and stimulated with Wnt3a or control media (ESC basal media) for 24 h. Then cells were prepared as described above and analyzed using a FACSFortessa system (BD Biosciences). The same gating strategy was used, followed by an SSC-FITC (GFP+ cells) gate. FACS was also used to analyze Nanog-Venus expression levels in ESCs or pESCs, or to sort LRP6-eGFP+, N-cadherin–eGFP+, or E-cadherin–mCherry+ cells. Cell sorting was performed as described previously, and cells were used immediately for experiments. The gating strategy for these experiments is described in Fig. S5. For all experiments, analysis was performed using FlowJo software (FlowJo).
Live-cell imaging
To measure ESC and pESC interaction with TSCs, 1,500 ESCs or pESCs were co-cultured with 1,500 TSCs in a well of a tissue culture–treated, imaging-grade 96-well plate, in ESC basal media (no Wnt3a). In some experiments, ESCs or pESCs were pretransfected with pEGFP-N3-LRP6, E-cadherin–mCherry, or N-cadherin–eGFP plasmids and sorted before the experiment (described above). For ESC and pESC interaction with Wnt3a beads, 3,000 ESCs or pESCs plus 0.3 µg Wnt3a or control beads were seeded per well in ESC basal media. Plates were incubated for 1 h at 37°C, 5% CO2, to allow cell attachment. Cells were transferred to a Nikon Eclipse Ti Inverted Spinning-Disk confocal (equipped with a Yokogawa CSU-1 disk head and an Andor Neo sCMOS camera) with an incubation system at 37°C, 5% CO2. 15–25 positions were selected, laser intensity was adjusted as required, and three z-positions were defined. Cells were imaged using a Plan Apo VC 20×/0.75 dry objective for differential interference contrast (DIC) and the corresponding fluorescent channel (GFP and/or RFP) every 10 min for 12 h, using the NIS Elements software (Nikon). Analysis was performed using Fiji (ImageJ) to analyze the number of reactive and nonreactive interactions between ESCs or pESCs and TSCs or beads (as described in Fig. 3). The distance measurement tool in Fiji was used to measure the distances between cells or between cells and beads (as described in Figs. S2 and S3).
To measure FZD1-GFP polarization, 3,000 ESCs or pESCs transfected with FZD1-GFP plus 0.3 µg Wnt3a beads were seeded per well in ESC basal media. Imaging was performed as described above. Analysis was performed using the “Plot profile” tool in Fiji.
To analyze the percentage and characteristics of the cytonemes in ESCs or pESCs, 3,000 cells were seeded per well in ESC basal media supplemented with increasing concentrations of colcemid (demecolcine; cat. no. D7385; Sigma-Aldrich) or cytochalasin D (cat. no. PHZ1063; Thermo Fisher Scientific) and H2O or DMSO as controls, respectively. Cells were transferred to a Zeiss inverted Axio Imager (equipped with a CoolSNAP HQ2 camera) at 37°C, 5% CO2. Cells were imaged every 15 min for 4 h using a Plan-Neofluar 20×/0.8 dry objective, with the Zen software (Blue edition; Zeiss). After imaging, cells were fixed and stained (see below) and the percentage of cells with cytonemes as well as the dynamics of the cytonemes were measured using Fiji.
Immunofluorescence
For immunofluorescence analysis of single cells, ESCs or pESCs were seeded at 3,000 cells/well with or without 0.3 µg Wnt3a beads and incubated at 37°C, 5% CO2. When required, 10 µM CNQX or 100 µM kainate was added to the media. Cells were fixed with 2% paraformaldehyde in PBS plus 0.05% Triton X-100 for 8 min at RT. A blocking-permeabilization step was performed by incubation with 0.3% BSA and 0.05% Triton X-100 in PBS for 1 h at RT. Primary antibody solution was prepared in blocking solution, added to the cells, and incubated overnight at 4°C. After incubation, cells were washed four times in PBS plus 0.1% Tween 20 and incubated in Alexa Fluor (AF) 488–, AF555-, or AF647-tagged secondary antibodies diluted in blocking buffer for 1 h at RT. Sometimes, AF488-phalloidin (cat.no. A12379; Thermo Fisher Scientific) was added to the secondary antibody incubation. Cells were washed four times in PBS plus 0.1% Tween 20 plus DAPI and mounted with ProLong Gold Antifade Mountant (cat. no. P36935; Thermo Fisher Scientific) or kept in PBS for imaging. Cells were imaged in a Nikon Eclipse Ti Inverted Spinning-Disk confocal (equipped with a Yokogawa CSU-1 disk head and an Andor Neo sCMOS camera) using a Plan Apo VC 20×/0.75 dry, a Plan Apo lambda 40×/0.95 dry, or a Plan Apo lambda 100×/1.45 oil immersion objective, with type F oil, at 37°C, with the NIS Elements software.
LRP6, β-catenin, and iGluR polarization was measured using the “Plot profile” tool in Fiji. A 10 pixel-wide, 20-µm-long line was drawn from the position of the bead to the center of the cell (exemplified in Fig. S4). Intensity on that line was measured, as well as the background intensity next to the cell. Fluorescence intensity was processed by subtracting the background and normalizing to the maximum intensity value of the profile and was presented as a normalized intensity profile (ranging from 1 = maximum intensity to 0 = background intensity).
For analysis of ESC and pESC colonies, cells were grown in optical-grade, tissue culture–treated slides for 3 d prior to fixation and staining as described above. Stained colonies were imaged using a Leica SP8 confocal and a HC Plan Apo 20×/0.75 dry objective (using the LAS-X software). For analysis of ETS embryo structures, cells cultured for 4 d (96 h) were fixed and stained as described above. Correct ETS structures (as described in Fig. 2 and Fig. S1) were imaged using a Leica SP8 confocal and a HC Plan Apo 20×/0.75 dry objective (using the LAS-X software). Structure exterior size, as well as ESC/pESC compartment and TSC compartment structure and cavity size, were measured in Fiji. OCT3/4 and EOMES intensity was measured in the ESC/pESC and TSC compartments, respectively.
To verify protein overexpression, sorted cells expressing E-cadherin–mCherry, N-cadherin–eGFP, or LRP6-eGFP, or control ESCs or pESCs, were seeded in a tissue culture–treated, optical-grade, 96-well plate and incubated at 37°C, 5% CO2 overnight. Cells were fixed and stained as described above, and images were acquired in a Nikon Eclipse Ti Inverted Spinning-Disk confocal (equipped with a Yokogawa CSU-1 disk head and an Andor Neo sCMOS camera), using a Plan Apo VC 20×/0.75 dry objective at 37°C, with the NIS Elements software, and keeping the same laser intensities and exposure times for controls and experiments. Images were analyzed in Fiji and are presented at the same intensity range to allow comparison between panels.
Antibodies
The antibodies used were anti-α-tubulin (YL1/2; rat; ab6160; Abcam), anti–β-catenin (mouse; #610154; BD Transduction), anti-LRP6 (EPR2423(2); rabbit; ab134146; Abcam), anti-NANOG (rabbit; RCAB002P-F; Reprocell), anti-OCT3/4 (mouse; #611202; BD Transduction), anti-EOMES (rabbit; ab183991; Abcam), anti-GriA3 (mouse; MAB5416; Sigma-Aldrich), anti-GriA4 (rabbit; AB1508; Sigma-Aldrich), anti-GriK1 (rabbit; AGC-008; Alomone Laboratories), anti-GriK3 (rabbit; AGC-040; Alomone Laboratories), anti–N-cadherin (mouse; #33-3900; Thermo Fisher Scientific), anti–E-cadherin (DECMA-1; rat; ab11512; Abcam), anti-GFP (chicken; GFP-1020; Aves), anti-mCherry (goat; #200-101-379; Rockland), and AF488, AF555, or AF647-conjugated secondary antibodies (Thermo Fisher Scientific).
Imaging of whole-cell and cytoneme-localized Ca2+ transients
For cytoneme-localized Ca2+ analysis, ESCs or pESCs stably carrying pGP-cytomegalovirus (CMV)-GCaMP6s were seeded at a density of 4,500 cells/well in imaging plates (ibiTreat μ-Slide 8 well; cat. no. 80826; IBIDI) and incubated for ≥6 h in ESC basal media, at 37°C, 5% CO2. In some experiments, 100 µM kainate was added to the media. Cells were transferred to a Nikon Eclipse Ti Inverted Spinning-Disk confocal (equipped with a Yokogawa CSU-1 disk head and an Andor Neo sCMOS camera) with an incubation system at 37°C, 5% CO2, and beads were added in situ at a concentration of 0.6 µg beads/well. Cells were further incubated to allow the beads to precipitate for 15 min. Cells near beads were chosen. Images of GCaMP6s (GFP) and differential interference contrast (DIC) on the larger cytonemes were acquired every 6 s for ∼20 min, using a Plan Apo VC 20×/0.75 dry objective and the NIS Elements software (Nikon). The generation of localized calcium transients near the bead at the cytonemes was analyzed. Acquired time course images were analyzed using Fiji, and the GCaMP6s (GFP) signal was normalized to background.
For whole-cell Ca2+ analysis, GCaMP6s-expressing cells were seeded at 25,000 cells/well in imaging plates and incubated overnight at 37°C, 5% CO2. 10 min before the experiment, cell culture media were changed to bath solution for imaging (140 mM NaCl, 5 mM KCl, 2 mM MgCl2, 5 mM glucose, 10 mM Hepes, and 4 mM CaCl2, pH 7.3). Cells were transferred to a Nikon Eclipse Ti Inverted Spinning-Disk confocal (equipped with a Yokogawa CSU-1 disk head and an Andor Neo sCMOS camera) with an incubation system at 37°C, 5% CO2. To record changes in Ca2+ in response to kainate, a representative position was chosen, and GFP fluorescence images were taken every 2.5 s for 5 min, using a Plan Apo VC 20×/0.75 dry objective and the NIS Elements software (Nikon). Approximately 1 min after imaging start, a final concentration of 100 µM kainate (cat. no. 15467999; Thermo Fisher Scientific) or control was added as a single drop to the well. In some conditions, cells were pretreated with 10 µM CNQX (cat. no. C127; Sigma-Aldrich) for 10 min before imaging. Acquired time course images were analyzed using Fiji, and the GCaMP6s (GFP) signal was normalized to background and to preaddition basal intensity level (ΔF/F0).
RNA extraction and RT quantitative PCR (qPCR) analysis
For transcription analysis of ESCs or pESCs, cells were grown as described, and RNA was extracted using the RNeasy mini kit (cat. no. 74106; Qiagen). mRNA was retrotranscribed to cDNA using the QuantiTect Reverse Transcription Kit (cat. no. 205311; Qiagen). qPCR was performed using SYBR Green PCR Master Mix (cat. no. 4344463; Thermo Fisher Scientific) and primers against iGluR subunits (Table 1). Alternatively, qPCR was performed using TaqMan Fast Advanced Master Mix (cat.no. 444496; Thermo Fisher Scientific) and TaqMan probes against Otx2 (Mm00446859_m1; Thermo Fisher Scientific), Fgf5 (Mm03053745_s1; Thermo Fisher Scientific), Lrp6 (Mm00999795_m1; Thermo Fisher Scientific), Lrp5 (Mm01227476_m1; Thermo Fisher Scientific); and GAPDH (Mm99999915_g1; Thermo Fisher Scientific). To validate overexpression of LRP6, E-cadherin, or N-cadherin, sorted cells were lysed, and RNA was extracted and retrotranscribed as described before. RNA levels of the transgenes were assessed by TaqMan-based qPCR (Lrp6) or SYBR Green–based qPCR (Cdh1 and Cdh2), using the primers reported in Table 1. For all experiments, cycle threshold (Ct) values of targeted genes were normalized to housekeeping gene levels (DCt) and plotted as 2-DCt or as fold-change to control conditions (2-DDCt).
Table 1. Primers used for RT-qPCR.
| Gene | Forward primer sequence (5′ to 3′) | Reverse primer sequence (5′ to 3′) |
|---|---|---|
| GriA1 | GACAACTCAAGCGTCCAGAA | CGTCGCTGACAATCTCAAGT |
| GriA2 | GACCAGAACGGAAAACGAAT | TTCAAGCCCAGATGTGTCAT |
| GriA3 | CCTCCTGATCCTCCCAATG | CGCTCTCTATGGGGGACACC |
| GriA4 | AGAAGGACCCAGTGACCAAC | ATGCAGCCAGATTAGCAGTG |
| GriK1 | GCCCCTCTCACCATCACGTAT | TGGTCGATAGAGCCTTGGGCA |
| GriK2 | TTCCTGAATCCTCTCTCCCT | CACCAAATGCCTCCCACTATC |
| GriK3 | GGGTGTCAGCTGTGTCCTCT | GACAGAGCTTTGGGCATCAGT |
| GriK4 | CAAAGGCCTGGGAATGGAGAATA | CCGCCGCCTGGGATGGATA |
| GriK5 | CGACACCAAGGGCTACGGCAT | CCGCCACGAAGACAGCAATGA |
| β-Actin | CGTTGACATCCGTAAAGACCT | CAAAGCCATGCCAATGTTGTCTCT |
| Cdh1 | GGTTTTCTACAGCATCACCG | GCTTCCCCATTTGATGACAC |
| Cdh2 | ATCAACCCCATCTCAGGACA | CCATTCAGGGCATTTGGATC |
Statistical analysis
Data representation and statistical analysis were performed using Prism (GraphPad), as described in the figure legends. The statistical tests used were as follows: unpaired two-sided t test for Fig. 3, E and F; Fig. 4 A; Fig. 6, D and M; Fig. 7, C and D; and Fig. S1, C–G; one-way ANOVA with Tukey’s multiple comparison test for Fig. 1, C and F; Fig. 5 D; Fig. S2, C–H; and Fig. S3, C–F; one-way ANOVA with Šídák’s multiple comparison test for Fig. 5 A; two-way ANOVA with Tukey’s multiple comparison test for Fig. 4, C–F; and Fig. 7 G; two-way ANOVA with Šídák’s multiple comparison test for Fig. 1 D; Fig. 2, B and C; and Fig. S4 B; and one or multiple Fisher’s exact two-sided tests for Fig. 3, B and D; Fig. 5 C; Fig. 6, F and H–J; Fig. 7 F; Fig. S1 H; and Fig. S4 H. For all parametric tests, data distribution was assumed to be normal, but this was not formally tested. For all figures, symbols indicate statistical significance, as follows: #, P ∼ 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. We set the threshold for significance as P < 0.05, unless specified otherwise.
Online supplemental material
Fig. S1 shows immunofluorescence of NANOG and β-catenin in pESCs and quantification of ETS structures. Fig. S2 shows quantification of ESC or pESC interaction with TSCs. Fig. S3 shows quantification of ESC or pESC interaction with Wnt3a or control beads, and representative images of cytochalasin D or colcemid treatment. Fig. S4 shows that pESCs present functional iGluR receptors and similar Lrp5/6 levels to ESCs but exhibit impaired polarization of Wnt pathway components upon contact with a Wnt source. pESC polarization is recovered by kainate addition. Fig. S5 shows the gating strategy for FACS of ESCs or pESCs overexpressing tagged proteins.
Acknowledgments
We thank the Nikon Imaging Centre at King’s College London for help with light microscopy.
This work was supported by a Sir Henry Dale Fellowship (102513/Z/13/Z) to S.J. Habib.
The authors declare no competing financial interests.
Author contributions: S. Junyent planned and performed experiments, analyzed data, prepared figures, and wrote the manuscript; J. Reeves performed experiments and analyzed data; E. Gentleman provided critical review of the experiments and manuscript; and S.J. Habib conceived the idea, supervised the project, planned experiments, interpreted data, and wrote the manuscript.
References
- Alexandre, C., Baena-Lopez A., and Vincent J.-P.. 2014. Patterning and growth control by membrane-tethered Wingless. Nature. 505:180–185. 10.1038/nature12879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin, I., Dewi D.L., Hundshammer J., Erdmann G., Kerr G., and Boutros M.. 2017. Autocrine Wnt regulates the survival and genomic stability of embryonic stem cells. Sci. Signal. 10:eaah6829. 10.1126/scisignal.aah6829 [DOI] [PubMed] [Google Scholar]
- Bilic, J., Huang Y.-L., Davidson G., Zimmermann T., Cruciat C.-M., Bienz M., and Niehrs C.. 2007. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 316:1619–1622. 10.1126/science.1137065 [DOI] [PubMed] [Google Scholar]
- Boutros, M., and Niehrs C.. 2016. Sticking Around: Short-Range Activity of Wnt Ligands. Dev. Cell. 36:485–486. 10.1016/j.devcel.2016.02.018 [DOI] [PubMed] [Google Scholar]
- Bradley, A., Evans M., Kaufman M.H., and Robertson E.. 1984. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 309:255–256. 10.1038/309255a0 [DOI] [PubMed] [Google Scholar]
- Brons, I.G.M., Smithers L.E., Trotter M.W.B., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A., and Vallier L.. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 448:191–195. 10.1038/nature05950 [DOI] [PubMed] [Google Scholar]
- Cermola, F., D’Aniello C., Tatè R., De Cesare D., Martinez-Arias A., Minchiotti G., and Patriarca E.J.. 2019. Gastruloid development competence discriminates different states of pluripotency between naïve and primed. bioRxiv. 664920. (Preprint posted June 8, 2019) 10.1101/664920 [DOI] [PMC free article] [PubMed]
- Chen, B., Dodge M.E., Tang W., Lu J., Ma Z., Fan C.W., Wei S., Hao W., Kilgore J., Williams N.S., et al. 2009. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5:100–107. 10.1038/nchembio.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T.W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V., et al. 2013. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 499:295–300. 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching, W., Hang H.C., and Nusse R.. 2008. Lipid-independent secretion of a Drosophila Wnt protein. J. Biol. Chem. 283:17092–17098. 10.1074/jbc.M802059200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers, H., Loh K.M., and Nusse R.. 2014. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 346:1248012. 10.1126/science.1248012 [DOI] [PubMed] [Google Scholar]
- Farin, H.F., Jordens I., Mosa M.H., Basak O., Korving J., Tauriello D.V.F., de Punder K., Angers S., Peters P.J., Maurice M.M., and Clevers H.. 2016. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 530:340–343. 10.1038/nature16937 [DOI] [PubMed] [Google Scholar]
- Fuerer, C., and Nusse R.. 2010. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One. 5:e9370. 10.1371/journal.pone.0009370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin, C.L., and Habib S.J.. 2017. A Comparative Perspective on Wnt / β -Catenin Signalling in Cell Fate Determination. In Asymmetric Cell Division in Development, Differentiation and Cancer, Results and Problems in Cell Differentiation, vol. 61. Tassan J.-P., Kubiak J., editors. Springer International Publishing AG. 323–350. 10.1007/978-3-319-53150-2_15 [DOI] [PubMed] [Google Scholar]
- Gundry, R.L., Tchernyshyov I., Sheng S., Tarasova Y., Raginski K., Boheler K.R., and Van Eyk J.E.. 2010. Expanding the mouse embryonic stem cell proteome: combining three proteomic approaches. Proteomics. 10:2728–2732. 10.1002/pmic.201000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib, S.J., Chen B.-C., Tsai F.-C., Anastassiadis K., Meyer T., Betzig E., and Nusse R.. 2013. A localized Wnt signal orients asymmetric stem cell division in vitro. Science. 339:1445–1448. 10.1126/science.1231077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, S.E., Sozen B., Christodoulou N., Kyprianou C., and Zernicka-Goetz M.. 2017. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science. 356:eaal1810. 10.1126/science.aal1810 [DOI] [PubMed] [Google Scholar]
- Harrison, S.E., Sozen B., and Zernicka-Goetz M.. 2018. In vitro generation of mouse polarized embryo-like structures from embryonic and trophoblast stem cells. Nat. Protoc. 13:1586–1602. 10.1038/s41596-018-0005-x [DOI] [PubMed] [Google Scholar]
- Hayer, A., Shao L., Chung M., Joubert L.M., Yang H.W., Tsai F.C., Bisaria A., Betzig E., and Meyer T.. 2016. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat. Cell Biol. 18:1311–1323. 10.1038/ncb3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Osorno R., Tsakiridis A., and Wilson V.. 2012. In Vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep. 2:1571–1578. 10.1016/j.celrep.2012.10.022 [DOI] [PubMed] [Google Scholar]
- Ishiuchi, T., Ohishi H., Sato T., Kamimura S., Yorino M., Abe S., Suzuki A., Wakayama T., Suyama M., and Sasaki H.. 2019. Zfp281 Shapes the Transcriptome of Trophoblast Stem Cells and Is Essential for Placental Development. Cell Rep. 27:1742–1754.e6. 10.1016/j.celrep.2019.04.028 [DOI] [PubMed] [Google Scholar]
- Jamieson, P.R., Dekkers J.F., Rios A.C., Fu N.Y., Lindeman G.J., and Visvader J.E.. 2017. Derivation of a robust mouse mammary organoid system for studying tissue dynamics. Development. 144:1065–1071. 10.1242/dev.145045 [DOI] [PubMed] [Google Scholar]
- Jones, D.L., and Wagers A.J.. 2008. No place like home: anatomy and function of the stem cell niche. Nat. Rev. Mol. Cell Biol. 9:11–21. 10.1038/nrm2319 [DOI] [PubMed] [Google Scholar]
- Junyent, S., Garcin C.L., Szczerkowski J.L.A., Trieu T.-J., Reeves J., and Habib S.J.. 2020. Specialized cytonemes induce self-organization of stem cells. Proc. Natl. Acad. Sci. USA. 117:7236–7244. 10.1073/pnas.1920837117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, T., Wilder E., Klingensmith J., Zachary K., and Perrimon N.. 1996. The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes Dev. 10:3116–3128. 10.1101/gad.10.24.3116 [DOI] [PubMed] [Google Scholar]
- Kale, S., Biermann S., Edwards C., Tarnowski C., Morris M., and Long M.W.. 2000. Three-dimensional cellular development is essential for ex vivo formation of human bone. Nat. Biotechnol. 18:954–958. 10.1038/79439 [DOI] [PubMed] [Google Scholar]
- Karthaus, W.R., Iaquinta P.J., Drost J., Gracanin A., van Boxtel R., Wongvipat J., Dowling C.M., Gao D., Begthel H., Sachs N., et al. 2014. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 159:163–175. 10.1016/j.cell.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, M., Barber M., Mansfield W., Cui Y., Spindlow D., Stirparo G.G., Dietmann S., Nichols J., and Smith A.. 2020. Capture of mouse and human stem cells with features of formative pluripotency. bioRxiv. 2020.09.04.283218. (Preprint posted September 4, 2020) 10.1101/2020.09.04.283218 [DOI]
- Langton, P.F., Kakugawa S., and Vincent J.P.. 2016. Making, Exporting, and Modulating Wnts. Trends Cell Biol. 26:756–765. 10.1016/j.tcb.2016.05.011 [DOI] [PubMed] [Google Scholar]
- Lippert, A., Janeczek A.A., Fürstenberg A., Ponjavic A., Moerner W.E., Nusse R., Helms J.A., Evans N.D., and Lee S.F.. 2017. Single-Molecule Imaging of Wnt3A Protein Diffusion on Living Cell Membranes. Biophys. J. 113:2762–2767. 10.1016/j.bpj.2017.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes, M., Rotherham M., Price J.C., El Haj A.J., and Habib S.J.. 2016. Immobilized WNT Proteins Act as a Stem Cell Niche for Tissue Engineering. Stem Cell Reports. 7:126–137. 10.1016/j.stemcr.2016.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes, M., Junyent S., and Habib S.J.. 2017. Constructing cellular niche properties by localized presentation of Wnt proteins on synthetic surfaces. Nat. Protoc. 12:1498–1512. 10.1038/nprot.2017.061 [DOI] [PubMed] [Google Scholar]
- Merrill, B.J. 2012. Wnt pathway regulation of embryonic stem cell self-renewal. Cold Spring Harb. Perspect. Biol. 4:a007971. 10.1101/cshperspect.a007971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, K.M., Szczerkowski J.L.A., and Habib S.J.. 2017. Wnt ligand presentation and reception: from the stem cell niche to tissue engineering. Open Biol. 7:170140. 10.1098/rsob.170140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan, K.A., Fuerer C., Ching W., Fish M., Willert K., and Nusse R.. 2012. Secreted Wingless-interacting molecule (Swim) promotes long-range signaling by maintaining Wingless solubility. Proc. Natl. Acad. Sci. USA. 109:370–377. 10.1073/pnas.1119197109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano, K., Taoka M., Yamauchi Y., Itagaki C., Shinkawa T., Nunomura K., Okamura N., Takahashi N., Izumi T., and Isobe T.. 2005. Large-scale identification of proteins expressed in mouse embryonic stem cells. Proteomics. 5:1346–1361. 10.1002/pmic.200400990 [DOI] [PubMed] [Google Scholar]
- Neagu, A., van Genderen E., Escudero I., Verwegen L., Kurek D., Lehmann J., Stel J., Dirks R.A.M., van Mierlo G., Maas A., et al. 2020. In vitro capture and characterization of embryonic rosette-stage pluripotency between naive and primed states. Nat. Cell Biol. 22:534–545. 10.1038/s41556-020-0508-x [DOI] [PubMed] [Google Scholar]
- Neumann, C.J., and Cohen S.M.. 1997. Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development. 124:871–880. [DOI] [PubMed] [Google Scholar]
- Nichols, J., and Smith A.. 2009. Naive and primed pluripotent states. Cell Stem Cell. 4:487–492. 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- Niessen, C.M., Leckband D., and Yap A.S.. 2011. Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol. Rev. 91:691–731. 10.1152/physrev.00004.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka, S., Nishikawa-Torikai S., and Niwa H.. 2012. E-cadherin promotes incorporation of mouse epiblast stem cells into normal development. PLoS One. 7:e45220. 10.1371/journal.pone.0045220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuchi, Y., Reeves J., Ng S.S., Doro D.H., Junyent S., Liu K.J., El Haj A.J., and Habib S.J.. 2021. Wnt-modified materials mediate asymmetric stem cell division to direct human osteogenic tissue formation for bone repair. Nat. Mater. 20:108–118. 10.1038/s41563-020-0786-5 [DOI] [PubMed] [Google Scholar]
- Ortiz-Ramírez, C., Michard E., Simon A.A., Damineli D.S.C., Hernández-Coronado M., Becker J.D., and Feijó J.A.. 2017. GLUTAMATE RECEPTOR-LIKE channels are essential for chemotaxis and reproduction in mosses. Nature. 549:91–95. 10.1038/nature23478 [DOI] [PubMed] [Google Scholar]
- Pani, A.M., and Goldstein B.. 2018. Direct visualization of a native Wnt in vivo reveals that a long-range Wnt gradient forms by extracellular dispersal. eLife. 7:e38325. 10.7554/eLife.38325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivron, N.C., Frias-Aldeguer J., Vrij E.J., Boisset J.C., Korving J., Vivié J., Truckenmüller R.K., van Oudenaarden A., van Blitterswijk C.A., and Geijsen N.. 2018. Blastocyst-like structures generated solely from stem cells. Nature. 557:106–111. 10.1038/s41586-018-0051-0 [DOI] [PubMed] [Google Scholar]
- Rock, J.R., Onaitis M.W., Rawlins E.L., Lu Y., Clark C.P., Xue Y., Randell S.H., and Hogan B.L.M.. 2009. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA. 106:12771–12775. 10.1073/pnas.0906850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., and Clevers H.. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Shahbazi, M.N., and Zernicka-Goetz M.. 2018. Deconstructing and reconstructing the mouse and human early embryo. Nat. Cell Biol. 20:878–887. 10.1038/s41556-018-0144-x [DOI] [PubMed] [Google Scholar]
- Shahbazi, M.N., Scialdone A., Skorupska N., Weberling A., Recher G., Zhu M., Jedrusik A., Devito L.G., Noli L., Macaulay I.C., et al. 2017. Pluripotent state transitions coordinate morphogenesis in mouse and human embryos. Nature. 552:239–243. 10.1038/nature24675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, R., Satomi Y., Kurata T., Ueno N., Norioka S., Kondoh H., Takao T., and Takada S.. 2006. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev. Cell. 11:791–801. 10.1016/j.devcel.2006.10.003 [DOI] [PubMed] [Google Scholar]
- Takeichi, M. 2011. Self-organization of animal tissues: cadherin-mediated processes. Dev. Cell. 21:24–26. 10.1016/j.devcel.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Takeichi, M., Atsumi T., Yoshida C., Uno K., and Okada T.S.. 1981. Selective adhesion of embryonal carcinoma cells and differentiated cells by Ca2+-dependent sites. Dev. Biol. 87:340–350. 10.1016/0012-1606(81)90157-3 [DOI] [PubMed] [Google Scholar]
- Tanaka, S. 2006. Derivation and culture of mouse trophoblast stem cells in vitro. Methods Mol. Biol. 329:35–44. 10.1385/1-59745-037-5:35 [DOI] [PubMed] [Google Scholar]
- Tanaka, S., Kunath T., Hadjantonakis A.-K., Nagy A., and Rossant J.. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science. 282:2072–2075. 10.1126/science.282.5396.2072 [DOI] [PubMed] [Google Scholar]
- ten Berge, D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., Siu R.K., and Nusse R.. 2011. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 13:1070–1075. 10.1038/ncb2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar, P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., and McKay R.D.G.. 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 448:196–199. 10.1038/nature05972 [DOI] [PubMed] [Google Scholar]
- Thiery, J.P. 2002. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2:442–454. 10.1038/nrc822 [DOI] [PubMed] [Google Scholar]
- Tian, A., Duwadi D., Benchabane H., and Ahmed Y.. 2019. Essential long-range action of Wingless/Wnt in adult intestinal compartmentalization. PLoS Genet. 15:e1008111. 10.1371/journal.pgen.1008111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiridis, A., Huang Y., Blin G., Skylaki S., Wymeersch F., Osorno R., Economou C., Karagianni E., Zhao S., Lowell S., and Wilson V.. 2014. Distinct Wnt-driven primitive streak-like populations reflect in vivo lineage precursors. Development. 141:1209–1221. 10.1242/dev.101014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert, K., Brown J.D., Danenberg E., Duncan A.W., Weissman I.L., Reya T., Yates J.R. III, and Nusse R.. 2003. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 423:448–452. 10.1038/nature01611 [DOI] [PubMed] [Google Scholar]
- Wu, J., and Izpisua Belmonte J.C.. 2015. Dynamic Pluripotent Stem Cell States and Their Applications. Cell Stem Cell. 17:509–525. 10.1016/j.stem.2015.10.009 [DOI] [PubMed] [Google Scholar]
- Ying, Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., and Smith A.. 2008. The ground state of embryonic stem cell self-renewal. Nature. 453:519–523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca, M., Basler K., and Struhl G.. 1996. Direct and long-range action of a wingless morphogen gradient. Cell. 87:833–844. 10.1016/S0092-8674(00)81991-1 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Adileh M., Martin M.L., Klingler S., White J., Ma X., Howe L.R., Brown A.M.C., and Kolesnick R.. 2017. Establishing estrogen-responsive mouse mammary organoids from single Lgr5+ cells. Cell. Signal. 29:41–51. 10.1016/j.cellsig.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]