Abstract
Motivated by the lack of publicly available datasets of chest radiographs of positive patients with coronavirus disease 2019 (COVID-19), we build the first-of-its-kind open dataset of synthetic COVID-19 chest X-ray images of high fidelity using an unsupervised domain adaptation approach by leveraging class conditioning and adversarial training. Our contributions are twofold. First, we show considerable performance improvements on COVID-19 detection using various deep learning architectures when employing synthetic images as additional training set. Second, we show how our image synthesis method can serve as a data anonymization tool by achieving comparable detection performance when trained only on synthetic data. In addition, the proposed data generation framework offers a viable solution to the COVID-19 detection in particular, and to medical image classification tasks in general. Our publicly available benchmark dataset (https://github.com/hasibzunair/synthetic-covid-cxr-dataset.) consists of 21,295 synthetic COVID-19 chest X-ray images. The insights gleaned from this dataset can be used for preventive actions in the fight against the COVID-19 pandemic.
Keywords: Chest X-rays, COVID-19, Image synthesis, Deep learning, Image classification, Imbalanced data
Introduction
The World Health Organization (WHO) has declared COVID-19, the infectious respiratory disease caused by the novel coronavirus, a global pandemic due to the rapid increase in infections worldwide. This virus has spread across the globe, sending billions of people into lockdown, as many countries rush to implement strict measures in an effort to slow COVID-19 spread and flatten the epidemiological curve. Although most people with COVID-19 have mild to moderate symptoms, the disease can cause severe lung complications such as viral pneumonia, which is frequently diagnosed using chest radiography.
Recent studies have shown that chest radiography images such as chest X-rays (CXR) or computed tomography (CT) scans performed on patients with COVID-19 when they arrive at the emergency room can help doctors determine who is at higher risk of severe illness and intubation (Ai et al. 2020; Huang et al. 2020). These X-rays and CT scans show small patchy translucent white patches (called ground-glass opacities) in the lungs of COVID-19 patients. A chest X-ray provides a two-dimensional (2D) image, while a CT scan has the ability to form three-dimensional (3D) images of the chest. However, chest CT-based screening is more expensive, not always available at small or rural hospitals, and often yields a high false-positive rate. Therefore, the need to develop computational approaches for detecting COVID-19 via chest radiography imaging not only can save health care a tremendous amount of time and money, but more importantly, it can save more lives (Ng et al. 2020). By leveraging deep learning, several approaches for the detection of COVID-19 cases from chest radiography images have been recently proposed, including tailored convolutional neural network (CNN) architectures (Karim et al. 2020; Wang and Wong 2020) and transfer learning-based methods (Kassani et al. 2020; Narin et al. 2020; Li et al. 2020; Farooq and Hafeez 2020).
While promising, the predictive performance of these deep learning-based approaches depends heavily on the availability of large amounts of data. However, there is a significant shortage of chest radiology imaging data for COVID-19 positive patients, due largely to several factors, including the rare nature of the radiological finding, legal, privacy, technical, and data-ownership challenges. Moreover, most of the data are not accessible to the global research community.
In recent years, there have been several efforts to build large-scale annotated datasets for chest X-rays and make them publicly available to the global research community (Demner-Fushman et al. 2016; Johnson et al. 2019; Irvin et al. 2019; Wang et al. 2017; Bustos et al. 2019). At the time of writing, there exists, however, only one annotated COVID-19 X-ray image dataset (Cohen et al. 2020), which is a curated collection of X-ray images of patients who are positive or suspected of COVID-19 or other viral and bacterial pneumonia. This COVID-19 image data collection has been used as a primary source for positive cases of COVID-19 (Karim et al. 2020; Wang and Wong 2020; Kassani et al. 2020; Narin et al. 2020), where the detection of COVID-19 is formulated as a classification problem. While the COVID-19 image data collection contains positive examples of COVID-19, the negative examples were acquired from publicly available sources (Wang et al. 2017) and merged together for data-driven analytics. This fusion of multiple datasets results in predominantly negative examples with only a small percentage of positive ones, giving rise to a class imbalance problem (Demner-Fushman et al. 2016; Johnson et al. 2019; Irvin et al. 2019; Wang et al. 2017; Bustos et al. 2019). This in turn becomes a challenge of its own, as the merged data become highly imbalanced. In the context of a classifier training, the class imbalance problem in the training data distribution yields sub-optimal performance on the minority class (i.e., positive class for COVID-19).
In order to overcome the aforementioned issues, we present a domain adaptation framework by leveraging the inter-class variation of the data distribution for the task of conditional image synthesis by learning the inter-class mapping and synthesizing underrepresented class samples from the overrepresented ones using unpaired image-to-image translation (Zhu et al. 2017). The proposed framework combines class conditioning and adversarial training in a bid to synthesize realistic looking COVID-19 CXR images. The generated synthetic dataset contains 21,295 synthetic images of chest X-rays for COVID-19 positive cases.
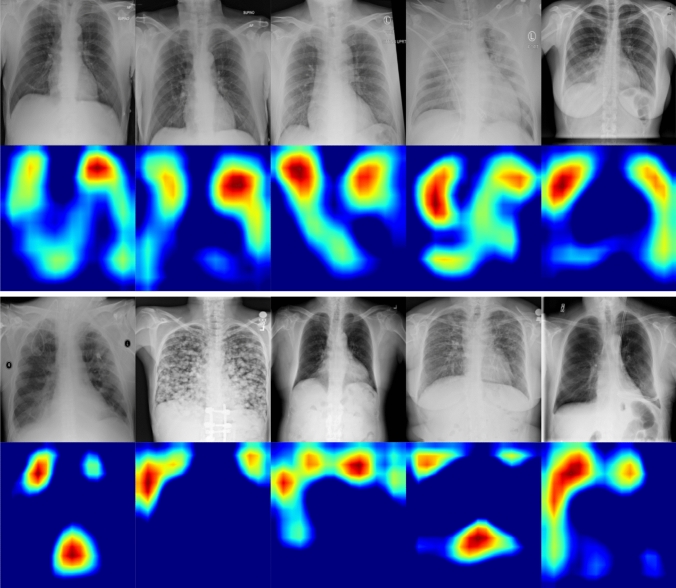
Understanding and interpreting the predictions made by a deep learning model provides valuable insights into the input data and the learned features learned so that the results can be easily understood by human experts. To visually explain the decisions made by the model in the sense that why an X-ray image is classified as COVID/Non-COVID, we use the gradient-weighted class activation map (Grad-CAM) to generate the saliency maps that highlight the most influential features affecting the predictions. Since the convolutional feature maps retain spatial information and that each pixel of the feature map indicates whether the corresponding visual pattern exists in its receptive field, the output from the last convolutional layer of the deep neural network shows the discriminative region in an image. To distinguish between the predicted COVID-19 and Non-COVID-19 images, we visualize the saliency maps for images that are correctly classified as COVID-19 and Non-COVID-19 (normal) by the proposed model. As shown in Fig. 1, the class activation maps for Non-COVID-19 (normal) demonstrate high activations for regions around the lungs, suggesting that there are no prediction features indicating that the disease is present. For most of the images that are correctly classified as COVID-19, the highlighted regions are within the lungs. Notice that in some cases, the model only highlights a specific part of the lung (e.g., left or right), which shows that COVID-19 features are present only on one side.
Fig. 1.
Saliency maps for the correctly classified COVID-19 (top two rows) and Non-COVID-19 (bottom two rows) images by the proposed model. Notice that for images that are classified as COVID-19, our model highlights the areas within the lungs, whereas for Non-COVID-19 images, the most important regions are around the lungs
In addition to demonstrating improved COVID-19 detection performance through the use of various deep convolutional neural network architectures on the synthetic data to boost training, we show how the proposed data generation and evaluation pipeline can serve as a viable data-driven solution to medical image analysis problems, and make our dataset publicly available, which is currently comprised of 21,295 synthetic images of chest X-rays for COVID-19 positive cases. The main contributions of this paper can be summarized as follows:
We present an integrated deep learning-based framework, which couples adversarial training and transfer learning to jointly address inter-class variation and class imbalance.
We synthesize chest X-ray images of COVID-19 to adjust the skew in training sets by over-sampling positive cases to mitigate the class imbalance problem, while training classifiers.
We demonstrate how the data generation procedure can serve as an anonymization tool by achieving comparable detection performance when trained only on synthetic data versus real data in an effort to alleviate privacy concerns.
The rest of this paper is organized as follows. In Sect. 2, we provide a brief overview of generative approaches for medical image synthesis. In Sect. 3, we present a generative framework, which couples adversarial training and transfer learning to jointly address inter-class variation and class imbalance. In Sect. 4, we present experimental results to demonstrate improved COVID-19 detection performance through the use of various deep convolutional neural network architectures on the generated data. Finally, we conclude in Sect. 5 and point out future work directions.
Related work
The advent of generative adversarial networks (GANs) (Goodfellow et al. 2014) has accelerated research in generative modeling and distribution learning. With the ability to replicate data distributions and synthesize images with high fidelity, GANs have bridged the gap between supervised learning and image generation. These synthetic images can then be used as input to improve the performance of various deep learning algorithms for downstream tasks, such as image classification and segmentation. GANs have not only been used in natural images’ settings, but have also been extensively employed in medical image analysis (Kazeminia et al. 2018), where labels are usually scarce or almost non-existent.
With the scarcity of annotated medical image datasets, there has been a surge of interest in developing efficient approaches for the generation of synthetic medical images. While several existing generative methods have addressed the translation between multiple imaging modalities CT-PET, CS-MRI, MR-CT, XCAT-CT (Ben-Cohen et al. 2017; Yang et al. 2017; Wolterink et al. 2017; Russ et al. 2019) based on distribution matching, other approaches have focused on the scarcity of labeled data in the medical field due in large part to the acquisition, privacy, and health safety issues. Conditional and unconditional image synthesis procedures, built on top of these generative models, have been proposed in retinal images (Costa et al. 2017; Dar et al. 2019) and MRI scans (Shin et al. 2018; Guibas et al. 2017; Korkinof et al. 2018). These models involve the training of paired data in both source and target domains to synthesize realistic, high-resolution images in order to aid in medical image classification and segmentation tasks.
Image synthesis methodologies have also been proposed in the context of chest X-rays (Teixeira et al. 2018). Our work is significantly different in the sense that we are specifically interested in synthesizing a particular class, whereas in Teixeira et al. (2018) X-rays are generated from surface geometry for landmark detection tasks. While some generative methods only require paired data in the source domain with target domain consisting of unlabeled examples, Cohen et al. (2018) have demonstrated that the phenomenon of hallucinating features (e.g., adding or removing tumors leading to a semantic change) leads to a high bias in these domain adaptation techniques. To overcome this issue, we have recently proposed a domain adaptation technique based on cycle-consistent adversarial networks in order synthesize high-fidelity positive examples to improve detection performance of melanoma from skin lesion images (Zunair and Hamza 2020).
Proposed method
In this section, we present the main building blocks of our proposed image synthesis framework using image-to-image translation, which is an increasingly popular machine learning paradigm that has shown great promise in a wide range of applications, including computer graphics, style transfer, satellite imagery, object transfiguration, character animation, and photo-enhancement. In an typical image-to-image translation problem, the objective is to learn a mapping that translates an image in one domain to a corresponding image in another domain using approaches that leverage paired or unpaired training samples. The latter is the focus of our work. While paired image-to-image translation methods use pairs of corresponding images in different domains, the paired training samples are, however, not always available. By contrast, the unpaired image-to-image translation problem, in which training samples are readily available, is more common and practical, but it is highly under-constrained and fraught with challenges. In our work, we build upon the idea that there exist no paired training samples showing how an image from one domain can be translated to a corresponding image in another domain. The task is to generate COVID-19 chest X-rays from chest X-ray images to address COVID-19 class imbalance problem. More specifically, our goal is to learn a mapping function between Non-COVID-19 images and COVID-19 in order to generate COVID-19 chest X-rays without paired training samples in an unsupervised fashion.
Chest X-ray image synthesis
We formulate the detection of COVID-19 as a binary classification problem. For the Normal vs. COVID-19 and Pneumonia vs. COVID-19 tasks, we train two translation models and synthesize COVID-19 images for each task in order to adjust the skew in the training data by over-sampling the minority class. For the sake of clarity and unless otherwise expressly indicated, we refer to the source domain of the two tasks as Non-COVID-19 instead of Normal and Pneumonia separately.
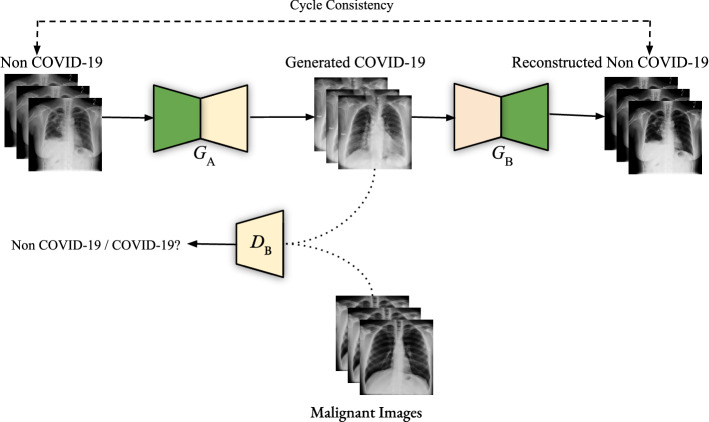
We adopt our unsupervised domain adaptation technique introduced in Zunair and Hamza (2020) to translate Non-COVID-19 images for each case (i.e., normal or pneumonia) to COVID-19. Given two image domains A and B denoting Non-COVID-19 and COVID-19, respectively, the goal is to learn to translate images of one type to another using two generators and , and two discriminators and , as illustrated in Fig. 2.
Fig. 2.
Illustration of the generative adversarial training process for unpaired image-to-image translation. Chest X-ray images are translated from Non-COVID-19 (i.e., Normal or Pneumonia) to COVID-19 and then back to Non-COVID-19 to ensure cycle consistency in the forward pass. The same procedure is applied in the backward pass from COVID-19 to Non-COVID-19
The generator (resp. ) translates images from Non-COVID-19 to COVID-19 (i.e., ), while the discriminator (resp. ) verifies how real an image of B (resp. A) looks. The overall objective function is defined as
| 1 |
which consists of two adversarial losses and a cycle consistent loss regularized by a hyper-parameter (Zhu et al. 2017). The first adversarial loss is given by
| 2 |
where the generator tries to generate images that look similar to COVID-19 images, while aims to distinguish between generated samples and real samples b. During the training, as generates a COVID-19 image, verifies if the translated image is actually a real COVID-19 image or a synthetic one. The data distributions of Non-COVID-19 and COVID-19 are and , respectively.
Similarly, the second adversarial loss is given by
| 3 |
where takes a COVID-19 image b from B as input and tries to generate a realistic image in A that tricks the discriminator . Hence, the goal of is to generate a Non-COVID-19 chest X-ray such that it fools the discriminator to label it as a real Non-COVID-19 image.
The third loss term is to enforce cycle consistency and is given by
| 4 |
which computes the difference between the input image and the generated one using the -norm.
Model optimization
The idea of the cycle consistency loss it to add a constraint such that and . In other words, the objective is to learn two bijective generator mappings by solving the following optimization problem:
| 5 |
For the generators and , the architecture is based on fully convolutional network (FCN). The discriminators and consist of a CNN classifier which verifies whether the image is real or synthetic.
Training procedure
The training for the generators and discriminators is carried out in the same way as in Zunair and Hamza (2020). First, we balance the inter-class data samples by performing undersampling. Then, we train CycleGAN to learn a function of the interclass variation between the two groups, i.e., we learn a transformation between Non-COVID-19 and COVID-19 radiographs. We apply CycleGAN to the overrepresented class samples in order to synthesize the target class samples (i.e., underrepresented class).
After training, we apply the generators and on the training datasets of Normal vs. COVID-19 and Pneumonia vs. COVID-19. We apply on the majority class of Normal vs. COVID-19, which consists of normal images in order to synthesize 16,537 COVID-19 images. We denote this synthesized dataset as , which consists of generated images by performing image-to-image translation from normal to COVID-19.
Similarly, for Pneumonia vs. COVID-19, we synthesize 4758 COVID-19 images by applying on the majority class consisting of pneumonia images and we denote the synthesized dataset as , which is comprised of generated images by performing image-to-image translation from pneumonia to COVID-19. It is worth pointing out that for the sake of clarity, the discriminator is not depicted to Fig. 2, as our main is to generate COVID-19 images from Non COVID-19 images.
Experiments
In this section, we conduct extensive experiments to evaluate the performance of the proposed data generation framework on COVID-19 detection.
Datasets
We use two publicly available datasets of chest X-rays:
COVID-19 Image Data Collection. This dataset comprises 226 images of pneumonia cases with chest X-ray or CT images, specifically COVID-19 cases as well as MERS, SARS, and ARDS. Data are scraped from publications and websites such as Radiopaedia.org, Italian Society of Medical and Interventional Radiology1, and Figure1.com2. From this dataset, we discard the CT images and retain the 226 images positive for COVID-19 and their corresponding labels.
RSNA Pneumonia Detection Challenge. This dataset originated from a Kaggle challenge3 and consists of publicly available data from Wang et al. (2017). It is composed of 26,684 images, and each image was annotated by a radiologist for the presence of lung opacity; thereby providing a label for two classes. This label is included as both lung opacity and pneumonia.
Dataset splits and preprocessing
We partition the three classes from COVID-19 Image Data Collection and RSNA Pneumonia Detection Challenge into two sets, namely “Normal vs. COVID-19” and “Pneumonia vs COVID-19.” A patient level split is then applied using 80% as training set and the remaining 20% as test set to assess algorithm performance, and we follow the same evaluation protocol laid out in Shin et al. (2018), Zunair and Hamza (2020). We define the skew ratio as follows:
| 6 |
where represents a fully balanced dataset, shows that the negative samples are the majority, and represents positive sample dominance in the distribution.
The data distributions of Normal vs. COVID-19 and Pneumonia vs. COVID-19 are displayed in Fig. 3, which illustrates the class imbalance in the training dataset. For Pneumonia vs. COVID-19, the skew ratio is around 22.9, while the skew for Normal vs. COVID-19 is almost four times larger, indicating high imbalance in the classes.
Fig. 3.
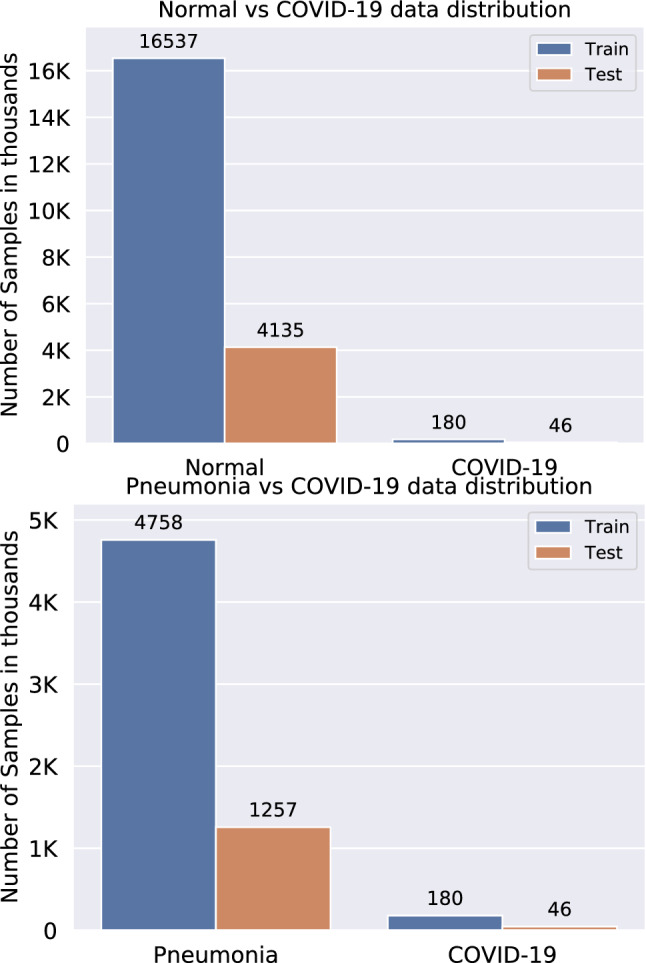
Data distributions of Normal vs. COVID-19 (top) and Pneumonia vs. COVID-19 (bottom) with skew ratios of 91.87 and 22.9, respectively
We also resize all images to pixels and scale the pixel values to [0, 1] for the training of classifiers. It is important to mention that when we use the term synthetic data, we refer to COVID-19 CXR images only.
Baselines
Since our aim is to provide a dataset to be used as a training set for the minority class, we test the effectiveness of several deep CNN architectures, including VGG-16 (Simonyan and Zisserman 2014), ResNet-50 (He et al. 2016) and DenseNet-102 (Huang et al. 2017), on the detection of the minority class. These pretrained networks were trained on more than a million images from the ImageNet database4. More specifically, we investigate the contribution of the synthetic datasets and , which consist of COVID-19 CXR images, to the overall performance of these deep learning models. The last layer of each of these models consists of a global average pooling (GAP) layer, which computes the average output of each feature map in the previous layer and helps minimize overfitting by reducing the total number of parameters in the model. The GAP layer turns a feature map into a single number by taking the average of the numbers in that feature map. Similar to max-pooling layers, GAP layers have no trainable parameters and are used to reduce the spatial dimensions of a 3D tensor. The GAP layer is followed by a single fully connected (FC) layer with a softmax function (i.e., a dense softmax layer of two units for the binary classification case), which yields the predicted classes’ probabilities that sum to one.
Evaluation metrics
Due to high class imbalance in the datasets, the choice of evaluation metrics plays a vital role in the comparison of classifiers. Threshold metrics such as accuracy and rank metrics (e.g., area under the ROC curve) may lead to a false sense of superiority and mask poor performance (Jeni et al. 2013), thereby introducing bias. Since we are interested in the detection of the minority class (COVID-19), we follow the recommendations provided in Brabec and Machlica (2018), Jeni et al. (2013) and perform quantitative evaluations using sensitivity and false negatives in the same vein as Kassani et al. (2020). Sensitivity is the percentage of positive instances correctly classified and is defined as
| 7 |
where TP, FP, TN and FN denote true positives, false positives, true negatives, and false negatives, respectively. TP is the number of correctly predicted malignant lesions, while TN is the number of correctly predicted benign lesions. A classifier that reduces FN (ruling COVID-19 out in cases that do have it) and FP (wrongly diagnosing COVID-19 where there is none) indicates a better performance. A false-negative COVID-19 result can be a serious problem due to the fact that we lose the benefits of early intervention. A false-positive result can also cause significant issues for both an individual and the community. Even from an epidemiologicial perspective, a high number of false positives can lead to a wrong understanding of the spread of COVID-19 in the community. Sensitivity, also known as recall or true-positive rate, indicates how often a classifier misses a positive prediction. It is one of the most common measures to evaluate a classifier in medical image classification tasks (Esteva et al. 2017). A larger value of sensitivity indicates a better performance of the classification model.
Implementation details
All experiments are performed on a Linux Workstation (CPU: AMD 2nd Gen Ryzen Threadripper 2950X, 16-Core, 64-Thread, 4.4GHz Max Boost; Memory: 64GB high-performance RAM; GPU: NVIDIA GeForce RTX 2080 Ti). We perform training/testing on both COVID-19 Image Data Collection and RSNA Pneumonia Detection Challenge. For training the models, we use the Adadelta optimization algorithm (Zeiler 2012) to minimize the binary cross-entropy loss function with a learning rate of 0.001 and batch size of 16. We initialize the weights using ImageNet and train all layers until the loss stagnates using an early stopping mechanism.
For each dataset, we follow the same evaluation protocol laid out in Shin et al. (2018), Zunair and Hamza (2020) for testing the contribution of newly added data. In this evaluation protocol, both training and test sets are used. The training set varies, as new data are added to each configuration. The deep CNN classifiers are trained on these data and evaluated on the held-out test set. For fair evaluation and comparison purposes, the size of the test set remains constant. It is important to mention that the test set does not contain any synthetic examples. Moreover, the hyper-parameters are not tuned and hence do not require a separate validation set.
Over-sampling with synthetic data
We demonstrate the effectiveness of the synthetic sets and in Tables 1 and 2 using four deep learning models, namely VGG-16 (Simonyan and Zisserman 2014), ResNet-50 (He et al. 2016), DenseNet-102 (Huang et al. 2017), and DenseNet-121 with a bagging tree classifier (DenseNet121 + BGT) (Kassani et al. 2020). For each task, we can observe that when is added, there is a significant increase in performance. While the addition of also results in an increase in performance, such an increase is not quite large compared to adding in some cases. We hypothesize that this is due to the number of COVID-19 examples in (16,537), which enables the models to learn better representations for COVID-19, whereas is comprised of only 4,758 COVID-19 examples. Further, an increase in performance using both metrics is observed when the skew in the training dataset decreases. The relative improvement seems to drop as the model complexity increases, which is in line with the findings in Raghu et al. (2019) due to the problem of over-parametrization. When synthetic data are used as additional training set, the detection performance significantly increases. However, the relative improvement drops when the architectural complexity of the model increases. Note that despite its simplicity, the VGG-16 network outperforms all the other baseline methods, while the “DenseNet121 + BGT” model yields the second best performance. For less complex models, we can see that using only synthetic dataset performs better than the original data. Moreover, Table 2 shows that with the exception of VGG-16, all models achieve sub-optimal performance when using synthetic data only.
Table 1.
COVID-19 detection performance results on Normal vs. COVID-19 test set when trained on real data; real + synthetic data; and only synthetic data (i.e., only is used for positive class examples in training each model)
| Model | Real | Real + | Real + + | Only Synthetic | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SEN (%) | FN | Skew | SEN (%) | FN | Skew | SEN (%) | FN | Skew | SEN (%) | FN | |
| VGG-16 | 19.56 | 37 | 91.87 | 54.34 | 21 | 0.98 | 63.04 | 17 | 0.79 | 50.00 | 23 |
| ResNet-50 | 32.61 | 31 | 91.87 | 41.30 | 27 | 0.98 | 43.47 | 26 | 0.79 | 10.86 | 41 |
| DenseNet-102 | 26.08 | 34 | 91.87 | 28.27 | 33 | 0.98 | 34.73 | 30 | 0.79 | 8.69 | 42 |
| DenseNet-121 + BGT | 36.95 | 29 | 91.87 | 45.65 | 25 | 0.98 | 52.17 | 22 | 0.79 | 21.73 | 36 |
SEN is short for sensitivity. Boldface numbers indicate the best performance
Table 2.
COVID-19 detection performance results on Pneumonia vs. COVID-19 test set when trained on real data; real + synthetic data; and only synthetic data (i.e., only is used for positive class examples in training each model)
| Model | Real | Real + | Real + + | Only Synthetic | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SEN (%) | FN | Skew | SEN (%) | FN | Skew | SEN(%) | FN | Skew | SEN (%) | FN | |
| VGG-16 | 8.69 | 42 | 22.9 | 29.50 | 24 | 0.95 | 52.17 | 22 | 0.19 | 39.13 | 28 |
| ResNet-50 | 21.73 | 36 | 22.9 | 36.95 | 29 | 0.95 | 41.30 | 27 | 0.19 | 13.04 | 40 |
| DenseNet-102 | 4.34 | 44 | 22.9 | 21.74 | 36 | 0.95 | 32.43 | 32 | 0.19 | 6.52 | 43 |
| DenseNet-121 + BGT | 32.60 | 31 | 22.9 | 41.30 | 27 | 0.95 | 47.82 | 24 | 0.19 | 32.60 | 31 |
Boldface numbers indicate the best performance
Training on anonymized synthetic data
We also evaluate the performance when the classifiers are trained on only synthetic COVID-19 images, as shown in Tables 1 and 2 for each dataset. Sub-optimal performance is achieved for both tasks for different CNNs, except for VGG-16 which shows performance improvement compared to when using the original COVID-19 examples. Since a new data sample is not attributed to an individual patient, but it is rather an instance which is conditioned on the training data, it does not entirely reflect the original data. This suggests that synthetic data alone cannot be used to achieve optimal performance. In other words, the synthetic data can be used as a form of pre-training, which often requires a small amount of real data to achieve comparable performance. In addition, the relatively large margin between the evaluation scores suggests that the observed difference between the models is actually real, and not due to a statistical chance.
Detecting target class with high confidence
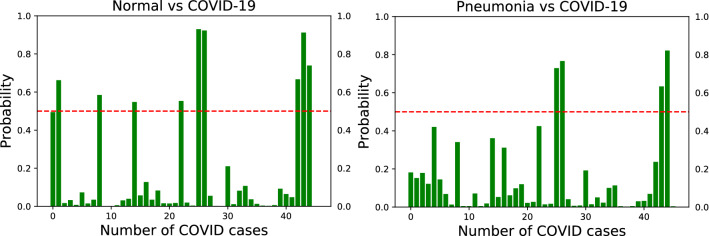
The output of the softmax function describes the probability (or confidence) of the learning model that a particular sample belongs to a certain class. The softmax layer takes the raw values (logits) of the last FC layer and maps them into probability scores by taking the exponents of each output and then normalize each number by the sum of those exponents so that all probabilities sum to one. Figure 4 shows the probability scores for the VGG-16 model on unseen test set of COVID-19 for the two binary classification tasks of Normal vs. COVID-19 and Pneumonia vs. COVID-19 using original data only. The red dashed line depicts the 0.5 probability threshold. Notice that Fig. 4(left) shows low confidence scores, while Fig. 4(right) shows sub-optimal performance for COVID-19 detection when using original training data only.
Fig. 4.
Confidence scores for the VGG-16 model on unseen test set of COVID-19 for the two binary classification tasks of Normal vs. COVID-19 (left) and Pneumonia vs. COVID-19 (right) using original data only
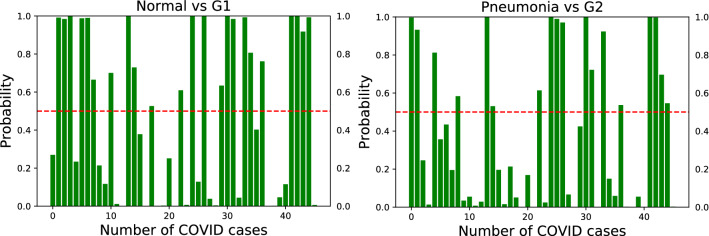
Figure 5 shows that synthetic data can be used without the original examples. When using synthetic data as additional training set, we observe that not only the number of correctly detected instances of COVID-19 increases, but also the predictions tend to improve, as demonstrated by the high probability scores.
Fig. 5.
Confidence scores for the VGG-16 model on unseen test set of COVID-19 for the two binary classification tasks of Normal vs. COVID-19 (left) and Pneumonia vs. COVID-19 (right) using synthetic data without the original examples. Notice that synthetic data increase the confidence scores
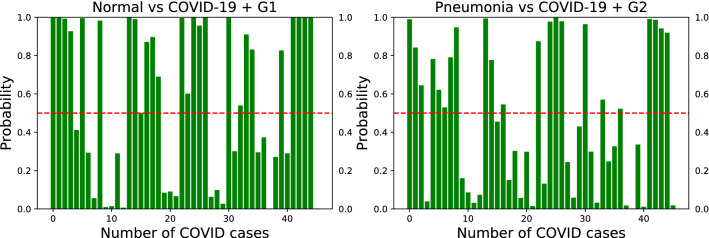
Figures 6 and 7 show improved detection performance when the synthetic data are used as additional training set. A similar trend was observed with the ResNet-50 and DenseNet-102 models.
Fig. 6.
Confidence scores for the VGG-16 model on unseen test set of COVID-19 for the two binary classification tasks of Normal vs. COVID-19 (left) and Pneumonia vs. COVID-19 (right) with synthetic data as additional training set. Left: adding 16,537 COVID-19 examples of to the original COVID-19 dataset. Right: adding 4758 COVID-19 examples of to the original COVID-19 dataset
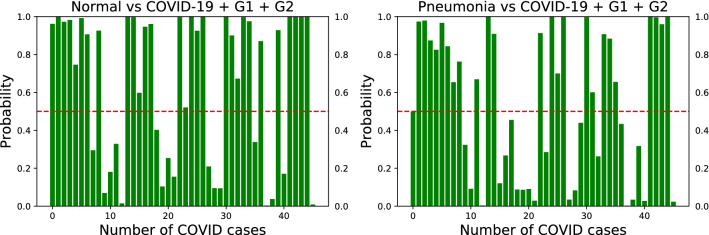
Fig. 7.
Confidence scores for the VGG-16 model on unseen test set of COVID-19 for the two binary classification tasks of Normal vs. COVID-19 (left) and Pneumonia vs. COVID-19 (right) with synthetic data as additional training set. Both and are added to the original COVID-19 dataset
Generating anonymized synthetic images with variation
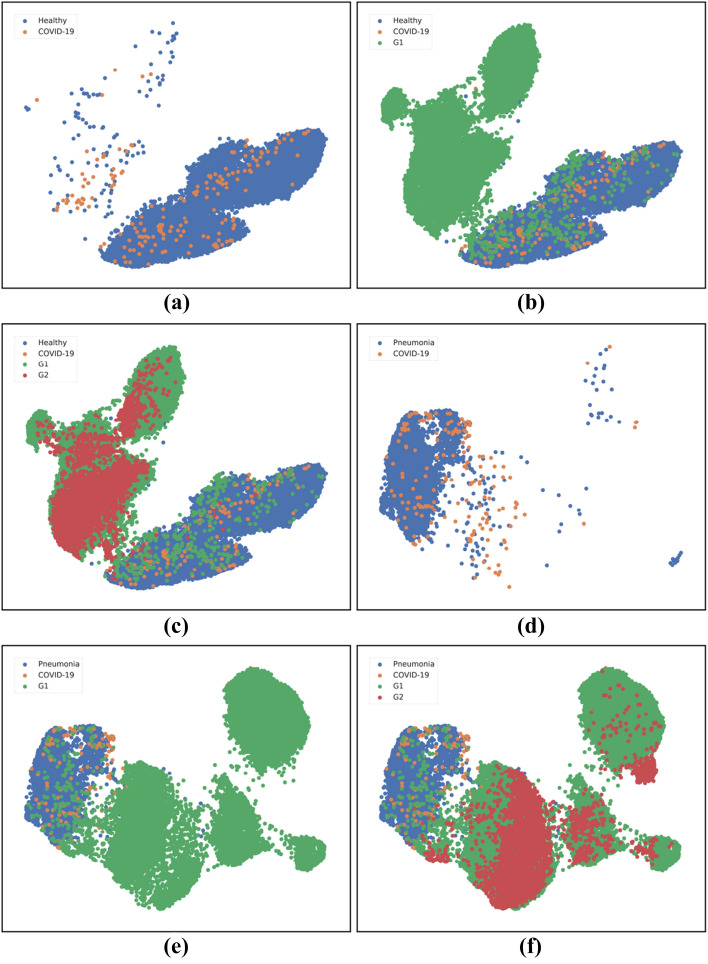
Data visualization based on dimension reduction plays an important role in data analysis and interpretation. The objective of dimension reduction is to map high-dimensional data into a low-dimensional space (usually 2D or 3D), while preserving the overall structure of the data as much as possible. A commonly used dimension reduction method is the Uniform Manifold Approximation and Projection (UMAP) algorithm, which is nonlinear technique based on manifold learning and topological data analysis. UMAP is capable of preserving both local and most of the global structure of the data when an appropriate initialization of the embedding is used. The two-dimensional UMAP embeddings of the features are shown in Fig. 8 to visualize the difference between the original and synthetic data. Notice that the synthetic samples are in a different distribution in the feature space, enabling a decision boundary between the classes. The original examples in Fig. 8a exhibit low inter-class variation and consist of outliers. In Fig. 8b, we can see that the synthetic examples of the dataset are in a different distribution in the feature space. While the UMAP embeddings may not be interpreted as a justification that the synthetic examples actually consist of COVID-19 symptoms from a clinical perspective, it is, however, important to note that the distribution of the synthetic images is significantly different than that of normal images, thereby enabling a proper decision boundary. A similar trend can be observed in Fig. 8d, e, f. The overlapping features for Pneumonia vs. COVID-19 can be explained by the fact that the findings of X-ray imaging in COVID-19 are not specific, and tend to overlap with other infections such as Pneumonia in this case.
Fig. 8.
Two-dimensional UMAP embeddings: (a) Normal vs. COVID-19; (b) Normal vs. COVID-19 + ; (c) Normal vs. COVID-19 + + ; (d) Pneumonia vs. COVID-19; (e) Pneumonia vs COVID-19 + ; (f) Pneumonia vs. COVID-19 + + . Here, G1 and G2 denote and , respectively
Discussion
Since the generative and classification models are trained to learn representations in the training data distribution, it is likely that a bias might occur toward that data. In light of the class imbalance problem, the generator is trained by under-sampling the majority class. This under-sampling process often leaves a relatively small number of data points (180 samples for each domain) to learn from. While a boost in performance is achieved when using the synthetic datasets, it is not conclusive enough to confirm whether our approach can be generalized across other COVID-19 datasets due largely to the lack of such benchmarks. While the improvements we have achieved using our proposed framework are encouraging, it is important to mention that a key objective of this work is not to claim state-of-the-art results, but rather to release an open source dataset to the research community in an effort to further improve COVID-19 detection.
Conclusion
In this paper, we presented an unsupervised domain adaptation approach by leveraging class conditioning and adversarial training to build an open database of synthetic COVID-19 chest X-ray images of high fidelity. This publicly available database comprises 21,295 synthetic images of chest X-rays for COVID-19 positive cases. The insights generated from applying recent deep learning approaches on this database can be used for preventive actions against the global COVID-19 pandemic, in the hope of containing the virus. We also demonstrated how the data generation procedure can serve as an anonymization tool by achieving comparable detection performance when trained only on synthetic data versus real data in an effort to alleviate data privacy concerns. Our experiments reveal that synthetic data can significantly improve the COVID-19 detection performance results, that as the amount of synthetic data is increased, sensitivity improves considerably and the number of false negatives decreases. We believe that the performance can be further improved by applying more application-specific preprocessing and exhaustive hyper-parameter tuning, as well as by leveraging ensemble methods, which we leave for future work.
Acknowledgements
This work was supported in part by and Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant Number N00929. This research was enabled in part by advanced computing resources provided by Compute Canada.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L (2020) Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology [DOI] [PMC free article] [PubMed]
- Ben-Cohen A, Klang E, Raskin SP, Amitai MM, Greenspan H (2017) Virtual PET images from CT data using deep convolutional networks: initial results. In: Proceedings of international workshop on simulation and synthesis in medical imaging, pp 49–57
- Brabec J, Machlica L (2018) Bad practices in evaluation methodology relevant to class-imbalanced problems, arXiv preprint arXiv:1812.01388
- Bustos A, Pertusa A, Salinas J-M, de la Iglesia-Vayá M (2019) PadChest: a large chest X-ray image dataset with multi-label annotated reports, arXiv preprint arXiv:1901.07441 [DOI] [PubMed]
- Cohen JP, Luck M, Honari S (2018) Distribution matching losses can hallucinate features in medical image translation. In: Proceedings of international conference on medical image computing and computer-assisted intervention, pp 529–536
- Cohen JP, Morrison P, Dao L (2020) COVID-19 image data collection, arXiv preprint arXiv:2003.11597,
- Costa P, Galdran A, Meyer MI, Abràmoff MD, Niemeijer M, Mendonça AM, Campilho A (2017) Towards adversarial retinal image synthesis, arXiv preprint arXiv:1701.08974 [DOI] [PubMed]
- Dar SU, Yurt M, Karacan L, Erdem A, Erdem E, Çukur T. Image synthesis in multi-contrast MRI with conditional generative adversarial networks. IEEE Trans Med Imaging. 2019;38(10):2375–2388. doi: 10.1109/TMI.2019.2901750. [DOI] [PubMed] [Google Scholar]
- Demner-Fushman D, Kohli MD, Rosenman MB, Shooshan SE, Rodriguez L, Antani S, Thoma GR, McDonald CJ. Preparing a collection of radiology examinations for distribution and retrieval. J Am Med Inform Assoc. 2016;23(2):304–310. doi: 10.1093/jamia/ocv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M, Hafeez A (2020) COVID-ResNet: a deep learning framework for screening of COVID19 from radiographs, arXiv preprint arXiv:2003.14395
- Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2014) Generative adversarial nets. In: Advances in neural information processing systems, pp 2672–2680
- Guibas JT, Virdi TS, Li PS (2017) Synthetic medical images from dual generative adversarial networks, arXiv preprint arXiv:1709.01872
- He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of IEEE conference on computer vision and pattern recognition, pp 770–778
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of IEEE conference on computer vision and pattern recognition, pp 4700–4708
- Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K et al (2019) CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. In: Proceedings of AAAI conference on artificial intelligence, vol 33, pp 590–597
- Jeni LA, Cohn JF, De La Torre F (2013) Facing imbalanced data–recommendations for the use of performance metrics. In: Proceedings of humaine association conference on affective computing and intelligent interaction, pp 245–251 [DOI] [PMC free article] [PubMed]
- Johnson AEW, Pollard TJ, Greenbaum NR, Lungren MP, ying Deng C, Peng Y, Lu Z, Mark RG, Berkowitz SJ, Horng S (2019) MIMIC-CXR-JPG: a large publicly available database of labeled chest radiographs, arXiv preprint arXiv:1901.07042 [DOI] [PMC free article] [PubMed]
- Karim M, Döhmen T, Rebholz-Schuhmann D, Decker S, Cochez M, Beyan O, et al (2020) DeepCOVIDExplainer: explainable COVID-19 predictions based on chest X-ray images, arXiv preprint arXiv:2004.04582
- Kassani SH, Kassasni PH, Wesolowski MJ, Schneider KA, Deters R (2020) Automatic detection of coronavirus disease (COVID-19) in X-ray and CT images: a machine learning-based approach, arXiv preprint arXiv:2004.10641 [DOI] [PMC free article] [PubMed]
- Kazeminia S, Baur C, Kuijper A, van Ginneken B, Navab N, Albarqouni S, Mukhopadhyay A (2018) GANs for medical image analysis, arXiv preprint arXiv:1809.06222 [DOI] [PubMed]
- Korkinof D, Rijken T, O’Neill M, Yearsley J, Harvey H, Glocker B (2018) High-resolution mammogram synthesis using progressive generative adversarial networks, arXiv preprint arXiv:1807.03401
- Li X, Li C, Zhu D (2020) COVID-MobileXpert: On-device COVID-19 screening using snapshots of chest X-Ray, carXiv preprint arXiv:2004.03042,
- Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks, arXiv preprint arXiv:2003.10849 [DOI] [PMC free article] [PubMed]
- Ng M-Y, Lee EY, Yang J, Yang F, Li X, Wang H, Lui MM, Lo CS-Y, Leung B, Khong P-L, et al. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiology. 2020;2(1):e200034. doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu M, Zhang C, Kleinberg J, Bengio S (2019) Transfusion: understanding transfer learning for medical imaging. In: Advances in neural information processing systems, pp 3342–3352
- Russ T, Goerttler S, Schnurr A-K, Bauer DF, Hatamikia S, Schad LR, Zöllner FG, Chung K. Synthesis of CT images from digital body phantoms using CycleGAN. Int J Comput Assist Radiol Surg. 2019;14(10):1741–1750. doi: 10.1007/s11548-019-02042-9. [DOI] [PubMed] [Google Scholar]
- Shin H-C, Tenenholtz NA, Rogers JK, Schwarz CG, Senjem ML, Gunter JL, Andriole KP, Michalski M (2018) Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In: Proceedings of international workshop on simulation and synthesis in medical imaging, pp 1–11
- Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556
- Teixeira B, Singh V, Chen T, Ma K, Tamersoy B, Wu Y, Balashova E, Comaniciu D (2018) Generating synthetic X-ray images of a person from the surface geometry. In: Proceedings of IEEE conference on computer vision and pattern recognition, pp 9059–9067
- Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of IEEE conference on computer vision and pattern recognition, pp 2097–2106
- Wang L, Wong A (2020) COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images, arXiv preprint arXiv:2003.09871, [DOI] [PMC free article] [PubMed]
- Wolterink JM, Dinkla AM, Savenije MH, Seevinck PR, van den Berg CA, Išgum I (2017) Deep MR to CT synthesis using unpaired data. In: Proceedings of international workshop on simulation and synthesis in medical imaging, pp 14–23
- Yang G, Yu S, Dong H, Slabaugh G, Dragotti PL, Ye X, Liu F, Arridge S, Keegan J, et al. DAGAN: deep de-aliasing generative adversarial networks for fast compressed sensing MRI reconstruction. IEEE Trans Med Imaging. 2017;37(6):1310–1321. doi: 10.1109/TMI.2017.2785879. [DOI] [PubMed] [Google Scholar]
- Zeiler MD (2012) ADADELTA: an adaptive learning rate method, arXiv preprint arXiv:1212.5701
- Zhu J-Y, Park T, Isola P, Efros AA (2017) Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of IEEE international conference on computer vision, pp 2223–2232
- Zunair H, Hamza AB. Melanoma detection using adversarial training and deep transfer learning. Phys Med Biol. 2020;65:135005. doi: 10.1088/1361-6560/ab86d3. [DOI] [PubMed] [Google Scholar]