Abstract
Retinoblastoma (RB) is the most common primary intraocular cancer type that occurs during retinal development in childhood. Previous studies have reported that long non-coding RNAs (lncRNAs) are involved in the development of RB. Therefore, the aim of the present study was to investigate the effects and underlying regulatory mechanisms of nuclear paraspeckle assembly transcript 1 (NEAT1) in RB. The expression levels of NEAT1, microRNA (miR)-24-3p and leucine-rich-α-2-glycoprotein (LRG1) were detected using reverse transcription-quantitative PCR (RT-qPCR). Moreover, the protein expression levels of LRG1, matrix metalloproteinase 9, N-cadherin and E-cadherin were detected via western blotting. Furthermore, cell migration and invasion abilities were evaluated via Transwell assays. The targeting relationships between miR-24-3p and NEAT1 or LRG1 were predicted using online software and confirmed via dual-luciferase reporter assay. In the present study, NEAT1 and LRG1 were upregulated, and miR-24-3p was downregulated in RB tissues and cells compared with the corresponding healthy tissues and cells. Moreover, miR-24-3p was identified as a target of NEAT and LRG1 was demonstrated to be a direct target gene of miR-24-3p. Knockdown of NEAT1 or LRG1 significantly suppressed RB cell migration and invasion ability, while the effects were reversed by an miR-24-3p inhibitor. In addition, the downregulation of LRG1 caused by miR-24-3p was restored following the overexpression of NEAT1 in RB cells. It was also demonstrated that NEAT1 knockdown inhibited the epithelial-to-mesenchymal transition (EMT) pathway by inhibiting the expression of LRG via targeting miR-24-3p. In conclusion, the present results suggest that silencing of NEAT1 suppresses cell migration, invasion and the EMT process by downregulating LRG1 expression via sponging miR-24-3p in RB, thus indicating that NEAT1 may be a potential candidate for RB treatment.
Keywords: retinoblastoma, long non-coding RNA nuclear paraspeckle assembly transcript 1, microRNA-24-3p, leucine-rich-α-2-glycoprotein, epithelial-to-mesenchymal transition
Introduction
Retinoblastoma (RB) is an aggressive eye cancer that occurs during retinal development in childhood, and it accounts for 2-4% of all childhood malignancies (1,2). Moreover, the occurrence of RB is a very complicated process and it can be present in a single eye or both eyes (3,4). Although the treatment methods have improved over the past few years, the mortality remains at ~70% in low- and middle-income countries (5). Therefore, it is important to improve the understanding of the pathogenesis of RB and develop more effective treatment strategies.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs (ncRNAs) with >200 nucleotides (nts), which have no protein-encoding ability (6). lncRNAs have been reported to serve crucial roles in the occurrence and progression of various cancer types (7,8). Furthermore, several lncRNAs have been revealed to participate in the regulation of RB. For example, knockdown of lncRNA Antisense ncRNA in the INK4 locus inhibits RB cell invasion and induces apoptosis via the ataxia-telangiectasia mutated/E2F transcription factor 1 pathway (9). Colon cancer associated transcript 1 has been demonstrated to promote RB cell proliferation via sponging microRNA (miRNA/miR)-218-5p (10). It has also been revealed that H19 imprinted maternally expressed transcript suppresses RB progression by targeting miR-17-92(11). Previous studies have reported that Nuclear paraspeckle assembly transcript 1 (NEAT1) plays an important role in different human cancer type progression (12). For example, NEAT1 promotes cell proliferation and invasion by regulating the miR-211/High mobility group AT-hook 2 axis in breast cancer (13). Moreover, NEAT1 promotes cell proliferation and invasion by targeting AKT/PI3K in cervical carcinoma (14). However, understanding of the role of NEAT1 in RB remains limited, and the potential effects and mechanisms of NEAT1 in RB require further investigation.
miRNAs are a group of small ncRNAs with 22-25 nts that exert their functions by binding to the 3'untranslated region (UTR) of their target gene (15). Previous studies have reported that miRNAs are aberrantly expressed in RB (16). Furthermore, miR-24 is downregulated in RB tissues compared with healthy retinas tissues (17). A recent study by Yu et al (18) reported that miR-24 suppresses cell proliferation, migration and invasion in RB. However, the mechanism underlying the role of miR-24-3p in the development of RB is not fully understood.
Leucine-rich-α-2-glycoprotein1 (LRG1) is a glycoprotein containing 312 amino acid residues (19). LRG1 has been reported to influence a variety of biological processes, including proliferation, apoptosis and invasion (20,21). In addition, previous studies have revealed that the dysregulation of LRG1 is associated with the progression of human cancer types, such as non-small cell lung cancer (22), bladder cancer (23) and ovarian cancer (24). Furthermore, Amer et al (25) demonstrated that LRG1 is elevated in RB, suggesting that LRG1 may be an important regulator of RB progression.
Therefore, the aim of the present study was to explore the function and underlying molecular mechanism of NEAT1 in RB through functional and mechanistic analysis.
Materials and methods
Patients and tissue samples
RB tissue samples and adjacent healthy retina tissue samples were collected from 20 patients with RB who underwent surgery at Renmin Hospital, Hubei University of Medicine (Shiyan, China) between October 2015 and March 2018. The patients were within the ages of 8 months to 9 years, including 9 girls and 11 boys. The patients were diagnosed as having RB by two experienced pathologists and none of the patients received any treatment before surgery. All tissue samples were stored in liquid nitrogen immediately after resection and kept at -80˚C until use. Written informed consent was obtained from every patient and the study was approved by the Ethics Committee of Renmin Hospital, Hubei University of Medicine.
Cell culture and transfection
Human RB cell lines (Y79 and WERI-RB1) and normal human retinal pigment epithelial cell line (ARPE-19) were obtained from the American Type Culture Collection. RB cell lines SO-Rb50 and HXO-Rb44 were obtained from the Cell Bank of Type Culture of Chinese Academy of Sciences. Y79, WERI-RB1, SO-Rb50 and HXO-Rb44 cells were cultured in RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.). ARPE-19 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.). All mediums were supplemented with 10% FBS (HyClone; GE Healthcare Life Sciences), 100 U/ml penicillin and 100 mg/ml streptomycin (HyClone; GE Healthcare Life Sciences). Cells were cultured at 37˚C in a humidified incubator containing 5% CO2.
Short hairpin RNA (shRNA) targeting NEAT1 (sh-NEAT1), shRNA targeting LRG1 (sh-LRG1) and their negative control (sh-NC), pcDNA3.1-NEAT1 overexpression vector (pcDNA-NEAT1), pcDNA3.1-LRG1 overexpression vector (pcDNA-LRG1) and their control pcDNA3.1 empty vector (pcDNA-NC) were purchased from Shanghai GenePharma Co., Ltd. miR-24-3p mimic (miR-24-3p; 5'-UGGCUCAGUUCAGCAGGAACAG-3' and 5'-GUUCCUGCUGAACUGAGCCAUU-3') and control mimic (miR-NC; 5'-UUCUCCGAACGUGUCACGUTT-3' and 5'-ACGUGACACGUUCGGAGAATT-3'), miR-24-3p inhibitor (anti-miR-24-3p; 5'-CUGUUCCUGCUGAACUGAGCCA-3') and its control (anti-miR-NC; 5'-CAGUACUUUUGUGUAGUACAA-3') were also purchased from Shanghai GenePharma Co., Ltd. Y79 and WERI-RB1 cells were seeded into 24-well plates at a density of 2.0x104 cells/well, following which 50 nM synthetic oligonucleotides or 2 µg vectors were transfected into the cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Cells were collected after 48 h for further experiments.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from tissues and cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. To analyze the expression of lncRNA and mRNA, the RNAs were reverse transcribed into cDNAs using PrimeScript™ RT reagent kit (Takara, Biotechnology Co., Ltd.) using the temperature protocol consisting of 50˚C for 15 min followed by 85˚C for 5 sec. To analyze the expression of miRNA, cDNAs were synthesized by All-in-One miRNA First strand cDNA Synthesis kit (GeneCopoeia, Inc.) using the temperature protocol consisting of 42˚C for 50 min followed by 70˚C for 15 min. qPCR was then performed using SYBR® Green qPCR Master Mix (Bio-Rad Laboratories, Inc.) with GAPDH (for lncRNA and mRNA) or U6 (for miRNA) as an endogenous control. The thermocycling conditions of the qPCR reaction were: Initial denaturation at 95˚C for 5 min, followed by 40 cycles of 95˚C for 30 sec and 60˚C for 45 sec. The relative expression levels of NEAT1, miR-24-3p and LRG1 were calculated via the 2-ΔΔCq method (26). The primers sequences were listed as follows: NEAT forward, 5'-GTGGCTGTTGGAGTCGGTAT-3' and reverse, 5'-TAACAAACCACGGTCCATGA-3'; miR-24-3p forward, 5'-GCCGAGTGGCTCAGTTCAGC-3' and reverse, 5'-CTCAACTGGTGTCGTGGA-3'; LRG1 forward, 5'-CCATGTCAGTGTGCAGATTC-3' and reverse, 5'-AAGAGTGAGAGGTGGAAGAG-3'; U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3'; and GAPDH forward, 5'-TCGGAGTCAACGGATTTGGT-3' and reverse, 5'-TTCCCGTTCTCAGCCTTGAC-3'.
Transwell assay
Cell migration and invasion abilities were evaluated using Transwell chambers (8-µm pore size; EMD Millipore). Cell suspension (2.0x105 cells/ml) was prepared in DMEM (Gibco; Thermo Fisher Scientific, Inc.) without serum. For the Transwell migration assay, ~500 µl cell suspension was seeded into the upper chamber and 300 µl DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone; GE Healthcare Life Science) was added into the lower chamber. After culturing for 24 h at 37˚C, cells that did not pass through the membrane were scraped using a cotton swab. Migrated cells that passed through the membrane were fixed in 90% ethyl alcohol for 15 min at room temperature, stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for 20 min at room temperature, observed and counted under a light microscope (Olympus Corporation) at x100 magnification.
For the Transwell invasion assay, the upper chamber was pre-coated with Matrigel (BD Biosciences) for 30 min at 37˚C, and the subsequent steps were the same as those in the cell migration assay.
Dual-luciferase reporter assay
The potential binding sites between NEAT1 and miR-24-3p, as well as miR-24-3p and LRG1 3'UTR were predicted using the online software starBase2.0 (http://starbase.sysu.edu.cn/index.php) and MiRcode 11 (http://mircode.org/), respectively. To assess the prediction, the sequences of NEAT1 and LRG1 3'UTR containing the potential wild-type (WT) or mutant (MUT) binding sites of miR-24-3p were amplified via PCR and cloned into pGL3-control luciferase reporter vectors (Promega Corporation), named as WT-NEAT1, MUT-NEAT1, LRG1 3'UTR-WT and LRG1 3' UTR-MUT. These procedures were conducted by Shanghai Sangon Co., Ltd. Y79 and WERI-RB1 cells (5.0x104 cells) were then seeded into 24-well plates before 50 nM miR-24-3p mimic or miR-NC and 100 ng WT-NEAT1, MUT-NEAT1, LRG1 3'UTR-WT or LRG1 3'UTR-MUT were co-transfected into Y79 and WERI-RB1 cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to manufacturer's instructions. After incubation for 48 h, firefly luciferase activity and Renilla luciferase activity were detected using a Dual Luciferase Reporter assay kit (Promega Corporation). Renilla luciferase activity was normalized to firefly luciferase activity.
Western blotting
Total protein was extracted from tissues and cells using RIPA lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.) and protein concentrations were measured using the bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology) according to the manufacturer's instructions. In total, 20 µg proteins were separated via 10% SDS-PAGE and transferred to PVDF membranes (EMD Millipore). The membranes were blocked in 5% non-fat milk for 1 h at room temperature. Subsequently, the membranes were immunoblotted with primary antibodies: E-cadherin (cat. no. ab231303; 1:2,000; Abcam), N-cadherin (cat. no. ab76011; 1:5,000; Abcam), matrix metalloproteinase (MMP)9 (cat. no. ab38898; 1:1,000; Abcam), LRG1 (cat. no. ab170953; 1:5,000; Abcam) or GAPDH (cat. no. ab9485; 1:2,500; Abcam) at 4˚C overnight. After the membranes were washed three times with TBS-1% Tween-20, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (cat. no. ab6721; 1:5,000; Abcam) for 1 h at room temperature. All antibodies used in the current study were purchased from Cell Signaling Technology, Inc.. The immunoreactive bands were detected using enhanced chemiluminescence (Pierce; Thermo Fisher Scientific, Inc.) and analyzed using ImageJ v1.8.0 software (National Institutes of Health). GAPDH was used as an internal reference.
Statistical analysis
All data in the present study were obtained from three independent experiments, analyzed by GraphPad Prism 7 software (GraphPad Software, Inc.) and are presented as the mean ± SD. Paired Student's t-test was used to analyze the difference between two groups, while one-way ANOVA followed by Sidak's test or Dunnett's test were used to analyze the differences between multiple groups. The correlations between NEAT1 and miR-24-3p, miR-24-3p and LRG1 mRNA expression levels were analyzed by Pearson's correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
NEAT1 is upregulated and miR-24-3p is downregulated in RB tissues and cell lines
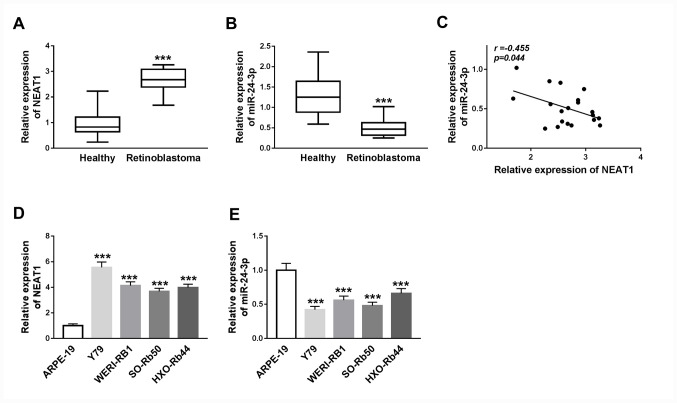
To investigate the potential roles of NEAT1 and miR-24-3p in RB progression, the expression levels of NEAT1 and miR-24-3p were detected in RB tissues via RT-qPCR. It was revealed that NEAT1 expression was significantly increased (P<0.001) and miR-24-3p expression was significantly decreased (P<0.001) in RB tissues compared with adjacent healthy tissues (Fig. 1A and B). Furthermore, Pearson's correlation analysis results suggested that the expression of miR-24-3p exhibited a moderate negatively correlation with the expression of NEAT1 in RB tissues (P<0.05; r=-0.455; Fig. 1C).
Figure 1.
NEAT1 expression is upregulated and miR-24-3p expression is downregulated in RB tissues and cells. (A) Expression levels of NEAT1 in RB tissues and adjacent healthy tissues were determined via RT-qPCR. ***P<0.001 vs. Healthy. (B) Expression levels of miR-24-3p in RB tissues and adjacent healthy tissues were determined via RT-qPCR. ***P<0.001 vs. Healthy. (C) Correlation between NEAT1 and miR-24-3p expression was analyzed using Pearson's correlation analysis. (D) NEAT1 expression in Y79, WERI-RB1, SO-Rb50, HXO-Rb44 and ARPE-19 cell lines was detected via RT-qPCR. (E) miR-24-3p expression in Y79, WERI-RB1, SO-Rb50, HXO-Rb44 and ARPE-19 cell lines was detected via RT-qPCR. ***P<0.001 vs. ARPE-19. RT-qPCR, reverse transcription-quantitative PCR; miR, microRNA; NEAT1, nuclear paraspeckle assembly transcript 1; RB, retinoblastoma.
The expression levels of NEAT1 and miR-24-3p were subsequently detected in RB cell lines (Y79, WERI-RB1, SO-Rb50 and HXO-Rb44) and the normal human retinal pigment epithelial cell line (ARPE-19) via RT-qPCR. NEAT1 expression was significantly upregulated, whilst miR-24-3p was significantly downregulated, in Y79, WERI-RB1, SO-Rb50 and HXO-Rb44 cells compared with that in ARPE-19 cells (all P<0.001; Fig. 1D and E). Thus, it was indicated that the dysregulation of NEAT1 and miR-24-3p may serve vital roles in RB progression.
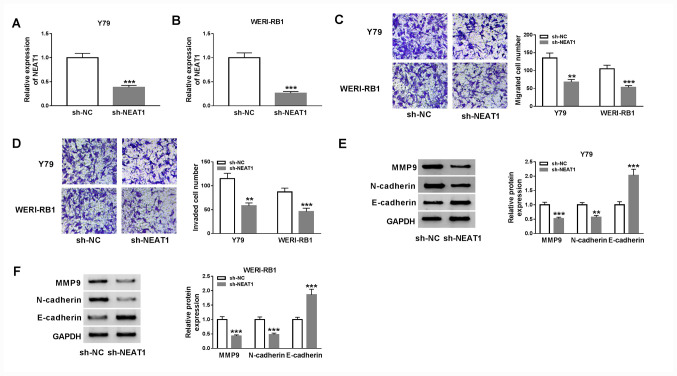
Subsequently, a Transwell assay was performed to examine cell migration and invasion. The results demonstrated that migration (Y79 cells, P<0.01; WERI-RB1 cells, P<0.001) and invasion (Y79 cells, P<0.01; WERI-RB1 cells, P<0.001) of Y79 and WERI-RB1 cells were significantly decreased after the knockdown of NEAT1 (Fig. 2C and D).
Figure 2.
NEAT1 knockdown significantly suppresses cell migration, invasion and EMT in RB cells. sh-NEAT1 or sh-NC were transfected into Y79 and WERI-RB1 cells. Expression of NEAT1 in (A) Y79 and (B) WERI-RB1 cells was examined via reverse transcription-quantitative PCR. (C) Migration and (D) invasion of Y79 and WERI-RB1 cells were evaluated via a Transwell assay. Protein expression levels of MMP9, N-cadherin and E-cadherin in (E) Y79 and (F) WERI-RB1 cells were detected via western blotting. **P<0.01 and ***P<0.001 vs. sh-NC. miR, microRNA; NEAT1, nuclear paraspeckle assembly transcript 1; RB, retinoblastoma; MMP, matrix metalloproteinase; sh, short hairpin RNA; NC, negative control; EMT, epithelial-to-mesenchymal transition.
It has been previously reported that the migratory and invasive phenotypes of tumor cells are gained via the process of epithelial-mesenchymal transition (EMT) (27,28). Therefore, the effects of NEAT1 on the protein expression levels of EMT markers, including MMP9, N-cadherin and E-cadherin, were examined via western blot analysis. The results indicated that NEAT1 knockdown resulted in a significant decrease in MMP9 (Y79 cells, P<0.001; WERI-RB1 cells, P<0.001) and N-cadherin (Y79 cells, P<0.01; WERI-RB1 cells, P<0.001) expression levels, and a significant increase in E-cadherin (Y79 cells, P<0.001; WERI-RB1 cells, P<0.001) expression in both Y79 and WERI-RB1 cells (Fig. 2E and F). Collectively, it was indicated that knockdown of NEAT1 may suppress cell migration, invasion and the EMT pathway in RB.
NEAT1 negatively regulates miR-24-3p expression via a direct interaction in RB cells
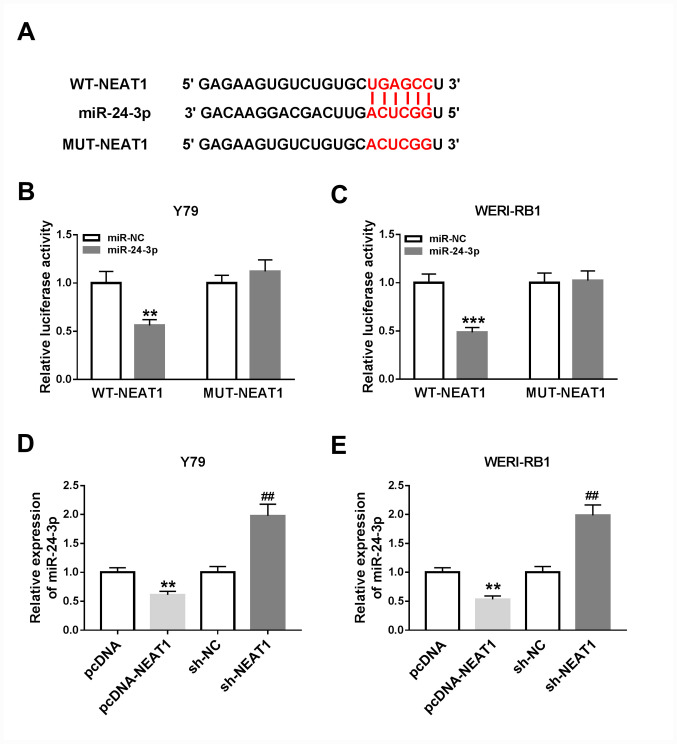
Based on the aberrant upregulation of NEAT1 and downregulation of miR-24-3p in RB cells, it was speculated that miR-24-3p may represent a target of NEAT1. According to the bioinformatics software starBase v2.0, NEAT1 was predicted to contain the putative binding sites of miR-24-3p (Fig. 3A), indicating that miR-24-3p may be targeted by NEAT1.
Figure 3.
NEAT1 directly targets miR-24-3p and negatively regulates miR-24-3p expression in RB cells. (A) Predicted binding sites between miR-24-3p and NEAT1. Luciferase activities in (B) Y79 and (C) WERI-RB1 cells co-transfected with miR-24-3p or miR-NC and WT-NEAT1 or MUT-NEAT1. **P<0.01 vs. miR-NC, ***P<0.001 vs. miR-NC. The expression level of miR-24-3p in (D) Y79 and (E) WERI-RB1 cells transfected with pcDNA, pcDNA-NEAT1, sh-NC or sh-NEAT1 was detected via reverse transcription-quantitative PCR. **P<0.01 vs. pcDNA, ##P<0.01 vs. sh-NC. miR, microRNA; NEAT1, nuclear paraspeckle assembly transcript 1; RB, retinoblastoma; sh, short hairpin RNA; NC, negative control; EMT, epithelial-to-mesenchymal transition; WT, wild-type; MUT, mutant.
To assess this prediction, a dual-luciferase reporter assay was conducted. It was revealed that the luciferase activities were significantly inhibited in Y79 and WERI-RB1 cells co-transfected with WT-NEAT1 and miR-24-3p, compared with cells co-transfected with WT-NEAT1 and miR-NC (Y79 cells, P<0.01; WERI-RB1 cells, P<0.001), while the luciferase activities were not affected in the MUT-NEAT1 groups (Fig. 3B and C).
It was demonstrated that pcDNA-NEAT1 transfection resulted in a significant increase in NEAT1 expression in Y79 and WERI-RB1 cells, suggesting that pcDNA-NEAT1 was successfully transfected (Fig. S1A). Subsequently, the expression of miR-24-3p in pcDNA-NEAT1-, pcDNA-, sh-NC- or sh-NEAT1-transfected Y79 and WERI-RB1 cells was detected via RT-qPCR. It was revealed that miR-24-3p expression was significantly decreased following NEAT1 overexpression and significantly increased following NEAT1 knockdown in both Y79 and WERI-RB1 cells (all P<0.01; Fig. 3D and E). Thus, it was speculated that NEAT1 negatively regulates miR-24-3p expression by directly interaction.
miR-24-3p inhibition restores the inhibitory effects of NEAT1 silencing on cell migration, invasion and the EMT process in RB cells
To further examine whether NEAT1 regulates RB cell migration, invasion and EMT, miR-24-3p mimic and anti-miR-24-3p were successfully transfected into Y79 and WERI-RB1 cells (Fig. S1B and C).
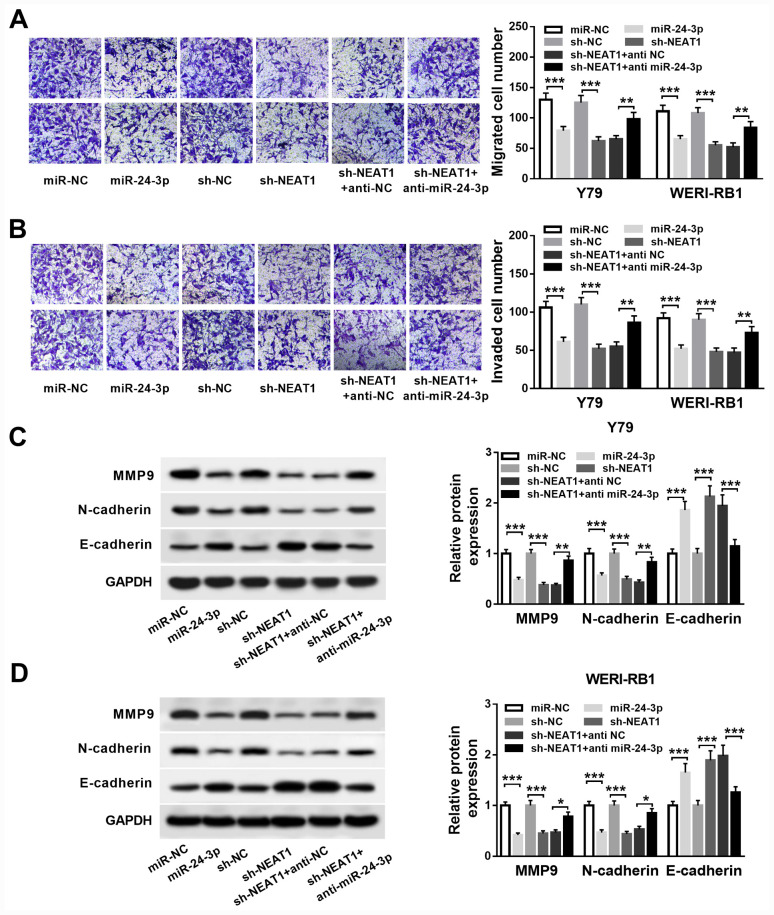
Next, Y79 and WERI-RB1 cells were transfected with either miR-24-3p, miR-NC, sh-NEAT1, sh-NC, sh-NEAT1 + anti-NC or sh-NEAT1 + anti-miR-24-3p. The Transwell assay results indicated that miR-24-3p overexpression or NEAT1 knockdown significantly suppressed migration and invasion abilities in Y79 and WERI-RB1 cells (all P<0.001). Moreover, inhibition of miR-24-3p could eliminate the inhibitory effects of NEAT1 knockdown on the migration and invasion of these cells (all P<0.01; Fig. 4A and B).
Figure 4.
NEAT1 represses RB cell migration, invasion and EMT via targeting miR-24-3p. Y79 and WERI-RB1 cells were treated with miR-24-3p, miR-NC, sh-NEAT1, sh-NC, sh-NEAT1 + anti NC or sh-NEAT1 + anti miR-24-3p. (A) Migrated and (B) invaded cell numbers of Y79 and WERI-RB1 cells were evaluated by Transwell assay. Protein expression levels of MMP9, N-cadherin and E-cadherin in (C) Y79 and (D) WERI-RB1 cells were detected by western blot analysis. *P<0.05, **P<0.01 and ***P<0.001. miR, microRNA; NEAT1, nuclear paraspeckle assembly transcript 1; RB, retinoblastoma; MMP, matrix metalloproteinase; sh, short hairpin RNA; NC, negative control; EMT, epithelial-to-mesenchymal transition.
Western blotting was then conducted to detect the protein expression levels of EMT markers MMP9, N-cadherin and E-cadherin. It was demonstrated that the expression levels of MMP9 and N-cadherin were significantly decreased (all P<0.001), while the expression of E-cadherin was significantly increased (all P<0.001) via miR-24-3p overexpression or NEAT1 knockdown in Y79 and WERI-RB1 cells. Furthermore, co-transfection with the miR-24-3p inhibitor reversed the effects of NEAT1 knockdown on MMP9 (Y79 cells, P<0.01; WERI-RB1 cells, P<0.05), N-cadherin (Y79 cells, P<0.01; WERI-RB1 cells, P<0.05) and E-cadherin (Y79 cells, P<0.001; WERI-RB1 cells: P<0.001) expression levels in Y79 and WERI-RB1 cells (Fig. 4C and D). Therefore, NEAT1 knockdown may suppress cell migration, invasion and the EMT process via targeting miR-24-3p in RB.
NEAT1 regulates LRG1 expression via sponging miR-24-3p in RB cells
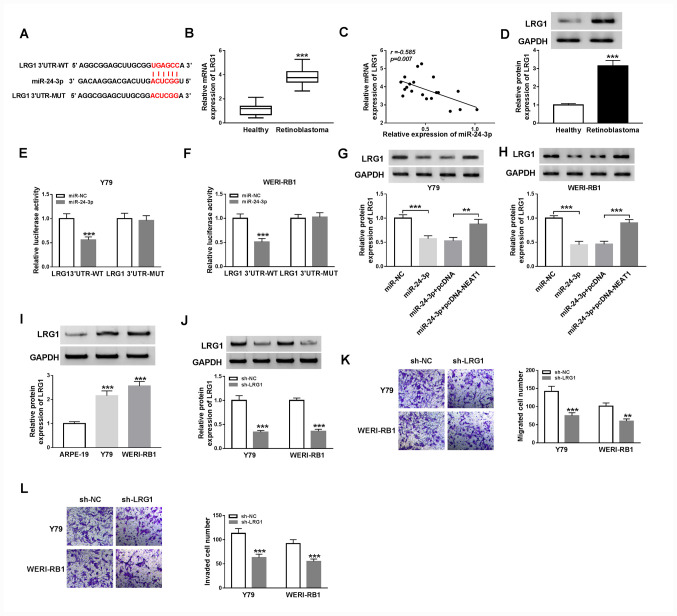
To investigate the potential mechanism of miR-24-3p in RB cell migration and invasion, an online software miRcode was used to predict the potential binding sites of miR-24-3p. LRG1 was predicted to be a target gene of miR-24-3p (Fig. 5A). Then, the mRNA expression level of LRG1 was detected via RT-qPCR and it was demonstrated that LRG1 mRNA was significantly upregulated in RB tissues compared with corresponding healthy tissues (P<0.001; Fig. 5B). Furthermore, there was an inverse correlation between LRG1 mRNA expression and miR-24-3p expression in RB tissues (P<0.01; r=-0.585; Fig. 5C). Western blotting results indicated that the protein expression level of LRG1 was also elevated in RB tissues compared with healthy tissues (P<0.001; Fig. 5D).
Figure 5.
NEAT1 upregulates LRG1 expression via miR-24-3p in RB cells. (A) Potential binding sites between miR-24-3p and the 3'UTR of LRG1. (B) mRNA expression levels of LRG1 in RB tissues and healthy tissues were determined via reverse transcription-quantitative PCR. ***P<0.001 vs. healthy. (C) Correlation between LRG1 and miR-24-3p was determined using Pearson's correlation analysis. (D) Protein expression levels of LRG1 in RB tissues and healthy tissues were detected via western blot analysis. ***P<0.001 vs. healthy. Luciferase activities were detected in (E) Y79 and (F) WERI-RB1 cells co-transfected with LRG1 3'UTR-WT or LRG1 3'UTR-MUT and miR-NC or miR-24-3p. ***P<0.001 vs. miR-NC. Protein expression levels of LRG1 in (G) Y79 and (H) WERI-RB1 cells treated with miR-24-3p, miR-24-3p + pcDNA-NEAT1 or the corresponding controls were measured via western blotting ***P<0.001 vs. miR-NC, **P<0.01 and ***P<0.001 vs. miR-24-3p+pcDNA. (I) Protein expression level of LRG1 in ARPE-19, Y79 and WERI-RB1 cells was detected via western blotting. ***P<0.001 vs. ARPE-19. (J) LRG1 protein expression in Y79 and WERI-RB1 cells transfected with sh-NC or sh-LRG1. ***P<0.001 vs. sh-NC. (K) Migratory and (L) invasive abilities of Y79 and WERI-RB1 cells were evaluated via Transwell assays. **P<0.01 and ***P<0.001 vs. sh-NC. miR, microRNA; NEAT1, nuclear paraspeckle assembly transcript 1; RB, retinoblastoma; sh, short hairpin RNA; NC, negative control; WT, wild-type; MUT, mutant; LRG1, leucine-rich α-2-glycoprotein; UTR, untranslated region.
To assess the targeting association between miR-24-3p and LRG1, a dual-luciferase reporter assay was performed by transfecting miR-24-3p or miR-NC and LRG1 3'UTR-WT or LRG1 3'UTR-MUT into Y79 and WERI-RB1 cells. The luciferase activities were significantly suppressed in Y79 and WERI-RB1 cells co-transfected with LRG1 3'UTR-WT and miR-24-3p compared with LRG1 3'UTR-WT and miR-NC (both P<0.001), while luciferase activities were not significantly affected in the LRG1 3'UTR-MUT groups (Fig. 5E and F).
Subsequently, the protein expression levels of LRG1 in Y79 and WERI-RB1 cells transfected with miR-24-3p, miR-24-3p + pcDNA-NEAT1 or the corresponding controls was examined via western blot analysis. It was demonstrated that miR-24-3p overexpression significantly suppressed the expression of LRG1 (both P<0.001), while NEAT1 overexpression reversed this effect (Y79 cells, P<0.05; WERI-RB1 cells, P<0.001; Fig. 5G and H). Moreover, the protein expression level of LRG1 was elevated in Y79 and WERI-RB1 cells compared with ARPE-19 cells (both P<0.001; Fig. 5I). sh-LRG1 was then transfected into Y79 and WERI-RB1 cells before transfection efficiency was evaluated by western blotting. sh-LRG1 transfection led to a significant reduction in LRG1 protein expression in Y79 and WERI-RB1 cells compared with that in the sh-NC groups (both P<0.001; Fig. 5J). In addition, it was discovered that knockdown of LRG1 caused a significant decrease in the migration (both P<0.001) and invasion (Y79 cells, P<0.01; WERI-RB1 cells, P<0.001) abilities of Y79 and WERI-RB1 cells (Fig. 5K and L). Thus, it was indicated that NEAT1 may positively regulate LRG1 expression via the targeting of miR-24-3p in RB cells.
NEAT1 modulates RB cell migration, invasion and EMT by upregulating LRG1 expression via sponging miR-24-3p
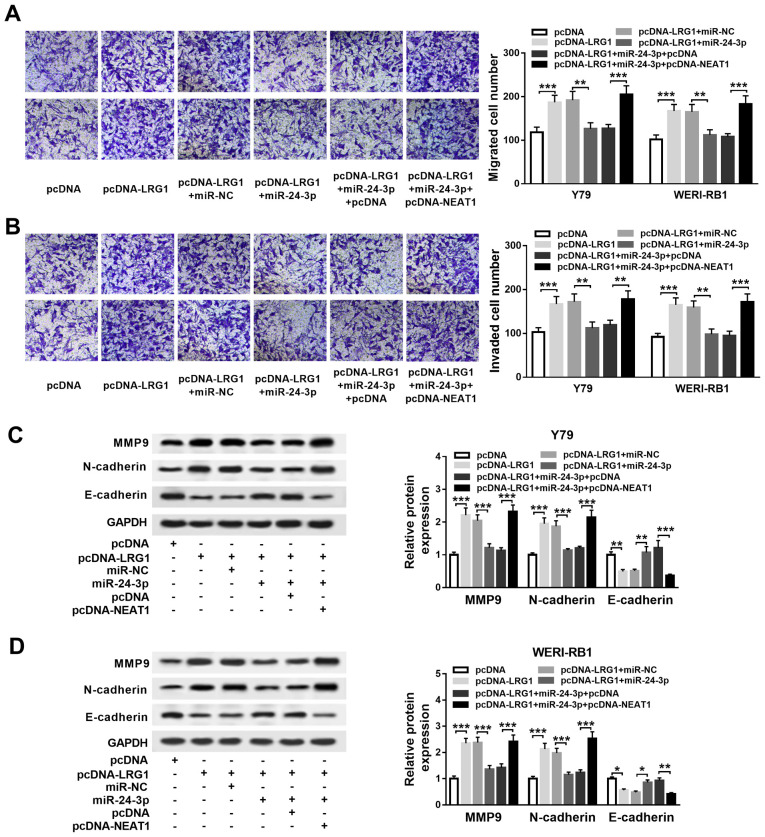
In order to examine the regulatory effects of NEAT1, miR-24-3p and LRG1 in RB progression, pcDNA-LRG1, pcDNA-LRG1 + miR-24-3p, pcDNA-LRG1 + miR-24-3p + pcDNA-NEAT1 or their matched controls were transfected into Y79 and WERI-RB1 cells. The transfection efficiency of pcDNA-LRG1 was determined via western blotting, and it was identified that the protein expression level of LRG1 was significantly increased in Y79 and WERI-RB1 cells, which demonstrated the success of transfection (Fig. S1D).
Transwell assay results suggested that the migratory (both P<0.001) and invasive (both P<0.001) abilities of Y79 and WERI-RB1 cells were significantly enhanced following pcDNA-LRG1 transfection, and the effects were reversed following miR-24-3p overexpression (all P<0.01). However, the effects of pcDNA-LRG1 + miR-24-3p on cell migration (both P<0.001) and invasion (Y79 cells, P<0.05; WERI-RB1 cells, P<0.05) were reversed by the overexpression of NEAT1 (Fig. 6A and B).
Figure 6.
NEAT1 affects RB cell migration, invasion and EMT by upregulating LRG1 expression via the sponging of miR-24-3p. Y79 and WERI-RB1 cells were divided into six groups and transfected with the following: pcDNA-LRG1, pcDNA, pcDNA-LRG1 + miR-24-3p, pcDNA-LRG1 + miR-NC, pcDNA-LRG1 + miR-24-3p + pcDNA-NEAT1 or pcDNA-LRG1 + miR-24-3p + pcDNA. (A) Migration and (B) invasion of Y79 and WERI-RB1 cells were evaluated via a Transwell assay. MMP9, N-cadherin and E-cadherin protein levels in (C) Y79 and (D) WERI-RB1 cells were determined via a western blot assay. *P<0.05, **P<0.01 and ***P<0.001. miR, microRNA; NEAT1, nuclear paraspeckle assembly transcript 1; RB, retinoblastoma; MMP, matrix metalloproteinase; sh, short hairpin RNA; NC, negative control; EMT, epithelial-to-mesenchymal transition; LRG1, leucine-rich α-2-glycoprotein.
The expression levels of MMP9, N-cadherin and E-cadherin were then determined via western blotting. It was indicated that MMP9 (both P<0.001)and N-cadherin expression levels were significantly elevated (both P<0.001), while E-cadherin expression was significantly reduced (Y79 cells, P<0.01; WERI-RB1 cells, P<0.05) in pcDNA-LRG1-treated Y79 and WERI-RB1 cells; these effects on MMP9 (both P<0.001), N-cadherin (both P<0.001) and E-cadherin (Y79 cells, P<0.01; WERI-RB1 cells, P<0.05) levels were restored in pcDNA-LRG1 + miR-24-3p-treated cells. Furthermore, NEAT1 overexpression decreased the impact of pcDNA-LRG1 + miR-24-3p on the expression levels of MMP9 (both P<0.001), N-cadherin (both P<0.001) and E-cadherin (Y79 cells, P<0.001; WERI-RB1 cells, P<0.01; Fig. 6C and D). Collectively, the results indicated that NEAT1 regulates cell migration, invasion and EMT via the miR-24-3p/LRG1 axis in RB.
Discussion
RB is an aggressive eye cancer associated with poor prognosis in infants and children (2). Previous studies have reported that lncRNAs serve important roles in RB progression (10,29,30). However, understanding of the role lncRNA NEAT1 in RB is limited. The present results suggest that NEAT1 is significantly upregulated in RB tissues and cells, compared with corresponding healthy tissues and cells. Furthermore, it was demonstrated that NEAT1 promotes RB cell migration, invasion and EMT by upregulating LRG1 expression via the sponging of miR-24-3p.
It has been revealed that the aberrant expression of NEAT1 regulates the development of different cancer types. For instance, Liu et al (31) reported that NEAT1 was significantly increased in renal cell carcinoma (RCC), and that NEAT1 knockdown suppressed RCC cell proliferation, migration and invasion. Moreover, Ji et al (32) reported that there is a high expression of NEAT1 in osteosarcoma and that NEAT1 overexpression accelerates osteosarcoma cell proliferation and inhibits cell apoptosis. Wang et al (33) revealed that there is a high expression of NEAT1 in RB tissues compared with healthy retinal tissues, and NEAT1 silencing represses RB cell proliferation, while inducing cell cycle arrest and apoptosis. In line with the findings from Wang et al (33), the present results indicate that NEAT1 expression is elevated in RB tissues and cell lines. Furthermore, two cell lines (Y79 and WERI-RB1) were selected to investigate the roles of NEAT1 in RB. It was demonstrated that NEAT1 knockdown significantly suppresses the migratory and invasive abilities of these cells. Thus, it was speculated that NEAT1 may function as an oncogene in RB.
Previous studies have demonstrated that lncRNAs exert their biological functions via serving as miRNA sponges (34). To the best of our knowledge, the present study was the first to identified that NEAT1 binds miR-24-3p in Y79 and WERI-RB1 cells. miR-24 is downregulated and function as a tumor suppressor in various cancer types, such as osteosarcoma (35), gastric (36) and bladder cancer (37). Moreover, miR-24 expression is decreased in RB and its overexpression represses RB cell proliferation, migration and invasion abilities (17,18). In line with these previous findings, the present results suggested that miR-24-3p expression was significantly decreased and negatively modulated by NEAT1 in RB. Furthermore, overexpression of miR-24-3p suppressed RB cell migration and invasion, and inhibition of miR-24-3p restored the inhibitory effects of NEAT1 knockdown on cell migration and invasion in Y79 and WERI-RB1 cells. Therefore, it was indicated that miR-24-3p serves as a tumor inhibitor in RB, and that NEAT1 regulates cell migration and invasion via sponging miR-24-3p in RB.
It has been revealed that LRG1 is aberrantly upregulated in several types of cancer, including RB (25). In addition, the present results were consistent with these previous findings, and demonstrated that LRG1 expression was significantly elevated in RB tissues and cells. It was also demonstrated that LRG1 is a target of miR-24-3p, and the mRNA expression level of LRG1 was negatively correlated with miR-24-3p expression. Furthermore, the roles of LRG1 in RB were examined and it was identified that LRG1 overexpression promoted the migration and invasion of Y79 and WERI-RB1 cells, while these effects were reversed by miR-24-3p overexpression.
The EMT process results in increased mobility and invasiveness of cancer cells, which is considered as a key early step in cancer progression and metastasis (38,39). Moreover, Li et al (40) reported that NEAT1 induces EMT in breast cancer. Lu et al (41) also reported that overexpression of NEAT1 is associated with the EMT process via the miR-204/zinc finger E-box binding homeobox 1 axis in nasopharyngeal carcinoma. The present results suggest that knockdown of NEAT1 may suppress MMP9 and N-cadherin expression, as well as promote E-cadherin expression; however, these effects were reversed by miR-24-3p knockdown. Furthermore, the effects of LRG1 overexpression on EMT markers was reversed by co-transfection with an miR-24-3p mimic, while the effects were further strengthened by the overexpression of NEAT1 in RB cells. Therefore, the present results indicated that NEAT1 facilitates the EMT pathway by upregulating LRG1 via sponging miR-24-3p in RB cells. However, the present study did not conduct in vivo experiments on animals, and thus these will be performed in future studies through transfecting sh-NEAT1 into nude mice and then monitoring the volume and weight of xenograft tumors.
In conclusion, the present results suggested that NEAT1 expression is elevated in RB. It was also demonstrated that NEAT1 knockdown represses RB progression via suppressing cell migration, invasion and the EMT process. Moreover, the present study established a novel regulatory network; the NEAT1/miR-24-3p/LRG1 axis, which was revealed to regulate RB progression via functional and mechanism analysis. Collectively, the findings indicated that NEAT1 may represent a potential therapeutic target for RB treatment.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LaL conceived and designed the study, and drafted the first draft of the manuscript. All experiments were completed by all authors. QH, YW, LuL and JL analyzed and collated the results. All authors reviewed and critiqued the manuscript, and agreed to the final submission of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from every patient and the study was approved by the Ethics Committee of Renmin Hospital, Hubei University of Medicine (Shiyan, Hubei).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dimaras H, Kimani K, Dimba EA, Gronsdahl P, White A, Chan HS, Gallie BL. Retinoblastoma. Lancet. 2012;379:1436–1446. doi: 10.1016/S0140-6736(11)61137-9. [DOI] [PubMed] [Google Scholar]
- 2.Kivelä T. The epidemiological challenge of the most frequent eye cancer: Retinoblastoma, an issue of birth and death. Br J Ophthalmol. 2009;93:1129–1131. doi: 10.1136/bjo.2008.150292. [DOI] [PubMed] [Google Scholar]
- 3.Cimino PJ, Robirds DH, Tripp SR, Pfeifer JD, Abel HJ, Duncavage EJ. Retinoblastoma gene mutations detected by whole exome sequencing of merkel cell carcinoma. Mod Pathol. 2014;27:1073–1087. doi: 10.1038/modpathol.2013.235. [DOI] [PubMed] [Google Scholar]
- 4.Khan AA, Bukhari MH, Mehboob R. Association of retinoblastoma with clinical and histopathological risk factors. Nat Sci. 2013;5:437–444. [Google Scholar]
- 5.Abramson DH, Shields CL, Munier FL, Chantada GL. Treatment of retinoblastoma in 2015: Agreement and disagreement. JAMA Ophthalmol. 2015;133:1341–1347. doi: 10.1001/jamaophthalmol.2015.3108. [DOI] [PubMed] [Google Scholar]
- 6.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan Q, Yang L, Zhang X, Peng X, Wei S, Su D, Zhai Z, Hua X, Li H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018;414:107–115. doi: 10.1016/j.canlet.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 8.Gutschner T, Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Peng XW. The silencing of long non-coding RNA ANRIL suppresses invasion, and promotes apoptosis of retinoblastoma cells through the ATM-E2F1 signaling pathway. Biosci Rep. 2018;38(BSR20180558) doi: 10.1042/BSR20180558. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Zhang H, Zhong J, Bian Z, Fang X, Peng Y, Hu Y. Long non-coding RNA CCAT1 promotes human retinoblastoma SO-RB50 and Y79 cells through negative regulation of miR-218-5p. Biomed Pharmacother. 2017;87:683–691. doi: 10.1016/j.biopha.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Zhang A, Shang W, Nie Q, Li T, Li S. Long non-coding RNA H19 suppresses retinoblastoma progression via counteracting miR-17-92 cluster. J Cell Biochem. 2018;119:3497–3509. doi: 10.1002/jcb.26521. [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Li Z, Zheng H, Chan MT, Wu WK. NEAT 1: A novel cancer-related long non-coding RNA. Cell Prolif. 2017;50(e12329) doi: 10.1111/cpr.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Wang S, Li Z, Long X, Guo Z, Zhang G, Zu J, Chen Y, Wen L. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int J Biol Macromol. 2017;105:346–353. doi: 10.1016/j.ijbiomac.2017.07.053. [DOI] [PubMed] [Google Scholar]
- 14.Guo H, Yang S, Zhao S, Li L, Yan M, Fan M. LncRNA NEAT1 regulates cervical carcinoma proliferation and invasion by targeting AKT/PI3K. Eur Rev Med Pharmacol Sci. 2018;22:4090–4097. doi: 10.26355/eurrev_201807_15400. [DOI] [PubMed] [Google Scholar]
- 15.Lü J, Qian J, Chen F, Tang X, Li C, Cardoso WV. Differential expression of components of the microRNA machinery during mouse organogenesis. Biochem Biophys Res Commun. 2005;334:319–323. doi: 10.1016/j.bbrc.2005.05.206. [DOI] [PubMed] [Google Scholar]
- 16.Zhao JJ, Yang J, Lin J, Yao N, Zhu Y, Zheng J, Xu J, Cheng JQ, Lin JY, Ma X. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst. 2009;25:13–20. doi: 10.1007/s00381-008-0701-x. [DOI] [PubMed] [Google Scholar]
- 17.To KH, Pajovic S, Gallie BL, Thériault BL. Regulation of p14ARF expression by miR-24: A potential mechanism compromising the p53 response during retinoblastoma development. BMC Cancer. 2012;12(69) doi: 10.1186/1471-2407-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu F, Pang G, Zhao G. ANRIL acts as onco-lncRNA by regulation of microRNA-24/c-Myc, MEK/ERK and Wnt/β-catenin pathway in retinoblastoma. Int J Biol Macromol. 2019;128:583–592. doi: 10.1016/j.ijbiomac.2019.01.157. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi N, Takahashi Y, Putnam FW. Periodicity of leucine and tandem repetition of a 24-amino acid segment in the primary structure of leucine-rich alpha 2-glycoprotein of human serum. Proc Natl Acad Sci USA. 1985;82:1906–1910. doi: 10.1073/pnas.82.7.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serada S, Fujimoto M, Terabe F, Iijima H, Shinzaki S, Matsuzaki S, Ohkawara T, Nezu R, Nakajima S, Kobayashi T, et al. Serum leucine-rich alpha-2 glycoprotein is a disease activity biomarker in ulcerative colitis. Inflamm Bowel Dis. 2012;18:2169–2179. doi: 10.1002/ibd.22936. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Abraham S, McKenzie JAG, Jeffs N, Swire M, Tripathi VB, Luhmann UFO, Lange CAK, Zhai Z, Arthur HM, et al. LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature. 2013;499:306–311. doi: 10.1038/nature12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakajima M, Miyajima M, Ogino I, Watanabe M, Miyata H, Karagiozov KL, Arai H, Hagiwara Y, Segawa T, Kobayashi K, Hashimoto Y. Leucine-rich α-2-glycoprotein is a marker for idiopathic normal pressure hydrocephalus. Acta Neurochir (Wien) 2011;153:1339–1346. doi: 10.1007/s00701-011-0963-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindén M, Lind SB, Mayrhofer C, Segersten U, Wester K, Lyutvinskiy Y, Zubarev R, Malmström PU, Pettersson U. Proteomic analysis of urinary biomarker candidates for nonmuscle invasive bladder cancer. Proteomics. 2012;12:135–144. doi: 10.1002/pmic.201000810. [DOI] [PubMed] [Google Scholar]
- 24.Andersen JD, Boylan KL, Jemmerson R, Geller MA, Misemer B, Harrington KM, Weivoda S, Witthuhn BA, Argenta P, Vogel RI, Skubitz AP. Leucine-rich alpha-2-glycoprotein-1 is upregulated in sera and tumors of ovarian cancer patients. J Ovarian Res. 2010;3(21) doi: 10.1186/1757-2215-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amer R, Tiosano L, Pe'er J. Leucine-rich α-2-glycoprotein-1 (LRG-1) expression in retinoblastoma. Invest Ophthalmol Vis Sci. 2018;59:685–692. doi: 10.1167/iovs.17-22785. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Wang JX, Yang Y, Li K. Long noncoding RNA DANCR aggravates retinoblastoma through miR-34c and miR-613 by targeting MMP-9. J Cell Physiol. 2018;233:6986–6995. doi: 10.1002/jcp.26621. [DOI] [PubMed] [Google Scholar]
- 30.Hu C, Liu S, Han M, Wang Y, Xu C. Knockdown of lncRNA XIST inhibits retinoblastoma progression by modulating the miR-124/STAT3 axis. Biomed Pharmacother. 2018;107:547–554. doi: 10.1016/j.biopha.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Liu F, Chen N, Gong Y, Xiao R, Wang W, Pan Z. The long non-coding RNA NEAT1 enhances epithelial-to-mesenchymal transition and chemoresistance via the miR-34a/c-Met axis in renal cell carcinoma. Oncotarget. 2017;8:62927–62938. doi: 10.18632/oncotarget.17757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji S, Wang S, Zhao X, Lv L. Long noncoding RNA NEAT1 regulates the development of osteosarcoma through sponging miR-34a-5p to mediate HOXA13 expression as a competitive endogenous RNA. Mol Genet Genomic Med. 2019;7(e673) doi: 10.1002/mgg3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Yang D, Tian R, Zhang H. NEAT1 promotes retinoblastoma progression via modulating miR-124. J Cell Biochem. 2019;120:15585–15593. doi: 10.1002/jcb.28825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P, Ning S, Zhang Y, Li R, Ye J, Zhao Z, Zhi H, Wang T, Guo Z, Li X. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res. 2015;43:3478–3489. doi: 10.1093/nar/gkv233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song L, Yang J, Duan P, Xu J, Luo X, Luo F, Zhang Z, Hou T, Liu B, Zhou Q. MicroRNA-24 inhibits osteosarcoma cell proliferation both in vitro and in vivo by targeting LPAATβ. Arch Biochem Biophys. 2013;535:128–135. doi: 10.1016/j.abb.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Duan Y, Hu L, Liu B, Yu B, Li J, Yan M, Yu Y, Li C, Su L, Zhu Z, et al. Tumor suppressor miR-24 restrains gastric cancer progression by downregulating RegIV. Mol Cancer. 2014;13(127) doi: 10.1186/1476-4598-13-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S, Zhang C, Liu W, Zheng W, Zhang Y, Wang S, Huang D, Liu X, Bai Z. MicroRNA-24 upregulation inhibits proliferation, metastasis and induces apoptosis in bladder cancer cells by targeting CARMA3. Int J Oncol. 2015;47:1351–1360. doi: 10.3892/ijo.2015.3117. [DOI] [PubMed] [Google Scholar]
- 38.Bates RC, Mercurio AM. The epithelial-mesenchymal tansition (EMT) and colorectal cancer progression. Cancer Biol Ther. 2005;4:371–376. doi: 10.4161/cbt.4.4.1655. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Li W. Epithelial-mesenchymal transition in human cancer: Comprehensive reprogramming of metabolism, epigenetics, and differentiation. Pharmacol Ther. 2015;150:33–46. doi: 10.1016/j.pharmthera.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Wang S, Li Z, Long X, Guo Z, Zhang G, Zu J, Chen Y, Wen L. NEAT1 induces epithelial-mesenchymal transition and 5-FU resistance through the miR-129/ZEB2 axis in breast cancer. FEBS Lett. 2017;591(570) doi: 10.1002/1873-3468.12474. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y, Li T, Wei G, Liu L, Chen Q, Xu L, Zhang K, Zeng D, Liao R. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016;37:11733–11741. doi: 10.1007/s13277-015-4773-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.