Abstract
Background
Long non-coding RNAs (lncRNA) are reported to influence colorectal cancer (CRC) progression. Currently, the functions of the lncRNA ZNF561 antisense RNA 1 (ZNF561-AS1) in CRC are unknown.
Methods
ZNF561-AS1 and SRSF6 expression in CRC patient samples and CRC cell lines was evaluated through TCGA database analysis, western blot along with real-time PCR. SRSF6 expression in CRC cells was also examined upon ZNF561-AS1 depletion or overexpression. Interaction between miR-26a-3p, miR-128-5p, ZNF561-AS1, and SRSF6 was examined by dual luciferase reporter assay, as well as RNA binding protein immunoprecipitation (RIP) assay. Small interfering RNA (siRNA) mediated knockdown experiments were performed to assess the role of ZNF561-AS1 and SRSF6 in the proliferative actives and apoptosis rate of CRC cells. A mouse xenograft model was employed to assess tumor growth upon ZNF561-AS1 knockdown and SRSF6 rescue.
Results
We find that ZNF561-AS1 and SRSF6 were upregulated in CRC patient tissues. ZNF561-AS1 expression was reduced in tissues from treated CRC patients but upregulated in CRC tissues from relapsed patients. SRSF6 expression was suppressed and enhanced by ZNF561-AS1 depletion and overexpression, respectively. Mechanistically, ZNF561-AS1 regulated SRSF6 expression by sponging miR-26a-3p and miR-128-5p. ZNF561-AS1-miR-26a-3p/miR-128-5p-SRSF6 axis was required for CRC proliferation and survival. ZNF561-AS1 knockdown suppressed CRC cell proliferation and triggered apoptosis. ZNF561-AS1 depletion suppressed the growth of tumors in a model of a nude mouse xenograft. Similar observations were made upon SRSF6 depletion. SRSF6 overexpression reversed the inhibitory activities of ZNF561-AS1 in vivo, as well as in vitro.
Conclusion
In summary, we find that ZNF561-AS1 promotes CRC progression via the miR-26a-3p/miR-128-5p-SRSF6 axis. This study reveals new perspectives into the role of ZNF561-AS1 in CRC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13046-021-01882-1.
Keywords: Colorectal cancer, ZNF561-AS1, SRSF6, ceRNA
Background
Colorectal cancer (CRC) ranks 3rd among the most frequent cancers globally [1]. Despite remarkable advances, the molecular basis of CRC progression is not fully understood and more effective therapeutic strategies are needed [2].
Long non-coding RNAs (lncRNAs) are composed of > 200 nucleotides reported to influence various disorders [3–5], including cancers [6–8], by adsorbing miRNAs [9], mediating DNA interactions [10], and binding to proteins as decoys [11]. Many lncRNAs are show abnormal expression in CRC which affects its progression and drug resistance [12–14]. For instance, lncRNA GLCC1 promotes CRC tumorigenesis by stabilizing c-Myc [15]. By regulating the activities of PKM2, actions of FEZF1-AS1 leads to accelerated cell proliferation and metastasis [16]. Exosomal lncRNA H19 confers CRC cells stemness and chemo-resistance [17]. Since lncRNAs influence CRC progression, their identification and understanding of their biological functions in CRC are imperative as they have anti-CRC therapeutic value.
ZNF561-AS1 is a novel lncRNA whose function in CRC is unknown. Here, we find that ZNF561-AS1 overexpressed in CRC samples and this promotes the proliferative rates and survival of CRC cells in vivo, as well as in vitro. Mechanically, ZNF561-AS1 upregulates SRSF6 levels by sponging miR-26a-3p and miR-128-5p. Additionally, reduced SRSF6 levels upon ZNF561-AS1 depletion were restored by miR-26a-3p or/and miR-128-5p inhibition. Exogenous SRSF6 expression rescued proliferation of ZNF561-AS1-depleted CRC cells. The present results underscores the potential of ZNF561-AS1 to be targeted as an anti-CRC target.
Materials and methods
Clinical tissues
Six human colorectal cancer tissues along with matched adjacent normal tissues were collected from the Second Affiliated Hospital of Harbin Medical University with patients’ consents. The ethics committee of Harbin Medical University approved the study. Matched pre-treated, post-treated and recurrent samples from one patient, in total six patients were collected. The samples obtained at surgery refers to pre-treated samples. Post-treatment samples were collected after finishing chemotherapy by colonoscope. In 1–2 years after chemotherapy, the patients were suffered from relapsed tumors which refer to recurrent samples. Upon collection, tissues were snap-frozen in liquid nitrogen for further use.
Patients information is shown on Supplementary Table S1.
Cell culture
The following human CRC cell lines were used in the experiments: HCT-116, HT-29, SW620, LoVo, SW480, and SW48, all obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). These cells were inoculated in Dulbecco’s modified Eagle medium (DMEM, Invitrogen, Carlsbad, CA, USA), containing 10% fetal bovine serum (FBS; Invitrogen), 50 μg/mL streptomycin and 50 U/mL penicillin (Invitrogen). CCD841 CoN, a normal human colonic epithelial cell line, was supplied by the American Type Culture Collection (ATCC, Manassas, VA, USA) and grown in Eagle’s Minimum Essential Medium (EMEM, Lonza, Basel, Switzerland), supplemented with 10% FBS. All cells were maintained in an incubator with humidified conditions at 37 °C, 5% CO2.
Cell transfection
ZNF561-AS1 siRNA, as well as negative control siRNA were constructed by GenePharma, Shanghai, China. MiR-26a-5p and miR-128-3p mimics, inhibitor NC, mimics NC, miR-26a-5p and miR-128-3p inhibitors and were purchased from Ambion (Austin, TX, USA). ZNF561-AS1 and SRSF6 expression vectors were purchased from GeneCopoeia, Guangzhou, China. To transfect the cells with vectors, HCT-116 cells were first cultured in 6 cm dishes for 24 h. The cells were transfected using lipofectamine 2000 for 5 h, and then the transfection media was replaced with complete media. The transfected cells were harvested at 48 h after transfection.
Measurement of mRNA expression of ZNF561-AS1
Total RNA for Real-time PCR was isolated by lysing the cells with TRIzol (Invitrogen,). cDNA was generated from the RNA using reverse transcription kit. Real time PCR for ZNF561-AS1 expression was performed following routine procedures using the Power SYBR Green PCR master Mix (Thermo Fisher Scientific, Waltham, USA). Relative mRNA expression was determined using the 2-ΔΔCt method. Experiments were run in triplicate and average values were obtained. The primers used to measure ZNF561-AS1 expression were:
ZNF561-AS1-F: 5′- ACCAAGACCTCCCACAACTCTCC -3′;
ZNF561-AS1-R: 5′- CAGGATCTGGCTTCACTGCTCTTC -3′.
GAPDH-F: 5′- CTGGGCTACACTGAGCACC -3′;
GAPDH-R: 5′- AAGTGGTCGTTGAGGGCAATG -3′.
GAPDH was used as reference gene.
To assess miR-26a-5p and miR-128-3p expression levels, 1 μg of total RNA was retrotranscribed using miR-26a-5p or miR-128-3p specific stem-loop primers (Genepharma, Shanghai, China). RT-qPCR analysis of miR-26a-5p or miR-128-3p was done with the Power SYBR Green PCR master Mix. The stem loop primers and PCR primers for miR-26a-5p or miR-128-3p used in the reverse transcription and qPCR were:
miR-26a-5p specific stem loop primer: GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAGCCTATC.
miR-26a-5p-F: 5′- GGCTTCAAGTAATCCAGG − 3′.
miR-26a-5p-R: 5′- ATTGCGTGTCGTGGAGTCG − 3′.
miR-128-3p specific stem loop primer: GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTTTCTCTG.
miR-128-3p-F: 5′- GGGTCACAGTGAACCGGTC − 3′.
miR-128-3p-R: 5′- ATTGCGTGTCGTGGAGTCG -3′.
U6-F: 5′- GCTTCGGCAGCACATATACTAAAAT -3′.
U6-R: 5′- CGCTTCACGAATTTGCGTGTCAT − 3′.
U6 was served for normalization.
Western blot analysis
A RIPA buffer supplemented with protease inhibitors cocktail (Roche, Switzerland) was used to extract protein samples from cells. Equal amounts of proteins were resolved on SDS-PAGE and electro-transferred to PVDF membranes (Thermo Scientific, USA) and then blocked with 5% skim mile. After blocking, the blots were probed with following primary antibodies to SRSF6 (1:1000, Abcam, Cambridge, MA, USA), PCNA (1:1000), Cyclin D1 (1:1000), CDK4 (1:1000), cleaved Caspase-3 (1:1000) and p21 (1:1000) (Cell signaling technology, USA) and Actin (1:1000, Santa Cruz, USA). After washing and inoculating with HRP-conjugated rabbit or mouse, secondary antibodies (1:5000, Cell signaling Technology), signal was then developed using ECL reagent (GE healthcare, USA).
Cell viability assays
ZNF561-AS1 siRNA or control siRNA transfected HCT-116 cells were seeded in 96-well plates with 4 × 103 cells in each well for 4 days. The viability of cells was evaluated using a Cell Counting Kit-8 (CCK-8, Dojindo, Japan) at day 0, 2, and 4.
Colony formation assays
800 HCT-116 cells transfected with ZNF561-AS1 siRNA or control siRNA were counted and seeded into 6 cm dishes and cultured for 14 days. The cells were stained with 0.1% crystal violet in 20% methanol for 20 min, and the number of colonies formed were photographed and counted.
Immunofluorescence staining
HCT-116 cells transfected with ZNF561-AS1 siRNA or control siRNA respectively were seeded onto coverslips. 48 h after transfection, cells were fixed with 4% PFA at room temperature. This was followed by incubation with mouse anti-Ki-67 primary antibody (1:100, Cell Signaling Technology, USA) at room temperature for 1 h. Then, cells were washed with PBS and incubated at room temperature for 20 min with of anti-mouse Alexa-488 secondary antibody (1:1000, Cell Signaling Technology, USA) and counterstained with DAPI. Stained coverslips were mounted using prolong® diamond antifade mountant (Applied Biosystems, USA). Images were captured using Zeiss Axiovert 200 microscope.
Acridine orange/ethidium bromide (AO/EB) fluorescence staining
After transfection with ZNF561-AS1 siRNA or control siRNA for 48 h, HCT-116 cells were treated with AO/EB reagent (Solarbio Biotechnology, China) for 5 min. The morphology of cells after treatment was imaged with a fluorescence microscope at 200X magnification. The percentage of cell apoptosis was determined as follows: apoptotic rate(%) = number of apoptotic cells/number of all cells counted.
TUNEL assay
Click-iT TUNEL Alexa Fluor Imaging Assay Kit (Invitrogen, USA) was employed to explore cells apoptosis. In brief, control siRNA or ZNF561-AS1 siRNA transfected HCT-116 cells were fixed with 4% PFA for 15 min and permeabilized with 0.25% Triton X-100 for 10 min at room temperature. Then cells were washed and incubated with TdT reaction buffer for 10 min and incubated with Click-iT reaction cocktail for 30 min at room temperature. Cell nuclei were counterstained with DAPI after washing with PBS. Stained coverslips were mounted using prolong® diamond antifade mountant (Applied Biosystems, USA). Cells were imaged using a Zeiss Axiovert 200 microscope.
Nude mice experiments
Athymic BALB/c nude mice (6-weeks old) were purchased from Vital River Laboratory, Beijing, China to establish tumor xenograft models. Mice were maintained and housed under specific pathogen-free conditions and handled using aseptic procedures.
HCT-116 cells (5 × 106) transfected with ZNF561-AS1 siRNA or control siRNA were then subcutaneously administered into the right shank of the mice (3 mice/group). Tumor size (length; L, and width; W) determined with a caliper every 7 days and tumor volume given by the formula: ½LW2. Three weeks later, mice were killed. Isolated xenograft tumors were weighed and subjected to the further experiments.
Immunohistochemistry staining
Harvested tumors were fixed in 4% PFA for 24 h at room temperature, followed by permeabilization with PBST for 20 min and inactivation of endogenous peroxidases by incubating tissues in 0.3% H2O2 for 20 min. After washing, tissues were blocked with 5% normal goat serum (Invitrogen, USA) for 30 min and incubated with anti-Ki-67 antibody (Cell Signaling Technology, USA) overnight at 4 °C. Subsequently, tissues were rinsed and probed with a biotin-labeled secondary antibody for 30 min and washed. After washing, the specimens were developed with 0.05% DAB (Sigma-Aldrich, Oakville, ON, Canada) and 0.03% H2O2, and counterstained with hematoxylin, dehydrated in increasing ethanol concentrations, cleared with xylene. Images of stained specimens were captured using an Olympus BX51 microscope. Digital images were analyzed using Image-Pro Plus 6.0 software.
RNA binding protein immunoprecipitation (RIP)
RIP assay was done using the EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) as described by the manufacturer protocol. Briefly, HCT-116 cells were lysed in complete RIP lysis buffer. 100 μl whole cell lysate from each group were incubated with RIP buffer containing magnetic beads conjugated to mouse anti-Ago2 antibody (Millipore, USA), or negative control normal mouse IgG (Millipore, USA). Next, proteins were digest by incubated with proteinase K and immunoprecipitated RNA was harvested. The RNA concentration was quantified on NanoDrop spectrophotometer (Thermo Scientific, USA). Purified RNA was reverse transcribed into cDNA and subjected to qPCR to examine the presence of ZNF561-AS1, miR-26a-3p, and miR-128-5p using indicated primers.
Dual luciferase reporter assay
MiR-26a-3p and miR-128-5p binding sites on ZNF561-AS1 and SRSF6 were searched on bioinformatics platforms. Wild-type binding site and flanking sequences (~ 300 bp) were amplified and cloned into SpeI and HindIII sites of the pMIR-REPORT Luciferase vector (Ambion, Austin, TX, USA) and named ZNF561-AS1-WT and SRSF6-WT. To generate mutant miR-26a-3p and/or miR-128-5p binding sites in the luciferase vector, several point mutations were introduced into ZNF561-AS1-WT and SRSF6-WT using Phusion Site-Directed Mutagenesis Kit (Thermo Fisher Scientific, USA), and named ZNF561-AS1-26aMut or ZNF561-AS1-128Mut, and SRSF6-26aMut or SRSF6-128Mut (single mutant) or ZNF561-AS1-26a/128Mut or SRSF6-26a/128Mut (double mutant). To determine if ZNF561-AS1 and SRSF6 are direct miR-26a-3p and miR-128-3p targets, HCT-116 cells were seeded in 6-well plates and co-transfected with the indicated vectors using the Invitrogen Lipofectamine 2000 (Invitrogen, USA) for 48 h. Luciferase activity was analyzed using Promega Dual Luciferase-reporter 1000 assay system (Promega, Madison, WI, USA) and normalized to Renilla luciferase activity.
Data analysis
For biological experiments, data were obtained from at least 3 independent experiments and presented as the mean ± SD. The data were subjected to F-test, and then subjected to two-tailed Student’s t test (comparison between two groups) or ANOVA (Comparisons among three groups or more) using Statistical Program for Social Sciences (SPSS) version 17.0 (Chicago, IL, USA). GEPIA web server was utilized to analyze the data of expression of ZNF561-AS1 and SRSF6 in CRC tumor and adjacent normal tissues from TCGA database. The data were subjected to ANOVA analysis for differentially expressed genes based on the comparison of tumors and matched normal samples, and Pearson correlation analysis for correlation between ZNF561-AS1 and SRSF6.
Results
Overexpression of ZNF561-AS1 in CRC positively regulates SRSF6 expression
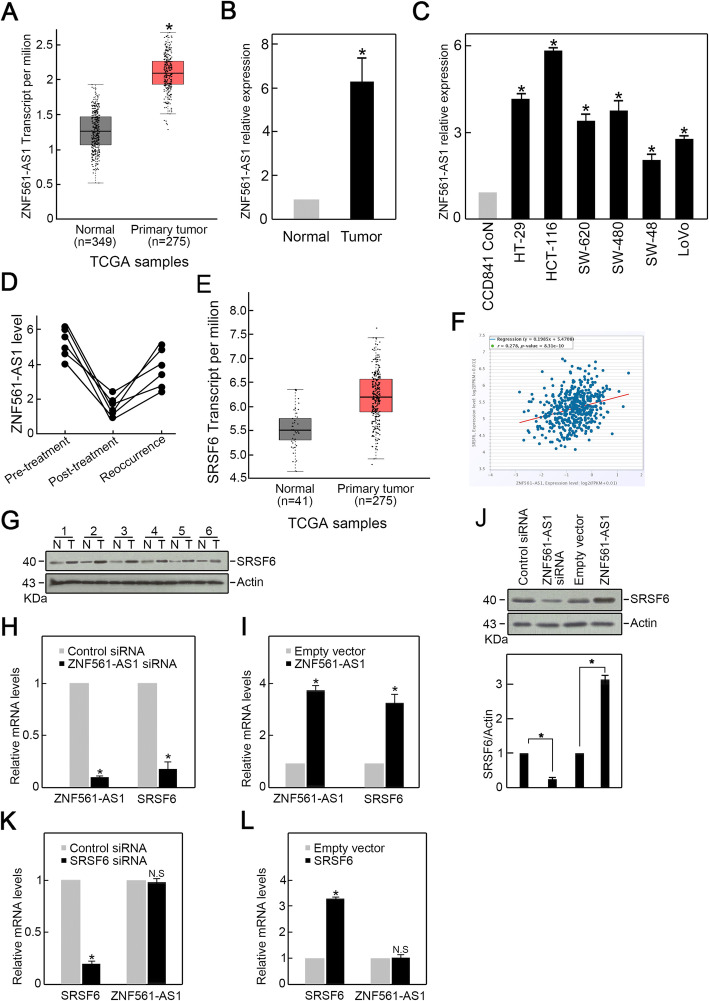
To examine the ZNF561-AS1 expression in CRC, analysis of TCGA CRC datasets revealed an elevated ZNF561-AS1 levels in CRC patients (Fig. 1a). Consistently, significantly higher ZNF561-AS1 level was found in our collected CRC patient samples compared to their matched adjacent normal tissue samples (Fig. 1b). Analysis of ZNF561-AS1 level in various CRC cell lines consisting of SW620, HCT-116, HT-29, SW480, SW48 and LoVo cells showed that ZNF561-AS1 expression was increased in all CRC cell lines compared to non-malignant human colon epithelial cell line, CCD841 CoN (Fig. 1c), with HCT-116 expressing the highest ZNF561-AS1 levels. Moreover, we found that compared to CRC samples from untreated patients, samples from patients treated with chemotherapy exhibited lower ZNF561-AS1 levels (Fig. 1d). However, ZNF561-AS1 rose in recurrent CRC samples (Fig. 1d). Together, these data suggest a critical role for ZNF561-AS1 in CRC.
Fig. 1.
Elevated ZNF561-AS1 positively regulates SRSF6 expression in CRC. a ZNF561-AS1 expression in CRC patients. Data from TCGA data resource. *p < 0.05. b ZNF561-AS1 expression is elevated in six patient CRC samples. *p < 0.05. c ZNF561-AS1 level is elevated in various CRC cell lines relative to CCD841 CoN normal human colonic epithelial cell line. n = 3, *p < 0.05. d ZNF561-AS1 expression was reduced by chemotherapy but restored in recurrent CRC samples. e SRSF6 expression in CRC patients. Data from TCGA data resource. f Positive correlation between ZNF561-AS1 and SRSF6 in CRC patients. Data from TCGA data resource. g Increased SRSF6 expression was observed in CRC patients tissue samples. h ZNF561-AS1 silencing suppressed SRSF6 mRNA levels in HCT-116 cells. n = 3, *p < 0.05. i ZNF561-AS1 overexpression elevated SRSF6 mRNA levels in HCT-116 cells. n = 3, *p < 0.05. j ZNF561-AS1 overexpression enhanced SRSF6 proteins level in HCT-116 cells. Actin was employed as loading control. Representative blots are shown (left panel). Ratios of level of SRSF6 vs Actin were computed after densitometric analysis of blot images on Image J (right panel). Data were from 3 independent experiments. k SRSF6 knockdown did not alter ZNF561-AS1 expression. n = 3, *p < 0.05. l SRSF6 overexpression did not alter ZNF561-AS1 expression. n = 3, *p < 0.05
Surprisingly, TCGA CRC dataset analysis found that Serine and arginine-rich splicing factor 6SRSF6 (SRSF6), an oncogenic factor, was also elevated in CRC and showed a positive association with ZNF561-AS1 expression (Fig. 1e-f). Elevated SRSF6 levels were also detected in CRC patients samples (Fig. 1g). To assess the correlation between ZNF561-AS1 and SRSF6 expression in CRC, we used siRNA to silence ZNF561-AS1 or SRSF6 in HCT-116 and then evaluated SRSF6 or ZNF561-AS1 expression. Interestingly, we found that compared to control group, ZNF561-AS1 silencing correlated with reduced SRSF6 mRNA and protein levels (Fig. 1 h, j). While ZNF561-AS1 overexpression correlated with increased SRSF6 mRNA and protein level (Fig. 1 i, j). However, SRSF6 knockdown and overexpression did not affect ZNF561-AS1 expression (Fig. 1 k-l). These data suggested that ZNF561-AS1 was an upstream positive regulator of SRSF6 in CRC cells.
ZNF561-AS1 sponges miR-26a-5p and miR-128-3p to promote SRSF6 expression
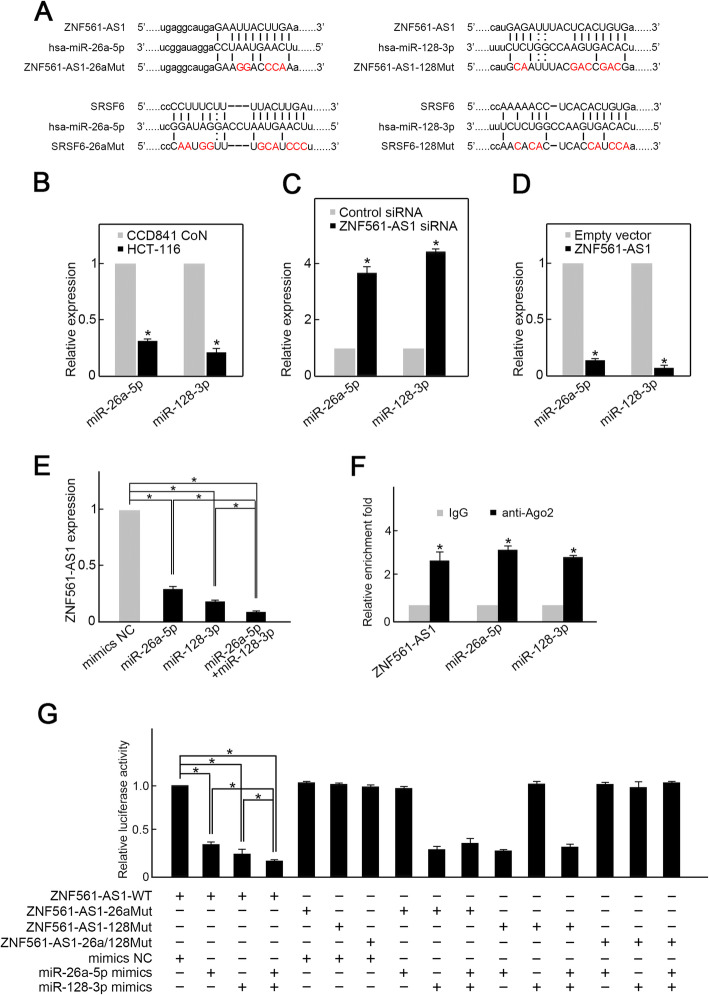
LncRNAs modulate gene transcription by functioning as miRNA sponges [18]. Using Starbase and Targetscan bioinformatics analysis, we surprisingly found that ZNF561-AS1 and SRSF6 bear potential binding sites for miR-26a-5p, as well as miR-128-3p (Fig. 2a). Thus, we hypothesize that ZNF561-AS1 might enhance SRSF6 expression by acting as a sponge of miR-26a-5p and miR-128-3p. To test this possibility, we first examined miR-26a-5p and miR-128-3p expression in HCT-116 cells and demonstrated that the levels of miR-128-3p and miR-26a-5p were remarkably reduced in HCT-116 cells in comparison of CDD841 CoN cells (Fig. 2b). Moreover, ZNF561-AS1 silencing upregulated levels of miR-26a-5p and miR-128-3p, while its overexpression reduced levels of miR-26a-5p and miR-128-3p (Fig. 2 c-d). In contrast, miR-26a-5p mimics and/or miR-128-3p mimics suppressed ZNF561-AS1 expression (Fig. 2e). These observations indicating a reciprocal regulation between ZNF561-AS1 and miR-26a-5p/miR-128-3p. Next, we used RIP analysis to determine whether ZNF561-AS1 and miR-26a-5p/miR-128-3p occupy the same RISC complex. To this end, HCT-116 lysates were immunoprecipitated using anti-Ago2 antibody and enrichment of ZNF561-AS1, miR-26a-5p, and miR-128-3p in immunoprecipitates were assessed by RT-qPCR. We found that the levels of ZNF561-AS1, miR-26a-5p, and miR-128-3p were all remarkably higher in Ago2 pellets relative to IgG pellets (Fig. 2f). Next, luciferase reporter assays to determine if ZNF561-AS1 was a direct target of miR-26a-5p and miR-128-3p demonstrated that miR-26a-5p or/and miR-128-3p mimics remarkably reduced ZNF561-AS1-WT luciferase activity but not that of single or double binding site mutants (Fig. 2g). Altogether, these data suggest that ZNF561-AS1 is a ceRNA for miR-26a-5p and miR-128-3p.
Fig. 2.
ZNF561-AS1 acts as a ceRNA for miR-26a-5p and miR-128-3p in CRC cells. a Schematic of potential miR-26a-5p and miR-128-3p binding sites on ZNF561-AS1. b Both miR-26a-5p and miR-128-3p were decreased in HCT-116 cells. n = 3, *p < 0.01. c ZNF561-AS1 silencing elevated miR-26a-5p and miR-128-3p levels in HCT-116 cells. n = 3, *p < 0.05. d ZNF561-AS1 overexpression suppressed miR-26a-5p and miR-128-3p levels in HCT-116 cells. n = 3, *p < 0.05. e Transfection of miR-26a-5p or/and miR-128-3p mimics suppressed ZNF561-AS1 expression. n = 3, *p < 0.05. f Relative enrichments of ZNF561-AS1, miR-26a-5p, as well as miR-128-3p in RISC was examined by RIP assay using anti-Ago2 antibody. n = 3, *p < 0.01. g Luciferase reporter assay. HCT-116 cells were transfected as indicated. Relative luciferase activity was assessed 48 h post transfection. n = 3, *p < 0.01
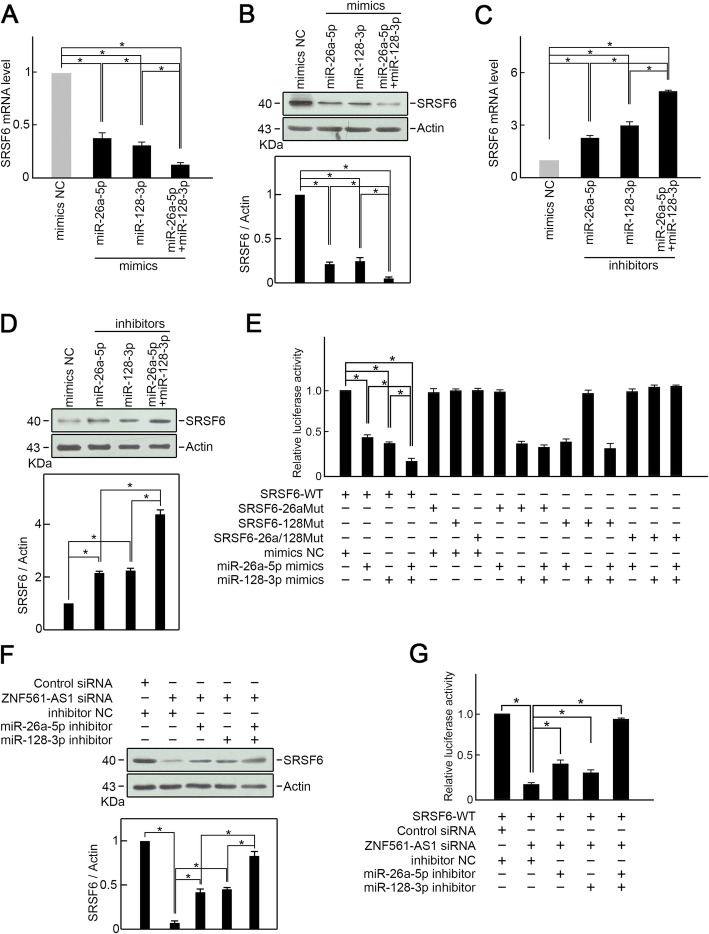
Next, we examined whether miR-26a-5p and miR-128-3p suppress SRSF6 expression through potential binding sites. Overexpression of miR-26a-5p or miR-128-3p suppressed SRSF6 expression (Fig. 3 a-b). Moreover, simultaneous overexpression of miR-26a-5p and miR-128-3p suppressed SRSF6 expression even more strongly than miR-26a-5p or miR-128-3p alone (Fig. 3 a-b). MiR-26a-5p and/or miR-128-3p silencing enhanced SRSF6 expression (Fig. 3 c-d). Luciferase reporter assay illustrated that miR-26a-5p and/or miR-128-3p mimics remarkably suppressed SRSF6-WT luciferase activity but not that of single or double binding site mutants (Fig. 3e).
Fig. 3.
ZNF561-AS1 promotes SRSF6 expression by sponging miR-26a-5p and miR-128-3p. a Transfection of miR-26a-5p or/and miR-128-3p mimics suppressed SRSF6 mRNA levels. n = 3, *p < 0.01. b Transfection of miR-26a-5p or/and miR-128-3p mimics suppressed SRSF6 protein level. Actin was employed as loading control. Representative blots are shown (upper panel). Ratios of SRSF6 level vs Actin were determined after densitometric analysis of blot images on Image J (lower panel). Data were from 3 independent experiments. *p < 0.01. c Knockdown of miR-26a-5p or/and miR-128-3p increased SRSF6 mRNA level. n = 3, *p < 0.01. d Knockdown of miR-26a-5p or/and miR-128-3p increased SRSF6 protein level. Actin was served as loading control. Representative blots were shown (upper penal). Ratios of level of SRSF6 vs Actin were calculated after densitometric analysis of blot images using NIH Image J 1.61 (lower panel). Data were from three individual experiments. *p < 0.05. e Luciferase reporter assay. HCT-116 cells were transfected as indicated. Relative luciferase activity was explored 48 h post-transfection. n = 3, *p < 0.05. f MiR-26a-5p or/and miR-128-3p silencing restored SRSF6 expression in ZNF561-AS1 depleted HCT-116 cells. Actin served as loading control. Representative blots are shown (upper panel). Ratios of SRSF6 vs Actin levels were computed after densitometric analysis of blot images on Image J (lower panel). Data were from 3 independent experiments. *p < 0.01. g Luciferase reporter assay. MiR-26a-5p or/and miR-128-3p silencing restored SRSF-WT luciferase activity in ZNF561-AS1-depleted HCT-116 cells. n = 3, *p < 0.01
Since we had already demonstrated that ZNF561-AS1 is an upstream SRSF6 regulator, we then assessed if ZNF561-AS1 regulates SRSF6 expression via miR-26a-5p and/or miR-128-3p. Evaluation of the expression of SRSF6 in HCT-116 cells co-transfected with ZNF561-AS1 siRNA and miR-26a-5p and/or miR-128-3p inhibitors found that in ZNF561-AS1 depleted cells, single miR-26a-5p inhibitor or miR-128-3p inhibitor partially reversed SRSF6 expression (Fig. 3f), whereas co-transfection with miR-26a-5p and miR-128-3p inhibitors almost fully rescued SRSF6 expression (Fig. 3f). Similar results were obtained from luciferase reporter assays. ZNF561-AS1 silencing markedly suppressed SRSF6-WT luciferase activity (Fig. 3g). However, miR-26a-5p and/or miR-128-3p inhibitors could partially or fully rescue luciferase activity in ZNF561-AS1 depleted cells (Fig. 3g). Taken together, these data show that ZNF561-AS1 promotes SRSF6 expression in CRC cells by sponging miR-26a-5p and miR-128-3p.
ZNF561-AS1 is critical for CRC cells proliferation and survival
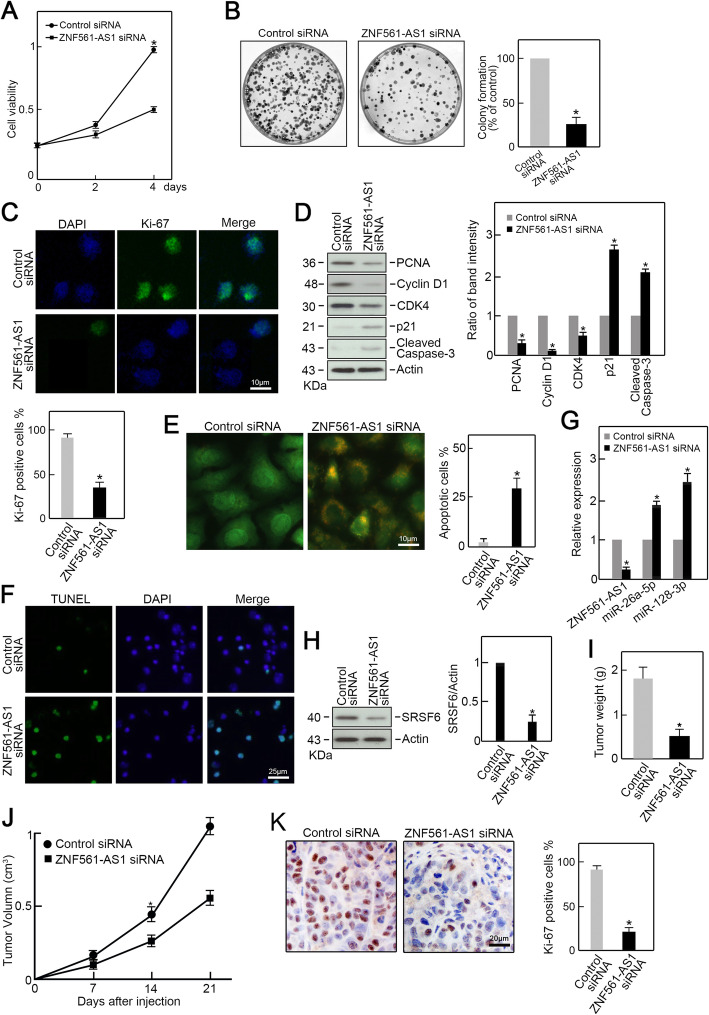
Since we found that ZNF561-AS1 is dramatically increased in CRC and it positively regulates SRSF6 expression, we next examined its biological effects on CRC cells. As expected, we found ZNF561-AS1 silencing significantly suppressed HCT-116 growth and colony formation compared to controls (Fig. 4 a-b). Ki-67 Immunofluorescence staining exhibited significantly lower Ki-67 signal in ZNF561-AS1 depleted HCT-116 cells compared to controls, indicating that ZNF561-AS1 silencing suppressed HCT-116 cells proliferation (Fig. 4c). Additionally, we found that expression of the cell cycle promoting factors, PCNA, CDK4, and Cyclin D1 was reduced markedly in ZNF561-AS1 deficient HCT-116 cells (Fig. 4d). However, p21, a strong cell cycle inhibitor, was elevated (Fig. 4d). Interestingly, the level of cleaved caspase-3, an apoptotic factor, was increased upon ZNF561-AS1 knockdown (Fig. 4d). This observation led us to examine whether ZNF561-AS1 silencing would trigger cell apoptosis in HCT-116 cells. AO/EB staining revealed that knockdown of ZNF561-AS1 increased the proportion of apoptotic cells in ZNF561-AS1 depleted HCT-116 cells compared to control cells (Fig. 4e). This phenotype was further confirmed by TUNEL staining (Fig. 4f).
Fig. 4.
ZNF561-AS1 silencing suppresses CRC progression in vitro and in vivo. a ZNF561-AS1 silencing inhibited HCT-116 growth. n = 3, *p < 0.01. b Colony formation assay. ZNF561-AS1- or mock-silenced HCT-116 cells were planted in 6 cm dishes and colonies counted after 12 days. Formed colonies were stained with crystal violet and counted. n = 3, *p < 0.05. c Ki-67 level was significantly reduced in ZNF561-AS1 depleted cells compared to controls assessed by immunofluorescence staining. n = 3, *p < 0.01. d PCNA, Cyclin D1, CDK4, p21, and cleaved caspase-3 levels in ZNF561-AS1- and mock-silenced HCT-116 cells. Actin served as loading control. Representative blots are shown (left panel). Ratios of PCNA, Cyclin D1, CDK4, p21, and cleaved caspase-3 vs Actin levels were computed after densitometric analysis of blot images on Image J (right panel). Data are 3 independent experiments. *p < 0.05. e Knockdown of ZNF561-AS1 induced cell apoptosis assessed by AO/EB staining. n = 3, *p < 0.01. f Knockdown of ZNF561-AS1 induced cell apoptosis assessed by TUNLE staining. n = 3. g The expression of ZNF561-AS1, miR-26a-5p and miR-128-3p in xenograft tumors. n = 3, *p < 0.05. h Levels of SRSF6 in xenograft tumors. Actin was served as loading control. Representative blots were shown (left penal). Ratios of level of SRSF6 vs Actin were calculated after densitometric analysis of blot images using NIH Image J 1.61 (right panel). Data were from three individual experiments. *p < 0.01. i Knockdown of ZNF561-AS1 reduced xenograft tumors weight. n = 3, *p < 0.05. j Knockdown of ZNF561-AS1 reduced xenograft tumors volume. n = 3, *p < 0.05. k Ki-67 immunohistochemical staining in xenograft tumor specimens. n = 3, *p < 0.05
To study the role of ZNF561-AS1 in vivo, ZNF561-AS1 siRNA or control siRNA transfected HCT-116 cells were subcutaneously injected into the flanks of nude mice. After 3 weeks, the mice were sacrificed, and xenograft tumors were isolated. RT-qPCR analysis confirmed ZNF561-AS1 depletion in tumors from ZNF561-AS1 depleted cells (Fig. 4g), which also exhibited increased miR-26a-5p and miR-128-3p levels and reduced SRSF6 levels relative to controls (Fig. 4 g-h). Tumors from ZNF561-AS1 depleted cells were smaller in size and lower in weight compared to control tumors. (Fig. 4 i-j). Immunohistochemical staining revealed lower Ki-67 levels in ZNF561-AS1 depleted tumors (Fig. 4k). Together, these data indicate that ZNF561-AS1 is crucial for CRC cell proliferation, as well as survival both in vitro and in vivo.
ZNF561-AS1 promotes CRC cells proliferation via SRSF6
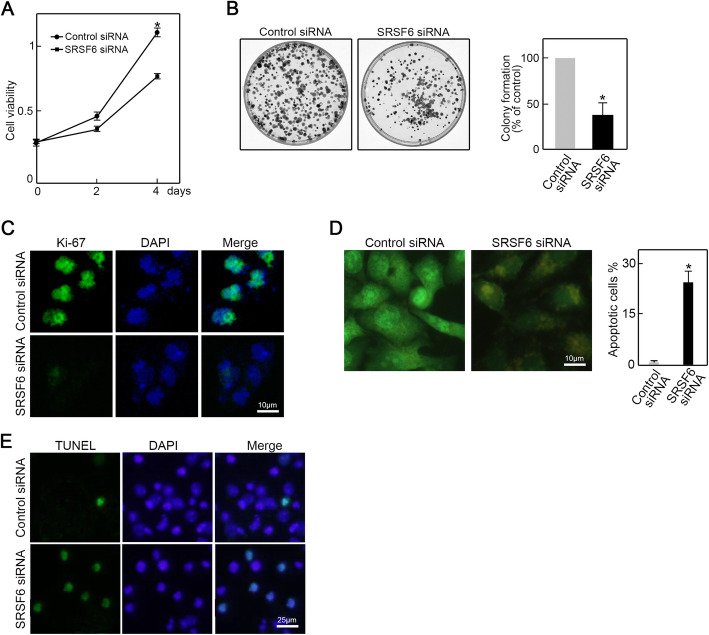
SRSF6 is associated with the progression of various cancers. Here, we find that SRSF6 is essential for CRC cell proliferation and survival as its depletion phenocopies ZNF561-AS1 knockdown as revealed by CCK-8 analysis, colony formation, and Ki-67 immunofluorescence staining (Fig. 5 a-c). Moreover, AO/EB and TUNEL assay demonstrated that SRSF6 depletion triggered apoptosis in HCT-116 cells (Fig. 5 d-e). These data show that SRSF6 is crucial for CRC proliferation and survival. Depletion of SRSF6 induced similar phenotypes as ZNF561-AS1 knockdown.
Fig. 5.
RSF6 silencing repressed cell proliferation and triggered apoptosis in CRC cells. a Knockdown of SRSF6 inhibited HCT-116 cells growth. n = 3, *p < 0.01. b Colony formation assay. HCT-116 cells transfected with SRSF6 siRNA or control siRNA were seeded in 6 cm dishes for 12 days culture. Formed colonies were stained with crystal violet and counted. n = 3, *p < 0.05. c Proliferation capabilities of ZNF561-AS1 siRNA or control siRNA transfected HCT-116 cells was examined by Ki-67 immunofluorescence staining. n = 3. d Knockdown of SRSF6 induced cell apoptosis assessed by AO/EB staining. n = 3, *p < 0.01. e Knockdown of SRSF6 induced cell apoptosis assessed by TUNLE staining. n = 3
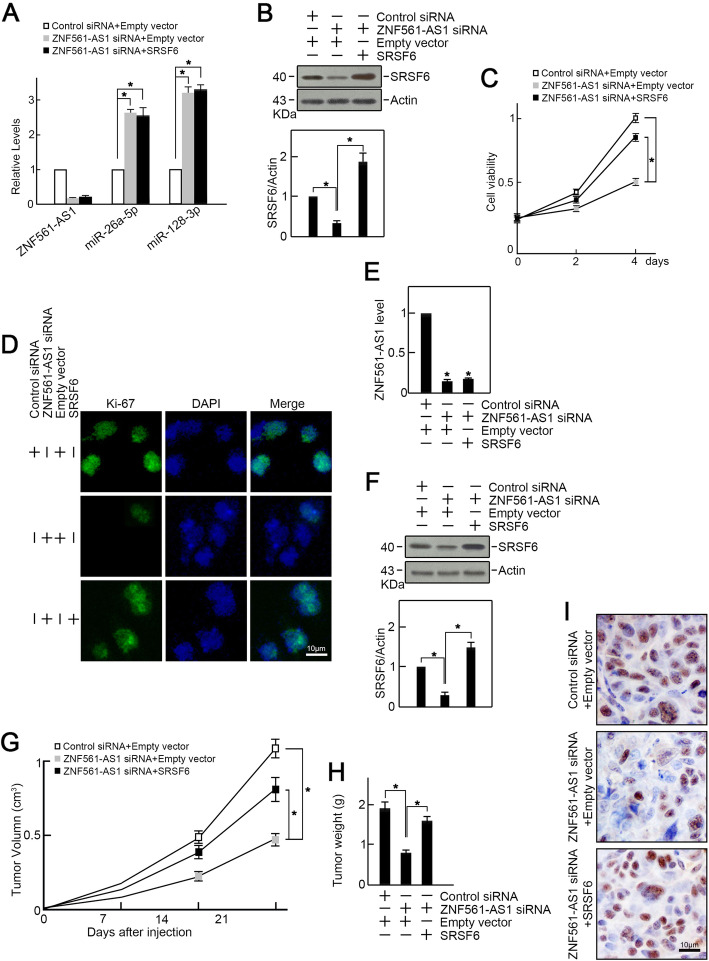
To further test whether ZNF561-AS1 promoted CRC cell proliferation via SRSF6, we overexpressed SRSF6 in ZNF561-AS1 depleted HCT-116 cells. We found that ZNF561-AS1 expression was suppressed by ZNF561-AS1 siRNA transfection (Fig. 6a) and the levels of miR-26a-5p and miR-128-3p were increased (Fig. 6a) while the level of SRSF6 was decreased (Fig. 6b). Co-transfection of SRSF6 expression vector restored SRSF6 expression in ZNF561-AS1 depleted HCT-116 cells and as expected, rescued proliferation in ZNF561-AS1-depleted cells as revealed by CCK-8 and Ki-67 assays (Fig. 6 c-d).
Fig. 6.
Exogenous SRSF6 expression rescued proliferation of ZNF561-AS1-depleted CRC cells in vitro and in vivo. HCT-116 cells were co-transfected as indicated. a RT-qPCR analysis of ZNF561-AS1, miR-26a-5p, and miR-128-3p levels. n = 3, *p < 0.05. b SRSF6 expression was examined by immunoblotting. Actin served as loading control. Representative blots were shown (upper penal). Ratios of level of SRSF6 vs Actin were calculated after densitometric analysis of blot images using NIH Image J 1.61 (lower panel). Data were from three individual experiments. *p < 0.01. c Enforced expression of SRSF6 restored cell growth capabilities in HCT-116 cells depleted of ZNF561-AS1. n = 3, *p < 0.05. d Enforced expression of SRSF6 restored Ki-67 expression in HCT-116 cells depleted of ZNF561-AS1. n = 3. e ZNF561-AS1 expression in xenograft tumors. n = 3, *p < 0.01. f Co-transfection of SRSF6 expression vector restored SRSF6 level in xenograft tumors. Actin served as loading control. n = 3, *p < 0.01. g Exogenous expression of SRSF6 rescued tumor volume. n = 3, *p < 0.05. h Exogenous expression of SRSF6 rescued tumor weight. n = 3, *p < 0.05. i SRSF6 overexpression restored Ki-67 levels in xenograft tumors. n = 3
Furthermore, HCT-116 cells depleted of ZNF561-AS1 were co-transfected with SRSF6 and subcutaneously injected into the flanks of nude mice. After 3 weeks, all mice were sacrificed, and xenograft tumors were isolated. qRT-PCR demonstrated that ZNF561-AS1 siRNA suppressed the level of ZNF561-AS1 in xenograft tumors (Fig. 6e). SRSF6 overexpression restored SRSF6 expression in ZNF561-AS1 depleted tumors (Fig. 6f). ZNF561-AS1 depleted tumors were smaller in size and lower in weight, compared to controls (Fig. 6 g-h). But ectopic SRSF6 expression rescued this phenotype (Fig. 6 g-h). Consistently, immunohistochemical staining analysis revealed reduced Ki-67 levels in ZNF561-AS1 depleted tumors compared to controls, which was restored by SRSF6 overexpression (Fig. 6 i). Collectively, these data indicate that ZNF561-AS1 promotes CRC cells proliferation by modulating SRSF6 expression.
Discussion
Herein, we investigated the role of ZNF561-AS1 in CRC and the reasonable mechanism. ZNF561-AS1 was upregulated in CRC compared to matched adjacent normal tissues and ZNF561-AS1 silencing suppressed CRC cells proliferation and survival, suggesting an oncogenic role in CRC. In contrast with our findings, another study found that ZNF561-AS1 downregulation promoted migration and invasion of laryngeal cancer cells (LSCC), indicating tumor suppressor function [19]. This difference was mostly considered from the different histological types [20]. The disparity in ZNF561-AS1 function in LSCC vs CRC suggests tissue-specific or cancer-specific effects.
Increasing evidence shows that numerous lncRNAs are dysfunctional in CRC, acting as oncogenes or tumor suppressors [21, 22]. LncRNA CCAL regulates CRC progression by targeting the activator protein 2α (AP-2α), which activates Wnt/β-catenin signaling [23]. LncRNA DLEU1 contributes to CRC progression by activating KPNA3 via SMARCA1 recruitment, which is an essential subunit of the NURF chromatin remodeling complex [24]. LncRNA SNHG5 enhances CRC cell survival via counteracting STAU1-induced mRNA destabilization [25]. Our data indicate that ZNF561-AS1 modulates CRC cell proliferation along with survival. In vitro and in vivo studies indicate that ZNF561-AS1 sequesters miR-26a-5p and miR-128-3p, thereby enhancing SRSF6 expression and promoting CRC cell proliferation and survival.
SRSF6 is a member of the serine-arginine-rich splicing factor family. These proteins are essential for pre-mRNA splicing and mRNA stability, export, and translation [26]. SRSF6 is a proto-oncogene often overexpressed in human skin cancer [27]. By controlling exon skipping, SRSF6 is critical for leukemia cells survival [28]. SRSF6 is also overexpressed in CRC [29]. Evidence show that SRSF6-regulated alternative splicing of ZO-1 promotes CRC progression by directly binding its motif to ZO-1 exon23 [30]. This alternative splicing effect relies on SRSF6’s RRM2 domain. Here, we find that ZNF561-AS1 promotes SRSF6 expression by acting as sponge of miR-26a-5p and miR-128-3p. SRSF6 overexpression rescued proliferation and survival in ZNF561-AS1 depleted cells. Indicating that ZNF561-AS1 regulates CRC cells proliferation and survival by modulating SRSF6 expression. Because SRSF6 knockdown did not alter ZNF561-AS1 expression, while ZNF561-AS1 knockdown suppressed SRSF6 expression, we speculate that ZNF561-AS1 is an upstream post-transcriptional regulator of SRSF6.
Conclusion
In summary, our study revealed an essential role of over-expression of ZNF561-AS1 for CRC cells proliferation and survival. ZNF561-AS1 servers as a ceRNA, competitively binding to miR-26a-5p and miR-128-3p to enhance the expression of SRSF6, an alternative splicing factor. These findings provide novel insight into our understanding of CRC progression and highlight ZNF561-AS1 as a potential therapeutic target against CRC.
Supplementary Information
Additional file 1: Table S1. CRC patient information.
Acknowledgments
We thank Department of colorectal cancer surgery, The Second Affiliated Hospital of Harbin Medical University for providing the CRC patients tissue samples and matched anonymous clinical information.
Abbreviations
- CRC
Colorectal cancer
- ZNF561-AS1
ZNF561 antisense RNA 1
- RIP
RNA binding protein immunoprecipitation
- TCGA
The Cancer Genome Atlas
- LncRNA
Long non-coding RNA
- TUNEL
Terminal deoxynucleotidyl transferase dUTP nick end labeling
- SRSF6
Serine and arginine-rich splicing factor 6
- ANOVA
Analysis of variance
Authors’ contributions
ZS performed proliferation assay and qRT-PCR; LY performed in vivo experiments and western blotting; HJ analyzed gene expression in patients’ samples. ZS, LY and LW performed the statistical analyses; XDW designed the work and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Foundation of Heilongjiang province (QC2017018) and Heilongjiang dostdoctoral funding (LBH-Z16164).
Availability of data and materials
The data used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Experimental protocols for human tissues and animal were approved by the Ethics Committee at Harbin Medical University (KY2016–036).
Consent for publication
Not applicable.
Competing interests
The authors report no conflicts of interest in this work.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zizhen Si and Lei Yu contributed equally to this work.
References
- 1.Schreuders EH, Ruco A, Rabeneck L, Schoen RE, Sung JJ, Young GP, et al. Colorectal cancer screening: a global overview of existing programmes. Gut. 2015;64(10):1637–1649. doi: 10.1136/gutjnl-2014-309086. [DOI] [PubMed] [Google Scholar]
- 2.Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14(4):235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 3.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 4.Devaux Y, Zangrando J, Schroen B, Creemers EE, Pedrazzini T, Chang CP, et al. Long noncoding RNAs in cardiac development and ageing. Nat Rev Cardiol. 2015;12(7):415–425. doi: 10.1038/nrcardio.2015.55. [DOI] [PubMed] [Google Scholar]
- 5.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127(3):761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitt AM, Chang HY. Long noncoding RNAs in Cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 10.Kajino T, Shimamura T, Gong S, Yanagisawa K, Ida L, Nakatochi M, et al. Divergent lncRNA MYMLR regulates MYC by eliciting DNA looping and promoter-enhancer interaction. EMBO J. 2019;38(17):e98441. doi: 10.15252/embj.201798441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y, Gu L, Lin X, Cui K, Liu C, Lu B, et al. LINC00265 promotes colorectal tumorigenesis via ZMIZ2 and USP7-mediated stabilization of beta-catenin. Cell Death Differ. 2020;27(4):1316–1327. doi: 10.1038/s41418-019-0417-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16(1):9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rigoutsos I, Lee SK, Nam SY, Anfossi S, Pasculli B, Pichler M, et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol. 2017;18(1):98. doi: 10.1186/s13059-017-1224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang J, Yan T, Bao Y, Shen C, Yu C, Zhu X, et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat Commun. 2019;10(1):3499. doi: 10.1038/s41467-019-11447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bian Z, Zhang J, Li M, Feng Y, Wang X, Zhang J, et al. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal Cancer by regulating PKM2 signaling. Clin Cancer Res. 2018;24(19):4808–19. doi: 10.1158/1078-0432.CCR-17-2967. [DOI] [PubMed] [Google Scholar]
- 17.Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8(14):3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyu K, Li Y, Xu Y, Yue H, Wen Y, Liu T, et al. Using RNA sequencing to identify a putative lncRNA-associated ceRNA network in laryngeal squamous cell carcinoma. RNA Biol. 2020;17(7):977–989. doi: 10.1080/15476286.2020.1741282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan C, et al. The lncRNA NEAT1 activates Wnt/beta-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. 2018;11(1):113. doi: 10.1186/s13045-018-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m (6) a reader YTHDF3. Mol Cancer. 2019;18(1):143. doi: 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2016;65(9):1494–1504. doi: 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- 24.Liu T, Han Z, Li H, Zhu Y, Sun Z, Zhu A. LncRNA DLEU1 contributes to colorectal cancer progression via activation of KPNA3. Mol Cancer. 2018;17(1):118. doi: 10.1186/s12943-018-0873-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damas ND, Marcatti M, Come C, Christensen LL, Nielsen MM, Baumgartner R, et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. 2016;7:13875. doi: 10.1038/ncomms13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anko ML. Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin Cell Dev Biol. 2014;32:11–21. doi: 10.1016/j.semcdb.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 27.Jensen MA, Wilkinson JE, Krainer AR. Splicing factor SRSF6 promotes hyperplasia of sensitized skin. Nat Struct Mol Biol. 2014;21(2):189–197. doi: 10.1038/nsmb.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y, Han C, Wang E, Lorch AH, Serafin V, Cho BK, et al. Posttranslational regulation of the exon skipping machinery controls aberrant splicing in leukemia. Cancer Discov. 2020;10(9):1388–1409. doi: 10.1158/2159-8290.CD-19-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen-Eliav M, Golan-Gerstl R, Siegfried Z, Andersen CL, Thorsen K, Orntoft TF, et al. The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J Pathol. 2013;229(4):630–639. doi: 10.1002/path.4129. [DOI] [PubMed] [Google Scholar]
- 30.Wan L, Yu W, Shen E, Sun W, Liu Y, Kong J, et al. SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut. 2019;68(1):118–129. doi: 10.1136/gutjnl-2017-314983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. CRC patient information.
Data Availability Statement
The data used and analyzed during the current study are available from the corresponding author on reasonable request.