Abstract
Telomere biology disorders, largely characterized by telomere lengths below the first centile for age, are caused by variants in genes associated with telomere replication, structure, or function. One of these genes, ACD, which encodes the shelterin protein TPP1, is associated with both autosomal dominantly and autosomal recessively inherited telomere biology disorders. TPP1 recruits telomerase to telomeres and stimulates telomerase processivity. Several studies probing the effect of various synthetic or patient-derived variants have mapped specific residues and regions of TPP1 that are important for interaction with TERT, the catalytic component of telomerase. However, these studies have come to differing conclusions regarding ACD haploinsufficiency. Here, we report a proband with compound heterozygous novel variants in ACD (NM_001082486.1)—c.505_507delGAG, p.(Glu169del); and c.619delG, p.(Asp207Thrfs*22)—and a second proband with a heterozygous chromosomal deletion encompassing ACD: arr[hg19] 16q22.1(67,628,846-67,813,408)x1. Clinical data, including symptoms and telomere length within the pedigrees, suggested that loss of one ACD allele was insufficient to induce telomere shortening or confer clinical features. Further analyses of lymphoblastoid cell lines showed decreased nascent ACD RNA and steady-state mRNA, but normal TPP1 protein levels, in cells containing heterozygous ACD c.619delG, p.(Asp207Thrfs*22), or the ACD-encompassing chromosomal deletion compared to controls. Based on our results, we conclude that cells are able to compensate for loss of one ACD allele by activating a mechanism to maintain TPP1 protein levels, thus maintaining normal telomere length.
Keywords: abnormality of B cell number, bone marrow hypocellularity, fingernail dysplasia, microcephaly, oral leukoplakia, reticulated skin pigmentation, toenail dysplasia
INTRODUCTION
Telomeres, the specialized structures that cap the ends of chromosomes, are essential for protecting genomic integrity and supporting cell proliferation. At birth, human telomere length ranges between 8 and 14 kilobasepairs (kb), depending on the cell type (Harley et al. 1990; Vaziri et al. 1994). As cells divide, the constraints of the semiconservative DNA replication machinery cause telomere shortening (Allsopp and Harley 1995). A few cell types, including embryonic cells and stem cells, express telomerase, a specialized reverse transcriptase that counteracts telomere shortening by catalyzing the de novo addition of telomeric repeats onto telomere ends (Wright et al. 1996). In contrast, telomerase expression is repressed in somatic cells after embryonic development (Kim et al. 1994). When telomeres become too short, senescence is induced by a DNA damage response, thereby preventing replicative immortality (d'Adda di Fagagna et al. 2003; Herbig et al. 2004). If senescence is bypassed (e.g., by loss of p53), critically short telomeres induce genomic instability, which can promote tumorigenesis (Artandi et al. 2000; Herate and Sabatier 2020). Conversely, abnormally long telomeres may also promote tumorigenesis by removing the constraint that age-related telomere shortening exerts on cell proliferation. Thus, maintaining telomeres within an age-appropriate physiologic range is important to prevent inappropriate senescence or apoptosis and cancer development.
The telomere biology disorders (TBDs) are a collection of disorders that arise as a consequence of abnormally short telomeres (Barbaro et al. 2016; Dodson and Bertuch 2018; Savage 2018). Dyskeratosis congenita (DC), the prototypical TBD, is classically defined by a mucocutaneous triad of nail dystrophy, reticulated skin pigmentation, and oral leukoplakia. Affected individuals are at high risk of bone marrow failure and a multitude of other medical conditions, including but not limited to pulmonary fibrosis, cirrhosis, hepatopulmonary syndrome, and specific cancers (e.g., squamous cell carcinoma of the tongue) (Kirwan and Dokal 2008; Alter et al. 2009, 2010; Calado et al. 2009; Giri et al. 2019). Hoyeraal–Hreidarsson syndrome (HHS) is another TBD and is generally recognized as a severe form of DC. HHS presents in infancy with clinical features of intrauterine growth restriction, microcephaly, cerebellar hypoplasia, and immunodeficiency, particularly profound B-cell deficiency (Hoyeraal et al. 1970; Hreidarsson et al. 1988; Berthet et al. 1994; Glousker et al. 2015). Other TBDs may become apparent in adulthood, with presentations such as isolated pulmonary fibrosis, bone marrow failure (also known as aplastic anemia), cirrhosis, or combinations thereof (Armanios et al. 2005; Yamaguchi et al. 2005; Calado et al. 2009; Parry et al. 2011). Thus, the spectrum of the TBDs ranges from multisystem, early childhood onset to more restricted phenotypic expression and adult onset.
Despite having a range of phenotypes and age of onset, the TBDs share a defining characteristic of telomere lengths below the first centile for age across cell populations, measured most often in lymphocyte subsets (Alter et al. 2007). In ∼80% of affected individuals, this can be attributed to a variant in one of 15 genes that impact telomere biology (Savage 2018). These include genes that encode core telomerase subunits (TERT, TERC); proteins involved in various aspects of telomere biology, including telomerase RNA subunit metabolism, telomerase biogenesis, duplex telomere replication, or telomere end structure formation (PARN, ZCCHC8, DKC1, NAF1, NHP2, NOP10, WRAP53, CTC1, STN1, RTEL1); and components of shelterin, the protein complex that coats telomeres and promotes proper structure and function (TINF2, ACD, POT1) (ZCCHC8 reported in Gable et al. [2019]; all others reviewed in Dodson and Bertuch [2018] with disease-associated mutations compiled in the Telomerase Database [Podlevsky et al. 2008; http://telomerase.asu.edu/ accessioned 4/18/2020]). Haploinsufficiency is a known mechanism for several of these genes, including TERT and TERC, with telomere shortening and disease arising from heterozygous loss-of-function (LOF) variants or deletions (Vulliamy et al. 2001; Armanios et al. 2005). In addition to short telomere disorders, variants in the genes encoding shelterin components ACD (Fig. 1A), POT1, TERF2IP, and TINF2 have also been reported in association with certain types of familial cancer and long telomeres (Shi et al. 2014; Aoude et al. 2015; Calvete et al. 2015; Speedy et al. 2016; Wong et al. 2019; Gong et al. 2020; He et al. 2020; Schmutz et al. 2020).
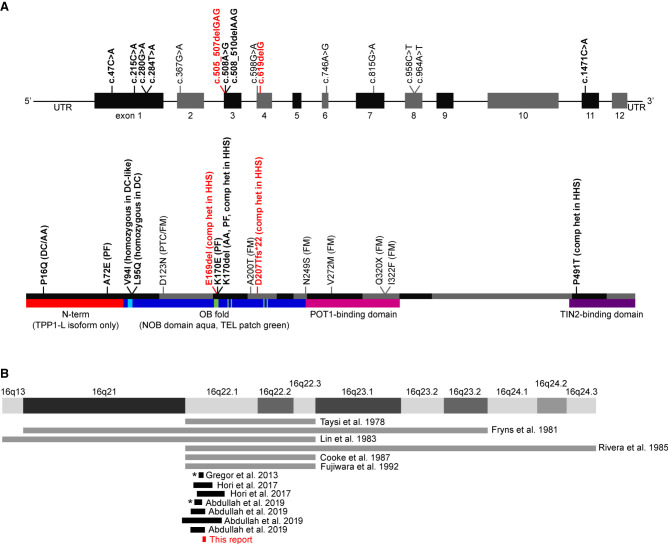
Figure 1.
Previously reported ACD variants and Chr 16q22 deletions. (A) Reported ACD variants. Nucleotide positions are depicted in the upper graphic, whereas amino acid positions are depicted in the lower graphic. Red text indicates variants described in this report. Bold text indicates variants associated with short telomeres. Non-bold text indicates variants found in cancer-prone families and associated with long telomeres. (DC) Dyskeratosis congenita, (AA) aplastic anemia, (PF) pulmonary fibrosis, (PTC) papillary thyroid carcinoma, (FM) familial melanoma, (HHS) Hoyeraal–Hreidarsson syndrome, (comp het) compound heterozygous variant found in the context of another disease-causing variant. (B) Reported Chromosome 16q22.1 deletions. Gray lines indicate deletions for which specific nucleotide information is not available. Asterisks indicate deletions that do not encompass ACD.
This report focuses on ACD, which encodes the shelterin protein TPP1. The gene name is derived from the adrenocortical dysplasia associated (ACD) phenotype in mice, and the protein name evolved from combination of the original monikers TINT1 (TIN2-interacting protein), PTOP (POT1- and TIN2-organizing protein), and PIP1 (POT1-interacting protein 1) given by three research groups who discovered the protein simultaneously (Houghtaling et al. 2004; Liu et al. 2004; Ye et al. 2004). The initial classification of ACD as a TBD-causing gene came from the identification of two unrelated families in which TBD features and short telomeres cosegregated with a heterozygous ACD (NM_001082486.1) c.508_510del, p.(Lys170del) variant (Fig. 1A; Guo et al. 2014; Kocak et al. 2014). Telomere lengths of individuals heterozygous for the p.(Lys170del) variant in these families as well as in a subsequent report were typically at or below the first centile for age (Guo et al. 2014; Kocak et al. 2014; Hoffman et al. 2019). The proband in one of these families also had a second ACD variant in trans, c.1471C > A, p.(Pro491Thr), very short telomeres, and an HHS phenotype (Kocak et al. 2014). Individuals from two unrelated families with homozygous deleterious variants in ACD (c.280C > T, p.(Val94Ile), and c.284T > A, p.(Leu95Gln)), short telomeres, and TBD clinical features were subsequently reported (Tummala et al. 2018). Consequently, ACD is considered to be associated with both autosomal dominantly and autosomal recessively inherited TBDs (for review, see Dodson and Bertuch 2018). The frequencies at which the previously reported ACD variants are present in the Genome Aggregation Database (gnomAD; https://gnomad.broadinstitute.org/; accessioned 12/3/20) (Karczewski et al. 2020) are shown in Supplemental Table 1.
TPP1 has several important functions. In addition to its role in the recruitment of telomerase to telomeres (Xin et al. 2007; Nandakumar et al. 2012; Zhong et al. 2012), TPP1 stimulates the ability of telomerase to add multiple telomeric repeats onto the chromosome end during a single binding event, known as repeat addition processivity (Wang et al. 2007; Latrick and Cech 2010). TPP1, along with its binding partner POT1, also contributes to the protection of telomere ends by inhibiting ATR-mediated resection (Kibe et al. 2016). Extensive mutagenesis screens have identified several TPP1 residues required for its interaction with TERT, through which it impacts telomerase. Two of the most prominent are Glu169 and Glu171 in the TEL patch, a region of acidic residues within the oligonucleotide/oligosaccharide-binding (OB) fold of TPP1, which is critical for telomerase recruitment (Nandakumar et al. 2012; Sexton et al. 2012; Zhong et al. 2012). The TPP1 p.[Glu169Ala;Glu171Ala] synthetic mutation has a detrimental effect on TPP1–TERT interaction and stimulation of telomerase processivity, as well as on telomere length, when overexpressed, with Glu169 specifically being important for these activities (Nandakumar et al. 2012; Sexton et al. 2012). The TBD-associated Lys170 also lies within the TEL patch, and although it is not predicted to interact directly with TERT, its deletion alters the position of Glu169 and impairs TPP1–TERT interaction and telomerase recruitment (Bisht et al. 2016).
Further studies probed the effect of TPP1 site-directed mutagenesis on cell phenotypes. Sexton et al. replaced TEL patch amino acids 166–172 with a GSSG amino acid linker in human embryonic stem cells (hESCs) (Sexton et al. 2014). Although this variant abrogated interaction with telomerase, heterozygous clones exhibited no telomere length decrease up to 80 days postediting (Sexton et al. 2014). However, Bisht et al. (2016) found that triploid HEK293T clones CRISPR-edited to express the TBD-associated TPP1 p.(Lys170del) allele, a LOF indel allele, and a wild-type (WT) allele exhibited telomere shortening compared to clones containing a silent mutation instead of the p.(Lys170del) variant, but at a slower rate than clones with two alleles disrupted by indels. Thus, these data were consistent with ACD dosage sensitivity. There are several possible explanations for the differences between the two studies, including differences in the mutations and cell lines. Regardless of the reasons for the differences, the question remains: Do TBD-associated ACD variants contribute to disease through ACD haploinsufficiency, as observed for TERT and TERC?
Arguing against this is the ACD nonsense, splice acceptor, and splice donor LOF variant observed/expected (o/e) ratio of 0.82 (90% confidence interval = 0.58–1.19) in the Genome Aggregation Database (https://gnomad.broadinstitute.org/; accessioned 12/3/20) (Karczewski et al. 2020), which suggests that loss of one allele is tolerated. Moreover, ACD deletions exist among a subset of individuals with 16q22.1 microdeletion syndrome (Fig. 1B; Taysi et al. 1978; Fryns et al. 1981; Lin et al. 1983; Rivera et al. 1985; Cooke et al. 1987; Fujiwara et al. 1992; Gregor et al. 2013; Hori et al. 2017; Abdullah et al. 2019). These individuals had clinical features including failure to thrive, microcephaly, congenital heart defects, feeding difficulties, developmental delay, autistic behavior, muscular hypotonia, craniofacial dysmorphism, and nail anomalies. Some of these features, such as failure to thrive, microcephaly, developmental delay, and nail anomalies, are also found in patients with DC and HHS, but other common TBD symptoms such as oral leukoplakia, abnormal blood cell counts, and immunodeficiency have not been reported in 16q22.1 microdeletion syndrome. Additionally, not all individuals with 16q22.1 microdeletion syndrome have a deletion encompassing ACD (indicated by asterisks in Fig. 1B). Together, these data suggest that ACD is not a primary contributor to 16q22.1 microdeletion syndrome and that hemizygosity of ACD does not produce a clinical phenotype. Notably, of the seven germline ACD variants that have been reported in individuals with TBDs to date (Fig. 1A, bold black text), none are LOF (Guo et al. 2014; Kocak et al. 2014; Tummala et al. 2018; Arias-Salgado et al. 2019; Hoffman et al. 2019).
Whether inactivation of one copy of ACD results in a defect of telomere maintenance has important clinical implications, as it is expected that widespread clinical exome and genome sequencing will result in discovery of additional ACD LOF variants and this knowledge will influence risk counseling and medical management. If a single ACD LOF variant is sufficient to cause disease, health-care providers might recommend family studies and screening for TBD-associated conditions. However, if a single ACD LOF variant is not sufficient to cause disease, providers may choose not to reveal the finding or to instead counsel about the probability of a child with disease following an autosomal recessive inheritance pattern.
Here, we describe three novel ACD variants in two families. By analyzing the consequences of these variants, we investigate the question of ACD haploinsufficiency and find that LOF variants in ACD do not result in a telomere length defect. We conclude that the absence of such an effect may be the result of a homeostatic mechanism that maintains physiologic levels of TPP1 protein.
RESULTS
Clinical Histories
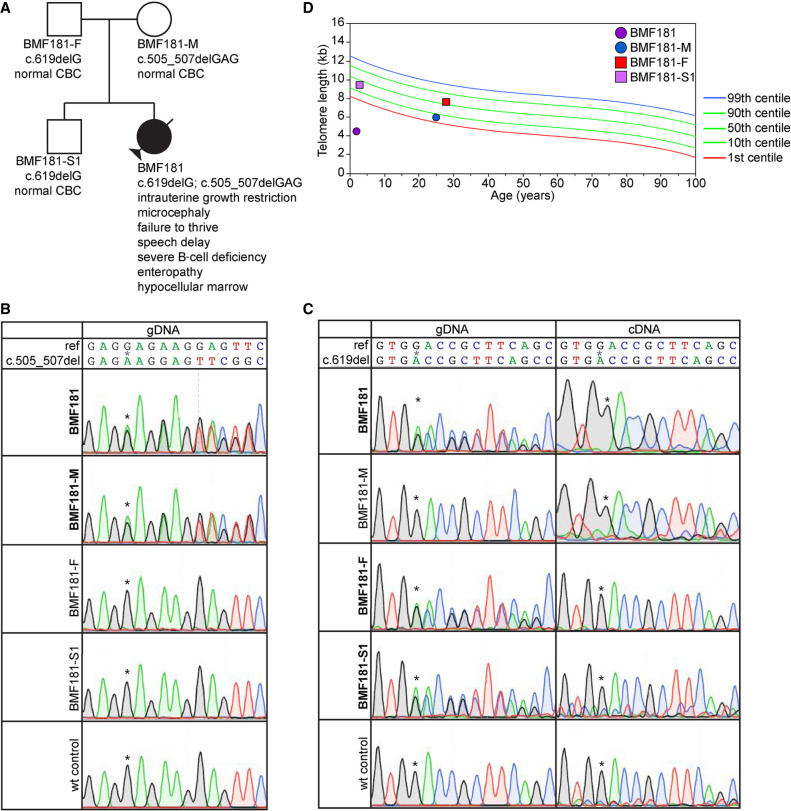
Variants described in this study are listed in Table 1. Proband BMF181 presented at 2 yr of age with clinical features that included intrauterine growth restriction, microcephaly, failure to thrive, speech delay, severe B-cell deficiency with associated life-threatening infections, severe enteropathy, and hypocellular bone marrow (Fig. 2A). Her mother (BMF181-M), father (BMF181-F), and brother (BMF181-S1) were reportedly healthy and had normal complete blood counts (CBCs) at 25, 28, and 3 yr of age, respectively. Clinical trio (BMF181 and her parents) exome sequencing (Baylor Genetics, Supplemental Table 2) showed that the proband had compound heterozygous variants in ACD. The maternal variant, c.505_507delGAG, p.(Glu169del), encoded an in-frame deletion of amino acid Glu169, notably mapping adjacent to the previously reported p.(Lys170del) variant (Figs. 1A and 2B). The paternal variant, c.619delG, p.(Asp207Thrfs*22), which was also inherited by the proband's sibling (BMF181-S1), encoded a frameshift in exon 4 and premature termination codon in exon 5 of 12 (Figs. 1A and 2C). Neither variant was present in gnomAD, although a c.617dupT, p.(Asp207Glyfs*30), variant was reported in a single read (https://gnomad.broadinstitute.org/; accessioned 12/3/20). Focused inspection of genes known or with potential to influence telomere biology (Supplemental Table 3) showed no other variants that may have contributed to disease.
Table 1.
Variants described in this report
| Variant | Gene | Chromo some | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | ClinVar ID | Genotype |
|---|---|---|---|---|---|---|---|---|
| c.505_507delGAG, p.(Glu169del) | ACD | 16 | NG_042874.1 | NP_001075955.2 | Deletion | Single-amino acid deletion | SCV001450738 | Heterozygous |
| c.619delG, p.(Asp207Thrfs* 22) | ACD | 16 | NG_042874.1 | NP_001075955.2 | Deletion | Frameshift | SCV001450737 | Heterozygous |
| arr[hg19] 16q22.1(67,628,846-67,813,408)x1 | ACD and surrounding genes | 16 | NG_042874.1 | NP_001075955.2 | Copy-number variation | Whole-gene deletion | SCV001450739 | Heterozygous |
Figure 2.
The ACD c.619delG variant is subject to nonsense-mediated decay but is not associated with telomere shortening. (A) Pedigree for proband BMF181. (B) Genomic DNA (gDNA) Sanger sequence chromatograms surrounding nucleotides c.505_507. Asterisks denote the location of the first deleted G in BMF181 and BMF181-M. (C) gDNA and complementary DNA (cDNA) Sanger sequence chromatograms surrounding nucleotide c.619. Asterisks denote the location of the deleted G in BMF181, BMF181-F, and BMF181-S1. (D) Lymphocyte telomere lengths as measured by telomere flow FISH for BMF181 and family members.
Given the biallelic ACD variants in BMF181, telomere flow cytometry fluorescence in situ hybridization (flow FISH) analysis was performed (RepeatDx, Inc.), which demonstrated telomere lengths ∼4.0–4.5 kb in all five measurable leukocyte populations (the B-cell number was too low for analysis), well below the first centile for age (Fig. 2D; Supplemental Fig. 1A). The clinical features, genetics, and telomere lengths led to a diagnosis of HHS. BMF181's numerous CBCs were most often characterized by moderate anemia requiring transfusion despite reticulocytosis, normal neutrophil counts, and normal platelet counts, except in the final 2 mo of life when she became critically ill. Her bone marrow was hypocellular, with an overall cellularity of 30%–40%, trilineage hematopoiesis with maturation and without dysplasia, and adequate number of megakaryocytes. She ultimately succumbed to multiorgan failure following a prolonged hospitalization for severe protein losing enteropathy and infection shortly after the age of 3 yr.
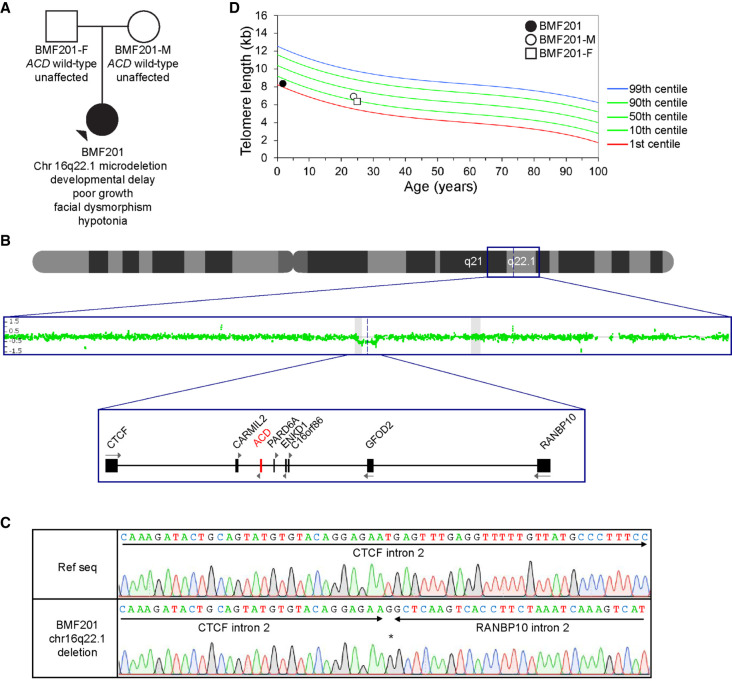
A second proband, BMF201, presented at 22 mo of age with developmental delay, poor growth, facial dysmorphism, and hypotonia (Fig. 3A). A clinical chromosomal microarray (Quest Diagnostics) on the proband and subsequently on the parents showed that BMF201 had a de novo 185-kb chromosomal deletion, arr[hg19] 16q22.1(67,628,846-67,813,408)x1 (Figs. 1B and 3B), which established a diagnosis of 16q22.1 microdeletion syndrome. Given the resulting hemizygosity of ACD, which we confirmed by sequencing the deletion junction (Fig. 3C), telomere flow FISH was performed, and telomere lengths were found to be between the first and 10th centiles for age in the five lymphocyte populations (Fig. 3D; Supplemental Fig. 1B). Multiple CBCs obtained between the ages of 16 mo and 4 yr were normal.
Figure 3.
Deletion of one ACD allele is not associated with telomere shortening. (A) Pedigree for proband BMF201. (B) Chromosomal microarray (CMA) indicating a deletion encompassing ACD and neighboring genes. Scanning from the top down, the graphics depict a schematic of Chromosome 16, the CMA data indicating deletion of 185 kb in Chr 16q22.1, and a schematic of the genes deleted or disrupted in BMF201. (C) Sanger sequence chromatograms confirming the heterozygous deletion of Chr 16q22.1(67,628,846-67,813,408) in BMF201. The asterisk denotes a single inserted G. (D) Lymphocyte telomere lengths as measured by telomere flow FISH for BMF201 and parents.
Heterozygous ACD c.505_507delGAG Variant Was Associated with Telomere Shortening, and the TPP1 p.169del Protein Had Reduced Stimulation of Telomerase Activity
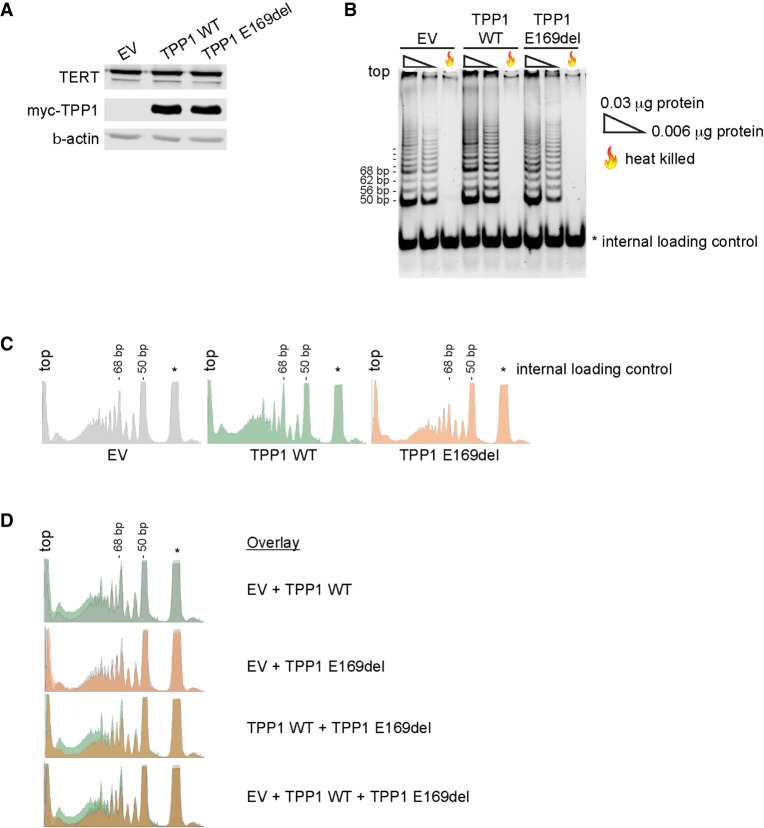
We next assessed lymphocyte telomere lengths in BMF181's parents and sibling. BMF181-M, heterozygous for the c.505_507delGAG, p.(Glu169del), variant (Fig. 2B), had short telomeres, with lengths between the first and 10th centiles for age in three of four lymphocyte subsets (Fig. 2D; Supplemental Fig. 1A). This was predicted given the known impact of in-frame deletion of neighboring Lys170 (Guo et al. 2014; Kocak et al. 2014; Hoffman et al. 2019) and the importance of Glu169 for TPP1–TERT interaction (Nandakumar et al. 2012; Sexton et al. 2012). To further test this, we performed the telomerase repeated amplification protocol (TRAP) on 293T lysates prepared from cells cotransfected with plasmids expressing TERT, hTR, and either empty vector (EV), myc-epitope tagged WT TPP1, or TPP1 p.Glu169del (Fig. 4A). Previous work has established that the interaction of TPP1 with TERT, in addition to mediating the recruitment of telomerase to telomeres, is important for activating telomerase (Grill et al. 2019) and stimulating telomerase processivity (Wang et al. 2007; Latrick and Cech 2010). As expected, we found that lysates prepared from cells transfected with WT TPP1 had increased telomerase activity, particularly with respect to higher-molecular-weight products indicative of stimulation of telomerase processivity, compared to cells transfected with EV (Fig. 4B–D). However, this increase was abrogated in cells expressing TPP1 p.Glu169del, indicating the ACD c.505_507delGAG variant is deleterious.
Figure 4.
TPP1 p.Glu169del is defective for telomerase stimulation. Shown are representative images for five independent experiments. (A) Western blot of lysates of 293T cells transiently transfected with plasmids expressing TERT, hTR, and either empty vector (EV), wild-type TPP1 (TPP1 WT), or TPP1 p.Glu169del (TPP1 E169del) used for the telomerase repeated amplification protocol (TRAP) assay shown in B. β-actin was used as a loading control. (B) TRAP assays performed with 0.03 µg protein, 0.006 µg protein, or 0.03 µg of heat-killed protein. Internal control is denoted by *. (C) Densitometry traces of the 0.03 µg TRAP reactions shown in B. The top of each lane aligns with the left of the densitometry trace. Internal control is denoted by *. Traces were generated using ImageJ software. (D) Overlay images of the densitometry traces shown in C.
Heterozygous ACD Frameshift or Whole-Gene Deletion Were Not Associated with Telomere Shortening
Interestingly, BMF181-F and BMF181-S1, which are heterozygous for the c.619delG, p.(Asp207Thrfs*22), variant (Fig. 2C), had pan-lymphocyte telomere lengths around the 50th centile for age and between the 10th and 50th centiles for age, respectively (Fig. 2D). Similar findings were observed for three of four lymphocyte subsets (Supplemental Fig. 1A), suggesting cells do not suffer from deleterious effects of ACD haploinsufficiency.
Large cohort studies have concluded that telomere length of individuals is determined largely by the lengths of their parents’ telomeres (Broer et al. 2013). Therefore, to better interpret BMF201's telomere lengths, we examined the telomere lengths of her parents. We found that, although BMF201's lymphocyte telomere lengths were between the first and 10th centiles across subsets, so were her father's (BMF201-F) in four of six leukocyte populations and her mother's (BMF201-M), with lengths just above the first centile for granulocytes and at or marginally above the 10th centile in the pan-lymphocyte, memory T-cell, and B-cell populations (Fig. 3D; Supplemental Fig. 1B). Because neither parent had the ACD gene deletion, we conclude BMF201's telomere lengths were reflective of her parents’, and the heterozygous ACD deletion had little, if any, impact.
ACD Transcript Levels Are Reduced in Lymphoblastoid Cell Lines (LCLs) Containing Either Heterozygous ACD c.619delG or Chromosomal Deletion Encompassing ACD
Given the novel ACD frameshift and chromosomal deletion variants identified, the BMF181 and BMF201 families provided a unique opportunity to investigate the question of ACD haploinsufficiency beyond the previously published studies using TEL patch-edited hESCs and HEK293T cells (Sexton et al. 2014; Bisht et al. 2016). The finding of telomere lengths well within normal range in BMF181-F and BMF181-S1 was surprising because the ACD c.619delG mutant transcript bore a premature termination codon in exon 5 of 12 and was therefore expected to undergo nonsense-mediated decay (NMD). To test this prediction, we generated Epstein–Barr virus (EBV)-transformed LCLs from BMF181-F, BMF181-S1, and BMF181-M to compare to three normal control LCLs. We were unable to generate LCLs from BMF181, likely because of the paucity of B cells and their very short telomeres. Consistent with our prediction of NMD, Sanger sequencing of genomic DNA (gDNA) from BMF181 (peripheral blood), BMF181-F (LCLs), and BMF181-S1 (LCLs) at position c.619 (noted by asterisks in Fig. 2C) clearly demonstrated the reference (G) and variant (A) bases, whereas the variant base (A) was markedly reduced in the complementary DNA (cDNA), with similar differences observed for downstream bases. In addition, we did not detect truncated TPP1 protein in BMF181-F and BMF181-S1 LCLs that would have indicated translation of the frameshift transcript (Supplemental Fig. 2B). Thus, we conclude that ACD c.619delG, p.(Asp207Thrfs*22), is a LOF variant.
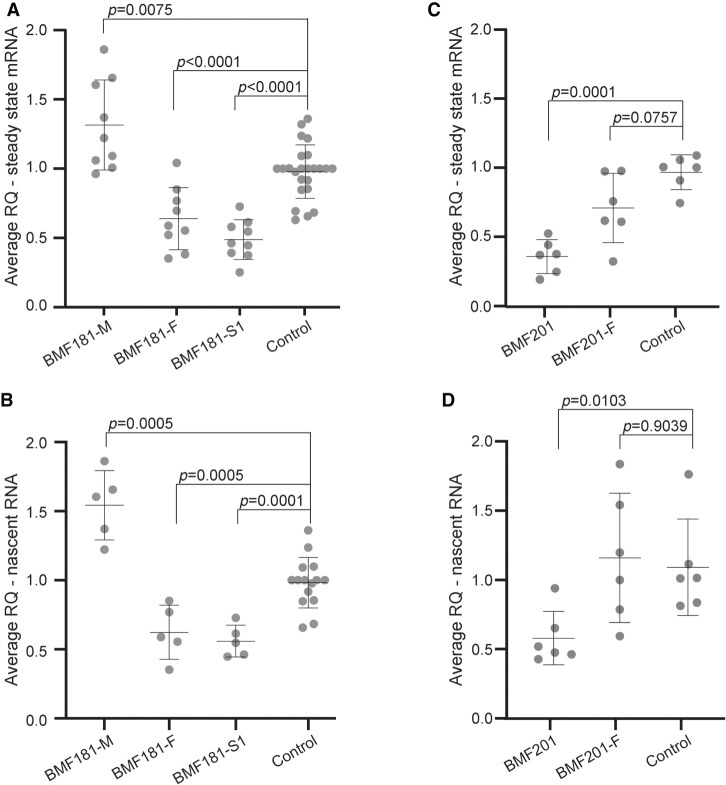
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) of cDNA generated from the polyadenylated RNA of LCLs demonstrated decreased ACD mRNA in BMF181-F and BMF181-S1 and increased ACD mRNA in BMF181-M compared to controls (Fig. 5A). Analysis of nascent ACD RNA using random hexamers for cDNA synthesis and forward and reverse primers within exon 1 and intron 1, respectively, for RT-qPCR showed similar results (Fig. 5B). Thus, despite a reduction in ACD nascent RNA and steady-state mRNA, telomere length was preserved within the normal range for age, arguing against haploinsufficiency of ACD.
Figure 5.
ACD steady-state mRNA and nascent transcript levels are reduced in the presence of the c.619delG variant. (A) Reverse transcriptase quantitative PCR (RT-qPCR) analysis of steady-state ACD mRNA expression relative to GAPDH control in BMF181 family LCLs. Three independent control LCL lines were used and grouped together for analysis. Control 2 was used to normalize values across experiments. Mean and standard deviation are shown. Results were log-transformed and then analyzed using one-way ANOVA followed by the Holm–Sidak test to adjust for multiple comparisons. N = 19 for control, N = 9 for all other samples. (RQ) Relative quantification, fold change compared to calibrator sample. (B) RT-qPCR analysis of nascent ACD RNA relative to GAPDH control in BMF181 family LCLs. Three independent control LCL lines were used and grouped together for analysis. Nascent transcripts were specifically amplified using primers spanning the exon 1-intron 1 junction. Results analyzed as in A. N = 15 for control, N = 5 for all other samples. (C) RT-qPCR analysis of steady-state ACD mRNA expression relative to GAPDH control in BMF201 family LCLs. A single control LCL line was used for comparison. Results analyzed as in A. N = 6. (D) RT-qPCR analysis of nascent ACD RNA relative to GAPDH control in BMF201 family LCLs. A single control LCL line was used for comparison. Results analyzed as in A. N = 6.
We predicted that BMF201 should resemble the phenotype of BMF181-F and BMF181-S1, with a decrease in both nascent and steady-state mRNA expression due to the loss of one ACD allele. Indeed, RT-qPCR of steady-state and nascent ACD transcripts revealed the expected reduction in BMF201 LCLs to ∼50% the level observed in a control LCL (Fig. 5C,D), supporting the argument against ACD haploinsufficiency.
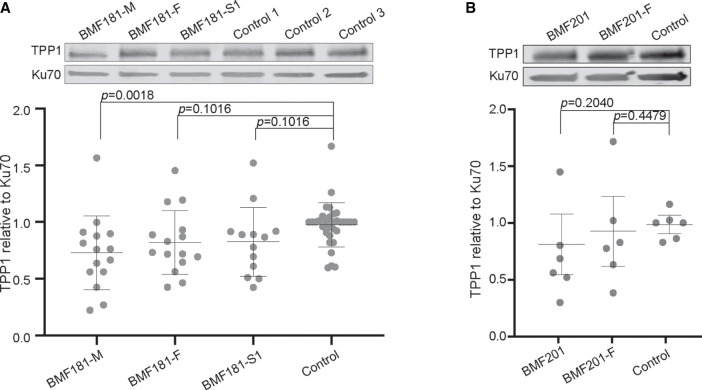
TPP1 Protein Levels Were Maintained in LCLs Containing Either Heterozygous ACD c.619delG or Chromosomal Deletion Encompassing ACD
The finding that telomere lengths were unaffected by the ACD frameshift and chromosomal deletion variants, in addition to the RT-qPCR data indicating that both nascent and steady state ACD mRNA levels were reduced in these cells, suggested that reduction in TPP1 by these variants was tolerated. However, we found that despite the presence of these variants, TPP1 protein levels were not significantly different in BMF181-F, BMF181-S1, or BMF201 LCLs relative to control (Fig. 6A,B; Supplemental Fig. 2C,D). Similarly, BMF181-M did not exhibit higher TPP1 protein levels, despite an increase in both nascent and steady-state mRNA (Fig. 6A; Supplemental Fig. 2C). In fact, statistical analysis showed BMF181-M TPP1 levels to be significantly lower than control; however, this might have been due to the specific TPP1 antibody we used (Bethyl A303-069A-M), which may have recognized an epitope inclusive of Glu169 (Supplemental Fig. 2G). To ensure that inherent variability in TPP1 expression did not affect our analysis, we conducted several western blots measuring TPP1 protein levels over time in the LCLs generated from BMF181 family members as well as one control line (Supplemental Fig. 2C). These experiments indicated that TPP1 levels are fairly stable in these LCLs, lending credibility to our results.
Figure 6.
TPP1 protein levels are maintained in the presence of heterozygous ACD c.619delG. (A) Representative western blot and quantification of TPP1 in BMF181 family LCLs. Ku70 was used as a loading control. Three independent control LCL lines were used and grouped together for analysis. Control 3 was used to normalize values across experiments. Mean and standard deviation are shown. Results were log-transformed and then analyzed using one-way ANOVA followed by the Holm–Sidak test to adjust for multiple comparisons. N = 15 for BMF181-M, BMF181-F, and BMF181-S1. N = 4 for controls 1 and 2. N = 24 for control 3. See Supplemental Figure 2C for compilation of western blots used for analysis. (B) Representative western blot and quantification of TPP1 in BMF201 family LCLs. A single control LCL line was used for comparison. Results analyzed as in A. No statistically significant difference was detected between the cell lines. N = 6. See Supplemental Figure 2D for compilation of western blots used for analysis.
DISCUSSION
Haploinsufficiency underlies abnormally short telomere lengths and TBD clinical features in individuals bearing pathogenic variants in TERT and TERC (Vulliamy et al. 2001; Armanios et al. 2005), highlighting the exquisite sensitivity of cells to sufficient telomerase levels. In contrast, the telomere lengths of the members of the two families described here argue against ACD haploinsufficiency, because the individuals with one WT and one LOF allele did not display marked telomere shortening. Although the number of individuals examined is small, the results are consistent with the lack of major TBD-associated clinical features reported for individuals with 16q22.1 microdeletion syndrome, the majority of which are hemizygous for ACD (Fig. 1B; Taysi et al. 1978; Fryns et al. 1981; Lin et al. 1983; Rivera et al. 1985; Cooke et al. 1987; Fujiwara et al. 1992; Gregor et al. 2013; Hori et al. 2017; Abdullah et al. 2019). Our results are also consistent with gnomAD data, which, as noted above, suggests ACD is not intolerant to LOF variants (o/e ratio 0.82, 90% confidence interval = 0.58–1.19). This is in stark contrast to TERT, which has a LOF o/e ratio of 0.16 (90% confidence interval = 0.09–0.29), indicating intolerance of LOF variants (Karczewski et al. 2020). It should also be noted that gnomAD contains 20 ACD frameshift variants (allele frequencies ranging from 4.58 × 10−5 to 4.57 × 10−6), indicating that such LOF variants can be found, however rarely, in the general population.
Although TPP1 is required for telomerase recruitment and processivity, tolerance of ACD LOF variants might be attributed to the relative abundance of TPP1 compared to telomerase (Fagerberg et al. 2014). However, the results of our studies instead suggest that the absence of telomere shortening secondary to a heterozygous ACD LOF variant is due to maintenance of TPP1 protein levels. We note that it is possible the results presented here, using EBV-transformed lymphocytes (LCLs), may produce slightly different results than if we had used primary cells, because LCLs have been known to show variability within and between cell lines as a result of the EBV transformation process. However, experiments conducted using peripheral blood mononuclear cells (PBMCs) stimulated with phytohemagglutinin A (PHA) (Supplemental Fig. 2E,F) produced highly variable results in our hands which did not lend themselves to reliable analysis. Given the reproducibility of our results using LCLs, we are satisfied that these are representative of the molecular workings of normal cells.
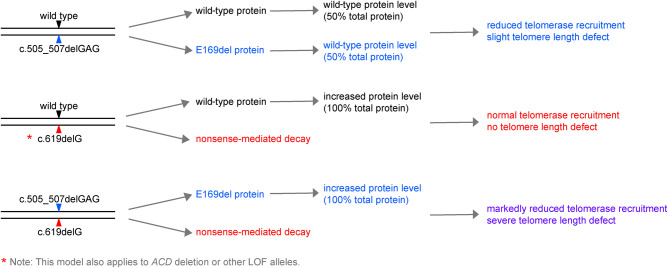
Application of the American College of Medical Genetics variant classification criteria (Richards et al. 2008) to the novel ACD c.505_507delGAG and c.619delG variants identified in BMF181 yields both as pathogenic (Supplemental Table 4). Our proposed model of the effect of these variants alone or in combination is outlined in Figure 7. In the case of heterozygosity of the ACD c.505_507delGAG, p.(Glu169del), allele, we predict that cells express the WT and variant allele in equal quantities, thus producing approximately equal amounts of the TPP1 WT and p.(Glu169del) proteins. Equal recruitment of the WT and variant proteins to telomeres would then result in reduced telomerase recruitment due to impaired interaction of TPP1 p.(Glu169del) with TERT, leading to reduced telomere extension and a slight telomere length defect as observed in BMF181-M. In the case of the ACD c.619delG, p.(Asp207Thrfs*22), variant, which is targeted for NMD, only the WT protein is expressed. This protein is subject to homeostatic regulation, resulting in normal telomerase recruitment and telomere length maintenance, as observed in BMF181-F and BMF181-S1. The Chr 16q22.1 microdeletion encompassing ACD in BMF201 would elicit the same effect as the NMD-targeted c.619delG variant. When compound heterozygous for c.505_507delGAG, p.(Glu169del) and c.619G, p.(Asp207Thrfs*22) variants, NMD of the frameshift allele would result in only the p.(Glu169del) variant protein being expressed and homeostatically regulated. Because the c.505_507delGAG, p.(Glu169del), variant exhibited reduced interaction with telomerase, as measured by reduced telomerase activity in vitro, this would lead to severely reduced telomere extension and clinical symptoms, as seen in BMF181.
Figure 7.
Working model for the impact of the BMF181 family and BMF201 ACD variants on TPP1 protein expression and telomere length. Presented is a schematic for our working model to explain the results described. According to our model, BMF181-M (top schematic) would express TPP1 WT and p.Glu169del in equal amounts, leading to reduced telomerase recruitment due to the interaction defect of TPP1 p.Glu169del with TERT, thus leading to telomere shortening. When combined with a second variant in trans, as in BMF181 (bottom schematic), the absence of WT TPP1 would result in rapid and dramatic telomere shortening. However, our model predicts that variants resulting in NMD, as in BMF181-F and BMF181-S1, or whole-gene deletions, as in BMF201, would result in increased WT protein to compensate for loss of a second functional allele, thus allowing normal telomerase recruitment (middle schematic).
The mechanism by which homeostatic regulation of TPP1 protein levels occurs requires further investigation. However, based on the RT-qPCR results, it is unlikely that ACD mRNA is being stabilized, because this would cause BMF181-F, BMF181-S1, and BMF201 steady-state mRNA levels to be comparable to the control (Fig. 5A,C). We can also conclude based on analysis of the nascent RNA (Fig. 5B,D) that ACD transcription is not being up-regulated. It is possible that translation is up-regulated or that TPP1 protein is stabilized, because either of these mechanisms would maintain protein levels without maintenance of WT RNA levels. Another possibility is that transport of ACD mRNA from the nucleus to the cytoplasm is affected, resulting in a higher number of transcripts being actively translated into protein.
Based on our clinical and in vitro findings, we surmise that the symptoms displayed by BMF201 were a result of hemizygosity of CTCF, which is involved in several cellular processes, including regulation of chromatin structure and transcription (Braccioli and de Wit 2019; Lazniewski et al. 2019), and is uniformly deleted in Chr 16q22.1 microdeletion syndrome, as illustrated in Figure 1B (Taysi et al. 1978; Fryns et al. 1981; Lin et al. 1983; Rivera et al. 1985; Cooke et al. 1987; Fujiwara et al. 1992; Gregor et al. 2013; Hori et al. 2017; Abdullah et al. 2019). We also note that the compound heterozygous nature of BMF181's ACD variants was a key factor in her severe clinical symptoms, because any modulation of TPP1 levels would affect only the impaired TPP1 p.(Glu169del) protein (Figs. 4 and 7). In summary, we conclude that ACD is not haploinsufficient and that tolerance of LOF variants is due to an as yet unknown mechanism by which TPP1 protein levels are maintained in the presence of diminished ACD mRNA.
METHODS
Telomere Length Analysis
Telomere flow FISH analysis was performed on peripheral blood leukocytes by RepeatDx as previously described (Baerlocher et al. 2006).
Generation and Culture of Lymphoblastoid Cell Lines (LCLs)
LCLs were generated by the tissue culture core laboratory within the Department of Molecular and Human Genetics, Baylor College of Medicine. LCLs were cultured in RPMI 1640 medium containing l-glutamine (Invitrogen) and 10% fetal bovine serum in 5% CO2.
DNA Sequencing
Clinical exome sequencing was conducted on BMF181 proband and parents according to standard protocols at Baylor Genetics. For sequence validation, genomic DNA was isolated from whole blood (BMF181) or LCLs (BMF181-M, BMF181-F, BMF181-S1, WT control LCLs) using the DNeasy Blood and Tissue kit (QIAGEN), and the region surrounding ACD c.505_507 and c.619 was PCR-amplified (Supplemental Table 5). The PCR products were analyzed by Sanger sequencing (Eurofins Genomics).
Chromosome Microarray Analysis (CMA) and Validation
CMA analysis was performed on a clinical basis by Quest Diagnostics. To validate the CMA data, genomic DNA was isolated from whole blood (BMF201, BMF201-F) and a WT LCL and PCR amplified using primers recognizing either CTCF intron 2 for the WT allele or regions flanking the chromosomal deletion (Supplemental Table 5). The PCR products were analyzed by Sanger sequencing (Eurofins Genomics).
Telomerase Repeated Amplification Protocol (TRAP)
293T cells were cotransfected with plasmids expressing TERT, hTR, and either empty vector, wild-type TPP1, or TPP1 p.Glu169del in a 1:1:2 ratio using Lipofectamine transfection reagent (Invitrogen 18324-012) according to manufacturer instructions. Cells were harvested at 72 h, and TRAP was conducted using the Millipore TRAPeze Telomerase Detection Kit (Millipore S7700) according to manufacturer's instructions. Briefly, cells were suspended in CHAPS buffer supplemented with protease inhibitors (Millipore 539134) and RNase inhibitors (Promega N2111), incubated on ice 30 min to lyse, then cleared by centrifuging at 12,000g for 20 min at 4°C. Protein concentration was determined using the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific 23225) according to manufacturer instructions. Three PCR reactions (total volume 25 μL) were set up for each sample, using 0.03 μg protein, 0.006 μg protein, and 0.03 μg protein that had been heat inactivated for 10 min at 95°C. Control reactions included telomerase positive cells (1:10 dilution of telomerase positive cells provided with TRAPeze kit and lysed via kit instructions, active and heat-killed), a PCR positive control provided with the TRAPeze kit, and negative controls using CHAPS buffer or water. Reactions cycled as follows: 1 cycle for 30 min at 30°C, 1 cycle for 2 min at 95°C, 34 cycles for 15 sec at 94°C, for 30 sec at 59°C, for 1 min at 72°C. A 12.5% nondenaturing PAGE gel was used to separate the products as described by the manufacturer. Products were visualized by staining in 1 μg/mL ethidium bromide for 1 h, and then destaining in distilled deionized water for 1 h before imaging. Results were analyzed using the densitometry function in ImageJ (Schneider et al. 2012; https://imagej.nih.gov/ij/index.html).
Reverse Transcriptase Quantitative PCR (RT-qPCR)
RNA was isolated using the RNeasy kit (QIAGEN 74106) according to manufacturer instructions and quantified using a NanoDrop 2000 spectrophotometer. One microgram RNA was reverse transcribed using the qScript Flex cDNA synthesis kit (Quantabio 95049) according to manufacturer instructions. Briefly, RNA was combined with the primer (oligo-dT for mRNA or random hexamer for total RNA), denatured for 5 min at 65°C and annealed on ice. 5× qScript Flex Reaction Mix and qScript RTase were added, then incubated for 90 min at 42°C, for 5 min at 85°C, and held at 4°C. Products were diluted with nine volumes of RNase-free water. RT-qPCR was conducted using PowerUp SYBR Green reaction mix (Applied Biosystems A25742) according to manufacturer instructions. Four microliters of the diluted cDNA (∼20 ng) were used per reaction. Samples were analyzed on a QuantStudio 6 Flex thermocycler with the following cycle conditions: one cycle for 2 min at 50°C, one cycle for 2 min at 95°C, 40 cycles for 15 min at 95°C and then 1 min at 60°C. Each step ramped at a rate of 1.6°C/sec. Melt curve analysis was conducted as follows: 1.6°C/sec ramp from 60°C to 95°C, hold 15 sec; 1.6°C/sec ramp to 60°C, hold 1 min; 0.15°C/sec ramp to 95°C, hold 15 sec. The relative changes in RNA abundance were calculated by comparative ΔCt method with normalization to GAPDH. Independent biological replicates were collected for each experiment from continuously growing LCL cultures. A single RNA preparation was used to generate cDNA corresponding to mRNA or total RNA, as indicated, for each sample. Three independent control LCLs were used for each experiment with BMF181 family LCLs, resulting in a higher number of control data points compared to each of the experimental LCLs. A single control LCL was used for the BMF201 family experiments.
Western Blot Analyses
Cells were pelleted, washed in PBS, and resuspended in RIPA lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% Igepal [Sigma-Aldrich CA-630], 0.5% sodium deoxycholate, 0.1% SDS, 5 mM EDTA, 1 mM PMSF, 1× protease inhibitors [Millipore 539134]) for 10 min on ice. Lysed cells were sonicated in a Diagenode Bioruptor UCD-200 on high for 5 min (30 sec on, 30 sec off), centrifuged, and the supernatant transferred to a new tube. Protein concentration was quantified using Pierce BCA protein assay kit (Thermo Scientific 23225) according to manufacturer instructions. Proteins were separated on a 4%–20% gradient gel (BioRad 456-1094) and transferred to Immobilon-FL PVDF membrane (Millipore IPFL00010). Membranes were probed with TPP1 primary antibody (Bethyl A303-069A-M, diluted 1:500) or Ku70 primary antibody (Lab Vision/NeoMarkers MS-329-P1, diluted 1:1000). Independent biological replicates were collected for each experiment from continuously growing LCL cultures. Three independent control LCLs were used for each experiment with BMF181 family LCLs, with the exception of blots investigating TPP1 expression variability over time, resulting in a higher number of control data points compared to each of the experimental LCLs. A single LCL was used for the BMF201 family experiments. The identity of the TPP1 band was verified by siRNA knockdown (Supplemental Fig. 2A). Imaging was conducted on Odyssey and Odyssey CLx systems, and bands were quantified using Image Studio Lite software. For TRAP experiments, membranes were probed with primary antibody against TERT (Abcam ab32020, diluted 1:1000), myc (Sigma-Aldrich M4439, diluted 1:1000), or β-actin (Sigma-Aldrich a5441, diluted 1:1000). For antibody characterization experiments, membranes were probed with primary antibodies against TPP1 (Bethyl A303-069A-M, diluted 1:500), myc (Sigma-Aldrich M4439, diluted 1:1000), neomycin phosphotransferase II (Millipore AC113 1:1000), and Ku70 (Lab Vision/NeoMarkers MS-329-P1, diluted 1:1000).
Statistical Analyses
Western and RT-qPCR data were analyzed using GraphPad Prism. All data sets were log transformed and then analyzed using one-way ANOVA followed by the Holm–Sidak test for multiple comparisons. A P-value < 0.05 was considered statistically significant.
ADDITIONAL INFORMATION
Data Deposition and Access
The BMF181 family ACD (NM_001082486.1) variants c.619delG, p.(Asp207Thrfs*22) and c.505_507delGAG, p.(Glu169del) were deposited to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under the accession numbers SCV001450737 and SCV001450738, respectively. Patient consent was not granted to deposit WES data. BMF201's chromosomal deletion, arr[hg19] 16q22.1(67,628,846-67,813,408)x1, was deposited to ClinVar under accession number SCV001450739.
Ethics Statement
This investigation was conducted according to the Declaration of Helsinki Principles. Informed written consent was received from participants (or their parents, if minors) before inclusion in the study according to protocol H-7698 Genetic and Biological Determinants of Bone Marrow Failure approved by the Institutional Review Board for Baylor College of Medicine and affiliated hospitals.
Acknowledgments
We thank the BMF181 and BMF201 families for their participation in this study as well as the healthy individuals who served as controls.
Author Contributions
G.H. and A.A.B. designed the experiments. G.H. and C.L.W. conducted the experiments. P.L. provided assistance in analyzing exome sequence data. G.H. and A.A.B. wrote the manuscript.
Funding
This study was funded by National Institutes of Health award R01HL131744 to A.A.B. and T32GM008231 program training grant to the Integrative Molecular and Biomedical Sciences graduate program, Baylor College of Medicine.
Competing Interest Statement
Baylor College of Medicine (BCM) and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of Baylor Genetics (BG), which performs genetic testing and derives revenue. Pengfei Liu is an employee of BCM and derives support through a professional services agreement with BG.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Abdullah S, Helal M, Dupuis L, Stavropoulos DJ, Louro P, Ramos L, Mendoza-Londono R. 2019. 16q22.1 microdeletion and anticipatory guidance. Am J Med Genet A 179: 1287–1292. 10.1002/ajmg.a.61155 [DOI] [PubMed] [Google Scholar]
- Allsopp RC, Harley CB. 1995. Evidence for a critical telomere length in senescent human fibroblasts. Exp Cell Res 219: 130–136. 10.1006/excr.1995.1213 [DOI] [PubMed] [Google Scholar]
- Alter BP, Baerlocher GM, Savage SA, Chanock SJ, Weksler BB, Willner JP, Peters JA, Giri N, Lansdorp PM. 2007. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood 110: 1439–1447. 10.1182/blood-2007-02-075598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP, Giri N, Savage SA, Rosenberg PS. 2009. Cancer in dyskeratosis congenita. Blood 113: 6549–6557. 10.1182/blood-2008-12-192880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP, Giri N, Savage SA, Peters JA, Loud JT, Leathwood L, Carr AG, Greene MH, Rosenberg PS. 2010. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol 150: 179–188. 10.1111/j.1365-2141.2010.08212.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoude LG, Pritchard AL, Robles-Espinoza CD, Wadt K, Harland M, Choi J, Gartside M, Quesada V, Johansson P, Palmer JM, et al. 2015. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J Natl Cancer Inst 107: dju408 10.1093/jnci/dju408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Salgado EG, Galvez E, Planas-Cerezales L, Pintado-Berninches L, Vallespin E, Martinez P, Carrillo J, Iarriccio L, Ruiz-Llobet A, Catala A, et al. 2019. Genetic analyses of aplastic anemia and idiopathic pulmonary fibrosis patients with short telomeres, possible implication of DNA-repair genes. Orphanet J Rare Dis 14: 82 10.1186/s13023-019-1046-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M, Chen JL, Chang YP, Brodsky RA, Hawkins A, Griffin CA, Eshleman JR, Cohen AR, Chakravarti A, Hamosh A, et al. 2005. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci 102: 15960–15964. 10.1073/pnas.0508124102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. 2000. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406: 641–645. 10.1038/35020592 [DOI] [PubMed] [Google Scholar]
- Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. 2006. Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat Protoc 1: 2365–2376. 10.1038/nprot.2006.263 [DOI] [PubMed] [Google Scholar]
- Barbaro PM, Ziegler DS, Reddel RR. 2016. The wide-ranging clinical implications of the short telomere syndromes. Intern Med J 46: 393–403. 10.1111/imj.12868 [DOI] [PubMed] [Google Scholar]
- Berthet F, Caduff R, Schaad UB, Roten H, Tuchschmid P, Boltshauser E, Seger RA. 1994. A syndrome of primary combined immunodeficiency with microcephaly, cerebellar hypoplasia, growth failure and progressive pancytopenia. Eur J Pediatr 153: 333–338. 10.1007/BF01956413 [DOI] [PubMed] [Google Scholar]
- Bisht K, Smith EM, Tesmer VM, Nandakumar J. 2016. Structural and functional consequences of a disease mutation in the telomere protein TPP1. Proc Natl Acad Sci 113: 13021–13026. 10.1073/pnas.1605685113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braccioli L, de Wit E. 2019. CTCF: a Swiss-army knife for genome organization and transcription regulation. Essays Biochem 63: 157–165. 10.1042/EBC20180069 [DOI] [PubMed] [Google Scholar]
- Broer L, Codd V, Nyholt DR, Deelen J, Mangino M, Willemsen G, Albrecht E, Amin N, Beekman M, de Geus EJ, et al. 2013. Meta-analysis of telomere length in 19,713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur J Hum Genet 21: 1163–1168. 10.1038/ejhg.2012.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado RT, Regal JA, Kleiner DE, Schrump DS, Peterson NR, Pons V, Chanock SJ, Lansdorp PM, Young NS. 2009. A spectrum of severe familial liver disorders associate with telomerase mutations. PLoS ONE 4: e7926 10.1371/journal.pone.0007926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete O, Martinez P, Garcia-Pavia P, Benitez-Buelga C, Paumard-Hernández B, Fernandez V, Dominguez F, Salas C, Romero-Laorden N, Garcia-Donas J, et al. 2015. A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li–Fraumeni-like families. Nat Commun 6: 8383 10.1038/ncomms9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke A, Tolmie J, Darlington W, Boyd E, Thomson R, Ferguson-Smith MA. 1987. Confirmation of a suspected 16q deletion in a dysmorphic child by flow karyotype analysis. J Med Genet 24: 88–92. 10.1136/jmg.24.2.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198. 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- Dodson L, Bertuch AA. 2018. Dyskeratosis congenita and the telomere biology disorders. In Bone marrow failure (ed. Kupfer GM, Reaman GH, Smith FO), pp. 111–135. Springer International Publishing, Cham. [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13: 397–406. 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryns JP, Proesmans W, Van Hoey G, Van den Berghe H. 1981. Interstitial 16q deletion with typical dysmorphic syndrome. Ann Genet 24: 124–125. [PubMed] [Google Scholar]
- Fujiwara M, Yoshimoto T, Morita Y, Kamada M. 1992. Interstitial deletion of Chromosome 16q: 16q22 is critical for 16q- syndrome. Am J Med Genet 43: 561–564. 10.1002/ajmg.1320430311 [DOI] [PubMed] [Google Scholar]
- Gable DL, Gaysinskaya V, Atik CC, Talbot CC, Kang B, Stanley SE, Pugh EW, Amat-Codina N, Schenk KM, Arcasoy MO, et al. 2019. ZCCHC8, the nuclear exosome targeting component, is mutated in familial pulmonary fibrosis and is required for telomerase RNA maturation. Genes Dev 33: 1381–1396. 10.1101/gad.326785.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri N, Ravichandran S, Wang Y, Gadalla SM, Alter BP, Fontana J, Savage SA. 2019. Prognostic significance of pulmonary function tests in dyskeratosis congenita, a telomere biology disorder. ERJ Open Res 5: 00209-2019 10.1183/23120541.00209-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glousker G, Touzot F, Revy P, Tzfati Y, Savage SA. 2015. Unraveling the pathogenesis of Hoyeraal–Hreidarsson syndrome, a complex telomere biology disorder. Br J Haematol 170: 457–471. 10.1111/bjh.13442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Stock AJ, Liu Y. 2020. The enigma of excessively long telomeres in cancer: lessons learned from rare human POT1 variants. Curr Opin Genet Dev 60: 48–55. 10.1016/j.gde.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor A, Oti M, Kouwenhoven EN, Hoyer J, Sticht H, Ekici AB, Kjaergaard S, Rauch A, Stunnenberg HG, Uebe S, et al. 2013. De novo mutations in the genome organizer CTCF cause intellectual disability. Am J Hum Genet 93: 124–131. 10.1016/j.ajhg.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S, Bisht K, Tesmer VM, Shami AN, Hammoud SS, Nandakumar J. 2019. Two separation-of-function isoforms of human TPP1 dictate telomerase regulation in somatic and germ cells. Cell Rep 27: 3511–3521 e3517. 10.1016/j.celrep.2019.05.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Kartawinata M, Li J, Pickett HA, Teo J, Kilo T, Barbaro PM, Keating B, Chen Y, Tian L, et al. 2014. Inherited bone marrow failure associated with germline mutation of ACD, the gene encoding telomere protein TPP1. Blood 124: 2767–2774. 10.1182/blood-2014-08-596445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345: 458–460. 10.1038/345458a0 [DOI] [PubMed] [Google Scholar]
- He H, Li W, Comiskey DF, Liyanarachchi S, Nieminen TT, Wang Y, DeLap KE, Brock P, de la Chapelle A. 2020. A truncating germline mutation of TINF2 in individuals with thyroid cancer or melanoma results in longer telomeres. Thyroid 30: 204–213. 10.1089/thy.2019.0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herate C, Sabatier L. 2020. Telomere instability initiates and then boosts carcinogenesis by the butterfly effect. Curr Opin Genet Dev 60: 92–98. 10.1016/j.gde.2020.01.005 [DOI] [PubMed] [Google Scholar]
- Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. 2004. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol Cell 14: 501–513. 10.1016/S1097-2765(04)00256-4 [DOI] [PubMed] [Google Scholar]
- Hoffman TW, van der Vis JJ, van der Smagt JJ, Massink MPG, Grutters JC, van Moorsel CHM. 2019. Pulmonary fibrosis linked to variants in the ACD gene, encoding the telomere protein TPP1. Eur Respir J 54: 1900809 10.1183/13993003.00809-2019 [DOI] [PubMed] [Google Scholar]
- Hori I, Kawamura R, Nakabayashi K, Watanabe H, Higashimoto K, Tomikawa J, Ieda D, Ohashi K, Negishi Y, Hattori A, et al. 2017. CTCF deletion syndrome: clinical features and epigenetic delineation. J Med Genet 54: 836–842. 10.1136/jmedgenet-2017-104854 [DOI] [PubMed] [Google Scholar]
- Houghtaling BR, Cuttonaro L, Chang W, Smith S. 2004. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol 14: 1621–1631. 10.1016/j.cub.2004.08.052 [DOI] [PubMed] [Google Scholar]
- Hoyeraal HM, Lamvik J, Moe PJ. 1970. Congenital hypoplastic thrombocytopenia and cerebral malformations in two brothers. Acta Paediatr Scand 59: 185–191. 10.1111/j.1651-2227.1970.tb08986.x [DOI] [PubMed] [Google Scholar]
- Hreidarsson S, Kristjansson K, Johannesson G, Johannsson JH. 1988. A syndrome of progressive pancytopenia with microcephaly, cerebellar hypoplasia and growth failure. Acta Paediatr Scand 77: 773–775. 10.1111/j.1651-2227.1988.tb10751.x [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581: 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibe T, Zimmermann M, de Lange T. 2016. TPP1 blocks an ATR-mediated resection mechanism at telomeres. Mol Cell 61: 236–246. 10.1016/j.molcel.2015.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015. 10.1126/science.7605428 [DOI] [PubMed] [Google Scholar]
- Kirwan M, Dokal I. 2008. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet 73: 103–112. 10.1111/j.1399-0004.2007.00923.x [DOI] [PubMed] [Google Scholar]
- Kocak H, Ballew BJ, Bisht K, Eggebeen R, Hicks BD, Suman S, O'Neil A, Giri N, Maillard I, Alter BP, et al. 2014. Hoyeraal–Hreidarsson syndrome caused by a germline mutation in the TEL patch of the telomere protein TPP1. Genes Dev 28: 2090–2102. 10.1101/gad.248567.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrick CM, Cech TR. 2010. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J 29: 924–933. 10.1038/emboj.2009.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazniewski M, Dawson WK, Rusek AM, Plewczynski D. 2019. One protein to rule them all: the role of CCCTC-binding factor in shaping human genome in health and disease. Semin Cell Dev Biol 90: 114–127. 10.1016/j.semcdb.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Lowry RB, Snyder FF. 1983. Interstitial deletion for a region in the long arm of Chromosome 16. Hum Genet 65: 134–138. 10.1007/BF00286649 [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. 2004. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol 6: 673–680. 10.1038/ncb1142 [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. 2012. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492: 285–289. 10.1038/nature11648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry EM, Alder JK, Qi X, Chen JJ, Armanios M. 2011. Syndrome complex of bone marrow failure and pulmonary fibrosis predicts germline defects in telomerase. Blood 117: 5607–5611. 10.1182/blood-2010-11-322149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. 2008. The telomerase database. Nucleic Acids Res 36: D339–D343. 10.1093/nar/gkm700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE, Molecular Subcommittee of the ALQAC. 2008. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med 10: 294–300. 10.1097/GIM.0b013e31816b5cae [DOI] [PubMed] [Google Scholar]
- Rivera H, Vargas-Moyeda E, Moller M, Torres-Lamas A, Cantu JM. 1985. Monosomy 16q: a distinct syndrome. Apropos of a de novo del(16) (q2100q2300). Clin Genet 28: 84–86. 10.1111/j.1399-0004.1985.tb01223.x [DOI] [PubMed] [Google Scholar]
- Savage SA. 2018. Beginning at the ends: telomeres and human disease. F1000Res 7: F1000 Faculty Rev-524 10.12688/f1000research.14068.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz I, Mensenkamp AR, Takai KK, Haadsma M, Spruijt L, de Voer RM, Choo SS, Lorbeer FK, van Grinsven EJ, Hockemeyer D, et al. 2020. TINF2 is a haploinsufficient tumor suppressor that limits telomere length. Elife 9: e61235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AN, Youmans DT, Collins K. 2012. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem 287: 34455–34464. 10.1074/jbc.M112.394767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AN, Regalado SG, Lai CS, Cost GJ, O'Neil CM, Urnov FD, Gregory PD, Jaenisch R, Collins K, Hockemeyer D. 2014. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev 28: 1885–1899. 10.1101/gad.246819.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Yang XR, Ballew B, Rotunno M, Calista D, Fargnoli MC, Ghiorzo P, Bressac-de Paillerets B, Nagore E, Avril MF, et al. 2014. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet 46: 482–486. 10.1038/ng.2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speedy HE, Kinnersley B, Chubb D, Broderick P, Law PJ, Litchfield K, Jayne S, Dyer MJS, Dearden C, Follows GA, et al. 2016. Germ line mutations in shelterin complex genes are associated with familial chronic lymphocytic leukemia. Blood 128: 2319–2326. 10.1182/blood-2016-01-695692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taysi K, Fishman M, Sekhon GS. 1978. A terminal long arm deletion of Chromosome 16 in a dysmorphic infant: 46,XY,del(16) (q22). Birth Defects Orig Artic Ser 14: 343–347. [PubMed] [Google Scholar]
- Tummala H, Collopy LC, Walne AJ, Ellisen LW, Cardoso S, Aksu T, Yarali N, Aslan D, Akata RF, Teo J, et al. 2018. Homozygous OB-fold variants in telomere protein TPP1 are associated with dyskeratosis congenita like phenotypes. Blood 132: 1349–1353. 10.1182/blood-2018-03-837799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM. 1994. Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci 91: 9857–9860. 10.1073/pnas.91.21.9857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. 2001. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413: 432–435. 10.1038/35096585 [DOI] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. 2007. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510. 10.1038/nature05454 [DOI] [PubMed] [Google Scholar]
- Wong K, Robles-Espinoza CD, Rodriguez D, Rudat SS, Puig S, Potrony M, Wong CC, Hewinson J, Aguilera P, Puig-Butille JA, et al. 2019. Association of the POT1 germline missense variant p.I78T with familial melanoma. JAMA Dermatol 155: 604–609. 10.1001/jamadermatol.2018.3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. 1996. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet 18: 173–179. [DOI] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. 2007. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature 445: 559–562. 10.1038/nature05469 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS. 2005. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med 352: 1413–1424. 10.1056/NEJMoa042980 [DOI] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. 2004. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev 18: 1649–1654. 10.1101/gad.1215404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. 2012. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150: 481–494. 10.1016/j.cell.2012.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The BMF181 family ACD (NM_001082486.1) variants c.619delG, p.(Asp207Thrfs*22) and c.505_507delGAG, p.(Glu169del) were deposited to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under the accession numbers SCV001450737 and SCV001450738, respectively. Patient consent was not granted to deposit WES data. BMF201's chromosomal deletion, arr[hg19] 16q22.1(67,628,846-67,813,408)x1, was deposited to ClinVar under accession number SCV001450739.