Abstract
Since the outbreak of SARS-CoV-2, the etiologic agent of the COVID-19 pandemic, the viral genome has acquired numerous mutations with the potential to increase transmission. One year after its emergence, we now further analyze emergent SARS-CoV-2 genome sequences in an effort to understand the evolution of this virus.
Since the outbreak of SARS-CoV-2, the etiologic agent of the COVID-19 pandemic, the viral genome has acquired numerous mutations with the potential to increase transmission. One year after its emergence, we now further analyze emergent SARS-CoV-2 genome sequences in an effort to understand the evolution of this virus.
Main text
The global coronavirus disease 2019 (COVID-19) pandemic has incurred over 100 million confirmed cases and more than 2 million fatalities since December of 2019 (https://covid19.who.int/). As an RNA virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has a relatively high mutation rate that results in abundant variations within its genome. In the past year, over 20,000 mutations (https://bigd.big.ac.cn/ncov/variation/annotation) and some insertion/deletions have been detected in SARS-CoV-2 strains. Many of these are located in the viral spike (S) protein that engages the host receptor ACE2 for target cell entry. Notably, viral strains with the D614G mutation in S protein and three recent variants (501Y.V1, 501Y.V2, and 501Y.V3) with the shared N501Y mutation in the S protein have raised global concerns and been extensively studied. The D614G mutation alters the S protein to an ACE-2-binding, fusion-competent conformation, thereby increasing viral transmission by enhancing viral replication in the upper respiratory tract of COVID-19 patients (Plante et al., 2020; Yurkovetskiy et al., 2020). Position 501 in the S protein has been identified as one of six residues comprising the receptor binding domain (RBD), and the N501Y mutation has been shown to enhance the binding affinity of SARS-CoV-2 to human ACE2 in vitro (Starr et al., 2020). Although mutations that enhance viral infectivity or transmissibility have been detected, the majority of mutations likely negatively affect viral fitness (Grubaugh et al., 2020). Therefore, heritable mutations or mutations that recurrently appear in viral populations should be given our utmost attention, because these mutations may have a positive effect on viral fitness and indicate future evolutionary directions. Deletions are of particular interest because they escape the proofreading function of the coronavirus RNA-dependent RNA polymerase, potentially accelerating coronaviral evolution. Recent evidence has demonstrated the existence of recurrent deletion regions (RDRs) that map to defined antibody epitopes, and deletions in these regions appear to emerge independently in a parallel, convergent pattern of viral antigenic evolution that may confer resistance to neutralizing antibodies (McCarthy et al., 2020). Here, we identify the mutations and deletions accumulated throughout the past year within representative genome sequences of SARS-CoV-2, explore the possible epidemiological patterns of potentially parallel mutations, and evaluate the impact of existing and potential mutations on the efficacy of monoclonal antibodies and vaccines.
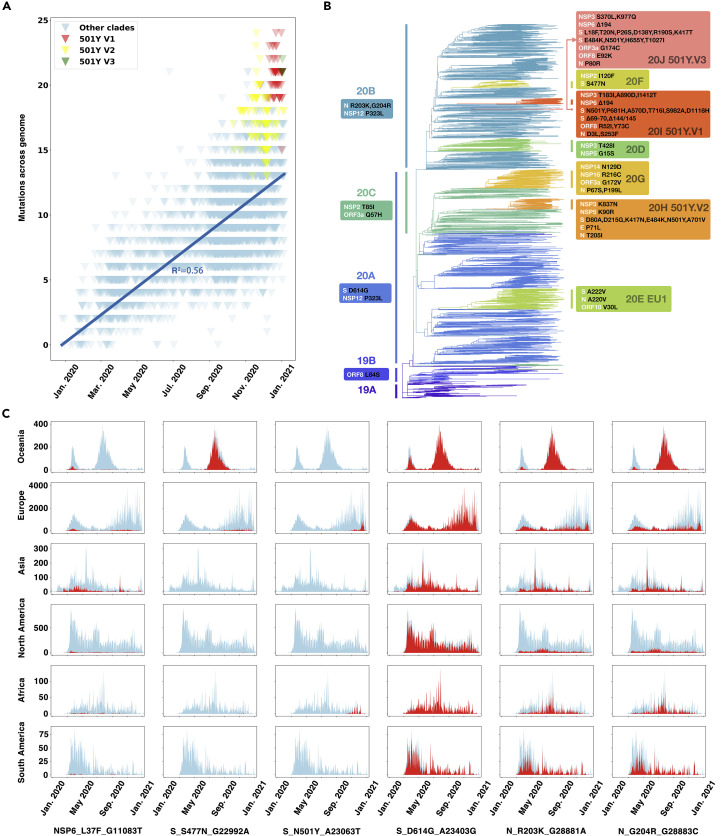
As of January 11th, 2021, a total of 355,067 SARS-CoV-2 genome sequences were available in GISAID (Shu and McCauley, 2017), an invaluable resource for detecting the evolution of SARS-CoV-2 and tracking its transmission. By comparing 3,823 representative viral genomes (referring to the result of Nextstrain, https://nextstrain.org/ncov/global, as of January 11th, 2021) to the early reference strain (EPI_ISL_402125), we found that SARS-CoV-2 accumulated about 0.035 amino acid mutations per day on average within the past year (Figure 1 A), while S proteins showed a nonlinear variation pattern, which might be the result of different selection pressures on the whole genome compared to the S protein. The three recently identified SARS-CoV-2 variants N501Y.V1, N501Y.V2, and N501Y.V3 harbor a relatively large number of mutations (Figures 1A and 1B). The heritable amino acid mutations in the major strain clades identified by Nextstrain (Hadfield et al., 2018) are shown hierarchically in the phylogenetic tree (Figure 1B). Some of these mutations have been given considerable attention. Specifically, all viral strains in clade 20A have the featured mutation D614G in the S protein, while the viral strains in clade 20B feature two additional mutations, R203K and G204R, in the nucleocapsid (N) protein. Within the S protein, the 20A subclade 20H (501Y.V2) has the heritable mutations D80A, K417N, E484K, N501Y, D614G, and A701V (Figure 1B); the 20B subclade 20J (501Y.V3) is featured with ten substitutions including K417T, E484K, and N501Y, while 20I (501Y.V1) has the heritable mutations N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H as well as deletions at 69–70 and 144/145 (Figure 1B).
Figure 1.
Phylogenetic pathway and spatiotemporal distribution of accumulated mutations in the SARS-CoV-2 genomes
(A) The accumulated mutations in SARS-CoV-2 strains compared with early reference strain EPI_ISL_402125 since January of 2021. The 501Y.V1, 501Y.V2, and 501Y.V3 sub-clades were colored as red, yellow, and green, respectively. The linear regression line was shown and labeled.
(B) Phylogenetic tree with fixed amino acid mutations for representative SARS-CoV-2 strains selected by Nextstrain until January 11th, 2021. Fixed mutations detected in each cluster were displayed in boxes. Viral strains were divided into hierarchical clusters as those in Nextstrain, including four big clusters (19A, 19B, 20A, and 20B) and eight small clusters (20C, 20D, 20E, 20F, 20G, 20H, 20I, and 20J).
(C) The spatiotemporal distribution of SARS-CoV-2 genomes collected in GISAID with the six representative mutations during January 2020 to January 2021. In the histogram for each continent/mutation pairing, the x axis represents the collection date of the sequenced viruses and the y axis represents the number of sequences with the indicated mutation. The number of mutated sequences is shown in red, with the total number of all strains shown in light blue for comparison.
Collectively, we detected a total of 130 nucleotide mutations acquired by SARS-CoV-2 genomes in the past year; of these, 75 are heritable, non-synonymous mutations (Figure S1). Viral evolution studies have indicated that parallel mutations and independently recurrent mutations have higher associations with viral adaptation (van Dorp et al., 2020). Thus, from the 75 non-synoymous mutations, we further identified 24 heritable mutations, including the two well-known mutations D614G and N501Y, that potentially arose in parallel (Table S1A). It should be noted that the potentially parallel mutations detected here were based on representative sequences and thus could be highly under-estimated. To investigate the occurrence and transmission of these potentially parallel mutations, we plotted the spatiotemporal distribution of sequences from GISAID with these mutations in the past year as an indicator of their possible epidemiological distribution (Figures 1C and S2). Significant epidemiological patterns were observed for these mutations in the SARS-CoV-2 genomes (Figures 1C and S2). Of them, the D614G mutation is notable for having raised global concern over its rapid transmission and dominance. The L37F mutation in nonstrucutral protein 6 (NSP6) protein has appeared frequently in different clades and across continents. The N501Y mutation in the S protein is closely related to the recent outbreak in the United Kingdom, South Africa, and Brazil. The S477N mutation was likely responsible for causing the epidemic from July to September of 2020 in Oceania. Strikingly, two potentially parallel mutations observed in the Ser/Arg (SR)-rich linker region of the N protein (R203K and G204R) co-occurred across six continents (Figure 1C).
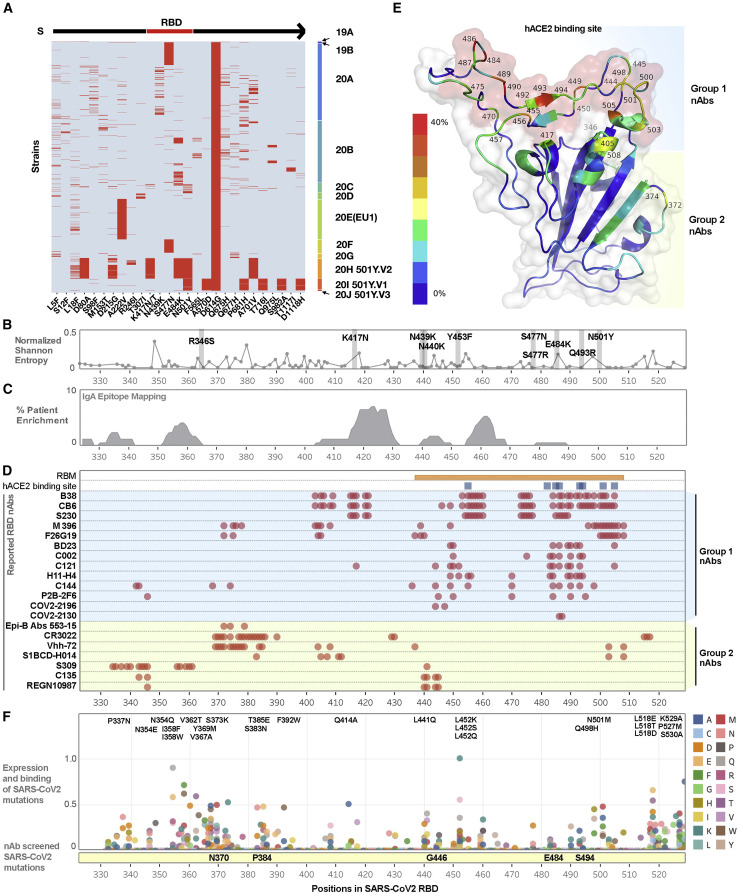
To evaluate the current status of therapeutic antibodies against mutations in the S protein of SARS-CoV-2, we first identified the most prevalent mutations in the S protein, as illustrated by the heatmap (Figure 2 A). There are ten key mutations located in the RBD of the S protein. We further analyzed the phylogenetic pattern of these mutations (Figure S3) and evaluated their positional relationship to the distribution of genetic diversity score across RBD residues (Figure 2B). Based on the reported epitope mapping of COVID-19 patients by Shrock et al. (Figure 2C), despite most COVID-19 patients producing antibodies against SARS-CoV-2 proteins, the levels of neutralizing antibodies (nAbs) vary among individual patients and typically correspond to the severity of the infection (Shrock et al., 2020). Therefore, it is not clear whether these patients, especially those with mild disease, will produce sufficient amounts of nAbs against SARS-CoV-2 to prevent reinfection (Bošnjak et al., 2020). Recent studies have already indicated that mutations or indels in the RBD region may impact the neutralization efficacy of nAbs (Wang et al., 2021). To evaluate the current state of therapeutic antibodies, we determined the frequency of epitopes corresponding to 20 reported nAbs (listed in Figure 2D and Table S1B) and visualized epitope frequency within the RBD structure (Figure 2E). Based on these analyses, we classified these nAbs into two groups. Group 2 nAbs bind epitopes closer to the N terminus of the RBD (330–430), whereas group 1 nAbs bind epitopes mostly within the receptor binding motif (RBM).
Figure 2.
Systematic analysis of SARS-CoV-2 RBD mutations and neutralizing antibodies
(A) Recurrent (appeared more than 20 times or >0.52% in the representative sequences) amino acid mutations in the S protein of SARS-CoV-2. To more clearly display the varied mutations in each strain, we show only those viral strains with the D614G mutation in the S protein. Each row in the y axis of the heatmap represent a viral strain; these strains were grouped by clade and plotted in the order on the right. Each red line represents a mutation that occurred with the pattern annoted in the x axis. Among them, four mutations, K417N, N439K, S477N, and N501Y, are located in the RBD of the S protein.
(B) Genetic diversity across residues of the RBD using representative SARS-CoV-2 strains selected by Nextstrain updated on January 11th, 2021. Ten recently reported mutations were labeled and highlighted.
(C) Reported enriched SARS-CoV-2 antigenic sites as identified by unbiased screening using phage immunoprecipitation (IP) with serum immunoglobulin A (IgA) of infected cohorts.
(D) Epitope mapping of currently reported neutralizing monoclonal antibodies that target the RBD.
(E) SARS-CoV-2 RBD monomer structure (PDB:7BZ5). Ribbon is colored by the frequencies of epitopes in the nAbs listed in Figure 2C. The red color indicates the highest frequency while the dark blue indicates the lowest (0 in 20). Known human ACE2 binding site is shown using surface view and is colored in red.
(F) Reported unbiased mutations of each amino acid in the RBD shown as a combination of mutation expression levels and human ACE2 binding affinity. Mutants with the highest combined scores are listed, and the reported experimentally determined escape mutants have been highlighted in the yellow bar below.
Due to the highly plastic nature of SARS-CoV-2, we further evaluated potential mutations based on previously reported data from Starr et al. (Figure 2F). Here, we integrated the expression and binding data from the unbiased screening of all potential mutations, revealing several potential mutations especially in the 330–370 and 518–530 regions (Starr et al., 2020). A recent study by Greaney et al. further demonstrated selection of escape mutations using a combination of nAbs (Greaney et al., 2021). It’s unclear whether these mutations will occur in nature and how they would impact our immune protection.
Numerous efforts have now been made to study the effects of mutations on the efficacy of vaccines and monoclonal antibodies (Poland et al., 2020). Mutations that affect viral replication do not necessarily correlate with mutations that escape immune protection. As an example, although they all share the same D614G mutation, the E484K-mutation-containing 501Y.V2 and 501Y.V3 but not the 501Y.V1 varients resisted nAbs against the RBD, as D614G and E484K mutations may contribute to the evolution of SARS-CoV-2 by enhancing viral infectivity and reducing immune protection, respectively (Korber et al., 2020; Plante et al., 2020; Wang et al., 2021). Given that most of the neutralization assays were carried out in vitro with S protein or pseudoviruses, more in vivo and especially human studies with intact viruses are needed to accurately evaluate whether current or future SARS-CoV-2 mutants may escape from monoclonal antibody treatments or vaccine protection that contains both antibody and cellular immunity.
We should be concerned about the rapid growth and spread of various SARS-CoV-2 mutants. The SARS-CoV-2 population has accumulated over 75 heritable mutations in only a year since the initial outbreak, a short time on the evolutionary scale. Our analysis of mutations, both of those naturally occurring in COVID-19 patients within the past year and of those experimentally generated within the laboratory, leads us to infer the likely generation of SARS-CoV-2 strains with even greater infectivity and pathogenicity within the coming year. Time will tell whether recovered COVID-19 patients or vaccinated individuals will have enough immunity to be protected from future infection with new strains and whether we can provide sufficiently broad-spectrum antibodies or drug treatments against the rapidly evolving SARS-CoV-2 virus.
Methods
Data collection and mutation/deletion identification
The multiple sequence alignments (MSAs) of >320,000 quality-checked genome sequences as of January 11th, 2021 were downloaded from GISAID after access was granted. By comparing with the reference genome EPI_ISL_402125 (Wu et al., 2020), we identified the mutations and deletions in these genomes. The insertions or ambiguous nucleotides were ignored when counting the mutations and deletions. We used the same site-numbering scheme as the reference genome. The open reading frame (ORF) and protein annotation of the genome were inferred from NCBI RepSeq NC_045512 (Wu et al., 2020).
Identifying heritable mutations and parallel events
The global sampling density of sequences in GISAID varied greatly across different regions. Therefore, to ensure a representative dataset with an equitable and balanced spatiotemporal sequence distribution, we used the selected sequences and phylogenetic tree generated by Nextstrain (https://nextstrain.org/ncov/global) as of January 11th, 2021 for further analysis. 196 tree tips with low-quality sequences and 3 tips without corresponding available sequences were trimmed before analysis, sampled at different dates, and were evenly distributed across all six regions and had no effect on the representative nature of the subsample. We plotted the distribution of all the amino acid and nucleotide mutations in viral genomes with ggtree (http://bioconductor.org/packages/release/bioc/html/ggtree.html). Mutations that could be inherited in at least one small cluster were defined as heritable mutations. Mutations with the same pattern in different clusters with featured heritable mutations were considered to be potentially parallel events.
Spatiotemporal distribution of the mutations
Mutations were found by comparison to the reference genome. The number of mutations (excluding insertions or ambiguous amino acids/nucleotides) was plotted against the sequence collection date using matplotlib library (https://matplotlib.org/stable/index.html). The tips in two N501Y clusters were highlighted in either yellow or red. The geo-distribution of selected mutations was also visualized using matplotlib library. The number of mutations per day was plotted against the total number of sequences collected as a background for each continent.
Mutations on the spike protein
A total of 3,823 representative sequences were selected to profile the amino acid mutations of the S protein. Those mutations within the S protein recurring in more than 20 sequences were selected and plotted as a heatmap using pheatmap library (https://cran.r-project.org/web/packages/pheatmap/index.html). The Nextstrain clade scheme was used to group the sequences.
RBD structure mapping
The residues in the SARS-CoV-2 RBD monomer structure (PDB:7BZ5) were colored according to the frequency of epitopes in the nAbs listed in Figure 2D. The frequency was calculated using Python v3.7. The structural illustration was generated using PyMol (https://pymol.org/2/).
Deep mutational scanning and escape mutants
The data analyzed in Figure 2F were obtained from Jesse Bloom’s Lab GitHub repository (https://github.com/jbloomlab/SARS-CoV-2-RBD_DMS). For purposes of clarity, we have combined viral expression level and human ACE2 binding data. Antibody escape screening data were obtained from https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_clinical_Abs/. The most significant escape mutants were highlighted in yellow.
Acknowledgments
This project is supported by the National Key Plan for Scientific Research and Development of China (2016YFD0500301), the National Natural Science Foundation of China (32070678), research funds from Chinese Academy of Medical Sciences (2016-I2M-1-005, 2019-I2M-1-003, and 2019XK310002), US National Institutes of Health funds (AI069120, AI158154, and AI149718), the UCLA AIDS Institute and UCLA David Geffen School of Medicine – Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Award Program, Microbial Pathogenesis Training Grant (AI7323-31), and AWS Diagnostic Development Initiative.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2021.02.017.
Supplemental information
References
- Bošnjak B., Stein S.C., Willenzon S., Cordes A.K., Puppe W., Bernhardt G., Ravens I., Ritter C., Schultze-Florey C.R., Gödecke N., et al. Low serum neutralizing anti-SARS-CoV-2 S antibody levels in mildly affected COVID-19 convalescent patients revealed by two different detection methods. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-00573-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Petrone M.E., Holmes E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020;5:529–530. doi: 10.1038/s41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Natural deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. bioRxiv. 2020 doi: 10.1101/2020.11.19.389916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. MGH COVID-19 Collection & Processing Team Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370 doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Richard D., Tan C.C.S., Shaw L.P., Acman M., Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020;11:5986. doi: 10.1038/s41467-020-19818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. bioRxiv. 2021 doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.