Abstract
Summary
Light microscopes can now capture data in five dimensions at very high frame rates producing terabytes of data per experiment. Five-dimensional data has three spatial dimensions (x, y, z), multiple channels (λ) and time (t). Current tools are prohibitively time consuming and do not efficiently utilize available hardware. The hydra image processor (HIP) is a new library providing hardware-accelerated image processing accessible from interpreted languages including MATLAB and Python. HIP automatically distributes data/computation across system and video RAM allowing hardware-accelerated processing of arbitrarily large images. HIP also partitions compute tasks optimally across multiple GPUs. HIP includes a new kernel renormalization reducing boundary effects associated with widely used padding approaches.
Availability and implementation
HIP is free and open source software released under the BSD 3-Clause License. Source code and compiled binary files will be maintained on http://www.hydraimageprocessor.com. A comprehensive description of all MATLAB and Python interfaces and user documents are provided. HIP includes GPU-accelerated support for most common image processing operations in 2-D and 3-D and is easily extensible. HIP uses the NVIDIA CUDA interface to access the GPU. CUDA is well supported on Windows and Linux with macOS support in the future.
Over the past decade novel signal collection methods, combined with increased sensitivity of sensors, have enabled the observation of phenomena beyond what was previously thought possible. This is especially true with light microscopy. Conventional confocal microscopes have been able to create large static volumes by tiling sub-volumes together. These microscopes can also capture live time-lapse sequences but they have low-temporal resolution, and can be harmful to living cells. The introduction of light sheet techniques has allowed for much longer image sequences. Modern image processing software are not equipped to process data of this size and complexity. Current challenges include: extending 2-D operations to 3-D, speeding up existing 3-D operations and efficient memory management. While there has been some progress in each of these areas (Eklund et al., 2013), no tool has done this in a comprehensive manner, nor effectively leveraged hardware acceleration for the broad range of processing requirements. Here we present a new software toolset for performing GPU accelerated image processing operations from easy-to-use scriptable programming environments including MATLAB and Python in a manner that is faster, more accurate and more scalable for large datasets.
Hydra image processor (HIP) is a library created from the ground up to be extensible, accelerate image processing operations and handle processing of large datasets. Many MATLAB image processing functions have GPU-acceleration support, but with important limitations. There is also a lack of GPU-accelerated functionality for Python and OpenCV. HIP addresses these limitations as follows. First, support for operations not available at all in 3-D include the broadly useful Laplacian of Gaussian and Wiener smoothing filters (full list on website). Second, HIP allows datasets that do not fit in GPU memory to be automatically partitioned for sequential processing on a single GPU or distributed among all available GPUs. HIP is only constrained by available virtual memory; MATLAB can only process images that fit in GPU memory. Third, HIP uses a kernel renormalization at the boundary pixels to accurately compute kernel response at the edges while MATLAB offers only zero-padding or mirroring as approximate solutions. See website for further discussion on kernel renormalization. HIP is broadly useful, fast, accurate and more efficient with system resources compared to other available approaches.
This versatile library adjusts at runtime to balance computational load across all system GPUs, images are partitioned spatially when larger than available vRAM and across channels and time, where appropriate. HIP can accelerate computations upwards of 100 times compared to a CPU implementation. In the case that multiple GPUs are present, there is a near linear speedup based on the number of GPU devices. This is particularly powerful when HIP is run on GPU cluster nodes. However, even laptops with CUDA-capable GPUs will see a considerable speedup over the same operation running on the CPU. Hardware configurations are natively supported without any user intervention. The addition or improvement of GPU hardware will automatically increase processing speed.
Graphic processing units have specialized memory units that enable rapid memory access from GPU cores. However, due to limited space and power available to GPUs, this memory is difficult and expensive to produce. This means that often there is not enough memory available on the GPU to process an entire dataset at once. When necessary, data is automatically partitioned into overlapping sections (see website for details). The optimal dimension to partition across is chosen to minimize redundant processing. The overlap is calculated based on the filter support size, allowing each chunk to be operated on independently. These overlapping sections provide enough information to ensure that there are zero artifacts between chunks during reconstruction. The chunks are automatically distributed either sequentially on a single GPU (if only one exists on the system) or amongst all of the available GPUs. Additional GPUs provide a near linear speedup in computation when image chunking is required.
HIP was created to simplify both the developer experience when writing new algorithms and the end user experience when using the library from scripting languages. HIP function calls within MATLAB are simple and consistent with MATLAB standards. Functions have similar parameters to those of the native MATLAB processing calls and follow a standard parameter layout. Users need not worry about the capability of their GPU when writing new processing pipelines. When CUDA-capable hardware is available, the user can expect a speedup regardless of the specific hardware class (e.g. GTX, GTXm, Tesla, Quadro). If a CUDA-capable device is not present, HIP will fall back to a software implementation based on approximate solutions. Compute clusters with MATLAB installed will experience considerable speedup (nearly linear over single GPU based on the number of devices per node, see stars in Fig. 1). Integration with MATLAB provides a powerful use case for this library. Not only do many institutions have MATLAB licenses, many other software tools (e.g. ImageJ, Imaris, ICY) have interfaces that can directly interact with MATLAB. Python wrappers are also included.
Fig. 1.
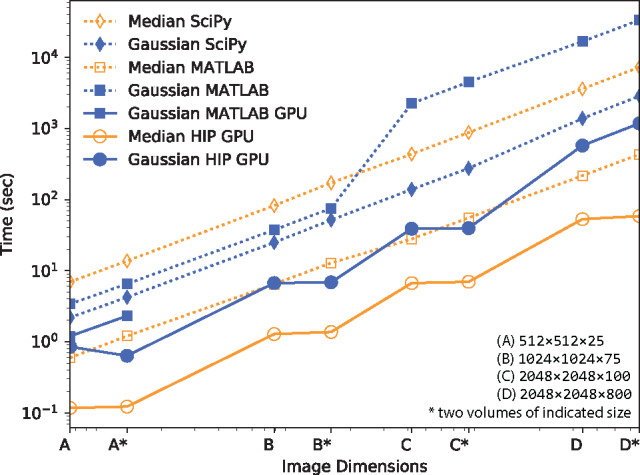
Comparison between MATLAB, SciPy and HIP runtime. Total number of voxels for each test images is shown on a log scale on the X axis and runtime on the Y. MATLAB Gaussian filter can be run on the GPU but runs out of vRAM after the second image. MATLAB does not have a 3-D median filter on the GPU. HIP distributes across multiple GPUs when images are bigger than available vRAM or more than three dimensions*. Adding additional GPUs would further reduce execution times. Computer used had dual Xeon E5-2697 CPUs (32 cores each and 512 GB of RAM total) and dual P6000 GPUs (3840 cores and 24 GB of vRAM each)
The HIP library also includes support utilities written to assist in the tasks that invariably accompany biological image processing. The first task when processing images is to read microscope data, soon followed by a need to write out results. HIP is released with a set of standard input/output utilities that support common microscope and data processing formats. Reader and writer helper functions are provided that support data import using the Bioformats library (Linkert et al., 2010), as well as input and output from HDF5 and KLB files (Amat et al., 2015). Effective visualization is critical in working with multichannel 2-D and 3-D images (Royer et al., 2015). Here, visualization support is provided using a 5-D texture renderer originally developed for the LEVER program (Winter et al., 2016) to support 2-D phase contrast movies (Winter et al., 2015) and 3-D multichannel movies (Wait et al., 2014). This viewer interactively displays data from 2-D to 5-D images and can embed polygonal mesh data such as segmentation and tracking results. The viewer additionally supports loading multiple versions for fast comparison of high-dimensional processing results.
HIP is designed for usability and extensibility. C++ templated classes simplify syntax for creating new operations making it as easy as copying a template, and changing a handful of lines unique to the new operation (see website for template). This template is designed to remove the burden of partitioning data onto devices and memory management concerns on both the CPU and GPU. Helper routines also reduce the code complexity of common image processing tasks like memory access and iterating over a neighborhood. In addition, a simple self-documenting Mex template is provided as an example of providing the MATLAB interface to utilize a HIP operation. HIP is weakly typed, preserving the input image class across all operations in order to best conserve system resources.
Microscope technology will continue to improve, acquiring denser and ever larger datasets. It is already becoming intractable to run current processing pipelines on these large datasets. HIP is a hardware-accelerated processing library that provides new functionality and significantly improves image processing speeds. The simplicity of writing new operations for HIP and the ease of inclusion in other software tools will allow for quick adoption. HIP is released free and open source under the permissive BSD 3-clause license to encourage widespread use. HIP is continually being updated by the authors and significant changes will be announced on the website. The image processing community is encouraged to request changes through the website forum and contribute code. Current compiled versions and source code are available.
Funding
Portions of this research were supported by the NIH NIA (R01AG041861).
Conflict of Interest: none declared.
References
- Amat F. et al. (2015) Efficient processing and analysis of large-scale light-sheet microscopy data. Nat. Protoc., 11, 1679–1696. [DOI] [PubMed] [Google Scholar]
- Eklund A. et al. (2013) Medical image processing on the GPU—past, present and future. Med. Image Anal., 17, 1073–1094. [DOI] [PubMed] [Google Scholar]
- Linkert M. et al. (2010) Metadata matters: access to image data in the real world. J. Cell Biol., 5, 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer L. et al. (2015) ClearVolume: open-source live 3D visualization for light-sheet microscopy. Nat. Methods, 6, 480–481. [DOI] [PubMed] [Google Scholar]
- Wait E. et al. (2014) Visualization and correction of automated segmentation, tracking and lineaging from 5-D stem cell image sequences. BMC Bioinform., 15, 328.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E. et al. (2016) LEVER: software tools for segmentation, tracking and lineaging of proliferating cells. Bioinformatics, 32, 3530–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter M.R. et al. (2015) Computational image analysis reveals intrinsic multigenerational differences between anterior and posterior cerebral cortex neural progenitor cells. Stem Cell Reports, 5, 609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]


