Abstract
Background
In generalized epidemic settings, strategies are needed to prioritize individuals at higher risk of human immunodeficiency virus (HIV) acquisition for prevention services. We used population-level HIV testing data from rural Kenya and Uganda to construct HIV risk scores and assessed their ability to identify seroconversions.
Methods
During 2013–2017, >75% of residents in 16 communities in the SEARCH study were tested annually for HIV. In this population, we evaluated 3 strategies for using demographic factors to predict the 1-year risk of HIV seroconversion: membership in ≥1 known “risk group” (eg, having a spouse living with HIV), a “model-based” risk score constructed with logistic regression, and a “machine learning” risk score constructed with the Super Learner algorithm. We hypothesized machine learning would identify high-risk individuals more efficiently (fewer persons targeted for a fixed sensitivity) and with higher sensitivity (for a fixed number targeted) than either other approach.
Results
A total of 75 558 persons contributed 166 723 person-years of follow-up; 519 seroconverted. Machine learning improved efficiency. To achieve a fixed sensitivity of 50%, the risk-group strategy targeted 42% of the population, the model-based strategy targeted 27%, and machine learning targeted 18%. Machine learning also improved sensitivity. With an upper limit of 45% targeted, the risk-group strategy correctly classified 58% of seroconversions, the model-based strategy 68%, and machine learning 78%.
Conclusions
Machine learning improved classification of individuals at risk of HIV acquisition compared with a model-based approach or reliance on known risk groups and could inform targeting of prevention strategies in generalized epidemic settings.
Clinical Trials Registration
Keywords: clinical prediction rule, HIV risk score, HIV prevention, PrEP, SEARCH Study
Using population-based testing data on 75 558 adults from rural East Africa, we developed and validated a machine learning human immunodeficiency virus (HIV) risk score and found it improved targeting of intensified HIV prevention services compared with model-based and risk group approaches.
(See the Editorial Commentary by Krakower and Marcus on pages 2334–5.)
Despite rapid scale-up in diagnosis and access to antiretroviral therapy (ART), an estimated 800 000 new human immunodeficiency virus (HIV) infections occurred in eastern and southern Africa in 2017 [1]. Identifying who remains at risk of HIV acquisition is crucial to guiding the application of more intensive prevention interventions such as preexposure prophylaxis (PrEP). In generalized epidemic settings, a focus on known risk groups, such as serodiscordant spouses and young women, can effectively reach many high-risk individuals and align with guidelines for identifying persons “at substantial risk for HIV”[1–3]. This approach, however, may miss less well-recognized or easily described subgroups who face elevated risk [4] and may not expend resources most efficiently [5].
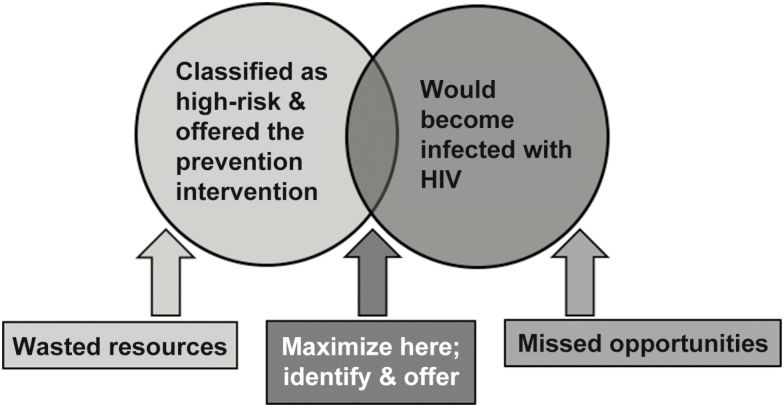
Self-assessment provides one means of identifying individuals at elevated risk despite absence of a known risk factor. However, an individual’s risk perception can depend on their HIV-related knowledge [6] and may fail to capture unanticipated or uncontrolled exposures. Data-driven tools offer an alternative and potentially complementary approach to efficiently and effectively identify persons who would most benefit from intensified prevention interventions (Figure 1).
Figure 1.
Schematic representation of a targeted prevention strategy with the goal of maximizing the intersection of the population offered intensified prevention (light gray) with the population at risk of seroconversion (medium-gray). Sensitivity is the proportion of individuals at high risk who are correctly identified by the strategy: the number in the dark-gray intersection divided by the number in the medium-gray circle. The rate of positive predictions is the proportion of the population targeted: the number in the light-gray circle divided by the total population size. The number needed to target (equal to 1/positive predictive value) is the number classified as high risk per seroconversion identified: the number in the light-gray circle divided by the number in the dark-gray intersection. Abbreviation: HIV, human immunodeficiency virus.
In eastern and southern Africa, a number of HIV risk scores have been developed and validated to predict HIV seroconversion within known risk groups, including serodiscordant couples [7], African women [8, 9], and men-who-have-sex-with-men (MSM) [10, 11]. These risk scores were constructed using standard approaches for clinical prediction rules, which assign a point value to each predictor based on regression model coefficients [12]. Machine learning, which uses computational and statistical algorithms to flexibly learn complex relationships from data, might improve risk classification by relaxing the modeling assumptions made by standard approaches. Ensemble methods such as Super Learner are particularly promising due to their ability to combine multiple approaches, including regression models, known risk groups, and more flexible data-adaptive algorithms [13]. Despite its promise, however, the application of machine learning to predict HIV acquisition has been limited [14–18].
We used population-level HIV incidence data from 3 regions of rural Kenya and Uganda and applied Super Learner [13] to construct a machine learning risk score. We hypothesized that this machine learning risk score would classify future seroconversions as “high-risk” and eligible for intensified prevention more efficiently (fewer persons targeted to achieve a fixed sensitivity) and with higher sensitivity (under a fixed number of persons targeted) compared with either a model-based risk score constructed using standard methods [7–12] or an approach based on known risk groups (eg, young women, having a spouse living with HIV).
METHODS
Study Setting and Population
HIV risk scores were developed and evaluated using data from 16 communities in the intervention arm of the SEARCH Study, a cluster-randomized test-and-treat trial conducted in rural Uganda and Kenya in 2013–2017 [19]. Following a household census, 90% of community residents aged ≥15 years were tested at study baseline using multidisease health fairs combined with home visits [20]. Baseline HIV prevalence varied by region: 4% in Uganda-East, 7% in Uganda-West, and 19% in Kenya [21]. Three subsequent rounds of repeat HIV testing reached 78%, 76%, and 82% of residents, inclusive of in-migrants and newly 15 year-olds. All persons with a negative HIV test followed by a repeat test 1 year later were included in analyses, providing 3 annual incidence cohorts.
Measures
HIV serostatus was determined using country-standard rapid HIV antibody algorithms [20]. The following predictors were assessed with self-report at baseline and year 3: age, sex, marital status, education, occupation, mobility, relationship to the head of the household, alcohol use, family planning, and male circumcision (Supplementary Table 1). We assumed these factors were stable over a 1-year period and imputed interim (years 1 and 2) values. Household predictors included socioeconomic status [20] and summaries of household HIV testing behavior and outcomes. Serodiscordant relationships and characteristics of partners living with HIV were determined by linking HIV testing data between heads of households and their spouses.
Risk Score Development
We generated 3 scores to predict the 1-year risk of HIV acquisition. First, we considered an approach based on belonging to a known “risk group”: women aged 15–24 years, individuals with spouses who were living with HIV, alcohol users, widow(er)s, and persons employed in transportation, bars, or fishing [1, 22–25]. The score was calculated be summing the groups to which an individual belonged.
Second, we generated a “model-based” risk score using standard methods for constructing clinical prediction rules [7–12]. Specifically, we reduced the set of candidate predictors (Supplementary Table 1) based on univariate associations with the outcome (P < .05), applied forward and backward stepwise logistic regression to the remaining predictors, and normalized the fitted coefficients (divided by the smallest coefficient and rounded) to generate a point value for the final predictors.
Third, we used Super Learner to build a “machine learning” risk score. Super Learner is an ensemble method that uses internal sample-splitting to build a weighted combination of algorithm-specific predictions generated from a library of candidate algorithms (Supplementary Materials) [13]. Our library included penalized logistic regression, generalized additive models, main terms logistic regression using known risk factors, and stepwise logistic regression after screening based on univariate associations with the outcome. The weighted combination was chosen using the negative log-likelihood loss and 5-fold sample splitting, stratified on the individual.
Risk Score Evaluation
For the model-based and machine learning approaches, we constructed cross-validated risk scores to evaluate performance on data not used in their development (Supplementary Figure 1). Specifically, individuals were partitioned into 5 mutually exclusive and exhaustive “folds.” Data from 4/5 folds were used to derive the model-based and machine learning risk scores. The resulting algorithms were then applied to predict HIV seroconversion among participants in the remaining fold. By rotating through the folds, we obtained cross-validated scores for each participant. We plotted receiver operating characteristic curves and estimated the areas under the curve (AUC).
Evaluation of Targeting Strategies
We used the cross-validated risk scores to evaluate alternative strategies for targeting intensified prevention. If an individual’s risk score was greater than or equal to a score-specific cutoff, we classified that individual as “high-risk.” First, we selected the minimum score-specific cutoff to achieve at least {50%,60%,70%,80%} sensitivity (proportion of seroconversions correctly classified as high-risk). For the selected cutoffs, we evaluated efficiency by comparing the resulting rate of positive predictions (proportion of the population flagged as high-risk) and number of persons targeted per seroconversion correctly classified (“number needed to target” [NNT]; Figure 1). Second, we selected the maximum score-specific cutoff that would not exceed a rate of positive predictions of {20%,30%,40%,45%}. Under these cutoffs, we calculated sensitivity and NNT.
We pooled across regions when selecting cutoffs and also selected region-specific cutoffs. We evaluated each strategy within strata defined by sex and age. Analyses were completed in R-v3.5.1 with the SuperLearner package [26].
RESULTS
Study Population
A total 75 558 persons who were followed for 166 723 person-years (PY) had at least 1 negative HIV test with a repeat test 1 year later and were included in analyses (250 806 total tests). At baseline, 39% of participants were aged 15–24 years, 44% were male, 3% had a spouse living with HIV, and 15% used alcohol (Table 1). A total of 519 HIV seroconversions were observed (incidence rate, 0.31/100 PY). There were 212 seroconversions among 57 296 persons in the first year (0.37/100 PY), 158 seroconversions among 57 284 persons in the second year (0.29/100 PY), and 149 seroconversions among 60 668 persons in the third year (0.27/100 PY).
Table 1.
Characteristics of 3 Annual Human Immunodeficiency Virus Incidence Cohorts
| Characteristic | Year 0 to Year 1 | Year 1 to Year 2 | Year 2 to Year 3 |
|---|---|---|---|
| N = 57 296 (%) | N = 57 284 (%) | N = 60 668 (%) | |
| Region | |||
| Uganda-East | 21 451 (37) | 22 062 (39) | 22 514 (37) |
| Uganda-West | 18 853 (33) | 17 897 (31) | 20 065 (33) |
| Kenya | 16 992 (30) | 17 325 (30) | 18 089 (30) |
| Sex | |||
| Female | 32 072 (56) | 32 221 (56) | 34 271 (56) |
| Male | 25 224 (44) | 25 063 (44) | 26 397 (44) |
| Age, y | |||
| 15–24 | 22 320 (39) | 21 956 (38) | 22 631 (37) |
| 25–34 | 11 441 (20) | 10 997 (19) | 11 920 (20) |
| 35–44 | 8434 (15) | 8625 (15) | 9232 (15) |
| 45–54 | 6156 (11) | 6236 (11) | 6698 (11) |
| 55+ | 8945 (16) | 9470 (17) | 10 187 (17) |
| Marital statusa | |||
| Single | 16 902 (30) | 14 257 (27) | 15 247 (26) |
| Married | 33 395 (59) | 32 732 (62) | 36 305 (63) |
| Widowed | 4425 (8) | 4384 (8) | 4858 (8) |
| Divorced, separated | 1576 (3) | 1489 (3) | 1676 (3) |
| Educationb | |||
| Less than primary | 38 966 (68) | 40 540 (71) | 38 074 (63) |
| Primary completed | 7836 (14) | 7442 (13) | 9328 (15) |
| Some secondary or beyond | 10 494 (18) | 9302 (16) | 13 266 (22) |
| Occupationc | |||
| Transportation | 550 (1) | 511 (1) | 867 (1) |
| Bar | 124 (0) | 116 (0) | 94 (0) |
| Fishing | 959 (2) | 933 (2) | 908 (1) |
| Serodiscordant spoused | 1011 (3) | 1096 (3) | 1177 (3) |
| Any alcohol usee | 8165 (15) | 7483 (15) | 7109 (12) |
| Human immunodeficiency virus seroconversion by end of risk period | 212 (0) | 158 (0) | 149 (0) |
The cohorts consisted of individuals with a negative human immunodeficiency virus test and a repeat test 1 year later, measured between 2013 and 2017 in 16 communities in rural Kenya and Uganda.
aMissing data on 998 (2%) at year 0, 4422 (8%) at year 1, and 2582 (4%) at year 2.
bMissing data on 90 (0%) at year 0, 124 (0%) at year 1, and 2600 (4%) at year 2.
cMissing data on 1002 (2%) at year 0, 4426 (8%) at year 1, and 2576 (4%) at year 2.
dMissing data on 21 520 (38%) at year 0, 23 028 (40%) at year 1, and 26 068 (43%) at year 2.
eMissing data on 4072 (7%) at year 0, 6411 (11%) at year 1, and 144 (0%) at year 2.
AUC of Risk Scores
Machine learning more accurately ranked individuals who acquired HIV as higher-risk than those who did not (AUC, 0.73; 95% confidence interval [CI], 0.71–0.76; Supplementary Figure 2) compared with both the model-based score (AUC, 0.70; 95% CI, 0.68–0.73, P = .03) and the risk group score (AUC, 0.59; 95% CI, 0.55–0.62, P < .001).
Efficiency for a Fixed Sensitivity
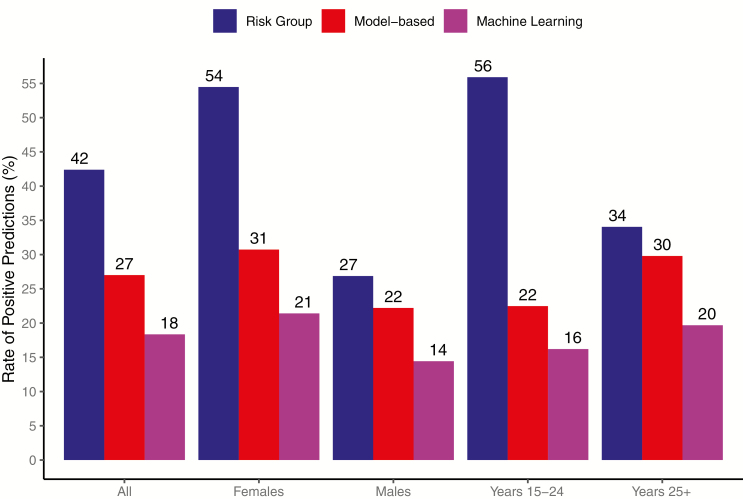
A risk group strategy targeting persons with at least 1 known risk factor would have fallen short of 60% sensitivity. For comparison, we therefore focus our discussion on achieving 50% sensitivity, while noting machine learning improved efficiency at all thresholds (Table 2).
Table 2.
Cross-Validated Efficiency, Defined as the Rate of Positive Predictions (Proportion of the Population Flagged as High Risk) to Achieve a Fixed Sensitivity for Correct Classification of Seroconversions (Top); and Cross-Validated Sensitivity That Would Have Been Achieved When Fixing the Rate of Positive Predictions (Bottom)
| Rate of Positive Predictions, % | |||
|---|---|---|---|
| Needed to meet a minimum sensitivity, % | Risk Groupa | Model-based | Machine Learning |
| 50 | 42 | 27 | 18 |
| 60 | NA | 39 | 26 |
| 70 | NA | 51 | 37 |
| 80 | NA | 63 | 48 |
| Sensitivity Achieved, % | |||
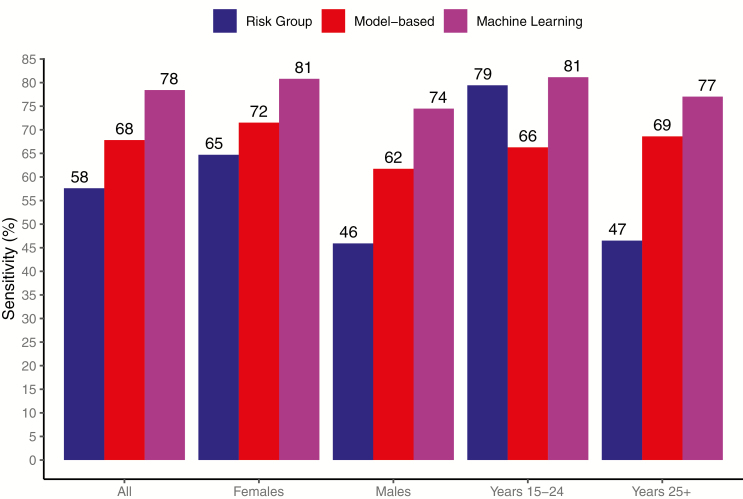
| Limiting the rate of positive predictions, % | Risk Groupb | Model-based | Machine Learning |
| 20 | 8 | 40 | 52 |
| 30 | 8 | 55 | 65 |
| 40 | 8 | 68 | 74 |
| 45 | 58 | 68 | 78 |
Abbreviation: NA, not applicable.
aA strategy to target all persons with at least 1 known risk factor (score ≥ 1) would have offered intensified prevention to 42% of the population; a lower threshold (score ≥ 0) is NA.
bA strategy to target all persons with at least 2 known risk factors (score ≥ 2) would have achieved 8% sensitivity, while a strategy to target all persons with at least 1 known risk factor (score ≥ 1) would have achieved 58% sensitivity.
To correctly classify at least 50% of seroconversions as high-risk, a risk group strategy targeting persons with at least 1 known risk factor (score≥1) would have identified 42% of the population for intensified prevention (Figure 2). To achieve the same sensitivity, the model-based strategy would have targeted 27% of the population and machine learning would have targeted 18%. Machine learning provided relative efficiency improvements of 2.3 and 1.5 compared with the risk group and model-based approaches, respectively. Within age–sex strata, machine learning resulted in 2.6-times fewer women, 1.9-times fewer men, 3.5-times fewer younger adults, and 1.7-times fewer older adults targeted than the risk group approach. Compared with the model-based strategy, efficiency gains from machine learning within age–sex strata were smaller but still present. Overall and within strata, the NNT using machine learning was 1.8- to 2.4-times lower than the risk group strategy and 1.3- to 1.5-times lower than the model-based strategy (Supplementary Figure 3).
Figure 2.
Cross-validated efficiency of each candidate targeting strategy, defined as the proportion of the population that would have been classified as high risk (rate of positive predictions) to achieve 50% sensitivity for correct classification of seroconversions.
Machine learning also improved efficiency within each region (Supplementary Table 2). To correctly classify 50% of seroconversions as high-risk in Uganda-West, 43% of the regional population would be targeted by the risk group strategy, 34% by the model-based strategy, and 20% by machine learning (efficiency improvement, 1.7–2.1 from machine learning). To correctly classify half of seroconversions in Uganda-East, the risk group and model-based strategies would have targeted 44% of the regional population and machine learning would have targeted 26% (efficiency improvement, 1.7 from machine learning). Finally, to correctly classify 50% of seroconversions in Kenya, 40% of the regional population would be targeted by the risk group strategy, 31% by the model-based strategy, and 24% by machine learning (efficiency improvement, 1.3–1.7 from machine learning).
When fixing the sensitivity within each region, machine learning also reduced the proportion of women and younger and older adults targeted compared with either of the other strategies (Supplementary Table 2). For example, across the 3 regions, 51%–59% of younger adults would be targeted by the risk group strategy, 25%–36% by the model-based strategy, and 19%–22% by machine learning. Results varied for men. Machine learning resulted in 16% fewer men targeted than the risk group approach in Uganda-West and 7% fewer men targeted in Uganda-East, but equal proportions of men targeted in Kenya. Nonetheless, among men in each region, the NNT from machine learning was 1.2- to 2.1-times lower than the risk group strategy and 1.2- to 1.6-times lower than the model-based strategy.
Sensitivity for a Fixed Proportion Targeted
A risk group strategy of targeting persons with at least 2 risk factors (score≥2) would have flagged 3% of the population as high-risk, but with limited sensitivity of 8%. For comparison, we focus on strategies with a rate of positive predictions that is ≤45%, corresponding to the proportion flagged as high-risk under a risk group strategy of targeting persons with at least 1 risk factor (score≥1), while noting machine learning improved sensitivity at all thresholds (Table 2).
With the proportion targeted fixed at 45%, the risk group strategy would have covered 58% of seroconversions, the model-based strategy would have covered 68%, and machine learning would have covered 78% (Figure 3); machine learning thus provided 20% and 10% higher sensitivity than the risk group and model-based strategies, respectively. Compared with the risk group strategy, machine learning correctly classified 16% more seroconversions among women, 28% more among men, and 30% more among older adults. Absolute gains in sensitivity compared with the model-based strategy were smaller (8%–15%) but still present. Among younger adults, the model-based strategy achieved 66% sensitivity, while the risk group strategy achieved 79% and machine learning achieved 81%.
Figure 3.
Cross-validated sensitivity for correct classification of seroconversions that would have been achieved by targeting 45% of the overall population.
Fixing the number targeted within each region, machine learning also correctly classified more region-specific seroconversions than either alternative (Supplementary Table 3). When targeting 45% of the population in Uganda-West, the risk group strategy covered 56% of regional seroconversions, the model-based strategy covered 60%, and machine learning covered 78% (sensitivity improvement, 18%–22% from machine learning). When targeting the same proportion of the population in Uganda-East, the risk group strategy correctly classified 54% of seroconversions, the model-based strategy correctly classified 64%, and machine learning correctly classified 68% (sensitivity improvement, 4%–14% from machine learning). When targeting 45% of the population in Kenya, the risk group strategy correctly classified 60% of seroconversions, the model-based strategy correctly classified 69%, and machine learning correctly classified 74% (sensitivity improvement, 5%–14% from machine learning).
Within each region, machine learning also resulted in the highest sensitivity among women, men, and older adults (Supplementary Table 3). In Uganda-West, for example, 42% of seroconversions among older adults would be identified by the risk group strategy, 62% by the model-based strategy, and 78% by machine learning. The risk group strategy had the highest sensitivity among younger adults in each region but at the price of higher NNT: 807 in Uganda-East, 222 in Uganda-West, and 156 in Kenya.
DISCUSSION
In this population-based study conducted in 3 generalized epidemic settings in rural East Africa, we used demographic data to build risk scores for HIV acquisition based on known risk groups; stepwise regression, a model-based approach; and machine learning. We compared strategies for targeting individuals for intensified prevention based on these risk scores and found that machine learning substantially improved efficiency compared with the other strategies. When maintaining the same sensitivity, machine learning reduced the number of individuals who would have been targeted by 33% and 57% compared with the model-based and risk group approaches, respectively. Within age–sex subgroups, machine learning also reduced the number targeted by 25%–50% compared with the alternatives. Efficiency gains were seen for both the population as a whole (allowing for reallocation of prevention resources across regions) and within each region (offering more efficient allocation within-region).
Across regions, machine learning resulted in notable gains in sensitivity when controlling the rate of positive predictions. For a fixed global constraint on the number targeted, machine learning achieved 10% higher coverage of seroconversions than the model-based strategy and 20% higher coverage than the risk group strategy. Improvements in sensitivity were also observed within region: machine learning improved coverage by 4%–18% compared with the model-based strategy and by 14%–22% compared with the risk group strategy. Coverage gains were also seen within most age–sex strata. However, the sensitivity of machine learning among youth, while 10%–22% higher than the model-based strategy, was lower than the risk group strategy in all regions. Similar age-related challenges have been observed when applying the VOICE score for African women to other trials’ data [27–29]. Future work could consider a modified approach to ensure minimum coverage among key groups such as youth. Specifically, selecting age-specific cutoffs would have resulted in superior sensitivity from machine learning (Supplementary Table 4).
This work builds on existing HIV risk scores from eastern and southern Africa in several ways [4,7–11,27–30]. First, it provides evidence that data-driven tools can improve characterization of HIV seroconversion risk at a population-level in the setting of universal ART eligibility, the current standard of care [31], and high population-level viral suppression [21], likely to become more common as HIV testing coverage and ART access expand. In this context, population-based HIV risk stratification may become both more challenging and more crucial for cost-effective targeting of services.
Second, we developed and evaluated risk scores using demographic predictors in a general East African population. Prior risk scores have largely focused on specific subpopulations (eg, African women, serodiscordant couples, MSM, and individuals who report recent sexual activity) [4,7–11,27–30]. Applying risk scores after screening may improve the specificity of risk classification but may also miss individuals at risk of HIV acquisition despite absence or underreporting of a known predictor. We were unable to directly compare our population-level risk scores with this approach because measures of sexual behavior and symptoms used by existing scores were unavailable. Interestingly, our machine learning score achieved similar performance despite reliance on demographic data (eg, when applied to other prevention trials, the VOICE risk score for African women achieved AUCs of 0.56–0.70) [8,28,29]. Nonetheless, incorporation of sexual behavior and risk self-assessment might further improve the performance of the strategies considered.
Our results provide evidence of the utility of machine learning for constructing population-level HIV risk scores. Recent work among US clinic-based populations supports this finding. In particular, Krakower et al applied machine learning to electronic health records (EHRs) and found penalized regression differentiated well between prevalent HIV cases and controls [17]. In a similar setting, Feller et al applied natural language processing to unstructured clinical notes and identified keywords related to sexual orientation (eg, “msm”) and drug use (eg, “methamphetamine”), incorporation of which improved classification of prevalent HIV cases [15]. Most recently, Marcus et al applied penalized regression to EHR data and demonstrated improved ability to predict incident HIV compared with risk group approaches based on MSM status and sexually transmitted infection positivity [18].
There are several limitations to our study. First, with the goal of predicting incident seroconversion over 1 year, our risk algorithms were built and evaluated using data on individuals with at least 2 repeat HIV tests. While testing coverage was >75% annually, individuals not tested may have risk profiles that are distinct from those analyzed [21]. Second, the values of certain demographic factors were imputed during interim years. This imputation, however, should adversely impact the performance of all algorithms equally and thus not change their comparative performance. Finally, our data on spousal discordance was limited to heads of households and their partners and thus was missing for many participants. This missingness, however, should affect all algorithms equally, and, if available, all algorithms could be updated to include serodiscordance as assessed via targeted testing.
Open questions remain regarding the generalizability of risk scores. In a sensitivity analysis, the inclusion of community indicators as candidate risk factors did not yield meaningful gains in performance, providing preliminary evidence of generalizability to other communities in these regions. However, risk score performance outside these regions, as well in the same regions as the epidemic evolves, requires further study.
Open questions also remain regarding the feasibility and acceptability of using machine learning to prioritize individuals for HIV prevention services. Preliminary data suggest implementation of a machine learning risk score is feasible in rural East Africa. During community-wide HIV testing in SEARCH (2016–2017), we screened 69 121 individuals not living with HIV and referred 7256 for PrEP based on a point-of-contact Super Learner risk score [14,16]. Other community-based or facility-based testing programs could incorporate machine learning in practice to identify high-risk individuals [17,18]. Going forward, multidisciplinary teams, including experts in data science, implementation science, behavioral science, social science, and costing, will be required to implement, evaluate, and update these approaches.
Improved data-driven HIV risk prediction provides one potential means to prioritize individuals for intensified prevention services. The results of this study suggest that in generalized epidemic settings with varying HIV prevalence, the use of machine learning methods such as Super Learner may improve the identification of individuals at high risk for HIV infection, a first step toward more efficient and effective approaches for targeted prevention delivery.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the Ministry of Health of Uganda and of Kenya; the research teams and administrative teams in San Francisco, Uganda, and Kenya; collaborators and advisory boards; and especially all the communities and participants involved in the study.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases and the National Institute of Mental Health (NIMH) at the National Institutes of Health (grant numbers U01AI099959, UM1AI068636, R01AI074345, and K23MH114760); the President’s Emergency Plan for AIDS Relief; and Gilead Sciences, which provided Truvada.
Potential conflicts of interest. G. C. reports grants from the Bill & Melinda Gates Foundation outside the submitted work. C. A. K. reports grant support to her institution from the Gilead Research Scholars Program in HIV outside the submitted work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Joint United Nations Programme on HIV/AIDS \. Global AIDS Update 2018: Miles To Go. Geneva, Switzerland; 2018. Available at: https://www.unaids.org/en/20180718_GR2018. Accessed 15 July 2019. [Google Scholar]
- 2. World Health Organization. Who expands recommendation on oral preexposure prophylaxis of HIV infection (PrEP) 2015. Available at: https://www.who.int/hiv/pub/prep/policy-brief-prep-2015/en/. Accessed 15 July 2019.
- 3. Centers for Disease Control and Prevention. Preexposure prophylaxis for the prevention of HIV infection in the United States—2017 Update 2017. Available at:https://www.cdc.gov/hiv/pdf/risk/prep/cdc-hiv-prep-guidelines-2017.pdf. Accessed 15 July 2019.
- 4. Kagaayi J, Gray RH, Whalen C, et al. Indices to measure risk of HIV acquisition in Rakai, Uganda. PLoS One 2014; 9:e92015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cambiano V, Miners A, Phillips A. What do we know about the cost-effectiveness of HIV preexposure prophylaxis, and is it affordable? Curr Opin HIV AIDS 2016; 11:56–66. [DOI] [PubMed] [Google Scholar]
- 6. Maughan-Brown B, Venkataramani AS. Accuracy and determinants of perceived HIV risk among young women in South Africa. BMC Public Health 2017; 18:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kahle EM, Hughes JP, Lingappa JR, et al. An empiric risk scoring tool for identifying high-risk heterosexual HIV-1-serodiscordant couples for targeted HIV-1 prevention. J Acquir Immune Defic Syndr 2013; 62:339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balkus JE, Brown E, Palanee T, et al. An empiric HIV risk scoring tool to predict HIV-1 acquisition in African women. J Acquir Immune Defic Syndr 2016; 72:333–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pintye J, Drake AL, Kinuthia J, et al. A risk assessment tool for identifying pregnant and postpartum women who may benefit from preexposure prophylaxis. Clin Infect Dis 2017; 64:751–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wahome E, Fegan G, Okuku HS, et al. Evaluation of an empiric risk screening score to identify acute and early HIV-1 infection among MSM in coastal Kenya. AIDS 2013; 27:2163–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wahome E, Thiong’o AN, Mwashigadi G, et al. An empiric risk score to guide PrEP targeting among MSM in coastal Kenya. AIDS Behav 2018; 22:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laupacis A, Sekar N, Stiell IG. Clinical prediction rules. A review and suggested modifications of methodological standards. JAMA 1997; 277:488–94. [PubMed] [Google Scholar]
- 13. van der Laan MJ, Polley EC, Hubbard AE. Super Learner. Stat Appl Genet Mol Biol 2007; 6:Article 25. [DOI] [PubMed] [Google Scholar]
- 14. Zheng W, Balzer L, van der Laan M, Petersen M, the SEARCH Collaboration Constrained binary classification using ensemble learning: an application to cost-efficient targeted PrEP strategies. Stat Med 2018; 37:262–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feller DJ, Zucker J, Yin MT, Gordon P, Elhadad N. Using clinical notes and natural language processing for automated HIV risk assessment. J Acquir Immune Defic Syndr 2018; 77:160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koss CA, Ayieko J, Mwangwa F, et al. ; SEARCH Collaboration Early adopters of human immunodeficiency virus preexposure prophylaxis in a population-based combination prevention study in rural Kenya and Uganda. Clin Infect Dis 2018; 67:1853–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krakower DS, Gruber S, Hsu K, et al. Development and validation of an automated HIV prediction algorithm to identify candidates for pre-exposure prophylaxis: a modelling study. Lancet HIV 2019; EPubJul. Available at: https://www.thelancet.com/journals/lanhiv/article/PIIS2352-3018(19)30139-0/abstract. Accessed 12 September 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marcus JL, Hurley LB, Krakower DS, Alexeeff S, Silverberg MJ, Volk JE. Use of electronic health record data and machine learning to identify candidates for HIV pre-exposure prophylaxis: a modelling study. Lancet HIV 2019; EpubJuly. Available at: https://www.thelancet.com/journals/lanhiv/article/PIIS2352-3018(19)30137-7/abstract. Accessed 12 September 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perriat D, Balzer LB, Hayes R, et al. Comparative assessment of five large-scale studies of universal HIV testing and treatment in sub-Saharan Africa. J Int AIDS Soc 2018; 21:e25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chamie G, Clark TD, Kabami J, et al. A hybrid mobile approach for population-wide HIV testing in rural East Africa: an observational study. Lancet HIV 2016; 3:e111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Havlir DV, Balzer LB, Charlebois ED, et al. HIV testing and treatment with the use of a community health approach in rural Africa. N Engl J Med 2019; 38:219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uganda Ministry of Health and ICF International. 2011 Uganda AIDS Indicator Survey: Key Findings. Maryland, USA, 2012. [Google Scholar]
- 23. National AIDS and STI Control Programme. Kenya AIDS Indicator Survey 2012: Final Report. Nairobi, NASCOP, 2014. [Google Scholar]
- 24. Kissling E, Allison EH, Seeley JA, et al. Fisherfolk are among groups most at risk of HIV: cross-country analysis of prevalence and numbers infected. AIDS 2005; 19:1939–46. [DOI] [PubMed] [Google Scholar]
- 25. Lindan CP, Anglemyer A, Hladik W, et al. ; Crane Survey Group High-risk motorcycle taxi drivers in the HIV/AIDS era: a respondent-driven sampling survey in Kampala, Uganda. Int J STD AIDS 2015; 26:336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Polley E, LeDell E, Kennedy C, van der Laan M. SuperLearner: Super Learner Prediction 2018. Available at: http://CRAN.R-project.org/package=SuperLearner. Accessed 15 July 2019.
- 27. Burgess EK, Yende-Zuma N, Castor D, Karim QA. An age-stratified risk score to predict HIV acquisition in young South African women. In: Conference on Retroviruses and Opportunistic Infections. Boston, MA, 2018. Available at: http://www.croiconference.org/sessions/age-stratified-risk-score-predict-hiv-acquisition-young-south-african-women. Accessed 10 June 2019. [Google Scholar]
- 28. Balkus JE, Brown ER, Palanee-Phillips T, et al. Performance of a validated risk score to predict HIV-1 acquisition among African women participating in a trial of the dapivirine vaginal ring. J Acquir Immune Defic Syndr 2018; 77:e8–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burgess EK, Delany-Moretlwe S, Pisa P, et al. Validation of a risk score for HIV acquisition in young African women with FACTS 0001. In: Conference on Retroviruses and Opportunistic Infections. Seattle, WA, 2017. Available at: http://www.croiconference.org/sessions/age-stratified-risk-score-predict-hiv-acquisition-young-south-african-women. Accessed 10 June 2019. [Google Scholar]
- 30. Pintye J, Singa B, Wanyonyi K, et al. Preexposure prophylaxis for human immunodeficiency virus (HIV) prevention among HIV-uninfected pregnant women: estimated coverage using risk-based versus regional prevalence approaches. Sex Transm Dis 2018; 45:e98–e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. World Health Organization. Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection. Geneva, Switzerland; 2016. Available at: http://www.who.int/hiv/pub/arv/arv-2016/en/. Accessed 15 July 2019. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.