Abstract
Sudden cardiac death results from arrhythmias commonly caused by channelopathies and cardiomyopathies, often due to several genetic factors. An emerging concept is that these disease states may in fact overlap, with variants in traditionally classified ‘cardiomyopathy genes’ resulting in ‘channelopathies phenotypes’. Another important concept is the influence of both genetic and non-genetic factors in disease expression, leading to the utilization of systems biology approaches, such as genomics/epigenomics, transcriptomics, proteomics, metabolomics, lipidomics, and glycomics, to understand the disease severity and progression and to determine the prognosis and the best course of treatment. In fact, our group has discovered significant differences in metabolites, proteins, and lipids between controls and Brugada syndrome patients. Omics approaches are useful in overcoming the dogma that both channelopathies and cardiomyopathies exist as Mendelian disorders (caused by a mutation in a single gene). This shift in understanding could lead to new diagnostic and therapeutic approaches.
Keywords: Sudden cardiac death, Brugada syndrome, Channelopathies, Cardiomyopathy, Genetics
Introduction
Sudden cardiac death (SCD) in young, otherwise healthy individuals is often due to ventricular tachyarrhythmias, resulting from a variety of diseases, traditionally grouped into the categories of channelopathies and cardiomyopathies. Channelopathies are primarily electrical disorders affecting ion channels, while cardiomyopathies affect sarcomeric proteins, desmosomes, the cytoskeleton, and the nuclear envelope. Examples of diseases traditionally classified as channelopathies include Brugada syndrome (BrS), long-QT syndrome (LQTS), short-QT syndrome (SQTS), and catecholaminergic polymorphic ventricular tachycardia. Examples of genetic cardiomyopathies are hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), left ventricular non-compaction (LVNC), arrhythmogenic right ventricular cardiomyopathy (ARVC), and restrictive cardiomyopathy.
Identifying patients at risk of arrhythmic events is challenging, as SCD may be the first symptom of such conditions. However, risk stratification is not completely reliable yet in most of these conditions, making the clinical management quite problematic. Often the implantable cardioverter-defibrillator (ICD) is the only reliable prophylactic measure. However, ICD implantation comes with many pitfalls, including inappropriate shocks, the need for battery replacement, potential infection or broken leads, and psychological consequences. Besides, ICD aims to treat, rather than prevent, the potential fatal arrhythmias.
An emerging concept is that channelopathies and cardiomyopathies may overlap, as patients harbouring variants in genes associated with channelopathies may develop ‘cardiomyopathy phenotypes’, and patients with genetic cardiomyopathies often develop ‘arrhythmic phenotypes’ resembling channelopathies. While several reports have already been published in this field,1–3 this line of research is still in its infancy. Another emerging concept is the interaction between both genetic and non-genetic factors in the disease expression, relative to arrhythmogenesis and myocardial dysfunction. In order to assess the mechanisms behind overlap and interaction of these two disease types, a new approach is required, encompassing whole genome sequencing (genomic), the RNA landscape (transcriptomic), the protein compounds derived from RNA (proteomic), and the characterization of lipid molecules (lipidomics). Such systems biology approach is defined as ‘omics’ (Table 1).
Table 1.
Definitions of omics
| Genomics | Application of molecular biology techniques to the complete DNA sequencing in a given organism. |
| Epigenomics | Study of the functional interaction among coding and non-coding regions and regulatory genes. |
| Transcriptomics | Application of molecular biology techniques to study RNA produced from DNA. |
| Metabolomics | Large-scale study of small molecules produced by living cells. |
| Proteomics | Application of biochemical techniques to study the complete set of proteins produced by an organism. |
| Lipidomics | Full characterization of lipid molecular species compared to proteins. |
| Glycomics | Comprehensive study of glycans that a cell or tissue produces under specified conditions of time, location, and environment. |
| Glycoproteomics | The systems-level analysis of glycoproteins, including their protein identities, sites of glycosylation, and glycan structures. |
Omics are providing the instruments to shed a new light onto the source of the wide phenotypic variability, even in the presence of similar genotypes. In cardio-genetics, omics may assist to overcome the dogma that both channelopathies and cardiomyopathies behave as Mendelian disorders, and may help to better understand the disease course and prognosis, improving the diagnostic approach and therapeutic management.
The interaction between genetic background and environmental factors is complex, and often it is difficult to discern the relative contribution of genetic and environmental factors that may result in a certain disease condition. Many examples exist of environmental factors (including pharmaceutical treatments) that may influence the clinical phenotype of either channelopathies or cardiomyopathies. For example, LQTS or BrS may become overt only after being exposed to certain drugs or stressful conditions, or post-chemotherapy dilative cardiomyopathy may be triggered especially in the presence of a genetic background that predisposes a patient to this effect of the therapy. In this view, omics can contribute to understand the mechanisms, to discover the links, and to identify new biomarkers that may lead to innovative diagnostic and therapeutic strategies.
Genomics/epigenomics
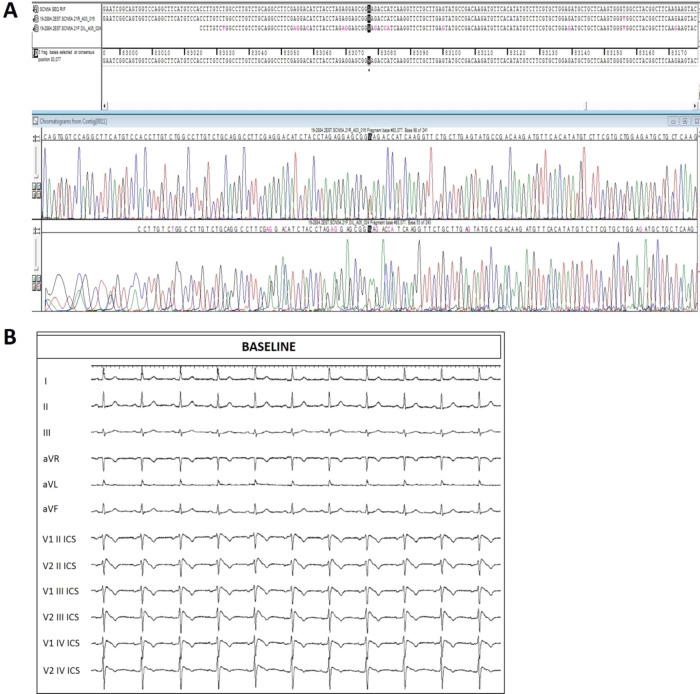
Early genetic studies by Sanger were sequencing screened patients for variants in candidate genes thought to be causative for the clinical phenotypes (Figure 1). Variants in several genes, including KCNQ1, SCN5A, KCNH2, RYR2, PKP2, DSP, and MYBPC34 were discovered over time in patients with personal and familial history of SCD, but, nonetheless, channelopathies and cardiomyopathies were still considered two completely separate conditions.
Figure 1.
(A) Representative Sanger sequencing for the confirmation of the SCN5A heterozygous mutation (NM_198056.2):c.3697A>T found in a BrS patient. (B) Spontaneous 12-lead ECG demonstrating the type 1 BrS pattern in the patient described in Figure 1.
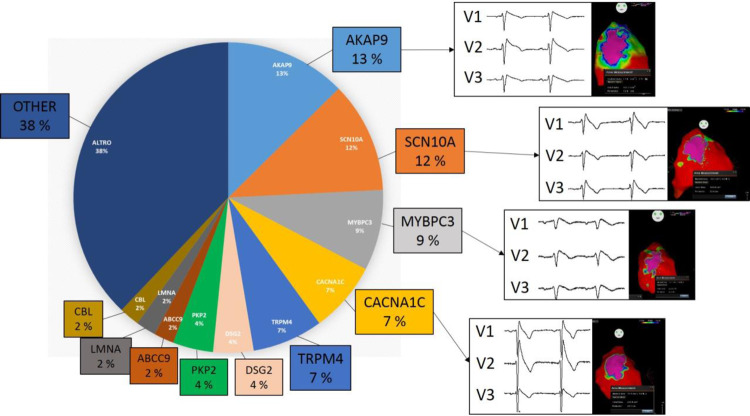
With the development of next-generation sequencing, moving towards the study of the whole DNA sequence, this paradigm became obsolete, since genomic studies provided increasing evidence of cardiomyopathy gene mutations in patients bearing a channelopathy phenotype, and vice versa. At our centre, out of 200 BrS patients who tested positive during genetic testing, 95 did not harbour variants in the SCN5A gene, but rather in an array of other genes (Figure 2). After SCN5A, variants were most commonly found in the AKAP9 gene (13%), followed by SCN10A (12%), MYBPC3 (9%), CACNA1C (7%), TRPM4 (7%), DSG2 (4%), PKP2 (4%), ABCC9 (2%), LMNA (2%), and CBL (2%). Regardless of the genotype, these patients exhibited similar arrhythmogenic substrates, demonstrating that variants in an array of genes are associated with BrS. This data demonstrates that the BrS phenotype cannot currently be explained by variants in a single gene, but rather, BrS is associated with variants in genes encoding for a wide variety of proteins, including signalling, channel, sarcomeric, and desmosomal proteins.
Figure 2.
At our centre, out of 200 BrS patients who tested positive during genetic testing, 95 did not harbour variants in the SCN5A gene, but rather in an array of other genes. After SCN5A, variants were most commonly found in the AKAP9 gene (13%), followed by SCN10A (12%), MYBPC3 (9%), CACNA1C (7%), TRPM4 (7%), DSG2 (4%), PKP2 (4%), ABCC9 (2%), LMNA (2%), and CBL (2%).
The complex functional interaction among coding and non-coding regions and regulatory genes is defined as epigenomics. Recent evidence suggests that mutations in regulatory genes may influence the clinical expression of gene mutations associated with channelopathies or cardiomyopathies. For example, experimental evidence suggested that MYBPC3 pathogenic mutations, known to be associated with DCM, can be modulated by epigenetic factors.5 Similarly, a recent study suggests a role for the differential methylation and imprinting of KCNQ1 (a gene involved in type 1 LQTS) in the risk for symptomatic prolonged QT interval.6 Moreover, the reversible modifications occurring in DNA or histones modulates gene expression even without altering the DNA sequence. The ways by which these complex processes occur are yet incompletely understood but can be considered part of epigenomics. However, epigenomics alone cannot entirely explain the complex link between channelopathies and cardiomyopathies. Therefore, the latest studies employ the whole genomic approach to clarify the contribution of additional coding and non-coding regions to these disease conditions. For example, HCM or DCM are thought to be inherited only in an autosomal dominant manner,7 but about 5% of affected patients have been reported to harbour more than one mutation, consistent with the hypothesis of an oligogenic inheritance. Additionally, some recent exome studies provided evidence of a possible high false positive rate in genetic testing for both HCM and DCM.8
Along these lines, it is likely that channelopathies and cardiomyopathies are not monogenic disorders and that they are possibly influenced, or even caused by, an interaction between genomic factors, including modifier genes, and environmental factors. This would also explain the pleomorphic phenotypes that can emerge even within a single patient, for example, a spontaneous type 1 BrS electrocardiogram pattern that occurs only transiently, or the fact that patients can live for decades completely asymptomatic, then suddenly experience cardiac arrest, often under very specific conditions, such as while sleeping or during a febrile state, something that appears to also be influenced by age and gender.2
There have been numerous studies linking several genes to SCD, ranging from sodium channel, calcium channel, potassium channel, desmosomal, and sarcomeric genetic variants. In spite of this, the genetics of such conditions still remains elusive, without any known disease-causing mutation found in approximately 20% of families meeting clinical diagnostic criteria for LQTS or in approximately 60% of BrS patients.9,10
Most genetic studies on SCD have focused on the SCN5A gene, which encodes for the alpha subunit of the cardiac sodium channel. SCN5A heterozygous mutations are considered causative for a variety of both channelopathies and cardiomyopathy phenotypes, including LQTS, BrS, ARVC, LVNC, DCM, idiopathic ventricular fibrillation, sick sinus syndrome, and progressive heart block.2
In BrS, due to the low prevalence of molecular confirmation, many clinical genomic studies have been performed in search of candidate genes, many of those suggesting that other sodium channel genes, for example, SCN1B and SCN10A, might play a role in the pathogenesis of BrS.2,11 However, a recent report underlined that, so far, only the SCN5A gene can be considered as causative for BrS, since it is the only BrS-associated gene that has withstood a systematic, evidence-based evaluation supporting the genotype–phenotype correlation.12
Other ionic channels besides sodium channels are implicated in the genesis of SCD. The function of calcium channels, including L-type calcium channels, ryanodine receptors, the sodium/calcium exchanger, the sarco/endoplasmic reticulum Ca2+-ATPase, and phospholamban, is central to excitation–contraction coupling, and the dysregulation of any of these proteins can lead to fatal arrhythmias. The most relevant genes implicated in these pathways are CACNA1C, CACNB2, CACNA2D1, RYR22 (associated with LQTS), and CACNA1C9 (associated with BrS).
Other genes involved in the genesis of SCD are those encoding potassium channels, such as KCNQ1 (associated with type 1 LQTS) and KCNH2 (associated with many different arrhythmic phenotypes, including type 2 LQTS, SQTS, and BrS).
Of note, heterozygous mutations in SCN5A, the major gene associated with cardiac channelopathies, were observed in patients with cardiomyopathies. Indeed, the finding of right ventricular dysfunction is not rare in BrS patients with SCN5A mutations. Therefore, it is now a well-established concept that SCN5A is a pleiotropic gene, causative of both electrical and structural phenotypes. However, the mechanisms behind such SCN5A pleiotropism are not yet understood, and, so far, genomic approaches have provided limited clues relating to this issue.
Several channelopathy-associated ion channel genes have been associated also with cardiomyopathies, including SCN5A and KCNQ1 with DCM, the KCNQ1, RYR2, and HCN4 genes with LVNC, and RYR2 with ARVC.15 However, these findings are less common than those observed with SCN5A. Therefore, it is possible that other omics approaches, beyond genomics and epigenomics, will explain the mechanisms of gene pleiotropism and overlapping syndromes between channelopathies and cardiomyopathies.
From genomic/epigenomic to other omics approaches
The complex scenario of cardio-genetic conditions can be studied with other omics approaches, such as proteomics, transcriptomics, lipidomics, metabolomics, and glycomics. So far, such methods are still underutilized in cardio-genetics, even though the role of environmental and metabolic variations in determining the phenotype of channelopathies and cardiomyopathies has long been recognized.
Omics approaches could also be utilized to better differentiate BrS from Brugada phenocopies that can result from a number of non-genetic factors, including acute myocardial ischaemia, pulmonary embolism, electrolyte abnormalities, or adverse drug reactions, such as beta-receptor blocker or cocaine or marijuana use, in the absence of an identifiable genetic background.
As examples of proteomic changes, in the plasma of BrS patients, levels of apolipoprotein E, prothrombin, vitronectin, complement-factor H, vitamin-D-binding protein, voltage-dependent anion-selective channel protein 3, and clusterin have been reportedly increased, while levels of alpha-1-antitrypsin, fibrinogen, and angiotensinogen were decreased when compared to control subjects.13 Additionally, our group recently discovered significant differences in circulating metabolites, proteins, and lipids between controls and BrS patients with a type 1 pattern expressed either spontaneously or after ajmaline administration, providing evidence that BrS may be a metabolomic disease (unpublished data). Such changes might affect the ionic channel traffic, explaining dynamic variation in BrS phenotypes.
The protein encoded by SCN5A (called the NaV1.5 protein) undergoes several post-translational modifications, such as phosphorylation and sialylation. Both these processes have been implicated in BrS pathogenesis.14 Following these findings, proteomic changes may be interpreted as a ‘fingerprint’ of the disease process. For example, we recently observed that BrS patients displayed post-translational modification of Nav1.5, compared to controls. This may provide a new diagnostic method and may be a new prognostic tool for the management of BrS patients.
Other omics, such as transcriptomic, metabolomics, lipidomics, and glycomics are still in an early phase due to methodological issues. Our group is pioneering their application both to BrS and to other related conditions.
Conclusions
Cardio-genetic diseases were always considered pure Mendelian disorders, in spite of much contradicting evidence. The technical improvements in biosciences have enabled the omics approach to illuminate that cardio-genetic diseases follow a more complex pathogenesis than a ‘one mutation in a single gene’ mechanism. This is true, especially for conditions without a well-established genetic background, such as BrS and some forms of cardiomyopathies. Omics approaches can be used in these conditions to understand disease severity, natural progression, prognosis, and overlap between seemingly distinct phenotypes. Moreover, omics approaches can impact the clinical management and treatment plan with positive consequences for affected patients.
Conflict of interest: none declared.
References
- 1. Pappone C, Monasky M, Ciconte G.. Epicardial ablation in genetic cardiomyopathies: a new frontier. Eur Heart J Suppl 2019;21:B61–B66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Monasky MM, Pappone C, Piccoli M, Ghiroldi A, Micaglio E, Anastasia L.. Calcium in Brugada syndrome: questions for future research. Front Physiol 2018;9:1088.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Monasky MM, Ciconte G, Anastasia L, Pappone C.. Commentary: next generation sequencing and linkage analysis for the molecular diagnosis of a novel overlapping syndrome characterized by hypertrophic cardiomyopathy and typical electrical instability of Brugada syndrome. Front Physiol 2017;8:1056.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Osman J, Tan SC, Lee PY, Low TY, Jamal R.. Sudden cardiac death (SCD)—risk stratification and prediction with molecular biomarkers. J Biomed Sci 2019;26:39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tabish AM, Arif M, Song T, Elbeck Z, Becker RC, Knoll R, Sadayappan S.. Association of intronic DNA methylation and hydroxymethylation alterations in the epigenetic etiology of dilated cardiomyopathy. Am J Physiol Heart Circ Physiol 2019;317:H168–H180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coto E, Calvo D, Reguero JR, Morís C, Rubín JM, Díaz-Corte C, Gil-Peña H, Alosno B, Iglesias S, Gómez J.. Differential methylation of lncRNA KCNQ1OT1 promoter polymorphism was associated with symptomatic cardiac long QT. Epigenomics 2017;9:1049–1057. [DOI] [PubMed] [Google Scholar]
- 7. Raghow R. An ‘omics’ perspective on cardiomyopathies and heart failure. Trends Mol Med 2016;22:813–827. [DOI] [PubMed] [Google Scholar]
- 8. Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, Minikel EV, MacArthur DG, Farrall M, Cook SA, Watkins H; Exome Aggregation Consortium. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med 2017;19:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alders M, Bikker H, Christiaans I.. Long QT syndrome. In Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, eds. GeneReviews((R)). Seattle (WA: ); 1993. [Google Scholar]
- 10. Monasky MM, Micaglio E, Ciconte G, et al. Genotype/phenotype relationship in a consanguineal family with Brugada syndrome harboring the R1632C missense variant in the SCN5A gene. Front Physiol 2019;10:666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Monasky MM, Micaglio E, Vicedomini G, Locati ET, Ciconte G, Giannelli L, Giordano F, Crisà S, Vecchi M, Borrelli V, Ghiroldi A, D'Imperio S, Di Resta C, Benedetti S, Ferrari M, Santinelli V, Anastasia L, Pappone C.. Comparable clinical characteristics in Brugada syndrome patients harboring SCN5A or novel SCN10A variants. Europace 2019;21:1550–1558. [DOI] [PubMed] [Google Scholar]
- 12. Hosseini SM, Kim R, Udupa S, Costain G, Jobling R, Liston E, Jamal SM, Szybowska M, Morel CF, Bowdin S, Garcia J, Care M, Sturm AC, Novelli V, Ackerman MJ, Ware JS, Hershberger RE, Wilde AAM, Gollob MH; on behalf of the National Institutes of Health Clinical Genome Resource Consortium. Reappraisal of reported genes for sudden arrhythmic death. Circulation 2018;138:1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Domenico M, Scumaci D, Grasso S, Gaspari M, Curcio A, Oliva A, Ausania F, Di Nunzio C, Ricciardi C, Santini AC, Rizzo FA, Romano Carratelli C, Lamberti M, Conti D, La Montagna R, Tomei V, Malafoglia V, Pascali VL, Ricci P, Indolfi C, Costanzo F, Cuda G.. Biomarker discovery by plasma proteomics in familial Brugada Syndrome. Front Biosci (Landmark Ed) 2013;18:564–571. [DOI] [PubMed] [Google Scholar]
- 14. Valdivia CR, Ueda K, Ackerman MJ, Makielski JC.. GPD1L links redox state to cardiac excitability by PKC-dependent phosphorylation of the sodium channel SCN5A. Am J Physiol Heart Circ Physiol 2009;297:H1446–H1452. [DOI] [PMC free article] [PubMed] [Google Scholar]