Abstract
Background and Aims
Ileal strictures are the major indication for resective surgery in Crohn’s disease [CD]. We aimed to define ileal gene programmes present at diagnosis and linked with future stricturing behaviour during 5-year follow-up, and to identify potential small molecules to reverse these gene signatures.
Methods
Antimicrobial serologies and pre-treatment ileal gene expression were assessed in a representative subset of 249 CD patients within the RISK multicentre paediatric CD inception cohort study, including 113 that are unique to this report. These data were used to define genes associated with stricturing behaviour and for model testing to predict stricturing behaviour. A bioinformatics approach to define small molecules which may reverse the stricturing gene signature was applied.
Results
A total of 19 of the 249 patients developed isolated B2 stricturing behaviour during follow-up, while 218 remained B1 inflammatory. Using deeper RNA sequencing than in our previous report, we have now defined an inflammatory gene signature including an oncostatin M co-expression signature, tightly associated with extra-cellular matrix [ECM] gene expression, in those who developed stricturing complications. We further computationally prioritise small molecules targeting macrophage and fibroblast activation and angiogenesis which may reverse the stricturing gene signature. A model containing ASCA and CBir1 serologies and a refined eight ECM gene set was significantly associated with stricturing development by Year 5 after diagnosis {AUC (area under the curve) (95th CI [confidence interval]) = 0.82 [0.7–0.94)}.
Conclusions
An ileal gene programme for macrophage and fibroblast activation is linked to stricturing complications in treatment of naïve pediatric CD, and may inform novel small molecule therapeutic approaches.
Keywords: Paediatric Crohn disease, ileum, small molecule, surgery, transcriptome
1. Introduction
Most Crohn’s disease [CD] patients present with inflammatory behaviour, and many progress to a stricturing complication.1 Increased use of anti-tumour necrosis factor [TNF] therapy has not resulted in a population-wide reduction in bowel resections.2 The CALM study demonstrated lower rates of surgeries with medication intensification guided by C-reactive protein [CRP] and faecal calprotectin.3 However, heterogeneous CD biology may also be associated with anti-TNF non-response and disease complications.4–7 A microarray study identified colon genes associated with anti-TNF response; ileal genes could not be defined due to low rates of ileal healing.8 A mechanism of anti-TNF non-response involving expression of oncostatin M [OSM] by colon stromal cells, leading to pro-inflammatory myeloid cell cytokine expression, was reported in adult UC.9 Subsequently, an inflammatory mononuclear phagocyte gene expression module was defined using single-cell RNASeq in adult CD ileal resections.10 As was previously published, when applied to our paediatric CD ileal bulk RNASeq data, higher pre-treatment expression of macrophage and fibroblast activation genes within this module was associated with anti-TNF non-response.10 Whether this or other molecular mechanisms could be associated with progression to ileal strictures in paediatric CD patients is not known.
We have conducted the RISK paediatric CD inception cohort study to identify host, microbial, and therapeutic factors associated with disease complications.4,11 We reported that ASCA and CBir1 serologies, and ileal extra-cellular matrix [ECM] gene expression, were associated with future stricturing complications.4 Data suggest that macrophages produce pro-inflammatory cytokines leading to myofibroblast activation and ECM production in CD.9,12 Whether a specific myeloid gene signature would be linked to ECM production and the stricturing outcome was not known. To test this, we have conducted a more extensive RNA sequencing of ileal samples in a RISK replication and validation sub-cohort. These data have identified a novel inflammatory gene set involving oncostatin M [OSM], antimicrobial signalling pathways, and activated macrophages linked to ECM production, and have begun to define small molecules which may reverse the stricturing gene signature.
2. Methods
2.1. Clinical cohort
RISK4 is a paediatric CD inception cohort study including 28 sites in the USA and Canada; 913 CD patients younger than 18 years, with inflammatory disease behaviour [B1] at diagnosis and during the first 3 months of follow-up, were enrolled between 2008 and 2012 [ClinicalTrials.gov identifier NCT00790543], and served to define baseline factors associated with progression to disease complications during 5-year follow-up. We excluded patients with diagnoses other than CD, those with incomplete information on disease location, and those without at least one follow-up visit. Patients were managed according to their physicians. The disease behaviour classification was based upon an audit conducted at each of the RISK sites through 60-month follow-up. Stricturing disease [B2] was defined as persistent luminal narrowing with pre-stenotic dilatation as shown by small bowel contrast imaging. For patients who required surgery, the presence of a stricture was confirmed by review of the pathology report. Internal penetrating disease [B3] was defined as intra-abdominal fistulising disease resulting in intra-abdominal or pelvic abscesses or fistulas to an adjacent organ [excluding the vagina or perianal region]. B1 refers to an uncomplicated inflammatory disease state.13,14 In case of B2 and B3 co-occurrence, those were categorised separately for the current report.
Subsequently, central reading of the initial magnetic resonance enterography [MRE] study has been completed for 167 [18%] of the 913 RISK CD participants classified with inflammatory [B1] behaviour at diagnosis. Of those 167, seven were classified as B2 stricturing [4%] based on the central read of the baseline MRE. These were each initially classified as B1 inflammatory in the RISK clinical metadata, and were each later classified as progression to B2 stricturing after 98, 169, 236, 434, 512, 1797, and 2013 days. Those who had the central read of the baseline MRE completed included 15% [37 of 249] of the CD subjects included here, with one CD patient [3%] classified as B2 stricturing at diagnosis. This subject had evolved to B2 stricturing after 169 days from diagnosis. We therefore estimate a potential 4% misclassification of the initial B1 inflammatory status, although in all cases examined the CD patients ultimately were classified as B2 stricturing during follow-up.
2.1.1. Sub-cohort for gene expression analysis
The reduction from 913 participants in the overall cohort to 249 participants for gene expression analysis was based upon: 1] availability of ileal biopsies for research purposes; 2] high=quality ileal RNA for sequencing; and 3] due to cost constraints, we examined all available B2, B2 + B3, and B3 patients [n = 31] and a representative group of B1 patients [n = 218], aiming to include equal numbers of previously sequenced and new samples for replication and validation, respectively.
2.2. Ethical considerations
The institutional review board [IRB] at each site reviewed and approved the protocol, and informed written consent was obtained from all parents or guardians and assent was provided from patients [Cincinnati Children’s IRB was 2008-0764].
2.3. Patient and public involvement statement
The RISK cohort study was designed with active collaboration from the Crohn’s and Colitis Foundation [CCF] patient advocacy group, where disease complications were viewed as a high priority by patient groups.
2.4. Serological assays
Serological determination4,7 of anti-Saccharomyces cerevisiae antibodies [ASCA] IgG, ASCA IgA, and anti-CBir1, was performed using baseline blood samples at Cedars-Sinai Hospital [Los Angeles, CA, USA].15 Antibody levels were determined and results are expressed as ELISA units [EU/ml], which are relative to a Cedars-Sinai Laboratory standard, derived from a pool of CD patient sera with well-characterised disease found to have reactivity to this antigen; ASCA IgA is considered positive above 20 EU/ml, ASCA IgG above 40 EU/ml, and anti-CBir1 above 25 EU/ml.
2.5. RNA isolation and sequencing
RNA was isolated from pre-treatment ileal biopsies as described.5 NEBNext Ultra RNA Library Prep Kit [NEB, Ipswich, MA, USA] was used for RNAseq libraries using Paired end [PE] 150 base pair chemistry [on the HiSeq system by GENEWIZ, South Plainfield NJ]. Reads (median coverage of 35.4M and interquartile range [IQR] 32.2M, 39.4M]) were quantified by kallisto,16 using Gencode v24 as the reference genome and transcripts per million [TPM] as an output, by three independent analysts [YH, RK, TB]. We included 14 370 protein-coding genes with TPM above 1 in 20% of samples. Differentially expressed genes between CD B2 and B1, with fold change differences [FC] >= 1.5 and using false-discovery rate correction [FDR; <0.05] were determined in parallel in both GeneSpring® software and using R package DESeq2 version 1.24.0, and importing and summarising the kallisto output files to gene level with R package tximport version 1.12.3. Euclidean distance metric and Ward’s linkage rule were used for unsupervised hierarchical clustering. The top quartile of co-expressed genes [Pearson correlation] with either OSM or COL1A2 were recorded with only six genes overlapping between the two. Those gene sets were used for ToppGene17 and ToppCluster18 functional annotation enrichment analyses with visualisation using Cytoscape.v3.0.2.19 The RNASeq data is deposited in the SRA database SUB6656230.
Principal component analysis [PCA] distills large gene sets into component variables that are indicative of overarching biological themes among the original gene sets, and we focus on the first component, which explained the greatest amount of variability. PCA plots PC1 were used for Pearson correlations and two-way Student’s t tests to test for statistically significant differences between ileal samples with subsequent B2 and B1 behaviour. In addition, for validation, we assessed the discriminative capacity of previously published4 gene sets indicative of: 1] inflammatory processes [GO:0006954, n = 569 genes, INFL]; and 2] fibrosis [GO:0005201, n = 69 genes, ECM], using PCA PC1 results.
2.6. Perturbagen analysis
To discover potential anti-stricture agents, we used the functional enrichment analysis from ToppGene Suite.17 Enrichment p-values were calculated using a hypergeometric test. We then used the differentially expressed genes [DEGs] enriched for angiogenesis, ECM, or macrophage-related processes and pathways, to query the NIH’s LINCS Library.20 We used the LINCS cloud web tool20 to identify small molecules from this library. We focused on gene-expression signatures located in the ‘Touchstone’ dataset because it represents a set of thoroughly annotated perturbagens. The LINCS cloud web tool compares queried signatures with gene expression profiles from the Touchstone library, comprising gene expression signatures from more than 2000 compounds including Food and Drug Administration [FDA]-approved drugs. Compounds whose signatures are in opposition to the disease signature are assumed to have therapeutic potential. We also performed direct comparison of each of the prioritised compound signatures with the B2 gene signature associated with progression to stricturing behaviour [differentially expressed pre-treatment genes between CD patients who progressed to B2 stricturing behaviour and CD patients who maintained B1 inflammatory behaviour] identified using the RISK ileal RNASeq data. Overlaying genes that were reciprocally connected [ie, upregulated in the B2 gene signature and downregulated in compound treatment] were defined and gene set enrichment analysis was performed. Additionally, for the select compounds, known targets were downloaded from the DrugBank21 and the Drug Repurposing Hub22 and used for enrichment analysis using the ToppGene Suite. Visualisation of enrichment network was done using Cytoscape.19
2.7. Sirius red immunohistochemistry
Sirius Red [Sigma-Aldrich, catalogue number: 365548, Saint Louis, MO, USA] for collagen type I and III protein immunohistochemistry23 and Image J24 were used to determine the extent of collagen infiltration into the sub-cryptal space in formalin-fixed, paraffin-embedded [FFPE] biopsies of patients who exhibited B1 behaviour at diagnosis and later developed B2 stricturing [n = 22] or those who maintained B1 inflammatory behaviour [n = 35]. The area containing collagen was captured [Olympus microscope] and measured with the rotated rectangle tool in ImageJ. Thickness was calculated as:
where:
and width is fixed at 50 μm, in five technical replicates per patient.
2.8. Analytical approach
We assessed differential distribution of categorical variables between patient subgroups [B2 vs B1 by 5 years] using Fisher’s exact test [significance at p <0.05] and of continuous variables using the non-parametric Wilcoxon rank-sum test [p <0.05]. Baseline variables were assessed for association with the stricturing phenotype with a univariate logistic regression analysis with an odds ratio (with 95% confidence interval [CI]) calculated for each variable. These baseline variables included the PC1 from the previously published INFL [GO:0006954] and ECM [GO:0005201] gene sets, and of preselected eight gene panels from those gene sets. Preselection of eight INFL gene [ADAMTS12, AOX1, FN1, IL24, KRT16, MASP1, NOX4, SELE] and eight ECM gene [ASPN, COL11A1, COL12A1, COL1A1, ELN, ITGA11, LRRC15, MFAP5] panels involved prioritisation based on high TPM [mean of above 5], lower corrected p-value, and higher fold change between B2 and B1 in the previously reported dataset.4
In order to minimise model overfitting, multivariate logistic regression models for the prediction of B2 stricturing by 5 years were built using an iterative, stepwise process, where variables with univariate significance of p <0.01 were tested as possible predictors. In addition, with only 16 B2 stricturing events, we limited our final model to three variables. Model fit was assessed using Akaike’s information criteria [AIC], which applies a penalty for increased model complexity, model significance, variable significance, and area under the curve [AUC]. The final logistic model was fitted using a sequential method of starting with a single predictor and adding subsequent predictors to achieve the maximum likelihood and minimise overfitting by calculating McFadden’s adjusted pseudo-R2 at each step. Model comparison included the likelihood ratio test and DeLong’s test [to compare AUC]. Collinearity of potential predictors was also assessed using Pearson’s correlation test, where we observed a strong linear association between PC1 of the inflammatory [INFL] and fibrosis [ECM] gene sets [R = 0.89, p <0.0001]. Logistic regression and receiver operating characteristic [ROC] curves were performed with R version 4.0.0 [R Development Core Team, Vienna].
3. Results
3.1. Clinical and demographic characteristics of the RISK sub-cohort included for molecular characterisation
Of the 913 paediatric CD patients in the RISK cohort with B1 inflammatory behaviour at diagnosis and within 3 months of diagnosis, 80 patients [9%] developed stricturing behaviour [B2] and 797 remained B1 inflammatory during 5-year follow-up. Clinical and demographic characteristics of the overall RISK cohort, and the 249 sub-cohort included for molecular characterisation, are shown in Tables 1 and 2 stratified by disease behaviour through 60 months of follow-up. The 249 sub-cohort did not vary in age, sex, race, anthropometrics, disease location, or activity compared with the overall cohort [Table 2]. As expected, the frequency of ASCA or CBir1 sero-positivity was increased in those who developed B2 behaviour. Here we used this representative 249 sub-cohort of CD patients with ileal mucosal biopsies to explore the biology at diagnosis which is linked to later development of stricturing complications, including 124 also used in the previous report for replication4 and 113 unique to this report for an additional validation [Table 2]. The B2 validation group had fewer females and more perianal disease, but otherwise did not vary in clinical, demographic, or serological characteristics.
Table 1.
Clinical and demographic characteristics of the RISK study sub-cohort stratified by disease behaviour by 60 months after diagnosis.
| B1 [n = 218] | B2 [n = 19a] | B2 + B3 [n = 7] | B3 [n = 5] | |
|---|---|---|---|---|
| Demographics and follow-up | ||||
| Age at diagnosis [years] | 12.4 [10.5, 14.6] | 12.8 [11, 15.1] | 14.4 [11, 15.5] | 13.9 [11.2, 16.1] |
| Female sex [%] | 80 [37%] | 6 [32%] | 3 [43%] | 2 [40%] |
| African American or mixed race | 37 [17%] | 2 [11%] | 1 [29%] | 1 [20%] |
| Duration of follow-up [months] median[IQR] | 59 [44, 65] | 63 [45, 70] | 58 [55, 58.5] | 52 [43, 63] |
| Time to behaviour change [days] | - | 478 [193, 851] | 681 [496, 1131] | 678 [161, 1494] |
| Disease activity and treatment exposures | ||||
| Moderate-to-severe disease activity [%] | 106 [49%] | 10 [53%] | 4 [57%] | 3 [60%] |
| Height Z-score | -0.2 [-0.8, 0.6] | -0.5 [-0.9, 0.2] | -1 [-1.5, -0.4] | -1.3 [-1.5, -1.2] |
| BMI Z-score | -0.6 [-1.7, 0.3] | -0.9 [-1.7, 0.2] | -0.9 [-1.5, -0.5] | -2.5 [-2.9, -0.5] |
| Anti-TNFα within 90 days | 48 [22%] | 3 [16%] | 2 [28%] | 1 [20%] |
| Immunomodulator within 90 days | 86 [40%] | 9 [47%] | 4 [47%] | 2 [40%] |
| Small bowel imaging within 6 months | 164 [75%] | 17 [89%] | 7 [100%] | 5 [100%] |
| Disease location | ||||
| Isolated terminal ileum | 43 [20%] | 5 [26%] | 3 [43%] | 0 [0%] |
| Isolated colonic | 52 [24%] | 1 [5%] | 2 [29%] | 0 [0%] |
| Ileo-colonic | 123 [56%] | 13 [69%] | 3 [29%] | 5 [100%]b |
| Perianal | 40 [18%] | 3 [16%] | 1 [14%] | 1 [20%] |
| Serological reactivity status at diagnosis | ||||
| ASCA IgA above 20 EU/ml | 43 [21%] | 7 [37%] | 5 [71%] b | 1 [20%] |
| ASCA IgG above 40 EU/ml | 40 [20%] | 9 [47%] b | 6 [71%] b | 0 [0%] |
| CBir1 above 25 EU/ml | 72 [36%] | 11 [58%] b | 6 [86%] b | 3 [60%] |
IQR, interquartile range; BMI, body mass index; TNF, tumour necrosis factor.
aThree B2 had complications at diagnosis; those were excluded from the stricturing complication regression model.
b p <0.05 versus B1 group.
Table 2.
Clinical and demographic characteristics of the overall RISK cohort and the sub-cohort used for mRNASeq gene expression analysis.
| Overall RISK cohort with 5-year B1 or B2 behaviour [n = 877] | RISK sub-cohort with mRNASeq data and 5-year B1 or B2 behaviour [n = 237] | Replication group with mRNASeq data [n = 124] | Validation group with mRNASeq data [n = 113] | |||||
|---|---|---|---|---|---|---|---|---|
| Inflammatory [B1] [n = 797] | Stricturing [B2] [n = 80] | Inflammatory B1 [n = 218] | Stricturing B2 [n = 19*] | Inflammatory B1_Repl [n = 115] | Stricturing B2_Repl [n = 9] | Inflammatory B1_Val [n = 103] | Stricturing B2_Val [n = 10a] | |
| Demographics and follow-up | ||||||||
| Age at diagnosis [years] | 12.3 [9.92, 14.6] | 12.5 [10.5, 14.4] | 12.4 [10.5, 14.6] | 12.8 [11, 15.1] | 12.4 [10.7, 14.7] | 12.1[8.8, 12.8] | 12.4 [10.2, 14.2] | 13.9[12.5, 15.9] |
| Female sex [%] | 297 [37.3%] | 34 [42.5%] | 80 [37%] | 6 [32%] | 42 [37%] | 5 [55%] | 38 [37%] | 1 [10%]^ |
| African American or mixed race | 97 [12.2%] | 14 [17.5%] | 37 [17%] | 2 [11%] | 17 [15%] | 0 [0%] | 20 [19%] | 2 [20%] |
| Duration of follow-up [months] median[IQR] | 60 [42, 60] | 60 [45, 66] | 59 [44, 65] | 63 [45, 70] | 58 [40, 65] | 63 [40, 68] | 59 [44, 65] | 64 [52, 71] |
| Time to behaviour change [days] | - | 637 [388, 1095] | - | 478 [193, 851] | - | 427 [254, 584] | - | 606 [42, 1321] |
| Disease activity and treatment exposures | ||||||||
| Moderate-to-severe disease activity [%] | 367 [46%] | 38 [47.5%] | 106 [49%] | 10 [53%] | 61 [53%] | 5 [56%] | 45 [44%] | 6 [60%] |
| Height Z-score | -0.25 [-0.93, 0.45] | -0.38 [-1.17, 0.23] | -0.2 [-0.8, 0.6] | -0.5 [-0.9, 0.2] | -0.2 [-0.7, 0.6] | -0.7 [-1.2, 0.2] | -0.2 [-1.1, 0.5] | -0.3 [-0.8, 0.2] |
| BMI Z-score | -0.64 [-1.61, 0.15] | -0.91 [-1.83, 0.33] | -0.6 [-1.7, 0.3] | -0.9 [-1.7, 0.2] | -0.5 [-1.6, 0.3] | -0.8 [-1.7, 0.4] | -0.7 [-1.8, 0.2] | -1.3 [-1.7, 0.4] |
| Anti-TNFα within 90 days | 169 [21.2%] | 15 [18.8%] | 48 [22%] | 3 [16%] | 19 [17%] | 1 [11%] | 29 [28%] | 2 [20%] |
| Immunomodulator within 90 days | 364 [45.7%] | 33 [41.3%] | 86 [40%] | 9 [47%] | 52 [45%] | 6 [66%] | 34 [33%] | 3 [30%] |
| Small bowel imaging within 6 months | 579 [72.6%] | 65 [81.3%] | 164 [75%] | 17 [89%] | 87 [76%] | 9 [100%] | 77 [75%] | 8 [80%] |
| Disease location | ||||||||
| Isolated terminal ileum | 156 [19.6%] | 21 [26.3%] | 43 [20%] | 5 [26%] | 19 [17%] | 1 [11%] | 24 [23%] | 4 [40%] |
| Isolated colonic | 205 [25.7%] | 12 [15%] | 52 [24%] | 1 [5%] | 28 [24%] | 0 [0%] | 24 [23%] | 1 [10%] |
| Ileo-colonic | 436 [54.7%] | 47 [58.8%] | 123 [56%] | 13 [69%] | 68 [59%] | 8 [90%] | 55 [53%] | 5 [50%] |
| Perianal | 111 [13.9%] | 9 [11.3%] | 40 [18%] | 3 [16%] | 24 [21%] | 0 [0%] | 16 [16%] | 3 [30%]b |
| Serological reactivity status at diagnosis | ||||||||
| ASCA IgA | 167 [21%] | 30 [38%]c | 43 [21%] | 7 [37%] | 28 [24%] | 3 [33%] | 15 [15%] | 4 [40%]b |
| ASCA IgG | 169 [21%] | 27 [34%b] | 40 [20%] | 9 [47%b] | 24 [21%] | 5 [56%] b | 16 [16%] | 4 [40%]b |
| CBir1 | 270 [34%] | 48 [60%]c | 72 [36%] | 11 [58%]b | 43 [37%] | 6 [67%] | 29 [28%] | 5 [50%] |
There were 36 B3 in the overall CD RISK cohort of 913 participants.
IQR, interquartile range; BMI, body mass index; TNF, tumour necrosis factor; Repl: replication group included in the Lancet 2017 report; Val: validation group unique to this report.
aThreeB2 had complications at diagnosis; those were excluded from the complication regression model.
b p <0.05 and cp <0.01 B2 versus B1 group in each dataset.
3.2. Inflammatory and tissue remodelling pathways enriched in the ileum of patients who developed stricturing complications
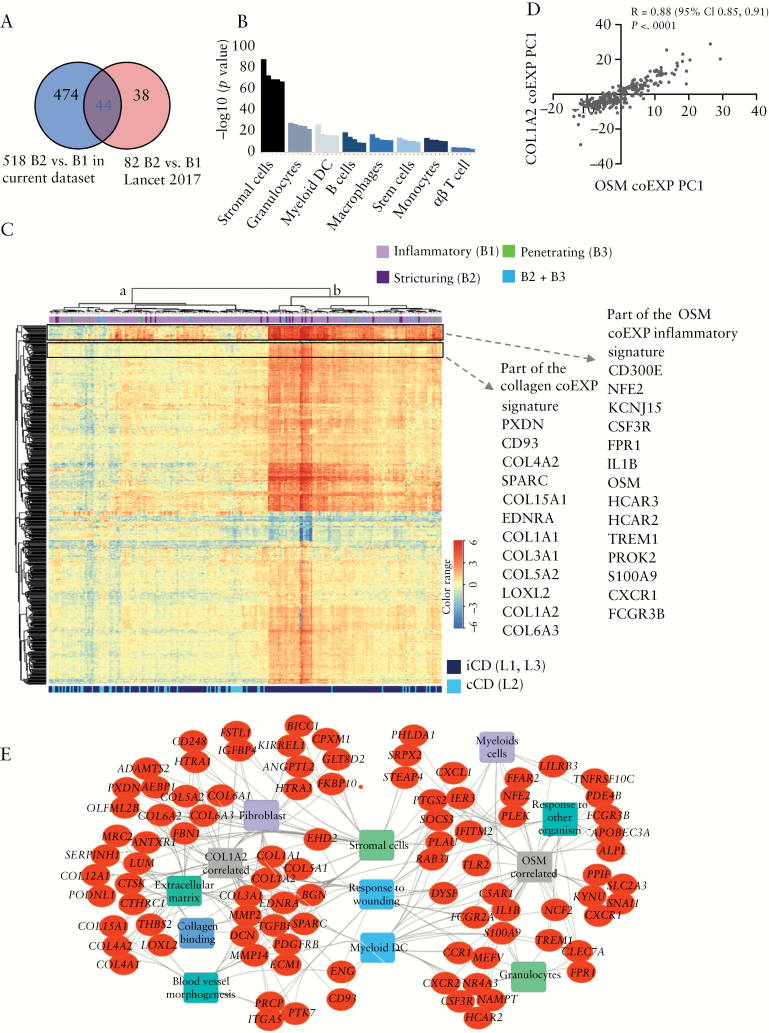
When comparing CD patients who developed stricturing complications [B2, n = 19] with CD patients who maintained inflammatory disease behaviour during follow-up [B1, n = 218] using 3-fold deeper level of RNA sequencing in the current study, we defined 518 B2 genes that were differentially expressed in the ileum at diagnosis [FC ≥1.5, FDR <0.05, Supplementary dataset 1, available as Supplementary data at ECCO-JCC online, and Figure 1]. We were underpowered to perform differential expression analysis between the B3 internal penetrating or B2 + B3 combined stricturing/internal penetrating groups and the B1 inflammatory group, but included the mean TPM expression per group in the Supplementary dataset 1 for the 518 B2 genes. Those 518 genes overlapped with 44 of the 82 previously reported ileal genes linked to B2 behaviour4 [Figure S1] but also added a substantial number of 474 novel genes [Figure 1A]. These 518 B2 stricturing genes were notable for up-regulation of: OSM, implicated in anti-TNF non-response; NCF2 and CSF3R, implicated in myeloid cell activation; ANTXR1; collagen genes; and TGFBI, implicated in tissue fibrosis.
Figure 1.
Tissue remodelling linked to inflammatory pathways is enriched in the ileum of patients who developed stricturing complications. A. Venn diagram comparing the 518 genes differentially expressed in the ileum between patients with B2 stricturing [n = 19] and B1 inflammatory [n = 218] behaviour [Supplementary dataset 1] in the current dataset in comparison with the previous dataset.4 B. Cell type enrichment analysis of the 468 up-regulated genes differentially expressed in the ileum between patients with B2 stricturing and [n = 19] and B1 inflammatory [n = 218] behaviour [Supplementary dataset 1], using the Immunological Genome Project data series as a reference through ToppGene.17 Enrichment for a given stromal or immune cell class is illustrated by coloured bars on the × axis, with the significance for each individual cell subtype within the class shown as the –log10[p-value] on the y axis. DC, dendritic cells. C. Hierarchical clustering of the 518 genes differentially expressed in the ileum between patients with B2 stricturing [n = 19] and B1 inflammatory [n = 218] behaviour is visualised as a heatmap. Above the heatmap, individual Crohn’s disease [CD] patient behaviour during 5-year follow-up is indicated [218 B1 in light purple, 19 B2 in dark purple, 5 B3 in green, 7 B2 + B3 in blue]. Dendrogram main branches are marked in a [left] and b [right]. The top and bottom frames highlight part of the OSM and collagen co-expression [coEXP] inflammatory and collagen [COL1A2] signatures, respectively, with specific genes indicated on the right of the heatmap. D. Samples loading PC1 values of the COL1A2 coEXP were plotted against the PC1 loading of the oncostatin M [OSM] coEXP and Pearson R correlation was calculated. E. Functional annotation enrichment analyses of the OSM and collagen [COL1A2] coEXP signatures using ToppGene,17 ToppCluster,18 and Cytoscape.19 Detailed functional annotation enrichment analyses are summarised in Supplementary dataset 1.
3.3. Characterisation of inflammatory pathways linked to ECM production and subsequent stricturing behaviour
Previous studies have supported a mechanism of fibrosis in CD involving activation of ECM-producing stromal cells, including myofibroblasts, by pro-inflammatory myeloid cells. Consistent with this, ileal genes up-regulated in B2 patients were enriched in stromal cells [p = 6.6E-89] and pro-inflammatory granulocytes [p = 2.1E-28], myeloid DC [p = 5E-27], B cells [p = 2.2E-19], and macrophages [p = 1.6E-17, Figure 1B]. Unsupervised hierarchical clustering analysis using the 518 genes grouped a larger fraction of B1 that evolved to B2 [n = 12] on the right dendrogram cluster [with 90 B1 that remained B1] and seven B1 that evolved to B2 and 128 persistent B1 clustered on the left dendrogram cluster [Fisher’s exact test p = 0.037, Figure 1C]. Ontologies identified in up-regulated ileal genes of B2 patients included pro-inflammatory cytokine and growth factor pathways [p = 1.6E-22], vasculature development and angiogenesis [p = 3.2E-40], and ECM [p = 4.5E-46] organisation. Suppressed genes were associated with response to metal ions and zinc [p = 1.5E-16] and mineral absorption [p = 5E-11] [Supplementary dataset 1].
Strong linkage to gut inflammation, response to anti-TNF therapy, and B2 complications prompted us to specifically examine the top quartile genes out of the 518 that were co-expressed with OSM or COL1A2, using Pearson correlation, across all CD patients in the cohort. Some of those gene subsets are highlighted in the heatmap in Figure 1C, where the upper panel includes the inflammatory signature linked with OSM and includes TREM1, CXCR1, IL1B, S100A9, and HCAR2/3. The lower highlighted panel is linked to the fibrosis signature and includes collagens I, III, IV, and VI. Although only six genes showed overlap between the OSM and COL1A2 co-expression signatures, the PCA PC1 values that summarised variation of those gene signatures showed a significant correlation [Figure 1D, Pearson r = 0.88, p <0.0001]. Functional annotation and network analyses of those co-expression signatures interestingly showed [Figure 1E] that response to wounding, myeloid dendritic cells, and gp38 + stromal cells are linked to both signatures. Extracellular matrix annotation, collagen binding, fibroblasts, and angiogenesis were more specific to the COL1A2 signature, and granulocytes, myeloid cells, and response to other organisms were more specific to the OSM co-expression signature.
Our earlier report4 showed enrichment for extra-cellular matrix [ECM, GO:0005201, 69 genes, Supplementary dataset 1] and inflammatory [INFL, GO:0006954, 569 genes, Supplementary dataset 1] terms in CD patients who developed subsequent B2 or B3 disease complications, respectively. Gene expression for these gene sets was reduced to PC1 for each participant to facilitate comparisons between groups. Here, we detected a significant increase in the ECM PC1 [accounting for 47% of the overall variance] and for the first time also the INFL gene signature PC1 [accounting for 34% of the variance] in the B2 CD ileum compared with the B1 CD ileum [Supplementary Figure S2A and B, available as Supplementary data at ECCO-JCC online], altogether and also separately in both the replication and validation subsets. Those INFL and ECM signatures were highly significantly correlated with each other, and varied between the B2 and B1 groups [Figure S2C]; 63 of the 569 INFL genes and 28 of the 69 ECM genes overlapped with the current 518 B2 gene set [Figure S2D]. Moreover, although we replicated enrichment of the ECM genes in patients who later developed stricturing complications,4 the enrichment of INFL genes in this sub-group was only detected in the current dataset [Figure S2D].
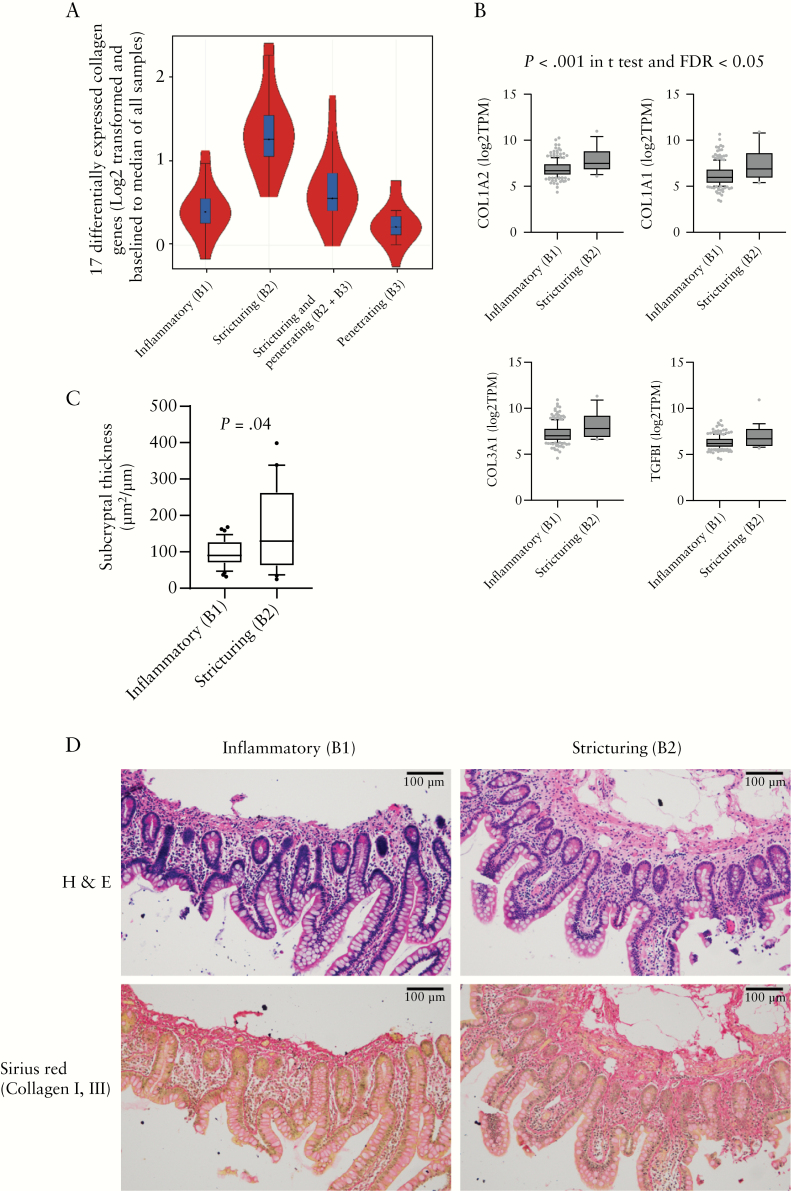
3.4. Collagen I and III are increased in the sub-cryptal space at diagnosis in the ileum of patients who later develop stricturing complications
Collagen I/III are the major types in the intestine under normal conditions, whereas in CD patients with strictures there is a significant increase in collagen I/III and V.25 We noted up-regulation of many collagen sub-type genes, including COL1A1/2, COL3A1, COL4A1/2, COL5A1/2/3, COL6A1/2/3, COL7A1, COL8A1, COL12A1, COL13A1, COL15A1, and COL27A1 specifically in B2 patients [Figure 2A, B], together with the pro-fibrogenic cytokine TGFBI. Increased deposition of collagen in both the sub-cryptal space [submucosa] and in the muscularis propria has recently been described in surgical resection samples from Crohn’s disease patients with ileal strictures.23,26 Here we tested for variation in submucosal collagen in ileal pinch biopsies. Sirius red staining confirmed an increase in sub-cryptal type I/III collagen protein in CD patients at diagnosis who progressed to B2 stricturing behaviour, in comparison with patients who remained B1 through 5-year follow-up [Figure 2C, D, and Figure S3 showing an example of the sub-cryptal area measurement].
Figure 2.
Collagen I and III are increased in the sub-cryptal space in the ileum of patients who developed stricturing complications. A. Violin plot of the 17 collagen genes that were significantly differentially expressed between B2 stricturing [n = 19] and B1 inflammatory [n = 218] behaviour Crohn’s disease [CD] patients including COL1A1/2, COL3A1, COL4A1/2, COL5A1/2/3, COL6A1/2/3, COL7A1, COL8A1, COL12A1, COL13A1, COL15A1, COL27A1, across B1 [n = 218], B2 [n = 19], B2 + 3 [n = 7], and B3 [n = 5] groups. B. Bar graph of COL1A2, COL1A1, COL3A1, and TGFBI RNAseq TPM values in 218 patients with an inflammatory B1 behaviour and in those 19 who developed stricturing B2 behaviour. C Sub-cryptal thickness measurement, as described in the Methods section, of ileal mucosal FFPE biopsy slides of patients who developed B2 stricturing behaviour [n = 22] or those who maintained B1 inflammatory behaviour [n = 35]. D. Haematoxylin and eosin [H&E] and SiriusRred stain of collagen I and II with representative images of C. A Student’s t test assessed the statistical difference.
3.5. Serological and molecular model linked to ECM production and subsequent stricturing behaviour
In univariate analyses, age, gender, and disease location were not linked with the development of Year 5 stricturing behaviour [Table S1]. Higher ASCA and CBir1 serologies and lower albumin were significantly linked to Year 5 stricturing behaviour. Importantly, both the previously reported INFL PC1 [accounting for 34% of variance, n = 569 genes] and ECM PC1 [accounting for 47% of variance, n = 69 genes] GO terms were associated with stricturing behaviour [Figure S2 and Table S1], as well as PC1 for the 518 B2 genes [accounting for 58% of variance] associated with future stricturing in the current analysis [Figure S4, and Table S1].
Moreover, INFL or ECM eight-gene panels [described in the Methods section] were also associated with B2 behaviour [Figure S4 and Table S1]. A multivariable model including the ileal ECM eight-gene PC1 [accounting for 44% of variance], and pre-treatment serum ASCA IgG and CBir1 serology provided excellent discriminant power [AUC:0.82] for predicting a subsequent stricturing outcome [Table 3 and Figure S5]. The likelihood ratio test confirmed that the model including the ECM PC1 and CBir1 and ASCA IgG serology was superior to models including the ECM PC1 alone, or the ECM PC1 and CBir1 serology [Table 3]. AUC of this model was similar to a model that included the ileal INFL eight-gene PC1 [AUC of 0.82 vs 0.81, Figure S5]. The discriminant power of models based on CBir1 serology alone (AUC: 0.68 [0.56–0.80]) or ASCA IgG alone (AUC: 0.69 [0.56–0.82]) was similar to model 1 based on ECM PC1 alone [Table 3]. A model including both CBir1 and ASCA IgG serology was superior to model 1 including only the ECM PC1 (AUC:0.79 [0.70–0.89], LR test: 7.4, p = 0.007].
Table 3.
Multivariate analyses for development of B2 stricturing complications.
| Model 1 | Model 2 | Model 3 | ||||
|---|---|---|---|---|---|---|
| Baseline predictor | Estimate | p-value | Estimate | p-value | Estimate | p-value |
| 8.ECM transcriptome | 1.5 [1.14–1.96] | 0.0039 | 1.45 [1.1–1.91] | 0.0067 | 1.4 [1.1–1.86] | 0.0145 |
| CBir [>25] | - | - | 4.2 [1.44–14] | 0.0116 | 3.4 [1.12–11.6] | 0.0364 |
| I gG.ASCA [>40] | - | - | - | - | 4.32 [1.43–13] | 0.0093 |
| Model characteristics | ||||||
| AUC | 0.73 [0.59–0.87] | 0.79 [0.68–0.9] | 0.82 [0.70–0.94] | |||
| AIC | 112.4 | 107.5 | 102.8 | |||
| R2 | 0.03 | 0.06 | 0.12 | |||
| Sensitivity | 75% [48–93] | 81% [54–96] | 88% [62–98] | |||
| Specificity | 67% [61–74] | 75% [69–80] | 70% [63–76] | |||
| PPV | 15% [11–42] | 19% [15–56] | 18% [14–66] | |||
| NPV | 97% [92–98] | 98% [94–99] | 99% [95–99] | |||
| Model comparison | ||||||
| Model 2 vs model 1 | Model 3 vs model 1 | |||||
| Likelihood ratio test | - | - | LR = 6.92 | 0.0085 | LR = 13.5 | 0.0012 |
| Comparison of AUC | - | - | 0.26 | 0.0103 | ||
| Model 3 vs model 2 | ||||||
| Likelihood ratio test | LR = 6.62 | 0.01 | ||||
| Comparison of AUC | 0.43 | |||||
Baseline predictors are shown with odds ratios with 95% confidence intervals [CIs] in parentheses. Model characteristics were based on a predicted probability of 0.5. Models comparisons were completed with likelihood ratio test and DeLong’s test [to compare AUC between models].
AUC, area under the ROC curve; AIC, Akaike’s information criteria; PPV, positive predictive value; NPV, negative predicitive value; LR, likelihood ratio.
With only 16 stricturing events, there is a risk of overfitting a model with >2 variables. To evaluate for overfitting, we calculated an adjusted pseudo R2 [McFadden statistic] for a base model including only the ECM PC1 [0.02] variable, and found a stable increase in the adjusted pseudo R2 with the addition of the CBir1 [0.062] and then ASCA IgG variables [0.102] suggesting that the model was not overfit until a fourth predictor was added. The final logistic regression model with the three variables [ECM PC1, CBir1, and ASCA IgG] had a McFadden statistic of 0.188. Collectively these data defined a model including baseline ileal ECM gene expression and antimicrobial serologies with very high negative predictive value for the 5-year stricturing outcome.
3.6. Prediction of small molecules likely to reverse the ileal gene signature associated with stricturing complications
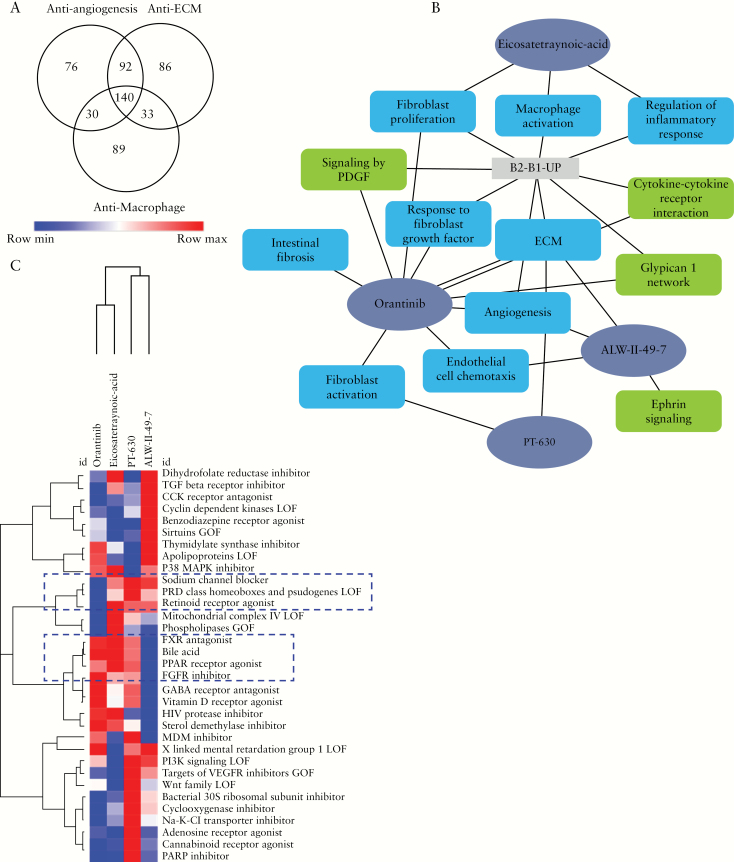
Finally, we asked which small molecules might reverse the ileal gene signature associated with stricturing behaviour. The Touchstone dataset contains a total of approximately 8400 perturbagens, including gene signature expression profiles for more than 2000 small molecules generated from testing in-vitro on a panel of nine cell lines. These cell lines include A375, A549, HEPG2, HCC515, HA1E, HT29, MCF7, PC3, and VCAP. Although the signatures are based on epithelial cell lines, we and others have shown that the signatures and the connectivity map approach can be leveraged for discovery of novel therapies for fibrotic disorders.27 The CMap connectivity score [tau] is a standardised parameter ranging from -100 to 100 as a measure of the connectivity between the queried gene expression signature and those of the other perturbagens in the database. A positive tau indicates a relative similarity between perturbagens signatures, and a negative score indicates relative opposing gene signatures,20 implying that this small molecule can potentially revert the pathogenic signature. We considered tau of + 90 or higher, and of -90 or lower, as strong scores for the current study. Perturbagen analysis identified a significant number of endogenous and pharmaceutical small molecules with summary score predicting a likely beneficial effect [Supplementary dataset 2, available as Supplementary data at ECCO-JCC online], implying that the perturbagen profile is opposite that of the pathological ileal gene signature. The summary score reflects the ability of a given small molecule to reverse a pathogenic gene expression signature, in this case the ileal B2 signature, across multiple epithelial, mesenchymal, and immune cell types.
Small molecules with the highest summary scores are shown in Figure 3A. CMap connectivity analysis using the angiogenesis, ECM, and macrophage signatures identified 345, 358, and 294 candidate small molecules with a tau score of -90 or lower. There were 140 small molecules that were common to all [Figure 3A]. Among these were alpha-linolenic-acid, ALW-II-49-7, clofibrate, eicosatetraynoic acid, liquiritigenin, orantinib, PT-630, sitagliptin, tipifarnib, and verapamil. Since our primary interest was to search for small molecules that had recognised mechanisms of action, strengthen previous findings, and support hypotheses for potential repurposing for stricturing, we focused on ALW-II-49-7 [ephrin inhibitor], eicosatetraynoic acid [ETYA, cyclooxygenase, and lipoxygenase inhibitor], orantinib [PDGFR tyrosine kinase receptor inhibitor, FGFR inhibitor, VEGFR inhibitor, and angiogenesis inhibitor], and PT-630 [dipeptidyl peptidase and fibroblast activation protein inhibitor]. To further elucidate the underlying mechanism of these compounds, we: [i] compiled compounds that induce transcriptionally similar profiles to the four select compounds; and [ii] performed functional enrichment analysis of the four compounds using their known targets [Figure 3B]. The heatmap output that is presented in Figure 3C gives the other compounds that are similar to the four select compounds. The similarity is based on the summarisation measure calculated across the nine cell lines in the LINCS signatures.
Figure 3.
Perturbagen analysis to discover and characterise potential anti-stricture agents. A. Venn diagram showing the comparison between candidate small molecules from CMap connectivity analysis using the angiogenesis, extraaa-cellular matrix [ECM], and macrophage signatures. B. Network representation of enriched biological processes and pathways for the four select repurposing candidates for stricturing. C. Heatmap representation of perturbagen classes that are similar to the select four repurposing candidates for structuring in the LINCS signatures.
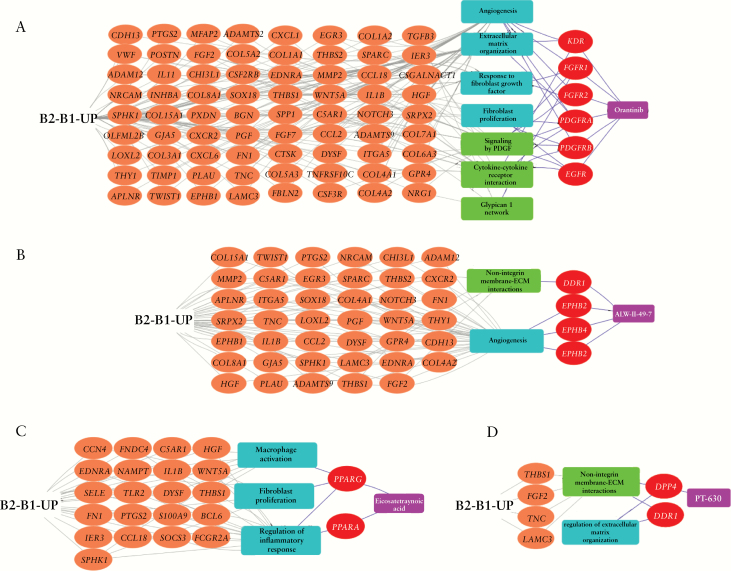
Moreover, we intersected differentially expressed genes by those compounds [genes that showed decreased expression using the compound] in vitro using the epithelial cell lines with genes that were increased in our B2 518 ileal mucosal biopsies gene set. Evaluations of those genes’ enriched functions are shown in Supplementary dataset 3, available as Supplementary data at ECCO-JCC online, highlighting ECM, angiogenesis, and wound healing. Most interestingly, gene enrichment analysis for specific cell types using single-cell data showed highest cell type enrichment for fibroblasts [p = 1.3E-9 for orantinib, p = 5E-5 for ETYA, p = 5E-7 for PT-630, and p = 7E-5 for ALW-II] To complement the Touchstone dataset approach, we also looked specifically into known targets of those compounds from DrugBank and Drug Repositioning Hub [see Methods] and linked those to pathways and genes from our B2 518 genes [Figure 4]. Orantinib’s targets include PDGF, PDGFRA, and PDGFRB, which are also up-regulated and are included in our 518 B2 gene set. Those targets and B2 genes are enriched for several common pathways as shown in Figure 4A. ETYA, on the other hand, targets PPARG and PPARA and is linked with both fibroblasts and the inflammatory component that is also linked to the B2 signature as shown in Figure 4C.
Figure 4.
Prioritised candidate compounds regulate biological processes and pathways in the B2 gene signature. Panels A, B, C, and D show the functional connectivity to B2 up-regulated genes for prioritised compounds orantinib, ETYA, PT-630, and ALW-II-49-7. Orange nodes are genes upregulated in the 518 B2 gene set and the red nodes are known targets of the four compounds. The turquoise and green rectangles are the enriched biological processes and pathways shared between the candidate compounds and the B2 up-regulated genes.
4. Discussion
Using deeper RNA sequencing than in the previous report,4 we have now defined for the first time an OSM co-expressed inflammatory gene signature tightly associated with ECM/collagen gene expression signatures linked with the development of B2 stricturing complications, through 60-month follow-up in a large multicentre paediatric CD inception cohort, RISK.4 These results emphasise for the first time that the OSM inflammatory signature, previously linked to anti-TNF non-response, is highly correlated with an ECM signature and with development of stricturing complications. We were able to capture at diagnosis the INFL and ECM signatures that are linked to later development of stricturing complications in 124 patients also included in the earlier report for replication,4 and 113 patients unique to this report for an additional validation [Table 2]. Sirius Red staining detected increased sub-cryptal collagen content in ileal pinch biopsies at diagnosis in patients who subsequently developed strictures. Perturbagen analysis defined for the first time a panel of small molecules which may reverse the ileal stricturing gene signature.
Beginning with the identification of NOD2,28,29 an important role for microbial signals in activating mononuclear phagocytes in CD pathogenesis has been suggested. The OSM cytokine produced by stromal cells has now also been shown to activate lamina propria monocytes and macrophages, leading to inflammatory cytokine and chemokine production9 and anti-TNF non-response.30 scRNASeq defined a mononuclear phagocyte: activated T cell: fibroblast module in adult CD ileal resection specimens,10 which when applied to the prior RISK bulk RNASeq dataset, was induced in patients who did not achieve durable steroid-free remission with anti-TNF. Using 3-fold deeper bulk RNASeq, we have now defined 518 genes that include induction of both OSM and COL1A2/ECM ileal co-expression signatures at diagnosis, linked to future stricturing behaviour in children with CD. These data support the role of pro-inflammatory myeloid cells in driving myofibroblast activation in CD, and may inform specific targets such as OSM to reduce inflammation and improve outcomes.31
The majority of patients in RISK have received anti-TNF during follow-up. Initiation of this medication within 3 months of diagnosis has been associated with higher 12-month rates of steroid-free remission,11 and lower 36-month rates of internal penetrating complications. However, a signal for lower rates of stricturing complications was not detected during the 36- and 60-month follow-up.4 This may reflect the need to increase anti-TNF exposure with tighter control of mucosal inflammation, as suggested by the CALM study,3 but may also support additional mechanisms requiring new therapeutic approaches. One attractive strategy would be to add therapies to anti-TNF in high-risk patients25 predicted to develop disease complications. We used bioinformatic perturbagen analysis to prioritise small molecules most likely to reverse the ileal gene signature for B2 stricturing. The Touchstone dataset consists of >8000 well-annotated compound and genetic perturbagens that have been profiled in a core set of nine cell lines. Although the signatures are based on epithelial cell lines, we27 and others have shown that the signatures and the connectivity map approach can be leveraged for fibrotic disorders.32,33 We employed a similar approach and dataset to discover preclinical therapeutic candidates for cystic fibrosis [https://www.biorxiv.org/content/10.1101/288324v1], which are currently in feasibility testing. Clearly, all candidate drugs including those identified by the studies using connectivity-based approaches and L1000 LINCs or Touchstone datasets, need to be validated in model systems before they go into clinical trials. Nevertheless, these compounds or hits can serve as leads for additional in-vitro or in-vivo assays.
Small molecules inhibiting macrophage and fibroblast activation and angiogenesis were predicted to have a substantial effect on pathways leading to stricturing. More specifically to the four compounds emphasised here, orantinib [SU6668] was shown to inhibit migration of intestinal subepithelial myofibroblasts and prevented myofibroblast migration into tumour cell clusters.34,35 Eicosatetraynoic acid [ETYA] is reported to block the platelet-induced proliferation of serum-starved subconfluent human fibroblasts,36 and decreased elastin gene transcription and alpha-smooth muscle actin mRNAs in cultured lipofibroblasts.37 ETYA was also shown to have anti-inflammatory effects on lipopolysaccharide [LPS]-stimulated macrophages,38 and through induction of PPARa in monocytes,39 and to inhibit pro-inflammatory signals by blocking NFKb translocation to the nucleus.40 Ephrin inhibition and PT-360 were less studied in comparison with the other two compounds.
However, targeting the ephrin system in an animal model of intestinal inflammation has demonstrated efficacy,41 and others have demonstrated its role in wound healing.42 PT-360 [L-glutamyl L-boroproline] was tested in vitro on rheumatoid arthritis synovial fibroblasts, with an effect on MMP levels.43 Collectively, these experimental studies have confirmed direct effects of compounds prioritised by our bioinformatic analysis in highly relevant cell types including activated myofibroblasts and inflammatory macrophages. These small molecules may now be prioritised for further study in preclinical model systems of inflammation and fibrosis, including transgenic mice in which over-expression of Tl1a leads to microbe-dependent ileitis and fibrosis.44
Our study has several strengths, but also some limitations. RISK is the largest CD inception cohort study to date linking pre-treatment clinical, demographic, serological, and molecular factors to patient outcomes captured in a prospective and highly audited manner during 60-month follow-up.4 Therapies were chosen by patient and physician preference, and so reflect real-world practice. Central reads of baseline MRE studies have suggested a very low rate of disease behaviour misclassification [4%]. However, these are also some limitations. The number of participants with both ileal gene expression data and 60-month stricturing outcomes was still relatively low, although key findings regarding the predictive ability of ASCA and CBir1 serology, and the ileal ECM gene signature, were replicated and validated in a new RNASeq dataset including approximately 50% of B1 and B2 participants not included in the previous report.4 However, as follow-up imaging was not obtained at regular intervals in all participants, we may have missed subclinical or mildly symptomatic strictures. It is possible that the baseline gene signature is maintained after the behaviour change; ongoing studies with follow-up samples will test this. Data are bulk RNASeq, and so cannot identify with certainty cell-type specific pathways as would be identified by scRNASeq. However, it should be noted that a mononuclear phagocyte module defined using scRNASeq in a small number of adult CD patients10 could be applied to the prior RISK bulk ileal RNASeq dataset, thereby classifying patients less likely to achieve durable remission with anti-TNF. An ongoing validation study will increase the number of participants with disease complications using bulk clinical FFPE ileal biopsies and will test the utility of obtaining such data from archived pathology samples.
In summary, we have defined OSM/INFL and ECM/collagen gene signatures specifically expressed in the pre-treatment ileum and linked with future stricturing disease complications in a large prospective CD cohort. Data support a critical role for activated macrophages in driving myofibroblast activation at an early stage of disease. We have further replicated and validated the previous ECM GO gene signature in the context of 60-month follow-up. A model including serum ASCA, CBir1 serology, and a refined ileal ECM eight-gene panel identified patients at higher risk for stricturing with excellent discriminant power, which would need to be validated in another cohort. If validated in another cohort or in the ongoing analysis of independent FFPE samples within RISK, this model may be used to enrich clinical trials for patients more likely to develop stricturing disease complications. This may include small molecules predicted to reverse the ileal gene signature for stricturing which, if validated in preclinical models, may provide an adjunct approach to anti-TNF therapy.
The RNASeq data are deposited in the SRA database, and the accession number is SUB6656230
Funding
This work was supported by: the Crohn’s and Colitis Foundation [LAD and SK]; the Gene Analysis and Integrative Morphology cores of the National Institutes of Health [NIH]-supported Cincinnati Children’s Hospital Research Foundation Digestive Health Center [1P30DK078392-01]; NIH grant T32 DK007727 [AT]; and NIH grant R01 DK098231 [LAD and SK].
Conflict of Interest
None. The authors have no financial arrangement[s] with a company whose product figures prominently in the submitted manuscript or with a company making a competing product.
Supplementary Material
Acknowledgements
The results published here are in whole or part based on data obtained from the IBD Plexus programme of the Crohn’s and Colitis Foundation. This work was supported by the Crohn’s and Colitis Foundation. We thank the Crohn’s and Colitis Foundation RISK study publication committee and Andres Hurtado-Lorenzo and Gerard Honig [CCF] for coordinating the gene expression and for critical review of this manuscript. We also thank Erin Bonkowski and Elizabeth Maier for technical support, and Kajari Mondal, Jarod Prince, and David Hercules for project and data management support.
Author Contributions
Study concept and design: LD, YH, BA, GG, SK. Acquisition of data: ST, DS, AT, BS, TW, RB, JN, JR, JM, JD, DM, AG, MH, SB, RK, DM, AP, AG, SJS, NL, AO, MOH, DZ, RG, DK, RG, SK, SG, SC, SS, MS, JD. Analysis and interpretation of data: YH, PM, RK, PJD, SG, ST, DS, BS, SV, TB, BA, GG, SK, AJ, LD. Drafting of the manuscript: YH, PM, RK, SG, ST, TB, AJ, LD. Critical revision of the manuscript for important intellectual content: YH, GG, MD, JH, SK, AJ, LD. Obtained funding: LD, SK. Administrative, technical, or material support: ST, DS, BS. Study supervision: LD, AJ, SK.
References
- 1. Vernier-Massouille G, Balde M, Salleron J, et al. Natural history of pediatric Crohn’s disease: a population-based cohort study. Gastroenterology 2008;135:1106–13. [DOI] [PubMed] [Google Scholar]
- 2. Murthy SK, Begum J, Benchimol EI, et al. Introduction of anti-TNF therapy has not yielded expected declines in hospitalisation and intestinal resection rates in inflammatory bowel diseases: a population-based interrupted time series study. Gut 2020;69:274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Colombel JF, Panaccione R, Bossuyt P, et al. Effect of tight control management on Crohn’s disease [CALM]: a multicentre, randomised, controlled phase 3 trial. Lancet 2018;390:2779–89. [DOI] [PubMed] [Google Scholar]
- 4. Kugathasan S, Denson LA, Walters TD, et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 2017;389:1710–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haberman Y, Tickle TL, Dexheimer PJ, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest 2014;124:3617–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haberman Y, BenShoshan M, Di Segni A, et al. Long ncRNA landscape in the ileum of treatment-naive early-onset Crohn disease. Inflamm Bowel Dis 2018;24:346–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haberman Y, Schirmer M, Dexheimer PJ, et al. Age-of-diagnosis dependent ileal immune intensification and reduced alpha-defensin in older versus younger pediatric Crohn Disease patients despite already established dysbiosis. Mucosal Immunol 2019;12:491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arijs I, Quintens R, Van Lommel L, et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm Bowel Dis 2010;16:2090–8. [DOI] [PubMed] [Google Scholar]
- 9. West NR, Hegazy AN, Owens BMJ, et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med 2017;23:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martin JC, Chang C, Boschetti G, et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 2019;178:1493–508.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walters TD, Kim M, Denson LA, et al. Increased effectiveness of early therapy with anti-tumor necrosis factor-alpha vs an immunomodulator in children with Crohn’s disease. Gastroenterology 2013;146:383‐91. [DOI] [PubMed] [Google Scholar]
- 12. Speca S, Giusti I, Rieder F, et al. Cellular and molecular mechanisms of intestinal fibrosis. World J Gastroenterol 2012;18:3635–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis 2011;17:1314–21. [DOI] [PubMed] [Google Scholar]
- 14. Silverberg MS, Satsangi J, Ahmad T, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol 2005;19[Suppl A]:5A–36A. [DOI] [PubMed] [Google Scholar]
- 15. Dubinsky MC, Kugathasan S, Mei L, et al. Increased immune reactivity predicts aggressive complicating Crohn’s disease in children. Clin Gastroenterol Hepatol 2008;6:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 17. Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009;37:W305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res 2010;38:W96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Subramanian A, Narayan R, Corsello SM, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 2017;171:1437–52 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledge base for drugs, drug actions and drug targets. Nucleic Acids Res 2008;36:D901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Corsello SM, Bittker JA, Liu Z, et al. The drug repurposing hub: a next-generation drug library and information resource. Nat Med 2017;23:405–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang X, Ko HM, Torres J, et al. Luminally polarized mural and vascular remodeling in ileal strictures of Crohn’s disease. Hum Pathol 2018;79:42–9. [DOI] [PubMed] [Google Scholar]
- 24. Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Graham MF, Diegelmann RF, Elson CO, et al. Collagen content and types in the intestinal strictures of Crohn’s disease. Gastroenterology 1988;94:257–65. [DOI] [PubMed] [Google Scholar]
- 26. Wagner M, Ko HM, Chatterji M, et al. Magnetic resonance imaging predicts histopathological composition of ileal Crohn’s disease. J Crohns Colitis 2018;12:718–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sontake V, Wang Y, Kasam RK, et al. Hsp90 regulation of fibroblast activation in pulmonary fibrosis. JCI Insight 2017;2:e91454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cho JH The Nod2 gene in Crohn’s disease: implications for future research into the genetics and immunology of Crohn’s disease. Inflamm Bowel Dis 2001;7:271–5. [DOI] [PubMed] [Google Scholar]
- 29. Cho JH, Abraham C. Inflammatory bowel disease genetics: Nod2. Annu Rev Med 2007;58:401–16. [DOI] [PubMed] [Google Scholar]
- 30. Haberman Y, Karns R, Dexheimer PJ, et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Commun 2019;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Verstockt S, Verstockt B, Vermeire S. Oncostatin M as a new diagnostic, prognostic and therapeutic target in inflammatory bowel disease [IBD]. Expert Opin Ther Targets 2019;23:943‐54. [DOI] [PubMed] [Google Scholar]
- 32. Karatzas E, Bourdakou MM, Kolios G, et al. Drug repurposing in idiopathic pulmonary fibrosis filtered by a bioinformatics-derived composite score. Sci Rep 2017;7:12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li L, Greene I, Readhead B, et al. Novel therapeutics identification for fibrosis in renal allograft using integrative informatics approach. Sci Rep 2017;7:39487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Iwanaga K, Okada M, Murata T, et al. Prostaglandin E2 promotes wound-induced migration of intestinal subepithelial myofibroblasts via EP2, EP3, and EP4 prostanoid receptor activation. J Pharmacol Exp Ther 2012;340:604–11. [DOI] [PubMed] [Google Scholar]
- 35. Walter-Yohrling J, Pratt BM, Ledbetter S, Teicher BA. Myofibroblasts enable invasion of endothelial cells into three-dimensional tumor cell clusters: a novel in vitro tumor model. Cancer Chemother Pharmacol 2003;52:263–9. [DOI] [PubMed] [Google Scholar]
- 36. Berg C, Hammarström S, Herbertsson H, et al. Platelet-induced growth of human fibroblasts is associated with an increased expression of 5-lipoxygenase. Thromb Haemost 2006;96:652–9. [PubMed] [Google Scholar]
- 37. McGowan SE, Jackson SK, Doro MM, Olson PJ. Peroxisome proliferators alter lipid acquisition and elastin gene expression in neonatal rat lung fibroblasts. Am J Physiol 1997;273:L1249–57. [DOI] [PubMed] [Google Scholar]
- 38. Chaudhri G Differential regulation of biosynthesis of cell surface and secreted TNF-alpha in LPS-stimulated murine macrophages. J Leukoc Biol 1997;62:249–57. [DOI] [PubMed] [Google Scholar]
- 39. Marx N, Mackman N, Schönbeck U, et al. PPARalpha activators inhibit tissue factor expression and activity in human monocytes. Circulation 2001;103:213–9. [DOI] [PubMed] [Google Scholar]
- 40. Stuhlmeier KM, Kao JJ, Bach FH. Arachidonic acid influences proinflammatory gene induction by stabilizing the inhibitor-kappaBalpha/nuclear factor-kappaB [NF-kappaB] complex, thus suppressing the nuclear translocation of NF-kappaB. J Biol Chem 1997;272:24679–83. [DOI] [PubMed] [Google Scholar]
- 41. Grandi A, Zini I, Palese S, et al. Targeting the Eph/Ephrin system as anti-inflammatory strategy in IBD. Front Pharmacol 2019;10:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Coulthard MG, Morgan M, Woodruff TM, et al. Eph/Ephrin signaling in injury and inflammation. Am J Pathol 2012;181:1493–503. [DOI] [PubMed] [Google Scholar]
- 43. Ospelt C, Mertens JC, Jüngel A, et al. Inhibition of fibroblast activation protein and dipeptidylpeptidase 4 increases cartilage invasion by rheumatoid arthritis synovial fibroblasts. Arthritis Rheum 2010;62:1224–35. [DOI] [PubMed] [Google Scholar]
- 44. Jacob N, Jacobs JP, Kumagai K, et al. Inflammation-independent TL1A-mediated intestinal fibrosis is dependent on the gut microbiome. Mucosal Immunol 2018;11:1466–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.