Fuentes-Fayos et al. show that the expression of spliceosome components and splicing factors is dysregulated in high-grade astrocytomas and glioblastomas. The splicing machinery could be a new source of diagnostic and prognostic biomarkers as well as therapeutic targets, with SRSF3 being a key splicing factor in glioblastomas.
Keywords: glioblastoma, splicing machinery, SRSF3, PDGFRB pathway, antitumour therapy
Abstract
Glioblastomas remain the deadliest brain tumour, with a dismal ∼12–16-month survival from diagnosis. Therefore, identification of new diagnostic, prognostic and therapeutic tools to tackle glioblastomas is urgently needed. Emerging evidence indicates that the cellular machinery controlling the splicing process (spliceosome) is altered in tumours, leading to oncogenic splicing events associated with tumour progression and aggressiveness. Here, we identify for the first time a profound dysregulation in the expression of relevant spliceosome components and splicing factors (at mRNA and protein levels) in well characterized cohorts of human high-grade astrocytomas, mostly glioblastomas, compared to healthy brain control samples, being SRSF3, RBM22, PTBP1 and RBM3 able to perfectly discriminate between tumours and control samples, and between proneural-like or mesenchymal-like tumours versus control samples from different mouse models with gliomas. Results were confirmed in four additional and independent human cohorts. Silencing of SRSF3, RBM22, PTBP1 and RBM3 decreased aggressiveness parameters in vitro (e.g. proliferation, migration, tumorsphere-formation, etc.) and induced apoptosis, especially SRSF3. Remarkably, SRSF3 was correlated with patient survival and relevant tumour markers, and its silencing in vivo drastically decreased tumour development and progression, likely through a molecular/cellular mechanism involving PDGFRB and associated oncogenic signalling pathways (PI3K-AKT/ERK), which may also involve the distinct alteration of alternative splicing events of specific transcription factors controlling PDGFRB (i.e. TP73). Altogether, our results demonstrate a drastic splicing machinery-associated molecular dysregulation in glioblastomas, which could potentially be considered as a source of novel diagnostic and prognostic biomarkers as well as therapeutic targets for glioblastomas. Remarkably, SRSF3 is directly associated with glioblastoma development, progression, aggressiveness and patient survival and represents a novel potential therapeutic target to tackle this devastating pathology.
Introduction
Astrocytomas are a subtype of malignant gliomas characterized by rapid growth and high diffusion through the brain (Louis et al., 2016). They are stratified from low to high aggressiveness (WHO criteria) (Louis et al., 2016) with low-grade astrocytomas (grades I and II) and high-grade astrocytomas (HGAs; grades III and IV), and grade IV [glioblastoma multiforme (GBM)] being the most aggressive and one of the most common malignant cancers in the brain (14.7% of total) and the CNS (Ostrom et al., 2018). Currently, HGAs/glioblastomas are detected by diagnostic imaging, with surgery as the first line of treatment, combined with chemotherapy and/or radiotherapy, depending on various clinical factors (type and degree of tumour, location and size, patient’s age and sex, etc.). However, about two-thirds of patients with HGAs, mainly glioblastomas, do not have a survival rate >2 years after diagnosis (Ozdemir-Kaynak et al., 2018).
Tumour pathologies, including HGAs, are known to share atypical presence, alteration or loss of relevant components of key molecular machineries regulating cell physiology, including the splicing machinery, which governs the splicing mechanism, that adequately processes the pre-mRNA to obtain mature mRNA (Chen and Manley, 2009; Pelechano et al., 2013). Splicing is an intricate process, tightly regulated and carried out by the spliceosome (Yan et al., 2019), a macromolecular machinery organized as a complex of RNA-protein multi-subunits, which act in a coordinated and sequential manner to develop its action in a highly dynamic way. Specifically, the spliceosome consists of a main core composed of five small nuclear ribonucleoproteins (SNRNPs), which is assisted by a large number of auxiliary core proteins and the so-called splicing factors (Jurica and Moore, 2003), which cooperate in the precise recognition of a multitude of target introns (Chen and Manley, 2009). In mammals there are two types of spliceosomes, the main or major spliceosome, also known as U2-dependent, and the minor, U12-dependent spliceosome (Chen and Manley, 2009; Matera and Wang, 2014).
Alternative splicing is not infrequent, as it is the mechanisms processing >90% of the human coding genes, and thereby provides an essential source of biological versatility. However, it is also a vulnerable regulatory point. Thus, growing evidence indicates that defects in the splicing process are frequent, leading to the appearance of altered spliceosome components, splicing factors and/or aberrant splicing variants (generated by alternative splicing), which are associated with the development, progression and aggressiveness of various types of cancer, such as lung, breast, prostate, colon, brain and pancreas (Stratton et al., 2009; Cordoba-Chacon et al., 2011; Guo et al., 2015; Kozlovski et al., 2017; Barbagallo et al., 2018; Song et al., 2019; Jimenez-Vacas et al., 2020). In fact, the study of the expression profile of splicing isoforms (‘splicing signature’) has been shown in some tumours to provide a more useful tool than the analysis of the genetic/omic profile (‘transcriptome signature’) to provide an accurate classification of tumours/patients in terms of diagnosis and treatment selection (Feero et al., 2010). Therefore, a better understanding of the regulation of splicing in normal and pathological tissues may help to identify novel diagnostic and prognostic biomarkers and therapeutic tools to target tumour pathologies.
However, to the best of our knowledge, no studies have reported hitherto a comprehensive analysis to ascertain whether the splicing machinery, i.e. the spliceosome and its auxiliary splicing factors, is altered in HGAs/glioblastomas. These data could provide new valuable avenues to develop novel strategies to tackle these terrible tumours. Thus, in this study we aimed to determine—for the first time—the expression profile of a representative set of spliceosome components and splicing factors and their relationship with key clinical and molecular features of glioblastoma samples/patients, as well as to determine the putative functional role and therapeutic potential of the most relevant components identified within this cellular machinery in this devastating disease.
Materials and methods
Reagents
Unless otherwise indicated, reagents and products were purchased from Sigma-Aldrich. Platelet-derived growth factor D (PDGFDD) (R&D systems, #1159-SB-025) was administered in vitro at 20 ng/ml based on a dose response experiment in glioblastoma cell lines (data not shown).
Patients and samples
Fresh HGAs/glioblastoma samples (AIII/IV; n = 29; Cohort-1) were obtained by intracranial surgery from patients previously diagnosed with HGAs/glioblastomas. Control brain tissue samples (CTs; n = 16) were obtained from four healthy donors [autopsy; four different brain areas (Brocca, Wernicke, cingulate and medial)] (Supplementary Table 1A). All samples were histologically studied by expert anatomopathologists to confirm as HGA/glioblastoma and control samples. Portions of each sample were rapidly frozen in liquid nitrogen and stored at −80°C until extraction for total RNA (see below) or formalin-fixed paraffin embedded (FFPE) for immunohistochemistry (IHC) analysis (see below). A validation cohort consisting of additional HGA/glioblastoma (AIII/IV; n = 46; Cohort-2) and control (n = 3; obtained by lobectomy surgery) samples were similarly obtained and analysed (Supplementary Table 1B). Demographic and clinical characteristics of both cohorts were collected to carry out clinical correlations. This study was approved by the Reina Sofia University Hospital Ethics Committee and was conducted in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from all individuals included in the study.
Primary patient-derived glioblastoma cell culture
Glioblastoma samples were collected within 15 min after intracranial surgery and immediately transported to the cell culture room in sterile cold Spinner’s modified Eagle medium (S-MEM) (Gibco, #11380-037) complemented with 0.1% bovine serum albumin (BSA) (#A2153), 0.01% l-glutamine (#G7513), 1% antibiotic-antimycotic solution (Gibco, #R01510) and 2.5% HEPES (#H3537). Glioblastoma samples were dispersed into single cells within the following 30 min by a mechanic/enzymatic protocol using 2% Collagenase IV (ThermoFisher-Scientific, #17104019) and 0.15% trypsin lyophilized powder (BD, #215310). Cell viability >90% was always obtained, as determined by the trypan-blue dye exclusion method. The single cells were cultured onto poly-l-lysine (#P1524-25MG) tissue culture plates in a 10% foetal bovine serum (FBS) (#F6765) containing Dulbecco’s modified Eagle medium (DMEM) (BI, #06-1055-09-1A) complemented as S-MEM medium.
Cell lines
Glioblastoma cell lines (U-87 MG and U-118 MG) were obtained from the American Type Culture Collection (ATCC, #HTB-14/#HTB-15, respectively) and cultured according to the supplier’s recommendations of passages <20. These cell lines were previously checked for mycoplasma contamination by PCR every month, as previously reported (Uphoff and Drexler, 2013).
PDGFA and PDGFB mouse models
All experimental procedures were carried out following the European Regulations for Animal Care, in accordance with guidelines and regulations, and under the approval of Ethics Committees (CEIyBA) from the Spanish National Cancer Research Centre (CNIO). Samples from two glioblastoma mouse models recently reported were used (PDGFA and PDGFB models; tumour and non-tumour samples) (Bejarano et al., 2017).
Electroporated mouse models
All experimental procedures were performed according to the Cedars-Sinai Institutional Animal Care and Use Committee. Paired-end bulk RNAseq datasets from glioma mouse models generated by postnatal lateral ventricle electroporation after a plasmid DNA mix injection with driver mutations, which had been identified as driver mutations in previous models or in patient tumours (Erbb2-V664E-EGFP/Hras-G12-EGFP/Kras-G12V-EGFP tumour versus EGFP control models) were used as previously described (Breunig et al., 2016).
RNA isolation, real-time qPCR and customized qPCR dynamic array based on microfluidic technology
Total RNA from fresh normal and tumour human and mouse tissue samples was extracted, followed by DNase treatment using the AllPrep DNA/RNA/Protein Mini Kit and the RNase-Free DNase set (Qiagen, #80004/#79254), respectively. Total RNA from glioblastoma cell lines was extracted with TRIzol® Reagent (ThermoFisher Scientific, #15596026). In all cases, total RNA concentration and purity were assessed by Nanodrop One Microvolume UV-Vis Spectrophotometer (ThermoFisher Scientific). For qPCR analyses, total RNA was retrotranscribed by using random hexamer primers and the RevertAid RT Reverse Transcription Kit (ThermoFisher Scientific, #K1691). Thermal profile and qPCR analysis to obtain absolute mRNA copy number/50 ng of sample of selected genes are reported elsewhere (Luque et al., 2013, 2015).
As recently reported (Jimenez-Vacas et al., 2019b, 2020), a qPCR dynamic array based on microfluidic technology (Fluidigm, #BMK-M-48.48) was implemented to determine the simultaneous expression of 48 transcripts in HGA/glioblastoma samples compared to control samples using the Biomark System and the Fluidigm® Real-Time PCR Analysis Software v.3.0.2 and Data Collection Software v.3.1.2 (Fluidigm). Specific primers for human and mouse transcripts including components of the major spliceosome (n = 13), minor spliceosome (n = 4), associated splicing factors (n = 28), PDGFRB pathway-related genes and three housekeeping genes were specifically designed with the Primer3 software (Supplementary Tables 2–4). To control for variations in the efficiency of the retrotranscription reaction, mRNA copy numbers of the different transcripts analysed were adjusted by a normalization factor, calculated with the expression levels of three housekeeping genes [β-actin (ACTB), hypoxanthine guanine phosphoribosyl-transferase (HPRT), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; only for human samples) and peptidylprolyl isomerase-A (Cyclophilin A; only for mouse samples)] (Supplementary Tables 2 and 3) and the GeNorm v.3.3 software as previously reported (Luque et al., 2015; Hormaechea-Agulla et al., 2017a; Jimenez-Vacas et al., 2019a).
Immunohistochemical analysis
IHC analysis of selected splicing factors was performed on FFPE samples obtained by intracranial surgery from patients diagnosed with HGAs/glioblastomas (Cohort-1) and control/non-pathological samples. Specifically, rabbit polyclonal antibodies against human SRSF3 and RBM22 (#ab73891 and #ab59157, respectively), goat polyclonal antibody against human PTBP1 (#ab5642) and mouse monoclonal antibody against human RBM3 (#ab211356) were used following the manufacturer’s instructions (Abcam). Specifically, deparaffinized sections were incubated with the antibody overnight at 4°C. Then, ImmPRESS® Anti-Mouse/Rabbit IgG Peroxidase (Vector Laboratories, #MP-7500-50) was used according to the supplier’s recommendations, except for PTBP1 in which an anti-Goat IgG was added to secondary solution at a concentration of 1:500. Finally, sections were developed with 3,39-diaminobenzidine (Envision system 2-Kit Solution DAB, ThermoFisher Scientific, #34065), contrasted with haematoxylin (#MHS128). As previously reported (Del Rio-Moreno et al., 2019; Jimenez-Vacas et al., 2020), the pathologists performed histopathological analysis of the samples following a blinded protocol. In the analysis, +, ++, +++ indicate low, moderate, and high intensities of tumour region staining compared with the normal control adjacent region.
Silencing of splicing factors by specific small interfering RNA
Pre-designed and validated specific small interfering RNA (siRNA) oligos for knockdown of endogenous SRSF3, RBM22 or RBM3 (#s12733, #s31273, #s142991, respectively; Silencer® Select siRNAs; ThermoFisher Scientific), and for PTBP1 (1:1 from B-C sequences; #SR303857; Trilencer-27 siRNA kit; Origene) were used. Cells (n = 200 000 cells/well) were transfected with 25 nM of each siRNA individually using Lipofectamine RNAiMax® (ThermoFisher Scientific, #13778-075) according to the manufacturer’s instructions. Silencer® Select Negative Control siRNA (ThermoFisher Scientific, #4390843) was used as a scramble control. After 48 h, cells were collected for validation of the transfection (qPCR and western blot) and seeded for different functional assays (see below).
Measurements of proliferation and migration rates
As previously described, cell proliferation (Hormaechea-Agulla et al., 2017b; Vazquez-Borrego et al., 2019) and migration (Hormaechea-Agulla et al., 2017a, b; Vazquez-Borrego et al., 2019) in response to SRSF3/RBM22/PTBP1/RBM3 silencing or after PDGFDD treatment was analysed using alamarBlue® Assay (3000 cells/well for cell lines and 10 000 cells/well for primary cell cultures) (Biosource International, #BUF012B) and the wound-healing technique (150 000 U-118 MG cells/well), respectively. For the migration assay, U-118 MG cells cultured under confluence were serum-starved for 24 h to achieve cell synchronization, and, the wound was then made using a 200 μl sterile pipette tip. Wells were rinsed and cells were incubated for 6 h and 24 h with supplemented medium without FBS. Wound-healing was compared with the area just after the wound was performed. Three pictures were randomly acquired along the wound per well to calculate the area by ImageJ software v.1.49 (Schneider et al., 2012).
Apoptosis analysis
As previously reported (Vazquez-Borrego et al., 2019), apoptosis induction in response to SRSF3/RBM22/PTBP1/RBM3 silencing in glioblastoma cell lines (5000 cells/well onto white-walled multi-well luminometer plates) was performed using Caspase-Glo® 3/7 Assay (Promega Corporation, #G8091). Cells were serum-starved for 24 h before the measurements, which were carried out following the manufacturer’s protocols.
Tumorsphere formation
This assay was carried out in SRSF3/RBM22/PTBP1/RBM3-silenced glioblastoma cell lines (100 cells/well) cultured in a Corning Costar ultra-low attachment plate (#CLS3473) with DMEM F-12 (Gibco, #11320033) with EGF (20 ng/µl) (#SRP3027) for 10 days (refreshed every 48 h), as previously reported (Jimenez-Vacas et al., 2019a). Briefly, an inverted microscope coupled to a digital camera was used to take photographs to visualize and measure cell morphology and area in order to calculate the number of tumorspheres after 10 days of incubation. Then, tumorspheres were trypsinized to determine the number of cells per tumorsphere. In addition, glioblastoma U-87 MG and U-118 MG cell lines were similarly analysed after 5 days of incubation with PDGFDD treatment (administered from time 0) combined with SRSF3 silencing.
VEGF secretion
The VEGF Human ELISA Kit (ThermoFisher Scientific, #KHG0112) was used to quantify vascular endothelial growth factor (VEGF) secretion in response to SRSF3/RBM22/PTBP1/RBM3 silencing in glioblastoma cell lines, following the manufacturer’s instructions. Briefly, silenced cells were seeded (150 000 cells/well) and incubated for 24 h in FBS medium. Then, cells were starved in medium without FBS for 1 h and incubated under the same conditions for 4 h and 24 h. Medium was collected, centrifuged, and stored for VEGF determination.
Western blotting
To determine protein levels, cells pellets were resuspended using pre-warmed SDS-DTT sample buffer [62.5 mM Tris-HCl (#10708976001), 2% SDS (#71726), 20% glycerol (#17904), 100 mM DTT (#D0632-5G) and 0.005% bromophenol blue (#B0126)] followed by sonication for 10 s and boiling for 5 min at 95°C. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes (Millipore, #1704270). Membranes were blocked with 5% non-fat dry milk (#T145.3), in Tris-buffered saline/0.05% Tween-20 (#93773) and incubated with the primary antibody [anti-SRSF3, anti-RBM22, anti-PTBP1, anti-RBM3, anti-AKT (CST, #4060S), anti-pAKT (CST, #9272S), anti-ERK1/2 (CST, #9102S), anti-pERK (CST, #4370S), anti-PDGFRB (CST, #3162) and anti-PIK3p110A (CST, #4249)], and their appropriate secondary antibodies [anti-rabbit IgG-HRP (CST, #7074), anti-mouse IgG-HRP (CST, #7076) or anti-goat IgG-HRP (#A9452)]. Proteins were developed using an enhanced chemiluminescence detection system (GE Healthcare) with dyed molecular weight markers. A densitometric analysis of the bands was carried out with ImageJ software v.1.49 (Schneider et al., 2012) using total protein loading (Ponceau staining, #P3504-10G) or total protein signal (in case of AKT and ERK) as normalizing factor, as previously reported (Jimenez-Vacas et al., 2020).
Preclinical mouse model
A preclinical xenograft mouse model with silenced SRSF3 in vivo was developed. Specifically, 5-week-old ATHYM-Foxn1nu/nu mice (n = 5; Janvier Labs) were injected subcutaneously with 3 × 106 U-87 MG cells in both flanks [in 100 μl of basement membrane extract (Trevigen, #3432-010-01)]. Once the tumour was clearly measurable, each mouse received an injection with SRSF3 siRNA into one flank and a negative control siRNA (used as a scramble control) into the other flank using AteloGene® reagent (KOKEN Co, #KKN1394) to transfect the siRNA molecule into cells by local administrations. Tumour growth was monitored every 4 days using a digital calliper. Sixteen days after injection, mice were sacrificed and each tumour was dissected, fixed, and sectioned for histopathological examination following haematoxylin and eosin staining. Examination of %KI67 and mitosis number was carried out by expert pathologists. Additional pieces from the tumour were in liquid nitrogen until RNA extraction using TRIzol reagent and protein extraction using SDS-DTT buffer, as previously reported (Jiménez-Vacas, 2019). These experiments were carried out according to the European Regulations for Animal Care under the approval of the university/regional government research ethics committees.
nCounter PanCancer signalling pathway analysis
As previously reported (Sarmento-Cabral et al., 2017), the nCounter PanCancer Pathways Panel kit (NanoString Technologies, #GXA-PATH1-12) was used and carried out at the Laboratory of Genetics at UCAIB (IMIBIC) to simultaneously examine the expression of 730 genes associated with cancer (i.e. 606 genes representing all major cancer pathways and 124 key cancer driver genes). Briefly, after analysing the quality of all samples using microelectrophoresis, RNA from four scramble-control and four SRSF3-silenced samples from the preclinical mouse model (samples with the best quality) were loaded in the plate provided in the nCounter Kit, and the experiment was run following the manufacturer’s protocol. The data were analysed using the nSolver Analysis Software v.4.0.66 from NanoString Technologies with the PanCancer Pathways Analysis Module using 36 genes as housekeeping genes. All specific target sequences and panel details are available on the manufacturer’s webpage.
Detection of splicing variants in response to SRSF3 silencing by end-point PCR
End-point PCR was developed using cDNA from glioblastoma cell lines in response to SRSF3 silencing versus scramble-control to detect different splicing variants of PDGFRB and TP73 (a PDGFRB transcription factor). Details of the end-point PCR to detect splicing events have previously been reported (Duran-Prado et al., 2009; Hormaechea-Agulla et al., 2016). Primer sequences are included in Supplementary Table 4.
Bioinformatics and statistical analysis
Several data-mining processes were carried out by bioinformatic experts (Supplementary Fig. 3A), ranging from the application of preprocessing methods to the construction of classification and clustering models for a better estimation of the relevance of specific factors. Data were evaluated for heterogeneity of variance using the Kolmogorov–Smirnov test. Statistical differences were assessed by t-test, Mann-Whitney U-test or by one-way ANOVA followed by Fisher’s correction exact test. Analysis of qPCR array results was carried out by a bioinformatic expert who used several tests, models and hierarchical cluster algorithms, which allowed us to make a fairly accurate assessment. Correlations were studied using the Pearson correlation test. All statistical analyses were performed using Prism software v.8.0 (GraphPad Software, La Jolla, CA, USA) except the clustering analyses which were performed with MetaboAnalyst Software v.4.0 (McGill University, Quebec, Canada). Paired-end bulk RNAseq data were aligned, analysed and normalized using Partek Flow® software (Partek Incorporated, St. Louis, MO, USA). For survival analysis, groups were selected based on the cut-off points determined by survminer R package or median. The computational methodology shown in Supplementary Fig. 3A was implemented in R language v.3.5. Values of P < 0.05 were considered statistically significant. Data represent median (interquartile range) or means ± standard error of mean (SEM). *P, 0.05; **P, 0.01; ***P, 0.001, significantly different from control conditions.
Data availability
Microfluidic array, RNAseq, nCounter and other data used in this paper are available upon request to the corresponding author. Chinese Glioma Genome Atlas RNAseq data (CGGA) and Murat microarray data were interrogated through the GlioVis Tools (http://gliovis.bioinfo.cnio.es) (Murat et al., 2008; Bowman et al., 2017).
Results
Splicing machinery is drastically dysregulated in high-grade astrocytomas/glioblastomas
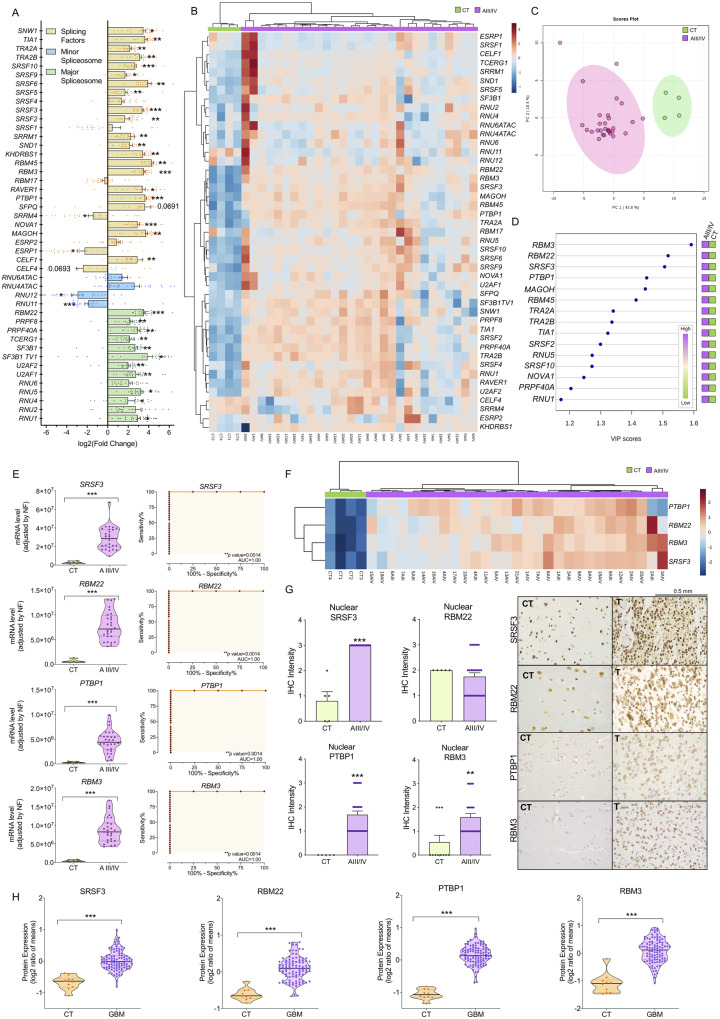
The expression levels of the components of the splicing machinery (spliceosome components and splicing factors) were analysed in HGAs/glioblastomas and control samples from a first cohort of patients (Supplementary Table 1A). An initial analysis in the four different brain areas obtained from healthy donors (control samples) revealed that the expression levels of all the splicing machinery components were completely homogenous across the different control brain areas (based on principal component analysis, PCA) (Supplementary Fig. 1A). Moreover, the individual expression level of these splicing machinery components did not vary across the four brain areas (based on qPCR analysis; data not shown). Similarly, PCA and individual mRNA expression comparisons also revealed that expression levels of these splicing machinery components were completely homogeneous between grade III versus IV HGAs (n = 8 and 21, respectively) (Supplementary Fig. 1B and C). Based on these results, further analyses were performed combining the expression levels of the four different control brain areas of each patient (control tissues, CTs; n = 4), and both HGA samples (III and IV-glioblastoma, HGAs/glioblastomas; n = 29).
Remarkably, we found a marked dysregulation in the expression levels of multiple components of the splicing machinery (spliceosome components and splicing factors) in HGAs/glioblastomas compared to the control tissues. Specifically, there was a significant upregulation of the 11 major spliceosome components (RBM22, PRPF8, PRPF40A, TCERG1, SF3B1, SF3B1TV1, U2AF2, U2AF1, RNU5, RNU4 and RNU1) and 20 splicing factors (SNW1, TIA1, TRA2A, TRA2B, SRSF10, SRSF9, SRSF6, SRSF5, SRSF3, SRSF2, SRRM1, SND1, KHDRBS1, RBM45, RBM3, RAVER1, PTBP1, NOVA1, MAGOH and CELF1), whereas two minor spliceosome components (RNU11 and RNU12) and two splicing factors (ESRP1 and SRRM4) were significantly downregulated (Fig. 1A and Supplementary Fig. 2). Interestingly, non-supervised hierarchical analysis based on the expression pattern of all spliceosome components and splicing factors was able to perfectly discriminate HGAs/glioblastomas and control tissues into two independent clusters (Fig. 1B), which was also confirmed by PCA (Fig. 1C). In fact, application of different bioinformatics analyses [i.e. five feature weighting and heuristic methods (Supplementary Fig. 3A), and variable importance in projection (VIP) score of partial least squares-discriminant analysis (PLS-DA; Fig. 1D)] revealed that SRSF3, RBM22, PTBP1 and RBM3 were always the highest score components, capable of discriminating between HGAs/glioblastomas and control tissues. Indeed, expression of these four components of the splicing machinery was elevated in HGAs/glioblastomas (Fig. 1E), and receiver operating characteristic (ROC) curve analyses corroborated their capacity to finely discriminate between HGAs/glioblastomas and controls samples, showing an area under the curve (AUC) of 1 (Fig. 1E). Moreover, as depicted in Fig. 1F, the heat map generated with only these four components perfectly discriminated HGAs/glioblastomas from control tissues, segregating them into two perfect clusters. Of note, overexpression of SRSF3, RBM22, PTBP1 and RBM3 levels in HGA/glioblastoma samples was further corroborated in a second cohort of patients collected in our hospital (validation cohort: three control tissues versus 46 HGAs/glioblastomas) (Supplementary Fig. 3B and Supplementary Table 1B), as well as in the well-characterized Murat and CGGA external dataset cohort available on the GlioVis data portal (Murat et al., 2008; Bowman et al., 2017). Furthermore, available data from the CGGA revealed that SRSF3, PTBP1 and RBM3, but not RBM22, mRNA levels were increased across glioma grades (III versus IV; n = 288; Supplementary Fig. 3D and E). Moreover, and similar to that previously found in the first cohort (Fig. 1A–F), the expression levels of SRSF3, RBM22, PTBP1 and RBM3 in this validation cohort again allowed the perfect discrimination between HGAs/glioblastomas and control tissues into two perfect clusters (Supplementary Fig. 3C). Based on these results, we selected SRSF3, RBM22, PTBP1 and RBM3 for further analyses.
Figure 1.
Splicing machinery is drastically dysregulated in HGAs/glioblastomas. (A) Individual fold-change of the expression of all the splicing machinery components analysed in control brain tissues (CT) and HGA/glioblastoma samples. (B) Hierarchical heat map generated using the expression levels of all the spliceosome components and splicing factors determined in control brain tissues from different brain areas [Controls (Brocca, Wernicke, cingulate and medial); n = 4] and HGA/glioblastoma samples (grades AIII/AIV; n = 29). (C) Principal components analysis of the mRNA expression levels of the splicing machinery components in the same sample set. (D) VIP scores obtained from partial least squares discriminant analysis (PLS-DA) of all of the splicing machinery components studies. (E) Levels of mRNA from selected splicing machinery components (SRSF3, RBM22, PTBP1 and RBM3) in control and tumour tissues and ROC curves analysis. (F) Hierarchical heat map generated using the expression levels of selected splicing machinery components (SRSF3, RBM22, PTBP1 and RBM3) in control brain tissues from different brain areas (CTs) and HGAs/glioblastomas (grades AIII/AIV). (G) IHC analysis of nuclear levels of SRSF3, RBM22, PTBP1 and RBM3 in FFPE samples from control and HGA/glioblastoma tissues (representative images are depicted). (H) Protein expression levels of SRSF3, RBM22, PTBP1 and RBM3 in GBM compared to non-tumour samples (GTEx tissues) using the proteomic glioblastoma CPTAC dataset. Data represent median (interquartile range) or means ± SEM. *P, 0.05; **P, 0.01; ***P, 0.001, significantly different from control conditions. See also Supplementary Figs 1–4.
Protein levels of SRSF3, RBM22, PTBP1 and RBM3 are elevated in HGA/glioblastoma samples
Consistent with the results of mRNA, IHC analyses using FFPE of all the samples from the first cohort of patients (Supplementary Table 1A) revealed that nuclear protein levels of SRSF3, PTBP1 and RBM3 were significantly elevated in HGA/glioblastoma samples compared to control samples; however, RBM22 staining was not elevated in these HGA/glioblastoma samples (Fig. 1G). Interestingly, we also had the possibility to compare the levels of these four splicing machinery components in a glioblastoma tumour tissue versus its non-tumour adjacent tissue, which confirmed that the protein levels of all of these factors, including RBM22, were clearly elevated in the glioblastoma tissue compared to the non-tumour area (Supplementary Fig. 4). In addition, low levels of cytoplasmic staining of SRSF3 were detected in HGA/glioblastoma tissues, although the score did not reach statistical significance (data not shown). Furthermore, SRSF3, RBM22, PTBP1 and RBM3 protein overexpression was corroborated in GBMs versus non-tumour samples (GTEx tissues) using the proteomic glioblastoma CPTAC dataset (Edwards et al., 2015) (Fig. 1H).
Dysregulation in splicing machinery is confirmed in different mouse models
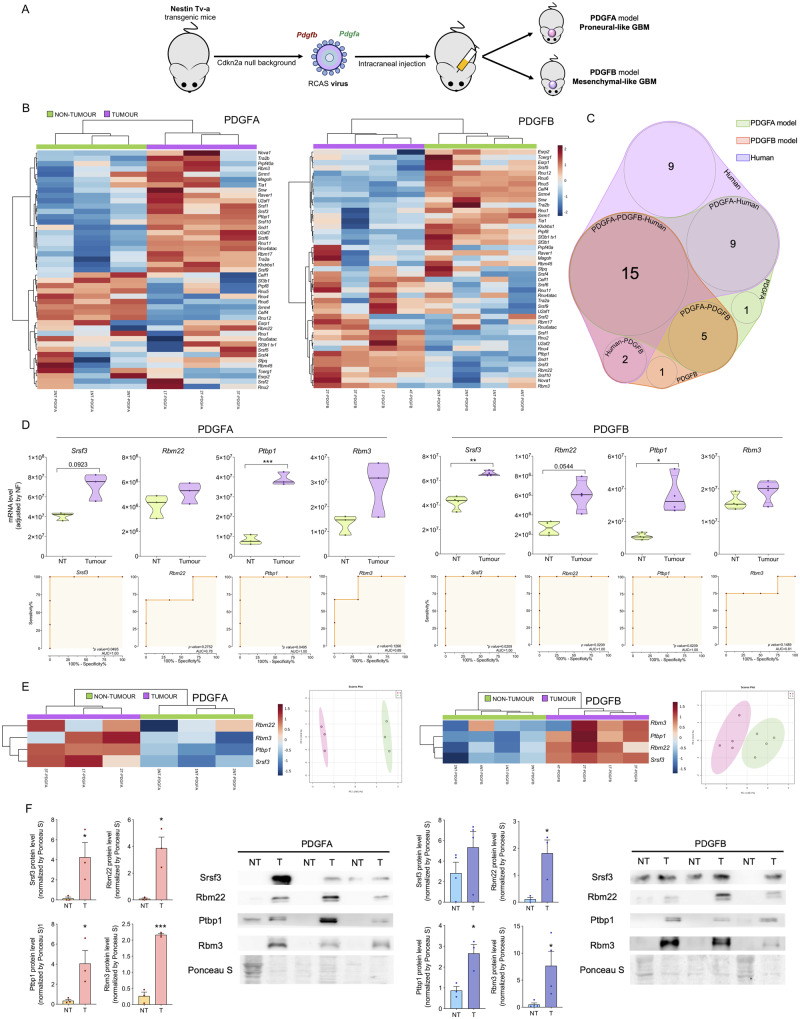
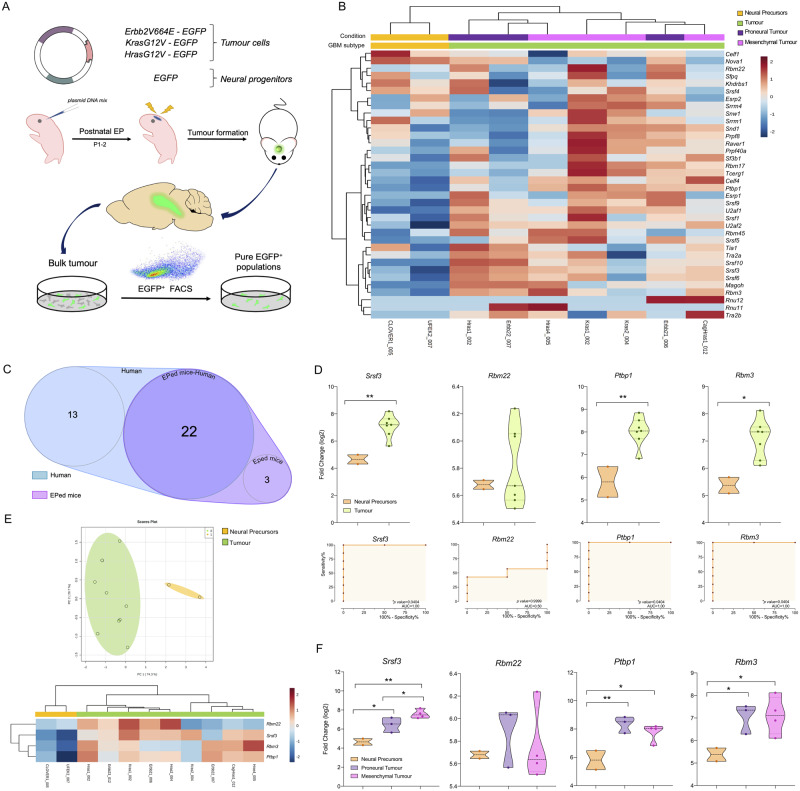
We also studied the expression levels of the 45 splicing machinery components (the same as previously analysed in human HGAs/glioblastomas) in two mouse models with proneural-like glioblastoma (PDGFA model) and mesenchymal-like glioblastoma (PDGFB model) recently published (Ozawa et al., 2014) (Fig. 2A), as well as in electroporated glioma mouse models with constitutively active oncogenes (Erbb2-V664E-EGFP/Hras-G12-EGFP/Kras-G12V-EGFP tumour versus EGFP-control model) as previously described (Breunig et al., 2016) (Fig. 3A).
Figure 2.
Dysregulation in the splicing machinery is also confirmed in two mouse models with proneural-like and mesenchymal-like glioblastoma. (A) Diagram of generation of mouse models of glioblastoma by overexpression of PDGFB or PDGFA in a Cdkn2a null background in Nestin-expressing cells (adapted from Bejarano et al., 2017). (B) Hierarchical heat map generated using the mRNA expression levels of spliceosome components and splicing factors in non-tumour and tumour samples from PDGFA/B-induced mouse models. (C) Venn diagram of altered splicing machinery components in PDGFA/B-induced glioblastoma mouse models and human HGA/glioblastoma samples. (D) Expression levels of mRNA and ROC curve analysis of selected splicing machinery components in PDGFA/B-induced mouse model samples. (E) Hierarchical heat map generated using the mRNA expression levels of previously selected splicing machinery components in the human study (Srsf3, Rbm22, Ptbp1 and Rbm3) and the PCA in the same set of murine samples. (F) Analysis and western blot images showing the validation of SRSF3, RBM22, PTBP1 and RBM3 protein overexpression in PDGFA/B-induced mouse tumours. Data represent median (interquartile range) or means ± SEM. *P, 0.05; **P, 0.01; ***P, 0.001, significantly different from control conditions.
Figure 3.
Dysregulation in the splicing machinery is also confirmed in electroporated glioma mouse models. (A) Diagram of the generation of mouse models of glioblastoma by plasmid DNA mix injection into the left lateral ventricle following mouse brain electroporation (EP) (adapted from Breunig et al., 2016). (B) Hierarchical heat map generated using the mRNA expression levels of spliceosome components and splicing factors in non-tumour and tumour samples classified in mesenchymal-like and proneural-like from electroporated models. (C) Venn diagram of altered splicing machinery components in electroporated (EPed) models and human HGA/glioblastoma samples. (D) mRNA expression levels and ROC curve analysis of selected splicing machinery components in the electroporated mouse model samples. (E) Hierarchical heat map generated using the mRNA expression levels of previously selected splicing machinery components in the human study (Srsf3, Rbm22, Ptbp1 and Rbm3) and the PCA in the same set of murine samples. (F) mRNA expression levels of selected splicing machinery components in the electroporated mouse model samples classified in proneural-like or mesenchymal-like phenotype versus control samples (neural precursors). Data represent median (interquartile range) or means ± SEM. *P, 0.05; **P, 0.01; ***P, 0.001, significantly different from control conditions.
Similar to that observed in humans, a marked dysregulation of the splicing machinery components was found in the PDGF mice and electroporated mice tumour samples (Figs 2B and 3B, respectively). Specifically, non-supervised hierarchical analysis based on the expression pattern of all these components was able to perfectly separate tumour and control samples into two independent clusters (Figs 2B and 3B). Fifteen of these components were commonly dysregulated in human and both mouse PDGF models (Fig. 2C), including the four key components (Srsf3, Rbm22, Ptbp1 and Rbm3) previously selected in human samples. Likewise, 22 components were also commonly dysregulated in human and the electroporated mouse models, including Srsf3, Ptbp1 and Rbm3, but not Rbm22 (Fig. 3C). In fact, ROC curve analyses of these four components corroborated their capacity to significantly discriminate between tumour and control tissues showing an AUC for Srsf3, Rbm22, Ptbp1 and Rbm3 of 1, 0.78, 1 and 0.89 in the PDGFA model, of 1, 1, 1 and 0.81 in the PDGFB-model, and of 1, 0.5, 1 and 1 in the electroporated glioma models, respectively (Figs 2D and 3D). Moreover, heat maps and PCA generated with these four components in PDGF and electroporated mouse models clearly showed that the expression pattern of just these four components is able to separate tumour and control tissues, segregating them into two perfect clusters/groups (Figs 2E and 3E).
In addition, western blot analyses using the PDGF samples revealed that protein levels of Srsf3, Rbm22, Ptbp1 and Rbm3 were significantly elevated in glioblastoma samples compared to their control tissues in both mouse models (Srsf33 expression tended to be elevated in the PDGFB model; P = 0.125; Fig. 2F). Moreover, analysis of the RNAseq dataset from the electroporated glioma models revealed that only Srsf3 expression was significantly higher in mesenchymal-like models in comparison with proneural-like models (Fig. 3F).
Silencing of SRSF3/RBM22/PTBP1/RBM3 expression decreases functional parameters in glioblastoma cells
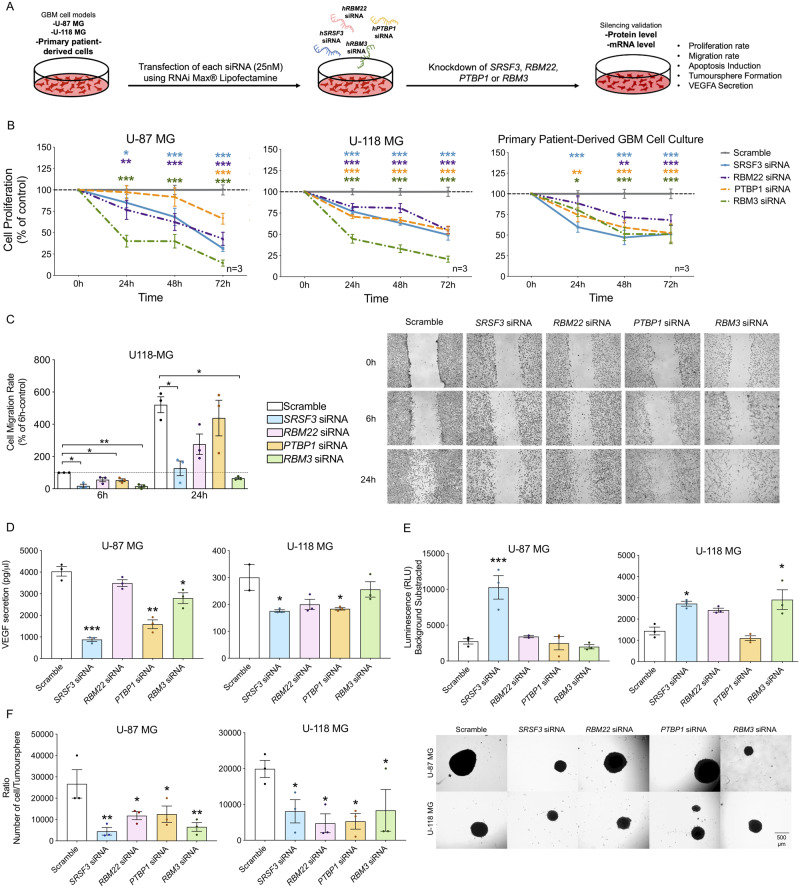
Two human glioblastoma cell line models (U-87 MG and U-118 MG) and/or primary patient-derived glioblastoma cell cultures were used to perform functional experiments (Fig. 4A). First, a heat map and a PCA generated with the expression pattern of all spliceosome components and splicing factors (Supplementary Fig. 5A and B) and also with only SRSF3, RBM22, PTBP1 and RBM3 (Supplementary Fig. 5A) were able to perfectly separate human control tissues into independent clusters/groups from human HGA/glioblastoma tissues and glioblastoma cell lines. In addition, data from available GBM cell lines (n = 34, including U-87 MG and U-118 MG) and primary GBM lines (n = 12) RNAseq were also analysed (Cancer Cell Line Encyclopedia) (Ghandi et al., 2019; Stringer et al., 2019), which revealed a similar expression profile of the spliceosome components and splicing factors across the different models, indicating that the U-87 MG and U-118 MG cell models used herein were appropriate glioblastoma models to study the functional role of the selected splicing machinery components (Supplementary Fig. 5C–E).
Figure 4.
Silencing of SRSF3, RBM22, PTBP1 and RBM3 expression decreases functional parameters of aggressiveness in glioblastoma cell lines and primary patient-derived glioblastoma cells. (A) Diagram showing the in vitro generation of SRSF3, RBM22, PTBP1 and RBM3 knockdown glioblastoma cells by silencing with specific siRNAs. (B) Proliferation rate of SRSF3, RBM22, PTBP1 and RBM3-silenced cells compared to control scramble-transfected cells (U-87 MG and U-118 MG models and primary patient-derived glioblastoma cells; n = 3). (C) Migration rate of SRSF3, RBM22, PTBP1 and RBM3-silenced U-118 MG cells compared to control scramble-transfected cells (representative images of the migration capacity are also included; n = 3). (D) VEGF secretion of SRSF3, RBM22, PTBP1 and RBM3-silenced cells compared to control scramble-transfected cells (U-87 MG and U-118 MG models; n = 3). (E) Apoptosis levels in SRSF3, RBM22, PTBP1 and RBM3-silenced cells compared to control scramble-transfected cells (U-87 MG and U-118 MG models; n = 3). (F) Number of cells per tumorsphere and representative images of formation of tumorspheres in SRSF3, RBM22, PTBP1 and RBM3-silenced cells compared to control scramble-transfected cells on cell morphology (U-87 MG and U-118 MG models; n = 3). Data represent means ± SEM. *P, 0.05; **P, 0.01; ***P, 0.001, significantly different from control conditions. See also Supplementary Fig. 5.
Silencing of SRSF3, RBM22, PTBP1 and RBM3 (confirmed at the mRNA and/or protein levels; Supplementary Fig. 5F) significantly decreased proliferation rate in a time-dependent manner in both glioblastoma cell lines and in primary patient-derived glioblastoma cell cultures (Fig. 4B). Similarly, silencing of these components reduced migration rate in a time-dependent manner in U-118 MG (Fig. 4C). Specifically, SRSF3, RBM22 and RBM3 silencing decreased migration capacity at 6 h and 24 h, but this difference did not reach statistical significance in the case of RBM22 (Fig. 4C). PTBP1 silencing significantly decreased migration only at 6 h (Fig. 4C). Moreover, a clear decrease in VEGF secretion was observed after SRSF3 and PTBP1 silencing in both cell lines, while RBM3 silencing decreased VEGF secretion only in U-87 MG cells, and a trend for statistical significance was found in response to RBM22 silencing in both cell lines (Fig. 4D). Furthermore, the use of the capase3/7 assay revealed that SRSF3 silencing induced apoptosis in both glioblastoma cells models, while RBM3 silencing only increased apoptotic rate in U-118 MG cells (Fig. 4E). Finally, application of a tumorsphere formation assay to quantify the proliferation capacity of cancer stem/progenitor cells revealed that SRSF3, RBM22, PTBP1 and RBM3 silencing decreased the number of glioblastoma stem/progenitor cells present in each tumorsphere in both cell lines (Fig. 4F).
Dysregulation of the splicing machinery is associated with relevant clinical parameters
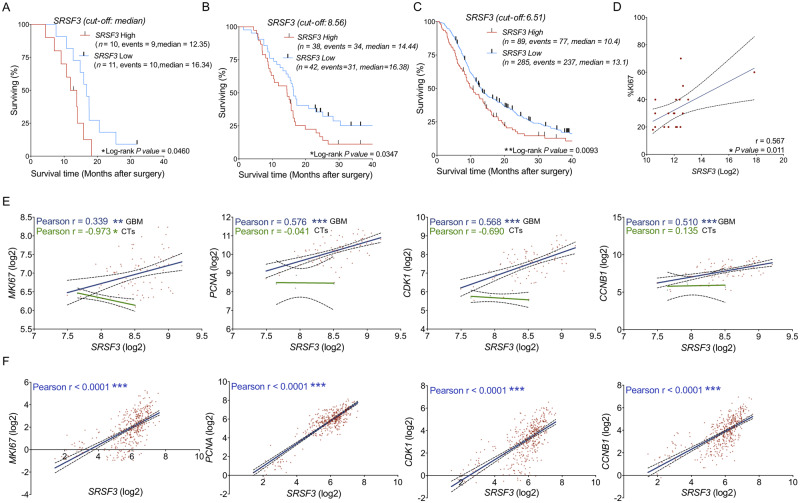
Low expression of SRSF3 (Fig. 5A), but not RBM22, PTBP1 and RBM3 (Supplementary Fig. 6A), was directly associated with better survival rate in glioblastoma patients from the first cohort. It should be noted that this analysis could not be implemented in our second cohort (validation cohort) as these patients were diagnosed near the end of the study. Nonetheless, we could also corroborate that SRSF3 expression was directly associated with a better survival rate in glioblastoma patients using the well-characterized Murat and CGGA dataset (Fig. 5B and C), this association being with SRSF3, but not with RBM22, PTBP1 and RBM3, consistent across the different datasets (Supplementary Fig. 6B and C). In line with this association, a higher expression of SRSF3 (Supplementary Fig. 6E; CGGA dataset), but not RBM22, PTBP1 and RBM3 (data not shown), was found in human mesenchymal GBM subtype (a poor survival GBM subtype) compared to proneural GBM subtype. Consistently, we found that the difference in mouse Srsf3 expression levels was more evident, in terms of P-values, when comparing mesenchymal GBM subtype versus control samples than in the case of proneural GBM subtype versus control samples in both mouse models analysed (PDGF and electroporated mice; Supplementary Fig. 6F). Additionally, we found a significant correlation between SRSF3 (Fig. 5D), but not RBM22, PTBP1 and RBM3 (Supplementary Fig. 6D), with the %KI67 index, a standard proliferative marker, in our cohort of patients. Likewise, a correlation between expression levels of SRSF3 and those of different key tumour progression markers (MKI67, PCNA, CDK1 and CCNB1) was also found in glioblastoma tissues (Murat and CGGA dataset), but not in the non-tumour samples (available only in Murat dataset) (Fig. 5E and F). Based on these and the previous results, SRSF3 was selected to perform further studies.
Figure 5.
The dysregulation of the splicing machinery is associated with relevant clinical parameters. Kaplan-Meier survival curves discerning between glioblastoma patients with high and low expression levels of SRSF3 from our cohort of patients (A), from the Murat dataset (B), and from the CGGA dataset (C). Correlation of SRSF3 mRNA expression levels and %KI67 in glioblastoma patients (D). Correlation of SRSF3 with different prognostic biomarkers in glioblastoma samples (blue regression line) and CTs (green regression line) using the Murat dataset (E). Correlation of SRSF3 with different prognostic biomarkers in glioblastoma samples using the CGGA dataset (F). See also Supplementary Fig. 6.
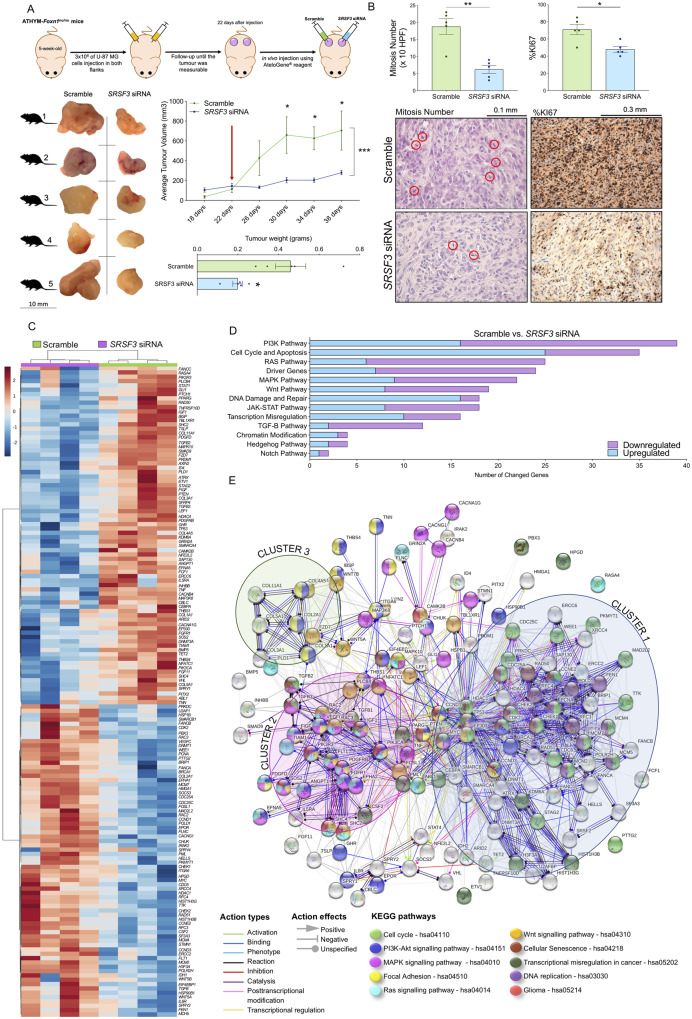
SRSF3 silencing prevents the progression of glioblastomas in vivo
SRSF3 silencing in vivo (validated at mRNA and protein levels; Supplementary Fig. 7A and B) reduced tumour volume and weight in the preclinical xenograft glioblastoma model (Fig. 6A). In fact, in vivo tumour volume across time clearly showed that glioblastoma progression was completely stopped in the SRSF3-silenced model (blue line) compared to the scramble-transfected tumours, which drastically continued their progression (green line; Fig. 6A). Moreover, the number of mitosis and %KI67 IHC staining was significantly decreased in the SRSF3-silenced model compared to the scramble-transfected (Fig. 6B) supporting the strong anti-tumour potential of SRSF3 silencing in glioblastoma cells previously observed in vitro (Fig. 4) and in vivo (Fig. 6A).
Figure 6.
SRSF3 silencing prevents the progression of glioblastomas and alters multiple key cancer related-pathways in vivo (U-87 MG-induced xenografts). (A) Diagram showing the geneneration of xenograft mouse models of glioblastoma by U-87 MG inoculation. SRSF3 in vivo silencing was carried out in one of the flanks of each mouse (n = 5). Average tumour volume and tumour weight of subcutaneous inoculated SRSF3 siRNA-transfected versus scramble-transfected tumours. Images of each tumour are shown. The red arrow indicates the treatment time with scramble or SRSF3-siRNA using AteloGene® reagent. (B) Mitosis number (×10 HPF) and %KI67 IHC analysis (n = 5/group) determined by experienced pathologists and representative images of haematoxylin staining of SRSF3 siRNA-transfected versus scramble-transfected tumours. (C) Hierarchical heat map generated with the mRNA expression levels of significantly altered genes (n = 149) measured by nCounter PanCancer in SRSF3 siRNA-transfected versus scramble-transfected tumours (n = 4 per group). (D) Number of hits of significantly altered genes in cancer-related pathways or processes. (E) Functional association network of the significantly altered genes in SRSF3 siRNA-transfected versus scramble-transfected tumours. These significantly altered genes were analysed using the STRING database, and they are marked according to their KEGG pathways analysis. See also Supplementary Figs 7 and 8.
SRSF3 silencing alters multiple key cancer-related pathways in vivo
SRSF3 silencing in vivo significantly altered the expression of 149 of 770 genes analysed with the nCounter PanCancer Pathways Panel kit compared to the scramble-transfected tumours (Fig. 6C). Specifically, the PI3K pathway was the most altered signalling pathway (i.e. 39 genes: 23 downregulated/16 upregulated), followed by the cell cycle and apoptosis pathways (i.e. 35 genes: 25 upregulated/10 downregulated; Fig. 6D). Additionally, other key signalling pathways were also altered (e.g. RAS, MAPK, Wnt, DNA-damage/repair, transcription misregulation, etc.) (Fig. 6D). Indeed, the heat maps generated with the genes involved in several of these pathways were able to perfectly discriminate between SRSF3-silenced versus scramble-transfected glioblastoma cells (Supplementary Fig. 8A).
We next used the STRING database to determine the potential functional association between the altered genes in response to SRSF3-silencing (Fig. 6E), which showed the existence of three clusters of altered genes strongly associated with the control of cell cycle/transcription/DNA replication (cluster-1: blue), PDGFR-activated pathway (cluster-2: magenta) and focal adhesion (cluster-3: green). An additional protein–protein interaction (PPI) functional association performance by Network Analyst software, revealed that the PDGFRB-activated pathway was the most relevant pathway altered (Supplementary Fig. 8B). In fact, a specific analysis of the glioma pathway using KEGG revealed a clear alteration in several genes involved in the PDGFRB pathway (i.e. PDGFRB, SOS2, SHC2, PIK3CR3 and PIK3CA) (Fig. 6E), which further supported the relevance of the PDGFRB in SRSF3-silenced cells.
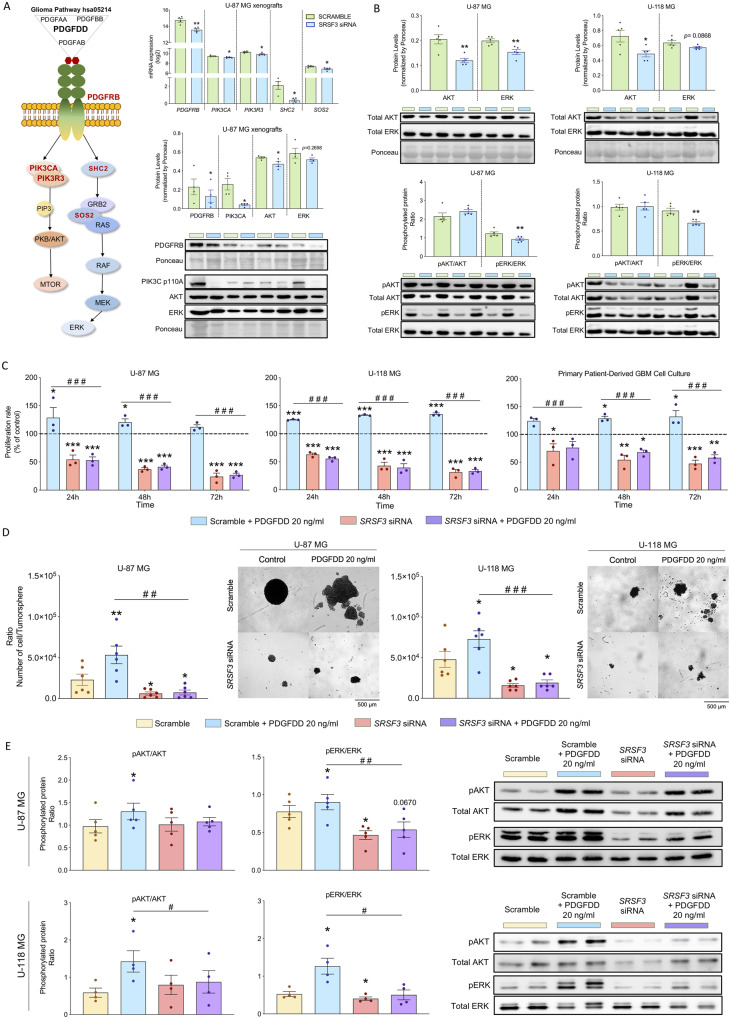
PDGFRB pathway inhibition as a driver of SRSF3 silencing-induced anti-glioblastoma tumour actions
mRNA levels of the glioma pathway components PDGFRB, SOS2, SHC2, PIK3CR3 and PIK3CA were significantly downregulated in SRSF3-silenced xenografted glioblastoma cells (Fig. 7A). Similarly, these changes were also observed in U-87 MG, U-118 MG and patient-derived primary glioblastoma cell cultures (Supplementary Fig. 9A). A protein downregulation of upstream (i.e. PDGFRB and PIK3 catalytic α-subunit) components of the glioma-pathway was also found in SRSF3-silenced xenografted glioblastoma cells (Fig. 7A), and also observed (i.e. PDGFRB) in U-87 MG and U-118 MG cells (Fig. 7B and Supplementary Fig. 9B). In addition, inhibition of total protein levels of the downstream components of the PDGF-PDGFRB pathway [i.e. AKT and ERK (classical end points of this pathway associated with cell survival, cell growth and cell proliferation)] was also observed in SRSF3-silenced xenografted glioblastoma cells (Fig. 7A) and/or SRSF3-silenced glioblastoma cells in vitro (U-87 MG and U-118 MG; Fig. 7B). Interestingly, we found that only the phosphorylation levels of ERK, but not AKT, were significantly downregulated in SRSF3-silenced glioblastoma cells in vitro (U-87 MG and U-118 MG; Fig. 7B). Remarkably, silencing of SRSF3 completely blocked the pro-oncogenic parameters (i.e. proliferation and number of glioblastoma stem/progenitor cells) induced in response to PDGFDD (specific ligand of PDGFRB receptor) in glioblastoma cells in vitro (U-87 MG and U-118 MG; Fig. 7C and D). Similar results in terms of proliferation rate were also observed in patient-derived glioblastoma primary cell cultures (Fig. 7C). Supporting these results is the fact that SRSF3-silencing blocked the stimulatory effects on AKT and ERK pathways induced by PDGFDD treatment in glioblastoma cells (U-87 MG and U-118 MG) (Fig. 7E).
Figure 7.
Inhibition of the PDGF-PDGFRB pathway as potential driver of the SRSF3 silencing-induced anti-glioblastoma tumour actions. (A) Expression levels (mRNA levels measured by nCounter PanCancer and protein levels measured by western blot) of different components of the PDGFRB activated pathways (in bold) in SRSF3 siRNA-transfected versus scramble-transfected U-87 MG-induced xenograft tumour samples (n = 4). (B) Phosphorylated protein ratios and total protein levels of AKT and ERK in SRSF3-silenced U-87 MG and U-118 MG cells compared to scramble-controls (n = 5) after 24 h of silencing. (C) Proliferation rates after treatment with PDGFDD homodimer (specific PDGFRB ligand) on scramble-transfected versus SRSF3-silenced U-87 MG, U-118 MG cells and primary patient-derived glioblastoma cells (n = 3). (D) Number of cells per tumoursphere and representative images of formation of tumorspheres after treatment with PDGFDD homodimer (specific PDGFRB ligand) on scramble-transfected and SRSF3-silenced U-87 MG and U-118 MG cells (n = 6). (E) Phosphorylated ratio of AKT/ERK after treatment with PDGFDD homodimer (specific PDGFRB ligand) on scramble-transfected U-87 MG cells versus SRSF3-silenced U-87 MG (n = 5) and U-118 MG cells (n = 4). Data represent means ± SEM. *P, 0.05; **P, 0.01; ***P, 0.001, significantly different from control conditions. #P, 0.05; ##P, 0.01; ###P, 0.001, significantly different from Scramble+PDGFDD condition. See also Supplementary Fig. 9.
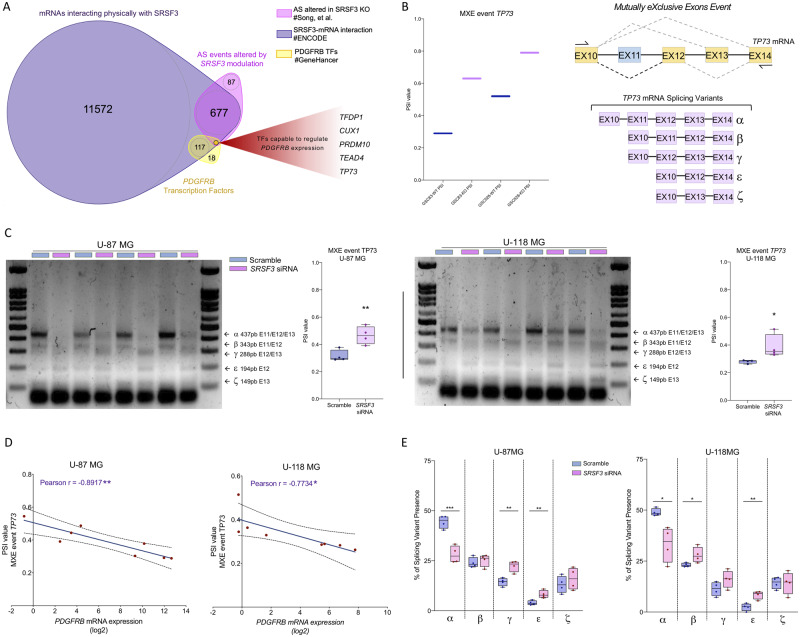
Changes in the expression of TP73 splicing variants as functional link between SRSF3-PDGF-PDGFRB pathway
Our results indicate that SRSF3 silencing does not alter the splicing of PDGFRB in GBM cell lines (Supplementary Fig. 10A) thus arguing against a direct action of this splicing factor on the receptor itself. Of note, our present results on SRSF3 silencing compare favourably with those in the Song et al. (2019) dataset, where SRSF3 knockout also downregulated PDGFRB (Supplementary Fig. 10B), while not altering PDGFRB splicing. Thus, to explore alternative mechanisms possibly linking SRSF3 and PDGF-PDGFRB pathway, we interrogated various datasets, aiming to ascertain the potential involvement of PDGFRB-controlling transcription factors whose splicing might be influenced by SRSF3. Specifically, we analysed: (i) the Song et al. (2019) dataset to define genes whose splicing is altered by SRSF3 knockout; (ii) ENCORI, Encyclopedia of RNA Interactomes (Ghandi et al., 2019), to identify mRNAs that physically interact with SRSF3; and (iii) GeneHancer database (Fishilevich et al., 2017), to predict transcription factors capable of regulating PDGFRB expression (Fig. 8A). Results from these analyses revealed five genes (TFDP1, CUX1, PRDM10, TEAD4 and TP73) that fulfil the three criteria and could thus represent a putative functional nexus between SRSF3 and PDGF-PDGFRB pathway (Fig. 8A). Indeed, analysis of PSI (Percent Spliced-in Index) of these five genes in the Song et al. (2019) dataset showed that SRSF3 knockout decreased the splicing events of TFDP1, CUX1 and PRDM10, and increased the splicing events of TEAD4 and TP73 (Fig. 8B and Supplementary Fig. 10C). Interestingly, from these transcription factors, only TP73 has already been experimentally shown to directly regulate PDGFRB expression (Hackzell et al., 2002; Uramoto et al., 2004; Wetterskog et al., 2009). Specifically, TP73 has seven splicing variants due to different inclusion of its C-terminal exons (Vikhreva et al., 2018), wherein only five could be experimentally detected (TP73α, β, γ, ε, ζ) (Fig. 8B). Some of the TP73 splicing variants appear to differentially impact oncogenesis (e.g. TP73α can play an oncogenic role) (Liu et al., 2004), and TP73β/γ variants can act as tumour suppressors (Jancalek, 2014; Vikhreva et al., 2018); while the function of TP73ε and TP73ζ is still unknown. The ability of SRSF3 to influence TP73 splicing in a GBM context is further substantiated by the altered PSI of TP73 found in our own experimental data on SRSF3-silenced GBM cell lines [i.e. an increased of the MXE (Mutually eXclusive Exon) splicing event in U-87 MG and U-118 MG; Fig. 8C]. In support of a link between the modulation of the expression of SRSF3, changes at the level of TP73 splicing and PDGF-PDGFRB pathway, an inverse correlation was found between MXE event PSI value of TP73 and PDGFRB expression in SRSF3-silenced glioblastoma cells (U-87 MG and U-118 MG; Fig. 8D). Moreover, a detailed analysis of the TP73 MXE event revealed that SRSF3 silencing in GBM cells distinctly altered the ratio of individual splicing variants with respect to the total amount of splicing variants detected (Fig. 8E). Specifically, while SRSF3 silencing decreased TP73α/TP73 total ratio, it increased the TP73β-γ-ε/TP73 total ratios (Fig. 8E).
Figure 8.
Changes in the expression profile of TP73 splicing variants as potential functional link between SRSF3 and PDGF-PDGFRB pathway. (A) Venn diagram of three levels of information derived from different sources: Song et al. (2019) dataset, ENCORI (Ghandi et al., 2019) and GeneHancer database (Fishilevich et al., 2017). (B) PSI (Percent Spliced-in Index) of TP73 from Song et al. (2019) dataset and diagram showing TP73 splicing variants detected and produced by a C-terminal MXE event. (C) PSI of TP73 in SRSF3 siRNA-transfected versus scramble-transfected U-87 MG and U-118 MG cells (n = 4). (D) Correlation of PDGFRB mRNA expression with PSI of TP73 in the same set of samples (n = 4). (E) Percentage of each splicing variant of TP73 identified in SRSF3 siRNA-transfected versus scramble-transfected U-87 MG and U-118 MG cells (n = 4). AS = alternative splicing. TFs = transcription factors.
Discussion
Glioblastoma is one of the most lethal human cancers, with late diagnosis and poor prognosis despite years of research, leading to a survival rate of 12–16 months from diagnosis (Singh and Eyras, 2017; Ostrom et al., 2018). As patients are often diagnosed at advanced stages, when cure is no longer possible, their quality of life is poor and worsens rapidly, while healthcare costs concomitantly increase. Indeed, despite extensive effort made in recent years to develop therapeutic approaches for tackling this pathology (Zanders et al., 2019), the current therapeutic strategies are not efficient at reducing tumour volume/growth or augmenting survival rate. This is likely due, in part, to the resistance acquired by tumours, particularly by stem cells progenitors, to current drugs (Noch et al., 2018). Thus, identification of new molecular diagnostic, prognostic and therapeutic tools to refine their detection, define tumour behaviour (from tumorigenesis and progression to metastasis mechanisms) and develop new treatments is crucial to combat this devastating pathology.
The splicing process is a highly coordinated mechanism, regulated and carried out by the spliceosome, that relies on a combination of multiple spliceosome components and splicing factors, intronic and exonic sequence elements, and temporal and spatial signalling pathways to adequately control gene expression, while increasing its complex versatility (Chen and Manley, 2009; Pelechano et al., 2013). Remarkably, increasing evidence over the last two decades has documented that the spliceosome and associated proteins are often altered in disease states, including metabolic diseases (Gahete et al., 2018; Del Rio-Moreno et al., 2019) and cancer (Dvinge et al., 2016; Jia et al., 2019; Jimenez-Vacas et al., 2019b, 2020), which augments pathobiological versatility through the generation of distinct/novel alternative splicing variants (Kozlovski et al., 2017; Singh and Eyras, 2017). Actually, every hallmark of cancer (Stratton et al., 2009) can be associated with several examples of proto-oncogenes, tumour-suppressor genes, or other genes whose splicing is altered to produce isoforms that contribute to the transformation process (Sveen et al., 2016). In fact, recent genomic characterization of different cancers has revealed recurrent copy number changes affecting genes encoding spliceosome components and splicing factors that contribute to cancer-relevant phenotype (Dvinge et al., 2016). Here, we demonstrate for the first time a drastic dysregulation of the expression profile of the splicing machinery in a well-characterized cohort of HGAs/glioblastomas, where a representative set of spliceosome components and splicing factors was markedly altered (77% and 79%, respectively). Moreover, bioinformatics analyses defined an expression-based molecular fingerprint of these spliceosome components and splicing factors able to perfectly discriminate between HGAs/glioblastomas versus control tissues from both human and mouse, which further reinforces our notion, suggesting that HGAs/glioblastomas have a global splicing dysregulation in different species. Further clustering and hierarchical bioinformatics analyses in two independent human sample cohorts revealed that SRSF3, RBM22, PTBP1 and RBM3 were the components of the splicing machinery with higher capacity to discriminate between human HGAs/glioblastomas and control tissues. Importantly, these results were confirmed in two independent external in silico cohorts of human glioblastomas (Murat/CGGA) and in different mouse glioma models analysed herein (PDGF/electroporated models). Moreover, data from the CGGA dataset also revealed that SRSF3, PTBP1 and RBM3 were significantly elevated across glioma grades (IV versus III versus II) supporting the diagnostic potential of these factors.
In this study, we evidenced an overall overexpression of SRSF3, RBM22, PTBP1 and RBM3, at both mRNA and protein level, in the different human cohorts and mouse models with gliomas analysed. Notably, silencing the expression of these splicing machinery components in glioblastoma cells induced marked reductions in aggressiveness features of glioblastomas (i.e. inhibition of proliferation, migration and VEGF secretion, and increase of apoptosis). Most notably, silencing of SRSF3, RBM22, PTBP1 and RBM3 strikingly decreased the number of glioblastoma stem/progenitor cells present in each tumorsphere, a relevant functional result that may help to explore how to overcome the well-known resistance of glioblastomas to current drugs (Noch et al., 2018). Thus, these results demonstrate that dysregulation of the splicing machinery can play a key pathophysiological role in glioblastoma cells, and some of its components (SRSF3, RBM22, PTBP1 and RBM3) could provide novel, useful tools as diagnostic biomarkers and potential therapeutic targets to tackle glioblastomas. The potential utility of SRSF3, RBM22, PTBP1 and RBM3 expression as prognostic biomarkers is further supported by the direct association found between their levels and relevant molecular features of aggressiveness (e.g. MKI67, PCNA, CDK1 and CCNB1 levels) in patients with glioblastomas but not in healthy patients, as well as between their levels and glioma grades and GBM subtypes (i.e. higher in IV versus III versus II grades, and in mesenchymal versus proneural GBM subtypes). Moreover, these observations suggested a causal link between dysregulation of these splicing machinery components and glioblastoma aggressiveness. While SRSF3, PTBP1 and RBM3, but not RBM22, have been associated with certain tumour pathologies (Guo et al., 2015; Barbagallo et al., 2018; Chen et al., 2019b; Jia et al., 2019; Melling et al., 2019; Song et al., 2019) and/or brain development (Vuong et al., 2016), to the best of our knowledge, this is the first report identifying a relevant functional role of these splicing machinery components in human glioblastomas, as well as in proneural/mesenchymal-like glioma mouse models with different prognosis.
Most importantly, this study reveals that expression of SRSF3 is directly associated with a better survival rate in HGA/glioblastoma patients, the main clinical problem in this pathology. This finding was corroborated in two external in silico cohorts of patients with glioblastomas (Murat and CGGA datasets), and is further supported by a recent study indicating that the knockout (KO) of SRSF3 extended overall survival of tumour-bearing animals (Song et al., 2019). In line with this, a higher SRSF3 expression was observed in human and mouse mesenchymal GBMs (with poor survival rate) compared to proneural GBM subtypes, which reinforced the prognostic value of SRSF3. The mechanisms underlying the link between SRSF3 and survival rate may relate to the direct association found between SRSF3 silencing and tumour progression, as well as with the noteworthy disruption in one of the main pro-oncogenic signalling pathways activated in GBM cells, i.e. the PDGFRB-glioma pathway (Liu et al., 2018). Specifically, we demonstrate here that SRSF3 is an effective target in glioblastomas in vivo, since silencing of this splicing factor effectively blocks glioblastoma progression of already established glioblastoma tumours in a preclinical mouse model of glioblastoma. In fact, SRSF3 silencing in glioblastoma in vivo markedly decreased tumour volume, tumour weight, mitosis number of glioblastoma cells and KI67 expression in all mice studied (100%), which, together with the previous in vitro data reported here using SRSF3-silencing glioblastoma cells, and with the extended overall survival observed in human studies, and in a recent study on KO-SRSF3 mice (Song et al., 2019), further demonstrate the clinico-pathophysiological relevance of the strong anti-tumour role of SRSF3 silencing in glioblastomas, and its potential value as a future therapeutic target in this devastating disease. In line with our results, previous studies have also indicated that the altered expression of specific components of the splicing machinery is tightly associated with aggressive disease and poor overall survival of patients with pancreatic cancer (Qiao et al., 2019) and in colorectal cancer (Sveen et al., 2011).
To determine the signalling mechanisms underlying the anti-tumour role of SRSF3 in glioblastomas, we explored an ample range of cancer-related signalling pathways in response to SRSF3 silencing in vivo and in vitro. Our data revealed, for the first time, a striking alteration in the PI3K pathway in different SRSF3-silenced glioblastoma cell models. This pathway has been reported to be closely associated with progression of glioblastomas and used as a target for the development of novel drugs (Massacesi et al., 2016; Mecca et al., 2018), as its inhibition or alteration causes anti-proliferative/invasive effects, and apoptosis (Mete et al., 2019; Xu et al., 2019). However, we found that the PDGFRB pathway, which is involved in widely known groups of glioma-related PI3K pathways (currently being used as therapeutic target) (Batchelor et al., 2017), was the most relevant pathway altered in SRSF3-silenced cells, since a clear alteration in several genes involved in this pathway (i.e. PDGFRB, SOS2, SHC2, PIK3CR3 and PIK3CA; all glioma pathway components) was observed. Moreover, a protein downregulation of upstream (i.e. PDGFRB and PIK3 catalytic α-subunit) and downstream (i.e. AKT) components of the glioma pathway representing classical end points for cell survival, growth and proliferation (Brennan et al., 2013), was also found in SRSF3-silenced xenografted glioblastoma cells. Most importantly, SRSF3 silencing blocked the stimulatory effects on AKT and ERK pathways induced by PDGFDD treatment, and completely blocked key pro-oncogenic parameters (i.e. proliferation and/or number of glioblastoma stem/progenitor cells) induced in response to PDGFDD in different glioblastoma cell models (U-87/U-118 cell lines and primary GBM cells). Therefore, these data provide original, compelling evidence that SRSF3 is functionally linked to these well-known pathophysiologically relevant pro-oncogenic pathways in gliomas (PDGFRB and PI3K), which further supports the potential clinico-pathophysiological importance of the strong anti-tumour role of SRSF3 silencing in glioblastomas in vivo and in vitro.
However, SRSF3 does not appear to control PDGFRB activity by directly modulating its splicing, in that no apparent changes in PDGFRB splicing variants were found in SRSF3-silenced (this report) or in SRSF3-knockout (Song et al., 2019) GBM cells. In contrast, SRSF3 could control the PDGFRB pathway indirectly, by modulating the alternative splicing of five potential regulators of PDGFRB expression such as TFDP1, CUX1, PRDM10, TEAD4 and TP73. Previously, altered TFDP1, CUX1 and PRDM10 splicing has been reported to generate variants that act as super enhancers of several oncogenes and cell cycle checkpoint genes (Rong Zeng et al., 2000; Sansregret and Nepveu, 2008; Cadieux et al., 2009; Park and Kim, 2010; Hulea and Nepveu, 2012; Ramdzan and Nepveu, 2014; Lu et al., 2016; Kaur et al., 2018; Zhang et al., 2018; Chen et al., 2019a; Zhou et al., 2020), while altered TEAD4 splicing events could result in VEGF inhibition (Appukuttan et al., 2012; Qi et al., 2016; Xu et al., 2018). Most interestingly, some splicing variants of TP73 (the only factor reported as a PDGFRB regulator from the five identified) can distinctly act as tumour suppressors, arresting the cell cycle and inducing apoptosis (Liu et al., 2004; Jancalek, 2014; Vikhreva et al., 2018). In this scenario, it is reasonable to posit that alterations in the alternative splicing of specific transcription factors (particularly TP73) induced by changes in SRSF3 expression may serve as the conduit linking SRSF3 to the PDGF-PDGFRB pathway, and thus the development/aggressiveness in glioblastoma. In support of this notion, our results indicate that SRSF3 silencing can increase the proportion of splicing variants such as TP73β and TP73γ, which have been linked to tumour suppressor roles (Liu et al., 2004) while decreasing that of the TP73α variant, which may play an oncogenic role (Jancalek, 2014; Vikhreva et al., 2018) in glioblastoma cells. Thus, modulation of TP73 MXE splicing event may serve as a putative mechanism linking SRSF3 and PDGFRB expression.
Taken together, our results unveiled new conceptual and functional avenues in glioblastomas, with potential therapeutic implications, by demonstrating for the first time a drastic dysregulation of the splicing machinery (spliceosome core and splicing factors; especially SRSF3/RBM22/PTPB1/RBM3) in HGAs/glioblastomas of different species. This is likely clinically relevant, because the dysregulation directly associates with development and aggressive features of glioblastomas. Moreover, we unveil a role of SRSF3 in crucial pathophysiological processes of glioblastomas, such as cell proliferation, migration, apoptosis, VEGF secretion and tumoursphere formation, which would underlie the relevant direct association of lower SRSF3 levels with reduced tumour progression and enhanced survival rate, observed herein in different glioblastoma human cohorts and animal models. These actions are likely mediated through the modulation of key signalling pathways (PDGFRB and PI3K) and may involve the distinct alteration of alternative splicing events of specific transcription factors controlling PDGFRB expression. Therefore, our study provides solid, convincing evidence demonstrating that SRSF3 has a functional role in the pathophysiology of glioblastomas, and invites suggestion that the development and use of SRSF3-targeting drugs could become a promising option to treat patients with this devastating pathology, offering a clinically relevant opportunity that should be tested for use in humans.
Acknowledgement
Special thanks to GraphicalAbstractDesign.com for assistance with graph and image design.
Funding
This work was funded by the Junta de Andalucía (CTS-1406, BIO-0139), the Spanish Ministry of Science, Innovation and Universities (FPU16/05059, FPU14/04290, PID2019-105564RB-I00), Instituto de Salud Carlos III, co-funded by European Union (ERDF/ESF, ‘Investing in your future’: PI16/00264, PI17/02287], Spanish Ministry of Economy and Competitiveness Projects (BFU2016-80360-R, TIN2017-83445-P) and CIBERobn. M.A.B. is funded by the Spanish Ministry of Economy and Competitiveness Projects (SAF2013-45111-R and SAF2015-72455-EXP), the Comunidad de Madrid Project (S2017/BMD-3770), the World Cancer Research (WCR) Project (16-1177), and the Fundación Botín (Spain). CIBER is an initiative of Instituto de Salud Carlos III, Spanish Ministry of Health, Social Services and Equality, Spain. J.J.B. is funded by the Samuel Oschin Comprehensive Cancer Institute (SOCCI), NIH grants (R33CA236687, and R03NS101529), American Cancer Society grant (RSG-16-217-01-TBG), and SOCCI Jack Mishkin Discovery, Prevention & Genetics, and Cancer Biology Awards.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Glossary
- GBM =
glioblastoma multiforme
- HGA =
high-grade astrocytoma
References
- Appukuttan B, et al. The related transcriptional enhancer factor-1 isoform, TEAD4(216), can repress vascular endothelial growth factor expression in mammalian cells. PLoS One 2012; 7: e31260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbagallo D, et al CircSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int J Mol Sci 2018; 19: 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor TT, et al. Feasibility, phase I, and phase II studies of tandutinib, an oral platelet-derived growth factor receptor-beta tyrosine kinase inhibitor, in patients with recurrent glioblastoma. Neuro Oncol 2017; 19: 567–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano L, et al. Inhibition of TRF1 telomere protein impairs tumor initiation and progression in glioblastoma mouse models and patient-derived xenografts. Cancer Cell 2017; 32: 590–607.e4. [DOI] [PubMed] [Google Scholar]
- Bowman RL, et al. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol 2017; 19: 139–41., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell 2013; 155: 462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig JJ, et al. Ets factors regulate neural stem cell depletion and gliogenesis in Ras pathway glioma. Cell Rep 2016; 17: 3407. [DOI] [PubMed] [Google Scholar]
- Cadieux C, et al. Mouse mammary tumor virus p75 and p110 CUX1 transgenic mice develop mammary tumors of various histologic types. Cancer Res 2009; 69: 7188–97. [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL.. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 2009; 10: 741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, et al. Transcriptional regulation of Bcl-2 gene by the PR/SET domain family member PRDM10. PeerJ 2019. a; 7: e6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, et al. RBM3 upregulates ARPC2 by binding the 3'UTR and contributes to breast cancer progression. Int J Oncol 2019. b; 54: 1387–97. [DOI] [PubMed] [Google Scholar]
- Cordoba-Chacon J, et al. Truncated somatostatin receptors as new players in somatostatin-cortistatin pathophysiology. Ann N Y Acad Sci 2011; 1220: 6–15. [DOI] [PubMed] [Google Scholar]
- Del Rio-Moreno M, et al. Dysregulation of the splicing machinery is associated to the development of nonalcoholic fatty liver disease. J Clin Endocrinol Metab 2019; 104: 3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Prado M, et al. Identification and characterization of two novel truncated but functional isoforms of the somatostatin receptor subtype 5 differentially present in pituitary tumors. J Clin Endocrinol Metab 2009; 94: 2634–43. [DOI] [PubMed] [Google Scholar]
- Dvinge H, et al. RNA splicing factors as oncoproteins and tum suppressors. Nat Rev Cancer 2016; 16: 413–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards NJ, et al. The CPTAC data portal: a resource for cancer proteomics research. J Proteome Res 2015; 14: 2707–13., [DOI] [PubMed] [Google Scholar]
- Feero WG, Guttmacher AE, Collins FS.. Genomic medicine–an updated primer. N Engl J Med 2010; 362: 2001–11. [DOI] [PubMed] [Google Scholar]
- Fishilevich S, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017; 2017: bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahete MD, et al. Changes in splicing machinery components influence, precede, and early predict the development of type 2 diabetes: from the CORDIOPREV study. EBioMedicine 2018; 37: 356–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019; 569: 503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Jia J, Jia R.. PTBP1 and PTBP2 impaired autoregulation of SRSF3 in cancer cells. Sci Rep 2015; 5: 14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackzell A, et al. p73 independent of c-Myc represses transcription of platelet-derived growth factor beta-receptor through interaction with NF-Y. J Biol Chem 2002; 277: 39769–76. [DOI] [PubMed] [Google Scholar]
- Hormaechea-Agulla D, et al. Ghrelin O-acyltransferase (GOAT) enzyme is overexpressed in prostate cancer, and its levels are associated with patient's metabolic status: potential value as a non-invasive biomarker. Cancer Lett 2016; 383: 125–34. [DOI] [PubMed] [Google Scholar]
- Hormaechea-Agulla D, et al. The oncogenic role of the In1-ghrelin splicing variant in prostate cancer aggressiveness. Mol Cancer 2017. a; 16: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormaechea-Agulla D, et al. The oncogenic role of the spliced somatostatin receptor sst5TMD4 variant in prostate cancer. FASEB J 2017. b; 31: 4682–96. [DOI] [PubMed] [Google Scholar]
- Hulea L, Nepveu A.. CUX1 transcription factors: from biochemical activities and cell-based assays to mouse models and human diseases. Gene 2012; 497: 18–26. [DOI] [PubMed] [Google Scholar]
- Jancalek R. The role of the TP73 gene and its transcripts in neuro-oncology. Br J Neurosurg 2014; 28: 598–605. [DOI] [PubMed] [Google Scholar]
- Jia R, et al. Oncogenic splicing factor SRSF3 regulates ILF3 alternative splicing to promote cancer cell proliferation and transformation. RNA 2019; 25: 630–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Vacas JM, et al. Spliceosome component SF3B1 as novel prognostic biomarker and therapeutic target for prostate cancer. Transl Res 2019. a; 212: 89–103. [DOI] [PubMed] [Google Scholar]
- Jimenez-Vacas JM, et al. Spliceosome component SF3B1 as novel prognostic biomarker and therapeutic target for prostate cancer. Transl Res 2019. b; 212: 89–103. [DOI] [PubMed] [Google Scholar]
- Jimenez-Vacas JM, et al. Dysregulation of the splicing machinery is directly associated to aggressiveness of prostate cancer. EBioMedicine 2020; 51: 102547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Vacas JM, , et al. Clinical utility of Ghrelin-O-Acyltransferase (GOAT) enzyme as a diagnostic tool and potential therapeutic target in prostate cancer. J Clin Med 2019; 8: 2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ.. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 2003; 12: 5–14. [DOI] [PubMed] [Google Scholar]
- Kaur S, et al. CUX1 stimulates APE1 enzymatic activity and increases the resistance of glioblastoma cells to the mono-alkylating agent temozolomide. Neuro Oncol 2018; 20: 484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlovski I, et al. The role of RNA alternative splicing in regulating cancer metabolism. Hum Genet 2017; 136: 1113–27. [DOI] [PubMed] [Google Scholar]
- Liu G, et al. DeltaNp73beta is active in transactivation and growth suppression. Mol Cell Biol 2004; 24: 487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, et al. PDGF-mediated mesenchymal transformation renders endothelial resistance to anti-VEGF treatment in glioblastoma. Nat Commun 2018; 9: 3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 2016; 131: 803–20. [DOI] [PubMed] [Google Scholar]
- Lu X, et al. Dysregulation of TFDP1 and of the cell cycle pathway in high-grade glioblastoma multiforme: a bioinformatic analysis. Genet Mol Res 2016; 15: gmr7646. [DOI] [PubMed] [Google Scholar]
- Luque RM, et al. A cellular and molecular basis for the selective desmopressin-induced ACTH release in Cushing disease patients: key role of AVPR1b receptor and potential therapeutic implications. J Clin Endocrinol Metab 2013; 98: 4160–9. [DOI] [PubMed] [Google Scholar]
- Luque RM, et al. Truncated somatostatin receptor variant sst5TMD4 confers aggressive features (proliferation, invasion and reduced octreotide response) to somatotropinomas. Cancer Lett 2015; 359: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massacesi C, et al. PI3K inhibitors as new cancer therapeutics: implications for clinical trial design. Onco Targets Ther 2016; 9: 203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Wang Z.. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 2014; 15: 108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecca C, et al. Targeting mTOR in glioblastoma: rationale and preclinical/clinical evidence. Dis Markers 2018; 2018: 9230479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melling N, et al. Prevalence and clinical significance of RBM3 immunostaining in non-small cell lung cancers. J Cancer Res Clin Oncol 2019; 145: 873–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mete M, et al. Punicic acid inhibits glioblastoma migration and proliferation via the PI3K/AKT1/mTOR signaling pathway. Anticancer Agents Med Chem 2019; 19: 1120–31. [DOI] [PubMed] [Google Scholar]
- Murat A, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clinc Oncol 2008; 26: 3015–24. [DOI] [PubMed] [Google Scholar]
- Noch EK, Ramakrishna R, Magge R.. Challenges in the treatment of glioblastoma: multisystem mechanisms of therapeutic resistance. World Neurosurg 2018; 116: 505–17. [DOI] [PubMed] [Google Scholar]
- Ostrom QT, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro Oncol 2018; 20: iv1–iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 2014; 26: 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir-Kaynak E, Qutub AA, Yesil-Celiktas O.. Advances in glioblastoma multiforme treatment: new models for nanoparticle therapy. Front Physiol 2018; 9: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JA, Kim KC.. Expression patterns of PRDM10 during mouse embryonic development. BMB Rep 2010; 43: 29–33. [DOI] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM.. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 2013; 497: 127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, et al. A splicing isoform of TEAD4 attenuates the Hippo-YAP signalling to inhibit tum proliferation. Nat Commun 2016; 7: ncomms11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L, et al. Identification of upregulated HNRNPs associated with poor prognosis in pancreatic cancer. Biomed Res Int 2019; 2019: 5134050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdzan ZM, Nepveu A.. CUX1, a haploinsufficient tum suppressor gene overexpressed in advanced cancers. Nat Rev Cancer 2014; 14: 673–82. [DOI] [PubMed] [Google Scholar]
- Rong Zeng W, et al. Exon/intron structure and alternative transcripts of the CUTL1 gene. Gene 2000; 241: 75–85. [DOI] [PubMed] [Google Scholar]
- Sansregret L, Nepveu A.. The multiple roles of CUX1: insights from mouse models and cell-based assays. Gene 2008; 412: 84–94. [DOI] [PubMed] [Google Scholar]
- Sarmento-Cabral A, et al. Metformin reduces prostate tumor growth, in a diet-dependent manner, by modulating multiple signaling pathways. Mol Cancer Res 2017; 15: 862–74. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW.. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9: 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Eyras E.. The role of alternative splicing in cancer. Transcription 2017; 8: 91–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, et al. SRSF3-regulated RNA alternative splicing promotes glioblastoma tumorigenicity by affecting multiple cellular processes. Cancer Res 2019; 79: 5288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA.. The cancer genome. Nature 2009; 458: 719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer BW, et al. A reference collection of patient-derived cell line and xenograft models of proneural, classical and mesenchymal glioblastoma. Sci Rep 2019; 9: 4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveen A, et al. Transcriptome instability in colorectal cancer identified by exon microarray analyses: associations with splicing factor expression levels and patient survival. Genome Med 2011; 3: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveen A, et al. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016; 35: 2413–27. [DOI] [PubMed] [Google Scholar]
- Uphoff CC, Drexler HG.. Detection of mycoplasma contaminations. Methods Mol Biol 2013; 946: 1–13. [DOI] [PubMed] [Google Scholar]
- Uramoto H, et al. p73 competes with co-activators and recruits histone deacetylase to NF-Y in the repression of PDGF beta-receptor. J Cell Sci 2004; 117: 5323–31. [DOI] [PubMed] [Google Scholar]
- Vazquez-Borrego MC, et al. A somatostatin receptor subtype-3 (SST3) peptide agonist shows antitumor effects in experimental models of Nonfunctioning Pituitary Tumors. Clin Cancer Res 2019; 26: 957–69. [DOI] [PubMed] [Google Scholar]
- Vikhreva P, Melino G, Amelio I.. p73 Alternative splicing: exploring a biological role for the c-terminal isoforms. J Mol Biol 2018; 430: 1829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong JK, et al. PTBP1 and PTBP2 serve both specific and redundant functions in neuronal Pre-mRNA splicing. Cell Rep 2016; 17: 2766–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterskog D, et al. Dysregulation of platelet-derived growth factor beta-receptor expression by DeltaNp73 in neuroblastoma. Mol Cancer Res 2009; 7: 2031–9. [DOI] [PubMed] [Google Scholar]
- Xu A, et al. Overexpression of TEAD4 correlates with poor prognosis of glioma and promotes cell invasion. Int J Clin Exp Pathol 2018; 11: 4827–35. [PMC free article] [PubMed] [Google Scholar]
- Xu PF, et al. PI3Kbeta inhibitor AZD6482 exerts antiproliferative activity and induces apoptosis in human glioblastoma cells. Oncol Rep 2019; 41: 125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Wan R, Shi Y.. Molecular mechanisms of pre-mRNA splicing through structural biology of the spliceosome. Cold Spring Harb Perspect Biol 2019; 11: a032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders ED, Svensson F, Bailey DS. Therapy for glioblastoma: is it working?. Drug Discov Today 2019; 24: 1193–201. [DOI] [PubMed] [Google Scholar]
- Zhang L, et al. Screening and function analysis of hub genes and pathways in hepatocellular carcinoma via bioinformatics approaches. Cancer Biomark 2018; 22: 511–21. [DOI] [PubMed] [Google Scholar]
- Zhou J, et al. Identification of chemoresistance-related mRNAs based on gemcitabine-resistant pancreatic cancer cell lines. Cancer Med 2020; 9: 1115–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microfluidic array, RNAseq, nCounter and other data used in this paper are available upon request to the corresponding author. Chinese Glioma Genome Atlas RNAseq data (CGGA) and Murat microarray data were interrogated through the GlioVis Tools (http://gliovis.bioinfo.cnio.es) (Murat et al., 2008; Bowman et al., 2017).