We review the effects of major abiotic stresses on the expression of pathogen virulence and plant defense genes, and conduct a metadata analysis of four multistress transcriptomic datasets.
Keywords: Abiotic stress, crosstalk, defense, plant–pathogen interaction, transcriptome, virulence
Abstract
Current environmental and climate changes are having a pronounced influence on the outcome of plant–pathogen interactions, further highlighting the fact that abiotic stresses strongly affect biotic interactions at various levels. For instance, physiological parameters such as plant architecture and tissue organization together with primary and specialized metabolism are affected by environmental constraints, and these combine to make an individual plant either a more or less suitable host for a given pathogen. In addition, abiotic stresses can affect the timely expression of plant defense and pathogen virulence. Indeed, several studies have shown that variations in temperature, and in water and mineral nutrient availability affect the expression of plant defense genes. The expression of virulence genes, known to be crucial for disease outbreak, is also affected by environmental conditions, potentially modifying existing pathosystems and paving the way for emerging pathogens. In this review, we summarize our current knowledge on the impact of abiotic stress on biotic interactions at the transcriptional level in both the plant and the pathogen side of the interaction. We also perform a metadata analysis of four different combinations of abiotic and biotic stresses, which identifies 197 common modulated genes with strong enrichment in Gene Ontology terms related to defense . We also describe the multistress-specific responses of selected defense-related genes.
Introduction
In crop production, non-optimal growth conditions, i.e. abiotic stress, and pathogens are two major factors that can negatively affect yield, potentially leading to huge losses. It has long been known that abiotic stresses affect plant disease, and these interactions can be very important. For example, in the case of the nitrogen-induced susceptibility of rice to the fungus Magnaporthe oryzae that follows nitrogen fertilization, which Vietnamese farmers have named Koe-imochi (Ballini et al., 2013). Current knowledge indicates that plant–pathogen interactions are affected during abiotic stress via the following factors: (i) plant metabolism, and hence nutrient availability for the pathogen; (ii) plant cell viability; (iii) signaling (for a review see Kissoudis et al., 2014); and (iv) the transcriptomic regulation of both the plant and the pathogen. We will briefly summarize the first two points in this Introduction, and then focus on the second two in the rest of this review.
Since pathogens need to find appropriate and sufficient nutrients when invading plant tissue, abiotic stress is likely to affect pathogen nutrition in planta (Lemaitre et al., 2008; Singh et al., 2019). In some cases the pathogens themselves manipulate plant primary metabolism to their advantage, as in the case of infection of susceptible tomatoes by the necrotrophic fungus Botrytis cinerea, which induces the expression of asparagine synthetase, leading to the accumulation of asparagine in the infected tissue (Seifi et al., 2014). Modifications to plant metabolism will affect pathogens to a greater or lesser degree depending upon their life cycle. Thus, biotrophs are generally thought to be more dependent on the metabolism of their host than necrotrophs (Ah-Fong et al., 2019). Understanding the precise impact on pathogen fitness in planta of the modifications to the accumulation of primary metabolites that are induced by abiotic stresses is a complex and rather overlooked field (reviewed in Fagard et al., 2014).
Abiotic stress can affect cell viability, and this in turn can affect the outcome of plant–pathogen interactions in many ways depending on the pathogen life cycle. For example, nitrogen-limitation favors the onset of senescence (Lemaitre et al., 2008), which is beneficial to some necrotrophic pathogens. However, such an effect on tissue senescence does not give the whole picture since some necrotrophic pathogens are more virulent in high-nitrogen conditions (Fagard et al., 2014).
Plant–pathogen interactions have been well studied and many key molecular factors have been identified both on the plant and the pathogen side (Gust et al., 2017). Upon perception of the pathogen by the plant through recognition of pathogen/microbe-associated molecular patterns (P/MAMPs) by pattern recognition receptors, the first layer of immunity, termed PAMP-triggered immunity (PTI), is activated. Adapted pathogens can overcome PTI by releasing protein effectors inside plant cells using a type 3 secreting system in the case of many gram-negative bacteria, or by secreting them in the apoplast in the case of fungi and oomycetes. In turn, plants that possess specific resistance genes of the NBS-LRR family can sense virulence effectors. This specific recognition triggers a powerful defense response termed effector-triggered immunity (ETI) (Deslandes and Rivas, 2012). Activation of both PTI and ETI involves signaling pathways that require MAPK-signaling, which are regulated by the major phytohormones salicylic acid (SA), jasmonic acid (JA), and ethylene (ET), and which in turn activate downstream responses via a large array of transcription factors (TFs). Generally, this defensive line culminates in a hypersensitive response at the site of infection, together with the synthesis of antimicrobial molecules such as phytoalexins and pathogenesis-related proteins (for review see Berens et al., 2017). Despite the extensive literature on plant–pathogen interactions, unfortunately little is known about how plant defense is affected by abiotic stresses. Furthermore, to the best of our knowledge, no review has addressed the subject of activation of virulence gene signals in planta and their modulation under different environmental constraints.
In this review, we focus on how abiotic stresses affect the expression of plant defense at the transcriptomic level, together with the expression of pathogen virulence. We focus on three major abiotic stresses: drought, extreme temperatures, and nitrogen starvation and other mineral deficits.
Temperature, hormones, and defense genes: multifaceted crosstalk
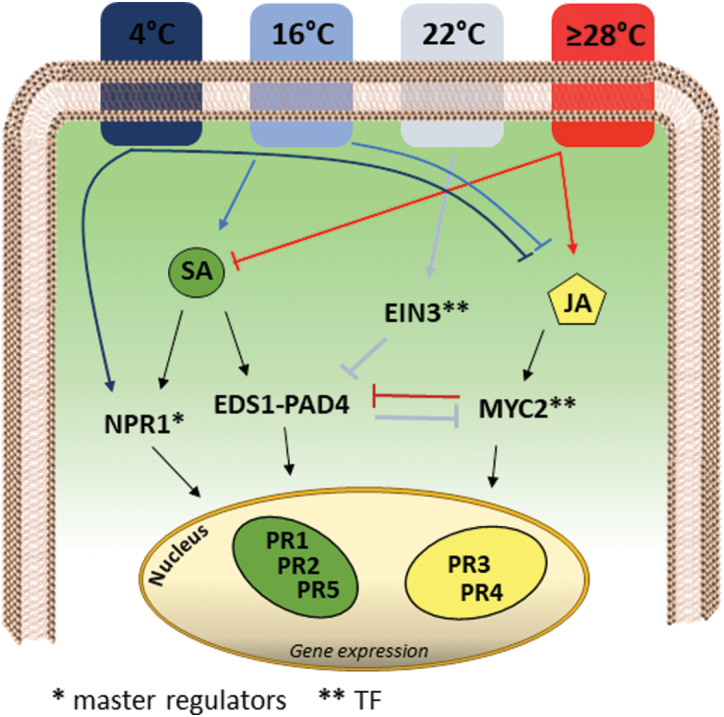
Under conditions favorable to growth, the activation of plant defenses is regulated by elaborate crosstalk among phytohormones such as SA, JA, and ET. In the present context of climate change, understanding how hormone-dependent gene expression is altered by ever-changing temperatures is of great interest. In the past few years, several reports have shown that the defensive responses mediated by SA during the interaction between Arabidopsis and Pseudomonas syringae are increased at low temperature (16 °C; Li et al., 2020), and compromised under high (28 °C; Wang et al., 2009) and extreme temperatures (37 °C and 42 °C; Janda et al., 2019). Interestingly, extreme temperatures compromise defense even when the exposure is short (Janda et al., 2019). An elegant study by Huot et al., (2017) reported that at an elevated temperature of 30 °C, Arabidopsis plants exposed to the synthetic SA analogue benzothiadiazole did not accumulate mRNA of the two SA-marker genes ICS1 and PR1 (see Supplementary Table S1 for a list of all Arabidopsis genes mentioned in this review). Moreover, several other positive regulators of SA biosynthesis and signaling, namely EDS1, PAD4, CBP60g, and SARD1, were negatively affected by elevated temperature. Interestingly, the inactivation of SA-responsive genes at 30 °C was unrelated to the inability of NPR1 to translocate to the nucleus. Instead, it appeared that elevated temperature affected SA-dependent gene expression through the activation of negative SA regulators. For example, MYC2, a master regulator gene of JA signaling and a negative regulator of SA signaling, showed higher expression levels at 30 °C than at 22 °C (Huot et al., 2017). Thus, at high temperatures, JA may confer susceptibility to P. syringae through negative regulation of PAD4 that is mediated by MYC2 and its homologs MYC3 and MYC4. Therefore, it appears that high temperature conditions lead to the suppression of SA responses due to the antagonist effect of JA signaling (Fig. 1). On the other hand, SA signaling is also known to antagonize JA signaling.
Fig. 1.
Modulation of plant-pathogen defense responses by cold and heat stress. Salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) are the major phytohormones involved in plant–pathogen interactions and they are modulated differently by cold and heat stress. Plants exposed to low temperature show high levels of resistance, and several reports have indicated that cold stress results in SA-related defense being enhanced while JA-dependent signaling is inhibited. SA responses occur independently of SA accumulation and of the EDS1-PAD4 complex depending on the intensity of the cold treatment, whereas NPR1 is a significant factor in signaling cold-induced gene expression whether the intensity is mild or strong (Cui et al., 2018; Olate et al., 2018; Li et al., 2020). An opposite scenario occurs at elevated temperature. SA biosynthesis is suppressed by the antagonist action of JA/ET. At 22 °C the transcription factor EIN3 blocks SA-dependent defense (Li et al., 2020) whilst the heat-induced JA responses may or may not be mediated via theMYC2 transcription factor (Huot et al., 2017; Mine et al., 2017).
A different scenario occurs when plants are grown at temperatures below their optimum. There is evidence indicating that cold stress confers increased disease resistance against hemi- and biotrophic pathogens. It has recently been reported that short-term cold stress at 4 °C positively modulates SA-dependent responses at the expense of the JA defensive pathway in Arabidopsis (Wu et al., 2019). In particular, the SA-marker genes PR2 and PR5 are up-regulated by cold treatment whereas the JA markers PR4 and MYC2 are inhibited by cold. Similarly, SA-dependent activation of PR1, PR2, and PR5 is observed in Arabidopsis exposed to long-term cold conditions (Seo et al., 2010). Moreover, the SA-dependent responses appear to play a key role in increasing the resistance to P. syringae even at a moderately low temperature of 16 °C (Li et al., 2020). By using transcriptomic and knock-out mutants for SA, JA, and ET signaling, the authors demonstrated that PAD4 and ICS1 are critical components of the SA-dependent responses in Arabidopsis at this temperature. On the other hand, the up-regulation of multiple SA-inducible genes, namely PR1, PR2, EDS1, WRKY18, and WRKY30, was shown to be negatively affected by EIN3, a master regulator of the ET-signaling pathway (Li et al., 2020). Thus, higher resistance to P. syringae at low temperature relies on SA–ET crosstalk that results in up-regulation of SA-dependent responses.
Taken together, it appears that at elevated temperatures, JA-dependent responses down-regulate SA-dependent signaling, leading to increased susceptibility to P. syringae, whereas cold temperatures mainly boost the SA-dependent response, leading to increased resistance to P. syringae (Fig. 1).
Recent data have opened up a new perspective on these processes. Olate et al. (2018) have shown that NPR1 can act as a hub in the molecular crosstalk between cold and biotic stresses, in an alternative regulatory mechanism to the canonical hormonal signaling network (Fig. 1). At low temperatures, NPR1 moves to the nucleus and regulates numerous genes associated with the responses to cold and pathogens (PR2, WRKY46, DMR6, NAC019) via interaction with the TF HSFA1 (Olate et al., 2018).
Temperature stress and virulence genes: the pathogen point of view
Plant pathogens usually only undergo gradual temperature changes during seasonal cycles and are generally not subjected to sudden temperature changes (Bocsanczy et al., 2014). However, due to climate change, extreme temperature events are predicted to occur more rapidly and more frequently. Extreme temperatures can directly affect pathogen physiology in different ways, which in turn can influence the outcome of plant–pathogen interactions. Several studies have described the adaptation of pathogen physiology to low or moderate temperatures (Ramos et al., 2001), including modifications of expression of virulence factors in planta, a key step in pathogenesis. However, the number of studies addressing the subject of virulence gene expression in plants grown at high temperatures is relatively low.
At low temperatures, a modification of virulence gene expression is observed for pathogens adapted to temperate climates. For example, Erwinia amylovora, the phytopathogenic bacterium responsible for fireblight in the Maloideae family, can adapt to lower temperatures (4 °C and 14 °C) by increasing the production of exopolysaccharides, which are involved in biofilm formation and in resistance to oxidative stress (Santander and Biosca, 2017). Another example of adaptation to low temperatures is found in another phytopathogenic bacterium Ralstonia solanacearum, a tropical pathogen, in which a few strains adapted to temperate climates do not show any reduction in their metabolism at a moderately low temperature of 18 °C (Bocsanczy et al., 2014). Interestingly, the differences in virulence are primarily explained by changes in temperature-dependent gene expression of the virulence regulators hrpB and hrpG. In addition, this study highlighted a role in virulence of a putative type 6 secretion system not previously associated with infection (Bocsanczy et al., 2014). Another study by Meng et al. (2015) focused on transcriptome responses in R. solanacearum to low temperature (4 °C) and showed an up-regulation of specific genes only in cold-adapted strains. Three of these genes (LecM, AidA, AidC) were required for full virulence, of which two (LecM, AidC) were present only in the genome of the adapted strains. Taken together, these studies point to a temperature-dependent regulation of virulence genes, whether they be known or novel, to explain the different virulence phenotypes observed at low temperatures.
High temperatures have also been shown to affect virulence in pathogens, and generally result in an increase. For example, translocation of P. syringae type III effectors increases at high temperature (Huot et al., 2017). In rice plants challenged with the fungus Magnaporthe oryzae, which causes rice blast, stronger necrotic symptoms are observed at high temperatures (Onaga et al., 2017), and transcriptome analysis has consistently indicated that many putative M. oryzae effector genes are more highly expressed in plants exposed to 35 °C than to 28 °C. A temperature rise could therefore increase the incidence and severity of rice blast, a serious threat that should not be underestimated in the present scenario of climate change (Onaga et al., 2017). Similar results have also been observed in the bacterium Dickeya solani, an emerging pathogen responsible for soft rot and blackleg in potato crops. At high temperatures, D. solani causes more severe symptoms than other Dickeya species, suggesting a temperature-dependent boost of virulence in adapted strains and species (Czajkowski et al., 2016). High temperatures have been shown to result in the up-regulation of 45 D. solani genes, four of which are required for biofilm production and virulence in potato. Interestingly, these key genes do not encode cell wall-degrading enzymes but a putative phospholipase (plcA), rhamnogalacturonase (rhiN), lysine aminomutase (yodO), and a regulatory protein (araC). The up-regulation of these loci could play a key role in the fitness of D. solani at high temperatures, a bad omen for potato crops given current climate change.
Some studies have reported a negative regulation of virulence under higher temperatures. A recent study by Saha et al. (2015) examined the effects of an array of temperatures between 18–37 °C on the phytopathogenic bacterium Pectobacterium carotovorum, which is responsible for bacterial soft rot in a wide range of plant species. The authors identified an optimal temperature of 33 °C for the production of the quorum-sensing signal molecule, acyl homoserine lactone, which regulates the bacterial population and virulence gene expression. Beyond this optimum, no accumulation of the quorum-sensing molecule and no disease were observed. A second example of a negative impact of high temperatures on virulence factors can be seen in P. syringae, in which the production of the phytotoxin coronatine is repressed at 28 °C compared to 18 °C (Ullrich et al., 1995).
Overall, most studies have found that high temperatures tend to favor pathogen virulence, while low temperatures tend to decrease virulence except in adapted strains. However, increasing temperatures beyond the optimal level for the expression of pathogen virulence will most likely decrease virulence, as seen in the example P. carotovorum.
Water stress: a positive or negative regulator of plant defense genes?
Drought stress is another major environmental factor that affects plant physiology, metabolism, and growth, and its occurrence is becoming increasingly worrying in many parts of the world. It can be caused by several phenomena, such as dehydration, salinity, high or low temperatures, and its effects depend on timing, severity, and the presence and types of interactions with other factors (Salehi-Lisar and Bakhshayeshan-Agdam, 2016). The whole plant defense system can be expected to be affected by water stress, but interestingly drought has been shown to cause both detrimental and beneficial effects on plant–pathogen interactions, both in terms of resistance and gene expression. Accumulation of abscisic acid (ABA) is very often observed in plants exposed to drought, and this leads to stomatal closure that prevents bacteria from entering through stomata, and also to other physiological responses with a putative role in plant–pathogen interactions (Melotto et al., 2017; Zarattini and Forlani, 2017). However, the precise role of ABA in plant–pathogen interactions is still a matter of debate. ABA can interact either synergistically or antagonistically with other defensive hormones such as SA, JA, and ET, thus affecting the outcome of biotic stress (reviewed in Cao et al., 2011).
As might be expected, there are frequent examples of plants being more susceptible to pathogens after a period of drought. For example, rice exposed to moderate drought conditions show higher susceptibility to M. oryzae (Bidzinski et al., 2016), which is due to lower expression of the defense marker genes PAL, PBZ1, POX22.3, and PR3. Water stress also appears to inhibit the immune system in forest trees. Transcriptomic and metabolomic analyses of seedlings of pine (Pinus koraiensis) challenged with Cenangium ferruginosum after experiencing water stress show that expression of defense genes is impaired (Ryu et al., 2018). In addition, reduced synthesis of specialized metabolites such as terpenoids, flavonoids, and phenolic acids is also observed whereas the levels of ABA are increased.
Drought can also have a positive effect on defense. For example, drought-stressed Arabidopsis and chickpea show enhanced resistance to the bacterial pathogens P. syringae DC3000 and P. syringae pv. phaseolicola, respectively (Gupta et al., 2016; Sinha et al., 2017). Drought-acclimated Nicotiana benthamiana plants show higher accumulation of mRNA of PR5 and PDF1.2 that leads to enhanced resistance to the fungus Sclerotinia sclerotiorum and the bacterium P. syringae pv. tabaci (Ramegowda et al., 2013). Cramer et al. (2006) found that drought increases the expression of the defense-related genes PR5, PR2 and Germin-like1.15 in grapevine. Comprehensive RNA-seq analysis of 2-year-old plants subjected to drought stress revealed that 72 genes encoding pathogenesis-related proteins were differentially expressed following drought (Haider et al., 2017); in particular, transcripts of several PR genes (PR1, PR2, PR3, PR5, PR10, PR14, and PR15) were positively modulated. Other studies have shown that application of PEG, sucrose, or salt to mimic the osmotic stress induced by drought can induce defense (Hatmi et al., 2014; Guan et al., 2018). For example, among the 35 WRKY genes induced by Fusarium udum infection in pigeonpea (Cajanus cajan), 11 were also induced by salt stress (Kumar et al., 2019).
Predicting a priori the impact of drought (as well as osmotic) stress on plant–pathogen interactions and plant defense therefore appears to be particularly difficult. Reduced water availability usually negatively affects plant physiology and growth; however, this is not always true when plants face a pathogen attack. The actual disease outcome is strongly dependent on the pathosystem that is being considered, and hence on how signaling pathways triggered by water and biotic stress interact to affect the expression of defense genes.
How pathogens respond to drought: is it possible to maintain or even increase virulence?
As outlined above, drought stress strongly affects plant defense but also induces various metabolic and physiological changes in the plant tissues. Phytopathogens that attack aerial organs often deal with the various stresses that they encounter on the surface of the host leaves by accumulating compatible osmolytes, and this includes the response to water limitation during the epiphytic phase (Bremer and Krämer, 2019). Such osmotic stress conditions not only interfere with the general metabolism and life cycle of phytopathogens but can also affect their pathogenic cycle. Thus, the pathogen’s capacity to cope with water limitation both in the phyllosphere and inside the leaf tissue will affect disease progression. Several studies have shown that salt stress, which also causes osmotic stress, can alter the expression of virulence genes. Although the question of whether plant pathogens alter their virulence when faced with water stress has not been extensively studied, several examples of induction of virulence genes by water limitation have been described.
The bacterial pathogen Xanthomonas citri subsp. citri spends part of its life cycle on the surface of citrus leaves, where it can face water limitation. By applying saline stress to mimic water limitation, Barcarolo et al. (2019) have identified proteins involved in bacterial tolerance to reactive oxygen species (ROS) that accumulate in vitro, including a putative NADPH dehydrogenase. Expression of the corresponding gene, Xac2229, is induced both by saline stress in vitro and during the plant–pathogen interaction. Interestingly, Xac2229 is required for virulence in planta but does not confer any advantage to X. citri under salt stress in vitro. This suggests that Xac2229 could be required for bacteria to cope with indirect effects of water limitation in planta rather than with water limitation/salt stress per se. Another study has examined Alternaria brassicicola, a necrotrophic fungus that causes important damage to cultivated Brassicaceae. Seed transmission is an important part of the life cycle of this fungus and this requires that the it can resist low water availability (N’Guyen et al., 2019). An in vitro transcriptome analysis of the fungus under water-limiting conditions resulted in the identification of a group of hydrophilin-encoding genes that show transcriptional activation under water limitation. Analysis of the corresponding knock-out mutants indicated that the genes are not involved in fungal virulence in plants grown in optimal conditions but that they are required for full transmission of the pathogen spores by Arabidopsis seeds (N’Guyen et al., 2019). Although these mutants show wild-type virulence under control conditions, whether the identified genes play a role in virulence in plants subject to water limitation remains an unanswered question.
Drought stress has also been shown to increase the aggressiveness of the fungus M. oryzae. RNA-seq analysis performed on fungal hyphae in planta revealed differences in the fungal gene expression profile between well-watered plants and plants exposed to drought conditions (Bidzinski et al., 2016). In particular, drought reduces the in planta expression of genes encoding effectors, such as Avr-PITA, and induces genes encoding cell wall-degrading enzymes. These results suggest that the fungus adapts its virulence program to the physiology of the stressed plants, probably through unknown signals produced by the plant and perceived by the fungus.
Although the literature on the subject remains limited, the current available data suggest that increases in the occurrence of drought stress will not only affect plants but will also increase the capacity of pathogens to express virulence through as yet unknown mechanisms.
Nutrient limitation: emerging implications for plant defense gene expression
Mineral depletion causes stress for plants and affects many different processes including defense. Several recent studies have described how mineral depletion affects the expression of genes associated with biotic stress. For example, genes involved in JA biosynthesis and signaling in barley (HvLOX2A, HvAOC, HvJIP60, HvJIP23, HvJIP37, and HvAOS) are induced by low potassium supply (Davis et al., 2018). Interestingly, this is correlated with an increase in resistance to Blumeria graminis, a JA-susceptible pathogen causing powdery mildew. In Arabidopsis, potassium deficiency induces the expression of JA-dependent downstream genes (Armengaud et al., 2010). These observations suggest that alongside the triggering of responses related to potassium depletion, such as up-regulation of the high-affinity K transporter HAK5) (Armengaud et al., 2004), plants also activate hormone-dependent defensive responses that are able to increase pathogen resistance. Surprisingly, opposite results have been observed in rice seedlings grown with low potassium, with decreased expression of two JA-dependent genes, OsLOX5 and Os12g14440, being observed (Shankar et al., 2013). However, it is currently difficult to know whether these differences are species-specific (rice versus barley and Arabidopsis) or whether they are due to differences in the experimental set-ups.
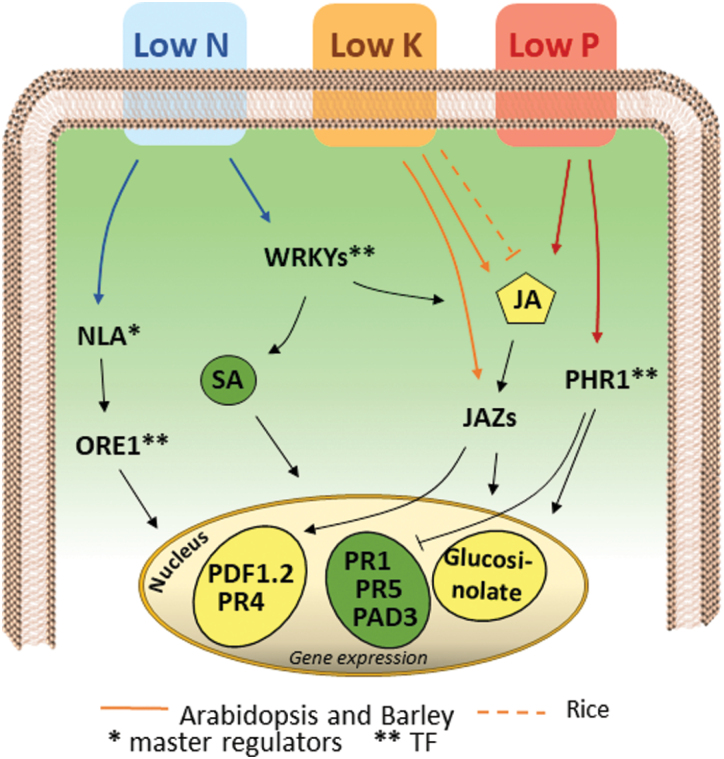
Phosphate and nitrogen limitations have also been shown to affect the expression of plant defense genes. Transcriptome analysis of roots of Medicago truncatula grown with a combination of low phosphate and low nitrogen has indicated induced expression of several stress-associated genes, including ones encoding NADPH oxidases (Bonneau et al., 2013). A recent study by Castrillo et al. (2017) in Arabidopsis examined the link between regulation of the immune system and the formation of microbiota under phosphate starvation. The authors concluded that the master regulator of phosphate starvation, PHR1, down-regulates SA-dependent responses while increasing the expression of JA-associated genes, mostly those related to glucosinolate biosynthesis (Fig. 2).
Fig. 2.
Modulation of plant-pathogen defense responses by mineral limitation. Increasing evidence indicates that nutritional status has an impact on plant defense. In Arabidopsis, phosphate limitation can modulate defense signaling either via the jasmonic acid (JA) pathway or via PHR1, a master regulator that governs responses to phosphate starvation. PHR1 has a dual function of modulating the plant immune system, either by inhibiting the expression of salicylic acid (SA)-dependent genes or by activating a subset of JA-responsive genes that are mainly involved in glucosinolate biosynthesis (Castrillo et al., 2017). Low potassium leads to opposite responses in Arabidopsis, barley, and rice (Armengaud et al., 2010; Shankar et al., 2013). Although low potassium leads to an increased level of JA and expression of JAZs genes in Arabidopsis and barley, a decreased level of JA occurs in rice. Upon low nitrate conditions, genes belonging to the WRKY transcription factor family are induced in Arabidopsis (Patterson et al., 2010), which in turn can regulate the SA/JA balance as well as hormone-related gene expression. Interestingly, a direct interaction between NLA and ORE1, two key regulators of nitrogen limitation and senescence, has recently been demonstrated. ORE1 is a NAC transcription factor (NAC092) that might modulate JA-dependent gene expression (Park et al., 2018).
Plants can use nitrogen in both oxidized and reduced forms, mainly as nitrate and ammonium. Arabidopsis roots grown in low nitrate or low ammonium show both common responses (a generic nitrogen-limitation response) as well as specific responses to the limitation in each nitrogen source (Patterson et al., 2010). In particular, this study showed that low ammonium triggered the expression of biotic-associated genes such as WRKY70, which regulates the SA/JA balance, and JA-responsive genes (Fig. 2). Another study has shown that growth in ammonium reduces resistance to an avirulent strain of P. syringae due to lower production of NO (Gupta et al., 2012). Ammonium triggered the accumulation of specialized metabolites, suggesting that nitrogen availability not only affects mineral homeostasis in the plant cells but can also activate defense at both the molecular and the biochemical levels.
Switching the nitrogen source to nitrate appears to have contrasting effects depending on the plant species considered. In tomato plants exposed to low nitrate, a principal component analysis showed that the transcriptional response clustered close to that of plants infected by Botrytis, strongly suggesting that low nitrate primes defense responses even in the absence of infection (Vega et al., 2015). Our own studies have shown that nitrogen limitation also affects the activation of transcriptional defense in Arabidopsis leaves in responses to bacterial infection (Farjad et al., 2018), fungal infection (Soulié et al., 2020), and to defense stimulators (Zarattini et al., 2017; Verly et al., 2020). In the absence of a pathogen or defense stimulator, several WRKY TFs are positively modulated when the nitrate source is limited, even if a weaker magnitude of expression is generally observed as compared to plants infected with pathogens (Fig. 3). Moreover, nitrate limitation alters the defense responses triggered by pathogens and defense stimulators. For example, in Arabidopsis low nitrate boosts the induction of PDF1.2a by the defense stimulator deoxycholic acid, a bile acid (Zarattini et al., 2017), and by B. cinerea, thus increasing resistance to this fungus (Soulié et al., 2020). Taken together, our data indicate that nitrate limitation strongly affects defense signaling pathways in response to a variety of biotic stimuli, emphasizing the importance of JA signaling in the integration of nutritional and defense cues.
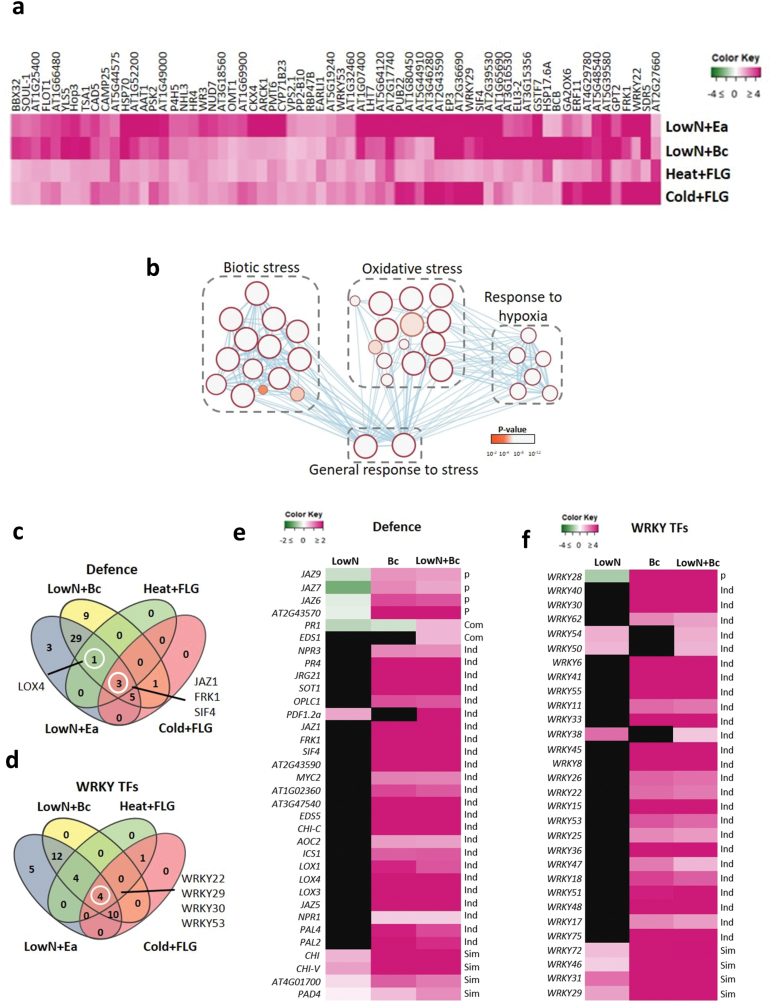
Fig. 3.
Meta-analysis of combined abiotic and biotic stress transcriptome data. Ten publicly available datasets were selected to study the modulation of defense gene expression in response to combined stresses. Transcriptome data for nitrate limitation (LowN), Botrytis cinerea (Bc), and their combination were obtained from Soulié et al. (2020), whilst the data for cold and flagellin (FLG) and for heat and FLG were extracted from the Gene Expression Omnibus repository (accession GSE41935; Rasmussen et al., 2013). (a) Expression of 66 commonly up-regulated genes in all four multistress conditions (fold change >1; see Supplementary Table S2). (b) Gene Ontology analysis was performed using the Cytoscape and g:Profiler software according to Reimand et al. (2019) using the total of 197 up- and down-regulated genes shared in all multistress conditions (fold-change >1 or <–1; Supplementary Table S2). (c. d) Venn diagrams showing (c) three defense genes and (d) four WRKYs commonly modulated by all the combined stress conditions. (e, f) Heat-maps showing expression of selected defense genes (e) and WRKY transcription factors (d), following single LowN stress, Bc infection, and their combinations. Black shading indicates genes not significantly modulated (P>0.05; Supplementary Tables S2, S3). The same genes reported in the heat-maps were also screened in datasets related to the combination of heat, cold, and flagellin (see Supplementary Table S3).
Mineral depletion represents a serious threat to agriculture since it affects plant growth and is a major cause of yield loss in crops. Molecular and transcriptomic studies indicate that stress linked to mineral depletion often primes defense responses; however, negative effects of mineral depletion on defense have been reported for multiple pathosystems. This should be taken into consideration for each crop when selecting cultivars and fertilization.
When nutrient limitation reaches the pathogen: a signal for virulence genes
Soil mineral depletion affects both plant metabolism and the chemistry of root and leaf exudates, which in turn affect the interactions of plants with their surrounding microbes, beneficial or not. Many studies have shown that plant growth conditions, in particular nitrogen availability, alter the capacity of nitrogen-fixing bacteria to establish symbiosis and alter the transcriptome of plant growth-promoting rhizobacteria in the soil (Carvalhais et al., 2013). However, little data exist concerning the effects of plant nutrient limitation on pathogen virulence. Many pathogens express their virulence factors specifically when infecting plants and not when grown in rich medium in vitro (Rico et al., 2011,Tan and Oliver, 2017). However, relatively few studies have addressed the actual metabolic environment encountered by pathogens in planta, and the signals that allow the expression of pathogen virulence genes in planta are not well known yet. On the other hand, several studies have shown that pathogens express their virulence factors in vitro when grown in limiting nutrient conditions (Bolton and Thomma, 2008; Tudzynski, 2014). For example, low nitrogen and low carbon both induce the Magnaporthe grisea gene Mgp1, and low nitrogen induces the avr9 gene in Cladosporium fulvum (Talbot et al., 1993; Van den Ackerveken et al., 1994). In Fusarium oxysporum, production of fusaric acid, a toxin required for disease, is greater in vitro with high nitrate (5 mM) than with low nitrate (1 mM) or ammonium (Zhou et al., 2017); the pathogen induces stronger disease symptoms when infecting plants grown with ammonium than with nitrate, indicating that there is no strict correlation between what is observed in vitro and in planta.
Taken together, in vitro studies have led to the hypothesis that nutrient limitation could represent a signal for the induction of virulence genes; however, in planta data to support this remain scarce and mostly indirect (Wilson et al., 2012). For example, it is well known that fungal secondary metabolism is affected by nitrogen sources, as shown for F. oxysporum (Tudzynski, 2014; Sharma and Jha, 2015). In Ustilago maidis, the Nit2 TF, which activates the fungal nitrogen catabolite repression process, has also been shown to regulate virulence since the nit2 mutant possesses reduced virulence (Horst et al., 2012). This would indicate that the source of nitrogen and its metabolic pathway not only modulates the pathogen biology but also its virulence, an aspect worth exploring to improve plant tolerance to biotic stresses.
Some rare studies have directly analysed the expression of virulence factors in plants grown with contrasting levels of fertilization. For example, M. oryzae expresses high levels of pathogenicity-related and effector genes in host plants grown under high nitrogen regimes (Huang et al., 2017). We have analysed virulence factors of E. amylovora and B. cinerea in plants grown on low or high nitrate and found that stronger symptoms are associated with higher expression of virulence factors and pathogenicity-related genes, which are observed under low nitrate for E. amylovora and under high nitrate for B. cinerea (Soulié et al., 2020; M. Farjad et al., unpublished results). Interestingly, among the highest expressed B. cinerea genes in high nitrate, we demonstrated for the first time the involvement in virulence of two genes that encode a protease (acp1) and a secondary metabolite biosynthesis enzyme (sm). SM encodes a putative oxydoreductase orthologous to the Cochliobolus heterostrophus gene RED1 that is involved in the synthesis of T-toxin. In addition, a third gene corresponds to the well-known bot2 that is involved in the biosynthesis of the toxin botrydial (Soulié et al., 2020).
Limitation of inorganic phosphate (Pi) can also be encountered by bacterial pathogens in the soil or in planta. Bacteria perceive Pi deficiency through the two-component PhoBR signal transduction system, which leads to activation of the Pho regulon, allowing Pi uptake and assimilation (Chekabab et al., 2014). Interestingly, several studies have shown that PhoBR also regulates bacterial virulence. This has mostly been studied in animal pathogens, but a few studies concerning phytopathogens exist (Petters et al., 2002). For example, in Agrobacterium tumefaciens, PhoB is essential for virulence, and low-Pi conditions induce biofilm formation and catalase-encoding genes that protect bacteria against oxidative stress (Mantis and Winans, 1993; Chekabab et al., 2014). In Xanthomonas oryzae, the pathogen of rice bacterial leaf blight, a PhoR loss-of-function mutant shows strongly reduced virulence (Zheng et al., 2018). Transcriptome analysis of this ΔphoR mutant shows that several hrp genes that are required for the synthesis of the type 3 secretion apparatus and effector proteins are down-regulated. However, this study also showed that the PhoBR regulon was not activated in planta, suggesting that the bacteria encountered Pi-rich conditions and that the main role of PhoBR could be during the nutrient-poor epiphytic stages of the bacterial life cycle.
Taken together, our current knowledge suggests that nutrient availability for plants affects the transcription of pathogenesis-related genes during infection; however, these effects seem to be pathogen-dependent and probably plant-pathogen dependent as well. Although this remains to be studied, it is likely that signals perceived by pathogens in planta are affected by plant metabolism, possibly in the form of secondary metabolites, which are themselves linked to mineral nutrition conditions.
Multistress signals orchestrate plant transcriptomic responses
In the past, most transcriptomic studies of abiotic and biotic stresses have examined them individually, a situation that rarely occurs under natural conditions. Analysis of data acquired in recent years, however, has led to the conclusion that abiotic and biotic stresses not only often occur simultaneously, but that the corresponding regulatory pathways can interact at several levels inside the plant. Recently, researchers have started to examine the transcriptomic responses of plants challenged with both biotic and abiotic stress (Table 1). Although the number of datasets remains limited, some lessons can be learned from their analysis. The first is that a very large number of genes responsive to combined stresses cannot be predicted from their responses to each single stress (Rasmussen et al., 2013; Farjad et al., 2018). The number of these genes that show a specific and non-predictable response to combined stresses varies from ~30% to ~60% of modulated genes depending both on the nature and the intensity of the combined stresses. These non-predictable genes show either a ‘prioritized’, ‘cancelled’, or ‘combinatorial’ response to stress combinations (as described below). Secondly, only a small percentage of genes are similarly modulated in their responses to numerous stress conditions, whether individual or combined (Prasch and Sonnewald, 2013). Thirdly, genotype plays an important role in the way plants integrate multistress signals. For example, high temperatures decrease the resistance to X. oryzae of rice carrying Xa4 resistance but increase resistance of rice carrying Xa7 resistance, and this is correlated with genotype-specific transcriptomic profiles under the multistress combination (Table 1; Dossa et al., 2020). The importance of genotype is supported by the involvement of PBS3, an actor in SA signaling, in the age-dependent trade-off between two abiotic stresses and immune responses in Arabidopsis (Berens et al., 2019): immune responses are reduced by drought and high salinity in older leaves, but not in younger leaves in which PBS3 antagonizes the trade-off. Finally, in a multistress combination one stress can outweigh another (Coolen et al., 2016; Davila Olivas et al., 2016). In particular, the response to sequential application of stresses most resembles the response to the last-occurring stress, although a signature of the first stress is present.
Table 1.
Summary of main conclusions drawn by studies analysing transcriptomic responses of plants to different abiotic–biotic multistress combinations
| Stress combination | Plant species | Key conclusions | Reference |
|---|---|---|---|
| Low N | |||
| Botrytis cinerea | Solanum lycopersicum | Nitrate limitation activates JA signaling and represses SA signaling in response to Bc | Vega et al. (2015) |
| B. cinerea | Arabidopsis thaliana | Nitrate limitation activates JA signaling and represses SA signaling in response to Bc; 182 A. thaliana and 22 B. cinerea genes specifically modulated by stress combination | Soulié et al. (2020) |
| Erwinia amylovora | A. thaliana | ~30% of modulated genes show a specific response to stress combination | Farjad et al. (2018) |
| Cold | |||
| Flagellin (flg22) | A. thaliana | ~50% of modulated genes show a specific response to stress combination | Rasmussen et al. (2013) |
| Drought | |||
| Pseudomonas syringae | A. thaliana | ~30% of modulated genes show a specific response to stress combination, among which 150 remain specifically modulated independently of the order of stress application | Gupta et al. (2016) |
| Magnaporthe oryzae | Oryza sativa | Strong modification of fungal virulence program by drought: repression of small secreted proteins, activation of cell wall-degrading enzymes. Repression of effector-triggered immunity under drought. | Bidzinski et al. (2016) |
| Botrytis cinerea | A. thaliana | Second stress is dominant in transcriptome response but contains the first-stress signature | Coolen et al. (2016) |
| High temperature | |||
| flg22 | A. thaliana | ~50% of modulated genes show a specific response to stress combination | Rasmussen et al. (2013) |
| SA-analog (BTH) | A. thaliana | Down-regulation of SA pathway | Huot et al. (2017) |
| flg22 | A. thaliana | ~50% of modulated genes show a specific response to stress combination | Rasmussen et al. (2013) |
| Xanthomonas oryzae | O. sativa | Up-regulation of ABA biosynthesis genes and down-regulation of SA pathway | Cohen et al. (2017) |
| X. oryzae | O. sativa | Down-regulation of cell wall biosynthesis genes in susceptible line; up-regulation of trehalose biosynthesis gene in resistant line | Dossa et al. (2020) |
| Drought × High temperature | |||
| Turnip mosaic virus | A. thaliana | 23 genes specifically modulated by triple stress combination; 11 genes modulated in all three stress conditions | Prasch and Sonnewald (2013) |
To further decipher the impact of different stress combinations, we selected 10 transcriptomic datasets for analysis (Supplementary Table S2). These comprise single cold, heat, and flagellin treatment and their combinations, together with single low-nitrate, E. amylovora, and B. cinerea stress and their combinations (Rasmussen et al., 2013; Farjad et al., 2018; Soulié et al., 2020). When comparing all 10 datasets, we identified only four genes that were significantly modulated among all single and combined stress conditions, which is consistent with previous observations made on other multistress combinations (Prasch and Sonnewald, 2013). Interestingly, these stress-robust genes comprise a putative kinase, a membrane glycoprotein, and a putative TIR-domain NBS-LRR resistance protein, none of which has yet been functionally characterized (Supplementary Table S2). We then focused on the four stress combinations and identified 197 genes that were modulated in all of them (Fig. 3a, Supplementary Table S2). Interestingly, these included several members of the WRKY and NAC TF families together with the defense-signaling kinase FRK1 and the leucine-rich repeat receptor-like kinase SIF4. Gene Ontology analysis performed on all 197 genes highlighted a strong enrichment in the defense-related terms ‘response to biotic stress’ (14 nodes), ‘oxidative stress’ (14 nodes), and ‘response to hypoxia’ (six nodes) (Fig. 3b). Taken together, these data indicate that multistress-robust responsive genes are not found among the genes that respond specifically to multistress (i.e. and not to single stresses) but among the genes that also respond to some single stresses and remain activated in response to a variety of multistress combinations. These multistress-robust genes can be considered to be generally stress-robust and are of great interest for future research.
In order to better understand the modulation of expression of defense genes in response to stress combinations, we checked the expression of a manually curated list of genes (~1300) covering different aspects of plant defense in our multistress transcriptomic data (Supplementary Table S3). This list contains genes related to defense as well as the family of WRKY TFs, which are known to play a key role in the response to both biotic and abiotic stresses. We first compared the modulation of these genes in the 10 datasets of our analysis and found three defense-related genes (Fig. 3c) and four WRKY TFs (Fig. 3d) to be modulated by all stress combinations, suggesting that they are robust stress-response genes. We then used previously defined categories depending on whether their response to stress combinations could be predicted from their response to both single stresses (independent and similar categories) or not (prioritized, combinatorial, and cancelled categories), thus revealing an interaction between the response to the stresses (Rasmussen et al., 2013; Farjad et al., 2018). The response of most WRKY TFs genes was simply additive, with the response to one or both stresses being maintained (Fig. 3f: independent and similar responses, respectively). However, WRKY28 showed a specific response to the multistress combination, with a prioritization of the response to B. cinerea over the response to nitrate limitation. The response of many defense-related genes to B. cinerea was maintained under nitrate limitation (Fig. 3e), as previously described (Farjad et al., 2018). Interestingly, for several genes related to JA signaling (JAZ 6, 7, 9), the response to B. cinerea was prioritized over the response to nitrate limitation, while two genes related to SA signaling (EDS1, PR1) showed an induction specifically in the multistress combination (Fig. 3e). Our analysis thus shows that the defense response to B. cinerea overtakes the response to an abiotic stress, in this case nitrate limitation. This is opposite to the effect of heat, which had a negative effect on resistance and for which several defense and WRKY genes were found to be cancelled (Supplementary Table S3). These differences are consistent with a previous meta-analysis that showed that each multistress combination generated a specific response (Zandalinas et al., 2019).
Taken together, analysis of multistress transcriptomic data point to a pivotal role in phytohormone signaling pathways in fine-tuning the plant’s response to multiple stresses. In our analysis, several genes involved the JA- and SA-dependent pathways showed a non-predictable response to multistress. Furthermore, several genes involved in phytohormone signaling were present in the first-stress signature in the response to the combined sequential stresses (Coolen et al., 2016), again highlighting the key role of phytohormones as general integrators of multistress responses.
Conclusions
Plants are constantly under the threat of both biotic and abiotic stresses. In this review, we have examined the literature to better understand how abiotic stresses affect the response of plants to biotic stress. This is of key importance, because abiotic stresses can have an impact early in the infection process as well as having implications for either the chemical and biological treatments used to prevent disease or the efficiency of genetic-based resistance.
The first way in which abiotic stress interacts with biotic stress is by directly activating or repressing genes that are known to be involved in responses to pathogens. Cold temperatures tend to repress JA-dependent genes and activate SA-dependent genes while high temperatures do the opposite (Fig. 1). Although genes associated with defense are generally modulated by abiotic stress at much lower levels compared to pathogen infections, their modulation by abiotic stress might affect the level of activation during a potential subsequent pathogen attack. Thus, a clear understanding of how abiotic stress impacts on the susceptibility of plants to pathogens is necessary and will require further investigation.
The second way by which abiotic stress affects biotic stress responses is by interfering with the signaling. The signaling crosstalk between biotic and abiotic stresses has been extensively described in the literature and we have not covered this aspect in the present review. In general, hormone signaling plays a key role in the integration of multistress signals. For example, the fine-tuning regulation of the SA/JA balance seems to be implicated in the integration of abiotic–biotic multistresses involving temperature and nutritional limitations. On the other hand, recent data indicate that the signaling of the abiotic-driven expression of defense genes might even occur independently of the accumulation of hormones, as in the response to cold, suggesting the existence of alternative signaling mechanisms (Fig. 1; Olate et al., 2018). Besides hormones, TFs are key regulatory elements governing different aspects of multistress signals. For example, PHR1, a TF that regulates responses to phosphate starvation, has been demonstrated to repress SA-dependent genes and to activate JA-dependent genes (Fig. 2; Castrillo et al., 2017). Metadata analysis, genome-wide TF-binding assays, and in silico modeling combined with technological advances, such as CRISPR systems, are examples of techniques that can potentially help to identify new regulatory genes implicated in the response of plants to multistress (Lai et al., 2018). Hence, although great advances have been made in recent decades, further analysis will be required to get a clearer picture of the gene regulatory network that occurs during combined biotic and abiotic stress conditions. Furthermore, increasing the number of studies will allow a better comparison of datasets by meta-analysis. Our metadata analysis allowed us to identify a list of stress-robust genes that could be of great interest for future research, among which were several WRKY TFs and important defense-signaling genes (FRK1, JAZ1, and SIF4; Fig. 3). Our analysis also confirmed that the defense response can overtake the response to nutritional limitation.
The third way by which abiotic stress impacts on biotic stress is by affecting pathogen fitness and virulence inside the host plant. Once inside the leaf tissue, the pathogen is completely dependent on plant metabolism to perform its life cycle. Most studies nowadays on pathogen development in planta are performed on plants grown in optimal conditions; however, several recent studies have shown that studying pathogen virulence in non-optimal conditions can unveil novel virulence genes that are not evident in optimal conditions (Barcarolo et al., 2020; Soulié et al., 2020). This suggests that pathogens can adapt to variations in the physiology and metabolism of their host plant, which is consistent with the ability of some pathogen species such as B. cinerea to adapt to an array of important hosts (Blanco-Ulate et al., 2014). Why do pathogens have virulence genes specifically expressed in plants undergoing abiotic stress? Further investigation is required to determine whether these virulence genes are unnecessary under optimal conditions or whether they allow adaptation to plants undergoing abiotic stress. However, the existing data should encourage us to look more closely into these conditions to identify new molecular actors and perhaps to understand part of the adaptability of pathogens. Finally, the data available, even though limited, suggests that pathogens perceive different plant signals in the leaf. Although studies of leaf pathogens are complicated since the analysis of leaf intercellular fluid is technically challenging, the current knowledge that we have suggests that an interesting development in the field of abiotic–biotic interactions would be to focus more on the plant-to-pathogen signaling that occurs in planta.
Supplementary data
The following supplementary data are available at JXB online.
Table S1. List of Arabidopsis genes mentioned in the review.
Table S2. Complete datasets used for the metadata analysis, and lists of genes common to all stress conditions or common to all multistress conditions.
Table S3. Heatmaps and multistress profiles of selected defense-related genes and WRKY TFs.
Acknowledgements
IJPB benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007). This work was supported by INRA-BAP grants Multipass and Nitropath (BAP2014_63). MZ was funded by Fonds de la Recherche Scientifique–FNRS (LUX: F.4502.19).
Author contributions
MZ and MFg were responsible for the conceptualization, methodology, investigations, and writing the original draft manuscript; MZ, MFj, AL, DC, MCS, GB, and MFg reviewed and edited the manuscript; and MFg supervised the work.
Data availability
The data that support the findings of this study are openly available in the Gene Expression Omnibus (GEO) repository, accessions GSE41935, GSE116135, and GSE97582.
References
- Ah-Fong AMV, Kagda MS, Abrahamian M, Judelson HS. 2019. Niche-specific metabolic adaptation in biotrophic and necrotrophic oomycetes is manifested in differential use of nutrients, variation in gene content, and enzyme evolution. PLoS Pathogens 15, e1007729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P, Breitling R, Amtmann A. 2004. The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiology 136, 2556–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P, Breitling R, Amtmann A. 2010. Coronatine-insensitive 1 (COI1) mediates transcriptional responses of Arabidopsis thaliana to external potassium supply. Molecular Plant 3, 390–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballini E, Nguyen TT, Morel JB. 2013. Diversity and genetics of nitrogen-induced susceptibility to the blast fungus in rice and wheat. Rice 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcarolo MV, Garavaglia BS, Gottig N, Ceccarelli EA, Catalano-Dupuy DL, Ottado J. 2020. A novel Xanthomonas citri subsp. citri NADPH quinone reductase involved in salt stress response and virulence. Biochimica et Biophysica Acta - General Subjects 1864, 129514. [DOI] [PubMed] [Google Scholar]
- Barcarolo MV, Garavaglia BS, Thomas L, Marondedze C, Gehring C, Gottig N, Ottado J. 2019. Proteome changes and physiological adaptations of the phytopathogen Xanthomonas citri subsp. citri under salt stress and their implications for virulence. FEMS Microbiology Ecology 95, fiz081. [DOI] [PubMed] [Google Scholar]
- Berens ML, Berry HM, Mine A, Argueso CT, Tsuda K. 2017. Evolution of hormone signaling networks in plant defense. Annual Review of Phytopathology 55, 401–425. [DOI] [PubMed] [Google Scholar]
- Berens ML, Wolinska KW, Spaepen S, et al. 2019. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proceedings of the National Academy of Sciences, USA 116, 2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidzinski P, Ballini E, Ducasse A, Michel C, Zuluaga P, Genga A, Chiozzotto R, Morel JB. 2016. Transcriptional basis of drought-induced susceptibility to the rice blast fungus Magnaporthe oryzae. Frontiers in Plant Science 7, 1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Ulate B, Morales-Cruz A, Amrine KC, Labavitch JM, Powell AL, Cantu D. 2014. Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Frontiers in Plant Science 5, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocsanczy AM, Achenbach UC, Mangravita-Novo A, Chow M, Norman DJ. 2014. Proteomic comparison of Ralstonia solanacearum strains reveals temperature dependent virulence factors. BMC Genomics 15, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton MD, Thomma BPHJ. 2008. The complexity of nitrogen metabolism and nitrogen-regulated gene expression in plant pathogenic fungi. Physiological and Molecular Plant Pathology 72, 104–110. [Google Scholar]
- Bonneau L, Huguet S, Wipf D, Pauly N, Truong HN. 2013. Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favorable for arbuscular mycorrhizal symbiosis in Medicago truncatula. New Phytologist 199, 188–202. [DOI] [PubMed] [Google Scholar]
- Bremer E, Krämer R. 2019. Responses of microorganisms to osmotic stress. Annual Review of Microbiology 73, 313–334. [DOI] [PubMed] [Google Scholar]
- Cao FY, Yoshioka K, Desveaux D. 2011. The roles of ABA in plant–pathogen interactions. Journal of Plant Research 124, 489–499. [DOI] [PubMed] [Google Scholar]
- Carvalhais LC, Dennis PG, Fan B, Fedoseyenko D, Kierul K, Becker A, von Wiren N, Borriss R. 2013. Linking plant nutritional status to plant–microbe interactions. PLoS ONE 8, e68555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo G, Teixeira PJ, Paredes SH, et al. 2017. Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekabab SM, Harel J, Dozois CM. 2014. Interplay between genetic regulation of phosphate homeostasis and bacterial virulence. Virulence 5, 786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SP, Liu H, Argueso CT, Pereira A, Vera Cruz C, Verdier V, Leach JE. 2017. RNA-seq analysis reveals insight into enhanced rice Xa7-mediated bacterial blight resistance at high temperature. PLoS ONE 12, e0187625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen S, Proietti S, Hickman R, et al. 2016. Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. The Plant Journal 86, 249–267. [DOI] [PubMed] [Google Scholar]
- Cramer GR, Ergül A, Grimplet J, et al. 2006. Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Functional & Integrative Genomics 7, 111–134. [DOI] [PubMed] [Google Scholar]
- Cui H, Qiu J, Zhou Y, Bhandari DD, Zhao C, Bautor J, Parker JE. 2018. Antagonism of transcription factor MYC2 by EDS1/PAD4 complexes bolsters salicylic acid defense in Arabidopsis effector-triggered immunity. Molecular Plant 11, 1053–1066. [DOI] [PubMed] [Google Scholar]
- Czajkowski R, Kaczyńska N, Jafra S, Narajczyk M, Lojkowska E. 2016. Temperature-responsive genetic loci in pectinolytic plant pathogenic Dickeya solani. Plant Pathology 66, 584–594. [Google Scholar]
- Davila Olivas NH, Coolen S, Huang P, et al. 2016. Effect of prior drought and pathogen stress on Arabidopsis transcriptome changes to caterpillar herbivory. New Phytologist 210, 1344–1356. [DOI] [PubMed] [Google Scholar]
- Davis JL, Armengaud P, Larson TR, Graham IA, White PJ, Newton AC, Amtmann A. 2018. Contrasting nutrient–disease relationships: potassium gradients in barley leaves have opposite effects on two fungal pathogens with different sensitivities to jasmonic acid. Plant, Cell & Environment 41, 2357–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Rivas S. 2012. Catch me if you can: bacterial effectors and plant targets. Trends in Plant Science 17, 644–655. [DOI] [PubMed] [Google Scholar]
- Dossa GS, Quibod I, Atienza-Grande G, Oliva R, Maiss E, Vera Cruz C, Wydra K. 2020. Rice pyramided line IRBB67 (Xa4/Xa7) homeostasis under combined stress of high temperature and bacterial blight. Scientific Reports 10, 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Launay A, Clément G, Courtial J, Dellagi A, Farjad M, Krapp A, Soulié MC, Masclaux-Daubresse C. 2014. Nitrogen metabolism meets phytopathology. Journal of Experimental Botany 65, 5643–5656. [DOI] [PubMed] [Google Scholar]
- Farjad M, Rigault M, Pateyron S, Martin-Magniette ML, Krapp A, Meyer C, Fagard M. 2018. Nitrogen limitation impacts the response of specific genes to biotic stress. International Journal of Molecular Sciences 19, 3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, Haider M, Khan N, Nasim M, Jiu S, Fiaz M, Zhu X, Zhang K, Fang J. 2018. Transcriptome sequence analysis elaborates a complex defensive mechanism of grapevine (Vitis vinifera L.) in response to salt stress. International Journal of Molecular Sciences 19, 4019–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Dixit SK, Senthil-Kumar M. 2016. Drought stress predominantly endures Arabidopsis thaliana to Pseudomonas syringae infection. Frontiers in Plant Science 7, 808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KJ, Brotman Y, Segu S, et al. 2012. The form of nitrogen nutrition affects resistance against Pseudomonas syringae pv. phaseolicola in tobacco. Journal of Experimental Botany 64, 553–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA, Pruitt R, Nürnberger T. 2017. Sensing danger: key to activating plant immunity. Trends in Plant Science 22, 779–791. [DOI] [PubMed] [Google Scholar]
- Haider MS, Zhang C, Kurjogi MM, Pervaiz T, Zheng T, Zhang C, Lide C, Shangguan L, Fang J. 2017. Insights into grapevine defense response against drought as revealed by biochemical, physiological and RNA-seq analysis. Scientific Reports 7, 13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatmi S, Trotel-Aziz P, Villaume S, Couderchet M, Clément C, Aziz A. 2014. Osmotic stress-induced polyamine oxidation mediates defence responses and reduces stress-enhanced grapevine susceptibility to Botrytis cinerea. Journal of Experimental Botany 65, 75–88. [DOI] [PubMed] [Google Scholar]
- Horst RJ, Zeh C, Saur A, Sonnewald S, Sonnewald U, Voll LM. 2012. The Ustilago maydis Nit2 homolog regulates nitrogen utilization and is required for efficient induction of filamentous growth. Eukaryotic Cell 11, 368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Nguyen Thi Thu T, He X, Gravot A, Bernillon S, Ballini E, Morel JB. 2017. Increase of fungal pathogenicity and role of plant glutamine in nitrogen-induced susceptibility (NIS) to rice blast. Frontiers in Plant Science 8, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B, Castroverde CDM, Velásquez AC, Hubbard E, Pulman JA, Yao J, Childs KL, Tsuda K, Montgomery BL, He SY. 2017. Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nature Communications 8, 1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda M, Lamparová L, Zubíková A, Burketová L, Martinec J, Krčková Z. 2019. Temporary heat stress suppresses PAMP-triggered immunity and resistance to bacteria in Arabidopsis thaliana. Molecular Plant Pathology 20, 1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissoudis C, van de Wiel C, Visser RG, van der Linden G. 2014. Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Frontiers in Plant Science 5, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G, Bajpai R, Sarkar A, Mishra RK, Kumar Gupta V, Singh HB, Sarma BK. 2019. Identification, characterization and expression profiles of Fusarium udum stress-responsive WRKY transcription factors in Cajanus cajan under the influence of NaCl stress and Pseudomonas fluorescens OKC. Scientific Reports 9, 14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X, Stigliani A, Vachon G, Carles C, Smaczniak C, Zubieta C, Kaufmann K, Parcy F. 2018. Building transcription factor binding site models to understand gene regulation in plants. Molecular Plant 12, 743–763. [DOI] [PubMed] [Google Scholar]
- Lemaitre T, Gaufichon L, Boutet-Mercey S, Christ A, Masclaux-Daubresse C. 2008. Enzymatic and metabolic diagnostic of nitrogen deficiency in Arabidopsis thaliana Wassileskija accession. Plant & Cell Physiology 49, 1056–1065. [DOI] [PubMed] [Google Scholar]
- Li Z, Liu H, Ding Z, Yan J, Yu H, Pan R, Hu J, Guan Y, Hua J. 2020. Low temperature enhances plant immunity via salicylic acid pathway genes that are repressed by ethylene. Plant Physiology 182, 626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantis NJ, Winans SC. 1993. The chromosomal response regulatory gene chvI of Agrobacterium tumefaciens complements an Escherichia coli phoB mutation and is required for virulence. Journal of Bacteriology 175, 6626–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Zhang L, Oblessuc PR, He SY. 2017. Stomatal defense a decade later. Plant Physiology 174, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Babujee L, Jacobs JM, Allen C. 2015. Comparative transcriptome analysis reveals cool virulence factors of Ralstonia solanacearum Race 3 Biovar 2. PLoS ONE 10, e0139090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine A, Nobori T, Salazar-Rondon MC, Winkelmüller TM, Anver S, Becker D, Tsuda K. 2017. An incoherent feed-forward loop mediates robustness and tunability in a plant immune network. EMBO Reports 18, 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N’Guyen GQ, Raulo R, Marchi M, et al. 2019. Responses to hydric stress in the seed-borne necrotrophic fungus Alternaria brassicicola. Frontiers in Microbiology 10, 1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olate E, Jiménez-Gómez JM, Holuigue L, Salinas J. 2018. NPR1 mediates a novel regulatory pathway in cold acclimation by interacting with HSFA1 factors. Nature Plants 4, 811–823. [DOI] [PubMed] [Google Scholar]
- Onaga G, Wydra KD, Koopmann B, Séré Y, von Tiedemann A. 2017. Elevated temperature increases in planta expression levels of virulence related genes in Magnaporthe oryzae and compromises resistance in Oryza sativa cv. Nipponbare. Functional Plant Biology 44, 358–371. [DOI] [PubMed] [Google Scholar]
- Park BS, Yao T, Seo JS, Wong ECC, Mitsuda N, Huang CH, Chua NH. 2018. Arabidopsis NITROGEN LIMITATION ADAPTATION regulates ORE1 homeostasis during senescence induced by nitrogen deficiency. Nature Plants 4, 898–903. [DOI] [PubMed] [Google Scholar]
- Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson AG, Escobar MA. 2010. Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant, Cell & Environment 33, 1486–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petters J, Göbel C, Scheel D, Rosahl S. 2002. A pathogen-responsive cDNA from potato encodes a protein with homology to a phosphate starvation-induced phosphatase. Plant & Cell Physiology 43, 1049–1053. [DOI] [PubMed] [Google Scholar]
- Prasch CM, Sonnewald U. 2013. Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiology 162, 1849–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramegowda V, Senthil-Kumar M, Ishiga Y, Kaundal A, Udayakumar M, Mysore KS. 2013. Drought stress acclimation imparts tolerance to Sclerotinia sclerotiorum and Pseudomonas syringae in Nicotiana benthamiana. International Journal of Molecular Sciences 14, 9497–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Gallegos MT, Marqués S, Ramos-González MI, Espinosa-Urgel M, Segura A. 2001. Responses of Gram-negative bacteria to certain environmental stressors. Current Opinion in Microbiology 4, 166–171. [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, Bones AM, Nielsen HB, Mundy J. 2013. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiology 161, 1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Isserlin R, Voisin V, et al. 2019. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nature Protocols 14, 482–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico A, McCraw SL, Preston GM. 2011. The metabolic interface between Pseudomonas syringae and plant cells. Current Opinion in Microbiology 14, 31–38. [DOI] [PubMed] [Google Scholar]
- Ryu M, Mishra RC, Jeon J, Lee SK, Bae H. 2018. Drought-induced susceptibility for Cenangium ferruginosum leads to progression of Cenangium-dieback disease in Pinus koraiensis. Scientific Reports 8, 16368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha ND, Chaudhary A, Singh SD, Singh D, Walia S, Das TK. 2015. Plant pathogenic microbial communication affected by elevated temperature in Pectobacterium carotovorum subsp. carotovorum. Current Microbiology 71, 585–593. [DOI] [PubMed] [Google Scholar]
- Salehi-Lisar SY, Bakhshayeshan-Agdam H. 2016. Drought stress in plants: causes, consequences, and tolerance. In: Hossain M, Wani S, Bhattacharjee S, Burritt D, Tran LS. Eds. Drought stress tolerance in plants, vol. 1. Cham, Switzerland: Springer International Publishing, 1–16. [Google Scholar]
- Santander RD, Biosca EG. 2017. Erwinia amylovora psychrotrophic adaptations: evidence of pathogenic potential and survival at temperate and low environmental temperatures. PeerJ 5, e3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifi H, De Vleesschauwer D, Aziz A, Höfte M. 2014. Modulating plant primary amino acid metabolism as a necrotrophic virulence strategy. The immune-regulatory role of asparagine synthetase in Botrytis cinerea-tomato interaction. Plant Signaling & Behavior 9, e27995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim J, Park CM. 2010. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. The Plant Journal 61, 661–671. [DOI] [PubMed] [Google Scholar]
- Shankar A, Singh A, Kanwar P, Srivastava AK, Pandey A, Suprasanna P, Kapoor S, Pandey GK. 2013. Gene expression analysis of rice seedling under potassium deprivation reveals major changes in metabolism and signaling components. PLoS ONE 8, e70321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B, Jha DK. 2015. Role of nitrogen sources in regulation of fungal secondary metabolism. In: Gupta VK, Mach RL, Sreenivasaprasad S. eds. Fungal biomolecules sources, applications and recent developments. Chichester, UK: Wiley Blackwell, 213–224. [Google Scholar]
- Singh AK, Dhanapal S, Yadav BS. 2019. The dynamic responses of plant physiology and metabolism during environmental stress progression. Molecular Biology Reports 47, 1459–1470. [DOI] [PubMed] [Google Scholar]
- Sinha R, Gupta A, Senthil-Kumar M. 2017. Concurrent drought stress and vascular pathogen infection induce common and distinct transcriptomic responses in chickpea. Frontiers in Plant Science 8, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulié MC, Mia Koka S, Floch K, et al. 2020. Plant nitrogen supply affects the Botrytis cinerea infection process and modulates known and novel virulence factors. Molecular Plant Pathology 21, 1436–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot NJ, Ebbole DJ, Hamer JE. 1993. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. The Plant Cell 5, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KC, Oliver RP. 2017. Regulation of proteinaceous effector expression in phytopathogenic fungi. PLoS Pathogens 13, e1006241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski B 2014. Nitrogen regulation of fungal secondary metabolism in fungi. Frontiers in Microbiology 5, 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich M, Peñaloza-Vázquez A, Bailey AM, Bender CL. 1995. A modified two-component regulatory system is involved in temperature-dependent biosynthesis of the Pseudomonas syringae phytotoxin coronatine. Journal of Bacteriology 177, 6160–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ackerveken GF, Dunn RM, Cozijnsen AJ, Vossen JP, Van den Broek HW, De Wit PJ. 1994. Nitrogen limitation induces expression of the avirulence gene avr9 in the tomato pathogen Cladosporium fulvum. Molecular & General Genetics 243, 277–285. [DOI] [PubMed] [Google Scholar]
- Vega A, Canessa P, Hoppe G, Retamal I, Moyano TC, Canales J, Gutiérrez RA, Rubilar J. 2015. Transcriptome analysis reveals regulatory networks underlying differential susceptibility to Botrytis cinerea in response to nitrogen availability in Solanum lycopersicum. Frontiers in Plant Science 6, 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verly C, Djoman ACR, Rigault M, Giraud F, Rajjou L, Saint-Macary ME, Dellagi A. 2020. Plant defense stimulator mediated defense activation is affected by nitrate fertilization and developmental stage in Arabidopsis thaliana. Frontiers in Plant Science 11, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bao Z, Zhu Y, Hua J. 2009. Analysis of temperature modulation of plant defense against biotrophic microbes. Molecular Plant-Microbe Interactions 22, 498–506. [DOI] [PubMed] [Google Scholar]
- Wilson RA, Fernandez J, Quispe CF, Gradnigo J, Seng A, Moriyama E, Wright JD. 2012. Towards defining nutrient conditions encountered by the rice blast fungus during host infection. PLoS ONE 7, e47392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Han S, Zhou H, Tuang ZK, Wang Y, Jin Y, Shi H, Yang W. 2019. Cold stress activates disease resistance in Arabidopsis thaliana through a salicylic acid dependent pathway. Plant, Cell & Environment 42, 2645–2663. [DOI] [PubMed] [Google Scholar]
- Zandalinas SI, Fritschi FB, Mittler R. 2019. Signal transduction networks during stress combination. Journal of Experimental Botany 71, 1734–1741. [DOI] [PubMed] [Google Scholar]
- Zarattini M, Forlani G. 2017. Toward unveiling the mechanisms for transcriptional regulation of proline biosynthesis in the plant cell response to biotic and abiotic stress conditions. Frontiers in Plant Science 8, 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarattini M, Launay A, Farjad M, Wénès E, Taconnat L, Boutet S, Bernacchia G, Fagard M. 2017. The bile acid deoxycholate elicits defences in Arabidopsis and reduces bacterial infection. Molecular Plant Pathology 18, 540–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Xue B, Shao Y, Yu H, Yao X, Ruan L. 2018. Activation of PhoBR under phosphate-rich conditions reduces the virulence of Xanthomonas oryzae pv. oryzae. Molecular Plant Pathology 19, 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang M, Sun Y, Gu Z, Wang R, Saydin A, Shen Q, Guo S. 2017. Nitrate increased cucumber tolerance to fusarium wilt by regulating fungal toxin production and distribution. Toxins 9, 100–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in the Gene Expression Omnibus (GEO) repository, accessions GSE41935, GSE116135, and GSE97582.