Abstract
Likely drug candidates which are identified in traditional pre-clinical drug screens often fail in patient trials, increasing the societal burden of drug discovery. A major contributing factor to this phenomenon is the failure of traditional in vitro models of drug response to accurately mimic many of the more complex properties of human biology. We have recently introduced a new microphysiological system for growing vascularized, perfused microtissues that more accurately models human physiology and is suitable for large drug screens. In this work, we develop a machine learning model that can quickly and accurately flag compounds which effectively disrupt vascular networks from images taken before and after drug application in vitro. The system is based on a convolutional neural network and achieves near perfect accuracy while committing potentially no expensive false negatives.
Keywords: Machine learning, computer vision
1. INTRODUCTION
THE total cost of bringing a new drug from discovery to approval has exhibited a steady, exponential rise over the past five decades [1]. One contributing factor to this phenomenon, dubbed Eroom’s law (Moore’s law backwards), appears to be the failure of traditional, pre-clinical models to accurately simulate many of the more complex features of their clinical successors. These pre-clinical, in vitro studies serve to quickly and cheaply identify compounds that exhibit promising effects for further study in vivo. However, traditional 2D monolayer culture systems (i.e., petri dishes) lack many features that are present in vivo, such as 3D cellular structure, heterogeneous cellularity, cell–cell interactions, the presence of a complex extracellular matrix (ECM), biomechanical forces (e.g., shear forces generated by fluid flow), and the presence of perfused vasculature [2]. Animal studies, on the other hand, are too complex to analyze and expensive to substitute for in vitro pre-screening, and often fail to identify potential human toxicity due to physiological differences between humans and the animal model [3]. In short, a compound that appears effective in traditional, pre-clinical studies may fail spectacularly in the human body, further contributing to the costly societal burden of failed clinical trials [4].
Microphysiological systems (MPSs), or “organ-on-a-chip” platforms, promise to help close the gap between in vitro and in vivo drug screens [5], [6], [7], and have seen rapid, recent development [8], [9], [10], [11], [12], supported in part through private-public partnerships fostered under the auspices of the National Center for Advancing Translation Science [13]. These MPSs make significant strides toward more accurately modeling the pertinent properties of in vivo biological environments for drug discovery, however many remain in a proof-of-concept stage and require complex peripheral equipment and accessories to operate and maintain.
We have demonstrated an MPS for growing vascularized, perfused microtissues [14], [15]. This platform produces highly robust and uniform vascular networks which are suitable for screening anti-tumor compounds [16] and in large-scale drug discovery studies [17], all while requiring little additional training for the user and no added equipment beyond a standard incubator. We have shown that the survival of these miniature tissues is dependent on nutrients delivered through living vasculature. Importantly, by accurately identifying drugs that target tumor cells, the vascular networks that supply them, or both, the system has proven much better at mimicking human drug responses than previous models. In our studies using FDA-approved or clinical trial compounds to target the vasculature, we have found that anti-angiogenic compounds such as sorafenib and axitinib induce regression on sprouting vessels, but do not have profound effect on mature, interconnected vascular networks. Therefore, they often show a milder effect on the vasculature. On the other hand, non-specific, anti-vascular compounds such as bortezomib and vincristine aggressively fragment the vascular network. In brief, this system exhibits exceptional potential for developing more targeted, effective anti-vascular and anti-angiogenic compounds to target the tumor vasculature without adverse effects on normal tissue.
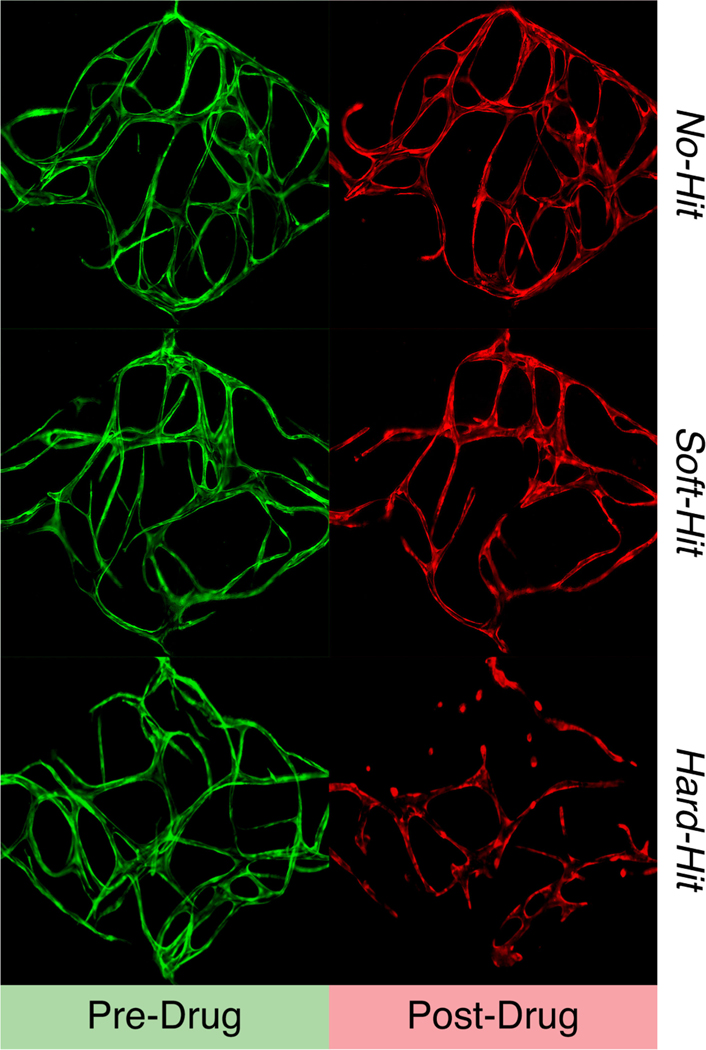
A remaining obstacle to deploying this system for truly large-scale anti-angiogenic and anti-vascular drug screening is the need to have human experts determine whether each compound is effective in targeting the vasculature network. Effects are categorized as no-hits (i.e., the compound had no effect on the vasculature network), soft-hits (i.e., the compound moderately disrupted the vasculature network or induced vascular regression), or hard-hits (i.e., the compound had a devastating effect on the vasculature network) from a primary screening (see Fig. 1). Once identified from the initial screen, soft-hit and hard-hit compounds can be further analyzed in a dose-response screen to identify the half maximal inhibitory concentration (IC50), optimized for molecular structure, and subsequently characterized for their pharmacokinetics in vivo. Soft-hit compounds are treated as anti-angiogenic while hard-hit compounds are treated as anti-vascular.
Fig. 1.
Example vessel images.
In the past, human raters have made this determination by manually analyzing each pair of before- and after-drug-application images and quantifying their total vessel length difference using AngioTool [18]. However, this workflow is imprecise—e.g., in its insensitivity to anti-angiogenic compounds that do not significantly affect total vessel length of a fully mature vascular network and its reliance on subjective human judgment—and low throughput—for its need to carefully tune several dataset-specific parameters in the software and the time it takes a human to look at each image.
Automatic classification of these images via machine learning could provide an attractive replacement to the slow and error-prone process of requiring human ratings. In this paradigm, a set of carefully hand-labeled images would be fed to a classifier which could “learn” to distinguish between classes.
A convolutional neural network is a type of machine learning model that is particularly suited to applications in computer vision. Not only do they offer state-of-the-art performance in general image classification tasks (e.g., [19]), they have also proven effective for biological applications, with past work demonstrating convolutional networks capable of detecting cardiovascular disease [20], spinal metastasis [21], and skin cancer [22] from medical images.
In this paper, we develop a convolutional neural network to automatically classify images of vasculature networks formed in our MPS into no-hit, soft-hit, and hard-hit categories. The accuracy of our best model is significantly better than our minimally-trained human raters and requires no human intervention to operate. This model is a first step toward automation of data analysis for high-throughput drug screening.
Alternative examples of applications of machine learning in drug discovery can be found in, for instance, [23], [24] and [25]. Most of these applications use machine learning models to predict drug-related properties of small molecules such as binding affinity, toxicity, and solubility.
2. METHODS
2.1. Data Collection
Drug studies were performed in the MPS as previously described [16], [17]. Briefly, the cell-ECM suspension was loaded into the platform and cultured for 7 days to allow the vascular network to develop inside the tissue chambers. Each tissue unit was exposed to various compounds obtained from the National Cancer Institute (NCI) Approved Oncology Compound Plate or purchased from Selleck Chemicals. Time course images of vascular network before and after drug treatment were taken using a Nikon Ti-E Eclipse epifluorescent microscope with a 4x Plan Apochromat Lambda objective. For close-up imaging of the tissue chambers, a 1.5x intermediate magnification setting was used.
2.2. Preprocessing
Each image in our dataset was between 1000 and 1300 pixels wide. Images of this size contain far more information than is needed for deep image classification (e.g., [26] classifies natural images taken from 1000 classes with 256×256 pixels images), so we downsampled images to create 4 separate constant-size datasets: one each of 128×128 px, 192×192 px, 256×256 px, and 320×320 px. Next, we z-normalized each image, subtracting the mean pixel intensity and dividing by the standard deviation of the pixel intensities within that image to obtain images with 0-centered pixel values and unitary standard deviation. This normalization helps our models to converge more quickly and uniformly across random initializations. After all this, we concatenated the pre-drug-application and post-drug-application images to obtain a single, 2-channel image.
2.2.1. Image Alignment
We would like the pre-drug-application and post-drug-application images to spatially align as closely as possible. If they do not, then our model swould be required to learn an extra invariance: that the channel images need not be aligned. Because the pre- and post-drug-application images were captured three days apart, it is not in general possible to ensure that the two images will be perfectly aligned (e.g., the later image might be shifted or rotated slightly compared to the original). To combat this effect, we implemented a rigid alignment preprocessing step to align the post-drug image to the pre-drug image using the warpAffine method in OpenCV3 [27]. For each image, we tried three sets of transformations:
A single Euclidean (translation + rotation) transformation on the full-resolution image.
A euclidean transformation on a smaller (32×32 px) copy of the image followed by a euclidean transformation on the full-resolution image.
A translation-only transformation on a smaller (32×32 px) copy of the image followed by a euclidean transformation on the full-resolution image.
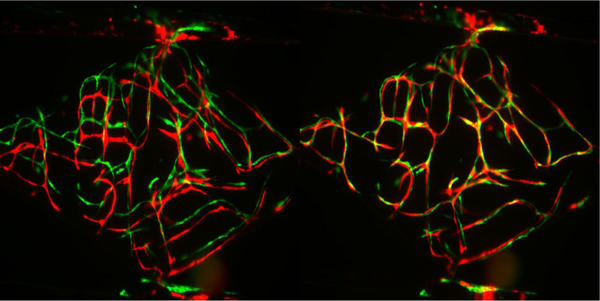
From these three, we selected the transformed version which yielded the highest possible correlation coefficient between the pre- and transformed post-drug image. See Fig. 2 for two examples of this alignment process in action.
Fig. 2.
A set of blood vessel images before (left) and after (right) alignment. The pre-drug-application images are placed in the image’s green channel and the post-drug-application images are placed in the red channel. The separate green and red vessels in the left image shows that the pre- and post-drug-application images are misaligned, the more pervasive yellow in the right image comes from the green and red channels being aligned on top of each other.
2.3. Human Ratings
Two human experts rated each of the 277 images, comparing disparate ratings where necessary to come to a consistent set of gold-standard ratings. 164 images were labeled as 0 or no-hit (59.2 percent), 52 were labeled as 1 or soft-hit (18.8 percent), while 61 were labeled as 2 or hard-hit (22.0 percent). These ratings are used throughout the remainder of this paper.
We also obtained ratings from 4 additional humans: undergraduate research assistants who were trained to recognize each image class and who had been assigned this task in the past. Raters were presented with the full set of 277 images in randomized order and were asked to provide an integer class assignment for each using the following instructions: “How much of an effect did the drug have? (0 for no effect, 1 for solid effect, 2 for devastating effect)”.
2.4. Loss Weighting
For the purposes of drug discovery, false negatives are potentially much costlier than false positives. A false positive (i.e., predicting that an image from an ineffective drug was actually effective) will result in secondary screening in which the ineffectiveness of the drug may be confirmed. A false negative (i.e., predicting that an image taken from an effective drug did not actually have any effect) may result in a potentially useful compound being overlooked in this and any future drug trials. To help control our model’s false-negative rate, we employed a weighted cross-entropy loss function of the form:
where i indexes over datapoints, c over classes, yic is an indicator variable that takes the value of 1 if the true class of datapoint i is c and 0 otherwise, citrue represents the true label of datapoint i (i.e., 0, 1, or 2), and the weights are drawn from the hand-tuned confusion weighting matrix shown in Table 1. Note that if all elements of this weight matrix were set to 1.0, then our weighted cross-entropy loss would reduce to standard cross-entropy.
TABLE 1.
Loss Function Weight Values
| Yipred = 0 | Yipred = 1 | Yipred = 2 | |
|---|---|---|---|
| Yitrue = 0 | 0.8 | 0.8 | 0.8 |
| Yitrue = 1 | 2.0 | 1.0 | 0.8 |
| Yitrue = 2 | 2.0 | 0.8 | 1.0 |
This loss function penalizes false negatives at twice the default value. In addition, it penalizes the treatment of all true no-hit images at 0.8 times the default value and reduces the penalties for confusing soft- and hard-hits to the same amount. We arrived at these weights through trial and error and use them for all experiments presented in this paper.
2.5. Training Procedure
We partitioned the full dataset of 277 images into a test set consisting of 25 percent of the images (69 images) and a training+validation set consisting of 75 percent of the images (208 images). We employed 4-fold cross validation on the training+validation set, training on 75 percent of its datapoints (156 images) and tracking validation loss on the remaining 25 percent (52 images). Unless otherwise noted, we trained on each fold for a total of 200 epochs. All deep neural networks presented in this paper were built in Keras [28] and trained on NVIDIA GPUs. We selected the model from each fold which attained the lowest validation-set loss value across all training epochs.
We combined the best models from each fold into a 4-model ensemble of models. We averaged the predictions across all 4 models in the ensemble to attain final predictions for each set of hyperparameters on the test dataset.
2.5.1. Data Augmentation
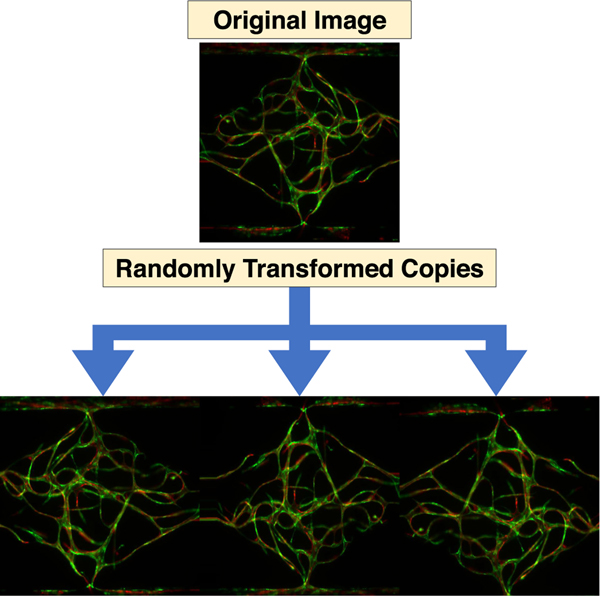
Since our training set is rather small, we employed random data augmentation during training. In each pass over the data, each training image was randomly rotated between −5 and 5 degrees clockwise, translated between −5 and 5 percent vertically and horizontally, zoomed in between 0 and 10 percent, and possibly flipped horizontally and vertically, with each transformation value selected uniformly at random from the legal range. Empty pixels that resulted from the random rotation and translation were filled with the values from their nearest existing neighbor pixel. Fig. 3 shows three randomly transformed versions of one training image. This random data augmentation scheme with continuous parameters yields an infinitude of variations for each 156-image training set and helps prevent our models from overfitting to the specific details of our training data.
Fig. 3.
Three examples of the data augmentation process used for training and inference. The top image is an actual training image, and the bottom three are randomly transformed copies of that image. Each time an image is visited during the training process, it is first randomly transformed in a way that simulates creating new images with respect to the true invariances of the training images (e.g., an image should have the same class as a copy of that image which is slightly shifted, rotated, or flipped). This random augmentation helps simulating a larger training set and prevent our model from overfitting.
At inference time, we randomly generated five versions of each validation or test image and averaged the model’s predictions for each image over all five of its randomly-generated copies.
2.6. Baseline Models
We trained a number of increasingly complex machine learning models on the data to use as comparison baseline: (1) logistic regression on the raw data; (2) logistic regression on a bag of words (BoW) representation of SIFT [29] or SURF features; and (3) RBF-kernel support vector machine classifiers (SVMs) trained on a BoW representation of SURF or SIFT features. We used the SVM-classifier and logistic regression implementations provided by scikit-learn.1
The logistic regression model was trained on images of sizes 128×128, 192×192, 256×256, and 320×320 with varying L2 regularization using the LBFGS optimizer, treating all concatenated pixels of both the pre- and post-drug application images as single input vector. We used OpenCV2 to extract SIFT and SURF features from the images at a resolution of 320×320, which yields a varying number of key-points/features per image. To build a bag of words representation we first clustered all SIFT/SURF descriptors of the training set with k-means. Then we mapped all descriptors to their nearest centroid (as found by k-means) and compute the histogram of these centroid mappings for each image separately. We experimented with two histogram normalization approaches: globally rescaling the bins to the 0–1 range or a binary representation that encodes whether at least one SIFT/SURF descriptor from the image was assigned to a given centroid. We treated pre- and post-drug application images separately and concatenated their BoW representations (the histograms) into one feature vector, as computing the BoW representation across SIFT/SURF features of both images together discards crucial discriminatory information and resulted in a reduced performance in preliminary experiments. We optimize the L2 regularization coefficient as well as the size of the BoW representation (number of clusters) for all models.
2.7. Convolutional Neural Network Models
Convolutional neural networks are based on a weight-sharing scheme in ‘convolutional’ layers [30]. These layers learn translation-invariant filters that are applied to e.g., all pixels of an image in the case of computer vision, and have lead to models achieving state-of-the-art classification performance on a variety of tasks [19], [31], [32].
Standard convolutional architectures for image classification include a series of convolutional layers followed by one or more fully connected layers [19], [26], [30]. Each convolutional and fully connected layer is followed by a rectified linear unit (ReLU) non-linearity [33] and max pooling layers are interspersed through some subset of the convolutional layers to repress non-maximal responses and reduce the number of parameters in subsequent layers. Dropout may also be used on some of the convolutional and fully connected layers to help prevent overfitting [34].
Overall, convolutional neural networks offer a well-established process for performing high-quality image classification.
2.8. Hyperparameter Search for Convolutional Architectures
Building a convolutional neural network requires specifying a large number of hyperparameters, such as the number of convolutional and fully-connected layers in the network, the size of each layer, dropout probabilities etc. The number of possible hyperparameter combinations grows exponentially with the number of hyperparameters, so a thorough grid search of hyperparameter combinations quickly becomes unwieldy.
Algorithm 1.
Outer-Loop Hyperparameter Optimization
| 1: HPO ← Hyperparameter Optimizer |
| 2: for Iteration i ← 1…K do |
| 3: Hyperparams αi ← HPO.NextHyperparams() |
| 4: Train model on training data with hyperparams αi |
| 5: Make predictions for validation data with model |
| 6: HPO.RecordValidationError() |
| 7: end for |
| 8: Best model is model with lowest validation error |
Instead, we employ a Gaussian-process-based meta-model which maps from a set of chosen hyperparameters to an estimate of the out-of-sample accuracy attained by a model trained with the given hyperparameters [35]. This meta-model of hyperparameter fitness is used in an outer-loop hyperparameter optimization process (see Algorithm 1). First, the meta-model proposes a hyperparameter set to try. For each hyperparameter set, we follow the same training procedure as detailed in Section 2.5, using 4-fold cross-validation on the training+validation set, building a 4-model ensemble from the best version of the model for each fold (across epochs and as judged by validation-set accuracy), and averaging each model’s validation- and test-set predictions over 5 randomly generated versions of each input image. At the end of training, we report the validation-set accuracy (averaged across all 4 folds) as the objective value attained for the given hyperparameter set. This objective value is used to update the meta-model of hyperparameter quality and the process repeats.
2.9. Pre-Trained Convolutional Architecture
Given the small size of our training dataset, we next tried a large convolutional architecture that had been pre-trained on a large, general purpose image recognition problem. For this purpose we picked the InceptionV3 architecture [36] as implemented in Keras [28] with weights that had been pre-trained on the ImageNet classification challenge [37]. The full convolutional portion of the InceptionV3 model contains 21,611,968 parameters and some 216 layers. We instantiated the model without including the final fully-connected layers, opting not to fine-tune its convolutional weights, but to train two fully connected and one 3-class softmax layer anew for our classification problem while using the convolutional portion of the InceptionV3 model as an elaborate, fixed computer vision preprocessing routine.
While fixing our convolutional architecture fixed many of the hyperparameters of our model, several still remained. These were: the input image size, the number of neurons in the fully connected layers, dropout probabilities for the dropout layers before and after the fully connected layers, the optimization batch size, the learning rate, and L1 and L2 regularization coefficients. Hyperparameters that control the amount of dropout, or the strength of the L1-, and L2- penalty terms have a regularizing effect and reduce the chances of overfitting the data, whereas the exact effect for other hyperparameters is in general more difficult to estimate. The exact ranges of hyperparameters that we optimized can be found in the supplementary material, which can be found on the Computer Society Digital Library at http://doi.ieeecomputersociety.org/10.1109/TCBB.2018.2841396.
2.10. Custom Convolutional Architecture
Though the Inception architecture employed in Section 2.9 has proven very useful for general-purpose image classification, the images of microscopic blood vessel networks used in this task have their own structure that does not necessarily match the constraints of general object recognition.3
For this purpose, we also trained a series of custom convolutional architectures specifically for this blood-vessel classification task. We constrained our architecture to contain several convolutional layers followed by two fully connected layers.
The hyperparameters that we optimized were: the input image size, the number of convolutional layers, number of convolutional filters, and number of neurons in fully connected layers in the model, the size of the max pooling receptive fields, the optimization batch size, and parameters related to model regularization: dropout probabilities and L1- and L2 penalty terms (see Supplement, available online, for detailed hyperparameter-ranges).
3. RESULTS
3.1. Human Rating Results
The four human raters found the vessel rating task difficult compared to the expert raters, matching the gold-standard ratings 72.9, 76.5, 69.3 and 83.0 percent of the time. The rounded average of all four raters’ ratings (i.e., 0, 1, or 2) matched the gold standard ratings 85.9 percent of the time. (See Table 2 and Section 4 for further details).
TABLE 2.
Test Set Confusion Matrix for Average of Four Human Raters
| Yipred = 0 | Yipred = 1 | Yipred = 2 | |
|---|---|---|---|
| Yitrue = 0 | 86% | 14% | 0 |
| Yitrue = 1 | 27% | 65% | 9% |
| Yitrue = 2 | 0 | 0 | 100% |
3.2. Baseline Models Results
The best logistic regression model trained on raw pixels obtained an average validation set accuracy of 79.6 percent across five repeated five-fold cross-validation experiments, using an input image size of 320 px ×320 px and an L2 regularization strength of 0.05. This model obtained an average three-class test accuracy of 73.3 percent, which is a notable improvement over guessing the majority class (62.3 percent). An even higher accuracy was reached by models using a bag of words (BoW) representation of SIFT or SURF features. The best such model was a support vector machine (SVM) using SURF features that were clustered into a binary feature vector of size 200, obtaining a validation accuracy of 84.4 percent and test-set accuracy of 78.0 percent. This is almost 5 percent better than the logistic regression model that was trained on raw pixels. A summary of best models for each category is given in Table 3; all BoW model results are averages over three repeated full cross-validations runs.
TABLE 3.
Average Test- and Validation Set Accuracies of Baseline Models
| Model | Features | Validation Acc. | Test Acc. |
|---|---|---|---|
| Logistic | raw pixels | 79.6% | 73.3% |
| BoW Logistic | SIFT | 82.7% | 77.7% |
| BoW Logistic | SURF | 81.3% | 77.6% |
| BoW SVM | SIFT | 82.0% | 76.0% |
| BoW SVM | SURF | 84.4% | 78.0% |
3.3. Pre-Trained Convolutional Neural Network Results
We explored a total of 100 hyperparameter sets for the pretrained convolutional architecture4 using the procedure explained in Section 2.8. The best model, as judged by three-way validation-set accuracy (87.0 percent), used 320 px×320 px input images, its first fully connected layer after the InceptionV3 convolutional stack contained 256 neurons, its second fully connected layer contained 1024 neurons, and the final dropout probability before the 3-way softmax layer was 0.27.
The optimization was completed with a batch size of 16, log10 of the learning rate of −1.24, a per-epoch learning rate decay factor of 0.98, log10 of L1 shrinkage of −9.0, and log10 of L2 shrinkage of −1.0.
A 4-model ensemble based on this architecture achieved a three-class accuracy value of 87.0 percent on the hitherto-unseen test (see the confusion matrix in Table 4 for details).
TABLE 4.
Test Set Confusion Matrix for Pre-Trained Convolutional Ensemble
| Yipred = 0 | Yipred = 1 | Yipred = 2 | |
|---|---|---|---|
| Yitrue = 0 | 98% | 2% | 0 |
| Yitrue = 1 | 45% | 36% | 18% |
| Yitrue = 2 | 0 | 7% | 93% |
3.4. Custom Convolutional Neural Network Results
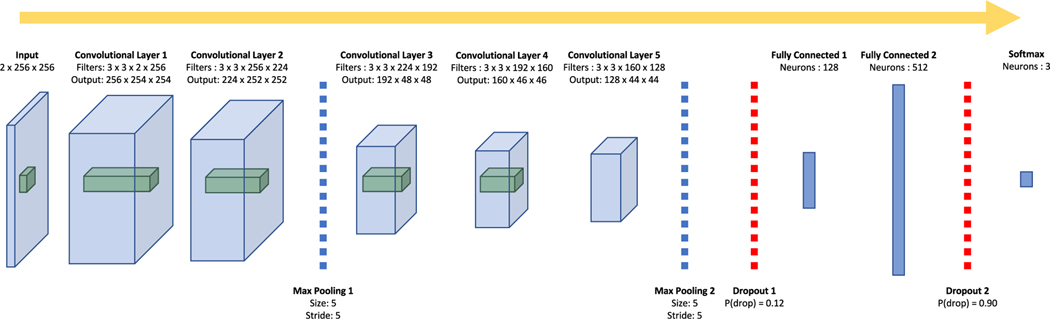
We explored a total of 1000 hyperparameter sets for our custom convolutional architecture, the best of which, as judged by three-class validation-set accuracy (96.6 percent), is a 21-layer convolutional neural network, the architecture for which is illustrated in Fig. 4.
Fig. 4.
The architecture for the best convolutional neural network we trained on these data. The blue prisms represent the 3-dimensional input images (two channels, width, and height) and the three dimensional output of each convolutional layer (filters, width, and height). The green prisms represent a sample receptive field for the subsequent convolutional layer.
The optimization was completed with a batch size of 1, a learning rate of 0.012, a per-epoch learning rate decay factor of 0.98, and both L1 and L2 coefficients at a value of 10−9.
A 4-model ensemble based on this architecture achieved a three-class accuracy value of 95.7 percent on the hitherto-unseen test set with no false negatives (see the confusion matrix in Table 5 for details).
TABLE 5.
Test Set Confusion Matrix for Custom Convolutional Ensemble
| Yipred = 0 | Yipred = 1 | Yipred = 2 | |
|---|---|---|---|
| Yitrue = 0 | 100% | 0 | 0 |
| Yitrue = 1 | 0 | 82% | 18% |
| Yitrue = 2 | 0 | 7% | 93% |
The data set (with 277 datapoints) is small in comparison to typical machine learning data sets, which raises concerns over potential overfitting of deep learning models. To shed light on whether overfitting occurs we plot the evolution of the validation accuracy for three, independently—randomly—initialized and trained, instantiations of our custom CNN model in Fig. 5. The curves are not smoothed and thus, as expected, relatively jagged due to the small size of the dataset and the various noise-injecting regularization techniques. Interestingly, we observe no evidence of overfitting within 200 epochs of training. Overfitting would have manifested as a decline in the average validation accuracy towards the end of training, but instead we only observe a reduction in the variance of validation accuracies. In short, we conclude that the employed model regularization techniques are very effective and that early stopping is, while still beneficial, not as crucial as initially expected.
Fig. 5.
Validation accuracy as a function of training progress for three different random initializations of the custom CNN model and different data cross-validation splits.
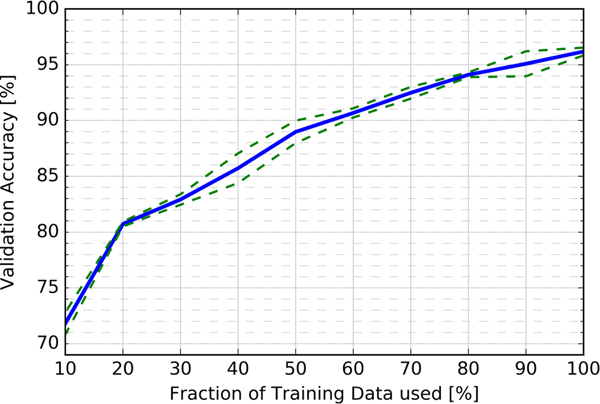
It is desirable to further estimate the sensitivity of the model to the number of samples in the training set. To this end we artificially and progressively reduce the amount of training data, while keeping all other factors identical (e.g., model architecture, hyperparamters, validation set). Fig. 6 presents results from training the custom CNN with ten different training set sizes in 10 percent increments, repeating the four-fold cross-validation training process four times for each training set size and averaging over these. As expected, decreasing the amount of training data directly reduces the validation accuracy. Interestingly, we also find that the CNN is able to match or outperform the best baseline model (an SVM trained on SURF features with a validation accuracy of 84.4 percent) when trained on only 40 percent of the original training data. Further, from extrapolating the graph beyond the 100 percent point, it seems virtually guaranteed that having access to more training data would enable us to train better models with accuracies beyond our current best result of 96.6 percent.
Fig. 6.
Average validation accuracy of the custom CNN as a function of training set size. (Blue line: average cross validation accuracy; green dashed lines: Delineate the empirical one-sigma confidence region)
4. DISCUSSION
In this paper, we present a new classification problem: to distinguish effective from ineffective drug compounds through automatic analysis of vascularization images.
This problem may appear to be simple in some cases, such as in Fig. 1, and solvable by merely counting the number of bright pixels in the pre- and post-treatment images. However, we find that a linear model obtains an overall test accuracy of 73.3 percent only, providing only a relatively small improvement over guessing the majority class (62.3 percent). The difficulty appears to be driven by the nuances of the classification problem, which cannot be captured in a simple linear decision boundary in pixel space. For example, the death of a bridge-to-nowhere vessel should be treated as less important than the death of a vessel on a major thoroughfare in the vasculature network. To further highlight its difficulty, even an ensemble of four trained human raters had some difficulty with this task (three-way accuracy: 85.9 percent).
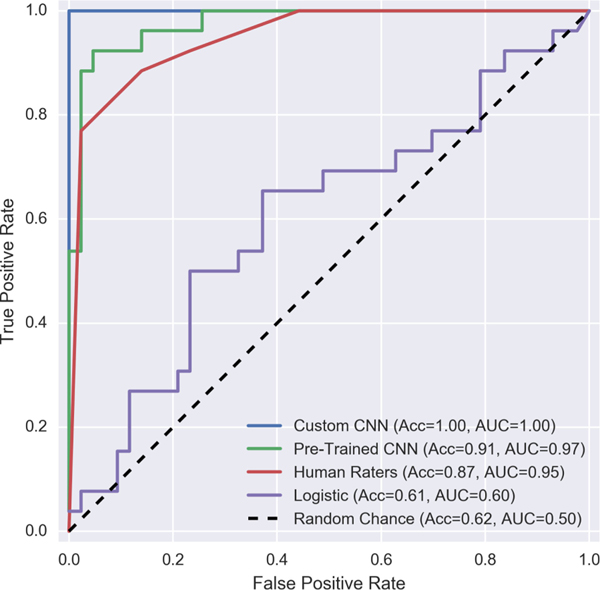
Convolutional neural networks significantly outperform the baseline models as well as human raters on this dataset. Where a cadre of four undergraduate raters achieved a three-way accuracy of 85.9 percent on this dataset, a convolutional ensemble based on the InceptionV3 architecture [36] and pre-trained on ImageNet data [37] achieved three-way accuracy of 87.0 percent (though it committed more false negatives than the human raters). A custom convolutional architecture, however, achieves a 95.7 percent three-way accuracy for drug-hit classification, while committing no false negatives. This pattern repeats itself if we reduce our 3-way classification problem to a binary problem by aliasing together the soft-hit and hard-hit categories (see Fig. 7).
Fig. 7.
Receiver operating characteristic curves for a binarized version of this classification problem (no-hit versus soft-hit or hard-hit). ROC-AUC scores range between 0.5 and 1.0, with 0.5 indicating performance at chance and 1.0 indicating perfect classification (a standard which the best custom convolutional neural network we tried achieves on this binarized problem).
The success of this convolutional model is driven in part by carefully tuning our loss function to discourage false negatives (see Section 2.4), but also by the steps taken to control overfitting in the model. One regularization strategy was to augment our limited training dataset to virtually infinite size via randomly transforming images during each training pass (see Section 2.5.1). Heavy use of dropout also contributed to the result. In fact, the hyperparameter optimization scheme that we used automatically picked a model with a large final layer (512 neurons) and a high dropout probability (0.90). Dropout can be interpreted as implicitly performing a geometric average over an ensemble of regularized subnetworks [38], so this model can be interpreted as implicitly averaging over a large ensemble of diverse sub-networks.
These regularization strategies were important, as our final network contained 2,485,827 learned parameters and 15 optimized hyperparameters, more than enough capacity to memorize the identity of 208 training+validation datapoints. However, our network still exhibits excellent generalization power, with test accuracy of 95.7 percent only barely lagging behind the hyperparameter optimized 96.6 percent validation accuracy which in turn closely follows the training accuracy of 98.1 percent. This tendency toward strong generalization performance is often seen in deep networks, and cannot yet be fully explained by any known regularization mechanism or learning theory[39].
5. CONCLUSION
In this paper, we have developed a convolutional neural network to improve the data analysis processes for high-throughput drug screening using our MPS. This network can classify new images near instantaneously and surpasses human accuracy on this task. A larger scale drug screening can be achieved by coupling this classifier and an automated microscope camera system to capture images before and after drug treatment.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Jerry C. Chen, Koyinsola B. Oloja, David E. Lewis, and Kim A. Lin for their work in rating images. This work was additionally supported in part by the following grants: National Science Foundation IIS-1550705, Defense Advanced Research Projects Agency D17AP00002, National Institutes of Health (NIH) R01 CA180122 (PQD5), NIH UH3 TR000481, and an NVDIA Corporation Hardware Award. P.B. receives support from a Google Faculty Research Award and C. C. W.H receives support from the Chao Family Comprehensive Cancer Center (CFCCC) through an NCI Center Grant award P30A062203.
Footnotes
For example, detecting eyes is very important for detecting the myriad animal types in ImageNet, but irrelevant for our task.
This model contained 21,611,968 fixed parameters that had been pre-trained on ImageNet data and 33,820,931 fully connected parameters that were trained on the vessel data.
Contributor Information
Gregor Urban, Department of Computer Science, University of California at Irvine, Irvine, CA 92697..
Kevin Bache, Department of Computer Science, University of California at Irvine, Irvine, CA 92697..
Duc T. T. Phan, Department of Molecular Biology and Biochemistry, University of California at Irvine, Irvine, CA 92697..
Agua Sobrino, Department of Molecular Biology and Biochemistry, University of California at Irvine, Irvine, CA 92697..
Alexander K. Shmakov, Department of Computer Science, University of California at Irvine, Irvine, CA 92697.
Stephanie J. Hachey, Department of Molecular Biology and Biochemistry, University of California at Irvine, Irvine, CA 92697.
Christopher C. W. Hughes, Department of Molecular Biology and Biochemistry, University of California at Irvine, Irvine, CA 92697.
Pierre Baldi, Department of Computer Science, University of California at Irvine, Irvine, CA 92697..
REFERENCES
- [1].Scannell JW, Blanckley A, Boldon H, and Warrington B, “Diagnosing the decline in pharmaceutical R&D efficiency,” Nature Rev. Drug Discovery, vol. 11, no. 3, pp. 191–200, 2012. [DOI] [PubMed] [Google Scholar]
- [2].Fabre KM, Livingston C, and Tagle DA, “Organs-on-chips (microphysiological systems): Tools to expedite efficacy and toxicity testing in human tissue,” Exp. Biol. Med, vol. 239, no. 9, pp. 1073–1077, 2014. [DOI] [PubMed] [Google Scholar]
- [3].Low LA and Tagle DA, “Tissue chips to aid drug development and modeling for rare diseases,” Expert Opinion Orphan Drugs, vol. 4, no. 11, pp. 1113–1121, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Arrowsmith J and Miller P, “Trial watch: Phase II and phase III attrition rates 2011–2012,” Nature Rev. Drug Discovery, vol. 12, no. 8, pp. 569–569, 2013. [DOI] [PubMed] [Google Scholar]
- [5].Esch M, King T, and Shuler M, “The role of body-on-a-chip devices in drug and toxicity studies,” Ann. Rev. Biomed. Eng, vol. 13, pp. 55–72, 2011. [DOI] [PubMed] [Google Scholar]
- [6].Esch EW, Bahinski A, and Huh D, “Organs-on-chips at the frontiers of drug discovery,” Nature Rev. Drug Discovery, vol. 14, no. 4, pp. 248–260, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sutherland ML, Fabre KM, and Tagle DA, “The national institutes of health microphysiological systems program focuses on a critical challenge in the drug discovery pipeline,” Stem Cell Res. Therapy, vol. 4, no. 1, 2013, Art. no. I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, and Ingber DE, “Reconstituting organ-level lung functions on a chip,” Sci., vol. 328, no. 5986, pp. 1662–1668, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schimek K, Busek M, Brincker S, Groth B, Hoffmann S, Lauster R, Lindner G, Lorenz A, Menzel U, Sonntag F, et al. , “Integrating biological vasculature into a multi-organ-chip microsystem,” Lab Chip, vol. 13, no. 18, pp. 3588–3598, 2013. [DOI] [PubMed] [Google Scholar]
- [10].Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, Marks N, Mandegar M, Conklin BR, Lee LP, et al. , “Human iPSC-based cardiac microphysiological system for drug screening applications,” Sci. Rep., vol. 5, 2015, Art. no. 8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brown JA, Pensabene V, Markov DA, Allwardt V, Neely MD, Shi M, Britt CM, Hoilett OS, Yang Q, Brewer BM, et al. , “Recreating blood-brain barrier physiology and structure on chip: A novel neurovascular microfluidic bioreactor,” Biomicrofluidics, vol. 9, no. 5, 2015, Art. no. 054124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Esch MB, Ueno H, Applegate DR, and Shuler ML, “Modular, pumpless body-on-a-chip platform for the co-culture of gi tract epithelium and 3D primary liver tissue,” Lab Chip, vol. 16, no. 14, pp. 2719–2729, 2016. [DOI] [PubMed] [Google Scholar]
- [13].Livingston CA, Fabre KM, and Tagle DA, “Facilitating the commercialization and use of organ platforms generated by the microphysiological systems (tissue chip) program through public–private partnerships,” Comput. Struct. Biotechnology J, vol. 14, pp. 207–210, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Moya ML, Hsu Y-H, Lee AP, Hughes CC, and George SC, “In vitro perfused human capillary networks,” Tissue Eng. Part C: Methods, vol. 19, no. 9, pp. 730–737, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang X, Phan DT, Sobrino A, George SC, Hughes CC, and Lee AP, “Engineering anastomosis between living capillary networks and endothelial cell-lined microfluidic channels,” Lab Chip, vol. 16, no. 2, pp. 282–290, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sobrino A, Phan DT, Datta R, Wang X, Hachey SJ, Romero-López M, Gratton E, Lee AP, George SC, and Hughes CC, “3D microtumors in vitro supported by perfused vascular networks,” Sci. Rep., vol. 6, 2016, Art. no. 31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Phan DT, Wang X, Craver BM, Sobrino A, Zhao D, Chen JC, Lee LY, George SC, Lee A, and Hughes C, “A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications,” Lab Chip, vol. 17, no. 3, pp. 511–520, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zudaire E, Gambardella L, Kurcz C, and Vermeren S, “A computational tool for quantitative analysis of vascular networks,” PloS One, vol. 6, no. 11, 2011, Art. no. e27385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Szegedy C, Ioffe S, Vanhoucke V, and Alemi AA, “Inception-v4, inception-resnet and the impact of residual connections on learning,” AAAI, 2017. [Google Scholar]
- [20].Wang J, Ding H, Azamian F, Zhou B, Iribarren C, Molloi S, and Baldi P, “Detecting cardiovascular disease from mammograms with deep learning,” IEEE Trans. Med. Imaging, vol. 36, no. 5, pp. 1172–1181, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang J, Fang Z, Lang N, Yuan H, Su M., and P. Baldi, “A multi-resolution approach for spinal metastasis detection using deep siamese neural networks,” Comput. Biol. Med, vol. 1, no. 84, pp. 137–146, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, and Thrun S, “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, pp. 115–118, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ramsundar B, Kearnes S, Riley P, Webster D, Konerding D, and Pande V, “Massively multitask networks for drug discovery,” arXiv:1502.02072, 2015. [Google Scholar]
- [24].Lusci A, Fooshee D, Browning M, Swamidass J, and Baldi P, “Accurate and efficient target prediction using a potency-sensitive influence-relevance voter,” J. Cheminform., vol. 7, no. 1, 2015, Art. no. 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gawehn E, Hiss JA, and Schneider G, “Deep learning in drug discovery,” Mol. Informat, vol. 35, no. 1, pp. 3–14, 2016. [DOI] [PubMed] [Google Scholar]
- [26].Krizhevsky A, Sutskever I, and Hinton GE, “Imagenet classification with deep convolutional neural networks,” in Proc. 25th Int. Conf. Neural Inf. Process. Syst, 2012, pp. 1097–1105. [Google Scholar]
- [27].Evangelidis GD and Psarakis EZ, “Parametric image alignment using enhanced correlation coefficient maximization,” IEEE Trans. Pattern Anal. Mach. Intell, vol. 30, no. 10, pp. 1858–1865, 2008. [DOI] [PubMed] [Google Scholar]
- [28].Chollet F, “keras,” 2015. [Online]. Available: https://github.com/fchollet/keras
- [29].Lowe DG, “Object recognition from local scale-invariant features,” in Proc. 6th IEEE Int. Conf. Comput. Vis, vol. 2, pp. 1150–1157, 1999. [Google Scholar]
- [30].LeCun Y, Bengio Y, et al. , “Convolutional networks for images, speech, and time series,” The Handbook of Brain Theory and Neural Networks. Cambridge, MA, USA: MIT Press, 1995. [Google Scholar]
- [31].Ren S, He K, Girshick R, and Sun J, “Faster r-cnn: Towards real-time object detection with region proposal networks,” in Proc. Advances Neural Inf. Process. Syst, 2015, pp. 91–99. [DOI] [PubMed] [Google Scholar]
- [32].Zhou B, Khosla A, Lapedriza A, Torralba A, and Oliva A, “Places: An image database for deep scene understanding,” CoRR, vol. abs/1610.02055, 2016. [Google Scholar]
- [33].Nair V and Hinton GE, “Rectified linear units improve restricted boltzmann machines,” in Proc. 27th Int. Conf. Mach. Learn, 2010, pp. 807–814. [Google Scholar]
- [34].Srivastava N, Hinton G, Krizhevsky A, Sutskever I, and Salakhutdinov R, “Dropout: A simple way to prevent neural networks from overfitting,” J. Mach. Learn. Res, vol. 15, no. 1, pp. 1929–1958, 2014. [Google Scholar]
- [35].Snoek J, Larochelle H, and Adams RP, “Practical bayesian optimization of machine learning algorithms,” in Proc. 25th Int. Conf. Neural Inf. Process. Syst, 2012, pp. 2951–2959. [Google Scholar]
- [36].Szegedy C, Vanhoucke V, Ioffe S, Shlens J, and Wojna Z, “Rethinking the inception architecture for computer vision,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit, 2016, pp. 2818–2826. [Google Scholar]
- [37].Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, et al. , “Imagenet large scale visual recognition challenge,” Int. J. Comput. Vis, vol. 115, no. 3, pp. 211–252, 2015. [Google Scholar]
- [38].Baldi P and Sadowski PJ, “Understanding dropout,” in Proc. Advances Neural Inf. Proc. Syst, 2013, pp. 2814–2822. [Google Scholar]
- [39].Zhang C, Bengio S, Hardt M, Recht B, and Vinyals O, “Understanding deep learning requires rethinking generalization,” ICLR, 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.