Glucocorticoid counteracts cellular mechanoresponses by a novel lncRNA-dependent mRNA decay machinery.
Abstract
Mechanical stimuli on cells and mechanotransduction are essential in many biological and pathological processes. Glucocorticoid is an important hormone, roles, and mechanisms of which in cellular mechanotransduction remain unknown. Here, we report that glucocorticoid counteracted cellular mechanoresponses dependently on a novel long noncoding RNA (lncRNA), LINC01569. Further, LINC01569 mediated glucocorticoid effects on mechanotransduction by destabilizing messenger RNA (mRNA) of mechanosensors including early growth response protein 1 (EGR1), Cbp/P300-interacting transactivator 2 (CITED2), and bone morphogenic protein 7 (BMP7) in glucocorticoid receptor–mediated mRNA decay (GMD) manner. Mechanistically, LINC01569 directly bound to the GMD factor Y-box–binding protein 1 (YBX1). Then, the LINC01569-YBX1 complex was guided to the mRNAs of EGR1, CITED2, and BMP7 through specific LINC01569-mRNA interaction, thereby contributing to the successful assembly of GMD complex and triggering GMD. Our results uncovered roles of glucocorticoid in cellular mechanotransduction and novel lncRNA-dependent GMD machinery and provided potential strategy for early intervention in mechanical disorder–associated diseases.
INTRODUCTION
The extracellular matrix (ECM) or neighboring cells surrounding living cells result in their constant exposure to mechanical stimuli (1). Cells use mechanotransduction to sense their physical environment. Mechanotransduction is a process in which biochemical signals are translated from mechanical forces. Consequently, alterations to mechanotransduction can disturb cellular or extracellular signaling in response to surrounding mechanical stretch, which has been implicated in the development of a wide array of diseases, such as cancer progression, cardiomyopathies, fibrosis, and muscular dystrophies (2). Determining how to alter cellular mechanotransduction will help develop preventive and therapeutic interventions for a multitude of pathological conditions.
Glucocorticoid, a kind of steroid hormone in the body, exerts diverse physiological functions via the glucocorticoid receptor (GR), which is expressed in almost all tissues (3, 4). Traditionally, once GR is bound by glucocorticoid, it translocates to the nucleus and functions as a transcription activator or suppressor for glucocorticoid-responsive genes (5). Recently, Park et al. (6) and Cho et al. (7) reported that the glucocorticoid-GR system also plays a role in the degradation of a subset of mRNAs independently of a translation event, which was named as GR-mediated mRNA decay (GMD). Park et al. (8) further identified that proline-rich nuclear receptor coregulatory protein 2 (PNRC2), ATM-mediated phosphorylation of up-frameshift suppressor 1 homolog (UPF1), and two GMD-specific factors, heat-responsive protein 12 (HRSP12) and Y-box–binding protein 1 (YBX1), are required for GMD, but how GMD-related factors are recruited to target mRNAs has not been described. Emerging evidence indicates glucocorticoid as one of the most important gene regulatory molecules. However, whether glucocorticoid affects cellular mechanotransduction and the underlying mechanisms remain unknown.
Long noncoding RNAs (lncRNAs) are a class of transcripts without protein coding potential, which are described longer than 200 nucleotides. Previous studies have revealed that lncRNAs can directly interact with various biomolecules including DNA, mRNA, lncRNA, small noncoding RNAs, proteins, and so on (9). Functionally, lncRNAs are classified in four different categories according to modes of actions: signal lncRNAs, decoy lncRNAs, guide lncRNAs, and scaffold lncRNAs (10). lncRNAs are being increasingly recognized as key molecules with the potential to provide mechanistic insight into many uncharacterized aspects of cellular, physiological, and pathological processes. Gong and Maquat (11) first reported that lncRNAs duplexed with 3′ untranslated regions (UTRs) via Alu elements and triggered staufen1 (STAU1)–mediated mRNA decay (SMD) for a subset of targets, indicating important roles of lncRNAs in mRNA metabolism process. GMD is a recently found mRNA decay mode that is different from traditional SMD, replication-dependent histone mRNA decay, and nonsense-mediated mRNA decay (NMD) (7, 12). However, roles of lncRNAs in glucocorticoid-regulated gene transcription or the GMD machinery have not been elucidated until now.
Hypertrophic scars, which occur in approximately 70% of patients with burn, can severely affect the patient’s quality of life (13). Traditionally, hypertrophic scars are characterized by excessive deposition of ECM in appearance. However, emerging evidence indicates that intrinsic cellular mechanical stretch is a primary drive force of hypertrophic scars, making it a cell model to study mechanotransduction (14). In the present study, we used a mechanically stretched hypertrophic scar fibroblast (HSFB) model to study roles of glucocorticoid and related lncRNAs in mechanotransduction. We found that glucocorticoid counteracted the cellular mechanoresponses including increased proliferation, decreased apoptosis, and excessive matrix production through a LINC01569-dependent GMD pathway. Upon glucocorticoid treatments, LINC01569 was up-regulated and bound to the GMD factor YBX1. Then, YBX1 was guided to the mRNAs of early growth response protein 1 (EGR1), Cbp/P300-interacting transactivator 2 (CITED2), and bone morphogenic protein 7 (BMP7) in an lncRNA-mRNA sequence–specific binding manner, resulting in triggering GMD and counteraction of cellular mechanoresponses. These findings provide new insights into roles of glucocorticoid in regulating mechanotransduction and the mechanisms by which lncRNAs affect GMD complex assembly for their target genes. In addition, we provide evidence that LINC01569-dependent GMD system might have potential therapeutic benefits in the prevention and treatment of mechanical disorder diseases.
RESULTS
Glucocorticoid counteracts the cellular mechanoresponses of HSFBs
Glucocorticoid is considered an important hormone and is a widely used drug involved in body development, metabolism, and immunity. Unfortunately, whether glucocorticoid affects cellular mechanotransduction and mechanical responses has never been determined until now. Hypertrophic scars can be produced by applied mechanical stress to a healing wound in mice that never produce scars naturally (15). Emerging evidence shows that the mechanoresponsive properties of fibroblasts have an evolutionarily conserved function in hypertrophic scar formation (16). Therefore, we treated the mechanical stretched HSFB model with triamcinolone acetonide (TA), a long-acting glucocorticoid drug, to investigate the effects of glucocorticoid in cellular responses to mechanical stimuli. HSFBs isolated from human hypertrophic scars were cultured for in vitro mechanical stretching assays (Fig. 1A).
Fig. 1. Glucocorticoid counteracts mechanical stretch–induced phenotypic transition.
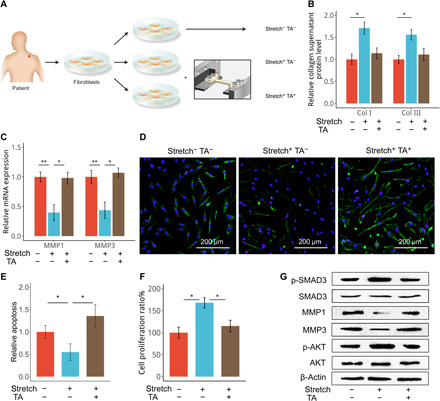
(A) Fibroblasts were isolated from hypertrophic scar tissues of patients and cultured in vitro. Cells were subjected to cyclic stretch for 48 hours and/or treated with TA (100 nM) as indicated. Cells cultured under static conditions served as a control. (B) Collagen I (Col I) and collagen III produced by cultured HSFBs were measured using enzyme-linked immunosorbent assay (ELISA). (C) Expression of matrix metalloproteinases (MMPs) by the cells described in (A) was determined using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). (D) Cells in (A) were subjected to terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) staining (green) with nuclei costained by 4′,6-diamidino-2-phenylindole (DAPI) (blue). (E) The relative apoptosis of cells in (A) were calculated. (F and G) Cells in (A) were subjected to the cholecystokinin 8 (CCK-8) assay (F) and Western blotting analysis (G). For (B), (C), (E), and (F), data are presented as means ± SD. *P < 0.05 and **P < 0.01, by analysis of variance (ANOVA) for more than two groups.
According to previously reports (17, 18), collagen synthesis, degradation, cell proliferation, and apoptosis are the molecular pathological mechanisms of scar formation; therefore, we analyzed these cellular biological events under mechanical stretch. As a result, stretch increased collagen production and down-regulated collagen-digesting enzymes, including matrix metalloproteinase 1 (MMP1) and MMP3, in the cultured HSFBs (Fig. 1, B and C). These effects were counteracted by treatment of cells with TA. Mechanical stretch also inhibited apoptosis and increased the proliferation of HSFBs, which were also relieved by TA treatment (Fig. 1, D to F, and fig. S1). In line with the changes of cellular behaviors, we observed that TA treatment reduced levels of MMPs and impaired the activation of the transforming growth factor–β pathway transcription factors SMAD family member 3 (SMAD3) and protein kinase B (PKB/AKT) (Fig. 1G), both of which are known members of intracellular mechanoresponsive pathways (19, 20). Thus, glucocorticoid treatment could counteract mechanical stretch–induced cellular responses including increased proliferation, decreased apoptosis, and excessive matrix production of HSFBs.
Counteraction of mechanoresponses by glucocorticoid involves down-regulation of mechanosensors in HSFBs
To investigate how glucocorticoid affects cellular responses of HSFBs to mechanical stretch and which genes and signaling pathways are involved, we profiled differentially expressed genes using a microarray analysis in stretched HSFBs treated with or without TA. TA treatment altered gene expression in HSFBs (Fig. 2A). We found the same altered expression of genes that were previously reported as glucocorticoid responsors, e.g., FKBP prolyl isomerase 5 (FKBP5) (21, 22), interleukin-8 (23), and the fos proto-oncogene (24) in the microarray. We further validated these genes using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis (fig. S2). This indicated that our microarray accurately represents the changes of gene expression after glucocorticoid treatment. Cytoscape analysis indicated that most TA-regulated genes were enriched in cell proliferation/apoptosis, responses to external stimulus or stress, and regulation of wound healing (Fig. 2B and fig. S3). Furthermore, gene ontology (GO) classification showed that the three most dysregulated biological processes were collagen synthesis, collagen degradation, and proliferation/apoptosis (Fig. 2C), all of which are involved in cellular mechanical responses of HSFBs. These bioinformatic data suggested that glucocorticoid affects cellular mechanical response–associated genes in HSFBs.
Fig. 2. Mechanosensors including EGR1, CITED2, and BMP7 participate in cellular mechanical responses.
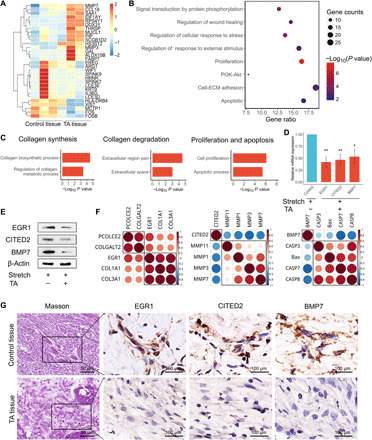
(A) HSFBs treated with TA were seeded in chambers and subjected to mechanical stretch. Paired HSFBs in the control groups were treated with solvent and subjected to mechanical stretch. Microarray analysis (GSE151240) of differentially expressed genes was performed. Red shading indicates increased gene expression, whereas blue shading indicates decreased expression. The mRNAs that were up-regulated by at least 2-fold (red bars) or down-regulated by at least 0.5-fold (blue bars) were included in the cluster analysis. (B) The bubble chart provides an overview of the most enriched pathways ranked as the top eight altered pathways according to Cytoscape analysis. Bubble size represents the number of genes in each pathway. The bubble color indicates the P value of each pathway. PI3K, phosphatidylinositol 3-kinase. (C) GO annotation using DAVID (Database for Annotation, Visualization, and Integrated Discovery) bioinformatic classification of gene categories from fig. S3. Biological process shows the signaling pathways that were most markedly changed by TA treatment of stretched HSFBs. (D and E) qRT-PCR (D) and Western blotting (E) analyses using HSFBs undergoing stretching and/or treatment with TA. The results of qRT-PCR were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression, and in the Western blotting analysis, β-actin was used as an internal loading control. Data are presented as means ± SD. *P < 0.05, by Student’s t test. (F) Correlation analysis of candidate genes involved in the indicated biological processes based on qRT-PCR assays of gene expression in (C). Red indicates positive correlations, and blue indicates negative correlations. The brightness is proportional to the strength of the correlation. (G) Masson’s trichrome staining for collagen using serial sections of clinical hypertrophic scar tissues before or after treatment with TA for 28 days. Immunohistochemical analysis of EGR1, CITED2, and BMP7 proteins in TA-treated hypertrophic scar tissues and paired control tissues is shown.
Given that cellular responses to mechanical stretch are dependent on mechanotransduction, which transforms physical cues into gene expression and signaling pathways, we paid more attention to candidate genes involved in mechanotransduction. Therefore, we selected three differentially expressed mechanosensor genes and further investigated, including EGR1 (25, 26), CITED2 (27), and BMP7 (28, 29), all of which act as mechanosensitive molecular switches and regulate transcription downstream of mechanical signals. qRT-PCR and Western blot assays verified that EGR1, CITED2, and BMP7 were significantly down-regulated by TA in stretched HSFBs (Fig. 2, D and E). Another glucocorticoid drug, dexamethasone (Dex), similarly impaired the mRNA and protein expression of these genes in stretched HSFBs (fig. S4, A and B).
In addition, previous studies reported that EGR1, CITED2, and BMP7 were key regulators in collagen synthesis (30, 31), matrix metabolism (27, 32–34), and apoptosis (35, 36), respectively, all of which are biological responses induced by mechanical stimulation in hypertrophic scar formation. Therefore, we analyzed the correlations between the three mechanosensors and the corresponding genes involved in collagen, matrix metabolism, and apoptosis. Quantitative analysis indicated that EGR1 expression correlated with collagen I and collagen III, CITED2 correlated with MMP1 and MMP3, and BMP7 correlated with apoptosis regulators Caspase-3, Caspase-7, and Caspase-8 and BCL2-associated X, apoptosis regulator (Bax) (Fig. 2F). Our data suggest that these documented mechanosensors (EGR1, CITED2, and BMP7) serve as downstream effectors of glucocorticoid to counteract cellular mechanical responses in HSFBs. Masson’s trichrome staining of hypertrophic scar tissues revealed that TA treatment restored regular and sparse distribution of collagens, which was accompanied by reduced protein and mRNA levels of EGR1, CITED2, and BMP7, as revealed by immunostaining (Fig. 2G) and in situ hybridization (ISH) (fig. S5). Therefore, down-regulation of mechanosensor proteins may be responsible for the counteraction of stretch-induced phenotypes of HSFBs by glucocorticoids.
LINC01569 mediates glucocorticoid-induced counteraction of cellular responses to mechanical stretch
Next, we sought to determine how TA affects mechanosensors in HSFBs under stretching environment. lncRNAs have emerged as an abundant and functionally diverse species of RNAs, which play important roles in regulating gene expression and are involved many biological processes (9, 10). Therefore, we screened for lncRNAs that participate in the regulation of mechanical stretched HSFBs upon TA treatment via expression profiles (Fig. 3A). qRT-PCR was used to verify that the expression levels of the lncRNAs were affected by TA treatment. lncRNA LINC01569, a validated ncRNA with gene ID (100507501 and NR_039999) in National Center for Biotechnology Information (NCBI), displayed the highest abundance among the lncRNAs altered by TA in stretched HSFBs (Fig. 3B). Consistently, Dex also significantly up-regulated LINC01569 in stretched HSFBs (fig. S6A). Further, LINC01569 expression was increased by both TA and Dex in HSFBs without mechanical stretch (fig. S6B). These results indicated that up-regulation of LINC01569 was dependent on glucocorticoid.
Fig. 3. LINC01569 is a candidate mediator of glucocorticoid-induced counteraction of cellular mechanical responses.
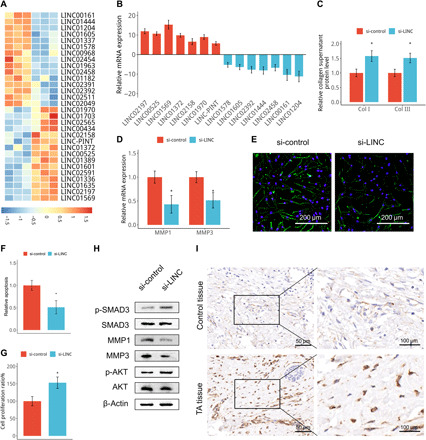
(A) Microarray analysis (GSE151153) of differentially expressed lncRNAs in TA-treated and untreated stretched HSFBs. Red shading indicates increased gene expression, and blue shading indicates decreased expression upon TA treatment. (B) lncRNA expression levels were assessed via qRT-PCR in TA-treated and untreated stretched HSFBs. Data were normalized to GAPDH expression. (C) HSFBs were transfected with scrambled (si-control) or a LINC01569-targeted small interfering RNA (siRNA) (si-LINC) and then subjected to TA and stretch treatment. The medium of cultured cells was harvested and analyzed using ELISA. (D to H) HSFBs were transfected and treated as described in (C), and cells were subjected to qRT-PCR for MMPs (D), TUNEL staining (green) for apoptosis assay (E and F), CCK-8 assay (G) and Western blotting analysis at 48 hours after transfection (H). (I) Sections of hypertrophic scar tissues treated with or without (control) TA were prepared and subjected to ISH for LINC01569. For (C), (D), (F), and (G), data are presented as means ± SD. *P < 0.05 and **P < 0.01, by Student’s t test.
Then, we hypothesized that LINC01569 may play important roles in glucocorticoid counteraction of mechanical responses. To investigate whether glucocorticoid counteraction of mechanical responses is attributed to LINC01569 up-regulation, we next performed a loss function assay of LINC01569 in HSFBs under TA and mechanical stretch treatment. In TA-treated and stretched HSFBs, knockdown of LINC01569 significantly promoted collagen synthesis and decreased MMP expression (Fig. 3, C and D, and fig. S7A); terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) staining and Western blotting of proapoptotic proteins including cleaved Caspase-3 and Bax showed that LINC01569 silencing inhibited apoptosis (Fig. 3, E and F, and fig. S7B). Cholecystokinin 8 (CCK-8) assay and EdU assay revealed that LINC01569 knockdown markedly rescue the proliferation impaired by TA in stretched HSFBs (Fig. 3G and fig. S7, C and D). These data suggested that LINC01569 up-regulation was required for glucocorticoid counteraction of mechanical responses including collagen synthesis, matrix metabolism, proliferation, and apoptosis. LINC01569 knockdown also significantly increased SMAD3 and AKT phosphorylation and impaired MMP expression in TA-treated and mechanically stretched HSFBs (Fig. 3H), suggesting that LINC01569 also mediated glucocorticoid regulation of signaling pathways in collagen synthesis, apoptosis, and matrix metabolism respectively involved in mechanical stretch. In addition, ISH analysis confirmed that LINC01569 expression was increased in hypertrophic scar tissues after TA treatment (Fig. 3I). Having demonstrated that glucocorticoid reversed cellular mechanoresponses of HSFBs by up-regulating LINC01569, we next investigated the LINC01569 expression in normal skin fibroblasts (NSFBs). LINC01569 expression was not statically different between NSFBs and HSFBs (fig. S8A). Mechanical stretch also did not change LINC01569 expression in NSFBs (fig. S8B). These results suggested that abnormal LINC01569 expression may not occur in hypertrophic scar formation. Together, our data revealed that LINC01569 is up-regulated by glucocorticoids and mediates glucocorticoid-induced repression of HSFBs responses to mechanical stretch.
LINC01569 mediates glucocorticoid effects on cellular mechanical responses via regulation of mechanosensors
Glucocorticoid down-regulates mechanosensors including EGR1, CITED2, and BMP7, all of which are important for cellular responses to mechanical stretch in HSFBs. In addition, the counteraction of cellular mechanical responses by glucocorticoid is LINC01569 dependent. Therefore, we hypothesized that LINC01569 might play a critical role in regulating EGR1, CITED2, or BMP7 expression in glucocorticoid-treated HSFBs under a stretch environment. To test this hypothesis, we investigated whether LINC01569 functions to down-regulate these mechanosensors in response to TA treatment in HSFBs. Actually, LINC01569 overexpression significantly reduced the levels of EGR1, CITED2, and BMP7, whereas knockdown of LINC01569 increased the expression of these genes in glucocorticoid (TA and Dex)–treated and stretched HSFBs (Fig. 4, A and B, and fig. S9). Consistently, the level of LINC01569 correlated inversely with those of the mechanosensors in hypertrophic scar tissues under TA treatment (Fig. 4C), which confirmed the regulation of mechanical sensors by LINC01569 under glucocorticoid treatment.
Fig. 4. LINC01569 mediates glucocorticoid repression of cellular mechanical responses via regulation of mechanosensors.
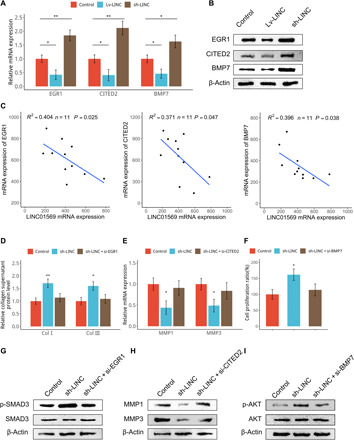
(A and B) HSFBs were infected with control lentiviruses or those expressing LINC01569 (Lv-LINC) or LINC01569-targeted short hairpin RNAs (shRNAs) (sh-LINC). Cells were subjected to mechanical TA and stretch treatment, followed by qRT-PCR (A) and Western blotting (B) analyses. (C) Hypertrophic scar tissues from patients were treated with TA and collected for qRT-PCR assay. The results were plotted and subjected to correlation analysis for the expression of the indicated genes. (D) HSFBs were infected with control or sh-LINC lentiviruses or further transfected with EGR1-targeted siRNAs. Collagen I and collagen III produced by cultured HSFBs were measured using ELISA. (E) HSFBs were infected with control or sh-LINC lentiviruses or further transfected with CITED2-targeted siRNAs. Expression of MMP1 and MMP3 were measured using qRT-PCR. (F) HSFBs were infected with control or sh-LINC lentiviruses or further transfected with BMP7-targeted siRNAs. Cells were then subjected to CCK-8 assays. (G to I) HSFBs were infected with control or sh-LINC lentiviruses and further transfected with the indicated siRNAs, followed by Western blotting analysis for indicated proteins. For (B), (E), and (F), data are presented as means ± SD. *P < 0.05 and **P < 0.01, by ANOVA for more than two groups. For (C), *P < 0.05, by Tukey’s post hoc test.
Given that EGR1, CITED2, and BMP7 play important roles in collagen synthesis, matrix metabolism and apoptosis, respectively, we wanted to know whether LINC01569 mediates glucocorticoid counteraction of cellular mechanical responses via these mechanosensors. In TA-treated and stretched HSFBs, silencing of LINC01569 promoted collagen synthesis; however, this was partially rescued by knockdown of EGR1 (Fig. 4D). Knockdown of LINC01569 decreased the mRNA levels of MMP1 and MMP3; however, this was partially rescued by further knockdown of CITED2 (Fig. 4E). LINC01569 depletion also promoted proliferation and suppressed apoptosis of HSFBs, which was partially rescued by knockdown of BMP7 (Fig. 4F and fig. S10). Knockdown efficacy of EGR1, CITED2, and BMP7 was showed in fig. S11. Furthermore, knockdown of LINC01569 reinforced SMAD3 signaling involved in collagen synthesis, repressed the expression of MMPs that participate in collagen degradation, and facilitated AKT activation involved in apoptosis and decreased proliferation, which were partially rescued by silencing EGR1 (Fig. 4G), CITED2 (Fig. 4H), or BMP7 (Fig. 4I), respectively. Together, these data provided further evidence that LINC01569 mediates glucocorticoid-induced mitigation of cellular mechanical responses via mechanosensors.
LINC01569 mediates glucocorticoid-induced destabilization of mechanosensor mRNAs through the GMD pathway
In Fig. 4, we demonstrated that LINC01569 mediates glucocorticoid regulation of mechanosensor expression, resulting in counteraction of cellular mechanical responses. GR is the unique GR that triggers glucocorticoid-induced signaling activation and biological effects. We next investigated whether LINC01569-mediated down-regulation of mechanosensors was dependent on GR. We observed that knockdown of GR in TA-treated stretched HSFBs significantly increased the mRNA levels of EGR1, CITED2, and BMP7 (Fig. 5A). However, knockdown or overexpression of LINC01569 in GR-silenced cells failed to affect the mRNA levels of these mechanosensors (Fig. 5A), suggesting that GR is required for LINC01569-mediated down-regulation of mechanosensors in TA-treated stretched HSFBs.
Fig. 5. LINC01569 mediates mechanosensor mRNA decay via GMD manner.
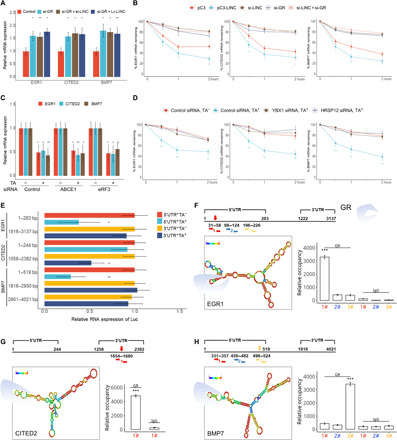
(A) Stretched HSFBs were treated with TA and transfected with control siRNAs or those targeting the indicated genes, followed by qRT-PCR. (B) Stretched HSFBs were transfected with a control (pC3) or LINC01569-overexpressing vector (pC3-LINC) and/or with the indicated siRNAs. Then, the cells were treated with actinomycin D (ACD) (5 μg/ml), and total cellular RNAs were extracted at the indicated time points for qRT-PCR to measure the half-lives of endogenous mechanosensors. The normalized level of each mechanosensor measured at 0 hours was set as 100%. The y axis represents a logarithmic scale of the levels of remaining mRNA (in percentage). (C) Stretched HSFBs were treated with or without TA and transfected with the indicated siRNAs, followed by qRT-PCR assay. (D) Stretched HSFBs were transfected with si-YBX1 or si-HRSP12 with or without TA treatment. The levels of remaining mRNA were analyzed as described in (B). (E) HSFBs were transfected with the luciferase (Luc) reporter constructs for the indicated mRNA regions of the mechanosensors. Cells were treated with or without TA and subjected to qRT-PCR analysis for luciferase mRNA levels. Data are normalized to GAPDH mRNA expression. (F to H) Top: The predicted RNA secondary structure of the hairpin formation in the indicated mRNAs. Red arrows indicate potential GR-binding regions. Bottom: The enrichment of GR-binding sequences on the indicated mRNAs, as assessed using PAR-CLIP (photoactivatable ribonucleoside–enhanced cross-linking and immunoprecipitation) quantitative PCR (qPCR). For (A) to (H), data are presented as means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001, by ANOVA for more than two groups.
First, we focused on GR function to investigate the down-regulation process of those mechanosensors mRNAs mediated by LINC01569. Traditionally, GR is considered to bind to glucocorticoid response elements (GREs) or negative GREs (nGREs) to regulate the transcription of target genes (37). Therefore, we scanned the promoter regions of the identified mechanosensors but found no GREs (5′-GGTACAnnnTGTTCT-3′) or nGREs (5′-CTCCn(0–2)GGAGA-3′). Previous studies report that GR also directly binds to a subset of mRNAs to affect their stability, acting as an RNA binding protein (38, 39). Therefore, we proposed that LINC01569 and GR might be involved in mRNA stability regulation of the mechanosensors. LINC01569 overexpression destabilized the mRNAs of the mechanosensors (Fig. 5B). Conversely, knockdown of either LINC01569 or GR stabilized these mRNAs, and combined LINC01569 and GR knockdown failed to further increase mRNA stability of the mechanosensors (Fig. 5B). These results suggested that both LINC01569 and GR participate in a process to destabilize the mRNAs of mechanosensors without synergistic effects.
How LINC01569 and GR affect mRNA stability of mechanosensors including EGR1, CITED2, and BMP7? Recently, Cho et al. (7) found that GR is involved in a newly described mRNA decay process named GMD. Thus, we hypothesized that destabilization of the mRNAs of the mechanosensors probably occurs in a GMD manner. To confirm our hypothesis, we investigated whether destabilization the mRNAs of EGR1, CITED2, and BMP7 has the characteristics of GMD. GMD is translation independent and relies on specific factors, including the RNA binding proteins YBX1 and HRSP12 (8), which is mechanistically distinct from NMD and SMD. Consistent with GMD, we found that knockdown of translation factors ABCE1 or eukaryotic release factor 3 (eRF3) had no significant effect on LINC01569-mediated mechanosensor mRNAs (Fig. 5C and fig. S12, A to C). However, knockdown of YBX1 and HRSP12 ablated TA-elicited decrease in the mRNA half-lives of EGR1, CITED2, and BMP7 (Fig. 5D and fig. S12, D and E), indicating that YBX1 and HRSP12 are required for LINC01569-mediated mRNA destabilization of the mechanosensors.
Besides the GMD characteristics described above, GR preloads on the UTRs before triggering mRNA degradation, which is important to elucidate GMD mechanisms for target mRNAs (7, 8). To identify the UTR sequences of the mechanosensors responsible for GMD, we first generated luciferase reporter constructs for different 5′UTR or 3′UTR regions of EGR1, CITED2, and BMP7. Introduction of these constructs into HSFBs showed that TA treatment reduced luciferase mRNA expression from the cassette containing specific 5′UTR segments of EGR1 or BMP7 and 3′UTR segments of CITED2 (Fig. 5E), suggesting that these UTR regions are critically involved in GMD of the mechanosensors. Next, we predicted the secondary structures of the UTR regions mentioned above by RNA-fold software. Ishmael et al. (39) first identified GR motif and the associated sequence and searched transcripts that bearing the GR motif sequence in the entire UniGene databases. On the basis of their identified GR motif, we searched potential GR-binding sequence against the corresponding UTR of EGR1, CITED2, and BMP7 by sequence alignment as described in Materials and Methods. We found that the EGR1, CITED2, and BMP7 candidate UTRs contained at least one region that could be potentially recognized and bound by GR (Fig. 5, F to H, top). The binding of GR to these UTR sequences was validated by PAR-CLIP [photoactivatable ribonucleoside–enhanced cross-linking and immunoprecipitation (IP)] combined with qRT-PCR (Fig. 5, F to H, bottom). Together, our results indicated that LINC01569 mediated the down-regulation of mechanosensor mRNAs through the GMD pathway.
LINC01569 directly interacts with YBX1 and promotes GMD of mechanosensors
Then, how do GR and LINC01569 work together to induce GMD of mechanosensors, including EGR1, CITED2, and BMP7 under glucocorticoid treatment? First, we investigated whether LINC01569 mediated GMD of mechanosensors via regulating or binding GR. We found that LINC01569 did not play a regulatory role in GR expression (fig. S13, A and B) nor did LINC01569 directly bind to GR (fig. S13C). Then, we investigated whether LINC01569 influences the enrichment of GR to the UTRs of target mechanosensors mRNA UTRs. We performed RNA IP (RIP) using an anti-GR antibody after ultraviolet (UV) cross-linking and found that knockdown of LINC01569 did not affect the enrichment of GR to the mRNA UTRs of mechanosensors UTR regions (Fig. 6A).
Fig. 6. LINC01569 functions in GMD through binding to YBX1.
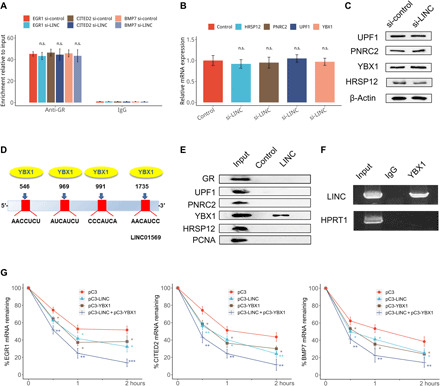
(A) Stretched HSFBs were treated with TA and transfected with the control or LINC01569 siRNAs. Forty-eight hours after transfection, cells were UV cross-linked, and the cytosolic fraction was isolated. IP was performed using an anti-GR antibody or a rabbit IgG as negative control. The immunoprecipitated RNA was extracted and subjected to reverse transcription. The mRNA levels of EGR1, CITED2, and BMP7 were evaluated. (B and C) Cells as described in (A) were subjected to qRT-PCR (B) and Western blotting (C) analyses. qRT-PCR data were normalized to GAPDH mRNA expression. (D) Schematic diagram of predicted YBX1 binding sites within the LINC01569 RNA. (E) Cell lysate of HSFBs in the absence of TA were subjected to an RNA pulldown assay using biotin-labeled LINC01569, followed by Western blotting analysis for the indicated proteins. Proliferating cell nuclear antigen (PCNA) was used as the negative control. (F) Cells as described in (A) were lysed and subjected to RIP using an anti-YBX1 antibody or IgG control. The precipitated RNAs were determined using qRT-PCR primers specific for LINC01569, with hypoxanthine-guanine phosphoribosyltransferase 1 (HPRT1) as the negative control. (G) HSFBs were transfected using empty vectors or those overexpressing LINC01569 and/or YBX1, followed by treatment with ACD (5 μg/ml). At the indicated time points, RNA was extracted, and mechanosensor expression was measured using qRT-PCR and normalized to the level of GAPDH. For (A), (B), and (G), data are presented as means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001, by Student’s t test for two groups or ANOVA for more than two groups. n.s., not significant.
Although GR is necessary for GMD of mechanosensors, LINC01569 is not directly associated with GR. Thus, we speculated that LINC01569 might regulate or interact with other GMD factors to mediate GMD of the mechanosensors. LINC01569 knockdown failed to affect the expression of GMD cofactors including UPF1, proline rich nuclear receptor coactivator 2 (PNRC2), YBX1, or HRSP12 (Fig. 6, B and C). Then, we want to know whether LINC01569 directly interacts with these GMD factors. To predict the physical interactions between LINC01569 and these factors, we searched for a common mRNA recognition motifs and putative protein-binding sites in LINC01569 using an online web server, RBPmap (22). Fortunately, we found that RNA binding protein YBX1 had an RNA signature motif that may directly bind to lncRNAs (fig. S13D). However, UPF1, PNRC2, and HRSP12, all of which own none RNA signature motif, were not included in the RBPmap database. In addition, LINC01569 was predicted to share several candidate putative binding sites for YBX1 (Fig. 6D). To validate the predicted interaction between LINC01569 and YBX1, we performed an RNA pulldown assay. Western blotting with an anti-YBX1 antibody confirmed the existence of YBX1 within pulldown samples of the LINC01569 probe (Fig. 6E). In contrast, GR, UPF1, PNRC2, HRSP12, and proliferating cell nuclear antigen (PCNA; a negative control) (40) were not associated with LINC01569. The results of RNA pulldown reflect RNA-protein interactions in vitro; therefore, we next performed an RIP assay to detect the direct interaction between YBX1 and LINC01569 in vivo in TA-treated and stretched HSFBs. As shown in Fig. 6F, LINC01569 was significantly enriched in samples immunoprecipitated using the anti-YBX1 antibody compared with samples immunoprecipitated using immunoglobulin G (IgG). The results demonstrate that LINC01569 directly binds with the YBX1 protein in vitro and in vivo but not with other GMD factors.
Furthermore, we analyzed both roles of LINC01569 and YBX1 in GMD of EGR1, CITED2, and BMP7. Data shown in Fig. 6G suggested that, consistently, overexpression of YBX1 or LINC01569 promoted the degradation of EGR1, CITED2, and BMP7 mRNA, which was further promoted by YBX1 and LINC01569 coexpression. Together, these results indicated that LINC01569 binds with YBX1, but not with other factors, to promote GMD of mechanosensors in glucocorticoid-treated HSFBs under mechanical stretch.
LINC01569 recruits YBX1 to target mechanosensors mRNAs through an lncRNA-mRNA interaction
We next sought to determine how the interaction between LINC01569 and YBX1 contributed to GMD of EGR1, CITED2, and BMP7. YBX1 is a key GMD factor, which binds to target mRNA/GR/UPF1-PNRC2 (GMD complex I) and completes the formation of GMD complex II, culminating in efficient mRNA degradation (8). However, how YBX1 is recruited to target mRNAs remains unknown. lncRNAs often serve as guide to recruit proteins to target DNA, mRNA, proteins, or other biomolecules, resulting in transcriptional or posttranscriptional gene regulation (41). We had the reason to speculate that LINC01569 might link YBX1 and target mRNAs to affect GMD cofactor assembly. To test this, we then performed RNA-IP assays in TA-treated stretched HSFBs and found that LINC01569 knockdown significantly impaired the enrichment of YBX1 on the mRNAs of EGR1, CITED2, and BMP7, but not on that of CXCL1 (C-X-C motif chemokine ligand 1), a proven positive binding partner of YBX1 (Fig. 7A) (42), thus sustaining the notion that LINC01569 mediates the recruitment of YBX1 to mRNAs of the mechanosensors.
Fig. 7. LINC01569 recruits YBX1 to target mRNAs in the GMD complex.
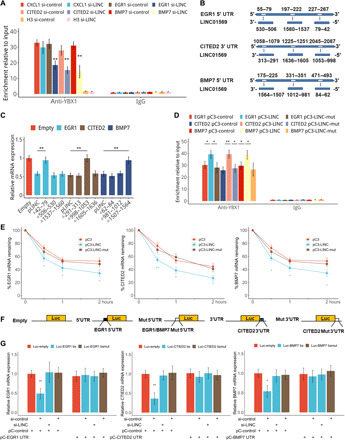
(A) Stretched HSFBs were treated with TA and transfected with the control or LINC01569 siRNAs. The cells were used in an IP experiment performed using an anti-YBX1 antibody (rabbit IgG as a negative control). EGR1, CITED2, and BMP7 mRNA levels were evaluated. (B) Schematic representation of the predicted binding sites of LINC01569 on the UTRs of EGR1, CITED2, and BMP7. (C) TA-treated and stretched HSFBs were transfected with empty vectors or those expressing wild-type or truncated LINC01569 and were further treated with ACD. The levels of indicated mRNAs were measured using qRT-PCR. (D) TA-treated and stretched HSFBs were transfected with the constructs for wild-type LINC01569 or LINC01569 harboring mutations (mut) in the sites responsible for binding to the corresponding mRNAs. RIP was performed using an anti-YBX1 antibody. The immunoprecipitated RNA was extracted and subjected to reverse transcription. (E) HSFBs were transfected with empty vectors or those overexpressing wild-type LINC01569 or LINC01569 harboring mutations in the mRNA binding sites, followed by treatment with ACD. The mRNAs of mechanosensors were measured using qRT-PCR. (F) Schematic representation of the UTR regions of the mechanosensors cloned into the luciferase plasmid. (G) Luciferase vectors containing wild-type or mutated LINC01569-binding sequences of EGR1, CITED2, and BMP7 UTRs and siRNAs were introduced into TA-treated and stretched HSFBs. pCDNA expressing EGR1, CITED2, and BMP7 UTRs were cotransfected as competitors on binding to LINC01569 with luciferase transcripts containing EGR1, CITED2, and BMP7 UTRs. Luciferase mRNA expression was determined by qPCR 48 hours after transfection. Data were normalized to GAPDH mRNA expression. For (A), (C), (D), (E), and (G), data are presented as means ± SD. *P < 0.05 and **P < 0.01, by Student’s t test for two groups or ANOVA for more than two groups.
We next explored the binding sites between LINC01569 and the target mRNAs of EGR1, CITED2, and BMP7. As demonstrated in Fig. 7B, the identified region in the 5′UTRs of EGR1 and BMP7 and the 3′-UTR of CITED2 was responsible for their TA-elicited GMD in HSFBs. Thus, from the regions above, we predicted the candidate-binding sequences for the interaction between LINC01569 and the target mRNA based on free energy calculations (Fig. 7B and see Supplementary Material Sequence).
Constructs for full-length and mutant LINC01569 were then generated and introduced into TA-treated and stretched HSFBs, followed by qRT-PCR assessment of the levels of the target mRNAs. As a result, we observed that the mutants lacking nucleotides 42 to 79, 998 to 1053, and 1507 to 1564 abrogated the ability of LINC01569 to mediate the down-regulation of EGR1, CITED2, and BMP7 mRNA, respectively, in TA-treated stretched HSFBs (Fig. 7C and see the Supplementary Materials). In addition, we validated that overexpression of full-length LINC01569, but not the aforementioned mutants, induced the enrichment of YBX1 on the mRNAs of the mechanosensors in stretched HSFBs (Fig. 7D), suggesting that these sequences in LINC01569 play critical roles in target mRNA recognition and YBX1 recruitment. Last, we assessed whether those biding sites are responsible for mRNA decay of EGR1, CITED2, and BMP7. Measurement of mRNA half-life indicated that overexpression of LINC01569 facilitated mRNA degradation of mechanosensors, whereas overexpression of the corresponding mutants of LINC01569 failed to have this effect (Fig. 7E). Overall, these results demonstrated that nucleotides 42 to 79, 998 to 1053, and 1057 to 1564 in LINC01569 were responsible for YBX1 recruitment to the target mRNAs and their decay.
To further verify the direct binding of LINC01569 to the mRNA UTRs of the mechanosensors, we generated luciferase reporter vectors for these UTRs and for those harboring mutations in the candidate LINC01569-binding sites (Fig. 7F). Compared with that of the control reporter construct, the ectopic expression of LINC01569 reduced luciferase mRNA from the EGR1, CITED2, and BMP7 UTR reporter constructs but not that from the mutant reporter constructs in TA-treated and stretched HSFBs. In addition, cotransfection of cells with plasmids expressing the corresponding UTRs of EGR1, CITED2, and BMP7 was able to restore luciferase mRNA expression from the corresponding wild-type UTR reporter constructs probably through competing with the luciferase transcript containing EGR1, CITED2, and BMP7 UTR in LINC01569 binding (Fig. 7G). These results demonstrated that LINC01569 recruited YBX1 to EGR1, CITED2, and BMP7 mRNAs and promoted their GMD in an lncRNA-mRNA sequence–specific binding manner. In conclusion, LINC01569 guided GMD cofactor YBX1 to the target mRNAs of mechanosensors, including EGR1, CITED2, and BMP7, to promote their GMD, resulting in counteraction of cellular responses to mechanical stretch under glucocorticoid treatment.
DISCUSSION
Aberrant cellular responses to mechanical stimuli underlie the pathology of many diseases, including myopathies, atherosclerosis, fibrosis, cancer, and some inherited diseases. However, little progress has been made in the development of preventions and treatments that target mechanotransduction pathways. Using mechanically stretched HSFBs as a cell model, we investigate whether and how glucocorticoid affected cellular mechanotransduction. We made the following primary observations: (i) We demonstrated that glucocorticoid counteracted the cellular mechanoresponses including increased proliferation, decreased apoptosis, and excessive matrix production in HSFBs; (ii) we identified the previously undescribed role of lncRNAs in GMD machinery, in which GMD of mechanosensors including EGR1, CITED2, and BMP7 were depended on the lncRNA LINC01569 in glucocorticoid-treated and stretched HSFBs; and (iii) our study elucidated how GMD factor YBX1 was recruited to target mRNA in GMD machinery. Mechanistically, LINC01569 directly interacts with YBX1 and guides it to the mRNAs of those mechanosensors through an lncRNA-mRNA interaction, thereby contributing to the successful assembly of the GMD complex and triggering GMD. In summary, our results showed that glucocorticoid counteracts cellular mechanoresponses via LINC01569-guided GMD of mechanosensors, providing a novel potential strategy to prevent or treat clinical disorders associated with mechanical stretch (Fig. 8).
Fig. 8. A schematic diagram showing that glucocorticoid counteracts mechanical cellular responses via LINC01569-guided GMD of mechanosensors.
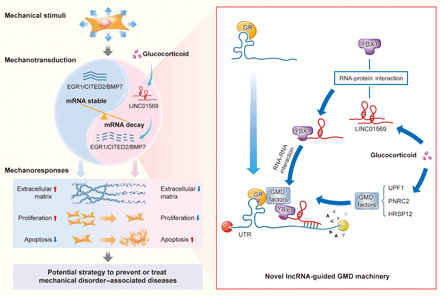
Mechanosensors including EGR1, CITED2, and BMP7 sense stretch and are stably expressed, leading to cellular mechanical responses of HSFBs, including increased collagen, increased proliferation, and decreased apoptosis. However, glucocorticoid up-regulates LINC01569, which mediates GMD of the mechanosensors, and counteracts the cellular mechanical responses of cells. Mechanistically, LINC01569 binds directly with the GMD protein factor YBX1 and guides it to the UTRs of EGR1, CITED2 and BMP7 mRNAs through lncRNA-mRNA interaction, thereby contributing to the successful assembly of the GMD complex that triggers GMD of the mechanosensors. Glucocorticoids and lncRNA-guided GMD system thus hold out promise for effective prevention and treatment of clinical disorders associated with mechanical stretch.
The importance of mechanical stimuli to control cell behaviors (e.g., migration, differentiation, and proliferation) in mechanical disorder diseases has only been acknowledged recently (43, 44). Unfortunately, the underlying mechanisms involved in mechanotransduction and how to target mechanical signaling cascades are largely unknown. Mechanical tension is associated with hypertrophic scar formation after trauma or burn (45). Even in murine wounds, which do not exhibit dermal scarring under normal circumstances, artificially applied mechanical stress is sufficient to produce excessive dermal scarring (46, 47). Emerging evidence indicates that mechanical stress drives hypertrophic scarring. Thus, stretched HSFBs are a good model to study the cellular mechanical responses and mechanotransduction. Glucocorticoid, a traditional anti-inflammatory drug, has not been reported to regulate cellular responses to mechanical stress. In this study, we describe a novel function of glucocorticoid in counteracting various mechanical responses, including increased collagen synthesis and cell proliferation, as well as decreased collagen degradation and cell apoptosis, by down-regulating a class of mechanosensors (EGR1, CITED2, and BMP7) and altering multiple pathways in hypertrophic scar cells. These findings indicated that glucocorticoid could be used in the attempt to prevent hypertrophic scar formation or other mechanical disorder–related diseases through suppressing the key mechanosensors.
Glucocorticoid hormones and their receptors (GRs) have been well established to orchestrate the transcription of a cohort of genes harboring GREs. In the present study, we found that the suppression of mechanosensors, including EGR1, CITED2, and BMP7, induced by glucocorticoid, is not dependent on GREs and nGREs but is dependent on LINC01569 and the GMD pathway. To the best of our knowledge, this is the first report to determine the biological roles of lncRNAs and GMD in mechanotransduction.
The GMD pathway shares protein factors, including UPF1 and PNRC2, with other pathways, such as NMD. However, unlike NMD, which requires an elongating ribosome to recognize a premature mRNA termination codon, GMD is translation independent and inducible by glucocorticoids. In GMD, GR is preloaded onto the target mRNA, and glucocorticoid binding initiates the recruitment of PNRC2 and UPF1. Recent studies by Park et al. (8) demonstrated that the active GMD complex also comprises HRSP12 and YBX1. HRSP12 is an endoribonuclease that contributes to the final cleavage of mRNAs, whereas the action mode of YBX1 in GMD remains unclear. In the present study, we found that LINC01569 binds directly with YBX1 but not with other GMD factors. This agreed with previous reports that YBX1, as a member of highly conserved double-stranded RNA binding protein family, dictates the stability of specific mRNAs via interaction with lncRNA(s) (48–50). Overexpression of LINC01569 or YBX1 promoted GMD of EGR1, CITED2, and BMP7, which was further accelerated by LINC01569 and YBX1 coexpression. Thus, the key role of lncRNAs in the GMD machinery was uncovered. However, further studies using a wider range of cell types are necessary to determine whether YBX1 and an adaptor lncRNA are fundamental components of the GMD complex.
Emerging studies have identified various lncRNA action modes, including interaction with RNA binding proteins, binding with mRNAs, scaffolding, repressing protein translation, and inhibiting microRNAs (miRNAs), in multiple biological events. lncRNA-mRNA interactions via base pairing are usually specific for target genes and have various consequences. For instance, lncRNAs termed half STAU1-binding site RNAs promote the decay of a series of specific target mRNAs (51). By contrast, lncRNA BACE1 antisense inhibits the degradation of its target transcript, BACE1 (52). We found that LINC01569 interacts with EGR1, CITED2, and BMP7 mRNA and promotes their degradation via GMD. Destruction of the lncRNA-mRNA interactions between LINC01569 and its target mRNA by mutating the binding sites repressed YBX1 recruitment to EGR1, CITED2, and BMP7 mRNA and resulted in impairment of GMD. Despite this finding, a question remains regarding how the specificity of GMD is determined. While GR interacts with a large quantity of mRNAs more frequently than with those subjected to degradation, we established here that GMD targets specific genes (EGR1, CITED2, and BMP7) as a result of LINC01569-YBX1 and LINC01569-mRNA interactions. LINC01569 can bind specifically to target mRNAs that were previously preloaded with GR; therefore, it can bind synchronously with YBX1, thus recruiting YBX1 and promoting the GMD of specific mechanosensors. These findings provide novel insights into the precision and efficiency of the GMD machinery.
In summary, our findings are of translational significance because they demonstrated that glucocorticoid could counteract cellular mechanoresponses by destabilizing mechanosensors including EGR1, CITED2, and BMP7 in HSFBs model. Mechanical stretch not only induces hypertrophic scarring but also induces lung fibrosis, hepatic fibrosis, heart fibrosis, and tubule interstitial fibrosis. Our findings suggested glucocorticoid and LINC01569-GMD as a novel strategy to prevent and treat mechanical disorder diseases. Our findings are also scientifically significant, because we not only revealed the critical role of lncRNAs in GMD machinery but also characterized GMD as an accurate mRNA decay mode. GMD accuracy relies on three specific interactions: the GR-mRNA interaction, the LINC01569-YBX1 interaction, and the LINC01569-mRNA interaction. However, further investigations are still required to understand the precise role of other molecules in GMD machinery, which will help develop preventions and treatments for mechanical stress–related diseases by targeting GMD efficiently.
MATERIALS AND METHODS
Patients and samples
This study was registered retrospectively in the Chinese clinical trial registry (registration number: ChiCTR-OOC-17011482). Ethical approval was obtained from the Ethics Committee of the First Affiliated Hospital of Fourth Military Medical University (registration number: KY20173012-1). Our protocol included patients aged 18 to 65 years with a scar on any body site that was present for less than 2 years. Patients were given local injections of glucocorticoid TA in the scar for more than 3 months. Patients who had not received TA treatment, or other drugs or lasers, were enrolled as controls. Hypertrophic scar tissues were collected in Xijing Hospital, which is affiliated with Fourth Military Medical University in Xi’an, China. The collected skin samples were preserved in 4% paraformaldehyde solution for ISH and immunohistochemistry.
Cell culture and stretch protocol
To separate fibroblasts, portions of wild-type hypertrophic dermal scars were minced and incubated in a solution of collagenase type I (0.1 mg/ml) (Novoprotein, Shanghai, China) for 3 hours at 37°C. The reaction mixtures were centrifuged, and the pelleted fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum (Gibco, Grand Island, NY, USA), streptomycin (100 U/ml), and penicillin (100 U/ml) in a 5% (v/v) CO2-humidified atmosphere at 37°C, according to our previously report (53). Subsequent experiments used cells between passages 3 and 6. The STB-140 STREX cell stretch system (STREX Co., Osaka, Japan) was used to subject cells to a uniaxial cyclic stretch force. Then, 2 ml of type I collagen (150 μg/ml; Atelo Cell KOKEN Co., Tokyo, Japan) was used to coat silicon resin chambers (STB-CH-10.0, STREX Co.), which were then dried overnight on a clean bench. The collagen-coated chambers (area, 10.24 cm2) were seeded with HSFBs at 1.5 × 106 cells and incubated for 24 hours in medium containing 10% fetal bovine serum (FBS). The medium was then replaced with fresh medium containing 5% exosome-depleted FBS. In the experimental groups, cells were subjected to cyclic stretch 20% elongation at 10 cycles/min for 48 hours (54). In the control groups, cells were cultured for 24 hours without cyclic stretch. Both groups of cells were grown in a humidified atmosphere (5% CO2). Thereafter, the silicon resin chambers were centrifuged at 1500 rpm for 15 min at 4°C, and the supernatant containing the cells was retained. TA or Dex at 100 nM was used to treat selected groups of cells. Paired HSFBs in the control groups were treated with solvent. The cells and their culture media were harvested, and the RNA and protein levels were analyzed at the indicated time points.
Microarray assay
HSFBs treated with TA were seeded in the chambers and subjected to mechanical stretch. Paired HSFBs in the control groups were treated with solvent and subjected to mechanical stretch. The RNAs of cells were extracted using TRIzol (Invitrogen, Waltham, MA, USA) and purified using the RNeasy Mini Kit (QIAGEN, Düsseldorf, Germany), according to the manufacturer’s instructions. RNAs were analyzed for gene expression using the PrimeView human gene expression array (Affymetrix, at Thermo Fisher Scientific, Waltham, MA, USA). lncRNA expression profiles were evaluated using a customized Agilent human microarray (OE Biotech human ceRNA microarray, design ID:086188, Agilent, Santa Clara, CA, USA). Gene expression profile and lncRNA expression data that support the findings of this study have been deposited in Gene Expression Omnibus (GEO) under the accession numbers of GSE151240 and GSE151153.
Bioinformatic analysis
The sequence of LINC01569 was downloaded from NCBI (gene ID: 100507501), from which a 2093–base pair (bp) sequence was extracted. The ClueGO + CluePedia tool in Cytoscape was used to perform cluster-based analysis of the biological processes dysregulated in TA-treated stretched HSFBs (55). The bubble chart is a scatter plot with an overview of the pathway categories in Cytoscape generated using the ggplot2 package in the R language. To further analyze functional categories, GO was implemented at the DAVID (Database for Annotation, Visualization, and Integrated Discovery) bioinformatic resources website (https://david.ncifcrf.gov/) (56). Subsequently, to analyze functional categories and the association between genes and the corresponding GO classifications, the R visualization package GOPlot was used to obtain a better visualization of the relationships between genes and the selected functional categories (57). The sequence regions were predicted at NCBI, and primer premier 5.0 software (PREMIER Biosoft International, San Francisco, CA, USA) was used to design primers to amplify the predicted regions for DNA fragment cloning and chromatin IP PCR. Ishmael et al. (39) first identified the GR-mRNA binding motif sequence and searched putative GR mRNA targets in the entire UniGene database. On the basis of their identified GR motif, we searched potential GR-binding sequence against the corresponding UTR of EGR1, CITED2, and BMP7 by sequence alignment via the COVE and COVELS software packages (58). RNA secondary structure prediction was conducted using online software (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) for hairpin formation in the UTR sequences of EGR1, CITED2, and BMP7 (59). The binding sites of RNA binding proteins on LINC01569 were predicted using the RBPmap website (http://rbpmap.technion.ac.il/) (60). The sites involved in the interactions between LINC01569 and the UTRs were predicted using the RNA-RNA binding prediction online software (http://rna.informatik.uni-freiburg.de/IntaRNA) with free energy calculation and potential interaction binding sequences (61).
Reagents
Glucocorticoid TA (CAS#: BP339), actinomycin D (ACD) (CAS#:129935), and Dex (CAS#:4902) were obtained from Sigma-Aldrich (St Louis, MO, USA).
Luciferase assay
Luciferase reporter constructs containing nucleotides 1 to 283 bp of the EGR1 5′UTR and 1916 to 3137 bp of the EGR1 3′UTR, 1 to 244 bp of the CITED2 5′UTR and 1058 to 2382 bp of the CITED2 3′UTR, 1 to 519 bp of the BMP7 5′UTR, 1816 to 2950 BMP7 3′UTR, and 2901 to 4021 BMP7 3′UTR, wild-type 5′UTR of the EGR1 5′UTR and mutants of 227 to 267 bp in the EGR1 5′ UTR binding site, wild-type 5′UTR of the CITED2 3′UTR and mutants of 2045 to 2087 bp in the CITED2 3′UTR binding site, and wild-type 5′UTR of the BMP7 5′UTR and mutants of 175 to 225 bp in the BMP7 5′ UTR binding site were cloned into PGL4-RLuc luciferase vector (Promega, Madison, WI, USA) or pMIR-REPORT firefly luciferase vector (Ambion, Austin, TX, USA). In luciferase reporter experiments, HSFBs were transfected with 400 ng of luciferase plasmid (containing the wild-type UTR binding site) or mutation luciferase plasmid (containing the corresponding UTR mutant) in a final volume of 0.2 ml using Lipofectamine 2000 (Invitrogen). At the same time, control or experiment vector/regents were cotransfected into HSFBs and treated with TA and stretch. The mRNA expression of luciferase was determined by qRT-PCR. The primers used for fragment cloning and PCR were listed in table S1.
PAR-CLIP quantitative PCR analysis
PAR-CLIP analysis was used to examine whether GR recognizes motif sequences in RNAs and affects the fate of the target transcript in specific directions (62, 63). In vitro, HSFBs were grown for 16 hours in medium containing 100 μM 4-thiouridine (Abcam) and washed with phosphate-buffered saline (PBS). To cross-link RNA to GR, a Spectrolinker XL-1500 UV cross-linker (Spectronics, Westbury, NY, USA) was used to irradiate the cells with 365-nm UV light (150 mJ/cm2). TA-treated stretched HSFBs were incubated for 16 hours in 200 μM 4-thiouridine. Then, to cross-link RNA to endogenous GR, the Spectrolinker XL-1500 UV cross-linker was used to irradiate the cells with 365-nm UV light (200 mJ/cm2). The cells were then collected, and three pellet volumes of NP-40 lysis buffer were used to lyse the cells. Tissue or cleared cell lysates were treated with RNase T1 (ribonuclease T1) (1 U/μl; Thermo Fisher Scientific, Fremont, CA, USA), and then a polyclonal anti-GR antibody (Thermo Fisher Scientific) attached to protein A/G Dynabeads was used to immunoprecipitate the endogenous GR protein. The RNA in the immunoprecipitates was then trimmed using RNase T1 (100 U/ml). Lysis buffer was used to wash the beads, and then deoxyribonuclease I (Zymo Research Corporation, Orange, CA, USA) and proteinase K (0.2 mg/ml; Thermo Fisher Scientific) were used to remove the DNA and proteins, respectively. Acidic phenol/chloroform extraction and ethanol precipitation were used to recover the RNA, which was then subjected to PCR analysis. Table S1 shows the primers used in PAR-CLIP quantitative PCR (qPCR).
Cell transfection
The jetPRIME reagent (Polyplus-transfection, Illkirch, France) was used for small interfering RNA (siRNA) transfection or DNA cotransfection (64). The final concentration of the siRNAs was 50 nM according to the manufacturer’s instruction for transfection. At 24 hours after transfection, the cell medium was changed, and at 48 hours after transfection, the cells were lysed. Polyethylenimide (PEI) (Polysciences Inc. Warrington, PA, USA) was used at a ratio of 1:2 (DNA:PEI) to perform DNA transfection. Serum-free DMEM, PEI, and plasmid DNA (10 to 15 μg for 10-cm dishes and 2 to 3 μg per well for six-well plates) were mixed, incubated for at room temperature for 10 min, and then added to the cells. For the luciferase reporter experiments, TA-treated HSFBs or untreated cells were grown in 48-well plates to 80 to 90% confluence in DMEM with 10% FBS. TA-treated and stretched HSFBs were cotransfected with 400 ng of luciferase reporter vectors for either the 3′UTR or 5′UTR of the mechanosensor. Cells were then subjected to qRT-PCR to measure mRNA levels (65).
Quantitative real-time reverse transcription polymerase chain reaction
Total cellular RNA was isolated from HSFBs using the TRIzol reagent (Invitrogen), and then random hexamer primers and RevertAid M-MuLV reverse transcriptase (Thermo Fisher Scientific) converted the RNA into complementary DNA (cDNA). The cDNA was subjected to qRT-PCR using gene-specific primers, and the SYBR green I master mix (FOREGENE, Chengdu, China) on the StepOnePlus System (Applied Biosystems, Foster City, CA, USA) was run in triplicate. Last, the relative quantities of miRNA and mRNA were calculated using the 2−ΔΔCT method after normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. The qRT-PCR primers are listed in table S1.
Measurement of mRNA half-life
Oligofectamine was used to transfect TA-treated stretched HSFBs with 100 nM siRNAs. Three days after transfection, the cells were treated with ACD (5 μg/ml) for the indicated times. The cells were then collected for total RNA extraction using the TRIzol reagent (Life Technologies) at the indicated time points. The levels of specific mRNAs were then assessed using qRT-PCR (66).
Oligonucleotides and constructs for gene knockdown and overexpression
siRNA oligonucleotides (listed in table S1) were synthesized by GenePharma (Shanghai, China) (67). The cDNAs of LINC01569 were synthesized and cloned into the pCDNA3 vector (Invitrogen, Waltham, MA, USA). LINC01569 mutation sequence fragments containing the putative binding site from the UTR were chemical synthesized and cloned into the pCDNA3 vector. The YBX1 cDNA was PCR amplified from the cDNA of HSFBs using primers shown in table S1 and then cloned into the pCDNA3 vector for stable expression. EGR1, CITED2, and BMP7 3′UTR or 5′UTR fragments were synthesized and cloned into the pCDNA3.1 vector. All of the constructs were named on the basis of the location of the promoter fragments relative to the 5′ UTR.
Construction and packaging of lentivirus vectors
The cDNA of full-length human LINC01569 was synthesized and ligated into the pMD-18 T vector (Takara, Dalian, China). DNA sequencing was used to confirm the construct. Next, the LINC01569 sequence was subcloned into pLenti-6.3, a lentivirus-based expression plasmid (Invitrogen). The lentiviral construct was then packaged using human embryonic kidney (HEK) 293T cells (American Type Culture Collection, Manassas, VA, USA). The complementary oligonucleotides coding LINC01569-targeted short hairpin RNA (shRNA) were synthesized. Oligonucleotides in annealing buffer were heated to 95°C for 5 min and then allowed to cool to room temperature, thus generating double-stranded DNA (dsDNA). The dsDNA oligonucleotides were ligated into pLKO.1, a lentiviral vector (plasmid 30323, Addgene, Cambridge, MA, USA). DNA sequencing was used to confirm the constructs. Constructed lentivirus plasmids were transiently transfected into HEK293T cells for packaging. Forty-eight hours later, the medium was collected and centrifuged at 3000 rpm for 20 min to purify the lentiviruses, which were then further evaluated for their multiplicity of infection. The recombinant lentiviruses were used to infect HSFBs according to the manufacturer’s instructions (68). The shRNA oligo sequences are listed in table S1.
RNA IP
TA-treated and stretched HSFBs were cross-linked and resuspended in cold PBS. For each sample, 50 million cells were fractionated into nuclear and cytoplasmic fractions using Nuclei EZ Lysis Buffer (Sigma-Aldrich). The volume of the cytoplasmic fraction was adjusted using 2× polysome buffer [20 mM tris-Cl (pH 7.4), protease inhibitor cocktail, SUPERase-In, 2 mM dithiothreitol, 1% Triton X-100, 2.5 mM MgCl2, and 200 mM NaCl]. Yeast RNA (0.1 mg/ml) and recombinant protein G agarose (Thermo Fisher Scientific) were used to preclear the lysates at 4°C for 30 min. One percent of the sample was reserved as the input. The rest of the sample was incubated with anti-GR or anti-YBX1 antibodies or rabbit serum IgG (I5006, Sigma-Aldrich) at 4°C overnight, followed by incubation with protein G Dynabeads (Invitrogen) to collect the RNA-antibody complexes. The samples were then subjected to four washes before treatment with proteinase K. RNA was then extracted and subjected to qRT-PCR analysis (8). Primers used for RIP are listed in table S1.
RNA pulldown assay
RNA pulldown was performed using the Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. LINC01569 was transcribed from the pC3-LINC01569 expression plasmid and then subjected to biotin-labeling using the Pierce RNA 3′End Desthiobiotinylation Kit (Thermo Fisher Scientific). Total cell lysates of HSFBs (without any treatments) were incubated with 50 pmol of the purified biotinylated transcripts at 4°C for 1 hour with rotation. Then, streptavidin-coupled T1 beads (Dynabeads) were used to retrieve the RNA-protein complexes, which were then rinsed five times using IP buffer, and eluted using Laemmli buffer. Western blotting assay for identifying the RNA interacted protein was performed lastly (69).
In situ hybridization
RNA hybridization was performed on thin (approximately 4 mm in thickness) tissue sections as described previously (70). Slides were rinsed twice in PBS and then dehydrated using a gradient series of ethanol (70, 90, and 100%). Slides were then incubated with oligonucleotide probes (1 to 2.5 nM of each oligonucleotide) in hybridization buffer [2× SSC, yeast transfer RNA (1 mg/ml), 10% dextran sulfate, and 25% formamide]. The samples were placed in a humidified chamber, and hybridization was conducted by incubation at 42°C for 3 to 4 hours. Specific ISH signals were identified as brown, punctate dots. A Nikon 90i microscope (Nikon, Tokyo, Japan) with a 60×, 1.4–numerical aperture VC objective lens, an Orca ER camera (Hamamatsu, Hamamatsu City, Japan), and Volocity software (PerkinElmer, Waltham, MA, USA) was used to perform the imaging. Slides were examined for ISH staining in morphologically intact cells and scored manually. The Primer3 website (https://primer3plus.com/) was used to design oligonucleotide probes recognizing LINC01569, EGR1, CITED2, and BMP7 and was selected manually. Single-stranded oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA, USA) and pooled, and amine-ddUTP was added using terminal transferase (New England Biolabs, Ipswich, MA, USA). The oligonucleotides were purified via phenol extraction and ethanol precipitation and were resuspended in 0.1 M sodium borate. N-hydroxysuccinimide–Cy3B (GE Healthcare, Chicago, IL, USA) or Alexa647 (Thermo Fisher Scientific) were used for dye conjugation overnight at room temperature. The dye-conjugated oligonucleotides were precipitated using ethanol, resuspended in diethyl pyrocarbonate–H2O, and then their quality was checked using a NanoDrop instrument (NanoDrop Technologies, Wilmington, DE, USA).
Immunohistochemistry
Paraffin-embedded TA-treated hypertrophic scar tissue and control tissue samples were immunostained with EGR1-, CITED2-, and BMP7-specific antibodies after deparaffinization and hydration. Immunohistochemical signal was amplified using a horseradish peroxidase (HRP) kit, and color was developed using a DAB substrate kit (PV-6000 and ZLI-9032, Beijing Zhongshan Golden Bridge Biotechnology Co. Ltd., Beijing, China). The immunostained sections were reviewed by a pathologist under a light microscope, and the MetaMorph image analysis software (Universal Imaging Corp., CA, USA) was used to quantify the staining results.
Western blotting analysis
HSFBs lysates were prepared, and total proteins (50 μg) were separated using SDS–polyacrylamide gel electrophoresis on 10% polyacrylamide gels and then transferred onto polyvinylidene fluoride (PVDF) membranes (Genergy Bio-Technology Co. Ltd., Shanghai, China). The PVDF membranes were then blocked using nonfat dry milk before incubation with primary antibodies (a 1:800 dilution). After washing, the blots were incubated with the appropriate HRP-conjugated anti-rabbit IgG secondary antibody (1:2000; Novoprotein, Shanghai, China). The membranes were then stripped and probed again using primary anti–β-actin antibodies. Antibodies recognizing the following proteins or peptides were used: EGR1 (1:500; #ab133695, Abcam, Cambridge, UK), CITED2 (1:400; #ab184145, Abcam), BMP7 (1:500; #ab129156, Abcam), GR (1:400; #ab3579, Abcam for Western blot; 1:100; #ab2768, Abcam for PAR-CLIP), UPF1 (1:200; #ab133564, Abcam), PNRC2 (1:100; #ab102068, Abcam), YBX1 (1:200; #D2A11, Cell Signaling Technology, Danvers, MA, USA; 1:200; #ab76149, Abcam for Western blot), ABCE1 (1:1000; #ab185548, Abcam), eRF3 (1:300; #14980, Cell Signaling Technology), HRSP12 (1:300;#PA531352, Thermo Fisher Scientific), cleaved Caspase-3 (1:500; #AB3623, Millipore Sigma, St. Louis, MO, USA), Bax (1:1000; #50599-2-Ig, Proteintech, Rosemont, IL, USA), PKB (AKT) (1:300; #ab179463, Abcam), p-AKT (1:400; #ab38449, Abcam), SMAD3 (1:300; #ab40854, Abcam), p-SMAD3 (1:250; #ab63403, Abcam), MMP1 (1:400; #ab137332, Abcam), MMP3 (1:300; #ab52915, Abcam), PCNA (1:300; #ab92552, Abcam), histone H3 (1:100; #ab8580, Abcam), and β-actin (#ab8227; 1:1500, Abcam).
Enzyme-linked immunosorbent assays
The levels of collagen I and collagen III secreted to the coculture medium were measured using enzyme-linked immunosorbent assay (ELISA) kits (#EKC33229 and #EKU03319, Biomatikt, Wilmington, DE, USA) according to the manufacturer’s instructions. The competitive ELISA procedure for collagen I and collagen III was as follows. A hundred microliter of biotinylated peptide was incubated in streptavidin-coated plates at 20°C for 30 min with shaking. The plates were then rinsed five times using wash buffer [20 mM tris and 50 mM NaCl (pH 7.2)]. Next, 20 μl of the sample, standard, or control was added to the wells, followed immediately by the addition of 100 μl of HRP-labeled monoclonal antibody and incubation at 20°C for 1 hour with shaking. Thereafter, the plates were rinsed five times using wash buffer. 3,3′,5,5′-Tetramethylbenzidine (100 μl) was then added and incubated for 15 min at 20°C in the dark. To stop the enzyme reaction of TMB, sulfuric acid (100 μl; 0.1%) was added, and then plate was placed in the ELISA reader and analyzed at 450 nm, with 650 nm as the reference. A four-parametric mathematical fit model was used to plot a standard curve. To monitor interassay variation, each ELISA plate incorporated kit controls. All the samples were quantified within the assay’s range. All samples with a value below the level of the lower limit of quantification were assigned the LLQQ value.
Cell proliferation and apoptosis assay
The EdU cell proliferation assay kit (CA1170, Solarbio, Beijing, China) was used for HSFB proliferation assay. Briefly, cells were incubated in 24-well plates with 50-μm EdU labeling medium at 37°C for 12 hours. After immobilization, coupled with 4′,6-diamidino-2-phenylindole (DAPI) staining, cells were observed under the fluorescence microscope. More than six random fields per well were captured, and the corresponding cells were counted. The CCK-8 assay (CK04, Dojindo Kumamoto, Japan) was used to assay cell proliferation. In 96-well plates, cells were seeded at 5 × 103 cells per well. A spectrophotometric plate reader set at 450 nm was used to measure cell viability. To count cells, the cells were seeded at day 0 and digested at day 7. Thereafter, we counted the live cells. The TUNEL assay using a Clik-iT TUNEL kit (Thermo Fisher Scientific) was performed to assess apoptosis according to the manufacturer’s protocol. Cells were costained with DAPI to visualize the nuclei.
Statistical analysis
Statistical analyses were performed with the R project for statistical computing software (https://r-project.org/, R version 3.5.3). All numerical data are presented as means ± SD from three or more independent replicates. Differences between two groups were assessed using a two-tailed Student’s t test. One-way analysis of variance (ANOVA) with Tukey’s posttest was used to evaluate data among more than two groups. The Mann-Whitney nonparametric test was used to evaluate associations between the product and the group parameters via univariate analysis. Statistical significance was accepted at P < 0.05.
Acknowledgments
We thank Y. Li for pathological diagnosis. Funding: This work was supported by the National Natural Science Foundation of China (81702554 to Y.L., 81971834 to J.L., 81802661 to W.B, and 81772071 to D.H.). Author contributions: H.Z. and W.B. conceived and designed the studies. Y.L. and J.L. performed the majority of the experiments. Z.Z., H.G., and H.W. performed the microarray assay. K.T. and J.L. performed histological and immunocytochemistry analyses. Y.W. and W.Z. performed plasmid constructs and luciferase activity assays. C.L. and J.L. gave technical support and conceptual advice. L.J. and D.H. analyzed the data and wrote the manuscript. All authors critically reviewed and approved the final manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Microarray data have been deposited in the GEO under accession numbers GSE151240 and GSE151153. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/9/eabd9923/DC1
REFERENCES AND NOTES
- 1.Cheng B., Wan W.-T., Huang G.-Y., Li Y.-H., Genin G. M., Mofrad M. R. K., Lu T.-J., Xu F., Lin M., Nanoscale integrin cluster dynamics controls cellular mechanosensing via FAKY397 phosphorylation. Sci. Adv. 6, eaax1909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyazaki T., Zhao Z., Ichihara Y., Yoshino D., Imamura T., Sawada K., Hayano S., Kamioka H., Mori S., Hirata H., Araki K., Kawauchi K., Shigemoto K., Tanaka S., Bonewald L. F., Honda H., Shinohara M., Nagao M., Ogata T., Harada I., Sawada Y., Mechanical regulation of bone homeostasis through p130Cas-mediated alleviation of NF-κB activity. Sci. Adv. 5, eaau7802 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kertser A., Baruch K., Deczkowska A., Weiner A., Croese T., Kenigsbuch M., Cooper I., Tsoory M., Ben-Hamo S., Amit I., Schwartz M., Corticosteroid signaling at the brain-immune interface impedes coping with severe psychological stress. Sci. Adv. 5, eaav4111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J.-C., Li J.-B., Cui X.-B., Li W.-B., Xue Y.-R., Shang P., Zhang H., Blocking glucocorticoid signaling in osteoblasts and osteocytes prevents mechanical unloading-induced cortical bone loss. Bone 130, 115108 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Weikum E. R., Knuesel M. T., Ortlund E. A., Yamamoto K. R., Glucocorticoid receptor control of transcription: Precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol. 18, 159–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park O. H., Do E., Kim Y. K., A new function of glucocorticoid receptor: Regulation of mRNA stability. BMB Rep. 48, 367–368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho H., Park O. H., Park J., Ryu I., Kim J., Ko J., Kim Y. K., Glucocorticoid receptor interacts with PNRC2 in a ligand-dependent manner to recruit UPF1 for rapid mRNA degradation. Proc. Natl. Acad. Sci. U.S.A. 112, E1540–E1549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park O. H., Park J., Yu M., An H.-T., Ko J., Kim Y. K., Identification and molecular characterization of cellular factors required for glucocorticoid receptor-mediated mRNA decay. Genes Dev. 30, 2093–2105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu L., Lou F., Sun Y., Sun L., Cai X., Liu Z., Zhou H., Wang H., Wang Z., Bai J., Yin Q., Zhang J., Chen L., Peng D., Xu Z., Gao Y., Tang S., Fan L., Wang H., A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci. Adv. 6, eaaz2059 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Täuber H., Hüttelmaier S., Köhn M., POLIII-derived non-coding RNAs acting as scaffolds and decoys. J. Mol. Cell Biol. 11, 880–885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong C.-G., Maquat L. E., lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470, 284–288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong K., Ryu I., Park J., Hwang H. J., Ha H., Park Y., Oh S. T., Kim Y. K., Staufen1 and UPF1 exert opposite actions on the replacement of the nuclear cap-binding complex by eIF4E at the 5′ end of mRNAs. Nucleic Acids Res. 47, 9313–9328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coentro J. Q., Pugliese E., Hanley G., Raghunath M., Zeugolis D. I., Current and upcoming therapies to modulate skin scarring and fibrosis. Adv. Drug Deliv. Rev. 146, 37–59 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Aarabi S., Bhatt K. A., Shi Y., Paterno J., Chang E. I., Loh S. A., Holmes J. W., Longaker M. T., Yee H., Gurtner G. C., Mechanical load initiates hypertrophic scar formation through decreased cellular apoptosis. FASEB J. 21, 3250–3261 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Dohi T., Padmanabhan J., Akaishi S., Than P. A., Terashima M., Matsumoto N. N., Ogawa R., Gurtner G. C., The interplay of mechanical stress, strain, and stiffness at the keloid periphery correlates with increased caveolin-1/ROCK signaling and scar progression. Plast. Reconstr. Surg. 144, 58e–67e (2019). [DOI] [PubMed] [Google Scholar]
- 16.Derderian C. A., Bastidas N., Lerman O. Z., Bhatt K. A., Lin S.-E., Voss J., Holmes J. W., Levine L. P., Gurtner G. C., Mechanical strain alters gene expression in an in vitro model of hypertrophic scarring. Ann. Plast. Surg. 55, 69–75 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Ku C.-H., Johnson P. H., Batten P., Sarathchandra P., Chambers R. C., Taylor P. M., Yacoub M. H., Chester A. H., Collagen synthesis by mesenchymal stem cells and aortic valve interstitial cells in response to mechanical stretch. Cardiovasc. Res. 71, 548–556 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Lehoux S., Tedgui A., All strain, no gain: Stretch keeps proliferation at bay via the NF-κB response gene iex-1. Circ. Res. 93, 1139–1141 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Subramanian A., Kanzaki L. F., Galloway J. L., Schilling T. F., Mechanical force regulates tendon extracellular matrix organization and tenocyte morphogenesis through TGFbeta signaling. eLife 7, e38069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borreguero-Muñoz N., Fletcher G. C., Aguilar-Aragon M., Elbediwy A., Vincent-Mistiaen Z. I., Thompson B. J., The Hippo pathway integrates PI3K-Akt signals with mechanical and polarity cues to control tissue growth. PLOS Biol. 17, e3000509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinars C. R., Cheung-Flynn J., Rimerman R. A., Scammell J. G., Smith D. F., Clardy J., Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a component of steroid receptor complexes. Proc. Natl. Acad. Sci. U.S.A. 100, 868–873 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bali U., Phillips T., Hunt H., Unitt J., FKBP5 mRNA expression is a biomarker for GR antagonism. J. Clin. Endocrinol. Metab. 101, 4305–4312 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Beaulieu E., Ngo D., Santos L., Yang Y.-H., Smith M., Jorgensen C., Escriou V., Scherman D., Courties G., Apparailly F., Morand E. F., Glucocorticoid-induced leucine zipper is an endogenous antiinflammatory mediator in arthritis. Arthritis Rheum. 62, 2651–2661 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Chen D. W.-C., Saha V., Liu J.-Z., Schwartz J.-M., Krstic-Demonacos M., Erg and AP-1 as determinants of glucocorticoid response in acute lymphoblastic leukemia. Oncogene 32, 3039–3048 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Herchenhan A., Dietrich-Zagonel F., Schjerling P., Kjaer M., Eliasson P., Early growth response genes increases rapidly after mechanical overloading and unloading in tendon constructs. J. Orthop. Res. 38, 173–181 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Gaut L., Robert N., Delalande A., Bonnin M.-A., Pichon C., Duprez D., EGR1 regulates transcription downstream of mechanical signals during tendon formation and healing. PLOS ONE 11, e0166237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun H. B., CITED2 mechanoregulation of matrix metalloproteinases. Ann. N. Y. Acad. Sci. 1192, 429–436 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Santos A., Bakker A. D., Willems H. M. E., Bravenboer N., Bronckers A. L. J. J., Klein-Nulend L., Mechanical loading stimulates BMP7, but not BMP2, production by osteocytes. Calcif. Tissue Int. 89, 318–326 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Miyashita S., Ahmed N. E. M. B., Murakami M., Iohara K., Yamamoto T., Horibe H., Kurita K., Takano-Yamamoto T., Nakashima M., Mechanical forces induce odontoblastic differentiation of mesenchymal stem cells on three-dimensional biomimetic scaffolds. J. Tissue Eng. Regen. Med. 11, 434–446 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Wu X.-Q., Cheng J.-Z., Li P., Yang M., Qiu S.-L., Liu P.-Q., Du J., Mechano-sensitive transcriptional factor Egr-1 regulates insulin-like growth factor-1 receptor expression and contributes to neointima formation in vein grafts. Arterioscler. Thromb. Vasc. Biol. 30, 471–476 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Hsieh Y.-P., Chen H.-M., Lin H.-Y., Yang H., Chang J. Z.-C., Epigallocatechin-3-gallate inhibits transforming-growth-factor-β1-induced collagen synthesis by suppressing early growth response-1 in human buccal mucosal fibroblasts. J. Formos. Med. Assoc. 116, 107–113 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Wang P., Yang L., You X., Singh G. K., Zhang L., Yan Y., Sung K. L. P., Mechanical stretch regulates the expression of matrix metalloproteinase in rheumatoid arthritis fibroblast-like synoviocytes. Connect. Tissue Res. 50, 98–109 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Yokota H., Goldring M. B., Sun H.-B., CITED2-mediated regulation of MMP-1 and MMP-13 in human chondrocytes under flow shear. J. Biol. Chem. 278, 47275–47280 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Kirwan R. P., Fenerty C. H., Crean J., Wordinger R. J., Clark A. F., O’Brien C. J., Influence of cyclical mechanical strain on extracellular matrix gene expression in human lamina cribrosa cells in vitro. Mol. Vis. 11, 798–810 (2005). [PubMed] [Google Scholar]
- 35.Wang N., Lin K. K., Lu Z., Lam K. S., Newton R., Xu X., Yu Z., Gill G. N., Andersen B., The LIM-only factor LMO4 regulates expression of the BMP7 gene through an HDAC2-dependent mechanism, and controls cell proliferation and apoptosis of mammary epithelial cells. Oncogene 26, 6431–6441 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Gautschi O. P., Cadosch D., Zellweger R., Joesbury K. A., Filgueira L., Apoptosis induction and reduced proliferation in human osteoblasts by rhBMP-2, -4 and -7. J. Musculoskelet. Neuronal Interact. 9, 53–60 (2009). [PubMed] [Google Scholar]
- 37.Surjit M., Ganti K. P., Mukherji A., Ye T., Hua G.-Q., Metzger D., Li M., Chambon P., Widespread negative response elements mediate direct repression by agonist liganded glucocorticoid receptor. Cell 145, 224–241 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Dhawan L., Liu B., Blaxall B. C., Taubman M. B., A novel role for the glucocorticoid receptor in the regulation of monocyte chemoattractant protein-1 mRNA stability. J. Biol. Chem. 282, 10146–10152 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Ishmael F. T., Fang X., Houser K. R., Pearce K., Abdelmohsen K., Zhan M., Gorospe M., Stellato C., The human glucocorticoid receptor as an RNA-binding protein: Global analysis of glucocorticoid receptor-associated transcripts and identification of a target RNA motif. J. Immunol. 186, 1189–1198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L., Yang Z.-H., Trottier J., Barbier O., Wang L., Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate Shp mRNA decay. Hepatology 65, 604–615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battistelli C., Sabarese G., Santangelo L., Montaldo C., Gonzalez F. J., Tripodi M., Cicchini C., The lncRNA HOTAIR transcription is controlled by HNF4α-induced chromatin topology modulation. Cell Death Differ. 26, 890–901 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon E., Todorova K., Wang J., Horos R., Lee K. K., Neel V. A., Negri G. L., Sorensen P. H., Lee S. W., Hentze M. W., Mandinova A., The RNA-binding protein YBX1 regulates epidermal progenitors at a posttranscriptional level. Nat. Commun. 9, 1734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaudhuri P. K., Low B. C., Lim C. T., Mechanobiology of tumor growth. Chem. Rev. 118, 6499–6515 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Feng Y., Tian X.-Y., Sun P., Cheng Z.-P., Shi R.-F., Simultaneous study of mechanical stretch-induced cell proliferation and apoptosis on C2C12 myoblasts. Cells Tissues Organs 205, 189–196 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Burke M., Scars. Can they be minimised? Aust. Fam. Physician 27, 275–278 (1998). [PubMed] [Google Scholar]
- 46.Martin P., Parkhurst S. M., Parallels between tissue repair and embryo morphogenesis. Development 131, 3021–3034 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Colwell A. S., Longaker M. T., Lorenz H. P., Fetal wound healing. Front. Biosci. 8, s1240–s1248 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Chen C. Y., Gherzi R., Andersen J. S., Gaietta G., Jürchott K., Royer H. D., Mann M., Karin M., Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 14, 1236–1248 (2000). [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang E., He X., Zhang C., Su J., Lu X., Si X., Chen J., Yin D., Han L., De W., A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 19, 154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng Z., Wang J., Shan B., Li B., Peng W., Dong Y., Shi W., Zhao W., He D., Duan M., Cheng Y., Zhang C., Duan C., The long noncoding RNA LINC00312 induces lung adenocarcinoma migration and vasculogenic mimicry through directly binding YBX1. Mol. Cancer 17, 167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J., Gong C., Maquat L. E., Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 27, 793–804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu T., Huang Y., Chen J., Chi H., Yu Z., Wang J., Chen C., Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1 AS expression. Mol. Med. Rep. 10, 1275–1281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu H.-Y., Bai W.-D., Wang H.-T., Xie S.-T., Tao K., Su L.-L., Liu J.-Q., Yang X.-K., Li J., Wang Y.-C., He T., Han J.-T., Hu D.-H., Peroxisome proliferator-activated receptor-γ agonist inhibits collagen synthesis in human keloid fibroblasts by suppression of early growth response-1 expression through upregulation of miR-543 expression. Am. J. Cancer Res. 6, 1358–1370 (2016). [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z., Maruyama K., Sakisaka Y., Suzuki S., Tada H., Suto M., Saito M., Yamada S., Nemoto E., Cyclic stretch force induces periodontal ligament cells to secrete exosomes that suppress IL-1β production through the inhibition of the NF-κB signaling pathway in macrophages. Front. Immunol. 10, 1310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang D.-W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walter W., Sánchez-Cabo F., Ricote M., GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 31, 2912–2914 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Eddy S. R., Durbin R., RNA sequence analysis using covariance models. Nucleic Acids Res. 22, 2079–2088 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gruber A. R., Bernhart S. H., Lorenz R., The ViennaRNA web services. Methods Mol. Biol. 1269, 307–326 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Paz I., Kosti I., Jr M. A., Cline M., Mandel-Gutfreund Y., RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 42, W361–W367 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mann M., Wright P. R., Backofen R., IntaRNA 2.0: Enhanced and customizable prediction of RNA–RNA interactions. Nucleic Acids Res. 45, W435–W439 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoon J.-H., De S., Srikantan S., Abdelmohsen K., Grammatikakis I., Kim J., Kim K. M., Noh J. H., White E. J., Martindale J. L., Yang X., Kang M.-J., Wood W. H. III, Noren H. N., Evans M. K., Becker K. G., Tripathi V., Prasanth K. V., Wilson G. M., Tuschl T., Ingolia N. T., Hafner M., Gorospe M., PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat. Commun. 5, 5248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moon B.-S., Bai J., Cai M., Liu C., Shi J., Lu W., Kruppel-like factor 4-dependent Staufen1-mediated mRNA decay regulates cortical neurogenesis. Nat. Commun. 9, 401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schopp I. M., Ramirez C. C. A., Debeljak J., Kreibich E., Skribbe M., Wild K., Béthune J., Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 8, 15690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sarrion-Perdigones A., Chang L., Gonzalez Y., Gallego-Flores T., Young D. W., Venken K. J. T., Examining multiple cellular pathways at once using multiplex hextuple luciferase assaying. Nat. Commun. 10, 5710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Damas N. D., Marcatti M., Côme C., Christensen L. L., Nielsen M. M., Baumgartner R., Gylling H. M., Maglieri G., Rundsten C. F., Seemann S. E., Rapin N., Thézenas S., Vang S., Ørntoft T., Andersen C. L., Pedersen J. S., Lund A. H., SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat. Commun. 7, 13875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu H.-Y., Li C., Bai W.-D., Su L.-L., Liu J.-Q., Li Y., Shi J.-H., Cai W.-X., Bai X.-Z., Jia Y.-H., Zhao B., Wu X., Li J., Hu D.-H., MicroRNA-21 regulates htert via pten in hypertrophic scar fibroblasts. PLOS ONE 9, e97114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang X., Zhang X., Hu S., Zheng M., Zhang J., Zhao J., Zhang X., Yan B., Jia L., Zhao J., Wu K., Yang A., Zhang R., Identification of miRNA-7 by genome-wide analysis as a critical sensitizer for TRAIL-induced apoptosis in glioblastoma cells. Nucleic Acids Res. 45, 5930–5944 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan X., Banerjee P., Guo H.-F., Ireland S., Pankova D., Ahn Y.-H., Nikolaidis I. M., Liu X., Zhao Y., Xue Y., Burns A. R., Roybal J., Gibbons D. L., Zal T., Creighton C. J., Ungar D., Wang Y., Kurie J. M., Epithelial-to-mesenchymal transition drives a pro-metastatic Golgi compaction process through scaffolding protein PAQR11. J. Clin. Invest. 127, 117–131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mao C., Lee M. Y., Jhan J.-R., Halpern A. R., Woodworth M. A., Glaser A. K., Chozinski T. J., Shin L., Pippin J. W., Shankland S. J., Liu J. T. C., Vaughan J. C., Feature-rich covalent stains for super-resolution and cleared tissue fluorescence microscopy. Sci. Adv. 6, eaba4542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carvalho C., L’Hôte V., Courbeyrette R., Kratassiouk G., Pinna G., Cintrat J.-C., Denby-Wilkes C., Derbois C., Olaso R., Deleuze J.-F., Mann C., Thuret J.-Y., Glucocorticoids delay RAF-induced senescence promoted by EGR1. J. Cell Sci. 132, jcs230748 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Shin D. H., Li S. H., Chun Y.-S., Huang L. E., Kim M.-S., Park J.-W., CITED2 mediates the paradoxical responses of HIF-1α to proteasome inhibition. Oncogene 27, 1939–1944 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Ohigashi M., Kobara M., Takahashi T., Toba H., Wada T., Nakata T., Pitavastatin suppresses hyperglycaemia-induced podocyte injury via bone morphogenetic protein-7 preservation. Clin. Exp. Pharmacol. Physiol. 44, 378–385 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Cho H., Kim K. M., Kim Y. K., Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol. Cell 33, 75–86 (2009). [DOI] [PubMed] [Google Scholar]
- 75.Trenkmann M., Brock M., Gay R. E., Michel B. A., Gay S., Huber L. C., Tumor necrosis factor α-induced microRNA-18a activates rheumatoid arthritis synovial fibroblasts through a feedback loop in NF-κB signaling. Arthritis Rheum. 65, 916–927 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Chu C., Liu X., Bai X., Zhao T., Wang M., Xu R., Li M., Hu Y., Li W., Yang L., Qin Y., Yang M., Yan C., Zhang Y., MiR-519d suppresses breast cancer tumorigenesis and metastasis via targeting MMP3. Int. J. Biol. Sci. 14, 228–236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poulain M., Frydman N., Duquenne C., N’Tumba-Byn T., Benachi A., Habert R., Rouiller-Fabre V., Livera G., Dexamethasone induces germ cell apoptosis in the human fetal ovary. J. Clin. Endocrinol. Metab. 97, E1890–E1897 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Fang Y.-Y., Li D., Cao C., Li C.-Y., Li T.-T., Glucocorticoid receptor repression mediated by BRCA1 inactivation in ovarian cancer. BMC Cancer 14, 188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L., Zhang M., Liu D.-X., Knock-down of ABCE1 gene induces G1/S arrest in human oral cancer cells. Int. J. Clin. Exp. Pathol. 7, 5495–5504 (2014). [PMC free article] [PubMed] [Google Scholar]
- 80.Malta-Vacas J., Aires C., Costa P., Conde A. R., Ramos S., Martins A. P., Monteiro C., Brito M., Differential expression of the eukaryotic release factor 3 (eRF3/GSPT1) according to gastric cancer histological types. J. Clin. Pathol. 58, 621–625 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li D., Chen T., Hu Y., Zhou Y., Liu Q., Zhou D., Jin X., Huang Z., An Ebola virus-like particle-based reporter system enables evaluation of antiviral drugs in vivo under non-biosafety level 4 conditions. J. Virol. 90, 8720–8728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crawford N. T., McIntyre A. J., McCormick A., D’Costa Z. C., Buckley N. E., Mullan P. B., TBX2 interacts with heterochromatin protein 1 to recruit a novel repression complex to EGR1-targeted promoters to drive the proliferation of breast cancer cells. Oncogene 38, 5971–5986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shin S.-H., Lee G. Y., Lee M., Kang J., Shin H.-W., Chun Y.-S., Park J.-W., Aberrant expression of CITED2 promotes prostate cancer metastasis by activating the nucleolin-AKT pathway. Nat. Commun. 9, 4113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li X., Chen T., Shi Q., Li J., Cai S., Zhou P., Zhong Y., Yao L., Angiopoietin-like 4 enhances metastasis and inhibits apoptosis via inducing bone morphogenetic protein 7 in colorectal cancer cells. Biochem. Biophys. Res. Commun. 467, 128–134 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Hashimoto Y., Hosoda N., Datta P., Alnemri E. S., Hoshino S.-I., Translation termination factor eRF3 is targeted for caspase-mediated proteolytic cleavage and degradation during DNA damage-induced apoptosis. Apoptosis 17, 1287–1299 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/9/eabd9923/DC1


