Abstract
We sought to determine whether donor-derived human herpesvirus (HHV) 6B–specific CD4+ T-cell abundance is correlated with HHV-6B detection after allogeneic hematopoietic cell transplantation. We identified 33 patients who received HLA-matched, non–T-cell–depleted, myeloablative allogeneic hematopoietic cell transplantation and underwent weekly plasma polymerase chain reaction testing for HHV-6B for 100 days thereafter. We tested donor peripheral blood mononuclear cells for HHV-6B–specific CD4+ T cells. Patients with HHV-6B detection above the median peak viral load (200 copies/mL) received approximately 10-fold fewer donor-derived total or HHV-6B–specific CD4+ T cells than those with peak HHV-6B detection at ≤200 copies/mL or with no HHV-6B detection. These data suggest the importance of donor-derived immunity for controlling HHV-6B reactivation.
Keywords: human herpesvirus 6, HHV-6, CD4+ T cell, hematopoietic cell transplant
We demonstrated that a higher number of donor-derived total and human herpesvirus (HHV) 6B–specific CD4+ T cells may reduce the risk for higher-level HHV-6B detection after allogeneic hematopoietic cell transplantation.
Reactivation of latent viruses poses a risk for treatment-related morbidity after allogeneic hematopoietic cell transplantation (aHCT) [1]. Human herpesvirus 6B (HHV-6B) is a ubiquitous virus that reactivates after aHCT in approximately 40% of patients. HHV-6B is the most common cause of post–hematopoietic cell transplantation (HCT) encephalitis and is associated with other complications [2]. HHV-6B typically reactivates within weeks after aHCT, when patients are still lymphopenic, suggesting the importance of virus-specific T-cell immunity.
CD4+ T-cell responses are known to be critical for control of herpesvirus infections [3]. After aHCT, this has been well characterized for cytomegalovirus (CMV), a virus closely related to HHV-6B. HCT recipients with CMV-seronegative donors have increased risk for CMV reactivation and delayed CMV-specific CD4+ T-cell reconstitution [4, 5]. Given that non–T-cell–depleted donor hematopoietic cell products contain donor-derived memory T cells in addition to stem cells, donor-derived virus-specific cells have a potential role in protection against infections after transplantation. Conversely, patients receiving umbilical cord blood stem cell products, which lack transferred memory T cells, have the highest risk for virus reactivation [1].
The relationship between HHV-6B–specific CD4+ T cells and HHV-6B detection after aHCT has not been previously studied. These data are relevant to ongoing efforts to develop immunotherapeutic treatments, such as virus-specific T cells, for HHV-6B and other viruses given toxic effects or the absence of currently available therapies [6, 7]. In the current study, we investigated whether donor-derived HHV-6B–specific CD4+ T-cell abundance and functionality is associated with the incidence and magnitude of HHV-6B detection after aHCT. We hypothesized that transfer of higher numbers of more polyfunctional virus-specific T cells would confer protection against HHV-6B detection.
METHODS
Study Participants
We identified patients who underwent HLA-matched (10 of 10 match for HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1), non–T-cell–depleted (whether in vitro or in vivo via ATG or alemtuzumab), non–cord blood, myeloablative aHCT who underwent weekly quantitative plasma polymerase chain reaction (PCR) testing for HHV-6B in plasma or serum for the first 100 days after HCT. Among these patients, we identified those for whom we had cryopreserved donor-derived peripheral blood mononuclear cells (PBMCs) and who had ≥5 HHV-6B PCR tests, with ≥2 samples obtained after HCT and before day 30. Patients who died before day 100 were included if they had ≥1 specimen obtained before day 30 and if the last specimen was obtained ≤30 days before their death. Patients in whom the donor or recipient had inherited chromosomally integrated HHV-6 were excluded based on previously described screening [8]. This study was approved by the Fred Hutchinson Cancer Research Center institutional review board. All participants provided informed consent in accordance with the Declaration of Helsinki.
HHV-6B Antigen Preparation
HHV-6B strain Z29 and control antigens were prepared as detailed in the Supplementary Material. HHV-6B Z29 was grown in SupT1 cells (National Institutes of Health AIDS Reagent Program) with Roswell Park Memorial Institute 1640 medium supplemented with 10% fetal calf serum, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L L-glutamine. Whole HHV-6B antigen was prepared by collecting Z29-SupT1 cells at 4+ cytopathic effect, sonicating, and UV irradiation for 30 minutes. Control preparations used uninfected SupT1 cells.
HHV-6B PCR
We used a quantitative PCR assay to detect HHV-6 with a lower level of quantitation of 50 copies/mL, as described elsewhere [9]. We used an additional PCR assay to distinguish between HHV-6 species A and B in approximately half the samples, as described elsewhere [9]. Given our findings and given that most HHV-6 reactivation is with HHV-6B [1], we refer to HHV-6 results as “HHV-6B” throughout.
Intracellular Cytokine Staining
HHV-6B specificity was tested by intracellular cytokine staining (ICS), as described elsewhere [10] and detailed in the Supplementary Material. Briefly, ex vivo PBMCs were thawed, counted, and incubated for 18 hours with mock or HHV-6B antigen (1:100 dilution), or 1.6 µg/mL phytohemagglutinin (PHA) positive control. Cells were permeabilized, fixed, and stained with fluorescently labeled antibodies to CD3, CD4, CD154, interferon γ, tumor necrosis factor α, and interleukin 2. Generally, each run included 8 samples corresponding to an equal proportion of aHCT recipients who did or did not have HHV-6B detection (operator blinded), as well as PBMCs from a healthy control with reactivity to HHV-6B. Data were analyzed using flow cytometry.
Statistical Analyses
For our principle measure, we defined HHV-6B–specific CD4+ T cells in donor PBMCs based on positivity for CD3, CD4, and ≥3 of the 4 activation markers. We had data for the total number of infused donor hematopoietic cells but not the composition of specific leukocyte subsets. To estimate absolute counts of donor-derived CD4+ T cells, we assumed that they accounted for 5% and 16% of the donor hematopoietic cell product obtained from bone marrow or granulocyte colony-stimulating factor (G-CSF)–mobilized peripheral blood stem cell (PBSC) sources, respectively [11]. We used Wilcoxon rank sum tests to compare the estimated numbers of donor-derived total and HHV-6B–specific CD4+ T cells infused per kilogram in patients with subsequent HHV-6B detection above the median peak viral load (200 copies/mL) with the numbers in patients with peak HHV-6B detection at ≤200 copies/mL or with no HHV-6B detection. We generated cumulative incidence curves of HHV-6B reactivation (at any level) after aHCT, stratified by the estimated infused donor HHV-6B–specific CD4+ T-cell counts. Curves were compared using log-rank tests.
In addition, we analyzed ICS data using the COMPASS package (RStudio) for combinatorial polyfunctionality analysis of antigen-specific T-cell subsets (described elsewhere [12]). The COMPASS package evaluates all possible functional T-cell subsets. The final readout is a single “polyfunctionality score” that summarizes the posterior probability of CD4+ T-cell antigen specificity based on distributions of T cells expressing all combinations of activation markers. A higher score indicates higher specific functionality. We used the Wilcoxon rank sum test to compare the polyfunctionality score between patients with HHV-6B detection above the median peak viral load (>200 copies/mL) compared with those with HHV-6B detection at ≤200 copies/mL or with no detection. SAS 9.43 software for Windows (SAS Institute) was used for analyses.
RESULTS
We identified 39 patients undergoing aHCT between 1998 and 2008 who met study criteria. For the subsequent analyses, three patients were excluded owing to poor live PBMC recovery (<1 × 104 live CD3+CD4+ cells in either mock- or HHV-6B–stimulated assays, or both), and three patients were excluded owing to <1% of CD4+ T cells responding to PHA (Supplementary Table 1). The final cohort of 33 patients is described in Table 1; characteristics of contemporaneous patients excluded owing to lack of HHV-6 testing or PBMC samples were similar (Supplementary Table 2). Ten patients (30%) had plasma HHV-6B detection after aHCT with a median peak viral load of 200 copies/mL (range, 57–3032 copies/mL). Patients with higher HHV-6B viral loads had more severe acute graft-vs-host disease. No patients had objective evidence of HHV-6B–associated end-organ disease.
Table 1.
Patient Demographic and Clinical Characteristics
| Characteristic | Patients, No. (%)a | P Valueb | ||
|---|---|---|---|---|
| No HHV-6B Detection (n = 23) | HHV-6B Detected at ≤200 Copies/mL (n = 5) | HHV-6B Detected at >200 Copies/mL (n = 5) | ||
| Donor age, median (range), y | 50 (10–63) | 49 (25–53) | 34 (22–42) | .08 |
| Patient age, median (range), y | 48 (40–55) | 51 (27–56) | 38 (17–42) | .08 |
| Male donor | 11 (48) | 4 (80) | 2 (40) | .66 |
| Male recipient | 14 (60) | 4 (80) | 3 (60) | >.99 |
| Related donor | 21 (91) | 5 (100) | 4 (80) | .4 |
| Hematopoietic cell source | ||||
| Peripheral blood stem cells | 17 (74) | 4 (80) | 1 (20) | .03 |
| Bone marrow | 6 (26) | 1 (20) | 4 (80) | |
| Year of transplantation, range | 1998–2008 | 1998–2007 | 1998–2007 | .07 |
| Donor or recipient CMV seropositive | 13 (57) | 3 (60) | 3 (60) | >.99 |
| Recipient with any CMV detection | 8 (35) | 1 (20) | 3 (60) | .33 |
| Underlying disease | ||||
| Acute leukemia | 11 (48) | 3 (60) | 0 (0) | .04 |
| Chronic leukemia | 5 (22) | 0 (0) | 3 (60) | |
| Other | 7 (30) | 2 (40) | 2 (40) | |
| Peak HHV-6B, median (range), copies/mL | 0 | 100 (57–150) | 550 (250–3032) | .008 |
| Acute graft-vs-host diseasec | ||||
| Grade 0 | 3 (13) | 1 (20) | 0 (0) | .86 |
| Grade 1–2 | 14 (61) | 3 (60) | 3 (60) | |
| Grade 3–4 | 6 (26) | 1 (20) | 2 (40) |
Abbreviations: CMV, cytomegalovirus; HHV, human herpesvirus.
aData represent no. (%) unless otherwise indicated. All patients underwent HLA-matched, non–T-cell–depleted, non–cord blood, myeloablative hematopoietic cell transplantation. HHV-6 species testing was performed in 4 of 10 patients and detected HHV-6 species B in all instances. The cutoff value of 200 copies/mL was the median peak viral load.
b P values determined with Fisher exact test for categorical variables or Wilcoxon rank sum test for continuous variables as appropriate, comparing the group with peak HHV-6B detection above the median (>200 copies/mL) with the other 2 detection groups.
cProphylaxis against graft-vs-host disease consisted of the following: Among patients with no HHV-6B detection, methotrexate and tacrolimus (n = 14), methotrexate and cyclosporine (n = 8), or mycophenolate mofetil and tacrolimus (n = 1); among patients with HHV-6B detection at ≤200 copies/mL, methotrexate and tacrolimus (n = 3) or methotrexate and cyclosporine (n = 2); and among patients with HHV-6B detection at >200 copies/mL, methotrexate and cyclosporine (n = 5).
Association Between Peak HHV-6B Detection and Total and HHV-6B–Specific CD4+ T Cells in Allografts
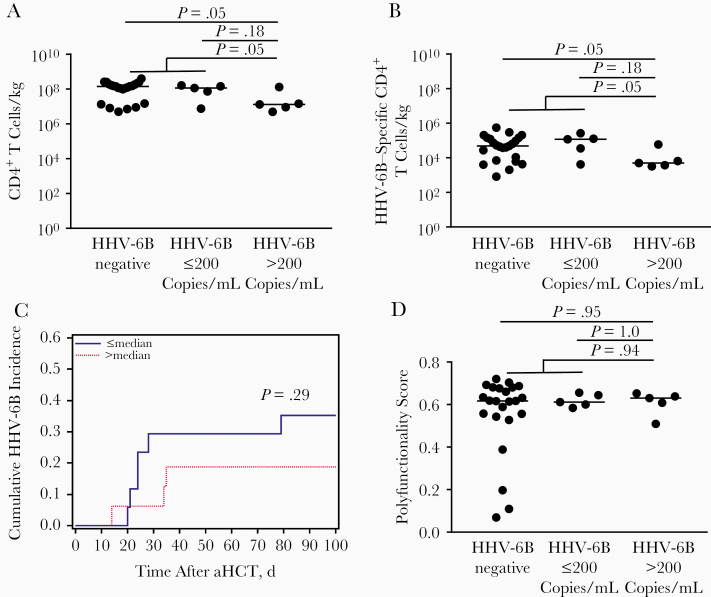
The median absolute count of infused hematopoietic cells was 7.4 × 108/kg (range, 9.9 × 107/kg to 2.5 × 109/kg). Based on these data, the estimated median number of infused CD4+ T cells was 8.9 × 106/kg from bone marrow and 1.5 × 109/kg from G-CSF–mobilized PBSCs. Patients with peak HHV-6B detection ≤200 copies/mL or with no HHV-6B detection received an estimated 10-fold more donor-derived total CD4+ T cells than those with peak HHV-6B detection >200 copies/mL (CD4+ T cells, 1.5 × 108/kg vs 1.3 × 107/kg, respectively; P = .047) (Figure 1A). Similar findings were seen among the patients excluded for poor PBMC viability or PHA response (Supplementary Table 1). We also demonstrated that patients with peak HHV-6B detection ≤200 copies/mL or without detection received an estimated 10-fold more donor-derived HHV-6B–specific CD4+ T cells than those with peak HHV-6B detection >200 copies/mL (5.1 × 104/kg vs 5.0 × 103/kg, respectively; P = .047) (Figure 1B).
Figure 1.
The quantity of donor-derived total or human herpesvirus (HHV) 6B–specific CD4+ T cells per kilogram, but not the polyfunctionality score, is correlated with reduced HHV-6B detection after allogeneic hematopoietic cell transplantation (aHCT). A, B, Patients with a peak HHV-6B viral load above the median (>200 copies/mL) after aHCT received fewer estimated total (A) and HHV-6B–specific (B) CD4+ T cells per kilogram in their hematopoietic cell products than patients with HHV-6B detected at ≤200 copies/mL or with no HHV-6B detection (P values based on Wilcoxon rank sum tests). C, Cumulative incidence plot demonstrates a numerically (but not significantly) lower incidence of HHV-6B in patients who received more than the median versus a lower number of donor-derived HHV-6B–specific CD4+ T cells per kilogram. D, Polyfunctionality scores for these T cells did not differ significantly between patient groups. Circles represent individual patients; horizontal lines, median values.
We also evaluated the cumulative incidence of time to any HHV-6B detection, which demonstrated that patients receiving more than the median number of donor-derived HHV-6B–specific CD4+ T cells per kilogram had a lower risk of HHV-6B detection than those receiving a lower number, but this difference did not reach statistical significance (Figure 1C).
COMPASS-Derived Polyfunctionality Score for Donor-Derived HHV-6B–Specific CD4+ T Cells in Patients With or Without HHV-6B Detection
Next, we used the COMPASS package to derive a polyfunctionality score for donor-derived HHV-6B–specific CD4+ T cells. This score did not discriminate between patients with and those without HHV-6B detection (Figure 1D). Interestingly, among patients with peak HHV-6B detection above the median viral load, the patient with the highest peak HHV-6B viral load had the highest number of infused total and HHV-6B–specific CD4+ T cells but the lowest polyfunctionality score. In an exploratory analysis in which we multiplied the polyfunctionality score by the estimated number of donor-derived CD4+ T cells to generate a composite index of transferred HHV-6B–specific immunity, this did not provide additional discrimination between patient groups than already provided by the absolute CD4+ T-cell counts (data not shown).
DISCUSSION
In the current study, we leveraged a unique cohort of aHCT recipients with weekly HHV-6B testing to demonstrate that a higher number of total and HHV-6B–specific CD4+ T cells in the hematopoietic cell product may reduce the risk for higher-level HHV-6B detection in the first 100 days after aHCT. The relative contribution of HHV-6B–specific versus total CD4+ T cells or other components of the stem cell product requires further study. We also applied a recently described analytic approach using the COMPASS package to derive polyfunctionality scores for donor-derived HHV-6B–specific CD4+ T cells, which has not been studied in the context of aHCT. Application of COMPASS has revealed cellular correlates of protection in the setting of HIV and other pathogens [12], suggesting that the character of T-cell response is functionally important. In our cohort, there was no correlation between CD4+ T-cell polyfunctionality and HHV-6B detection. Together, these data suggest that the absolute number of total and HHV-6B–specific donor-derived CD4+ T cells per kilogram are key factors for controlling the magnitude of HHV-6B detection after aHCT.
The correlation we demonstrate between donor-derived immunity and protection from higher magnitude detection is in accordance with findings pertaining to CMV reactivation after aHCT. CMV seropositive aHCT recipients have better CMV-specific T-cell recovery, fewer CMV-associated complications, and improved overall survival when their donor is also CMV seropositive [13]. These data also lend support to the observed clinical efficacy of donor-derived virus-specific T cells for treating refractory viral infections, including HHV-6, after aHCT [14].
Notably, 4 of the 5 patients with HHV-6B detection above the median peak viral load received hematopoietic cells from a bone marrow source. Bone marrow–derived stem cell products have fewer total cells compared with G-CSF-mobilized PBSCs [11], as demonstrated in our cohort. Thus, it is possible that the variable cell types from these sources may contribute to HHV-6B risk, in addition to the quantity of T cells.
The strengths of this study include the patient cohort in which we had donor PBMCs to directly test for HHV-6B–specific T cells coupled with weekly PCR assays for HHV-6B through 100 days after aHCT. The primary limitation was the small sample size, which limits aspects of our conclusions. We included only aHCT recipients receiving myeloablative conditioning and T-cell–replete allografts to focus on the role of donor-derived T-cell immunity. As such, the results are not generalizable to all HCT settings. Our cohort selection criteria likely contributed to a relatively narrow range of HHV-6B detection, which may have limited the discriminatory power of the HHV-6B–specific T-cell dose or the polyfunctionality score. In addition, our results are based on viral detection without objective evidence of HHV-6B end-organ disease, so caution is required in extrapolating these findings to more significant viral reactivation. Finally, we estimated absolute counts of donor-derived CD4+ T cells in the stem cell product, assays were limited to HHV-6B–specific T-helper 1 CD4+ T-cell immune markers, and we did not measure host-derived resting memory T cells that could contribute to viral control.
In conclusion, a higher number of donor-derived CD4+ T cells may reduce the magnitude of HHV-6B detection after aHCT. Further study is needed to confirm the relative contribution of HHV-6B–specific versus total donor-derived CD4+ T cells. These data provide a rationale for studies of pre- or posttransplantation pathogen-specific immune monitoring, such as those for CMV [15]. They also underscore the potential utility of adoptive cellular therapy to prevent HHV-6B reactivation and associated complications.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors extend thanks to Greg Finak, PhD, at the Fred Hutchinson Cancer Research Center for his consultation regarding the COMPASS package. Samples were obtained from the Infectious Disease Sciences Biospecimen Repository, Vaccine and Infectious Disease Division, and the Research Cell Bank, Fred Hutchinson Cancer Research Center.
Author contributions. D. J. H. performed experiments and collated data. H. X. and W. M. L. performed statistical analyses. D. J. H. and J. A. H. wrote the manuscript. D. M. Z., K. R. J., M. L. H., T. S. A., M. B., and D. M. K. reviewed and edited the manuscript.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grants T32AI118690 and K23 AI119133 to J. A. H.), the National Institutes of Health (grant 5P50GM115305-04), and the HHV-6 Foundation (Dharam Ablashi Research Fund grant to D. M. K.).
Potential conflicts of interest. M. B. has served as a consultant for and received research support from Chimerix, Allovir, and Gilead Sciences. J. A. H. has served as a consultant for Gilead Sciences and Allovir. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Hill JA, Mayer BT, Xie H, et al. The cumulative burden of double-stranded DNA virus detection after allogeneic HCT is associated with increased mortality. Blood 2017; 129:2316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hill JA Human herpesvirus 6 in transplant recipients: an update on diagnostic and treatment strategies. Curr Opin Infect Dis 2019; 32:584–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haberthur K, Engelmann F, Park B, et al. CD4 T cell immunity is critical for the control of simian varicella virus infection in a nonhuman primate model of VZV infection. PLoS Pathog 2011; 7:e1002367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gabanti E, Lilleri D, Ripamonti F, et al. Reconstitution of human cytomegalovirus-specific CD4+ T cells is critical for control of virus reactivation in hematopoietic stem cell transplant recipients but does not prevent organ infection. Biol Blood Marrow Transplant 2015; 21:2192–202. [DOI] [PubMed] [Google Scholar]
- 5. Ljungman P, Brand R, Hoek J, et al. ; Infectious Diseases Working Party of the European Group for Blood and Marrow Transplantation Donor cytomegalovirus status influences the outcome of allogeneic stem cell transplant: a study by the European Group for Blood and Marrow Transplantation. Clin Infect Dis 2014; 59:473–81. [DOI] [PubMed] [Google Scholar]
- 6. Hanson DJ, Hill JA, Koelle DM. Advances in the characterization of the T-cell response to human herpesvirus-6. Front Immunol 2018; 9:1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerdemann U, Keukens L, Keirnan JM, et al. Immunotherapeutic strategies to prevent and treat human herpesvirus 6 reactivation after allogeneic stem cell transplantation. Blood 2013; 121:207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hill JA, Magaret AS, Hall-Sedlak R, et al. Outcomes of hematopoietic cell transplantation using donors or recipients with inherited chromosomally integrated HHV-6. Blood 2017; 130:1062–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hill JA, Mayer BT, Xie H, et al. Kinetics of double-stranded DNA viremia after allogeneic hematopoietic cell transplantation. Clin Infect Dis 2018; 66:368–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dropulic LK, Oestreich MC, Pietz HL, et al. A randomized, double-blinded, placebo-controlled, phase 1 study of a replication-defective herpes simplex virus (HSV) type 2 vaccine, HSV529, in adults with or without HSV infection. J Infect Dis 2019; 220:990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang YT, Zhao XY, Zhao XS, et al. The impact of donor characteristics on the immune cell composition of mixture allografts of granulocyte-colony-stimulating factor-mobilized marrow harvests and peripheral blood harvests. Transfusion 2015; 55:2874–81. [DOI] [PubMed] [Google Scholar]
- 12. Lin L, Finak G, Ushey K, et al. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nat Biotechnol 2015; 33:610–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou W, Longmate J, Lacey SF, et al. Impact of donor CMV status on viral infection and reconstitution of multifunction CMV-specific T cells in CMV-positive transplant recipients. Blood 2009; 113:6465–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sutrave G, Gottlieb DJ. Adoptive cell therapies for posttransplant infections. Curr Opin Oncol 2019; 31:574–90. [DOI] [PubMed] [Google Scholar]
- 15. Chemaly RF, El Haddad L, Winston DJ, et al. Cytomegalovirus (CMV) cell-mediated immunity and CMV infection after allogeneic hematopoietic cell transplantation: the REACT study. Clin Infect Dis 2020. doi: 10.1093/cid/ciz1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.