Abstract
CD4 expression identifies a subset of mature T cells primarily assisting the germinal center reaction and contributing to CD8+ T-cell and B-cell activation, functions, and longevity. Herein, we present a family in which a novel variant disrupting the translation-initiation codon of the CD4 gene resulted in complete loss of membrane and plasma soluble CD4 in peripheral blood, lymph node, bone marrow, skin, and ileum of a homozygous proband. This inherited CD4 knockout disease illustrates the clinical and immunological features of a complete deficiency of any functional component of CD4 and its similarities and differences with other clinical models of primary or acquired loss of CD4+ T cells.
Keywords: CD4 deficiency, recurrent pneumonia, immunizations, double-negative T cells
The first inherited loss of any functional component of CD4, including soluble-CD4, is clinically distinct from any other congenital or acquired CD4 T-cell defect and characterized by compensatory changes in T-cell subsets and functional impairment of B-cells, monocyte and NK-cells.
(See the Editorial Commentary by Routy and Isnard, on pages 547–9.)
Adaptive immunological responses depend on the function of T cells and B cells expressing unique surface antigen-recognition receptors generated by somatic DNA rearrangements. In T cells, these clonal receptors (TCRs), along with the non–clonally distributed CD8 and CD4 co-receptors, determine which lymphocyte precursors will be positively or negatively selected based on the affinity for major histocompatibility complex (MHC) class I– or class II–bound peptides. Such dichotomous interactions define the selection of each TCRαβ + precursor and their distinct immunological functions: While MHC-II binding of CD4 extracellular domains defines the development of helper T cells, CD8 expression identifies cytotoxic T cells. In the periphery, similar mechanisms drive clonotypic expansion in response to individual antigen recognition through the interaction of CD4 or CD8 cytoplasmic tail with the signaling components of the TCR-CD3 complex. Shedding of extracellular CD4 domains by metalloproteinases as soluble CD4 may provide additional adaptive and innate regulatory functions [1] in viral infections and inflammatory or autoimmune disorders [2].
Although CD4 knockout mice [3, 4], as well as primary or acquired immunodeficient syndromes such as MHC-II deficiency [5] and human immunodeficiency virus (HIV)/AIDS [6], respectively, have provided insights into the consequences of CD4+ T-cell depletion in humans, the intertwined role of other viral and immunological factors prevents a more specific characterization of the function of CD4 co-receptor and the consequences of its absence in such clinical models. Similarly, a homozygous splicing variant abrogating the expression of the 2 carboxyl-terminal exons [7] of CD4 revealed some immunological correlates of the preserved expression of all extracellular and soluble CD4 domains in absence of its intracellular tail.
Herein, we present the first human case of a homozygous disruption of the CD4 translation-initiation codon, resulting in a complete multilineage loss of expression of any functional or structural component of CD4 and resulting in a novel primary immunodeficiency with distinct clinical and immunological consequences.
MATERIALS AND METHODS
Study Approval
All participants provided written informed consent and were enrolled in National Institute of Allergy and Infectious Diseases clinical protocols approved by the National Institutes of Health (NIH) institutional review board (ClinicalTrials.gov identifiers NCT00867269, NCT00001281, NCT00039689, and NCT00001316).
T Cells, Monocytes, B Cells, and Natural Killer Phenotypic Analyses
Surface and intracellular CD4 expression was evaluated using 4 different clones of fluorochrome-conjugated monoclonal antibodies: SK3-BUV737 (BD Biosciences), RPA-T4-BV605 (BD Biosciences), RPA-T4-BV605-FITC (BioLegend), MEM-241-APC-Cy7 (Sysmex), and 13B8.2-PC5 (Beckman Coulter). Additional fluorochrome-conjugated monoclonal antibodies used for T cells, monocytes, and natural killer (NK) phenotypic analysis are specified in the Supplementary Materials.
51Cr NK Cytotoxicity Assay
PBMCs were co-cultured with K562 target cells previously incubated with 100 µCi 51Cr for 4 hours in the presence or absence of 1000 U/mL interleukin 2 (IL-2) (Roche). The percentage of specific lysis was calculated as (sample – average spontaneous release) / (average total release – average spontaneous release) × 100.
Cytokine Production After In Vitro Stimulation
T cells’ intracellular cytokine production was investigated by flow cytometry upon stimulation with 2 µg/mL cytomegalovirus (CMV)–pp65 peptide-pool, or 25 µL/mL CMV grade 2 lysate (Microbix Biosystems) and costimulatory antibodies (αCD28, αCD49d, BD Biosciences). The description of the methodology used for detection of intracellular cytokines is specified in the Supplementary Materials.
Interleukin 7/STAT5 Signaling Pathway
Peripheral blood mononuclear cells (PBMCs) were either untreated or stimulated with 10 ng/mL interleukin 7 (IL-7) (Cell Signaling Technology) for 20 minutes at 37°C. Antibodies to stain surface antigens (anti-CD3-BUV395, anti-CD4-BUV737, and anti-CD8-APC) were added and cells were fixed and permeabilized with BD Phosflow Perm Buffer III before adding anti-pSTAT5 antibody (clone pY694, BD).
HIV-1-R5SF162 Infection of T Cells
T-cell subsets or unfractionated PBMCs were stimulated and then magneto-infected with HIVSF162 as reported previously [8]. HIV-1 infection was quantified by flow cytometry as frequencies of HIV-1-Gag p24+-CD3+ with anti-CD8-PerCP, anti-CD4-APC, anti-CD3-V450 (BD-Biosciences), and anti-p24/Kc57-RD1 (Kc57, Beckman Coulter).
Analysis of CD4 Messenger RNA and DNA Sequences
Total RNA was purified from PBMCs using TRIzol (Thermo Fisher Scientific) and reverse-transcribed into complementary DNA (cDNA) using SuperScript-III-First-Strand Synthesis (Thermo Fisher Scientific). Full-length CD4 cDNA was amplified using 5′CTCTCTTCATTTAAGCACGACTCTGCAGAA-3′ and 5′GTAAGTTTATTGTATTTTTATTTCAG-3′ as forward and reverse primers, respectively. Purified PCR product was sequenced with primers covering the full-length cDNA sequence as specified in the Supplementary Materials.
MHC-II Tetramer Staining
NIH Tetramer Core Facility provided DRB1*07:01-restricted Phycoerythrin and BV421-conjugated human cytomegalovirus (HCMV)–gB217–227|DYSNTHSTRYV (DYS); HCMV-pp65177–191| EPDVYYTSAFVFPTK (EPD); and Human-CLIP87–101| PVSKMRMATPLLMQA tetramers. One hundred microliters prediluted tetramer was added to PBMCs (final concentration 6.67 µg/mL) and analyzed by flow cytometry.
Immunohistochemistry
All biopsies were formalin-fixed, paraffin-embedded, and stained with hematoxylin-eosin. Immunohistochemical stains were performed using an automated immunostainer (Ultra, Roche) according to the manufacturer’s instructions with antibodies including CD20 (clone L26,, Roche), CD3 (clone 2GV6, Roche), CD4 (clone SP35, Roche, CD4 intracellular domain) CD8 (clone SP57, Roche), BCL6 (clone EP278, Cell Marque), CD21 (clone EP3093, Roche), CD279 (clone NAT105, Roche), and IgD (Dako) and were detected using Ultra-View kit (Roche) either with DAB or FAST-Red as chromogen.
Soluble CD4 Plasma Levels
Soluble CD4 plasma levels were measured from using a commercially available enzyme-linked immunoabsorbent assay (ELH-CD4-1, RayBiotech, Norcross, Georgia) according to the manufacturer’s instructions.
Statistical Considerations
Experimental data obtained from independent samples from each donor constituted the results of 1 experiment. Measures of central tendency (mean or median) and dispersion were calculated and presented as specified.
RESULTS
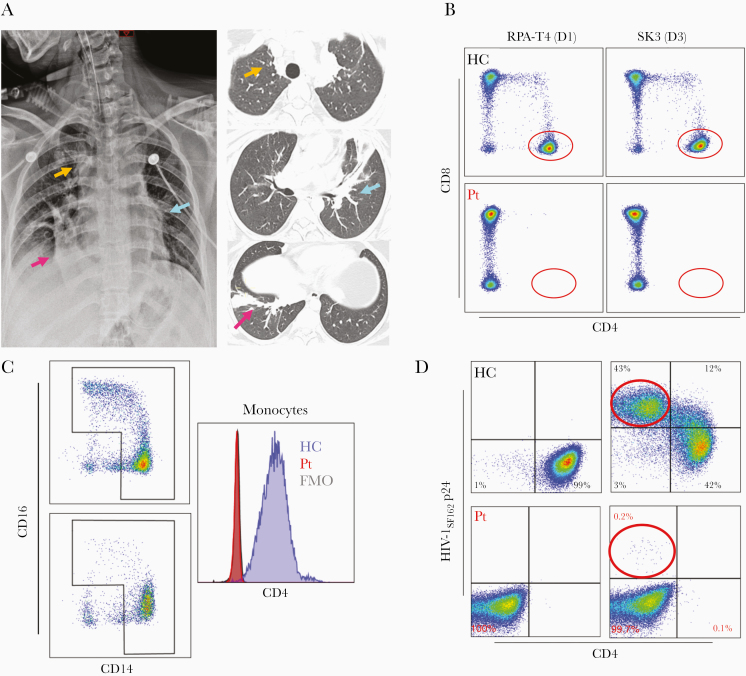
A 22-year-old white woman with a history of recurrent upper respiratory infections, otitis media, and multiple recalcitrant skin warts on trunk and extremities developed a multifocal pneumonia in September 2019 after 3 days of cough, chest pain, and wheezing (Figure 1A, Supplementary Table 1). Her symptoms, initially managed with bronchodilators and inhaled corticosteroids, rapidly progressed with development of hypoxic respiratory failure requiring mechanical ventilatory support. A nasopharyngeal molecular assay tested positive for rhinovirus and enterovirus, while additional microbiological evaluation on bronchoalveolar lavage was unrevealing. Lymphocyte subset analysis demonstrated a complete absence of CD4+ T cells, despite normal absolute numbers of CD3+ T cells (1315 cells/μL, 57% of lymphocytes) and CD8+ T cells (1011 cells/μL, 77% of CD3+ T cells), along with negative testing for HIV-1 antibodies/antigen and HIV-1 RNA. Complete clinical recovery was eventually achieved, but the expression of CD4 on T cells and monocytes remained undetectable by flow cytometry during hospital admission, at approximately 8 weeks and 4 months after the acute illness, as shown by both extracellular or intracellular staining with 4 antibodies targeting different epitopes within the extracellular domains of CD4 (Figure 1B and 1C, Supplementary Figure 1). Moreover, the proband T cells were not susceptible to HIV-1 R5SF162 infection ex vivo compared to a healthy control, further corroborating the lack of CD4 expression (Figure 1D).
Figure 1.
A, Multifocal pneumonia with acute bilateral dense airspace consolidations (colored arrows) on chest radiograph (left panel) and noncontiguous chest computed tomographic images (right panel). B, CD4+ and CD8+ T cells in healthy controls (upper panels) and in the proband (lower panels) using fluorescent-conjugated antibodies targeting 2 different epitopes on the extracellular domain D1 (RPA-T4) and D3 (SK3) of CD4. C, CD14- and CD16-expressing monocytes in a healthy control (left upper panel) and in the proband (left lower panel). Distribution of fluorescence intensity of CD4 staining in permeabilized monocytes from a healthy control (cyan histogram), proband (red histogram), and isotype control (gray histogram). D, Gating on CD3+ lymphocytes, surface CD4, and intracellular HIV-1 p24 expression in positively selected CD4+ T cells of a healthy control and unfractionated T cells of the proband upon anti-CD3/CD28 stimulation and HIV-1SF162 infection (healthy controls, upper panels; proband, lower panels). The left panels represent mock infection while the right panels represent HIV-1SF162 infection (healthy controls, upper panels; proband, lower panels). Abbreviations: FMO, fluorescence minus one; HC, healthy controls; HIV-1, human immunodeficiency virus type 1; Pt, proband.
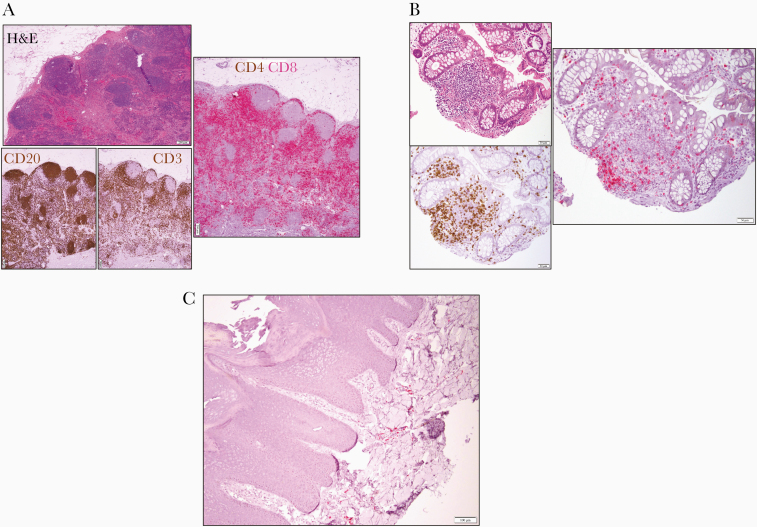
This loss of CD4 expression was further confirmed by immunohistochemical evaluation of CD4 expression on an inguinal lymph node excisional biopsy: No CD4 expression was noted despite a preserved general architecture and normal distribution of CD20+ B cells and CD3+CD8+ T cells in the follicular and parafollicular areas, respectively (Figure 2A).
Figure 2.
A, Excisional inguinal lymph node biopsy: hematoxylin and eosin (left upper panel) showing architectural preservation with open sinuses and regressed germinal centers confirmed by immunohistochemical staining for CD20 and CD3 (left lower panels, both in brown) and for combined CD4 (brown) and CD8 (red) staining (scale bar = 200 μm) demonstrating lack of CD4+ cells. B, Cecum biopsy hematoxylin and eosin (left upper panel), showing a lymphoid aggregate in the lamina propria. Immunohistochemical staining for CD3 (left lower panel, brown) and for combined CD4 (brown) and CD8 (red) staining revealing lack of CD4+ cells (scale bar = 50 μm). C, Immunohistochemical staining of lymphocytes in the superficial dermis of a left tibial verrucous plaque for CD4 (brown) and CD8 (red) (scale bar = 100 μm). Abbreviation: H&E, hematoxylin and eosin.
A similar immunohistochemical analysis extended to a terminal ileum/cecum biopsy, a large tibial human papillomavirus (HPV)–related verrucous plaque, and a bone marrow biopsy confirmed the complete absence of CD4 expression in different tissues and cell types (Figure 2, Supplementary Figure 2).
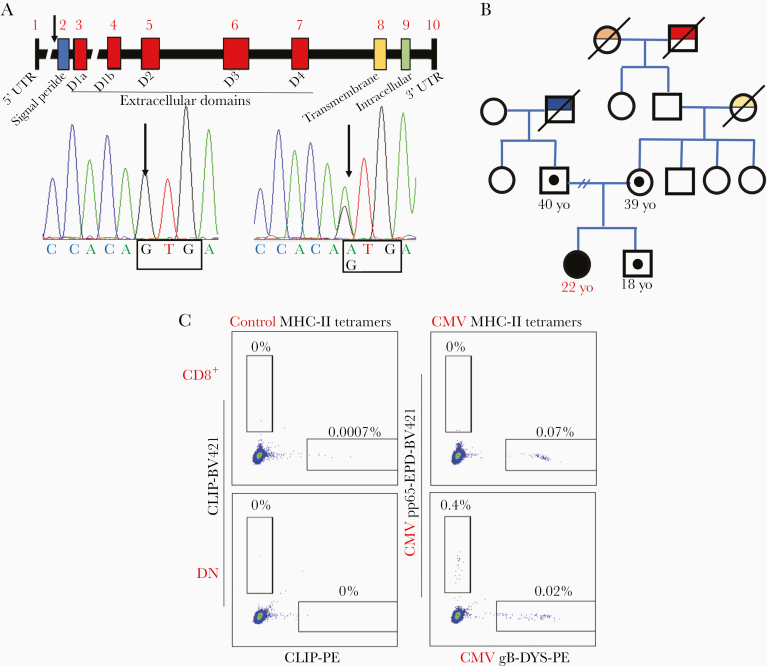
Such longstanding, complete, and multilineage loss of CD4 expression raised concern for an underlying primary immunodeficiency associated with either impaired development or selective depletion of CD4-expressing lymphoid and myeloid cells. Sequencing of full-length CD4 cDNA extracted from the proband’s PBMCs identified a homozygous missense variant in the translation-initiation codon, subsequently confirmed upon sequencing of the proband’s genomic CD4 DNA (c.1A > G, CD4:p.0, NM_000616), while the mother, father, and brother of the proband were heterozygous for the same variant (Figure 3A and 3B). This variant disrupts the CD4 start-codon preventing RNA translation and abrogating the expression of the entire CD4 protein including plasma-soluble CD4 (Supplementary Figure 3). The variant has a combined annotation-dependent deletion score of 22.7 (much higher than the mutation significance cutoff, which for CD4 is 3.313) and an allele frequency in gnomAD of 3.6 × 10−5 (n = 9, all heterozygous European non-Finnish) [9], underscoring both its predicted pathogenicity and rarity (Supplementary Figure 4).
Figure 3.
A, Diagram of the CD4 gene identifying untranslated regions (UTR), intron/exon structures, and protein domains. The arrow identifies the start-codon of the protein on exon 2 (blue). Sanger sequence of homozygous proband (left) and heterozygous mother (right). B, Pedigree with family history: Homozygous proband (full black circle) and heterozygous carriers who underwent genomic DNA sequencing are presented (small circle). Family members deceased with specific medical conditions are presented (red: peripheral T-cell lymphoma not otherwise specified [CD3+CD4+CD5+CD7+]; orange: lung cancer; yellow: breast cancer BRCA1/2 negative; blue: lung cancer, testicular cancer, and thyroid cancer). C, Evaluation of major histocompatibility complex class II tetramer-positive cytomegalovirus (CMV)–specific in CD8+ (upper panels) and double-negative T cells (lower panels) of the proband. Staining of DRB1*07:01 fluorescent-labeled tetramer loaded with nonspecific human peptide CLIP 87–101 PVSKMRMATPLLMQA is presented in the left upper and lower panels, while DRB1*07:01 Phycoerythrin-labeled tetramer loaded with CMV glycoprotein B (217–227)–DYSNTHSTRYV peptide and DRB1*07:01 BV-421–labeled tetramer loaded with CMV pp65 (177–191)–EPDVYYTSAFVFPTK is presented in the right upper and lower panels. Abbreviations: CLIP-PE, Class II-associated invariant chain peptide-Phycoerythrin; CMV, cytomegalovirus; DN, double-negative; gB, glycoprotein B; MHC-II, major histocompatibility complex class II; UTR, untranslated region; yo, years of age.
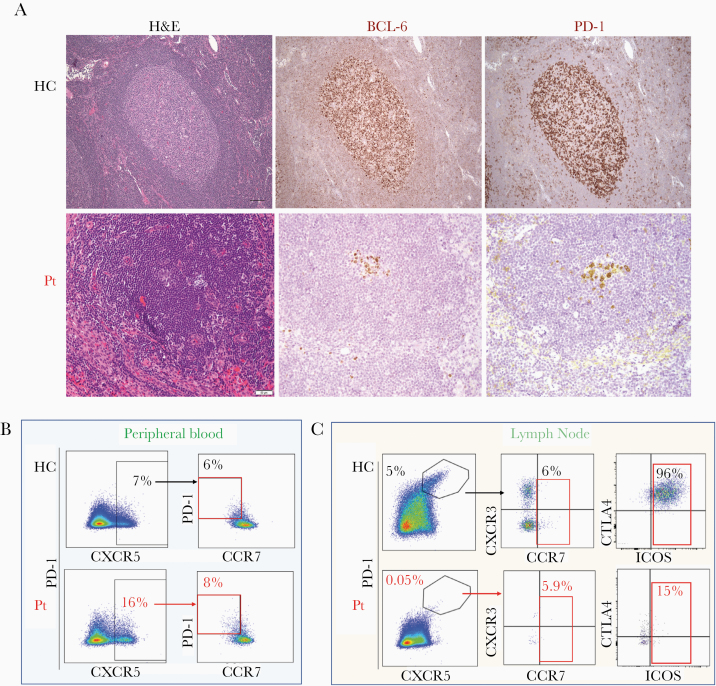
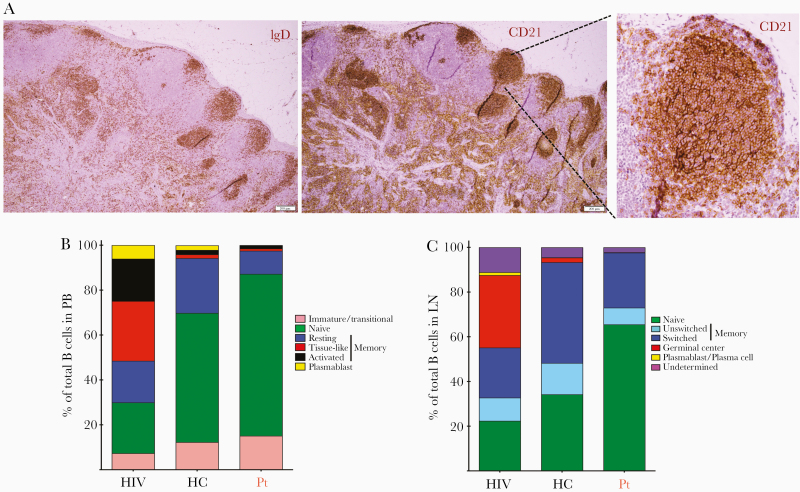
The immunological consequences of complete multilineage loss of CD4 were further investigated: The proband’s T cells were characterized by a relative expansion of CD4–CD8–TCRαβ + cells (double negatives [DNs]) compared to healthy subjects (50.8% vs 5.91%; Figure 1B), with preserved proportion of TCRγδ + and mucosal associated–invariant T cells (Supplementary Figure 5). Importantly, subsets of the proband’s DN T cells retained many phenotypic and functional characteristics of CD4+ T cells, including a distribution of naive/memory subsets that mirrored that of CD4+ T cells, expression of regulatory T-cell markers (FOXP3, CD25), expression of CD40 ligand upon activation, expression of interleukin 17 (IL-17) upon stimulation, IL-7 receptor α-chain expression (CD127), and brisk intracellular signaling in response to IL-7 stimulation similar to CD4+ T cells (Supplementary Figures 6 and 7). These findings suggest that DN T cells may perform certain specialized CD4+ T-cell functions, consistent with previous reports in knockout mice models [3, 4], in nonhuman primates [10], and in a case of aberrant CD4 expression from a splicing variant [7]. In addition, we found that the proband’s CD8+ T cells and/or DN T cells included MHC-II–restricted CMV-specific T cells (Figure 3C, Supplementary Figure 8), highlighting the importance of ontogenic adaptation of T-cell development and functions in the context of inherited CD4 deficiency compared to acquired CD4+ T-cell loss. The evidence of such compensatory MHC-II restriction in the proband DN T cells was further corroborated by the proliferation upon co-culture with allogenic monocytes in a 1-way mixed lymphocyte reaction and the suppression of such proliferation by incubation with an MHC-II blocking antibody (Supplementary Figure 9). The plasticity of T-cell development, however, did not appear to effectively compensate for all types of B-cell helper functions of CD4+ T cells in the proband [11]. In particular, the histological evaluation of an inguinal lymph node revealed diminutive and regressed germinal centers: The B-cell areas, enriched in IgD+ naive or unswitched B cells, contained primary follicles with a developed CD21+ follicular-dendritic mesh; they did not have well-developed secondary follicles with germinal centers but rather atretic follicles with small clusters of BCL-6– and PD-1–expressing cells (Figures 4A and 5A). Accordingly, while circulating T-follicular-helper PD-1+ cells (Tfh) were preserved in peripheral blood DN T-cell subsets, lymph node Tfh were severely reduced and expressed lower level of ICOS in the proband’s CD8+ and DN T-cell subsets compared to healthy subject subsets (Figure 4B and 4C, Supplementary Figures 10 and 11). Flow cytometric analysis of B-cell subsets in peripheral blood and lymph node further corroborated the relative expansion of naive B cells and the absence of germinal center B cells or plasmablast/plasma cells compared to healthy subjects (Figure 5B and 5C). Importantly, despite normal level of all immunoglobulin isotypes and preserved number and distribution of long-lived plasma cells in the bone marrow (Supplementary Figure 2C), the responses to some immunizations were impaired and hepatitis A and B, meningococcal, and Haemophilus influenzae vaccines resulted in a modest or a non-lasting response (Supplementary Table 2).
Figure 4.
A, Hematoxylin and eosin (H&E), BCL-6, and PD-1 staining of a typical secondary follicle with a germinal center in a lymph node of a healthy control subject is presented in the upper panels. H&E staining of the excisional inguinal lymph node biopsy of the proband highlighting a follicle and atrophic germinal center is presented in the left lower panel. The immunohistochemical staining in the right lower panels presents staining for BCL-6 and PD-1 in the same area (scale bar = 20 μm). B, T cells expressing phenotypic markers specific for T-follicular helper cells (CXCR5 and PD-1) within the CD4 or double-negative (DN) CD3+CD45RO+ subsets in peripheral blood of a healthy control (upper panels) and the proband (lower panels). The proportion of CCR7-negative CD4+CD45RO+CXCR5+PD-1+ T cells in a healthy subject is presented in the right upper panel, while the right lower panel presents the proportion of CCR7-negative DN CD45RO+CXCR5+PD-1+ T cells in the proband. C, T cells expressing phenotypic markers specific for T-follicular helper cells (CXCR5 and PD-1) within the CD4 or DN CD3+CD45RO+ subsets in the lymph node of a healthy control (upper panels) and proband (lower panels). The proportion of CCR7-expressing CD4+CD45RO+CXCR5+PD-1+ T cells in a healthy control is presented in the central upper panel, while the central lower panel presents the proportion of CCR7-expressing DN CD45RO+CXCR5+PD-1+ T cells in the proband. The proportion of ICOS-expressing CD4+CD45RO+CXCR5+PD-1+ T cells in a healthy control and proband is presented in the right upper and lower panels, respectively. Abbreviations: BCL-6, B-Cell Lymphoma-6; HC, healthy control; H&E, hematoxylin and eosin; PD-1, programmed cell death protein 1; Pt, proband.
Figure 5.
A, Immunohistochemical staining of an excisional inguinal lymph node biopsy: B cells are highlighted by the CD21 staining (left upper panel), while IgD staining highlights naive or unswitched memory B cells. Insert on right panel highlights the intricate CD21+ mesh of follicular dendritic cells within B-cell primary follicles (scale bar = 50 μm). B, Distribution of B-cell subsets in peripheral blood of HIV-1–infected individuals in the absence of antiretroviral treatment (HIV, average of 3 female donors, median CD4+ T-cell count 551 cells/μL, 27% of total T cells), healthy controls (average of 7 female donors, median CD4+ T-cell count 856 cells/μL, 55% of total T cells), and the proband. The proportion of immature/transitional B cells (CD10+CD27–) is in pink; in green is the proportion of naive B cells (CD21hiCD27–); in dark blue are resting memory B cells (CD21hiCD27+); in red are tissue-like memory B cells (CD21loCD27–); in black are activated memory B cells (CD21loCD27+); and in yellow is the proportion of plasmablasts (CD20–/CD21loCD27++). C, Distribution of B-cell subsets in lymph node of HIV-1–infected individuals in the absence of antiretroviral treatment (HIV, average of 3 female donors, median CD4+ T-cell count 551 cells/μL, 27% of total T cells), healthy controls (average of 7 female donors, median CD4+ T-cell count 856 cells/μL, 55% of total T cells), and the proband. The proportion of naive B cells (IgD+CD38–CD27–) is indicated in green, the proportion of unswitched memory B cells (IgD+CD38–CD27+) in light blue, switched memory B cells (IgD–CD38–CD27+) in dark blue, germinal center B cells (IgD–CD38+) in red, undefined or indeterminate B cells in purple, and plasmablast/plasma cells (IgD–CD38++) in yellow. Abbreviations: HC, healthy control; HIV, human immunodeficiency virus; LN, lymph node; PB, peripheral blood; Pt, proband.
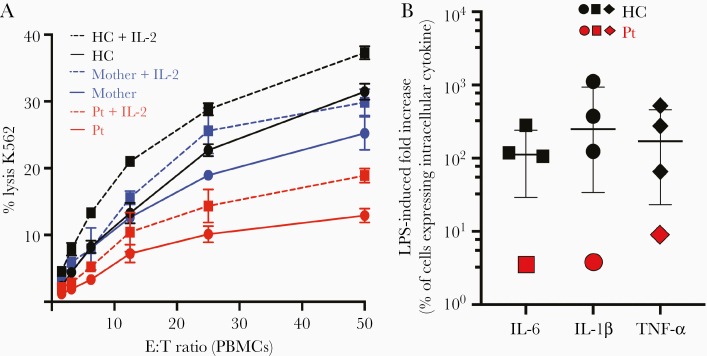
Since antigen-specific CD4+ T-cell function is also involved in NK cell activation [12, 13], we evaluated the NK cytotoxic function and found a significant impairment in lysis of MHC-I–deficient K562 cells in the proband that was only slightly improved by exogenous interleukin 2 (IL-2) stimulation (Figure 6A). Such functional impairment was not associated with changes in the number (Supplementary Table 1) and phenotype of NK cells, as no overt differences in NK cell maturation or expression of known activating and inhibitory killer immunoglobulin-like receptors, NKG2D, and perforin were detected (Supplementary Figure 12).
Figure 6.
A, Evaluation of natural killer cytotoxicity by a 4-hour 51Cr cytotoxicity assay in the presence (dashed line) or absence (solid line) of 1000 U/mL interleukin 2. Peripheral blood mononuclear cells from the proband (red), her mother (blue), or a healthy control (black) were used as effectors against K562 target cells at indicated ratios. Data shown from 1 biological replicate performed in triplicate; error bars indicate the standard deviation. B, Induction of intracellular cytokines upon lipopolysaccharide (LPS) stimulation in monocytes from healthy subjects (black symbols) and in the proband (red symbols) as measured by fold increase in the proportion of cells expressing intracellular cytokines compared to resting conditions. The median and interquartile range of the LPS-induced fold increase in the proportion of cells expressing intracellular interleukin 6 (squared), tumor necrosis factor–α (circles), and interleukin 1β (diamonds) for 3 different healthy controls are presented. Abbreviations: E:T, effector to target cell ratio; HC, healthy control; IL-1β, interleukin 1β; IL-2, interleukin 2; IL-6, interleukin 6; LPS, lipopolysaccharide; PBMCs, peripheral blood mononuclear cells; Pt, proband; TNF-α, tumor necrosis factor alpha.
Furthermore, as CD4 engagement by MHC-II has been previously associated with effect on monocyte differentiation/function [14], we investigated the phenotype and ex vivo response to lipopolysaccharide (LPS) stimulation of the proband’s freshly isolated peripheral blood monocytes. We found that a reduction in CD16-expressing subsets and in particular of the nonclassical/patrolling monocytes (CD14+CD16++), with relative expansion of the classical monocytes (CD14++CD16-), was accompanied with an increased expression of intracellular cytokines in resting conditions, and a blunted upregulation of interleukin 6 (IL-6), interleukin 1β (IL-1β), and tumor necrosis factor alpha (TNF-α), upon LPS stimulation (Supplementary Figure 13). In fact, while the median proportion of monocytes expressing IL-6, IL-1β, and TNF-α in the resting condition was 0.6%, 0.2%, and 0.2%, respectively, in healthy subjects (n = 3), it was 23%, 16%, and 2% in the proband’s monocytes. Upon LPS stimulation, the median proportions of monocytes expressing IL-6, IL-1β, and TNF-α increase to 76%, 74%, 50%, respectively, in healthy subjects (n = 3), and to 80%, 64%, and 19%, respectively, in the proband’s monocytes (Figure 6B, Supplementary Figure 13C).
DISCUSSION
The proband’s spectrum of clinical manifestations with increased frequency of upper respiratory infections and HPV-related skin lesions is consistent with defects in humoral immunity and innate antiviral immune responses. Collectively, the immunological studies revealed normal levels of total immunoglobulin isotypes with preserved long-living plasma cells in the bone marrow but impaired responses to some immunizations that reflected a paucity of memory B cells and plasmablasts/plasma cells and altered trafficking of Tfh in blood and lymph node, with accompanying impairment of innate immunological defense mechanisms related to the function of CD4+ T cells. Nonetheless, the patient has had a relatively modest infection history compared to other inborn errors of immunity and acquired immunodeficiencies associated with CD4+ T-cell depletion, such as HIV/AIDS.
In fact, the germline loss of CD4 expression, besides representing a novel genetic restriction factor for HIV-1 infection, was associated with development of CD4+ T-cell specialized functions by both CD8+ and DN T cells and brisk response to the homeostatic cytokine IL-7 by DN T cells. Such compensatory mechanisms allowed partial mitigation of the clinical phenotype with absence of opportunistic infections or autoimmune manifestations typically associated with primary or acquired loss of specific CD4+ T-cell subset or function [15–17]. Interestingly, even the conventional CD4-helper/CD8-cytotoxic dichotomy is surpassed in these circumstances; in fact, we document CMV-specific MHC-II-restricted responses in the proband’s CD8+ and DN T cells as well as MHC-II–restricted proliferative responses in a 1-way mixed lymphocyte reaction with allogeneic monocytes.
Moreover, the effect of the complete germline loss of CD4 on (1) distribution of B cells and Tfh cells in lymphoid tissue, (2) architecture of the germinal centers, (3) response to some immunogens and/or adjuvant formulations with stricter requirement for specific CD4 T helper cells [18–20], (4) NK cytotoxic function, and (5) monocyte maturation/function [14] suggests broader immunological consequences contributing to an earlier clinical onset and more several clinical manifestations compared to a previously reported case of inherited loss of surface CD4 expression [7]. Additional studies aimed to further disentangle primary defects from the compensatory/adaptive changes caused by loss of CD4 signaling in the current as well as other clinical models of CD4 deficiency can clarify the role of CD4 in different immunological functions including the extent and durability of humoral responses to different immunogens, the response to interleukin 16, or the maturation, function, and development of innate immune cells such as macrophages, eosinophils, and NK cells.
As for other primary immunodeficiencies caused by genetic defects intrinsic to the hematopoietic cells (HSCs), the definitive treatment of CD4 genetic defects would require replacement of HSCs carrying biallelic CD4 variants by hematopoietic stem cell transplantation with healthy donor HSCs or autologous genetically modified HSCs.
Nevertheless, the mitigated clinical phenotype observed in germline CD4 deficiency may suggest careful clinical and immunological monitoring along with primary prophylactic interventions rather than autologous or allogeneic stem cell transplantation.
Although a reduced ability to control some viral infections [21–24] and a possible impairment in the control of Pneumocystis murina (J. Kovacs, personal communication) have been reported in CD4 knockout mice models, the human clinical phenotype described herein is substantially different from such animal models as well as from any other human-acquired CD4 deficiency. We suggested close adherence to recommended immunizations with monitoring of the durability of antibody responses and repeat immunizations whenever nonprotective responses were identified (ie, hepatitis A and B and meningococcal vaccines). In addition, intermittent screening for serum cryptococcal antigen was recommended and sulfamethoxazole-trimethoprim and low-dose azithromycin were maintained in the proband for Pneumocystis jirovecii pneumonia and nontuberculous mycobacterial infection prophylaxis as well as to provide antimicrobial prophylaxis for recurrent respiratory infections in the context of a probable humoral immunity defect [25].
In conclusion, we report the first human with complete and ubiquitous inherited loss of expression of any functional component of CD4 in tissues as well as in peripheral blood, including plasmatic soluble CD4. Although persistent HPV-related skin disease has been observed in inherited or acquired CD4+ T-cell depletion, a concomitant defect in B-cell maturation, along with impaired NK cytotoxicity and monocyte maturation/function, is apparent in this novel and distinct inherited loss of any functional component of CD4. These additional immunological defects contributed to the proband’s recurrent respiratory infections with an episode of respiratory failure and may provide novel insights into the distinct role of lymphopenia vs defects in specific CD4+ T-cell functions in the pathogenesis of viral respiratory diseases. This case contextualizes in humans previous observations from animal knockout models, including the emergence of MHC-II–restricted CMV-specific DN and CD8+ T cells [3, 26], and disentangles CD4 co-receptor functions in the most definitive characterization of CD4 inherited deficiency.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. A. L., I. S., R. S. A., and P. Y. initiated the study, contributed to its conception and design, and wrote the manuscript. C. S. W., L. P., A. H., E. M., J. S. O., S. L., A. J. W., S. M., M. C., C. M. B., S. M., A. T., A. L. D., and S. P. acquired and analyzed data. A. L., I. S., M. V. A., P. O., and D. T. coordinated and provided clinical care. L. D. N. participated in clinical care, data interpretation, and drafting of the manuscript. All authors have approved the final manuscript for publication.
Acknowledgments. The authors thank Meera Patel for technical support in measurement of natural killer cytotoxic function, as well as Joie Davis, Catherine Rehm, and Ulisses Santamaria for clinical support. This article is dedicated to the memory of our colleague Benigno Rodriguez, a brilliant physician and a thoughtful and generous clinical researcher who died tragically in July 2020.
Financial support. This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases. NIH grant R01AI120989 provided financial support to J. S. O.
Potential conflicts of interest. J. S. O. disclosed personal fees from commercial entities which are not relevant to the content of the current work. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Goto H, Gidlund M. Soluble CD4: a link between specific immune mechanisms and non-specific inflammatory responses? Scand J Immunol 1996; 43:690–2. [DOI] [PubMed] [Google Scholar]
- 2. Yoneyama A, Nakahara K, Higashihara M, Kurokawa K. Increased levels of soluble CD8 and CD4 in patients with infectious mononucleosis. Br J Haematol 1995; 89:47–54. [DOI] [PubMed] [Google Scholar]
- 3. Pearce EL, Shedlock DJ, Shen H. Functional characterization of MHC class II-restricted CD8+CD4- and CD8-CD4- T cell responses to infection in CD4-/- mice. J Immunol 2004; 173:2494–9. [DOI] [PubMed] [Google Scholar]
- 4. Rahemtulla A, Fung-Leung WP, Schilham MW, et al. Normal development and function of CD8+ cells but markedly decreased helper cell activity in mice lacking CD4. Nature 1991; 353:180–4. [DOI] [PubMed] [Google Scholar]
- 5. Notarangelo LD. Partial defects of T-cell development associated with poor T-cell function. J Allergy Clin Immunol 2013; 131:1297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Okoye AA, Picker LJ. CD4(+) T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev 2013; 254:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fernandes RA, Perez-Andres M, Blanco E, et al. Complete multilineage CD4 expression defect associated with warts due to an inherited homozygous CD4 gene mutation. Front Immunol 2019; 10:2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Migueles SA, Weeks KA, Nou E, et al. Defective human immunodeficiency virus-specific CD8+ T-cell polyfunctionality, proliferation, and cytotoxicity are not restored by antiretroviral therapy. J Virol 2009; 83:11876–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020; 581:434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sundaravaradan V, Saleem R, Micci L, et al. Multifunctional double-negative T cells in sooty mangabeys mediate T-helper functions irrespective of SIV infection. PLoS Pathog 2013; 9:e1003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol 2015; 15:185–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bihl F, Pecheur J, Bréart B, et al. Primed antigen-specific CD4+ T cells are required for NK cell activation in vivo upon Leishmania major infection. J Immunol 2010; 185:2174–81. [DOI] [PubMed] [Google Scholar]
- 13. Jost S, Tomezsko PJ, Rands K, et al. CD4+ T-cell help enhances NK cell function following therapeutic HIV-1 vaccination. J Virol 2014; 88:8349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhen A, Krutzik SR, Levin BR, Kasparian S, Zack JA, Kitchen SG. CD4 ligation on human blood monocytes triggers macrophage differentiation and enhances HIV infection. J Virol 2014; 88:9934–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Puel A, Cypowyj S, Maródi L, Abel L, Picard C, Casanova JL. Inborn errors of human IL-17 immunity underlie chronic mucocutaneous candidiasis. Curr Opin Allergy Clin Immunol 2012; 12:616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McDonald DR. TH17 deficiency in human disease. J Allergy Clin Immunol 2012; 129:1429–35; quiz 36–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cepika AM, Sato Y, Liu JM, Uyeda MJ, Bacchetta R, Roncarolo MG. Tregopathies: monogenic diseases resulting in regulatory T-cell deficiency. J Allergy Clin Immunol 2018; 142:1679–95. [DOI] [PubMed] [Google Scholar]
- 18. Ko EJ, Lee YT, Kim KH, et al. Roles of aluminum hydroxide and monophosphoryl lipid A adjuvants in overcoming CD4+ T cell deficiency to induce isotype-switched IgG antibody responses and protection by T-dependent influenza vaccine. J Immunol 2017; 198:279–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martins KAO, Cooper CL, Stronsky SM, et al. Adjuvant-enhanced CD4 T cell responses are critical to durable vaccine immunity. EBioMedicine 2016; 3:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Catherine FX, Piroth L. Hepatitis B virus vaccination in HIV-infected people: a review. Hum Vaccin Immunother 2017; 13:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trautmann T, Kozik JH, Carambia A, et al. CD4+ T-cell help is required for effective CD8+ T cell-mediated resolution of acute viral hepatitis in mice. PLoS One 2014; 9:e86348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. von Herrath MG, Yokoyama M, Dockter J, Oldstone MB, Whitton JL. CD4-deficient mice have reduced levels of memory cytotoxic T lymphocytes after immunization and show diminished resistance to subsequent virus challenge. J Virol 1996; 70:1072–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Battegay M, Moskophidis D, Rahemtulla A, Hengartner H, Mak TW, Zinkernagel RM. Enhanced establishment of a virus carrier state in adult CD4+ T-cell-deficient mice. J Virol 1994; 68:4700–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science 2003; 300:337–9. [DOI] [PubMed] [Google Scholar]
- 25. Milito C, Pulvirenti F, Cinetto F, et al. Double-blind, placebo-controlled, randomized trial on low-dose azithromycin prophylaxis in patients with primary antibody deficiencies. J Allergy Clin Immunol 2019; 144:584–93.e7. [DOI] [PubMed] [Google Scholar]
- 26. Tyznik AJ, Sun JC, Bevan MJ. The CD8 population in CD4-deficient mice is heavily contaminated with MHC class II-restricted T cells. J Exp Med 2004; 199:559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.