Abstract
Background: The entity-attribute-value representation with classes and relationships (EAV/CR) provides a flexible and simple database schema to store heterogeneous biomedical data. In certain circumstances, however, the EAV/CR model is known to retrieve data less efficiently than conventionally based database schemas.
Objective: To perform a pilot study that systematically quantifies performance differences for database queries directed at real-world microbiology data modeled with EAV/CR and conventional representations, and to explore the relative merits of different EAV/CR query implementation strategies.
Methods: Clinical microbiology data obtained over a ten-year period were stored using both database models. Query execution times were compared for four clinically oriented attribute-centered and entity-centered queries operating under varying conditions of database size and system memory. The performance characteristics of three different EAV/CR query strategies were also examined.
Results: Performance was similar for entity-centered queries in the two database models. Performance in the EAV/CR model was approximately three to five times less efficient than its conventional counterpart for attribute-centered queries. The differences in query efficiency became slightly greater as database size increased, although they were reduced with the addition of system memory. The authors found that EAV/CR queries formulated using multiple, simple SQL statements executed in batch were more efficient than single, large SQL statements.
Conclusion: This paper describes a pilot project to explore issues in and compare query performance for EAV/CR and conventional database representations. Although attribute-centered queries were less efficient in the EAV/CR model, these inefficiencies may be addressable, at least in part, by the use of more powerful hardware or more memory, or both.
A problem that data modelers commonly encounter in the biomedical domain is organizing and storing highly diverse and heterogeneous data. For example, a single patient may have thousands of applicable descriptive parameters, all of which need to be easily accessible in an electronic patient record system. These requirements pose significant modeling and implementation challenges.
One increasingly popular solution that addresses these issues is to model data using the entity-attribute-value (EAV) approach. This model, which was historically seen first in LISP association lists,1 has been used in multiple applications, including the HELP system2,3 and the Columbia-Presbyterian Clinical Data Repository.4,5 The EAV approach offers many advantages, including its flexibility and ability to store heterogeneous data in a simple, easily maintained format. Our group has recently enhanced this representation to permit data modeling with classes and relationships (EAV/CR).6
Despite its significant benefits, EAV design has the potential to be less efficient than “conventional” database schemas when accessing data. In particular, attribute-centered queries, where the query criterion is based on the value of a particular attribute, are most likely to show impaired performance. This is especially true when query criteria combine one or more simple conditions in Boolean fashion. An example of such a query is “find all patients with blood cultures positive for either Streptococcus pneumoniae OR Candida albicans.” The reason for the potential performance degradation is that the relatively fast AND, OR, and NOT operations that would be required if we were operating on conventional schema tables must be converted to considerably slower set-based equivalents (set intersection, union, and difference, respectively) for EAV tables.
Attribute-centered queries are important for research questions; their performance is not critical for the care of individual patients. The contents of the production (patient-care) database serve, however, as the basis for any research databases that an institution must support, and the question is how such research databases must be implemented. There are two ways to do so:
The simplest solution is to periodically take a backup copy of the production database and restore it onto separate hardware, with only modest transformation, e.g., addition of indexes. (It is undesirable to run resource-intensive attribute-centric queries directly on the production system because they would take CPU cycles away from the simple but critical queries that assist management of individual patients.)
An alternative solution is to redesign the schema completely as numerous conventional tables on separate hardware and transform the production EAV data prior to loading it into these tables. The tables may reside in a single research database or in multiple, special-purpose databases. One could extract all the data or only a subset, focusing on the attributes of greatest clinical or epidemiologic interest. Such a procedure is followed at Intermountain Health Care, where multiple conventionally structured data marts are populated by export of data subsets from the HELP repository,17 whose contents are mostly in EAV form.
The first solution is easy to set up, but queries may not perform well. The second solution promises better performance but is more elaborate because of the data transformations involved. The required transformations can be extensive if one has to support multiple research efforts. To determine which solution is applicable in given circumstances, it would be useful to have performance metrics that compare the relative performance of attribute-centric queries on EAV schemas and equivalent queries on conventional schemas.
We have not found any published reports that quantify these performance differences systematically. Of related interest are the influences on query performance exerted by factors including database size and hardware configuration (e.g., amount of available physical memory). The questions addressed in this paper are:
Does degradation of query performance occur and, if so, how severely?
Can degradation, if present, be mitigated by database or query design strategies and, if so, to what extent?
Answers to these questions would provide valuable insights into the viability of the EAV approach for supporting large-scale databases.
In this paper, we explore issues in database performance for real-world microbiology data stored using an EAV/CR and conventional approach. In particular, we examine: 1) the differences in execution time for a set of clinically relevant database queries operating on an EAV/CR and conventional schema containing the same set of data, 2) the relative efficiency of different EAV/CR query strategies, and 3) the effect of database size and physical memory on database performance. Finally, we examine monitors of system performance and identify potential performance bottlenecks for the competing database schemas.
Background
The EAV model, which has been described in great detail elsewhere,7,8,9 can be visualized conceptually as a database table with three columns: 1) “Entity ID” or “Object ID”; 2) “Attribute,” or a pointer to a separate “Attributes” table; and 3) “Value,” containing the value corresponding to the particular entity and attribute. In an electronic patient record system, the entity ID usually refers to a specific patient-associated event stored as a patient ID and time stamp. The attribute and value columns correspond to a specific parameter (e.g., potassium) and its value, respectively. Each entity-attribute-value triplet occupies one row in the table. Since attributes are not represented as columns, this single table can store all values in the database.
EAV/CR enhances the basic EAV approach by using the object-oriented concepts of “classes” and “relationships” to facilitate data modeling. Classes in EAV/CR are data structures that store the attributes (possibly including other classes) associated with a particular type of entity. For example, a bacterial isolate from a blood culture could be modeled with the class “Bacterial Organism” that contains attributes “Organism_ID” (which references an organism name), “Quantity_cultured”, and “Antimicrobial_tested.” (The last attribute is multivalued, because multiple antimicrobials are typically tested against a single culture.) “Antimicrobial_tested,” in turn, is itself a class with the attributes “Antimicrobial_ID” (which references a drug name) and “Sensitivity_result.”
As in object-oriented modeling, objects represent instances of a class. Thus, in our previous example, every cultured bacterial isolate possesses an object ID as an instantiation of class “Bacterial Organism.” Relationships are used to describe inter-class interactions. Metadata, or information about the data contained in the database, is then applied to describe, maintain, update, and access the data. This overall approach allows the system to incorporate new attributes without altering the underlying physical database schema.
Conventional schemas, in contrast, model parameters as distinct columns. For example, the parameter “Organism_name” could be represented as a column in a table “Microbial_Organism.” As the database evolves to accommodate diverse data, the number of parameters grows and the number of columns and tables that are needed increases. Thus, with a rapidly evolving and expanding domain such as medicine, where new tests and concepts are commonplace, the underlying schema requires frequent modification. This is particularly true of clinical medicine, where there can be dozens of specialty-specific tables, each with a constantly growing list of fields.
The ability of the EAV approach to store diverse data in a single (or few) tables greatly simplifies the physical schema of the database. It provides an efficient way to access all data pertinent to a specific object (i.e., entity-centered queries). In an EMR, this often takes the form of “Select all data (clinical findings, lab tests, etc.) for patient X.” In a conventional schema with numerous tables, the user would need to search every table containing patient data to extract the relevant data for the patient, making query development laborious. Furthermore, not all these tables may contain data for a given patient; as a result, many tables may be searched unnecessarily.
As stated previously, it is the attribute-centered queries that are of concern to the developer who is focused on efficiency. To systematically quantify the performance of an EAV/CR schema compared with its conventional counterpart, we chose the domain of microbiology. This domain, which has been the subject of many previous database models,10,11 provides the potential to work with complex data structures where class attributes exist as both primitive types (e.g., strings) as well as other classes.
Methods
In this section, we describe our test database and schema and various aspects of the evaluation strategy that we devised. In each case we provide the rationale of our design decisions.
Data Description
Each specimen collected from a patient in the Veterans Administration (VA) system is assigned a unique ID. The ID of the ordering physician is also recorded against the specimen. One or more tests (e.g., Gram stain and culture) may then be run on each specimen. For each culture, a panel of antimicrobial sensitivity tests is performed. The data are stored in DHCP,12 an M language database that serves as the clinical patient repository used nationwide by VA Medical Center (VAMC) hospitals.
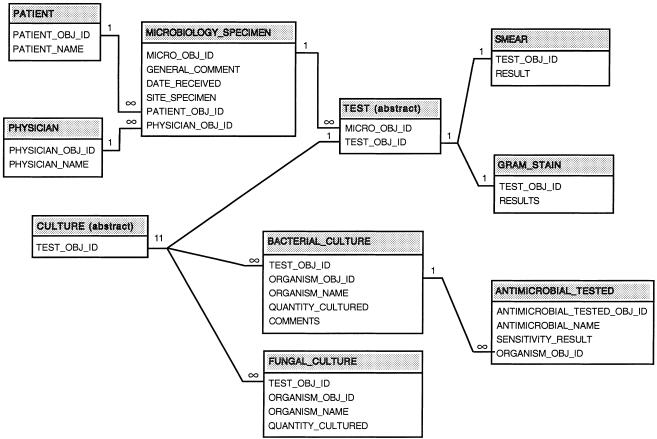
The entity-relationship diagram for the original data is shown in ▶ (details have been omitted for clarity). All classes/entities can be traced to the class “Microbiology Specimen,” which sits at the “top” of the class diagram. Classes contain either primitive types or other classes as attributes, which enable classes to relate to one another.
Figure 1.
Entity-relationship diagram for the microbiology data. For reasons of space, some tables, which are not relevant to the queries in the text, are not shown.
It should be noted that the entities “bacterial culture,” “fungal culture,” etc. shown in ▶ are represented in separate tables in DHCP. During the process of data transformation, we merged these into a single table, since the separate tables have similar structures.
The original, raw, antimicrobial sensitivity data had the names of antimicrobials (e.g., “streptomycin,” “chloramphenicol”) hard-coded as columns in a table. This represents poor table design. With each new antimicrobial introduced into the institution, the table structure needs updating, and all forms dependent on the table require redesign with each change. It may be possible to use this design in the M language, where all disk-based data structures are sparse by design. In relational database management systems (RDBMSs), however, such a design wastes much space, because NULL values of empty columns are still recorded. Further, RDBMSs have a limit on the maximum number of columns per table (e.g., 255), which might be exceeded in some circumstances. Even if space and column constraints are unimportant, necessitating alterations in the table structure and application code each time the number of objects changes is not a good system design approach.
Other parts of the DHCP schema are correctly designed. For example, in the Pharmacy subschema, names of medications are not hard-coded as columns but treated as data: the ID of the drug in the Orders table references a list of drugs in a Drugs table. Therefore, to make comparisons between EAV and conventional schemas more realistic, we first transformed the sensitivity data into the (correct) row-modeled form, where the antimicrobial ID was an attribute in a column and referenced a table of antimicrobial drug names.
Data Extraction
We extracted a dataset from VA Connecticut DHCP that contains all the available online data, which range from 5/1/87 to 9/26/98 and include results from more than 135,000 microbiology test specimens and more than 400,000 antimicrobial sensitivity tests for more than 28,000 patients. The extracted data were transformed for storage into a SQL Server 7.0 RDBMS. The extraction was done in two steps. We first created a conventional schema from the data (transforming the data where appropriate, as described above). We then created an EAV/CR schema from the conventional one by assigning a unique object ID to each instance of a particular class. In all, there were 965,529 instantiated objects in our database.
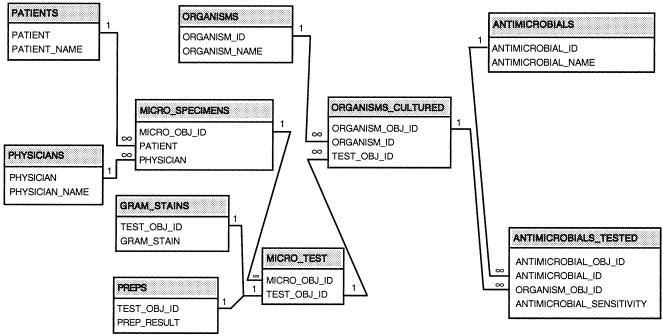
The Conventional Schema
The conventional schema is shown in ▶. All primary and foreign keys, as well as fields used for search (e.g., antimicrobial and organism names), are indexed.
Figure 2.
The conventional physical database schema. Fields in some tables are not shown, for reasons of space; for the most part, only fields linking to other fields are illustrated.
It should be noted that the culture and antimicrobial sensitivity tables can be considered special-purpose or homogeneous EAV tables (because each stores only a single class of data in row-modeled form). In contrast, “EAV” databases contain general-purpose tables that store heterogeneous data (across many classes).
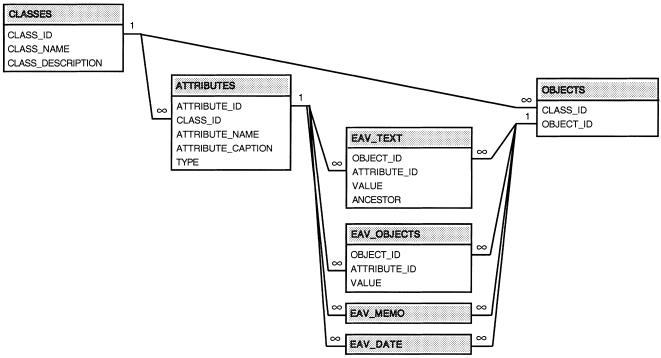
The EAV/CR Schema
The EAV/CR physical schema is shown in ▶. As in all EAV/CR schemas, there are two parts to the schema: metadata and data. (The EAV/CR metadata schema are not illustrated. They have been described in detail,6 and a summary is available on the Web at http://ycmi.med.yale.edu/nadkarni/eav_cr_frame.htm). Attributes are segregated by data type (e.g., string values are stored in “EAV_TEX,T” whereas dates are held in “EAV_DATE”).
Figure 3.
EAV physical database schema. Since all EAV tables share the same structure, the details of two tables have been omitted.
The table EAV_OBJECTS is of particular interest. It accommodates situations where one object can contain another object and permits objects to be associated with one another. In this table, the “Value” field is the ID of a “child” object. Thus, in our logical schema, tests are children of a specimen, cultures are children of a test, and antimicrobial sensitivity results are children of a culture. In other words, when data are hierarchic (as in consecutive one-to-many relationships), each record in EAV_OBJECTS represents one node of a tree. To traverse down an object hierarchy starting with a particular object, we locate that object's ID in the Object_ID column of EAV_OBJECTS. We then gather all Values (child Object IDs) for this entity. We then recurse, searching for these IDs in the Object_ID column, and so on.
One well-known method of minimizing the number of recursive queries when traversing an object hierarchy is to redundantly store the ID of the “ancestor” object against every record in a tree, so that all descendants of the ancestor can be retrieved in a single SQL statement.13 If a significant percentage of queries also use the “ancestor” class, the number of joins required to retrieve the data is also minimized. To allow us to explore this means of schema optimization and its impact on queries, we created an “ancestor” field containing the microbiology specimen ID associated with each test value in EAV_TEXT.
We indexed all object ID columns and created a compound index on attribute-value pairs for tables EAV_TEXT and EAV_DATE, since queries typically select data using both an “attribute ID” and “value” together as search criteria. In addition, a separate index was created for the Value column of EAV_OBJECTS, since the nature of a query might make it desirable to traverse an object hierarchy from descendant to ancestor.
Expanding the Size of the Data
To measure the effects of database size on performance, we replicated the contents of our initial two databases (EAV/CR-Small and Conventional-Small) and reassigned newly created, unique IDs for each new object. The replicated data were then appended onto the corresponding, original database to create “new” databases with twice the number of objects (EAV/CR-Medium and Conventional-Medium). This process was then repeated to yield databases four times larger than our originals (EAV/CR-Big and Conventional-Big). Since the underlying schemas did not change, queries did not require modification within a given representation. The “Big” data set represents the equivalent of 30 to 40 years' worth of microbiology data for all patients in VAMC Connecticut.
The benchmark timings for EAV/CR-Medium and Conventional-Medium were intermediate between the smallest and largest representations, with the data showing an approximately linear trend between the “small” and “big” databases. The purpose of studying three database sizes was to allow us to assess better how relative performance varied with size. To minimize information overload in the tables presented in the remainder of the paper, we omit further discussion of the two “medium” databases.
The EAV/CR representation consumed approximately four times the storage of our conventional schema. EAV/CR-Big consumed 1,177 Mb, with indexes accounting for more than 62 percent. Our largest conventional representation (Conventional-Big) was 301 MB, with approximately 32 percent accounted for by indexes.
It is true that EAV/CR is more space-efficient for sparse data. In this case, however, prior to importing the raw data, we had transformed the bulk of the original sparse data (e.g., the antimicrobial sensitivity results and the cultures) into dense, row-modeled facts in columns that are mostly IDs (long integers). It is well known that a row-modeled conventional table will always take significantly less space than the equivalent facts represented in EAV form. This is because a single set of facts is represented as a single row in a row-modeled table but as multiple rows in an EAV table, one per fact. For example, to fully describe a single antimicrobial sensitivity result, we store several related facts linked to the specimen ID: the micro-organism that was isolated, the antimicrobial tested, and the sensitivity of the former to the latter. In the EAV representation, each row has the extra overhead of Object ID and Attribute ID, plus accompanying indexes.
Query Benchmarking
To compare performance between the two competing physical representations, we developed several clinically oriented queries, as follows. The two attribute-centered queries were:
Query 1: Find all patients with cultures positive for Pseudomonas aeruginosa.
-
Query 2: Find all specimens that grew Streptococcus pneumoniae and Candida albicans.
The EAV/CR approach, as employed in electronic patient record systems, is generally known to be efficient for patient-centered queries (which correspond to queries based on the Object ID rather than on the Attribute and Value). However, we wanted to verify that this was indeed the case for our data set and determine the performance penalty, if any, in comparison with the conventional schema. Therefore, we also created two entity-centered queries:
Query 3: Find all microbiology tests run for a specific patient.
Query 4: Find all antimicrobials and sensitivities tested for a particular culture.
System Description: Memory Allocation
Tests were conducted on a Dell Dimension XPS D300 running Windows NT Server 4 with a 300-MHz Pentium II processor and 66-MHz system bus. Execution times were recorded using the SQL Server 7.0 Profiler tool.
We ran our benchmarks with 192 Mb and 256 Mb of physical RAM and allocated 64 Mb and 128 Mb, respectively, to SQL Server 7. We did this to ensure that the additional RAM was specifically allocated to the database. This left 128 Mb of RAM for the operating system and its associated tasks. The system employed a Western Digital UDMA, 9.5mS, 5400 RPM hard drive. We want to emphasize, however, that in our benchmarks it is the ratio of timings between the EAV/CR and conventional schemas that are important, rather than the absolute timings.
Alternative Formulation of EAV/CR Queries
Formulation of a complex query against an EAV/CR database is significantly more difficult and error-prone than is formulation of a functionally identical SQL query against an equivalent conventional schema. This is because the physical schema of an EAV/CR database—the actual tables holding the data —differs markedly from its logical schema, which specifies how the classes modeled in the database are related to each other. For example, when considering Query 1, the “culture,” “organism,” and “patients” tables do not physically exist in the EAV/CR schema. In a conventional schema, in contrast, logical and physical schemas are identical.
Queries against an EAV/CR system must, therefore, be formulated in terms of the semantics of the logical schema and must then be translated into operations on the tables in the physical schema. We present four ways to do this, the third and fourth being slight modifications of the second, as follows:
Method One
A single (large) SQL statement can be created, which extracts the desired data by joining the necessary tables. In an EAV/CR schema, however, a small set of physical tables is used to model many more logical tables. This results in the same tables being used repeatedly under different aliases, with “self-joins” being performed. In ▶, which illustrates this approach for Query 1, the table EAV_TEXT is used twice. As the logical schema becomes more complex and more classes have to be traversed, such SQL gets progressively harder to write and debug, because it often incorporates a large number of nested constructs.
Figure 4.
Database query developed using a single SQL statement. In this and the next three figures, attribute ID 20 refers to organism name and ID 9 refers to patient ID.
To complicate matters, when this SQL is sent to an RDBMS, the latter parses the code and tries to formulate an optimal strategy before executing it. Query optimization is a problem known to be factorial with respect to the number of tables or aliases participating in the join.14 (Factorial 8, or 8!, equals 8 × 7 × 6... × 2 × 1 = 40,320. An excellent discussion of the query optimization problem is posted at http://www.informix.com/informix/solutions/dw/redbrick/wpapers/star.html.)
Difficulties manifest, therefore, as the number of aliases increase. While DBMS optimizers are claimed by their vendors to use clever heuristics in such a situation, simply evaluating each of the numerous alternatives takes time. (In an experiment reported by Nadkarni and Brandt,9 an attempted 20-alias join that would have yielded only 50 rows in the result set caused an RDBMS to freeze.)
The only justifiable circumstances for using a single-statement approach for EAV/CR query are for queries with a modest number of aliases (as in ▶), or for “stored” queries, a facility available in many DBMSs. (In execution of stored queries, the parsing phase is bypassed, because the query has already been “compiled,” so to speak, with a plan of execution already worked out.)
Method Two
The second method uses a “divide and conquer” strategy. A series of simple queries are specified in terms of individual classes in the conceptual schema. Each query accesses one or two tables at a time to create a temporary table. The temporary tables are then combined with joins, using appropriate set operations (intersection, union, difference), if necessary. When the final result is obtained, temporary tables created along the way are deleted.
In this particular case, the DBMS optimizer has no problem with individual queries. In fact, the execution plan devised by optimizers often involves creating temporary tables, and we are explicitly telling the DBMS what to do at each step rather than relying on the DBMS to determine its own strategy. ▶ illustrates this method for Query 1.
Figure 5.
Database query developed using temporary tables created and deleted dynamically to store interim data.
It is, of course, possible that, if a DBMS spends enough time analyzing a complex query, it will arrive at a solution superior to what we have manually specified. However, for highly complex ad hoc queries (unlike stored procedures), the time required to devise an execution plan may be much greater than the time required to actually execute it.
Method Three
A slight modification of the second method is to repopulate and empty existing indexed temporary tables instead of creating and deleting them. The potential rationale is that if intermediate result sets created by a simple query are large, then, when the result sets are to be combined, the query optimizer will often create temporary indexes to speed up the join process. If we maintain several indexed tables for the sole purpose of holding intermediate results, then, when the tables are populated through INSERT queries, the indexes are also populated, and subsequent joins will be speeded up. ▶ illustrates this approach. Notice the very great similarity to the code in ▶. The only differences are that we use INSERT INTO... SELECT instead of SELECT... INTO, and TRUNCATE TABLE instead of DROP TABLE.
Figure 6.
Database query developed using previously created and indexed tables to store interim data.
This strategy, however, is double-edged. If intermediate results are small, then the DBMS may perform joins in memory rather than on disk. If so, we have created disk-based indexes for nothing. Index maintenance uses machine resources and time, because whenever rows are removed or added to a table, the indexes on the table must also be maintained.
Method Four
A minor modification of the third method is to use initially un-indexed temporary tables, which are indexed only after all records are inserted. Record insertion is expected to be faster because the indexes do not have to be maintained each time a new record is added to the corresponding table. The query is illustrated in ▶.
Figure 7.
Database query developed using un-indexed tables to store interim data, which are indexed only after data are inserted into the tables.
We applied all four methods to Query 1 operating on EAV/CR-Big with 128Mb RAM allocated to the database. We found that our second method (developing simple queries that created tables and later removed them) gave the best results.
Measuring Caching Effects
When the exact same query is sent to a DBMS multiple times, the execution times for the second and subsequent runs are often significantly shorter than for the first. This is because the DBMS uses caching, either in memory or on disk. The SQL string of a query is stored (so that the text of a new query may be compared with it), along with the execution plan. If the result set is small enough, it may also be cached. Therefore, in an extreme case, a repeated query will not cause any disk activity at all: the DBMS, having decided that it is identical to a previous one, will instantaneously return a stored result.
It is important to determine caching effects for conventional versus EAV/CR schemas, especially if queries on the latter consist of a batch rather than a single statement. (All the individual queries in a batch might not be cached.) To measure caching effects, each query was executed five times in succession immediately following system start-up for each combination of system memory and database size. We used five runs to control for “cold” database, or start-up effects.15
We report the timings for every query result in two ways: as initial query execution time and as cached execution time (the average of the four succeeding runs). The former probably simulates the real-world situation more closely than the latter, since it is less likely that two different users of the system will perform successive identical ad hoc queries.
Utilizing the Ancestor Field
We also compared execution times for queries that made use of the ancestor field construct with execution times for those that did not. As stated previously, the use of the ancestor field allows the composer of a query to “cheat” and shorten the traversal path in suitable circumstances. It was necessary to quantify the benefit obtained while recognizing that queries may not always be able to take advantage of this field.
Monitoring System Performance
Operating system performance was monitored with Windows NT performance monitor in conjunction with SQL Server Objects and Counters. We recorded the following parameters:
The number of physical database page reads per second
The number of disk reads per second
The percentage of time the hard disk was busy with read/write activity
The percentage of time the processor handled nonidle threads
Page read frequency is a surrogate measure of disk I/O (input/output) efficiency. A high percentage of hard disk activity and a large number of disk reads per second suggest a potential disk I/O bottleneck. Finally, a high degree of processor activity suggests a potential processor-related bottleneck.
We performed these evaluations for Query 1 during initial execution (when there was no data caching) and the fifth run of the sequence of five (where caching effects were presumed to be most significant). SQL Server 7 and Windows NT provide system measurements every second; we averaged these values over the duration of the query's execution to obtain summary measurements.
Results and Interpretation
Comparing Different EAV/CR Query Strategies for Attribute-centered Queries
As stated earlier, there are at least four different strategies to use when performing attribute-centered queries of EAV/CR data. Prior to contrasting the EAV/CR and conventional schemas for different memory configurations and database sizes, we had to decide which of these strategies was most viable. We report the comparative benchmarks with each approach for Query 1 (finding all patients with cultures positive for P. aeruginosa).
Initial execution times. The approach of developing multiple SQL statements to create temporary tables produced the quickest initial run (97.3 sec). The approach of creating a single large query took 108.5 sec, indicating that, even with a three-alias join, the time the optimizer takes to devise a plan is significant. (Disk activity was higher; this is expected because the DBMS optimizer must consult disk-based structures to make its decision.) The approach of multiple SQL statements reusing indexed temporary tables gave the longest time (126.2 sec), indicating that, at least in this particular case, pre-indexing reusable tables does not necessarily pay off. The approach of multiple SQL statements reusing initially un-indexed temporary tables took 114.7 sec. This was better than pre-indexing but not as good as de novo creation of temporary tables.
Cached execution times. Here, the single-statement approach gave results that were dramatically better than with multiple statements (14.3 sec versus 86.6 sec for temporary table creation, 95.9 sec for indexed temporary table reuse, and 89.9 sec for unindexed temporary table reuse). The operating system statistics showed zero disk activity for the single statement, indicating caching effects. We concluded that the DBMS does not efficiently cache batches of SQL statements; only the last-used statement appears guaranteed to be cached. In practice, as we have stated earlier, caching does not provide any real benefit for ad hoc queries, which are unlikely to be repeated twice in succession.
Similar results were observed when these strategies were applied to Query 2 (find all specimens with S. pneumoniae and C. albicans).
The time difference of 10 percent between the multiple-statement and single-statement approaches is not particularly dramatic. We hypothesized, however, that as the number of classes involved in a query increased, these differences would become increasingly pronounced. We tried out a new query, “Find all patients with cultures positive for P. aeruginosa that showed resistance to ceftazidime.” (This query also involves the “Antimicrobial_Tested” class and “Antimicrobial Name” attribute.) The initial execution times for the multiple-statement and single-statement queries were 194 sec compared with 338 sec, respectively. In other words, using multiple statements almost halved execution time. (Our experience is in line with that reported in a well-known database article,16 which stated that giant, “elegant” SQL statements are often too complex for the limited intelligence of a DBMS optimizer.)
Because of these results, we used the multiple statement approach, with creation of temporary tables, to benchmark EAV/CR query performance for the remaining analyses.
Comparing Performance of the EAV/CR and Conventional Representations for Entity-centered Queries
Queries on the conventional and EAV/CR schemas gave comparable execution times:
Query 3: Find All Microbiology Tests for a Single Patient. Initial times ranged from 1 to 2 sec for the EAV/CR schema, compared with 0.9 to 1.4 sec for the conventional schema. (The smaller and larger numbers in each range apply to the “Small” and “Big” versions of each database.) The corresponding cached times reduced to 0.6 and 0.5 sec irrespective of database size.
Query 4: Find All Antimicrobials and Sensitivities Tested for a Particular Culture. Initial times were 2.1 to 2.4 sec for the EAV/CR schema compared with 2.2 to 2.7 sec for the conventional, with cached timings for both EAV/CR and conventional schemas being 1.5 to 1.7 sec.
To summarize, increasing the volume of the data by a factor of four (in the “Small” versus “Big” databases) had little effect on entity-centered queries. This is expected, because for such queries, which use indexes well and return small amounts of data, search time tends to increase logarithmically rather than linearly with data size. The results indicate that, compared with the EAV/CR schema, the conventional schema may be marginally more efficient (if at all, since results for Query 4 are inconclusive), but the differences are not large enough to be of major practical importance.
Comparing Performance of the EAV/CR and Conventional Representations for Attribute-centered Queries
Query 1: Find All Patients with Cultures Positive for P. aeruginosa (▶). The conventional queries were three to five times faster than their EAV/CR counterparts in the initial run, and 2 to 12 times faster in the cached run. We analyze the results below.
Table 1.
Execution Times (in Seconds) for Query 1: “Find All Patients with Cultures Positive for Pseudomonas aeruginosa”
| EAV/CR-Small |
EAV/CR-Big |
|||||
|---|---|---|---|---|---|---|
| Anc | NoAnc | Conv-Small | Anc | NoAnc | Conv-Big | |
| 64-Mb RAM: | ||||||
| Initial query | 29.5 | 29.5 | 8.2 | 97.6 | 147 | 29.2 |
| Cached | 21.6 | 20.2 | 3.6 | 90.6 | 140 | 10.8 |
| 128-Mb RAM: | ||||||
| Initial query | 30.2 | 28.9 | 8.2 | 97.3 | 108 | 26.2 |
| Cached |
7.8 |
11.6 |
3.7 |
98.4 |
98.4 |
10.2 |
| Note: Times for EAV/CR schema are displayed with and without ancestor construct. Anc indicates ancestor; NoAnc, no ancestor. | ||||||
Increasing database size and system memory had little influence on the ratio for the initial run. With the smallest database, greater available RAM improved caching of EAV/CR queries, but this effect was lost for the largest database.
Using the ancestor field in the EAV/CR query provided little or no benefit for the smallest database but reduced the time for the large database. Here, the benefits were most pronounced with 64 Mb of RAM (with time reduced by a third), but less so (only one tenth) as more RAM was added.
The number of page reads per second was consistently larger in the EAV/CR schema, particularly as the database increased in size. This suggests that physical disk I/O is a likely bottleneck in this query operating on the EAV/CR model.
Query 2: Find All Specimens Positive for S. pneumoniae and C. albicans (▶). Here, the EAV/CR queries that used the “Ancestor” field were much faster than queries on the conventional schema: twice as fast with the initial query, six to eight times as fast when cached. Changing database size or increasing system memory had little effect on these ratios.
Table 2.
Execution Times (in Seconds) for Query 2: “Find All Specimens Positive for Both Pseudomonas aeruginosa and Candida albicans”
| EAV/CR-Small |
EAV/CR-Big |
|||||
|---|---|---|---|---|---|---|
| Anc | NoAnc | Conv-Small | Anc | NoAnc | Conv-Big | |
| 64-Mb RAM: | ||||||
| Initial query | 3.3 | 21.3 | 6.9 | 14.3 | 141 | 25.3 |
| Cached | 0.7 | 13.6 | 4.5 | 1.5 | 139 | 11.0 |
| 128-Mb RAM: | ||||||
| Initial query | 3.7 | 22.0 | 6.8 | 12.9 | 83.5 | 25.1 |
| Cached |
0.7 |
13.7 |
4.5 |
1.4 |
49.4 |
11.2 |
| Note: Times for EAV/CR schema are displayed with and without ancestor construct. Anc indicates ancestor; NoAnc, no ancestor. | ||||||
When the ancestor field was not used, the EAV/CR query took longer, with the difference in execution times widening for initial runs as database size increased. With 64 MB of RAM allocated, the ratios were 3 for the small databases and 5.6 for the large databases. As with Query 1, however, the ratios decreased (to 3.2 and 3.3, respectively) when 128 Mb of RAM was allocated.
Our results indicate that while EAV/CR is generally less efficient than its conventional equivalent, the inefficiency can be mitigated, at least in part, by additional memory. The dramatic benefits of the ancestor field (specimen ID) in Query 2, which uses this field specifically, should be put in perspective. Specifically, incorporation of this extra field in the schema amounts to deliberate denormalization, a standard space-for-time tradeoff.* (We could also have done this in the conventional schema, by adding a specimen ID column redundantly to the cultures and antimicrobial sensitivity tables. In this case, the altered conventional schema would almost certainly have outperformed the EAV/CR schema considerably.)
Our results do suggest that, if performance for certain types of commonly executed queries is critical in a particular database, judicious use of ancestor fields can exert more of an impact than the schema's underlying design (i.e., EAV/CR or conventional). In many electronic patient record systems, for example, almost every item of data is tagged with the ID of the patient to whom it directly or indirectly pertains, because a very large portion of queries relates to individual patients.
Discussion
We performed a pilot exploration of EAV/CR database efficiency issues that can help guide designers and programmers of EAV/CR databases, such as electronic patient record systems, as to the performance penalty they could expect if they tried to execute complex queries on the EAV/CR data directly, rather than exporting subsets of the data to a conventional database for querying purposes. However, the following limitations of our study must be noted.
We examined performance monitors for a single database engine running on a single operating system platform. We chose this configuration largely because of the available built-in system monitoring tools. The results generated here may not completely map to other database engines and operating systems. (Identical queries conducted on the two schemas using a Microsoft Access 97 database engine running on Windows 98, however, yielded qualitatively similar results.)
We cannot predict how performance would be affected by increasing the database to sizes larger than those tested here. Our results suggest, however, that the difference in performance appears to widen as database size increases for certain queries and that this can be offset, at least in part, by adding system memory.
We did not make use of the parallel processing capabilities of the SQL Server database engine, since we employed a single processor machine for testing.
Investigations are under way to study additional methods to optimize query generation and execution.
Our databases were populated with data from one specific domain, microbiology. We cannot predict how performance times would differ in a database populated from increasingly heterogeneous sources of data. We would expect, however, that the effort needed to develop queries would increase disproportionately in the conventional schema compared with the EAV/CR schema.
The logical schema of our test database may not be the best candidate for demonstrating the full potential value for an EAV/CR schema, since the data do not exhibit sparseness.
The Windows NT operating system performs certain background tasks for system maintenance that could conceivably affect specific performance runs. We tried to minimize these effects by generously dedicating 128 Mb of physical RAM to the operating system at all times.
These are the lessons we learned from this exercise:
For entity-centered queries, EAV/CR schemas are about as efficient as their conventional counterparts. Attribute-centered queries are distinctly slower for EAV/CR data than for conventional data (three to five times slower, for our test data). Adding more memory improves the ratio, especially for large volumes of data, as shown by performance monitor statistics.
The strategy of querying an EAV/CR database with simple statements run sequentially is a viable one. Each individual statement is simple to understand; in fact, it is simple enough to be created through a query generator that has knowledge of the EAV/CR logical schema. (A query generator for ACT/DB, an EAV/CR database for clinical studies, is described in Nadkarni and Brandt.9) We also found that using this query approach was more efficient than creating a single, large SQL query statement. The difference in efficiencies appeared to increase as queries became more complex. Additional studies are under way to examine this approach under a wider range of conditions.
The “ancestor” field yielded impressive benefits in improving efficiency for certain queries. Its primary limitations are that not all queries can make use of it and that certain domains may not possess “natural” candidate ancestor fields. (The existence of an ancestor field might also complicate the design of an automatic query generator considerably, because the generator must know the circumstances in which shortcuts can be taken.)
While attribute-centered EAV/CR queries are slower than queries on conventional schemas, this does not disqualify EAV/CR schemas from consideration for warehousing biomedical data. For a complex schema, the increased execution times are significantly offset by ease of database and query maintainability in the EAV/CR model. The conventional query's speed advantage (being three to five times faster) may appear to be discouraging for EAV/CR, but the fact is that the longest EAV/CR query in our fairly large data set took less than 3.5 min to complete. Continuing enhancements of CPU speed, increasing RAM capacity, and the availability of affordable multiprocessor machines will lower absolute times further, even though they would benefit conventional and EAV schemas equally.
In any case, long query execution times tend to be far less critical for non—real-time, attribute-centered queries, which are often submitted in batch mode. (Patient-centered queries typically submitted in an electronic patient record system, in contrast, demand quick response time.) Perhaps the most compelling justification for moving EAV/CR data subsets to specially designed, conventionally structured data marts is when users who demand response times of a few seconds repeatedly query these subsets.
Conclusions
This paper describes a pilot project to explore issues in query performance for EAV/CR and conventional database representations. Although we found that attribute-centered queries performed less efficiently in the EAV/CR model, we feel that many of the benefits inherent in the EAV/CR representation help offset this decrease in performance. Purchasing more memory, additional processors, or faster hardware, or a combination of these, may prove a very cost-effective approach to handling these potential inefficiencies, particularly if the alternative is to maintain two parallel versions of a database with different structures. We plan to continue building and optimizing query strategies to support this representation as well as testing its ability to handle increasingly complex databases for alternative biomedical domains.
This work was supported in part by NIH grants T15-LM07056 and G08-LM05583 from the National Library of Medicine and by grant U01-CA78266 from the National Cancer Institute.
Footnotes
Normalization refers to the database design principle of minimizing data redundancy by storing a fact, as far as possible, in only a single place. De-normalization refers to design that violates one or more of these principles.
References
- 1.Winston PH. Artificial Intelligence. 2nd ed. Reading, Mass: Addison-Wesley, 1984.
- 2.Huff SM, Berthelsen CL, Pryor TA, Dudley AS. Evaluation of an SQL model of the HELP patient database. Proc 15th Symp Comput Appl Med Care. 1991: 386-90. [PMC free article] [PubMed]
- 3.Huff SM, Haug DJ, Stevens LE, Dupont RC, Pryor TA. HELP the next generation: a new client-server architecture. Proc 18th Symp Comput Appl Med Care. 1994: 271-5. [PMC free article] [PubMed]
- 4.Friedman C, Hripcsak G, Johnson S, Cimino J, Clayton P. A generalized relational schema for an integrated clinical patient database. Proc 14th Symp Comput Appl Med Care. 1990: 335-9.
- 5.Johnson S, Cimino J, Friedman C, Hripcsak G, Clayton P. Using metadata to integrate medical knowledge in a clinical information system. Proc 14th Symp Comput Appl Med Care. 1990: 340-4.
- 6.Nadkarni PM, Marenco L, Chen R, Skoufos E, Shepherd G, Miller P. Organization of heterogeneous scientific data using the EAV/CR representation. J Am Med Inform Assoc. 1999; 6(6): 478-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niedner CD. The entity-attribute-value data model in radiology informatics. Proceedings of the 10th Conference on Computer Applications in Radiology. Anaheim, Calif: Symposia Foundation, 1990: 50-60.
- 8.Nadkarni PM, Brandt C, Frawley S, et al. Managing attribute-value clinical trials data using the ACT/DB client-server database system. J Am Med Inform Assoc. 1998;5(2): 139-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadkarni P, Brandt C. Data extraction and ad hoc query of an entity-attribute-value database. J Am Med Inform Assoc. 1998;5(6): 511-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nussbaum BE. Issues in the design of a clinical microbiology database within an integrated hospital information system. Proc 15th Annu Symp Comput Appl Med Care. 1991: 328-32. [PMC free article] [PubMed]
- 11.Delorme J, Cournoyer G. Computer system for a hospital microbiology laboratory. Am J Clin Pathol. 1980;74: 51-60. [DOI] [PubMed] [Google Scholar]
- 12.Department of Veterans Affairs. Decentralized Hospital Computer System Version 2.1: Programmer's Manual. San Francisco, Calif: Information Systems Center, 1994.
- 13.Celko J. SQL for Smarties: Techniques for Advanced SQL Programmers. San Mateo, Calif: Morgan Kaufman, 1996.
- 14.Sybase Corporation. Adaptive Server Anywhere (ASA) New Features and Upgrading Guide. (Chapter 4. Query Optimization Enhancements.) Available at: http://sybooks.sybase.come:7000/onlinebooks/group-aw/awg0603e/dbupen6/. Accessed Jul 29, 2000.
- 15.Zaniolo C, Ceri S, Faloutsos C, Snodgrass R, Subrahmanian V, Zicari R. Advanced Database Systems. San Francisco, Calif: Morgan Kaufmann, 1997.
- 16.Celko J. Everything you know is wrong. DBMS Mag. 1996;9(9): 18-20. [Google Scholar]
- 17.Wang P, Pryor TA, Narus S, Hardman R, Deavila M. The Web-enabled IHC enterprise data warehouse for clinical process improvement and outcomes measurement. AMIA Annu Fall Symp. 1997: 1028.