Abstract
Objective: The objective of this study is to predict in vivo lung mass density for patients with interstitial lung disease using different gradient boosting decision tree (GBDT) algorithms based on measurements from lung ultrasound surface wave elastography (LUSWE) and pulmonary function testing (PFT). Methods: Age and weight of study subjects (57 patients with interstitial lung disease and 20 healthy subjects), surface wave speeds at three vibration frequencies (100, 150, and 200 Hz) from LUSWE, and predicted forced expiratory volume (FEV1% pre) and ratio of forced expiratory volume to forced vital capacity (FEV1%/FVC%) from PFT were used as inputs while lung mass densities based on the Hounsfield Unit from high resolution computed tomography (HRCT) were used as labels to train the regressor in three GBDT algorithms, XGBoost, CatBoost, and LightGBM. 80% (20%) of the dataset was used for training (testing). Results: The results showed that predictions using XGBoost regressor obtained an accuracy of 0.98 in the test dataset. Conclusion: The obtained results suggest that XGBoost regressor based on the measurements from LUSWE and PFT may be able to noninvasively assess lung mass density in vivo for patients with pulmonary disease.
I. INTRODUCTION
Lung disease, the third-leading cause of mortality in the United States, is responsible for 4 00 000 deaths each year (Xu, 2010). While death rates from other major diseases such as cancer and cardiovascular disease are declining, mortality rate due to lung disease is increasing (American Lung Association, 2008). Many lung diseases, including interstitial lung disease (ILD), are associated with changes in the lung's biomechanical properties. ILD comprises of serious diseases in which the lung tissue is stiffened by fibrosis. Computed tomography (CT) is the clinical standard for diagnosing lung fibrosis (Mathieson et al., 1989; Verschakelen, 2010), but it substantially increases radiation exposure for patients, even when using various techniques to reduce the dose (Mayo, 2009).
Lung ultrasound imaging has recently attract attention for assessing lung disease (Demi et al., 2017; Mohanty et al., 2017; Demi et al., 2020; Soldati et al., 2019a; Soldati et al., 2019b; Mento et al., 2020). Given the COVID-19 epidemic, lung ultrasound has increasingly been used for disease diagnosis and early screening (Soldati et al., 2020). Safety issues related to using ultrasound imaging for assessing lung disease have been discussed (Miller et al., 2018). A deep learning framework has been proposed for classification and localization of COVID-19 markers in lung ultrasound (Roy et al., 2020). We have developed a lung ultrasound surface wave elastography (LUSWE) technique for noninvasively measuring surface wave speeds of superficial lung tissue for patients with ILD and found there is a statistical difference in the surface wave speed between patients and healthy controls (Zhang et al., 2011; Zhang et al., 2017a; Zhang et al., 2018a; Zhang et al., 2018b; Zhou et al., 2019). ILD is difficult to classify because>200 disorders have similar clinical, radiographic, physiologic, or pathologic manifestations, with limited consensus regarding the best management practices(Loscalzo, 2010). Because ILD has multiple causes, our work focuses on systemic sclerosis–associated ILD (SSc-ILD). This strategy has several advantages. First, patients with SSc have a high incidence of lung fibrosis (Steen and Medsger, 2000; Hudson et al., 2010). Second, lung involvement in patients with SSc may be predicted at early stages by evaluation of skin fibrosis because skin manifestation is an early and easily detectable marker of SSc disease activity. In this research, we do not study the analysis of lung mass density for different types of ILD.
This paper aims to develop LUSWE based artificial intelligence (AI) techniques to noninvasively analyze lung mass density. Lung mass density is an important biomarker that reflects lung health. However, there are currently no validated clinical imaging techniques to analyze lung mass density. A few studies have shown that the extent of emphysema measured by pathology scores in resected lung tissue is well correlated with lung density values obtained from preoperative computed tomography (CT) of the lung (Gould et al., 1988; Bankier et al., 1999). We first developed a method to relate the lung mass density with the Hounsfield unit (HU) of CT (Zhou et al., 2020a) These estimated lung mass densities were labeled for training the deep neural network (DNN) model. In a previous study (Zhou et al., 2020a), a DNN model was developed using the data of 57 ILD patients and 20 healthy controls. The accuracy was 0.84 and 0.89, respectively, for the rectified linear activation unit (ReLU) and exponential linear unit (ELU). The lung mass density of the ILD patients (317.68 ± 59.69 kg/m3) was statistically higher than that of the healthy subjects (218.75 ± 23.05 kg/m3).
Machine learning (ML) has increasingly been recognized in the field of medical imaging. Among ML algorithms, the gradient boosting decision tree (GBDT) algorithm is regarded as best-in-class for small-to-medium structured/tabular data. Gradient boosting is an ensemble technique where new models which predict the residuals of prior models via a gradient descent algorithm to minimize the loss are constructed and then combined together sequentially to make the final prediction until no further improvements can be achieved (Friedman, 2001, 2002). GBDT is a widely used ML algorithm due to its efficiency, accuracy and interpretability. XGBoost (short for extreme gradient boosting) is a decision-tree-based ensemble ML implementation that uses a gradient boosting framework (Chen et al., 2015; Chen and Guestrin, 2016). It has widely been used by data scientists to achieve state-of-the-art results on many ML challenges (Mangal and Kumar, 2016). CatBoost is a GBDT implementation that efficiently handles both categorical and numerical features (Prokhorenkova et al., 2018). LightGBM is also a GBDT implementation which uses gradient-base one-side sampling and exclusive feature bundling techniques to reduce the number of data instances and the number of features for the situations where the feature dimension is high and data size is large (Ke et al., 2017).
This study is a continuation of our previous investigation on the prediction of lung mass density based on LUSWE (Zhou and Zhang, 2018). The objective of this study is to predict in vivo lung mass density for patients with ILD using different GBDT algorithms based on measurements from LUSWE and pulmonary function testing (PFT). The forced volume capacity (FVC) level in PFT has traditionally been regarded as the measure of disease severity, and a decrease in FVC has been associated with increased mortality in ILD.
II. MATERIALS AND METHODS
A. Datasets
This was a retrospective study. From February 2016 to May 2017, there was a cumulative total of 91 patients (M/F: 51/40, mean age: 62.4 ±_12.9 years) and 30 healthy controls (M/F: 16/14, mean age: 45.4 ± _14.6 years) who underwent LUSWE; however, among them, HRCT and PFT data were available for 57 patients (M/F: 30/29, mean age: 60.9 ± 14.3 years) and 20 healthy subjects (M/F: 10/10, mean age: 44 ± 15.4 years). For LUSWE, a Verasonics ultrasound system with an L11-4 ultrasound probe with a central frequency of 6.4 MHz is used. The mechanical index of 0.15 was used in this study. In this study, the surface wave speed on the lung is measured at three frequencies of 100, 150, and 200 Hz (Clay et al., 2019; Wiley et al., 2019). The frequency ranges chosen in this study consider the wave motion amplitude, spatial resolution and wave attenuation (Zhou et al., 2020b; Zhou et al., 2020c). The lung is tested at total lung capacity when the subject takes a deep breath and holds for a few seconds. Both lungs of the subject are tested. Surface wave speeds at three frequencies, predicted forced expiratory volume (FEV1%pre), ratio of forced expiratory volume to forced vital capacity (FEV1% = FVC%), age, and weight of patients and healthy subjects were used as features for training ML models. A fivefold cross-validation was conducted by dividing the dataset into training (80%) and testing (20%) and repeated five times to calculate mean values and standard deviations so as to assess the performance of different algorithms.
B. GBDT algorithms
XGBoost, CatBoost, and LightGBM libraries all use decision trees as the base weak learner and gradient boosting to iteratively fit a sequence of such trees. LightGBM uses gradient-based one-side sampling to filter out the data instances for finding a split value while XGBoost uses pre-sorted algorithm or computing the best split. CatBoost has the flexibility of giving indices of categorical column using one-hot encoding. These algorithms were implemented in Python using the libraries XGBoost, CatBoost, and LightGBM in this study.
C. Feature importance assessment
In order to evaluate the importance of features used in training the models, a framework called SHAP (Shapley additive explanations) was used (Lundberg and Lee, 2017). SHAP is a unified approach to explain the result of any deep learning model, which connects the theory of games with local explanations joining several methods and representing the additive feature attribution method. SHAP assigns each feature an importance value for a particular prediction. These explain the model output as a sum of real values attributed to each input feature. Given the various GBDT algorithms used in this study, an implementation of Tree SHAP called TreeExplainer, which is a fast and exact algorithm to compute SHAP values for trees and ensembles of trees, was used (Lundberg et al., 2019).
III. RESULTS
Mean-square error (MSE) and correlation coefficients (R2) of the test dataset of three algorithms were shown in Table I. XGBoost obtained the best results. Predefined and predicted lung mass densities of three algorithms for the test dataset were shown in Table II. It showed that the prediction of XGBoost was in agreement with the predefined lung mass densities more than CatBoost or LightGBM.
TABLE I.
Comparison in metrics of the regressor in different GBDT libraries.
| R2 | MSE Loss | |
|---|---|---|
| XGBoost | 0.932 ± 0.112 | 3109.38 ± 104.5 |
| CatBoost | 0.817 ± 0.235 | 4166.54 ± 365.47 |
| LightGBM | 0.558 ± 0.247 | 3643.38 ± 433.56 |
TABLE II.
Predefined and predicted lung mass densities from XGBoost, CatBoost, and LightGBM in the test dataset.
| Predefined | XGBoost | CatBoost | LightGBM |
|---|---|---|---|
| 353 | 343 | 351 | 324 |
| 379 | 379 | 365 | 321 |
| 292 | 300 | 313 | 321 |
| 325 | 329 | 331 | 328 |
| 312 | 313 | 309 | 305 |
| 279 | 288 | 303 | 302 |
| 305 | 304 | 326 | 308 |
| 337 | 342 | 364 | 321 |
| 271 | 272 | 280 | 295 |
| 238 | 245 | 283 | 278 |
| 327 | 330 | 332 | 304 |
| 278 | 277 | 300 | 289 |
| 235 | 231 | 223 | 270 |
| 189 | 195 | 204 | 260 |
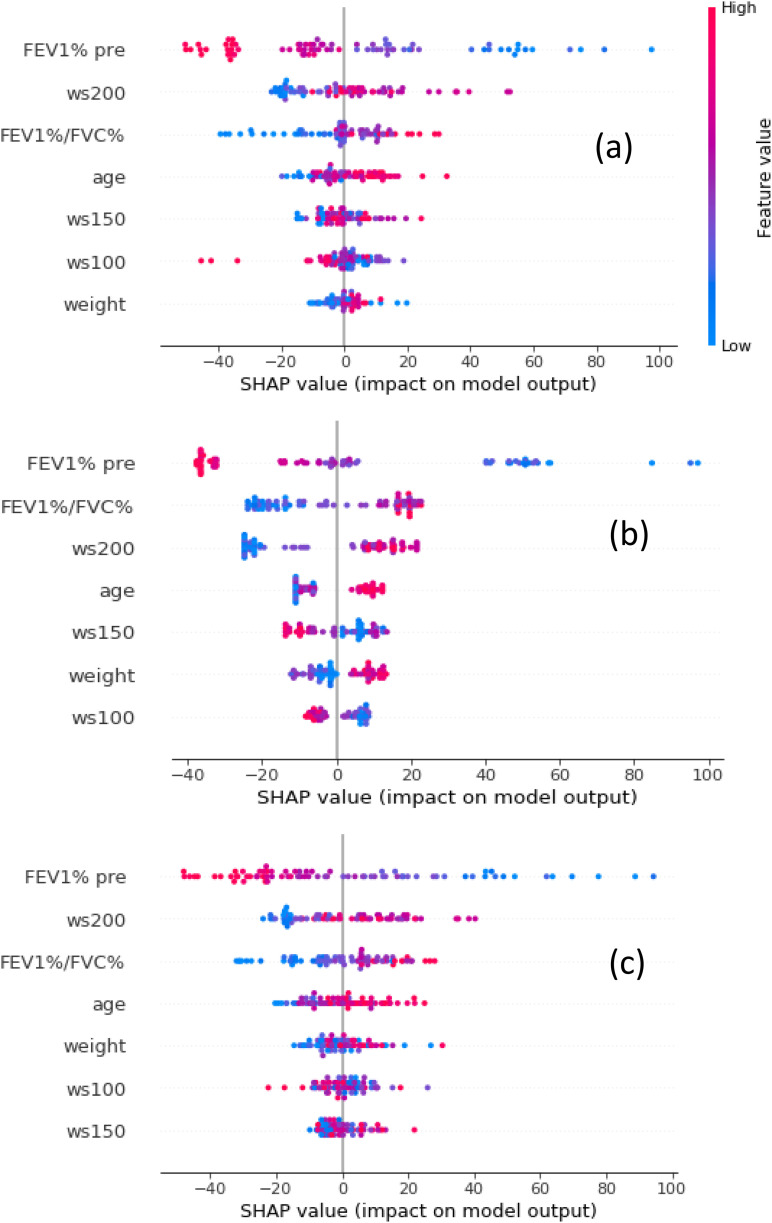
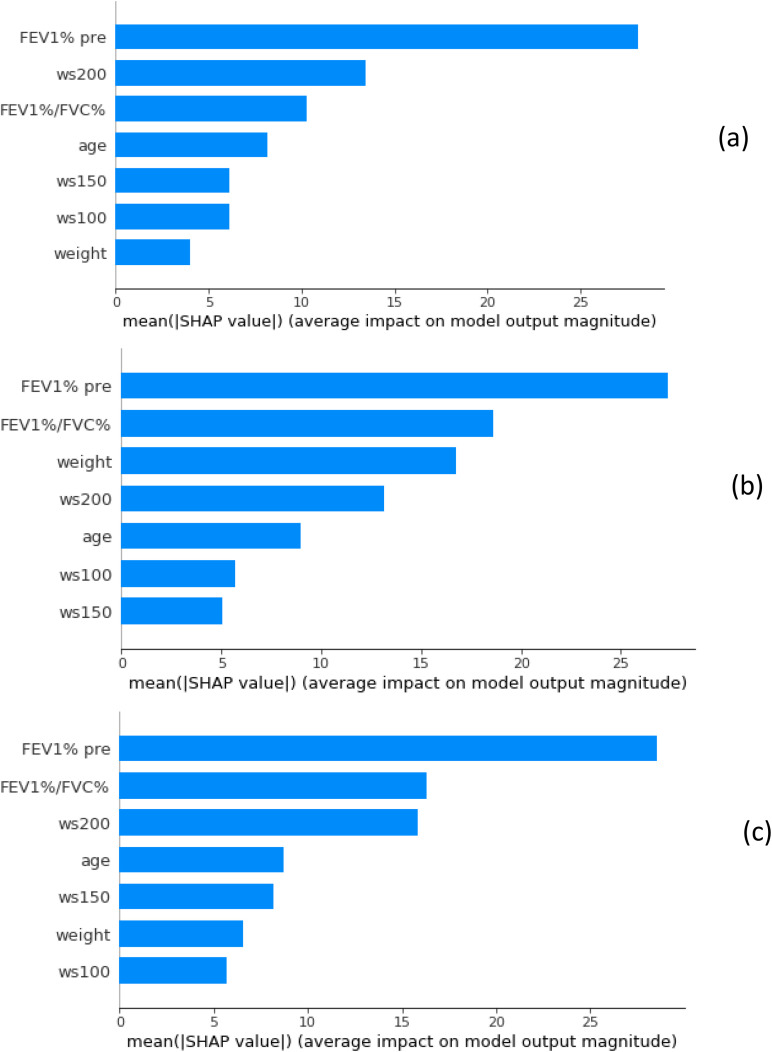
The importance of the seven features used in this study was shown in Figs. 1 and 2. In Fig. 1, features are sorted by the sum of the magnitudes of the SHAP values in all the samples, i.e., by their global impact. SHAP values are used to show the distribution of the impacts that each feature has on the model output. SHAP values are drawn horizontally. Each point represents a row of the dataset (FEV1% pre, FEV1%/FVC%, age, weight, ws100, ws150, ws200). The gradient color indicates the original value of that feature (high red, low blue). The average absolute value of the SHAP values for each feature was calculated to get a standard bar graph, Fig. 2. The SHAP values are represented along the horizontal axis and they quantify the change in log-odds.
FIG. 1.
(Color online) SHAP summary plots of the XGBoost (a), CatBoost (b), and LightGBM (c) models.
FIG. 2.
(Color online) The relative importance for each feature obtained by taking the average absolute value of the SHAP values from the XGBoost (a), CatBoost (b), and LightGBM (c) models.
IV. DISCUSSIONS
The aim of this study was to compare the performances of regressors from XGBoost, CatBoost, and LightGBM implementations to predict in vivo lung mass density based on age and weight of study subjects, as well as measurements obtained from LUSWE and PFT. The GBDT regressors were trained with surface wave speeds at three vibration frequencies, FEV1% pre and FEV1%/FVC% from PFT, age, weight, and predefined lung mass densities from HRCT. The obtained results show that XGBoost can be used to predict lung mass density with high accuracy.
It showed that the order of importance of the seven features changed with different algorithms, indicating that relative weights of features considering in XGBoost, CatBoost, and LightGBM. Moreover, FEV1% pre and FEV1%/FVC% are the top two features in all algorithms, suggesting good correlation between measurements from PFT to lung mass density predictions. This should be helpful for this technique combined with PFT to be translated in clinic. XGBoost is fast in execution and memory efficient compared with other GBDT algorithms and bagged decision trees. XGBoost is also dominant for structured or tabular datasets on classification and regression predictive modeling problems. In order to be used in gradient boosting algorithms, categorical features are preprocessed to convert categories to their target statistics which causes target leakage and prediction shift. Catboost uses an ordered boosting technique to avoid prediction shift caused by target leakage usually present in implementations of gradient boosting algorithms. CatBoost performs well only for categorical variables in the data. Since there are no categorical variables in our dataset, the CatBoost model cannot reach its maximum performance. LightGBM uses a novel technique of gradient-based one-side sampling to filter out the data instances for finding a split value while XGBoost uses pre-sorted algorithm and histogram-based algorithm for computing the best split. LightGBM can also handle categorical features by taking the input of feature names. It does not convert to one-hot coding and is much faster than one-hot coding. LGBM uses a special algorithm to find the split value of categorical features.
We used the seven features in this research. These features were obtained by clinical data and LUSWE measurements. It is interesting to know the contribution of each feature for predicting the lung mass density. We analyzed how SHAP values are attributed to the features globally in Figs. 1 and 2. In this case, all features are continuous and they are vertically sorted by their average impact on the predictions. We can see how FEV1% pre has the maximum impact of the prediction with high values of FEV1% pre associated with negative impacts on lung mass density predictions. High values of age are also significant and proportionally related to lung mass density predictions. FEV1%/FVC%, weight, age, and wave speeds at three frequencies, on the other hand, show a mixed pattern with high values associated both to high and low impacts on the predictions.
In this research, we use the CT data to label the lung mass density for training the DNN model. After the DNN models are developed, only the surface wave speeds and PFTs will be used to analyze lung mass density for patients. With known lung mass density, we can calculate two important biomarkers, lung elasticity, and viscosity, using LUSWE measurements. Thus, lung mass density, elasticity, and viscosity will be noninvasively analyzed for the first time. These novel biomarkers could prove useful for determining the clinical trajectory and prognosis of patients with ILD. CT is the clinical standard for lung assessment, but is costly, uses radiation, and typically is not performed often. In contrast, LUSWE can be used frequently to noninvasively evaluate the changes in lung mass and viscoelasticity. While conventional CT will remain the work-horse for initial lung disease diagnosis owing to its widespread availability and routine clinical adoption, we envision using LUSWE for follow up patient care so as to reduce potential radiation risk, mitigate the high costs associated with repeated CT.
Numerous deep learning techniques have been used in lung ultrasound imaging. This technique may be used to assess multiple lung disorders such as pulmonary edema when the alveoli are partly filled with blood or fluid. For congestive heart failure and inflammatory conditions, pulmonary edema is a basic characteristic. Extravascular lung water suggests worse development in critically ill patients and a higher risk of heart failure readmission or death (Gargani, 2015). In these “wet” lungs, ultrasound may penetrate deeper in the diseased tissue than healthy lung tissue, allowing imaging of deeper tissue (Zhou and Zhang, 2019). The development of a robust, quantitative method for lung mass density analysis has the potential to expand LUSWE in an almost exponential manner across a multitude of clinical applications. For example, quantitative LUSWE could be incorporated into the care of heart failure patients both in hospital practice and in outpatient clinics. In hospital practice, practitioners could perform daily LUSWE on patients undergoing diuresis for congestive heart failure and individually tailor medical therapy based on quantitative changes in LUSWE findings (Zhang et al., 2017b). Since each patient serves as their own reference standard, improvement in lung density through diuresis could be clearly documented and monitored. Outpatient providers could use quantitative LUSWE to monitor for increases in extravascular lung water and implement more timely changes in therapy.
Several limitations in this study should be taken into consideration when interpreting the results. First, this was a retrospective study on a dataset that was available at the time of the study. Further prospective investigation on the use of the GBDT in predicting in vivo lung mass density would be valuable. Second, the sample size used in this study is limited. The number of ILD patients and healthy subjects are not equal. Moreover, the age of ILD patients is statistically higher than that of healthy subjects. In the future, we plan to recruit more patients and older healthy subjects to enlarge the pool and further validate this technique. Third, lung density is dependent on pulmonary pressure. The measurements from LUSWE and PFT were obtained when the subject was taking a deep breath and holding. It could be difficult for critically patients with pulmonary fibrosis to hold their breath for more than one second. In this case, improvements need to be developed for LUSWE. In the future, we will investigate the relationship between lung mass density and pulmonary pressure using LUSWE, PFT, and GBDT.
V. CONCLUSIONS
In this study, we use different GBDT algorithms for in vivo lung mass density prediction based on measurements obtained from LUSWE and PFT, as well as subject age and weight information. The obtained results showed that given the tested algorithms and data, XGBoost outperforms the other two algorithms to predict in vivo lung mass density with high accuracy.
ACKNOWLEGEMENTS
This study is supported by NIH R01HL125234 from the National Heart, Lung and Blood Institute. We would like to thank Mrs. Jennifer Poston for editing this manuscript.
This paper is part of a special issue on Lung Ultrasound.
References
- 1.American Lung Association (2008). Lung Disease Data: 2008 ( American Lung Association, New York: ). [Google Scholar]
- 2. Bankier, A. A. , De Maertelaer, V. , Keyzer, C. , and Gevenois, P. A. (1999). “ Pulmonary emphysema: Subjective visual grading versus objective quantification with macroscopic morphometry and thin-section CT densitometry,” Radiology 211, 851–858. 10.1148/radiology.211.3.r99jn05851 [DOI] [PubMed] [Google Scholar]
- 3. Chen, T. , and Guestrin, C. (2016). “ Xgboost: A scalable tree boosting system,” in Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining (ACM), pp. 785–794. [Google Scholar]
- 4. Chen, T. , He, T. , Benesty, M. , Khotilovich, V. , and Tang, Y. (2015). “ XGBoost: Extreme gradient boosting,” R package version 0.4-2, pp. 1–4.
- 5. Clay, R. , Bartholmai, B. J. , Zhou, B. , Karwoski, R. , Peikert, T. , Osborn, T. , Rajagopalan, S. , Kalra, S. , and Zhang, X. (2019). “ Assessment of interstitial lung disease using lung ultrasound surface wave elastography: A novel technique with clinicoradiologic correlates,” J. Thoracic Imag. 34, 313–319. 10.1097/RTI.0000000000000334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Demi, L. , van Hoeve, W. , van Sloun, R. J. , Soldati, G. , and Demi, M. (2017). “ Determination of a potential quantitative measure of the state of the lung using lung ultrasound spectroscopy,” Sci. Rep. 7, 1–7. 10.1038/s41598-017-13078-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Demi, M. , Prediletto, R. , Soldati, G. , and Demi, L. (2020). “ Physical mechanisms providing clinical information from ultrasound lung images: Hypotheses and early confirmations,” IEEE Trans. Ultrasonics Ferroelectr. Freq. Control 67, 612–623. 10.1109/TUFFC.2019.2949597 [DOI] [PubMed] [Google Scholar]
- 8. Friedman, J. H. (2001). “ Greedy function approximation: A gradient boosting machine,” Ann. Stat. 29, 1189–1232. 10.1214/aos/1013203451 [DOI] [Google Scholar]
- 9. Friedman, J. H. (2002). “ Stochastic gradient boosting,” Comput. Stat. Data Anal. 38, 367–378. 10.1016/S0167-9473(01)00065-2 [DOI] [Google Scholar]
- 42. Gargani, L. (2015). “ Prognosis in heart failure: look at the lungs,” Eur. J. Heart Fail 17, 1086–1088. [DOI] [PubMed] [Google Scholar]
- 10. Gould, G. , MacNee, W. , McLean, A. , Warren, P. , Redpath, A. , Best, J. , Lamb, D. , and Flenley, D. (1988). “ CT measurements of lung density in life can quantitate distal airspace enlargement—An essential defining feature of human emphysema,” Am. Rev. Respiratory Disease 137, 380–392. 10.1164/ajrccm/137.2.380 [DOI] [PubMed] [Google Scholar]
- 11. Hudson, M. , Fritzler, M. J. , and Baron, M. (2010). “ Systemic sclerosis: Establishing diagnostic criteria,” Med. (Baltimore) 89, 159–165. 10.1097/MD.0b013e3181dde28d [DOI] [PubMed] [Google Scholar]
- 12. Ke, G. , Meng, Q. , Finley, T. , Wang, T. , Chen, W. , Ma, W. , Ye, Q. , and Liu, T.-Y. (2017). “ LightGBM: A highly efficient gradient boosting decision tree,” in Advances in Neural Information Processing Systems 30 (NIPS 2017), pp. 3146–3154. [Google Scholar]
- 13.Loscalzo, J. (2010). Harrison's Pulmonary and Critical Care Medicine ( Mc Graw Hill Medical, New York: ). [Google Scholar]
- 14. Lundberg, S. M. , Erion, G. , Chen, H. , DeGrave, A. , Prutkin, J. M. , Nair, B. , Katz, R. , Himmelfarb, J. , Bansal, N. , and Lee, S.-I. (2019). “ Explainable AI for trees: From local explanations to global understanding,” preprint arXiv:1905.04610. [DOI] [PMC free article] [PubMed]
- 15. Lundberg, S. M. , and Lee, S.-I. (2017). “ A unified approach to interpreting model predictions,” in Advances in Neural Information Processing Systems, pp. 4765n4774. [Google Scholar]
- 16. Mangal, A. , and Kumar, N. (2016). “ Using big data to enhance the Bosch production line performance: A Kaggle challenge,” in 2016 IEEE International Conference on Big Data (Big Data) (IEEE), pp. 2029–2035. [Google Scholar]
- 17. Mathieson, J. R. , Mayo, J. R. , Staples, C. A. , and Muller, N. L. (1989). “ Chronic diffuse infiltrative lung disease: Comparison of diagnostic accuracy of CT and chest radiography,” Radiology 171, 111–116. 10.1148/radiology.171.1.2928513 [DOI] [PubMed] [Google Scholar]
- 18. Mayo, J. R. (2009). “ CT evaluation of diffuse infiltrative lung disease: Dose considerations and optimal technique,” J. Thorac Imag. 24, 252–259. 10.1097/RTI.0b013e3181c227b2 [DOI] [PubMed] [Google Scholar]
- 19. Mento, F. , Soldati, G. , Prediletto, R. , Demi, M. , and Demi, L. (2020). “ Quantitative lung ultrasound spectroscopy applied to the diagnosis of pulmonary fibrosis: The first clinical study,” IEEE Trans. Ultrason. Ferroelectr. Freq. Control 67, 2265–2273. 10.1109/TUFFC.2020.3012289 [DOI] [PubMed] [Google Scholar]
- 20. Miller, D. L. , Dong, Z. , Dou, C. , and Raghavendran, K. (2018). “ Pulmonary capillary hemorrhage induced by different imaging modes of diagnostic ultrasound,” Ultrasound Med. Biol. 44, 1012–1021. 10.1016/j.ultrasmedbio.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mohanty, K. , Blackwell, J. , Egan, T. , and Muller, M. (2017). “ Characterization of the lung parenchyma using ultrasound multiple scattering,” Ultrasound Med. Biol. 43, 993–1003. 10.1016/j.ultrasmedbio.2017.01.011 [DOI] [PubMed] [Google Scholar]
- 22. Prokhorenkova, L. , Gusev, G. , Vorobev, A. , Dorogush, A. V. , and Gulin, A. (2018). “ CatBoost: Unbiased boosting with categorical features,” in Advances in Neural Information Processing Systems, pp. 6638–6648. [Google Scholar]
- 23. Roy, S. , Menapace, W. , Oei, S. , Luijten, B. , Fini, E. , Saltori, C. , Huijben, I. , Chennakeshava, N. , Mento, F. , Sentelli, A. , Peschiera, E. , Trevisan, R. , Maschietto, G. , Torri, E. , Inchingolo, R. , Smargiassi, A. , Soldati, G. , Rota, P. , Passerini, A. , Sloun, R. J. G. V. , Ricci, E. , and Demi, L. (2020). “ Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound,” IEEE Trans. Med. Imag. 39, 2676–2687. 10.1109/TMI.2020.2994459 [DOI] [PubMed] [Google Scholar]
- 24. Soldati, G. , Demi, M. , and Demi, L. (2019a). “ Ultrasound patterns of pulmonary edema,” Ann. Trans. Med. 7, S16. 10.21037/atm.2019.01.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soldati, G. , Demi, M. , Smargiassi, A. , Inchingolo, R. , and Demi, L. (2019b). “ The role of ultrasound lung artifacts in the diagnosis of respiratory diseases,” Expert Rev. Respir. Med. 13, 163–172. 10.1080/17476348.2019.1565997 [DOI] [PubMed] [Google Scholar]
- 26. Soldati, G. , Smargiassi, A. , Inchingolo, R. , Buonsenso, D. , Perrone, T. , Briganti, D. F. , Perlini, S. , Torri, E. , Mariani, A. , Mossolani, E. E. , Tursi, F. , Mento, F. , and Demi, L. (2020). “ Proposal for international standardization of the use of lung ultrasound for patients with COVID-19: A simple, quantitative, reproducible method,” J. Ultrasound. Med. 39, 1413–1419. 10.1002/jum.15285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Steen, V. D. , and Medsger, T. A., Jr. (2000). “ Severe organ involvement in systemic sclerosis with diffuse scleroderma,” Arthritis Rheum. 43, 2437–2444. [DOI] [PubMed] [Google Scholar]
- 28. Verschakelen, J. A. (2010). “ The role of high-resolution computed tomography in the work-up of interstitial lung disease,” Curr. Opin. Pulm. Med. 16, 503–510. 10.1097/MCP.0b013e32833cc997 [DOI] [PubMed] [Google Scholar]
- 29. Wiley, B. M. , Zhou, B. , Pandompatam, G. , Zhou, J. , Olgun Kucuk, H. , and Zhang, X. (2019). “ Application of lung ultrasound surface wave elastography for assessment of extravascular lung water in patients hospitalized with congestive heart failure,” J. Acoust. Soc. Am. 145, 1673–1674. 10.1121/1.5101139 [DOI] [Google Scholar]
- 30. Xu, J. , Kochanek, K. D. , Murphy, S. L. , and Tejada-Vera, B. (2010). “ Deaths: Final data for 2007,” Nat. Vital Stat. Rep. 58, 1–136. [PubMed] [Google Scholar]
- 31. Zhang, X. , Osborn, T. , Zhou, B. , Meixner, D. , Kinnick, R. , Bartholmai, B. , Greenleaf, J. , and Kalra, S. (2017a). “ Lung ultrasound surface wave elastography: A pilot clinical study,” IEEE Trans. Ultrason. Ferroelectr. Freq. Control 64, 1298–1304. 10.1109/TUFFC.2017.2707981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang, X. , Qiang, B. , Hubmayr, R. D. , Urban, M. W. , Kinnick, R. , and Greenleaf, J. F. (2011). “ Noninvasive ultrasound image guided surface wave method for measuring the wave speed and estimating the elasticity of lungs: A feasibility study,” Ultrasonics 51, 289–295. 10.1016/j.ultras.2010.09.005 [DOI] [PubMed] [Google Scholar]
- 33. Zhang, X. , Zhou, B. , Kalra, S. , Bartholmai, B. , Greenleaf, J. , and Osborn, T. (2018a). “ An ultrasound surface wave technique for assessing skin and lung diseases,” Ultrasound Med. Biol. 44, 321–331. 10.1016/j.ultrasmedbio.2017.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang, X. , Zhou, B. , Osborn, T. , Bartholmai, B. , Greenleaf, J. , and Kalra, S. (2017b). “ Assessment of interstitial lung disease using lung ultrasound surface wave elastography,” in 2017 IEEE International Ultrasonics Symposium (IUS), pp. 1–4. [Google Scholar]
- 35. Zhang, X. , Zhou, B. , Osborn, T. , Bartholmai, B. , and Kalra, S. (2018b). “ Lung ultrasound surface wave elastography for assessing interstitial lung disease,” IEEE Trans. Biomed. Eng. 66, 1346–1352. 10.1109/TBME.2018.2872907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou, B. , Bartholmai, B. J. , Kalra, S. , Osborn, T. G. , and Zhang, X. (2019). “ Lung US surface wave elastography in interstitial lung disease staging,” Radiology 291, 479–484. 10.1148/radiol.2019181729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou, B. , Bartholmai, B. J. , Kalra, S. , and Zhang, X. (2020a). “ Predicting lung mass density of patients with interstitial lung disease and healthy subjects using deep neural network and lung ultrasound surface wave elastography,” J. Mech. Behav. Biomed. Mater. 104, 103682. 10.1016/j.jmbbm.2020.103682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou, B. , Schaefbauer, K. J. , Egan, A. M. , Carmona Porquera, E. M. , Limper, A. H. , and Zhang, X. (2020c). “ An ex vivo technique for quantifying mouse lung injury using ultrasound surface wave elastography,” J. Biomech. 98, 109468. 10.1016/j.jbiomech.2019.109468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou, B. , and Zhang, X. (2018). “ Lung mass density analysis using deep neural network and lung ultrasound surface wave elastography,” Ultrasonics 89, 173–177. 10.1016/j.ultras.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou, B. , and Zhang, X. (2019). “ The effect of pleural fluid layers on lung surface wave speed measurement: Experimental and numerical studies on a sponge lung phantom,” J. Mech. Behav. Biomed. Mater. 89, 13–18. 10.1016/j.jmbbm.2018.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]