Abstract
Cord blood (CB) mononuclear cell populations have demonstrated significant promise in biomaterials-based regenerative therapies; however, the contributions of monocyte and macrophage subpopulations towards proper tissue healing and regeneration are not well understood, and the phenotypic responses of macrophage to microenvironmental cues have not been well-studied. In this work, we evaluated the effects of cytokine stimulation and altered substrate stiffness. Macrophage derived from CB CD14+ monocytes adopted distinct inflammatory (M1) and anti-inflammatory (M2a and M2c) phenotypes in response to cytokine stimulation (M1: lipopolysaccharide (LPS) and interferon (IFN-γ); M2a: interleukin (IL)-4 and IL-13; M2c: IL-10) as determined through expression of relevant cell surface markers and growth factors. Cytokine-induced macrophage readily altered their phenotypes upon sequential administration of different cytokine cocktails. The impact of substrate stiffness on macrophage phenotype was evaluated by seeding CB-derived macrophage on 3wt%, 6wt%, and 14wt% poly(ethylene glycol)-based hydrogels, which exhibited swollen shear moduli of 0.1, 3.4, and 10.3 kPa, respectively. Surface marker expression and cytokine production varied depending on modulus, with anti-inflammatory phenotypes increasing with elevated substrate stiffness. Integration of specific hydrogel moduli and cytokine cocktail treatments resulted in the differential regulation of macrophage phenotypic biomarkers. These data suggest that CB-derived macrophages exhibit predictable behaviors that can be directed and finely tuned by combinatorial modulation of substrate physical properties and cytokine profiles.
Keywords: Hydrogel, Cord blood, Macrophage, Cytokines
Graphical Abstract
1. Introduction
Cord blood (CB) mononuclear cells have shown significant promise as regenerative therapies for a variety of conditions, including reconstituting hematologic systems and repairing tissue damage in both pediatric and adult populations [1]. The successful use of CB cells for regenerative medicine has been attributed to the immunological immaturity of the cells, which allows increased transplant toleration between unrelated donors and recipients [2]. Moreover, CB mononuclear cell populations contain a high proportion of stem and progenitor cells with the capacity to regenerate functional tissue architecture when combined with instructive biomaterials [3]. Unfortunately, although more than 5.8 million units of unrelated CB are stored in public and private banks for potential use, the significant therapeutic promise of CB in clinical settings has not yet been fully realized [4–6]. Controlling the high degree of plasticity observed in CB-derived cells, demonstrated by their ability to differentiate towards immune, endothelial, cardiac, neuronal, hepatic, and epithelial lineages [7], remains a critical challenge, and importantly, there is a significant need to better understand the factors that influence CB-derived mononuclear cell activities in engineered regenerative therapies, particularly the context of myocardial repair, neuroregeneration, and revascularization.
Engineered microenvironments have been used ex vivo with CB-derived cells to condition the cells toward specific phenotypes [8], to propagate them [9, 10], and to build neo-tissues for implantation [11]. Hydrogel materials have also been used in conjunction with CB cell products in vivo [12]. Much emphasis has been placed on defining the role of CB hematopoietic stem cells, often defined as CD34+, and endothelial progenitor cells, currently demarcated as CD34+/CD133+/VEGFR2+, in promoting healing and tissue regeneration in these engineered contexts [3, 8]. However, little emphasis has been placed on understanding the critical roles of CB-derived CD14+ monocytes and macrophages, which are significant components of CB-derived populations [13] and which, in addition to CB stem cells, have been implicated in the regeneration of several tissue types [14, 15]. Although CB-derived macrophage fundamentally differ from macrophage derived from other sources [16], the regenerative capacities of CB-derived macrophage mimic those of macrophage in adult tissues, which are known to orchestrate wound healing, inflammation, angiogenesis, and tissue regeneration [17–19].
Proper tissue healing and regeneration are contingent on the spatiotemporal activation and dissipation of distinct macrophage sub-populations, with pro-inflammatory (M1) macrophages arising at early time points (1–3 days) and anti-inflammatory (M2) phenotypes arising at later stages in both wound healing and biomaterials-mediated inflammation [20]. Though the contribution of macrophage phenotype switching versus the recruitment of new macrophages exhibiting an altered phenotype to the site of healing remains unclear, it is known that activated macrophages are dynamic cells, and the adoption of M1 or M2 phenotypes is not permanent – macrophages can switch between phenotypes and even adopt hybrid phenotypes based on microenvironmental cues [21, 22]. In research models, macrophage phenotypes are conventionally induced from resting (Mϕ) cells by the addition of cytokines (M1: lipopolysaccharide (LPS) and interferon (IFN-γ); M2a: interleukin (IL)-4 and IL-13; M2c: IL-10).
In vivo and in biomaterials-based regenerative therapies, these cells encounter diverse microenvironments, and macrophage phenotypes are influenced by dynamic alterations in the physical properties of extracellular matrices within different tissues. Bioengineering studies have expanded our knowledge about how physical cues, such as matrix rigidity and topography, influence the functions of macrophage phenotypes among cells derived from human peripheral blood, murine bone marrow, or monocyte/macrophage-like cells lines. Alterations in substrate stiffness have been demonstrated to influence the attachment, morphology, migration, and proliferation of macrophages [23, 24]. Further, macrophage phenotypic indicators, including cell surface markers and secreted cytokines, can be influenced by substrate stiffness; however, generalizable mechanisms remain elusive, with multiple research groups reporting variable findings regarding the phenotypic responses of macrophage to altered substrate stiffness [25–27]. Critically, despite the potential therapeutic importance of CB, the effects of tissue substrate physical properties on CB-derived macrophage have not been well-characterized. Here, we examined pro- vs. anti-inflammatory modulation of macrophage by hydrogel modulus, which is a microenvironmental parameter that can be readily tuned to elicit desired cellular effects.
In this study, we utilized resting Mϕ macrophage from human CB-derived CD14+ monocytes and assessed the capacity of the cells to generate polarized M1, M2a, and M2c macrophage phenotypes in response to changes in the stiffness of simple poly(ethylene glycol) (PEG) substrates. PEG-based hydrogel platforms with substrate shear storage moduli between 0.1 and 10.3 kPa were employed, given the relevance of this range of modulus values in stromal tissues, as motivated by our and others’ previous work [8, 28, 29], and due to the facile deployment of modified PEG-based hydrogels in tissue regenerative approaches. Further, we investigated the response of CB-derived macrophage exposed to simultaneous alterations in substrate stiffness and cytokines directed at invoking phenotypic responses. The analysis of cell morphology, proliferation, surface markers, and secreted growth factors establishes associations between substrate physical properties and CB-derived cell behavior, suggesting the combinatorial influences of substrate stiffness and exogenous cytokines could be deployed to guide macrophage fate.
2. Materials and Methods
2.1. Monocyte Isolation
Fresh CB units were obtained through an IRB-approved agreement with the Carolinas Cord Blood Bank at Duke University (Durham, NC, USA). Units were collected with citrate-phosphate-dextrose anticoagulant and de-identified prior to shipping on ice. Cells were isolated using reagents and protocols from Miltenyi Biotec (Bergisch Gladbach, Germany) unless otherwise noted. In brief, CB samples were diluted 1:4 with cold autoMACS® Rinsing Solution, and mononuclear cells were isolated using a step gradient of Ficoll-Paque Premium (GE Healthcare, Pittsburgh, PA) at a specific gravity of 1.078 g/cc. Isopycnic centrifugation was performed at 400×g for 35 min. The mononuclear cell layer was collected, washed three times with autoMACS® Rinsing Solution, and the final cell pellet was resuspended in 500 μL of autoMACS® Rinsing Solution containing 3% BSA (3% BSA-MACS® buffer) prior to filtration through a 30 μm nylon mesh pre-separation filter. Magnetic beads coated with a monoclonal antibody specific for human CD14 were used to positively select for CD14+ cells. First, 20 μL of CD14 microbeads were added to every 80 μL cells and incubated for 15 min at 4°C. Cells were washed once by centrifugation, resuspended in 3% BSA-MACS® buffer, and CD14+ cells were then isolated using magnetic cell separation. Specifically, cells were applied to a MACS® LS column containing ferromagnetic spheres in the presence of a strong magnetic field and rinsed with 3 mL 3% BSA-MACS® buffer three times; the magnetic beads and bound cells were retained in the column. The column was removed from the magnetic field and the labeled cells were collected. Resulting monocytes were enumerated using a hemocytometer and cryopreserved in 90% FBS with 10% DMSO in liquid nitrogen until use. The yield of CD14+ monocytes, assessed by flow cytometry using a FITC-conjugated anti-CD14 (clone TÜK4, 1:11 dilution, Miltenyi Biotec) was ~95% (Table S1).
2.2. Preparation of Macrophage Phenotypes
Monocytes were cultured in ultra-low attachment polystyrene plates in RPMI 1640 medium (ATCC, Manassas, VA) with 10% heat-inactivated fetal bovine serum (Gibco, Gaithersburg, MD), 1% penicillin-streptomycin (ThermoFisher Scientific, Waltham, MA), and 20 ng/mL human monocyte colony stimulating factor (M-CSF; Peprotech, Rocky Hill, NJ) to differentiate the cells into macrophages (1x106 cells/mL, 2x105 cell/cm2). Culture medium was replaced on day 3. After 6 days of differentiation, macrophages were disaggregated using Accutase® (Sigma-Aldrich, St. Louis, MO), collected, and reseeded onto ultra-low attachment polystyrene plates (2x105 cells/mL, 5x104 cell/cm2; Nunc, Rochester, NY). Macrophage polarization was initiated by changing to complete medium supplemented with the following cytokines: 100 ng/mL lipopolysaccharide (LPS; Sigma-Aldrich) and 100 ng/mL interferon-gamma (IFNγ) for M1; 40 ng/mL IL-4 and 20 ng/mL IL-13 for M2a; and 40 ng/mL IL-10 for M2c (all cytokines obtained from Peprotech). Unactivated macrophages (Mϕ) were also cultured over the same time periods using complete medium. Polarization media were replenished on day 3. To characterize the ability of macrophages to switch phenotypes, a subset of experiments was carried out where the medium of M1 macrophages was switched to either M2a- or M2c-polarizing media, the medium of M2a macrophages was switched to either M1- or M2c-polarizing media, and the medium of M2c macrophages was switched to either M1- or M2a-polarizing media during the media change on day 3. After 3 or 6 days of culture, polarized and Mϕ cells were collected and analyzed by flow cytometry. Conditioned media samples were centrifuged at 400xg for 10min and frozen at −80°C until analysis.
2.3. Peptide Synthesis & Characterization
The peptide GRGDSPC was prepared using a Liberty Blue automated peptide synthesizer (CEM, Mathews, NC) with microwave assisted Fmoc-mediated solid phase synthesis, at 0.25 mmol scale using Rink-Amide resin (ChemPep Inc, Wellington, FL). The peptide was cleaved from the resin in cleavage cocktail (92.5% trifluoroacetic acid, 2.5% 1,2 ethanedithiol, 2.5% triisopropylsilane, and 2.5% H2O) at room temperature for 20 mins and precipitated in cold diethyl ether. Peptide purification was performed by reverse-phase high-performance liquid chromatography (Waters Corporation, Millford, MA), using a BEH130 Prep C18 10-μm column (XBridge, Waters Corporation). Crude peptides were dissolved in Milli-Q water containing 0.1% (v/v) TFA and were filtered (0.20-μm filter, Corning Inc.) before HPLC injection. The products were subjected to an elution gradient of 100% solvent A (0.1% (v/v) TFA in Milli-Q water) to 70% solvent A within 30 min; solvent B consisted of acetonitrile with 0.1% (v/v) TFA. Peptide fractions were detected using ultraviolet-visible detection at 214 nm (Waters 2489, Waters Corporation) and collected. Target product M/Z was verified using a Waters Xevo G2-XS QTof mass spectrometer (calculated exact mass= 689.7 Da, observed [M+H]+= 690.8) and was stored, under argon, as a lyophilized powder at −20°C until use.
2.4. Hydrogel Formation
Hydrogels were formed by crosslinking four-arm thiol-functionalized poly(ethylene glycol) (PEG-SH4; f=4, Mn 10,000 g/mol, JenKem Technology, Allen, TX) and four-arm maleimide-functionalized poly(ethylene glycol) (PEG-Mal4; f=4, Mn 10,000 g/mol, JenKem Technology). RGD peptides were incorporated into the hydrogels via reaction with the maleimide-functionalized PEG prior to hydrogel formation to provide cell-adhesive materials. Hydrogels were prepared at concentrations of 3wt%, 6wt%, and 14wt% (w/v%) by altering the concentrations of PEG-SH4 and PEG-Mal4, while the concentration of RGD was held constant at 1 mM, independent of hydrogel formulation. Hydrogels of a concentration of 3wt% comprised 1 mM PEG-SH4 and 2 mM PEG-Mal4, 6wt% comprised 3 mM PEG-SH4 and 3 mM PEG-Mal4, and 14wt% comprised 7 mM PEG-SH4 and 7 mM PEG-Mal4. To form hydrogels, RGD was pre-reacted with PEG-Mal4 in 10 mM citrate buffer, pH 4.5. PEG-SH4 was dissolved in 10 mM citrate buffer, pH 4.5, and both PEG solutions were sterilized through 0.2 μm very-low protein binding Durapore® PVDF filters (Millipore, Burlington, MA). Maleimide- and thiol-containing solutions were then mixed together via simple pipetting and deposited onto cell culture dishes (240 μL in a 24 well plate). Hydrogels were allowed to crosslink for 20 mins at 37°C. Following overnight storage in HBSS (pH 7.4; Gibco) at 4°C, hydrogels incubated in cell culture medium for 30 mins at 37°C before cell culture experiments.
2.5. Rheological Characterization
The mechanical properties of the swollen hydrogels were characterized via bulk oscillatory rheology (AR-G2, TA instruments), similar to previous reports [30–32]. Detailed methods are located in the supplementary materials. Briefly, the hydrogel precursor solutions were added to a cylindrical mold (30 μL; diameter = 4.6 mm, thickness = 1.8 mm) and allowed to crosslink for 20 mins at 37°C. Following gelation, hydrogels were carefully removed from the molds with a spatula, immersed in 1.5 mL of complete medium in a 24-well plate, and incubated at 37°C with 5% CO2 for 24 hrs. The swollen hydrogels were transferred to the rheometer, and elastic storage moduli of the swollen hydrogels were measured using a 20-mm diameter stainless steel, parallel plate geometry. Time sweep measurements were obtained within the linear viscoelastic regime using 2% constant strain and 2 rad/s angular frequency. A normal force of 0.2 N was applied to prevent slipping during measurement.
2.6. Culture of Macrophage on Hydrogels
To understand the effects of substrate stiffness in directing macrophage phenotype, CB monocytes were differentiated into macrophage with M-CSF for 6 days, as described above. After 6 days of differentiation, macrophages were collected using Accutase® and reseeded on the 240-μL PEG hydrogels (2x105 cells/mL, 5x104 cell/cm2) exhibiting various stiffnesses and cultured in complete medium for 6 days, with medium replenished on day 3. After 3 or 6 days of culture on hydrogels, macrophages were collected and analyzed by flow cytometry. Conditioned media samples were centrifuged at 400xg for 10min and frozen at −80°C until analysis. In a subset of experiments, macrophages were cultured for 3 days in complete medium supplemented with the following cytokines: 100 ng/mL IFNγ and 100 ng/mL LPS; 40 ng/mL IL-4 and 20 ng/mL IL-13; and 40 ng/mL IL-10. Culture medium was not replaced over the 3-day culture period.
2.7. Immunostaining
After 6 days of differentiation with M-CSF, Mϕ macrophages were fixed with 4% paraformaldehyde for 30 minutes, permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) for 15 min and blocked with 3% bovine serum albumin (Sigma-Aldrich) in D-PBS (Gibco) for 30 min. Samples were stained for 1 hr with primary antibodies (Table S2) at room temperature with shaking, rinsed, and incubated with secondary antibodies (Table S2) and the nuclear stain Hoechst 33258 (1:1000; Life Technologies) for 1 hr at room temperature with shaking. Cells were visualized using an EVOS® FL Auto Imaging System with a 20x LPlanFL 0.4 N.A. objective, controlled by EVOS® FL Auto software (version 1.6; ThermoFisher).
2.8. Characterization of Macrophage Phenotype
After 3 or 6 days of culture on ultra-low attachment polystyrene or PEG-based hydrogels, photomicrography was carried out using an EVOS® FL Auto Imaging System (ThermoFisher) with a 20x LPlanFL 0.4 N.A. objective, controlled by EVOS® FL Auto software (version 1.6; ThermoFisher). Morphological changes of macrophages were analyzed via ImageJ using images taken at selected time points. Area represents the total pixel area measured and converted to μm2 for analysis. Circularity was calculated using the following equation: 4π*area/perimeter2, where a value of 1 corresponds to a perfect circle, and a value approaching 0 indicates an increasingly elongated shape. Shape descriptors were calculated using >30 cells per treatment group.
Macrophages were then harvested for flow cytometry and conditioned media samples were centrifuged at 400xg for 10 min to remove debris and frozen at −80°C until analysis. Macrophages were collected via Accutase®, centrifuged at 300xg for 5 mins, and resuspended in 3% BSA-MACS® buffer. A small portion of the cells was used to determine viability by trypan blue dye exclusion (0.4% Trypan Blue Solution, Sigma-Aldrich), using a hemocytometer. To evaluate macrophage proliferation over time, the average number of cells counted in the hemocytometer was extrapolated to the average number of cells per well, which was normalized to number of cells initially seeded on day 0.
The remainder of the cells were triple-stained with phycoerythrin-conjugated anti-CCR7 (clone FR11–11E8), allophycocyanin (APC)-conjugated anti-CD163 (clone GHI/61.1), and APC-Vio770-conjugated anti-CD206 (clone DCN228) antibodies for 10 mins at 4°C, in accordance with the manufacturers’ instructions (all from Miltenyi Biotec and at a dilution of 1:11). Corresponding isotype controls were used as recommended by the manufacturer (Miltenyi Biotec; Table S2). The samples were then rinsed with 3% BSA-MACS® buffer, fixed with 2% paraformaldehyde in PBS (pH 7.4), and rinsed again with PBS. Labeled cells were analyzed using a Novocyte 3000 flow cytometer (ACEA Biosciences, San Diego, CA). Data were processed using NovoExpress Software (Version 1.2.4; ACEA Biosciences).
2.9. Secreted Protein Quantification Using ELISA
IL-6, TNF-α, IL-1β, and VEGF-A were measured with Meso Scale Discovery (MSD) electrochemiluminescence detection human U-PLEX kits, while TGF-β1 and TGF-β2 levels in collected medium were measured with an MSD human Cytokine Multiplex kit (Mesoscale Discovery, Rockville, MD). MMP-9 levels in the collected medium were quantified via Quantikine ELISA kits (R&D Systems, Minneapolis, MN). PDGF-BB levels in the collected medium were quantified via ELISA kits Development Kits (Peprotech).
2.10. Hierarchical Clustering Analysis
To better visualize complex sets of expression patterns, hierarchical clustering (HCL) analysis was utilized. In experiments where macrophages were sequentially exposed to cytokine cocktails, surface marker and cytokine data obtained for macrophages exposed to new cytokine cocktail conditions was normalized relative to their predecessors (e.g. M1→M2a relative to M1, etc.) and the normalized values were converted to log2 scale. Similarly, surface marker and cytokine data were converted to log2 fold changes for macrophages exposed to cytokine cocktails on hydrogels relative to macrophages on hydrogels alone. Hierarchical clustering (HCL) analysis, carried out using The Broad Institute’s online Morpheus software package (https://software.broadinstitute.org/morpheus), allowed biomarkers with similar expression patterns to be considered together. Clusters were identified for the phenotypic biomarkers analyzed, and a heat map was generated from log-transformed data to depict the relationship among surface markers or cytokines.
2.11. Statistics
Data are expressed as the mean ± standard error of the mean, unless otherwise noted. Details regarding the specific test utilized to evaluate each experiment are indicated in figure captions. In brief, assumptions of residual normality (Shapiro–Wilks test) and homoscedasticity (Bartlett’s test) were first verified for each group. After confirming that assumptions for parametric analysis were met, statistical significance was analyzed by performing either a one‐way or two‐way ANOVA, as appropriate, if the F‐test revealed significant statistical differences at the 0.05 level, pairwise comparisons were made using either Tukey HSD or Dunnett’s post‐hoc tests. Statistical interpretations were made using JMP Pro 14 (SAS Institute Inc.).
3. Results
3.1. Polarization of Cord Blood Derived Macrophage with Cytokines Leads to Specific and Discernible Phenotypes
Naïve macrophage (Mϕ) were generated by treating CB-derived CD14+ monocytes with M-CSF for 6 days on ultra-low attachment polystyrene (Figure 1A). After 6 days of treatment with M-CSF, adherent Mϕ cells expressed a mixture of morphologies, including highly refractive small cells and very large, flat, vesiculated cells (Figure S1A). Although various morphologies were observed following differentiation with M-CSF, immunocytochemistry experiments confirmed that Mϕ cells expressed CD11b and CD68, surface markers expressed by monocytes/macrophages (Figure S1B–C).
Figure 1.
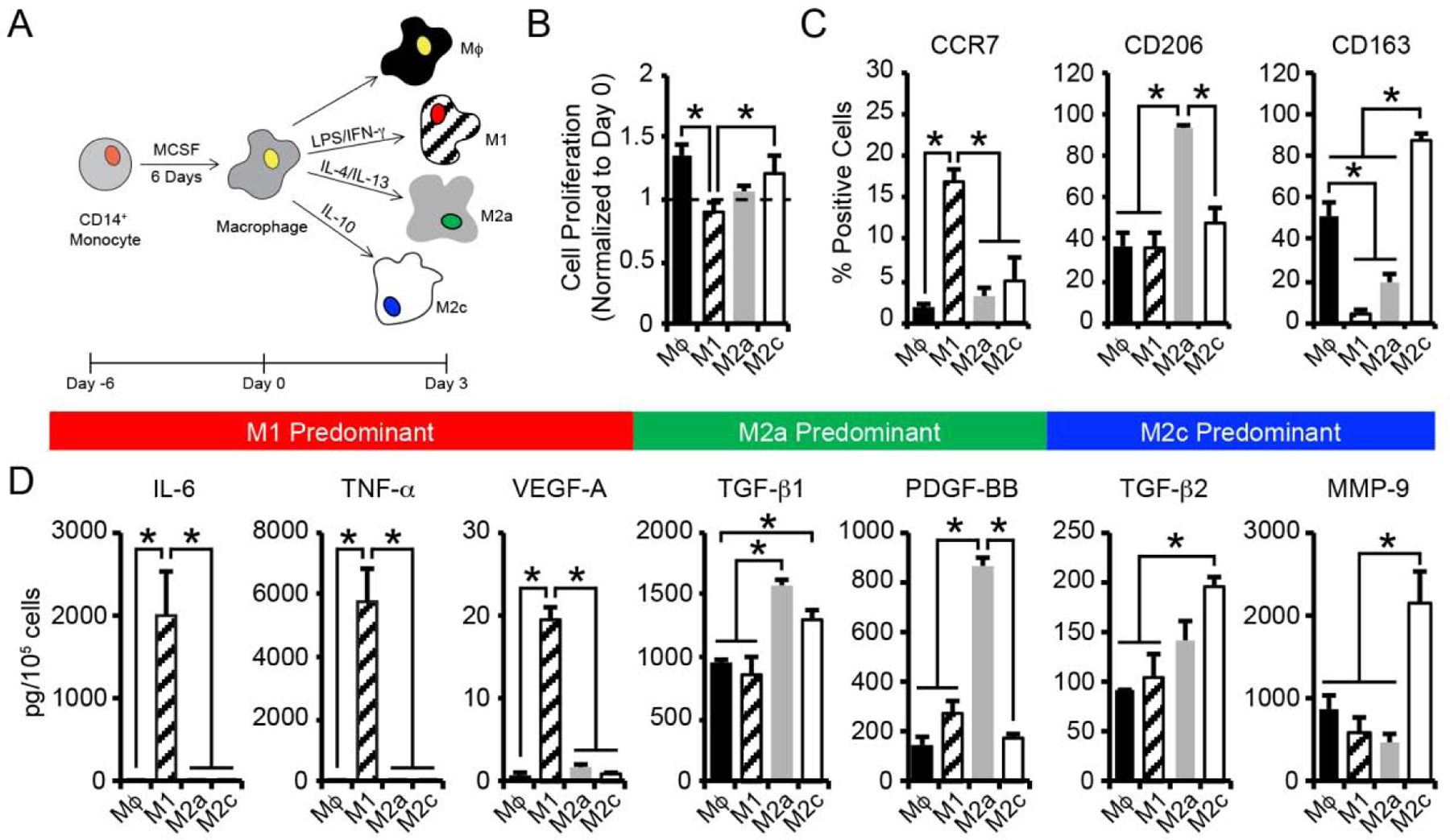
(A) CB monocytes were differentiated to macrophages (Mϕ) and polarized to 3 different phenotypes (M1, M2a, M2c) using established cytokine cocktails. (B) Macrophage cell proliferation was determined by enumerating the number cells present after 3 days of culture, normalized to the number of cells initially seeded on day 0. (C) Flow cytometric analysis of the percentage of macrophage staining positively for each phenotypic surface marker. (D) Quantification of secreted proteins from macrophage phenotypes. In B-D, data are represented as the mean ± SEM, with n = 3 human donors per condition. A one-way ANOVA, followed by a Tukey HSD post hoc test, was used to detect statistical significance, *p<0.05.
To determine their activation potential, CB-derived, naïve Mϕ were polarized to adopt either M1, M2a, or M2c phenotypes through the addition of cytokine cocktails (Figure 1A). Specifically, a cocktail of LPS and IFN-γ was used to invoke a pro-inflammatory M1 phenotype, IL-4 and IL-13 to stimulate M2a polarization, and IL-10 alone to promote an M2c phenotype. In addition, naïve Mϕ were maintained in culture for comparison. The morphologies of the polarized macrophage phenotypes were evaluated after 3 days. The naïve Mϕ elicited ovoid-shaped morphologies (circularity: 0.84 ± 0.01) with a spread area of 1863 ± 59 μm2 (Figure S2A–C). Cells treated with LPS/IFN-γ to stimulate an M1 phenotype formed loose clusters and exhibited an elongated morphology (circularity: 0.65 ± 0.04) and decreased cell area (1015 ± 77 μm2). In contrast, cultures stimulated to M2a polarization with an IL-4/IL-13 cocktail and cultures stimulated to M2c polarization with IL-10 retained ovoid-shaped morphologies (circularity: M2a: 0.74 ± 0.01, M2c: 0.79 ± 0.04) similar to naïve Mϕ macrophages. Interestingly, M2a macrophages exhibited increased area (2967 ± 99 μm2) compared to Mϕ, while M2c macrophages were of a similar area (2052 ± 116 μm2).
Proliferative activity among the various macrophage phenotypes was assessed by direct enumeration using a hemocytometer to estimate increases in the number of cells per well. A 1.4-fold increase in the number of naïve Mϕ was observed after 3 days of culture (Figure 1B). IL-10-induced M2c macrophages also proliferated but exhibited only a 1.2-fold increase in cell number. Significant proliferation was not observed for macrophage treated with either LPS/IFN-γ, to stimulate M1 polarization, or IL-4/IL-13 cocktails, to stimulate M2a polarization. Importantly, macrophage viability, determined via the trypan blue exclusion assay, remained high (>95%) for all macrophage phenotypes (Figure S2D).
To verify phenotypic changes in the macrophage populations, levels of cell surface receptors associated with polarized macrophage were characterized. As expected, flow cytometric analysis revealed that each distinct macrophage population expressed specific and quantifiable surface markers after 3 days of culture (Figure 1C, Figure S3). For example, the C-C chemokine receptor type 7 (CCR7) was expressed at higher frequency in the population of M1 macrophages (17 ± 1% positive cells), with minimal expression observed on Mϕ (1.9 ± 0.4%), M2a (3 ± 1%), and M2c (5 ± 3%) macrophages after 3 days of treatment. Over 94 ± 1% of M2a macrophages expressed the mannose receptor CD206, a putative M2a marker; however, CD206 expression was also detected in Mϕ (37 ± 7%), M1 (36 ± 7%), and M2c (48 ± 8%) phenotypes, albeit at lower levels. Expression of the hemoglobin scavenger receptor, CD163, was significantly increased in M2c macrophage (88 ± 3%) compared to Mϕ (52 ± 6%), M1 (5 ± 1%), and M2a macrophage (20 ± 4%).
We further examined protein secretion profiles of macrophage phenotypes using enzyme-linked immunosorbent assays (ELISA) to evaluate biomarkers associated with differential macrophage function in regulating inflammation, angiogenesis, and matrix remodeling during tissue regeneration. Secretion of pro-inflammatory cytokines, IL-6 and TNF-α, angiogenic growth factors, VEGF-A and PDGF-BB, anti-inflammatory cytokines, TGF-β1 and TGF-β2, and the matrix remodeling enzyme, MMP-9, were analyzed. Protein secretion profiles were dependent on macrophage subtype (Figure 1D). After 3 days of polarization, M1 macrophages secreted increased amounts of IL-6, TNF-α, and VEGF-A (IL-6: 2004 ± 525 pg/105 cells, TNF-α: 5817 ± 1058 pg/105 cells, VEGF-A: 20 ± 1 pg/105 cells), compared to Mϕ (IL-6: 0.9 ± 0.1 pg/105 cells, TNF-α: 2.8 ± 0.2 pg/105 cells, VEGF-A: 0.9 ± 0.2 pg/105 cells), M2a (IL-6: 0.9 ± 0.1 pg/105 cells, TNF-α: 4.2 ± 0.2 pg/105 cells, VEGF-A: 1.7 ± 0.1 pg/105 cells), and M2c (IL-6: 1.0 ± 0.1 pg/105 cells, TNF-α: 4 ± 1 pg/105 cells, VEGF-A: 0.8 ± 0.1 pg/105 cells) macrophages. TGF-β1 and PDGF-BB production was significantly increased in M2a macrophage (TGF-β1: 1571 ± 60 pg/105 cells, PDGF-BB: 870 ± 33 pg/105 cells) compared to Mϕ (TGF-β1: 963 ± 20 pg/105 cells, PDGF-BB: 151 ± 35 pg/105 cells), M1 (TGF-β1: 866 ± 133 pg/105 cells, PDGF-BB: 270 ± 59 pg/105 cells), and M2c (TGF-β1: 1310 ± 73 pg/105 cells, PDGF-BB: 173 ± 16 pg/105 cells) macrophages. M2c macrophage secreted increased amounts of TGF-β2 and MMP-9 (TGF-β2: 197 ± 9 pg/105 cells, MMP-9: 2140 ± 378 pg/105 cells) compared to Mϕ (TGF-β2: 91.0 ± 0.8 pg/105 cells, MMP-9: 872 ± 168 pg/105 cells), M1 (TGF-β2: 106 ± 22 pg/105 cells, MMP-9: 576 ± 186 pg/105 cells), and M2a (TGF-β2: 140 ± 21 pg/105 cells, MMP-9: 467 ± 110 pg/105 cells) macrophages. Combined with the observed differences in morphology and cell-surface receptor composition, these results demonstrate that discernable phenotypic differences exist between CB-derived Mϕ, M1, M2a, and M2c cell types following culture under standard, cytokine-dependent polarization conditions.
To evaluate whether the phenotypes of CB-derived macrophages remained stable over time, surface marker expression and cytokine production was re-assessed, via flow cytometry and ELISA, respectively, after 6 days of culture in the presence of polarizing stimuli. In general, surface marker expression was fairly stable through 6 days of culture (Figure S4), with the exception of CCR7 expression by M1 macrophages. The percentage of M1 cells expressing CCR7 was reduced by 2-fold after 6 days of culture with LPS/IFN-γ, compared to day 3 (Figure S5A). The population of M1 cells expressing CD206 or CD163 remained similar to the levels observed on day 3. Surface marker expression by M2a and M2c cells was more stable over time. The percentage of M2a macrophages expressing CD206 was similar between days 3 and 6 of culture with IL-4/IL-13, with little-to-no changes in CCR7+ or CD163+ cells observed. Similarly, M2c macrophages exhibited a stable surface marker expression pattern between days 3 and 6 of culture stimulated by IL-10, and little-to-no change in CCR7 or CD206 surface expression after 6 days.
Additional analysis of biomarkers associated with macrophage subtype function revealed altered levels of the M1 functional markers, IL-6, TNF-α, and VEGF-A, in M1 macrophages (i.e. LPS/IFN-γ-treated) over time (Figure S5B), where a 12-fold decrease in IL-6 and a 16-fold decrease in TNF-α was observed between days 3 and 6 of culture. VEGF-A production by these cells, on the other hand, increased by 3-fold between day 3 and 6 of culture. Importantly, production of IL-6, TNF-α, and VEGF-A by M1 cells remained increased compared to secretion of these markers by Mϕ, M2a, and M2c cells on day 6. Further, secretion of TGF-β1 and PDGF-BB, which are prototypical markers of M2a cells, as well as TGF-β2 and MMP-9, which are markers of M2c cells, remained stable between days 3 and 6, suggesting that M1 cells had not transdifferentiated.
Among M2a macrophages, TGF-β1 and PDGF-BB production were similar on days 3 and 6, and the production of TGF-β1 and PDGF-BB by M2a cells remained increased compared to by Mϕ, M1, and M2c cells on day 6. There were no significant changes in secretion of the M1 proteins, IL-6, TNF-α, and VEGF-A, or the M2c proteins, TGF-β2 and MMP-9. Likewise, for M2c cells, TGF-β2 and MMP-9 production was similar on day 6, compared to day 3, and the production of TGF-β2 and PDGF-BB by M2c cells remained increased compared to by Mϕ, M1, and M2a cells on day 6. No significant changes in secretion of the M1 proteins, IL-6, TNF-α, and VEGF-A, or the M2a proteins, TGF-β1 and PDGF-BB, observed between days 3 and 6 of culture. These results demonstrate that M2a and M2c cells retain discernable phenotypic differences following culture under standard, cytokine-dependent polarization conditions for up to 6 days.
3.2. Reversibility of the Polarized Macrophage Phenotypes
To determine whether CB-derived macrophages were able to switch their phenotypes in response to altered polarizing stimuli, macrophage were cultured with M1, M2a, or M2c-polarizing cytokines for 3 days, followed by culture for 3 additional days in medium supplemented with an alternative cytokine cocktail (Figure 2A). Surface marker expression and protein production were then determined via flow cytometry and ELISA, respectively. First, the effects M2a (IL-4/IL-13) or M2c (IL-10) polarizing stimuli (on macrophages previously stimulated with M1 cytokines were investigated. Following exposure to M2a-promoting stimuli, the M1 macrophage population shifted to express 4-fold less CCR7 and 1.5-fold more CD206 on day 6, while the percentage of CD163+ M1→M2a cells remained similar before and after IL-4/IL-13 exposure (Figure 2B). Likewise, M1 macrophages exposed to M2c-promoting stimuli on day 3 exhibited a 4-fold decrease in the percentage of CCR7+ cells on day 6. A corresponding 10-fold increase in the percentage of M1→M2c cells expressing CD163 was observed, while the M1→M2c population displayed similar levels of CD206 expression compared to their predecessors.
Figure 2.
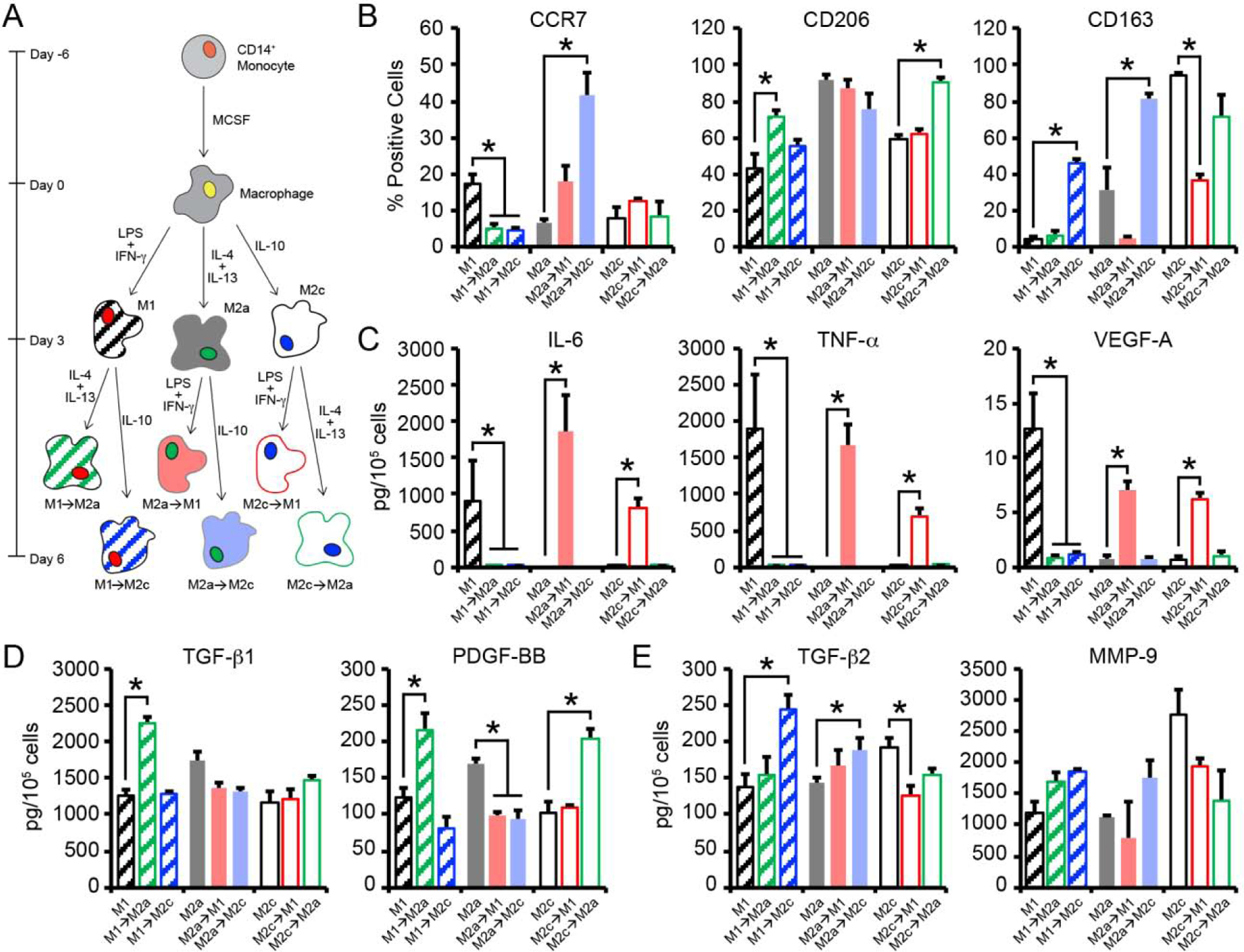
(A) CB macrophages were polarized to 3 different phenotypes (M1, M2a, M2c) using established cytokine cocktails for 3 days. Thereafter, macrophages were switched to medium supplemented with alternative polarizing stimuli (M1→M2a, M1→M2c, M2a→M1, M2a→M2c, M2c→M1, M2c→M2a) and cultured for 3 additional days. (B) Flow cytometric analysis of the percentage of macrophage staining positively for each phenotypic surface marker after 3 days of initial culture or on day 6, after switching. (C-E) Quantification of secreted (C) M1, (D) M2a, and (E) M2c related proteins from macrophages. In B-E, data are represented as the mean ± SEM, with n = 3 human donors per condition. A one-way ANOVA, followed by a Dunnett’s post hoc test, was used to detect statistical significance for repolarized macrophage populations compared to their predecessor, *p<0.05.
The impacts of M1 (LPS/IFN-γ) and M2c (IL-10) polarizing stimuli on macrophages previously treated with M2a cytokines were also evaluated. Both populations (M2a→M1 and M2a→M2c) maintained stable CD206 expression when compared to their predecessors. Interestingly, the population of M2a→M1 cells expressing CCR7 on day 6 was increased by 3-fold compared to M2a cells on day 3, while a 7-fold decrease in the percentage of CD163+ cells was observed for M2a→M1 cells. The percentage of M2a→M2c cells expressing CD163 increased by 3-fold and these cells further exhibited a 6-fold increase in the percentage of CCR7+ cells following IL-10 stimulation.
Mixed phenotypic plasticity was further observed for M2c macrophages following treatment with M1-promoting LPS/IFN-γ or M2a-promoting IL-4/IL-13. Both populations shifted to express less CD163 compared to their predecessor. Specifically, a 3-fold decrease in CD163 expression was observed for M2c→M1, while a 1.3-fold decrease in CD163 expression was observed for M2c→M2a. Differential regulation of surface markers associated with their predecessor was further observed. For example, the population of M2c→M1 cells expressing CCR7 increased by 1.5-fold, while a similar percentage of cells expressed CD206 compared to their predecessors. Conversely, the percentage of M2c→M2a cells expressing CD206 increased by 1.5-fold compared to their predecessors, while a similar percentage of cells expressed CCR7+ following IL-4/IL-13 stimulation. Taken together, these results suggest that polarized macrophages demonstrate plasticity following exposure to an alternative cytokine cocktail, where decreased expression of the surface marker associated with the predecessor cell type and increased expression in the surface marker associated with the newly induced phenotype were generally observed.
To confirm that CB-derived macrophages were able to switch their phenotypes, protein production was measured, via ELISA, following culture in alternative polarization media. Following exposure to M2a- and M2c-promoting stimuli, M1→M2a and M1→M2c macrophages exhibited decreased production of the M1 cytokines, IL-6 (M1→M2a: 250-fold decrease; M1→M2c: 130-fold decrease), TNF-α (M1→M2a: 140-fold decrease; M1→M2c: 160-fold decrease), and VEGF-A (M1→M2a: 17-fold decrease; M1→M2c: 12-fold decrease) on day 6, compared to their predecessors on day 3 (Figure 2C). M1→M2a cells also exhibited a 1.8-fold increase in TGF-β1 production and a 1.7-fold increase in PDGF-BB secretion on day 6, compared to M1 cells on day 3 (Figure 2D); however, no changes in TGF-β2 or MMP-9 production were observed (Figure 2E). On day 6 of culture, M1→M2c cells exhibited a 1.6-fold increase in TGF-β2 secretion and a 2-fold increase in MMP-9 secretion following stimulation with IL-10, while production of TGF-β1 and PDGF-BB remained stable when compared to production by M1 cells on day 3.
Altered cytokine production was also observed for M2a macrophages exposed to M1 and M2c-promoting stimuli. For example, M2a→M1 cells exhibited a 1.3-fold decrease in TGF-β1 production and a 1.7-fold decrease in PDGF-BB following stimulation with M1-promoting stimuli, compared to M2a cells on day 3. M2a→M1 cells were also observed to secrete more IL-6 (1600-fold increase), TNF-α (500-fold increase), and VEGF-A (9-fold increase) production compared to M2a cells after 3 days of stimulation; however, production of TGF-β2 and MMP-9 remained stable. M2a→M2c cells produced less TGF-β1 (1.4-fold decrease) and PDGF-BB (1.8-fold decrease) on day 6, compared to M2a macrophages on day 3. Further, secretion of TGF-β2 and MMP-9 by M2a→M2c macrophages was decreased compared to M2a macrophages; however, no changes in IL-6, TNF-α, or VEGF-A were observed.
Polarization of M2c macrophages with LPS/IFN-γ (i.e. M2c→M1) resulted in a 1.5-fold decrease in the expression of both MMP-9 and TGF-β2, while increased production of IL-6 (860-fold decrease), TNF-α (340-fold decrease), and VEGF-A (9-fold decrease) was detected. As expected, no changes in TGF-β1 and PDGF-BB were observed following polarization of M2c macrophages to M2c→M1. Conversely, polarization of M2c macrophages with IL-4/IL-13 (i.e. M2c→M2a) resulted in a 2-fold increase in PDGF-BB following stimulation with M2a-promoting stimuli, while no changes in TGF-β1 production were detected when compared to their predecessors. Importantly, production of TGF-β2 and MMP-9 concurrently decreased following treatment with IL-4/IL-13, while IL-6, TNF-α, or VEGF-A by M2c→M2a cells was similar to that produced by M2c macrophages on day 3. Overall, these results demonstrate that M1 conditions were able to downregulate expression of M2a or M2c biomarkers, while simultaneously upregulating the M1 phenotype. Similarly, M2a and M2c conditions were able to downregulate expression of M1 biomarkers, while simultaneously upregulating M2a and M2c phenotypes.
3.3. Hydrogel Fabrication and Characterization
To study the role of matrix stiffness in influencing CB-derived macrophage phenotype, hydrogels formed from polymer networks under physiological conditions without deleterious by-products were needed. Accordingly, thiol and maleimide end-functionalized PEG macromers were used to form stable crosslinks via Michael-type addition click reactions (Figure 3A) [30]. Cysteine-terminated RGD peptide was incorporated into the polymer network to facilitate integrin-mediated interactions between the macrophage and hydrogel. The viscoelastic properties of the hydrogels were tuned within physiologic ranges by varying the polymer concentration over a narrow range and validated via oscillatory shear rheology, using dynamic time sweep assays in the linear viscoelastic regime. Representative shear storage moduli (G’) and shear loss moduli (G”), which describe the elastic and viscous components of the hydrogel, respectively, were evaluated during hydrogel formation (Figure S6). These materials formed highly elastic networks within 158, 99, and 36 seconds for the 3wt%, 6wt%, and 14wt% hydrogels, respectively, as indicated by a crossover point between the storage and loss moduli, which is an indirect measure of gel point. As expected for PEG-based materials, concentration-dependent swelling varied with formulation, where swelling capacities of 97.3 ± 0.2%, 96.7 ± 0.2%, and 95.9 ± 0.3% were observed for 3wt%, 6wt%, and 14wt% hydrogels, respectively (Table S3). The equilibrium shear storage moduli of swollen 3wt%, 6wt%, and 14wt% hydrogels, measured after 24 h of incubation at 4°C in HBSS, were 0.1 ± 0.1, 3.4 ± 0.1, and 10.3 ± 0.4 kPa, respectively (Figure 3B).
Figure 3.
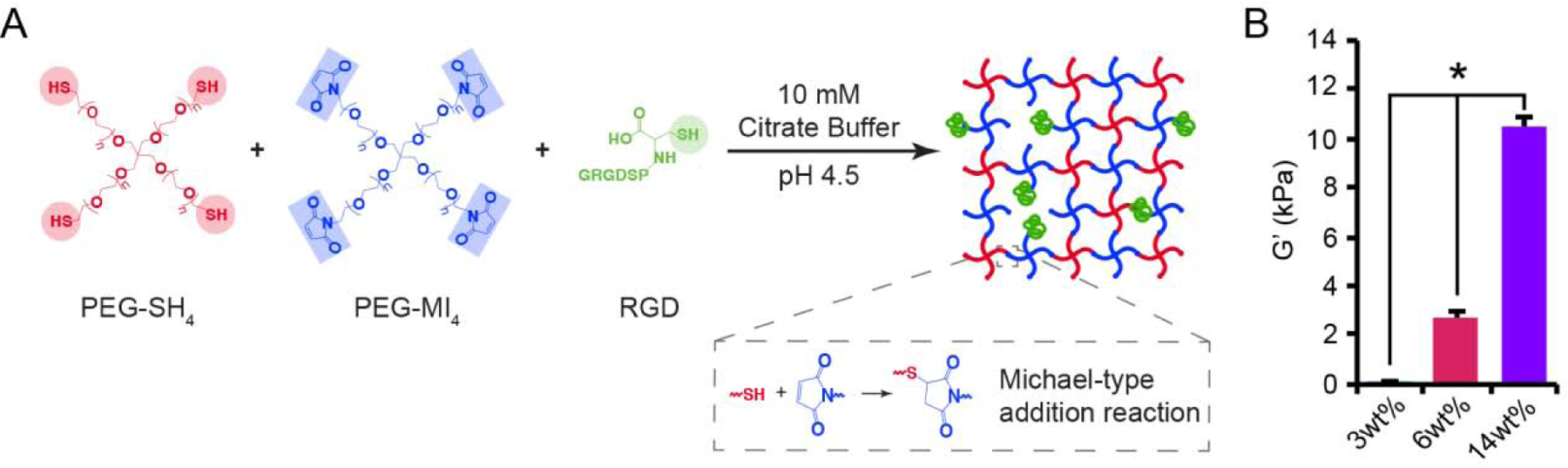
Formation and rheological characterization of PEG hydrogels. (A) Hydrogels were formed by reacting maleimide end-functionalized, four-arm PEG macromers (PEG-MI4) with thiol end-functionalized, four-arm PEG macromers (PEG-SH4) and cysteine-terminated RGD peptides using a Michael-type addition reaction. (B) Hydrogels were formed and incubated in cell culture medium prior to characterization via oscillatory shear rheology. Swollen 3wt%, 6wt%, and 14% hydrogels exhibited significantly different equilibrium shear storage moduli (G’) after 24 h. A one-way ANOVA, followed by a Tukey HSD post hoc test, was used to detect statistical significance, *p<0.05.
3.4. Hydrogel Stiffness Regulates Macrophage Phenotype
To investigate the impact of stiffness on CB-derived macrophage phenotype, CD14+ monocytes were first activated with M-CSF to generate Mϕ. Following differentiation, cells were gently lifted with Accutase® and re-seeded on swollen 3wt%, 6wt%, or 14wt% hydrogels (Figure 4A). After 3 days of culture, cell morphologies were evaluated via brightfield microscopy (Figure S7A). Macrophages on soft 3wt% hydrogels were highly refractive with small, ovoid morphology (circularity: 0.80 ± 0.03), with a spread area of 245 ± 16 μm2 (Figure S7B–C). Ovoid-shaped morphologies were also observed for macrophage cultured on 6wt% or 14wt% hydrogels (circularity: 6wt%: 0.79 ± 0.03, 14wt%: 0.78 ± 0.04); however, these cells exhibited larger areas (6wt%: 1224 ± 135 μm2, 14wt%: 1384 ± 176 μm2) after 3 days.
Figure 4.
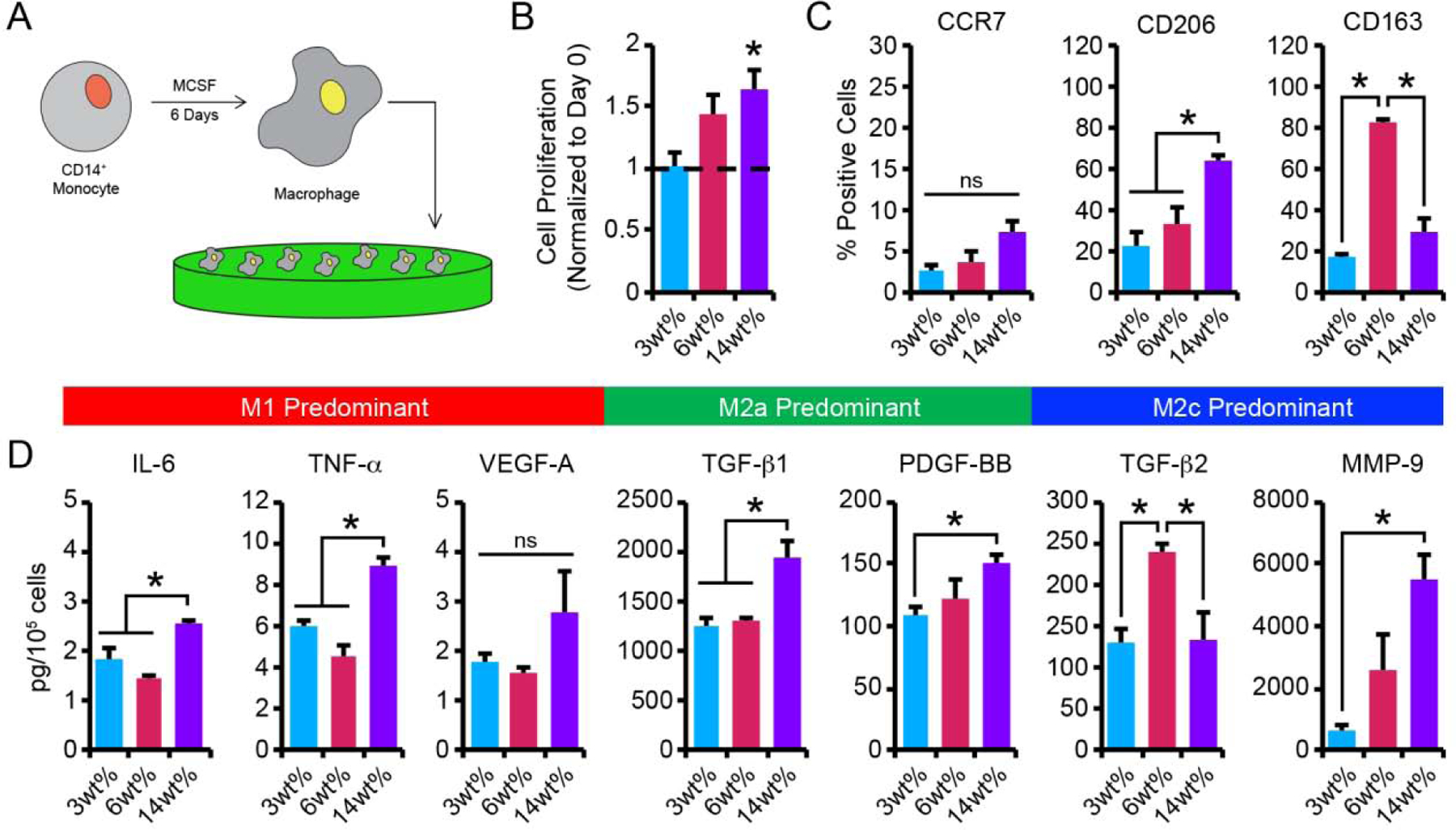
(A) CB monocytes were differentiated to macrophages (Mϕ) and seeded on 3wt%, 6wt%, and 14wt% hydrogels. (B) Macrophage cell proliferation was determined by enumerating the number cells present after 3 days of culture and normalizing to the number of cells initially seeded on day 0 (dashed line represents day 0) (C) Flow cytometric analysis of the percentage of macrophage staining positively for each phenotypic surface marker after 3 days of culture on hydrogels. (D) Quantification of secreted proteins from macrophage cultured on hydrogels for 3 days. In B-D, data are represented as the mean ± SEM, with n = 3 human donors per condition. A one-way ANOVA, followed by a Tukey HSD post hoc test, was used to detect statistical significance, *p<0.05, ns = no significance.
Macrophage proliferation in response to substrate stiffness was assessed by counting a small sample of cells on a hemocytometer, extrapolating the average number of cells per hydrogel, and normalizing the average number of cells on the hydrogel surface to the number of cells initially seeded on day 0. Macrophage proliferation increased with increased hydrogel stiffness (Figure 4B). On stiff 14wt% substrates, macrophages exhibited a 1.6-fold increase in cell number after 3 days of culture, while only a 1.4-fold increase in cell number was detected for macrophages on 6wt% hydrogels. In contrast, proliferation was not observed for macrophage cultured on soft 3wt% hydrogels, where a 0.9-fold change in cell number was observed after 3 days of culture. Importantly, macrophage viability, determined via the trypan blue exclusion assay, remained high (>95%) for macrophage cultured on hydrogels of varying stiffness (Figure S7D).
Flow cytometric analysis revealed three different patterns of surface marker expression by macrophage-cultured on hydrogels of three different stiffness (Figure 4C, Figure S8). After 3 days of culture on hydrogels, the percentage of CCR7+ macrophage was low (<10% CCR7+ cells), and no differences in the percentage of CCR7+ were observed across hydrogel formulations. Macrophage expression of CD206 increased with increased hydrogel stiffness, where significantly elevated CD206 expression was observed on 14wt% hydrogels (63.3 ± 3.0%) compared to 3wt% (23 ± 6%) and 6wt% (34 ± 8%) hydrogels. CD163, on the other hand, was relatively low on softer 3wt% hydrogels (17 ± 2%) and on stiffer 14wt% hydrogels (30 ± 6%), but significantly elevated in cultures on 6wt% hydrogel (82 ± 2%).
Cytokine secretion profiling for IL-6, TNF-α, VEGF-A, PDGF-BB, TGF-β1, TGF-β2, and MMP-9 indicated that patterns were dependent on substrate stiffness (Figure 4D). After 3 days of culture on hydrogels, macrophage on 14wt% hydrogels exhibited enhanced secretion of the M1 cytokines IL-6 (2.6 ± 0.1 pg/105 cells) and TNF-α (8.9 ± 0.4 pg/105 cells), compared to cells cultured on 3wt% (IL-6: 1.9 ± 0.2 pg/105 cells; TNF-α: 6.0 ± 0.3 pg/105 cells) or 6wt% hydrogels (IL-6: 1.4 ± 0.1 pg/105 cells; TNF-α: 5 ± 1 pg/105 cells). Interestingly, VEGF-A secretion, which was elevated in M1 macrophage polarized by cytokines, was low in all hydrogel formulations, with cells secreting 1.8 ± 0.2 pg/105 cells on 3wt%, 1.6 ± 0.1 pg/105 cells on 6wt%, and 3 ± 1 pg/105 cells on 14wt% hydrogels after 3 days of culture. In addition to secreting higher levels of M1 cytokines, macrophage on 14wt% hydrogels also exhibited elevated production of the M2a growth factors, TGF-β1 (1947 ± 156 pg/105 cells) and PDGF-BB (152 ± 5 pg/105 cells), compared to cells cultured on 3wt% (TGF-β1: 1240 ± 86 pg/105 cells; PDGF-BB: 108 ± 8 pg/105 cells) or 6wt% hydrogels (TGF-β1: 1314 ± 19 pg/105 cells; PDGF-BB: 123 ± 15 pg/105 cells). Production of the M2c-associated enzyme, MMP-9, was significantly increased by macrophage cultured on the stiffer 14wt% hydrogels (5488 ± 843 pg/105 cells), compared to the softer 3wt% and 6wt% hydrogels (3wt%: 680 ± 165 pg/105 cells, 2562 ± 1187 pg/105 cells). In contrast, TGF-β2 production was significantly increased in macrophage cultures grown on intermediate 6wt% hydrogels (239 ± 11 pg/105 cells) compared to 3wt% and 14wt% hydrogels (3wt%: 129 ± 17 pg/105 cells, 14wt%: 135 ± 33 pg/105 cells). These results, together with surface marker (CCR7, CD206, CD163) analysis, indicate that cells grown on the higher weight percent hydrogels exhibited cell surface marker expression indicative of anti-inflammatory M2a and M2c phenotypes but exhibited cytokine secretion profiles indicative of a mixed inflammatory/anti-inflammatory phenotype.
To evaluate the stability of macrophage phenotypes on hydrogels, surface marker expression and cytokine secretion were re-assessed after 6 days of culture. In general, surface marker expression by Mϕ macrophages cultured on hydrogels was stable through day 6, with similar patterns of intensity observed via flow cytometry for dual stained samples of cells on days 3 and 6 (Figure S9). Flow cytometric analysis confirmed that the percentage of CCR7+ macrophage remained low (<10% CCR7+ cells) across hydrogel formulations, with no differences in the percentage of CCR7+ observed across hydrogel formulations on day 6 (Figure S10A). Likewise, expression of CD206 and CD163 by macrophages cultured on 3wt%, 6wt%, or 14wt% hydrogels was similar between days 3 and 6.
Further analysis of biomarkers associated with macrophage subtype function revealed that production of M1 functional markers, IL-6 and TNF-α, remained similar over time for cells cultured on 3wt%, 6wt%, or 14wt% hydrogels (Figure S10B). Conversely, VEGF-A production by cells on 3wt% hydrogels increased 7-fold between days 3 and 6, while VEGF-A production by macrophage on 6wt% or 14wt% hydrogels was not altered over time. Importantly, VEGF-A secretion was significantly increased for macrophage on 3wt% hydrogels compared to 6wt% and 14wt% hydrogels on day 6, suggesting that substrates with lower moduli stimulate a pro-angiogenic phenotype over time. TGF-β1, TGF-β2, and PDGF-BB production by macrophages cultured on 3wt%, 6wt%, or 14wt% hydrogels were not altered on day 6, compared to day 3. MMP-9 production by macrophages on 3wt% hydrogels decreased by 3-fold on day 6, compared to day 3; however, MMP-9 secretion by macrophage cultured 6wt% or 14wt% hydrogels was not altered over time. Overall, surface markers and cytokine release profiles were similar on days 3 and 6, suggesting that hydrogel modulus-mediated effects on macrophage polarization were stable over time.
3.5. Activated Macrophage Respond Differentially to Hydrogel Moduli
As macrophage phenotypes are dynamic and influenced by complex and interacting factors, the relative contributions of substrate stiffness and exogenous cytokine stimuli were evaluated. As before, CD14+ monocytes were activated with M-CSF to generate Mϕ (Figure 5A). Following differentiation, cells were gently lifted with Accutase® and seeded onto 3wt%, 6wt%, or 14wt% hydrogels as above, except cultures were grown in medium supplemented with the LPS/IFN-γ, IL-4/IL-13, or IL-10 cytokine cocktails to induce polarization to M1, M2a, and M2c phenotypes, respectively. After 3 days of culture, macrophage morphologies, surface marker expression, and cytokine secretion were evaluated.
Figure 5.
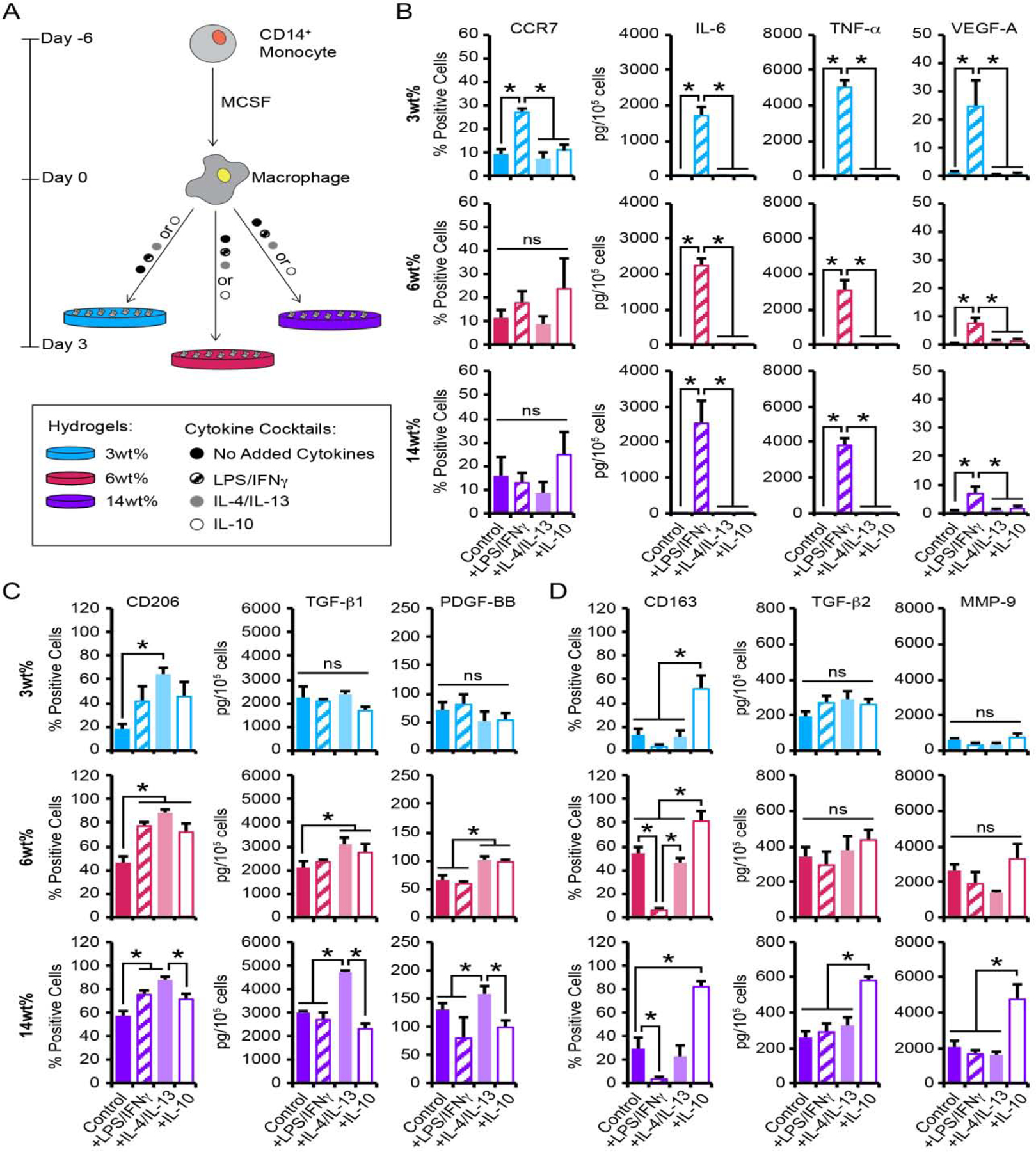
(A) CB monocytes were differentiated to macrophages (Mϕ), seeded on 3wt%, 6wt%, or 14wt% hydrogels, and cultured with or without the presence of pro- or anti-inflammatory cytokines. (B-D) Flow cytometric analysis of the percentage of macrophage staining positively for each phenotypic surface marker and quantification of secreted proteins from macrophage cultured on hydrogels for 3 days. Biomarkers for (B) M1, (C) M2a, and (D) M2c phenotypes were evaluated. B-D, data are represented as the mean ± SEM, with n = 3 human donors per condition. A one-way ANOVA, followed by a Tukey HSD post hoc test, was used to detect statistical significance, *p<0.05, ns = no significance.
In the absence of the cytokine cocktails, macrophage exhibited morphologies similar to those in previous experiments (Figure S7). Cells on 3wt% hydrogels were small and rounded (circularity: 0.76 ± 0.02; cell area: 283 ± 13 μm2), while macrophage cultured on 6wt% and 14wt% hydrogels were ovoid with increased surface area (circularity: 6wt%: 0.85 ± 0.02, 14wt%: 0.82 ± 0.03; cell area: 6wt%: 948 ± 51, 14wt% 869 ± 74 μm2; Figure S11A). LPS/IFN-γ treatment resulted in clustering of macrophage on all hydrogels. Aggregation increased with increasing substrate stiffness, with the average area of cell aggregates being 5,102 ± 557 μm2, 12,462 ± 1366 μm2, and 20,118 ± 2362 μm2 on 3wt%, 6wt%, and 14wt% hydrogels, respectively (Figure S11B). Treatment with IL-4/IL-13 or IL-10 did not induce cell clustering. Rather, exposure to exogenous IL-4/IL-13 or IL-10 resulted in macrophages that exhibited morphologies similar to those observed on hydrogels alone (Figure S11C). Interestingly, IL-4/IL-13 treated macrophage cultured on 3wt%, 6wt%, or 14wt% hydrogels exhibited a 1.3–1.6-fold increase in cell area compared to cells cultured on hydrogels alone; whereas differences in cell area were not observed between macrophage treated with or without IL-10 following culture on any hydrogel (Figure S11D).
Macrophage proliferation associated with substrate stiffness in the presence of exogenous cytokine stimuli was extrapolated from normalized direct cell counts. As expected from our prior results, in the absence of the cytokine cocktails, proliferation was not observed on 3wt% hydrogels, while macrophage cultured on 6wt% and 14wt% hydrogels exhibited 1.4- and 1.5-fold increases in cell number, respectively, after 3 days (Figure S12A). On soft 3wt% and stiff 14wt% hydrogels, macrophage proliferation was not altered by the addition of the cytokine cocktails. Further, no alterations in cell proliferation were observed following treatment with IL-4/IL-13 or IL-10 cocktails; however, a 1.8-fold reduction in proliferation was observed when macrophages were cultured on 6wt% hydrogels in the presence of LPS/IFN-γ, compared to control hydrogels. Importantly, macrophage viability remained high (>95%) for macrophage cultures in all treatment groups (Figure S12B).
Analysis of surface marker expression and growth factor secretion indicated that macrophage cultured on soft 3wt% hydrogels in the presence of LPS/IFNγ exhibited a 3-fold increase in CCR7 expression compared to macrophage cultured on 3wt% hydrogels alone (Figure 5B). Macrophage cultured on 3wt% hydrogels with LPS/IFNγ also exhibited enhanced production of the pro-inflammatory cytokines IL-6 (1100-fold increase), TNF-α (1200-fold increase), and VEGF-A (22-fold increase), compared to 3wt% hydrogels alone. Interestingly, CCR7 expression was not altered by LPS/IFNγ treatment on the stiffer 6wt% and 14wt% hydrogels, compared to hydrogels alone. LPS/IFNγ treatment on the 6wt% or 14wt% hydrogel cultures did, however, result in significantly increased production of the pro-inflammatory cytokines IL-6 (6wt%: 2000-fold increase; 14wt%: 1700-fold increase), TNF-α (6wt%: 1200-fold increase; 14wt%: 990-fold increase), and VEGF-A (6wt%: 15-fold increase; 14wt%: 9-fold increase), compared to macrophage cultured on hydrogels alone. As expected, CCR7 expression and pro-inflammatory cytokine production was not altered by the addition of IL-4/IL-13 on any substrate or following IL-10 treatment on soft 3wt% substrates. Conversely, IL-10-treated macrophage cultured on 6wt% or 14wt% hydrogels exhibited increased CCR7 expression, without any alterations in pro-inflammatory cytokine production. These findings suggest an uncoupling of M1 macrophage markers as the expression of CCR7 varied based on substrate, with elevation observed only for the softest substrate following LPS/IFNγ treatment or for the stiff substrates in the presence of IL-10, while the elevated production of the M1 cytokines, IL-6, TNF-α, and VEGF-A occurred independently of substrate stiffness following treatment with LPS/IFNγ.
The percentage of macrophage expressing CD206 following culture on 3wt% hydrogels was increased by 4-fold following treatment with IL-4/IL-13, compared to macrophage observed on 3wt% hydrogels alone. This effect occurred without significant changes in levels of TGF-β1 or PDGF-BB (Figure 5C). Similarly, CD206 expression by macrophage cultured on 6wt% hydrogels increased by 2-fold with IL-4/IL-13 treatment compared to control hydrogels. However, unlike 3wt% hydrogels, IL-4/IL-13 treatment resulted in a 1.5-fold increase in TGF-β1 and PDGF-BB production by macrophage on 6wt% hydrogels over controls. Macrophage cultured on the 14wt% hydrogels also exhibited a 1.5-fold increase in the percentage of CD206+ cells with IL-4/IL-13, as well as increased production of TGF-β1 (1.6-fold increase); however, PDGF-BB secretion was similar to control 14wt% hydrogels. Although treatment with IL-4/IL-13 increased CD206 expression as expected, CD206 was also increased by macrophages cultured on 3wt%, 6wt%, and 14wt% hydrogels following treatment with either LPS/IFNγ or IL-10. However, despite the increase in CD206+ cells following treatment with either the LPS/IFNγ or IL-10 cytokine cocktails, macrophage on the soft 3wt% and stiff 14wt% hydrogels did not elevate TGF-β1 or PDGF-BB levels. Interestingly, macrophage behaved differently on the intermediate stiffness 6wt% hydrogels, where IL-10 stimulated secretion of both TGF-β1 and PDGF-BB. These results indicate that although increases in the M2a surface marker CD206 on CB-derived macrophage following treatment with LPS/IFNγ, IL-10, or IL-4/IL-13 appeared to be independent of substrate stiffness, IL-4/IL-13-induced production of the M2a proteins, TGF-β1 and PDGF-BB, was dependent on substrate stiffness.
Exposure of hydrogel-cultured macrophage to M2c-inducing IL-10 also had differential effects across hydrogel formulations. As expected, macrophage cultured with IL-10 on 3wt% hydrogels exhibited a 4-fold increase in CD163 expression, compared to macrophage cultured on hydrogels alone (Figure 5D). However, IL-10 treatment of the 3wt% hydrogels did not result in the expected corresponding alterations in the production of TGF-β2 or MMP-9. The application of IL-10 to macrophages grown on 6wt% hydrogels resulted in increased expression of CD163 (1.5-fold increase) without corresponding changes in TGF-β2 and MMP-9, compared to macrophage cultured on hydrogels alone. The addition of IL-10 to macrophage cultured on 14wt% hydrogels also resulted in increased expression of CD163 (3-fold increase) compared to controls. Unlike the response observed on the softer 3wt% and 6wt% hydrogels, however, the addition of IL-10 to 14wt% hydrogel cultures resulted in increased production of TGF-β2 (2-fold increase) and MMP-9 (2-fold increase), compared to macrophage cultured on 14wt% hydrogels. Interestingly, LPS/IFNγ treatment of macrophage cultured on 3wt%, 6wt%, and 14wt% hydrogels resulted in significantly decreased CD163+ cells (4–10-fold decrease), while treatment with IL-4/IL-13 yielded CD163 expression that was similar to control hydrogels. Further, neither LPS/IFNγ nor IL-4/IL-13 altered production of TGF-β2 and MMP-9 in 3wt%, 6wt%, or 14wt% hydrogel cultures. Together, these findings suggest that IL-10 treatment enhances expression of the M2c surface marker, CD163, on CB-derived macrophage independent of substrate stiffness; however, IL-10-induced production of the M2c proteins, TGF-β2 and MMP-9, is dependent on substrate stiffness.
4. Discussion
CB-derived macrophage and other cells have significant clinical potential, but little is known about the fate of CB-derived cells in hydrogel-based, tissue regenerative approaches, which is a significant barrier to the development of new cell-based therapies. This study established that alterations in hydrogel modulus have significant differential effects on the phenotypic status of CB-derived macrophage in the presence or absence of exogenous biochemical signals. It is well-recognized that macrophage rapidly adapt to microenvironmental cues [33], with changes in cell morphology, surface marker expression, and growth factor production all reflecting altered macrophage polarization [34, 35]. Hydrogel substrate stiffness impacted each of these parameters, indicating that substrate stiffness may be a primary environmental driver of macrophage fate and suggesting that careful design of substrate stiffness in hydrogel-based interventions could be employed to drive desired cellular phenotypes.
We found that naïve Mϕ macrophage from CB exhibited ovoid morphologies and were CD11b+/CD68+ following initial differentiation on ultra-low attachment polystyrene (Figure S1). After cytokine-driven polarization, human CB-derived M1 macrophages exhibited elongated morphologies, while M2 subsets (both M2a and M2c) adopted a flat, ovoid morphology following induction during culture on ultra-low attachment polystyrene (Figure S2). The morphologies observed in this study are similar to those previously described for preparations of human macrophages obtained from peripheral blood [36–38], but diverge from reports for murine bone marrow derived macrophage (BMDM), which adopt a flat, circular morphology following stimulation with LPS/IFN-γ (M1) and elongated morphologies after exposure to IL-4/IL-13 (M2a) [34]. The differences among macrophage preparations suggest that morphological effects may vary with both the species and the tissue type from which the cells were isolated.
Surface marker and cytokine secretion patterns among polarized macrophage are indicative of phenotype but are also known to vary between species and tissue sources [39]. In the present study investigating human CB-derived cells, we found that M1 macrophages were readily identified in our initial validation studies on ultra-low attachment polystyrene, as these cells exhibited enhanced expression of the characteristic surface marker, CCR7, and increased production of pro-inflammatory cytokines (i.e., IL-6 and TNF-α) and the angiogenic growth factor, VEGF-A (Figure 1). Though we observed increased VEGF-A expression by M1 subtypes in this study, the extent of expression of this angiogenic growth factor by M1 vs. M2 subtypes remains controversial, with reports indicating that VEGF-A production is increased in M2 subtypes compared to M1 [40]. However, secretion of VEGF-A by M1 subtypes in this study is consistent with the defined role of pro-inflammatory M1 cells in initiating both inflammation and angiogenesis [41]. In this work, we were also able to distinguish CB-derived M2a and M2c phenotypes by evaluating the elevation of CD206 and CD163, respectively (Figure 1); these results were similar to published findings of M2 macrophage subsets generated from human peripheral blood [39]. CD206+ M2a macrophages in our study exhibited increased TGF-β1 and PDGF-BB production, while secretion of TGF-β2 and MMP-9 was enhanced in CD163+ M2c macrophage populations. Both M2a and M2c macrophages are known to play roles in resolving inflammation by modulating TGF-β [42]; however, the differential regulation of PDGF-BB and MMP-9 noted in these two phenotypes suggests that they have distinct functions related to regulating cell growth/survival versus matrix remodeling, respectively.
Though distinct phenotypes were observed 3 days after initial polarization of CB-derived macrophages, pro-inflammatory surface marker expression and cytokine production by M1 macrophages was not sustained over time (Figure S5). These results directly contrast previous findings for M1 macrophages derived from human peripheral blood monocytes, where pro-inflammatory surface marker expression and cytokine production was sustained over 6 days of culture with IFNγ treatment [21]. Interestingly, we observed increased VEGF-A production by macrophages cultured with LPS/IFN-γ after 6 days of culture. As previous reports have demonstrated that VEGF-A can skew macrophage differentiation toward the M2 phenotype [43], the loss of pro-inflammatory M1 phenotypic markers by cells may not be surprising. However, the loss of pro-inflammatory M1 phenotypic markers by cells cultured with LPS/IFNγ did not coincide with increased expression of markers associated with M2 subtypes, suggesting that the presence of an alternative or additional stimulus is required for phenotypic switching.
Phenotypic switching has been reported for macrophages derived from human peripheral blood monocytes and murine macrophages following sequential exposure to either pro- or anti-inflammatory cytokine cocktails [21, 44]. While the ability of individual macrophages to change phenotypes in vivo remains controversial, directing macrophage plasticity would be highly useful for resolving inflammation and/or fibrosis. To explore potential phenotypic switching by CB macrophage, we sequentially exposed Mϕ to the cytokine cocktails (LPS/IFNγ, IL-4/IL-13, or IL-10) that invoke canonical macrophage phenotypes (Figure 2). The complex set of expression patterns following sequential exposure to selected cytokine cocktails were better visualized following normalization of surface marker and cytokine data for macrophages exposed to new cytokine cocktail conditions relative to their predecessors (e.g. M1→M2a relative to M1, etc.). HCL analysis allowed biomarkers with similar expression patterns to be considered together. Clusters were identified for the phenotypic biomarkers analyzed, and a heat map was generated to depict the relationship among surface markers or cytokines (Figure 6). HCL analysis revealed that both M2a and M2c conditions were able to downregulate the M1 phenotype, while simultaneously upregulating M2a and M2c phenotypes, and vice versa. Together, these results suggest that (i) polarization assessed by cell surface marker expression does not implicate terminal differentiation and (ii) the reversibility of polarized CB-derived macrophage can be induced with specific combinations of stimuli resulting in the production of mixed phenotypes.
Figure 6.
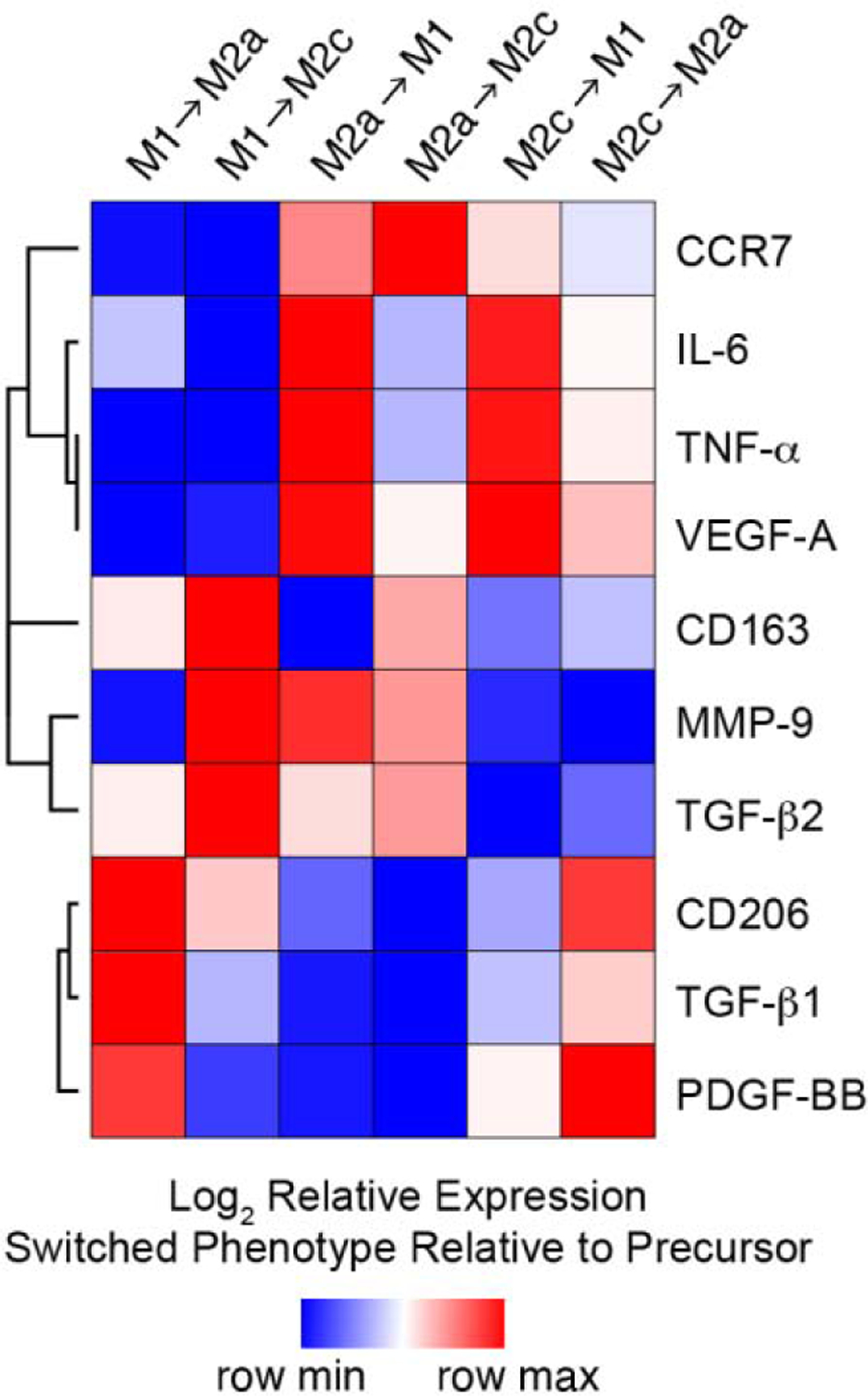
Hierarchical cluster analysis of surface markers and growth factor production data for macrophage polarized to 3 different phenotypes (M1, M2a, M2c) using established cytokine cocktails for 3 days and then cultured with alternative polarizing (M1→M2a, M1→M2c, M2a→M1, M2a→M2c, M2c→M1, M2c→M2a) stimuli for 3 additional days. Columns represent each culture condition; rows represent each surface marker or growth factor. Heat map colors represent the relative production of each biomarker normalized to the value of the predecessor (e.g. M1→M2a relative to M1, etc.), with blue indicating production that the value is in the lower half of the range across treatments for that biomarker and red indicating relatively elevated levels, as indicated in the scale bar (log2-transformed scale).
In regenerative therapy applications involving CB cells and biomaterials in combination, driving the desired remodeling of engineered scaffolds and controlling the high degree of plasticity characteristic of CB-derived cells are critical challenges, and there is significant need to better understand how the design of biomimetic materials can be used to influence CB-derived mononuclear cell activities. In particular, an understanding of how the biomechanical properties of hydrogel substrates contribute to CB-derived macrophage behavior is required. In the present study, we utilized PEG-based hydrogel platforms to investigate the impact of alterations in local tissue stiffness on CB-derived macrophage. Hydrogel formulations of 3wt%, 6wt%, and 14wt% (w/v%) were selected to give a range of physiologically relevant moduli, 0.1–10.3 kPa (Figure 3) for our studies, as motivated by previous work with materials of this composition from our group and others [8, 31, 45]. Prior reports establish that adherent cells generally exhibit rounded morphologies on soft substrates (~1 kPa) and spread morphologies on stiffer substrates (~35 kPa) [46]. CB-derived macrophages similarly responded to different hydrogel moduli, with small, ovoid cells observed on soft hydrogels and larger, spread cells detected on intermediate and stiff hydrogels. Interestingly, the average size of Mϕ macrophage on our 3wt% hydrogels were roughly the same as that reported for macrophages in suspension with little peripheral spreading [47], whereas cells on the stiffer hydrogels exhibited significantly greater interaction with the hydrogel surface. Our results are in agreement with other published work examining the impact of substrate stiffness on the morphology murine bone marrow macrophages, finding that macrophage area increased as the stiffness of either gelatin (1.9 to 29.2 kPa) or fibronectin-coated polyacrylamide (1 to 20 kPa) hydrogels increased [24, 48]. Similar stiffness-dependent morphological changes have also been reported for macrophages generated from the human myelomonocytic cell line (THP-1) and the murine macrophage cell line (RAW 264.7), but across a larger range of stiffer moduli (~10–840 kPa) [35, 48]. Interestingly, other studies with THP-1 cells describe macrophage being rounded and significantly aggregated following culture on collagen-coated polyacrylamide hydrogels with stiffnesses of 10 and 90 kPa [35]. The CB-derived macrophages utilized in the present study, on the other hand, were spread and not aggregated on 10.3 kPa hydrogels. In general, cells would be less prone to aggregate on collagen substrates versus PEG hydrogels; thus, the observed differences in cell morphology between studies may reflect differences due to cell-origin and suggest that CB-derived macrophage may be more sensitive to alterations in matrix mechanics in much softer tissues.
Substrate rigidity has been associated with alterations in cell phenotype for a large number of cell types, including macrophages obtained from human peripheral blood and murine bone marrow, as well as cell lines derived from monocyte- or macrophage-like cells [35]. However, predicting the response of macrophage to different substrate stiffnesses remains challenging with multiple research groups reporting sometimes conflicting findings regarding the phenotypic responses of macrophage to altered substrate stiffness [25–27]. For example, increases in substrate rigidity (over a range of 1.6–60.5 kPa) have been associated with increased inflammatory cytokine secretion by murine macrophages following culture in stiffer fibrin and gelatin hydrogels [26]; however, varying the mechanical properties of PEG-based materials minimally impacted the pro-inflammatory status of RAW 264.7 murine macrophage [27]. In the present study, we observed low expression of the pro-inflammatory surface receptor, CCR7, across the moduli tested in our PEG hydrogels (Figure 4). Although this result is perhaps not surprising as incorporation of RGD peptides into PEG-based matrices has been reported to reduce the inflammatory phenotype of bone marrow derived murine macrophages [49, 50]. In contrast, promotion of M2 phenotypes in CB-derived macrophages was induced by increased matrix stiffness, with M2c phenotypes (i.e., elevated CD163+) predominating on substrates with intermediate stiffness and M2a phenotypes (i.e., elevated CD206+) predominating on the stiffest substrates. The appearance of M2 sub-phenotypes on medium and high stiffness substrates is similar to effects seen in tumor-associated macrophages, which exhibit many similarities to M2 macrophages and appear to thrive in stiffer microenvironments [51]. Interestingly, macrophage on stiff 14wt% hydrogel substrates exhibited M2a surface marker patterns with increased production of the M2a-associated growth factors, TGF-β1 and PDGF-BB, but these cells also demonstrated elevated secretion of MMP-9, which we previously associated with M2c cells, and IL-6 and TNF-α, which are hallmarks of M1 cells. These combined findings indicate that substrates exhibiting shear storage moduli of 10.3 kPa promoted generation of hybrid macrophage phenotypes and underscore the sensitivity of CB-derived macrophage to substrate stiffnesses in this range.
Engineered microenvironments for tissue regenerative therapies provide platforms for regulating cell behavior via defined physical cues. Our results establish the influences of substrate modulus on CB-derived macrophage and highlight potential differences in responsiveness between cells derived from donor CB and cells derived from either peripheral blood or bone marrow. The design of materials to drive macrophage behaviors, however, can also leverage cell interactions with additional biological stimuli, including stimulatory cytokines from surrounding host tissue. An appropriately designed material could thus contribute to the dynamic alteration of macrophages that either arrive at the site of the implant from host circulation or arise from donor CB cell populations used to seed the regenerative biomaterial. Thus, to further explore the impacts of our modulus-tuned hydrogel platforms, we simultaneously exposed CB macrophages to substrates of selected moduli and to the cytokine cocktails that invoke canonical macrophage phenotypes. To invoke an M1/inflammatory phenotype, macrophages were cultured on hydrogels in the presence of LPS/IFNγ. This treatment resulted in the formation of large cell aggregates, which increased in size with increasing hydrogel modulus (Figure S11). Importantly, extensive cell clustering was not observed when cultured on hydrogels alone or on ultra-low attachment polystyrene in the presence of LPS/IFNγ. Macrophage cultured on hydrogels with anti-inflammatory cytokine cocktails (IL-4/IL-13 or IL-10) exhibited morphologies similar to those of cells cultured on hydrogels alone. Thus, our findings indicate that cell aggregation was due to a synergistic effect between hydrogel properties and pro-inflammatory cytokine stimuli and suggest that effects of exogenous stimuli on macrophages may differ on hydrogels compared to ultra-low attachment polystyrene. One possible explanation for this phenomenon is that local substrate stiffness-sensing is altered by exposure to IFNγ, which is known to modulate expression of cell adhesion molecules [52–54]. Previous reports have demonstrated that IFNγ induces clustering by regulating integrin activity in human THP-1 cells and in murine peritoneal exudate cells [55, 56]. Thus, in our study, IFNγ may induce more cluster formation on hydrogels compared to ultra-low attachment polystyrene due to differences in cellular pathways associated with stiffness-sensing – an interpretation that is supported by recent work from our group showing that fibroblasts alter MAPK pathways based on hydrogel substrate modulus [31].
To better visualize the complex set of expression patterns across hydrogel moduli in the presence of LPS/IFNγ, IL-4/IL-13, and IL-10 cytokine cocktails, HCL analysis, which allowed biomarkers, as well as hydrogel/cytokine cocktail combinations, with similar expression patterns to be considered together, was utilized and a heat map was generated depict to the relationship among surface markers or cytokines (Figure 7). HCL analysis revealed that hydrogel-cultured macrophages exhibited increased M1 biomarkers (CCR7, IL-6, TNF-α, and VEGF-A) in response to LPS/IFN-γ across all three hydrogel moduli examined, compared to cultures exposed to M2a-stimulating IL-4/IL-13 or to M2c-stimulating IL-10. HCL analysis also highlighted similar fold increases in TNF-α and IL-6 across hydrogel moduli after pro-inflammatory cytokine cocktail exposure; suggesting that LPS/IFN-γ-stimulation overwhelmingly influences secretion of pro-inflammatory factors by CB-derived macrophage regardless of substrate modulus. Interestingly, this finding contrasts with previous reports demonstrating increased TNF-α secretion by murine bone marrow macrophages on softer materials (1 kPa) in the presence of pro-inflammatory stimuli, compared to cultures on 20 kPa hydrogels [24]; however, the observed differences between studies may be attributed the use of LPS/IFN-γ in the present study, while LPS alone was utilized to generate M1 macrophage phenotypes in the prior work. Stiffness-dependent effects were observed for VEGF-A secretion and CCR7 expression in CB-derived macrophage cultured in the presence of LPS/IFN-γ. The concomitant decrease in the two markers with increased stiffness may be linked and further, is consistent with previous reports that CCR7 inhibition results in decreased VEGF-A production in in vivo models of retinopathy and colon cancer [57, 58]. Interestingly, although both markers showed decreasing expression with increasing modulus, the level of CCR7 expression in each case was similar to or below that of cells cultured on stiff hydrogels alone, while VEGF-A secretion was higher than that observed in all hydrogel-only controls, suggesting that CCR7-independent intracellular signaling pathways may also regulate VEGF-A production in CB-derived macrophage.
Figure 7.
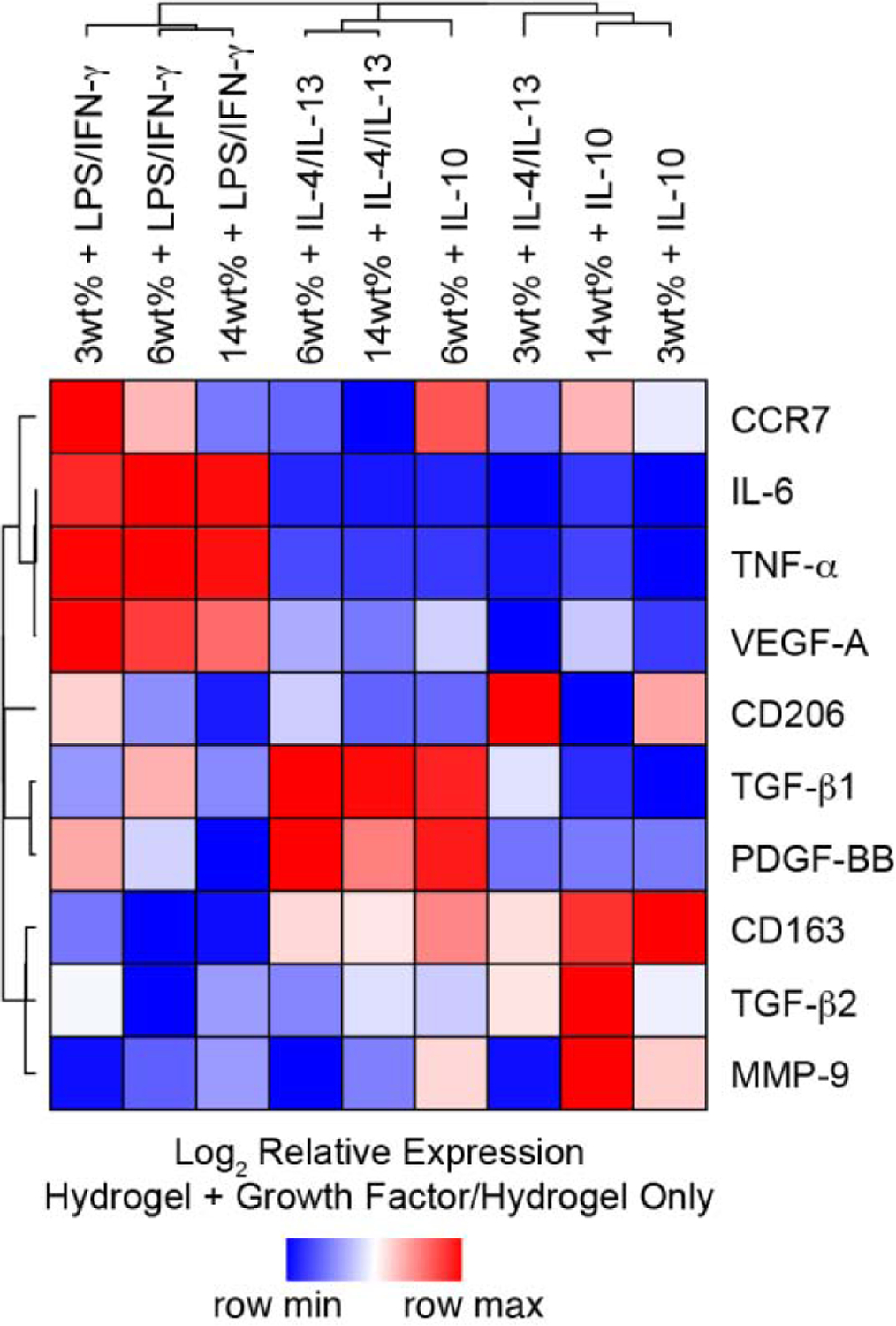
Hierarchical cluster analysis of surface markers and growth factor production data for macrophage cultured on 3wt%, 6wt%, and 14wt% hydrogels, and cultured with and without the presence of pro- or anti-inflammatory cytokines for 3 days. Columns represent each hydrogel/cytokine cocktail combination normalized to hydrogel alone; rows represent each surface marker or growth factor. Heat map colors represent the relative production of each biomarker normalized to the corresponding hydrogel-only control value, with blue indicating production that the value is in the lower half of the range across treatments for that biomarker and red indicating relatively elevated levels, as indicated in the scale bar (log2-transformed scale).
HCL analysis further highlighted the generation of hybrid macrophage phenotypes following exposure of CB Mϕ to multiple stimuli simultaneously. Specifically, a stiffness-dependent increase in CCR7, an M1 associated surface protein, was detected among IL-10 treated cells (Figure 7), which was not observed for macrophage cultured on ultra-low attachment polystyrene in the presence of IL-10. The stiff hydrogels employed in this study exhibit a swollen shear modulus of ~10 kPa, which, when converted to Young’s modulus (elastic modulus, E) using the formula E = 2G′(1 + ν) with ν = 0.5 [59], is equivalent to E≈30 kPa. As standard tissue culture polystyrene exhibits an E≈100,000 kPa, the differing responses observed for macrophage cultured on these two substrates is not surprising. In fact, numerous reports have demonstrated that the behavior of cells cultured on substrates exhibiting E≈30 kPa differs from the response observed on tissue culture polystyrene [45, 60], while others have shown that responses of cells across moduli in the range of E≈30 kPa are not necessarily monotonic functions [61, 62]. Importantly, the simultaneous expression of CCR7 and CD163 by IL-10-treated cells on stiffer hydrogels has been observed both in vitro and in vivo in environments when both M1- and M2- inducing cytokines are present [63, 64]; however, our results suggest potential regulation of CCR7 by non-classical stimuli. Interestingly, we also observed an unexpected increase in CCR7 expression following repolarization of M2a macrophage towards an M2c phenotype (Figure 2B, Figure 6). Together, our observations suggest that presentation of IL-10 to CB-derived macrophage exhibiting an M2a phenotype, induced either via substrate mechanical properties (Figure 4) or exogenous addition of IL-4/IL-13 (Figure 1), results in enhanced CCR7 expression. Interestingly, altered CCR7 expression associated with IL-10 treatment did not correspond to decreased M2c growth factor secretion or increased production of M1-associated growth factors, which is perhaps not surprising as IL-10 is known to inhibit production of IL-6 and TNF-α [65], and further suggests that CCR7 regulation is not cleanly tied to pro-inflammatory phenotype, substantiating previous reports [66–68].
One unexpected finding was that expression of CD206 by CB-derived macrophages also appeared to be regulated by non-classical stimuli on hydrogels, as treatment with LPS/IFN-γ, IL-4/IL-13, or IL-10 cocktails resulted in enhanced CD206 expression by Mϕ cultured across hydrogel moduli. Increased CD206 expression by Mϕ cultured on hydrogels in the presence of IL-10 is perhaps not surprising as delineation of M2a and M2c subsets by CD206 is often challenging and further, is dependent on cell source or specific culture conditions [39]. Conversely, induction of CD206 by Mϕ cultured on hydrogels in the presence of LPS/IFN-γ was unexpected in our hydrogel system; however, published reports suggest that both human and murine macrophages exposed to pro- and anti-inflammatory cytokine cocktails, either simultaneously or sequentially, may increase their production of both M1 and M2a biomarkers [21, 69]. In this work, stiff hydrogels evoked an M2a phenotype in CB-derived macrophage; thus, the simultaneous presentation of stiff matrices and LPS/IFN-γ to Mϕ may have contributed to the generation of hybrid macrophage phenotypes. Our results further highlight the complex associations between cell surface marker expression and pro- or anti-inflammatory cytokine release by CB-derived macrophage in response to altered microenvironmental cues.
Macrophages represent a critical subpopulation of CB cells that has received little research attention, and our results demonstrate of the stable polarization of these cord-blood derived cells marks an important step in the development of cell-based therapies. Moving forward, further functional studies exploring the phagocytic capacity of CB macrophage phenotypes and the interactions the CB macrophages with, as well as the ability of the phenotypes to regulate the behavior of, other cell types, will be paramount. Further, assessment of macrophage phenotype maintenance following hydrogel detachment may also be critical in pre-clinical and clinical development of CB cell-based therapeutics.
5. Conclusion
In conclusion, we demonstrate that Mϕ derived from CB CD14+ monocytes respond to exogenous cytokine cocktails (LPS/IFNγ, IL-4/IL-13, or IL-10) by adopting phenotypes that are similar to those seen observed for macrophage derived from peripheral blood or bone marrow. Macrophage readily altered their phenotypes following sequential administration of pro- and anti-inflammatory cytokine cocktails, demonstrating their inherent plasticity. CB-derived macrophage phenotypes were strongly influenced by substrate modulus, and culture on PEG-based materials with bulk moduli in the 0.1–10.3 kPa range dramatically influenced macrophage phenotype. Integration of hydrogel treatment and cytokine cocktail treatment effects resulted in differential regulation of macrophage phenotypic biomarkers. Our results suggest that CB-derived macrophages generally exhibit predicted behavior that can be further tuned by modulating cytokine profiles and emphasize the complex associations between cell surface marker expression and pro- and anti-inflammatory growth factor release, which are difficult to delineate for the CB-derived macrophage populations whose phenotypes are not yet well understood. Additional studies resolving the relationships between mechanosensitive signaling pathways, physicochemical stimuli, and cell behavior will be critical to our further understanding the response of CB-derived macrophage to engineered microenvironments, particularly in the development of advanced therapies for wound healing, neovascularization, and tissue regeneration.
Supplementary Material
Statement of Significance.
Cord blood (CB) mononuclear cells have elicited significant promise in biomaterials-based regenerative therapies; however, the phenotypic responses of CB-derived macrophage subpopulations towards altered microenvironmental cues are not well understood. Harnessing macrophage behaviors to drive desired phenotypes would tremendously benefit tissue engineering applications. We evaluated the effects of cytokine stimulation and alteration substrate stiffness on cord blood-derived macrophages. Administration of either specific cytokine cocktails or altered hydrogel stiffness to CB macrophage altered their inflammatory and anti-inflammatory phenotypes. Macrophage exposed simultaneously to specific hydrogel moduli and cytokine cocktail treatments resulted in differential regulation of macrophage behaviors. Thus, our research provides knowledge of the interactions between CB macrophages and microenvironmental cues, and offers a new perspective for combinatorial modulation for guiding macrophage response.
Acknowledgements
This research was made possible by funds from the Nemours Foundation and by support from the NHLBI (R01 HL108110, F32 HL127983). Instrumentation and other resources were provided by support from the NIGMS (P20 GM103446, P30 GM110758, U54 GM104941, and S10 OD016361).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Rizk M, Aziz J, Shorr R, Allan DS, Cell-Based Therapy Using Umbilical Cord Blood for Novel Indications in Regenerative Therapy and Immune Modulation: An Updated Systematic Scoping Review of the Literature, Biol Blood Marrow Transplant 23(10) (2017) 1607–1613. [DOI] [PubMed] [Google Scholar]
- [2].Gluckman E, Rocha V, Boyer-Chammard A, Locatelli F, Arcese W, Pasquini R, Ortega J, Souillet G, Ferreira E, Laporte J-P, Fernandez M, Chastang C, Outcome of Cord-Blood Transplantation from Related and Unrelated Donors, New England Journal of Medicine 337(6) (1997) 373–381. [DOI] [PubMed] [Google Scholar]
- [3].Peters EB, Christoforou N, Leong KW, Truskey GA, West JL, Poly(ethylene glycol) Hydrogel Scaffolds Containing Cell-Adhesive and Protease-Sensitive Peptides Support Microvessel Formation by Endothelial Progenitor Cells, Cell Mol Bioeng 9(1) (2016) 38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shearer WT, Lubin BH, Cairo MS, Notarangelo LD, Cord Blood Banking for Potential Future Transplantation, Pediatrics 140(5) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kurtzberg J, A History of Cord Blood Banking and Transplantation, Stem Cells Transl Med 6(5) (2017) 1309–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Brown KS, Rao MS, Brown HL, The Future State of Newborn Stem Cell Banking, J Clin Med 8(1) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].van de Ven C, Collins D, Bradley MB, Morris E, Cairo MS, The potential of umbilical cord blood multipotent stem cells for nonhematopoietic tissue and cell regeneration, Exp Hematol 35(12) (2007) 1753–65. [DOI] [PubMed] [Google Scholar]
- [8].Mahadevaiah S, Robinson KG, Kharkar PM, Kiick KL, Akins RE, Decreasing matrix modulus of PEG hydrogels induces a vascular phenotype in human cord blood stem cells, Biomaterials 62 (2015) 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ferreira MS, Jahnen-Dechent W, Labude N, Bovi M, Hieronymus T, Zenke M, Schneider RK, Neuss S, Cord blood-hematopoietic stem cell expansion in 3D fibrin scaffolds with stromal support, Biomaterials 33(29) (2012) 6987–97. [DOI] [PubMed] [Google Scholar]
- [10].Mousavi SH, Abroun S, Soleimani M, Mowla SJ, Expansion of human cord blood hematopoietic stem/progenitor cells in three-dimensional Nanoscaffold coated with Fibronectin, Int J Hematol Oncol Stem Cell Res 9(2) (2015) 72–9. [PMC free article] [PubMed] [Google Scholar]
- [11].Klontzas ME, Reakasame S, Silva R, Morais JCF, Vernardis S, MacFarlane RJ, Heliotis M, Tsiridis E, Panoskaltsis N, Boccaccini AR, Mantalaris A, Oxidized alginate hydrogels with the GHK peptide enhance cord blood mesenchymal stem cell osteogenesis: A paradigm for metabolomics-based evaluation of biomaterial design, Acta Biomaterialia 88 (2019) 224–240. [DOI] [PubMed] [Google Scholar]
- [12].Park YB, Ha CW, Kim JA, Han WJ, Rhim JH, Lee HJ, Kim KJ, Park YG, Chung JY, Single-stage cell-based cartilage repair in a rabbit model: cell tracking and in vivo chondrogenesis of human umbilical cord blood-derived mesenchymal stem cells and hyaluronic acid hydrogel composite, Osteoarthritis and Cartilage 25(4) (2017) 570–580. [DOI] [PubMed] [Google Scholar]
- [13].Normann E, Lacaze-Masmonteil T, Winkler-Lowen B, Guilbert LJ, Isolation of Non-Activated Monocytes from Human Umbilical Cord Blood, American Journal of Reproductive Immunology 63(1) (2010) 66–72. [DOI] [PubMed] [Google Scholar]
- [14].Saha A, Buntz S, Scotland P, Xu L, Noeldner P, Patel S, Wollish A, Gunaratne A, Gentry T, Troy J, Matsushima GK, Kurtzberg J, Balber AE, A cord blood monocyte–derived cell therapy product accelerates brain remyelination, JCI Insight 1(13) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Juhas M, Abutaleb N, Wang JT, Ye J, Shaikh Z, Sriworarat C, Qian Y, Bursac N, Incorporation of macrophages into engineered skeletal muscle enables enhanced muscle regeneration, Nature Biomedical Engineering 2(12) (2018) 942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jiang H, Van de Ven C, Satwani P, Baxi LV, Cairo MS, Differential Gene Expression Patterns by Oligonucleotide Microarray of Basal versus Lipopolysaccharide-Activated Monocytes from Cord Blood versus Adult Peripheral Blood, The Journal of Immunology 172(10) (2004) 5870–5879. [DOI] [PubMed] [Google Scholar]
- [17].van Amerongen MJ, Harmsen MC, van Rooijen N, Petersen AH, van Luyn MJ, Macrophage depletion impairs wound healing and increases left ventricular remodeling after myocardial injury in mice, Am J Pathol 170(3) (2007) 818–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Godwin JW, Pinto AR, Rosenthal NA, Macrophages are required for adult salamander limb regeneration, Proc Natl Acad Sci U S A 110(23) (2013) 9415–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Condeelis J, Pollard JW, Macrophages: obligate partners for tumor cell migration, invasion, and metastasis, Cell 124(2) (2006) 263–6. [DOI] [PubMed] [Google Scholar]
- [20].Troidl C, Jung G, Troidl K, Hoffmann J, Mollmann H, Nef H, Schaper W, Hamm CW, Schmitz-Rixen T, The temporal and spatial distribution of macrophage subpopulations during arteriogenesis, Curr Vasc Pharmacol 11(1) (2013) 5–12. [PubMed] [Google Scholar]
- [21].Spiller KL, Nassiri S, Witherel CE, Anfang RR, Ng J, Nakazawa KR, Yu T, Vunjak-Novakovic G, Sequential delivery of immunomodulatory cytokines to facilitate the M1-to-M2 transition of macrophages and enhance vascularization of bone scaffolds, Biomaterials 37 (2015) 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Agrawal H, Tholpady SS, Capito AE, Drake DB, Katz AJ, Macrophage phenotypes correspond with remodeling outcomes of various acellular dermal matrices, Open Journal of Regenerative Medicine Vol.01No.03 (2012) 9. [Google Scholar]
- [23].Adlerz KM, Aranda-Espinoza H, Hayenga HN, Substrate elasticity regulates the behavior of human monocyte-derived macrophages, European Biophysics Journal 45(4) (2016) 301–309. [DOI] [PubMed] [Google Scholar]
- [24].Gruber E, Heyward C, Cameron J, Leifer C, Toll-like receptor signaling in macrophages is regulated by extracellular substrate stiffness and Rho-associated coiled-coil kinase (ROCK1/2), Int Immunol 30(6) (2018) 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Previtera ML, Sengupta A, Substrate Stiffness Regulates Proinflammatory Mediator Production through TLR4 Activity in Macrophages, PLOS ONE 10(12) (2016) e0145813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Okamoto T, Takagi Y, Kawamoto E, Park EJ, Usuda H, Wada K, Shimaoka M, Reduced substrate stiffness promotes M2-like macrophage activation and enhances peroxisome proliferator-activated receptor γ expression, Experimental Cell Research 367(2) (2018) 264–273. [DOI] [PubMed] [Google Scholar]
- [27].Blakney AK, Swartzlander MD, Bryant SJ, The effects of substrate stiffness on the in vitro activation of macrophages and in vivo host response to poly(ethylene glycol)-based hydrogels, J Biomed Mater Res A 100(6) (2012) 1375–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Berger AJ, Linsmeier KM, Kreeger PK, Masters KS, Decoupling the effects of stiffness and fiber density on cellular behaviors via an interpenetrating network of gelatin-methacrylate and collagen, Biomaterials 141 (2017) 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen M, Zhang Y, Zhou P, Liu X, Zhao H, Zhou X, Gu Q, Li B, Zhu X, Shi Q, Substrate stiffness modulates bone marrow-derived macrophage polarization through NF-κB signaling pathway, Bioactive Materials 5(4) (2020) 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kharkar PM, Kiick KL, Kloxin AM, Design of thiol- and light-sensitive degradable hydrogels using Michael-type addition reactions, Polymer Chemistry 6(31) (2015) 5565–5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Scott RA, Kharkar PM, Kiick KL, Akins RE, Aortic adventitial fibroblast sensitivity to mitogen activated protein kinase inhibitors depends on substrate stiffness, Biomaterials 137 (2017) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sawicki LA, Ovadia EM, Pradhan L, Cowart JE, Ross KE, Wu CH, Kloxin AM, Tunable synthetic extracellular matrices to investigate breast cancer response to biophysical and biochemical cues, APL Bioeng 3(1) (2019) 016101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M, Macrophage plasticity and polarization in tissue repair and remodelling, The Journal of Pathology 229(2) (2013) 176–185. [DOI] [PubMed] [Google Scholar]
- [34].McWhorter FY, Wang T, Nguyen P, Chung T, Liu WF, Modulation of macrophage phenotype by cell shape, Proc Natl Acad Sci U S A 110(43) (2013) 17253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sridharan R, Ryan EJ, Kearney CJ, Kelly DJ, O’Brien FJ, Macrophage Polarization in Response to Collagen Scaffold Stiffness Is Dependent on Cross-Linking Agent Used To Modulate the Stiffness, ACS Biomaterials Science & Engineering 5(2) (2019) 544–552. [DOI] [PubMed] [Google Scholar]
- [36].Vogel DYS, Heijnen PDAM, Breur M, de Vries HE, Tool ATJ, Amor S, Dijkstra CD, Macrophages migrate in an activation-dependent manner to chemokines involved in neuroinflammation, Journal of Neuroinflammation 11(1) (2014) 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Eligini S, Crisci M, Bono E, Songia P, Tremoli E, Colombo GI, Colli S, Human monocyte-derived macrophages spontaneously differentiated in vitro show distinct phenotypes, Journal of Cellular Physiology 228(7) (2013) 1464–1472. [DOI] [PubMed] [Google Scholar]
- [38].Kurtzberg J, Buntz S, Gentry T, Noeldner P, Ozamiz A, Rusche B, Storms RW, Wollish A, Wenger DA, Balber AE, Preclinical characterization of DUOC-01, a cell therapy product derived from banked umbilical cord blood for use as an adjuvant to umbilical cord blood transplantation for treatment of inherited metabolic diseases, Cytotherapy 17(6) (2015) 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Spiller KL, Wrona EA, Romero-Torres S, Pallotta I, Graney PL, Witherel CE, Panicker LM, Feldman RA, Urbanska AM, Santambrogio L, Vunjak-Novakovic G, Freytes DO, Differential gene expression in human, murine, and cell line-derived macrophages upon polarization, Experimental Cell Research 347(1) (2016) 1–13. [DOI] [PubMed] [Google Scholar]
- [40].Zhao H, Kalish FS, Wong RJ, Stevenson DK, Hypoxia regulates placental angiogenesis via alternatively activated macrophages, Am J Reprod Immunol 80(3) (2018) e12989. [DOI] [PubMed] [Google Scholar]
- [41].Spiller KL, Anfang RR, Spiller KJ, Ng J, Nakazawa KR, Daulton JW, Vunjak-Novakovic G, The role of macrophage phenotype in vascularization of tissue engineering scaffolds, Biomaterials 35(15) (2014) 4477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Goodier HCJ, Carr AJ, Snelling SJB, Roche L, Wheway K, Watkins B, Dakin SG, Comparison of transforming growth factor beta expression in healthy and diseased human tendon, Arthritis Research & Therapy 18(1) (2016) 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li N, Qin J, Lan L, Zhang H, Liu F, Wu Z, Ni H, Wang Y, PTEN inhibits macrophage polarization from M1 to M2 through CCL2 and VEGF-A reduction and NHERF-1 synergism, Cancer Biol Ther 16(2) (2015) 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tarique AA, Logan J, Thomas E, Holt PG, Sly PD, Fantino E, Phenotypic, functional, and plasticity features of classical and alternatively activated human macrophages, Am J Respir Cell Mol Biol 53(5) (2015) 676–88. [DOI] [PubMed] [Google Scholar]
- [45].Robinson KG, Nie T, Baldwin AD, Yang EC, Kiick KL, Akins RE Jr., Differential effects of substrate modulus on human vascular endothelial, smooth muscle, and fibroblastic cells, Journal of Biomedical Materials Research Part A 100A(5) (2012) 1356–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D, Substrate Compliance versus Ligand Density in Cell on Gel Responses, Biophysical Journal 86(1) (2004) 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Krombach F, Münzing S, Allmeling AM, Gerlach JT, Behr J, Dörger M, Cell size of alveolar macrophages: an interspecies comparison, Environ Health Perspect 105 (1997) 1261–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhuang Z, Zhang Y, Sun S, Li Q, Chen K, An C, Wang L, van den Beucken JJJP, Wang H, Control of Matrix Stiffness Using Methacrylate–Gelatin Hydrogels for a Macrophage-Mediated Inflammatory Response, ACS Biomaterials Science & Engineering 6(5) (2020) 3091–3102. [DOI] [PubMed] [Google Scholar]
- [49].Lynn AD, Bryant SJ, Phenotypic changes in bone marrow-derived murine macrophages cultured on PEG-based hydrogels activated or not by lipopolysaccharide, Acta Biomater 7(1) (2011) 123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cha BH, Shin SR, Leijten J, Li YC, Singh S, Liu JC, Annabi N, Abdi R, Dokmeci MR, Vrana NE, Ghaemmaghami AM, Khademhosseini A, Integrin-Mediated Interactions Control Macrophage Polarization in 3D Hydrogels, Adv Healthc Mater 6(21) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, Chen YY, Liphardt J, Hwang ES, Weaver VM, Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration, Integr Biol (Camb) 7(10) (2015) 1120–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wang X, Michie SA, Xu B, Suzuki Y, Importance of IFN-gamma-mediated expression of endothelial VCAM-1 on recruitment of CD8+ T cells into the brain during chronic infection with Toxoplasma gondii, J Interferon Cytokine Res 27(4) (2007) 329–38. [DOI] [PubMed] [Google Scholar]
- [53].Russo E, Salzano M, Postiglione L, Guerra A, Marotta V, Vitale M, Interferon-γ inhibits integrin-mediated extracellular signal-regulated kinase activation stimulated by fibronectin binding in thyroid cells, J Endocrinol Invest 36(6) (2013) 375–8. [DOI] [PubMed] [Google Scholar]
- [54].Bartneck M, Heffels K-H, Pan Y, Bovi M, Zwadlo-Klarwasser G, Groll J, Inducing healing-like human primary macrophage phenotypes by 3D hydrogel coated nanofibres, Biomaterials 33(16) (2012) 4136–4146. [DOI] [PubMed] [Google Scholar]
- [55].Chandrasekar BS, Yadav S, Victor ES, Majumdar S, Deobagkar-Lele M, Wadhwa N, Podder S, Das M, Nandi D, Interferon-gamma and nitric oxide synthase 2 mediate the aggregation of resident adherent peritoneal exudate cells: implications for the host response to pathogens, PLoS One 10(6) (2015) e0128301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Dellacasagrande J, Ghigo E, Raoult D, Capo C, Mege J-L, IFN-γ-Induced Apoptosis and Microbicidal Activity in Monocytes Harboring the Intracellular Bacterium Coxiella burnetii Require Membrane TNF and Homotypic Cell Adherence, The Journal of Immunology 169(11) (2002) 6309–6315. [DOI] [PubMed] [Google Scholar]
- [57].Yuan LH, Chen XL, Di Y, Liu ML, CCR7/p-ERK1/2/VEGF signaling promotes retinal neovascularization in a mouse model of oxygen-induced retinopathy, Int J Ophthalmol 10(6) (2017) 862–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Xu Z, Zhu C, Chen C, Zong Y, Feng H, Liu D, Feng W, Zhao J, Lu A, CCL19 suppresses angiogenesis through promoting miR-206 and inhibiting Met/ERK/Elk-1/HIF-1α/VEGF-A pathway in colorectal cancer, Cell Death & Disease 9(10) (2018) 974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bryant KAS, Photopolymerization of Hydrogel Scaffolds. Scaffolding in Tissue Engineering., CRC Press; 2005. [Google Scholar]
- [60].Peyton SR, Putnam AJ, Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion, J Cell Physiol 204(1) (2005) 198–209. [DOI] [PubMed] [Google Scholar]
- [61].Chen M, Zhang Y, Zhou P, Liu X, Zhao H, Zhou X, Gu Q, Li B, Zhu X, Shi Q, Substrate stiffness modulates bone marrow-derived macrophage polarization through NF-κB signaling pathway, Bioact Mater 5(4) (2020) 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Liu F, Mih JD, Shea BS, Kho AT, Sharif AS, Tager AM, Tschumperlin DJ, Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression, J Cell Biol 190(4) (2010) 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kwiecień I, Polubiec-Kownacka M, Dziedzic D, Wołosz D, Rzepecki P, Domagała-Kulawik J, CD163 and CCR7 as markers for macrophage polarization in lung cancer microenvironment, Cent Eur J Immunol 44(4) (2019) 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tatano Y, Shimizu T, Tomioka H, Unique Macrophages Different from M1/M2 Macrophages Inhibit T Cell Mitogenesis while Upregulating Th17 Polarization, Scientific Reports 4(1) (2014) 4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kessler B, Rinchai D, Kewcharoenwong C, Nithichanon A, Biggart R, Hawrylowicz CM, Bancroft GJ, Lertmemongkolchai G, Interleukin 10 inhibits pro-inflammatory cytokine responses and killing of Burkholderia pseudomallei, Scientific Reports 7(1) (2017) 42791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mia S, Warnecke A, Zhang XM, Malmström V, Harris RA, An optimized protocol for human M2 macrophages using M-CSF and IL-4/IL-10/TGF-β yields a dominant immunosuppressive phenotype, Scand J Immunol 79(5) (2014) 305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kryczanowsky F, Raker V, Graulich E, Domogalla MP, Steinbrink K, IL-10–Modulated Human Dendritic Cells for Clinical Use: Identification of a Stable and Migratory Subset with Improved Tolerogenic Activity, The Journal of Immunology 197(9) (2016) 3607–3617. [DOI] [PubMed] [Google Scholar]
- [68].Fujiwara Y, Hizukuri Y, Yamashiro K, Makita N, Ohnishi K, Takeya M, Komohara Y, Hayashi Y, Guanylate-binding protein 5 is a marker of interferon-γ-induced classically activated macrophages, Clinical & Translational Immunology 5(11) (2016) e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Smith TD, Tse MJ, Read EL, Liu WF, Regulation of macrophage polarization and plasticity by complex activation signals, Integr Biol (Camb) 8(9) (2016) 946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


