Summary
SARS-CoV-2 infection results in viral burden in the respiratory tract, enabling transmission and leading to substantial lung pathology. The 1212C2 fully human monoclonal antibody was derived from an IgM memory B cell of a COVID-19 patient, has high affinity for the Spike protein receptor binding domain, neutralizes SARS-CoV-2, and exhibits in vivo prophylactic and therapeutic activity in hamsters when delivered intraperitoneally, reducing upper and lower respiratory viral burden and lung pathology. Inhalation of nebulized 1212C2 at levels as low as 0.6 mg/kg, corresponding to 0.03 mg/kg lung-deposited dose, reduced the viral burden below the detection limit and mitigated lung pathology. The therapeutic efficacy of an exceedingly low dose of inhaled 1212C2 supports the rationale for local lung delivery for dose-sparing benefits, as compared to the conventional parenteral route of administration. These results suggest that the clinical development of 1212C2 formulated and delivered via inhalation for the treatment of SARS-CoV-2 infection should be considered.
Keywords: SARS-CoV-2, COVID-19, B cell, IgM, monoclonal antibody, inhalation, neutralizing, Spike, RBD, hamsters
Graphical abstract

Highlights
The 1212C2 human monoclonal antibody potently neutralizes SARS-CoV-2
1212C2 mAb was isolated from an IgM memory B cell of a recovered COVID-19 patient
Inhaled 1212C2 mAb is rapidly distributed in the lungs
Inhaled 1212C2 mAb treatment reduces viral burden and lung pathology in hamsters
Piepenbrink et al. isolate a panel of potent SARS-CoV-2 neutralizing monoclonal antibodies and demonstrate that the 1212C2 mAb with high affinity for RBD is able to reduce the viral burden in SARS-CoV-2-infected hamsters. Inhaled 1212C2 enables rapid delivery to the lungs and therapeutic activity at lower doses compared with parenteral administration.
Introduction
The severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) global pandemic has infected >45 million people and has resulted in >1 million deaths so far. It is expected that new infections will continue for many more months, and the virus may persist endemically for years. Although severe infections and deaths have been reported in all ages and demographics, those older than 65 years and those with preexisting conditions are at highest risk for death.1, 2, 3, 4, 5 Moreover, in the United States, the number of coronavirus disease 2019 (COVID-19) patients who are hospitalized represent a small fraction (∼10%) of the active cases, which implies that the vast majority of COVID-19 symptomatic patients are not hospitalized and need treatment.6, 7, 8 Therefore, facilitating greater treatment coverage will be of importance to control transmissibility and healthcare burden.
Neutralizing antibodies (NAbs) induced either by natural infection or vaccination are likely to be critical for protection from SARS-CoV-2 infection and have been correlated with protection from SARS-CoV-2 in animal studies,9, 10, 11 and the passive transfer of neutralizing mAbs has demonstrated prophylactic and therapeutic activity against SARS-CoV-2 infection.12, 13, 14 Emerging results in humans treated with convalescent plasma with high titer NAbs suggest therapeutic activity.15,16 The primary target for SARS-CoV-2 neutralizing antibodies is the receptor binding domain (RBD), whereby antibodies are expected to inhibit the binding of the SARS-CoV-2 Spike (S) protein to the host angiotensin-converting enzyme 2 (ACE2), preventing viral attachment. Already several human mAbs (hmAbs) have been isolated from patients following SARS-CoV-2 infection that are specific for RBD, neutralize SARS-CoV-2, and have antiviral activity in animal models.12,17 RBD is used predominantly as the target in clinical stage vaccines and antibody candidates, with preliminary positive clinical responses reported.18,19
To obtain more precise resolution of the RBD-specific NAb response, a panel of RBD-specific hmAbs were isolated and their molecular features, reactivity profiles, and in vitro and in vivo antiviral activities were defined. Several high-affinity hmAbs with modest somatic hypermutation and potent SARS-CoV-2 neutralizing activity were isolated from immunoglobulin G (IgG), IgA, and IgM memory B cells. In this report, we show that the 1212C2 hmAb demonstrated substantial prophylactic and therapeutic activity in hamsters when delivered parenterally. Moreover, when delivered as inhaled liquid aerosols using a commercially available nebulizer, 1212C2 mediated the eradication of lung viral load at a substantially higher dosing efficiency than the parenteral route of administration. The combination of a potent SARS-CoV-2 mAb and inhaled, local lung delivery using widely available nebulizers could provide for a promising treatment option.
Results
Isolation of RBD+ B cells and hmAb screening
To identify and isolate RBD-specific B cells, recombinant RBD protein was expressed, biotinylated, and used to form streptavidin-conjugated RBD tetramers to various fluorochromes. Peripheral blood CD27+ memory B cells binding RBD were single cell sorted from convalescent SARS-CoV-2 patients and the Ig heavy- and light-chain variable regions of the B cells cloned to generate IgG1 recombinant hmAbs (Figure 1A). Twenty hmAbs were isolated that exhibited substantial binding to the SARS-CoV-2 RBD. These hmAbs all bound to recombinant SARS-CoV-2 RBD and S1 D614G proteins and exhibited varying reactivity SARS-CoV-2 S1S2 proteins (Figure 1B). As expected, minimal to no reactivity was observed in SARS-CoV-2 Nucleocapsid (N) protein, SARS-CoV-1, or HepG2 cell lysate for most hmAbs, indicating high specificity for SARS-CoV-2 RBD and minimal off-target binding. REGN10987, REGN10933, and CB6/JS016 hmAbs were synthesized and included as positive controls.17,20 To ascertain the avidity of the hmAbs for SARS-CoV-2 RBD, their binding stability was determined in the presence of the chaotropic agent 8 M urea. The hmAbs 1206D1, 1212C2, 1212F5, and 1215D1 retained at least 50% of their binding activity to SARS-CoV-2 RBD and S protein (Figure 1C). Binding affinity to RBD was further tested for a subset of the hmAbs by surface plasmon resonance (SPR), demonstrating a variety of high-affinity hmAbs exhibiting equilibrium dissociation constants (KD) ranging from 71 pM to ∼10 nM (Figures 1D and S1). Several of the hmAbs (1212C2, 1215D1, 1215D5) exhibited ∼8-fold or higher affinity to RBD than control hmAbs (REGN10987, REGN10933,20 and CB6/JS01617), predominantly due to their slower off rates (kd). These results indicate that this panel of hmAbs recognizes SARS-CoV-2 RBD specifically and with high affinity.
Figure 1.
Isolation of SARS-CoV-2 RBD-specific human monoclonal antibodies (hmAbs)
(A) Representative gating strategy for RBD+ B cell isolation. Initial plots are gated on live CD3−CD4− CD19+ annexin V-CD27+ B cells.
(B) hmAbs were tested at 10 and 1 μg/mL in duplicate by ELISA for binding to indicated protein; area under the curve (AUC) is indicated.
(C) hmAbs were tested at 10 μg/mL in duplicate for binding to indicated protein by ELISA in the presence or absence of 8 M urea treatment, mean and standard error presented.
(D) Binding to SARS-CoV-2 RBD was determined by surface plasmon resonance (SPR), mean and standard deviation presented.
Recognition and neutralization of SARS-CoV-2
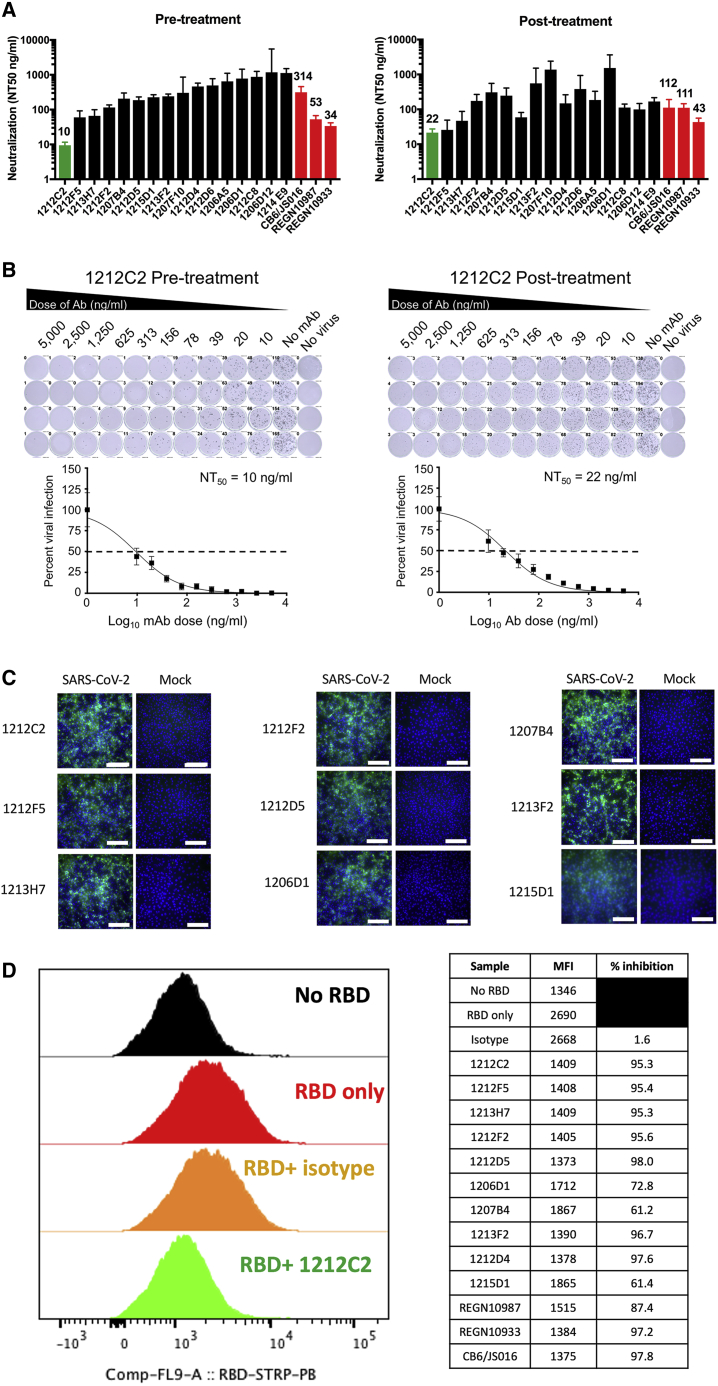
The process of entry into a susceptible host cell is an important determinant of infectivity and pathogenesis of viruses, including coronaviruses.21,22 SARS-CoV-2 relies on the ability of its S glycoprotein to bind to the ACE2 receptor through its RBD driving a conformational change that culminates in the fusion of the viral envelope with the host cell membrane and cell entry.23 The hmAbs were tested for the neutralization of live SARS-CoV-2 using a virus plaque reduction microneutralization (PRMNT) assay we previously described,24 with the hmAb and virus pre-incubated before culture with susceptible Vero E6 cells (pre-treatment) or allowing virus adsorption to Vero E6 cells to occur for 1 h before the addition of the hmAb (post-treatment). This gives an opportunity for the virus to initiate viral entry by binding to the cell surface receptor, potentially distinguishing the ability of hmAb to preferentially block the later steps of the virus entry into the cell or by inhibiting the cell-to-cell spread of the virus progeny. The panel of hmAbs neutralized SARS-CoV-2 in both pre- and post-treatment conditions at 50% neutralization (NT50) of ≤1 μg/mL, with 1212F5, 1212C2, and 1213H7 exhibiting the highest potency, with a NT50 of ≤100 ng/mL (Figure 2A). The hmAb 1215D1 was more effective in neutralizing in post-treatment (NT50 = 59 ng/mL) compared to pre-treatment (NT50 = 226 ng/mL). The 1212C2 hmAb was particularly potent (pre-treatment NT50 = 10 ng/mL, post-treatment NT50 = 22 ng/mL), with similar neutralizing activity to CB6/JS06, REGN10987, and REGN10933 (Figure 2B). In addition, testing of 1212C2 in a SARS-CoV-2 vesicular stomatitis virus (VSV) vectored pseudovirus assay confirmed its potent neutralizing activity (NT50 = 1.9 ng/mL) that was also similar to REGN10987 and CB6/JS016 (Table S1).
Figure 2.
Neutralization of SARS-CoV-2 infection and RBD binding to ACE2
(A) Indicated hmAbs were incubated with live SARS-CoV-2 (100 PFU/well) 1 h before (pre-treatment) or 1 h after (post-treatment) addition to Vero E6 cells. hmAbs were tested in quadruplicate cultures and NT50 and upper 95% confidence interval (CI) indicated.
(B) Representative titration curve of 1212C2 hmAb presented, mean and standard error presented.
(C) Binding of indicated hmAb to SARS-CoV-2 or mock-infected Vero E6 cells measured by immunofluorescence; scale bar, 100 μm.
(D) Indicated hmAb was incubated as single replicate with recombinant biotinylated RBD protein before incubation with HEK293-ACE2 cells measured by flow cytometry. Plot gated on 7-aminoactinomycin (7AAD)-ACE2+ cells.
The panel of hmAbs clearly recognized SARS-CoV-2-infected Vero E6 cells as seen via immunofluorescence (Figures 2C and S2). No background staining of mock-infected cells was evident with any of the hmAbs, which is consistent with their high-affinity specificity for SARS-CoV-2 S. The hmAbs also exhibited binding to SARS-CoV-2 viral lysate (Figure S3) and SARS-CoV-2 D614G-infected cells (Figure S4). The neutralizing activity of 1212C2 against SARS-CoV-2 D614G was confirmed (Figure S4). The ability of the hmAbs to directly inhibit the binding of RBD to ACE2 was determined using HEK293 cells overexpressing ACE2. All of the tested RBD-specific hmAbs inhibited the binding of recombinant SARS-CoV-2 RBD protein to the ACE2-expressing cells (Figure 2D). Inhibition was nearly complete, with the exceptions of 1206D1, 1207B4, and 1215D1. These results demonstrate the potent in vitro neutralizing and binding activity of these SARS-CoV-2 RBD-specific hmAbs.
Molecular features and clonal dynamics
The most potent hmAbs, 1212C2 and 1212F5, were both isolated from IgM+ B cells, belong to the same clonal lineage that uses the VH1-2 heavy-chain gene, and exhibited modest somatic hypermutation, with 1212C2 further mutated from the germline compared to 1212F5 (8.2% versus 6.1% amino acid VH) (Table 1). Most of the hmAbs were isolated from IgG1-expressing B cells, while 1215D1 and 1212C8 were isolated from IgA-expressing B cells. All hmAbs exhibited somatic hypermutation (2.0%–9.1% VH), suggesting that they arose from multiple rounds of germinal center reactions. VH3-66 and Vk1-9 gene usage was dominant among the hmAbs. Targeted VH-deep sequencing of the 1212C2/1212F5 clonal lineage from contemporary peripheral blood B cells identified numerous members (Figure 3). The lineage was dominated by IgM- and IgG-expressing B cells, with several expanded nodes of identical clones being expressed by both IgM- and IgG-expressing B cells.
Table 1.
Molecular characteristics of hmAbs
| mAb | VH | DH | JH | Isotype | % mutation AA | VL | JL |
% mutation AA |
| 1206D1 | IGHV1-2 | IGHD6-13 | IGHJ3 | IgG1 | 8.2 | IGLV2-8 | IGLJ2 | 8.2 |
| 1207F10 | IGHV1-2 | IGHD5-12 | IGHJ4 | IgG3 | 3.1 | IGLV6-57 | IGLJ3 | 4.1 |
| 1212C2 | IGHV1-2 | IGHD5-18 | IGHJ4 | IgM | 8.2 | IGLV2-23 | IGLJ2 | 5.2 |
| 1212F5 | IGHV1-2 | IGHD5-18 | IGHJ4 | IgM | 6.1 | IGLV2-23 | IGLJ2 | 3.1 |
| 1213H7 | IGHV1-58 | IGHD2-15 | IGHJ3 | IgG1 | 7.1 | IGKV3-20 | IGKJ1 | 3.2 |
| 1206A5 | IGHV3-9 | IGHD1-1 | IGHJ6 | IgG1 | 2.0 | IGLV2-14 | IGLJ2 | 5.1 |
| 1207B4 | IGHV3-9 | IGHD5-12 | IGHJ4 | IgG1 | 7.1 | IGLV1-44 | IGLJ2 | 3.1 |
| 1215D1 | IGHV3-9 | IGHD4-23 | IGHJ6 | IgA1 | 9.1 | IGLV6-57 | IGLJ2 | 3.1 |
| 1206G12 | IGHV3-15 | IGHD3-22 | IGHJ3 | IgG1 | 5.0 | IGKV1-39 | IGKJ4 | 6.3 |
| 1213F2 | IGHV3-53 | IGHD3-16 | IGHJ3 | IgG1 | 8.2 | IGKV1-9 | IGKJ3 | 4.3 |
| 1206D12 | IGHV3-66 | IGHD2-21 | IGHJ6 | IgG1 | 7.2 | IGKV1-33 | IGKJ2 | 2.1 |
| 1212C8 | IGHV3-66 | IGHD5-24 | IGHJ6 | IgA2 | 7.2 | IGKV1-12 | IGKJ4 | 4.3 |
| 1214E9 | IGHV3-66 | IGHD3-16 | IGHJ6 | IgG1 | 7.2 | IGKV1-9 | IGKJ2 | 0 |
| 1212D4 | IGHV3-66 | IGHD3-3 | IGHJ6 | IgG1 | 6.2 | IGKV1-9 | IGKJ2 | 6.4 |
| 1212F2 | IGHV3-66 | IGHD2-15 | IGHJ6 | IgG1 | 4.1 | IGKV1-9 | IGKJ5 | 3.2 |
| 1212D5 | IGHV3-66 | IGHD3-3 | IGHJ3 | IgG1 | 6.2 | IGKV1-5 | IGKJ2 | 4.2 |
| 1212D6 | IGHV3-66 | IGHD3-10 | IGHJ4 | IgG1 | 8.2 | IGKV1-9 | IGKJ3 | 2.1 |
Figure 3.
Phylogenic analysis of 1212C2 hmAb lineage
Lineage members defined as same heavy-chain V and J gene usage, HCDR3 length, and ≥85% HCDR3 similarity. Germline sequence is represented by the green diamond, the 1212C2 hmAb sequence is represented by the red circle, and the 1212F5 hmAb sequenced is represented by the orange circle. The size of the symbols is proportional to the number of identical sequences obtained of an individual lineage member (N = 2–50), with the exception of the germline and 1212C2 and 1212F5 hmAb sequences. The relationship between lineage members was determined by amino acid sequence similarity.
Epitope mapping
SPR epitope mapping was performed and used to cluster the hmAbs into five major epitopes (A–E, Figure 4). With one exception, all of the isolated hmAbs are part of the A epitope, which includes 1212C2. Thus, 1212C2 efficiently blocked all hmAbs from binding to RBD, except 1215B11 (E epitope), whose epitope overlaps with CR0322. Within the footprint of the A epitope, there are several sub-epitopes. In particular, we highlight hmAbs 1207B4/1215D1 (B epitope) and 1213H7 (C epitope), which exhibit essentially no overlap with one another, based on their ability to simultaneously bind RBD at >90% of their expected binding levels. Consistent with their distinct classification, B epitope hmAbs exhibit a reduced ability to block RBD attachment to ACE2-expressing cells (Figure 2D).
Figure 4.
SPR epitope mapping
(A) Representative sensorgram from the SPR competition assays used to subset hmAbs into distinct RBD binding epitopes. For each assay, a series of hmAbs were sequentially injected over immobilized SARS-CoV-2 RBD. In this example, 1212C2 was injected first, followed by a second injection of 1212C2, 2 injections of 1206D1, and the last injection was CR3022.
(B) Summary of all epitope mapping data, in which each block (first experiment from A in the red box) with a bold hmAb at the top represents a different experiment (10 experiments total). The bold hmAb is the “first” hmAb injected. The percentage of binding (100 = 100% binding and 0 = 0% binding) of subsequent hmAbs was recorded. mAbs were considered to have a different epitope (denoted by a distinct color) if they exhibited binding levels >30% in the presence of other mAbs. Thus, in the first experiment, CR3022 is defined as a new epitope (cyan), distinct from 1212C2.
(C) Schematic diagram of NmAb RBD epitopes defined in the mapping experiment. Five major epitopes (A–E) were identified, where the E epitope overlaps with control mAb CR3022 (cyan, epitope F). Four of the 5 epitopes (A–D) are located within the ACE2 binding site (purple), and all of the NmAbs are blocked by the 1212C2 epitope A (yellow). NmAbs with epitopes similar to B (orange) and C (green) are defined as B’ (light orange) and C’ (light green), respectively. The 1212C2 epitope A (yellow) blocks the binding of all NmAbs, with the exception of 1215B11, which occupies epitope E. Epitopes B and C are also blocked by epitope A NmAbs, but exhibit limited competition with each other.
Prophylactic and therapeutic activity in hamsters
The in vivo activity of 1212C2 and 1206D1 was evaluated in the golden Syrian hamster model of SARS-CoV-2 infection.25 1212C2 was chosen based on its potent in vitro neutralizing activity and high affinity, while 1206D1 was chosen based on its in vitro neutralizing activity and distinct affinity and reactivity profile from 1212C2.
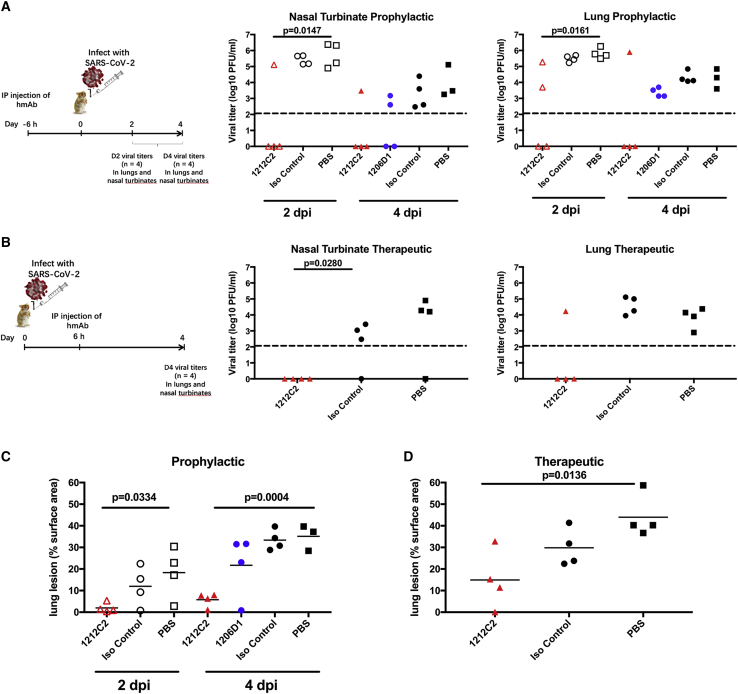
To test for prophylactic activity, 10 mg/kg hmAb was administered by intraperitoneal (i.p.) injection 6 h before intranasal (i.n.) challenge with SARS-CoV-2. At 2 days post-infection (dpi), all PBS control and isotype control hamsters had detectable live virus as measured by plaque assay in their nasal turbinates and lungs. In contrast, at 2 dpi, hamsters that received 1212C2 already started to exhibit meaningful viral load reduction in their nasal turbinate and lungs (Figure 5A). At 4 dpi, virus was detected in the nasal turbinates and lungs of all of the hamsters in the PBS and isotype control groups, although an overall decrease compared to 2 dpi, consistent with the viral dynamics of SARS-CoV-2 infection in hamsters.25 In comparison to the control groups at 4 dpi, prophylactically treated 1212C2 hamsters exhibited the eradication of viral loads in the nasal turbinates and lungs in 3 of 4 animals. Consistent with the viral load reduction, 1212C2 treated animals exhibited significantly less lung pathology compared to the PBS-treated group at 2 dpi (p = 0.0334) and 4 dpi (p = 0.0004), with this reduction also reaching significance compared to the isotype control group at 4 dpi (p < 0.0001) (Figure 5C). There was ∼80% reduction in lung pathology at 4 dpi when 1212C2 was given prophylactically. 1206D1 hmAb exhibited modest activity, noted by 50% of hamsters having no detectable virus in the nasal turbinates and all 1206D1 hamsters having detectable virus in the lungs, although trending to lower titers compared to PBS and isotype control groups. These in vivo results are consistent with the lower in vitro viral neutralization activity of 1206D1 in comparison to 1212C2 (Figure 2A). Overall, 1212C2 demonstrated substantial prophylactic activity as seen in sterilizing protection in 63% of the hamsters.
Figure 5.
Prophylactic and therapeutic activity of 1212C2 hmAb in SARS-CoV-2-infected hamsters
(A and B) Golden Syrian hamsters were (A) prophylactically treated i.p. with 10 mg/kg indicated hmAb or PBS 6 h before intranasal (i.n.) challenge with 2 × 105 PFU SARS-CoV-2 or (B) therapeutically treated i.p. with 25 mg/kg indicated hmAb or PBS 6 h following i.n. challenge with SARS-CoV-2. Virus present in nasal turbinates and lungs was determined by plaque assay. Dotted line indicates limit of detection.
(C and D) Distribution of pathologic lesion, including consolidation, congestion, and pneumonic lesions, was measured using ImageJ and represented as the percentage of the total lung surface area in prophylactically treated (C) and therapeutically treated (D) animals. Each symbol represents an individual animal. p values determined by 2-tailed t test.
Therapeutic activity was tested by treatment with 25 mg/kg of 1212C2 6 h following i.n. infection of hamsters with SARS-CoV-2 and evaluating viral burden at 4 dpi only due to the limited availability of hmAb. SARS-CoV-2 was detected in the nasal turbinates in 3 of 4 animals in the PBS and isotype control groups. No virus was detected in the nasal turbinates of the 1212C2 treated hamsters. SARS-CoV-2 was detected in the lungs of all PBS and isotype control treated hamsters, but only in 1 of 4 of the 1212C2 treated hamsters (Figure 5B). Overall, 1212C2 demonstrated substantial therapeutic activity, reducing virus to undetectable levels in 75% of the treated hamsters. SARS-CoV-2 infection in hamsters results in gross lung lesions, including consolidation, congestion, and atelectasis.25 Therapeutically, significantly less lung pathology was also observed in the 1212C2 treated hamster group compared to the PBS-treated group (p = 0.0136), and ∼58% reduction in lung pathology in the 1212C2 group compared to the control hamsters (Figure 5D). Importantly, these differences observed in lung lesions are consistent with the viral burden seen in the upper and lower respiratory tract (Figures 5A and 5B). Overall, these results demonstrate the ability of 1212C2 to substantially reduce the viral burden and lung pathology of SARS-CoV-2 infection when used either prophylactically or therapeutically.
Delivery of inhaled hmAb in hamsters
Therapeutic mAbs are typically delivered systemically through intravenous infusion or intramuscular injection. Such delivery typically results in <0.1% of the injected dose in the lung epithelial lining fluid at Cmax, thus highly inefficient in achieving optimal concentrations in the respiratory tract for the protection or treatment of respiratory infections, such as SARS-CoV-2.26
To evaluate the efficiency of lung-delivered dosing and clearance of an IgG1 hmAb when delivered as an i.p. bolus dose as compared to an inhaled nebulized aerosol, hamsters were administered the non-specific IgG used in the challenge studies and the serum and bronchoalveolar lavage (BAL) concentrations were analyzed at 30 min and 42 h post-dosing (Table 2). Intraperitoneal administration of ∼25 mg/kg hmAb resulted in high serum concentrations but little to no detectable hmAb in the BAL at 30 min post-treatment (Table 2). However, 42 h later, the hmAb was detectable in the BAL, suggesting that i.p.-injected hmAb gradually penetrates the lungs from the serum compartment. Inhalation (IH) administration of the hmAb using liquid aerosols delivered whole body to the animals from a commercially available nebulizer (Aerogen Aeroneb Solo nebulizer) showed that ∼1.7% of the inhaled dose was deposited in the BAL. The low percentage of the inhaled dose that is deposited in the lungs of hamsters is in line with prior lung uptake studies for the ∼4-μm diameter droplets produced by the nebulizer used here (volume mean diameter measured by laser diffraction 4.1 μm), and is a result of a higher degree of inertial impaction of liquid aerosols in the upper airways of small animals.27 Nevertheless, these lung-deposited (BAL) doses are still substantially higher at both time points than the BAL concentration achieved with the i.p. route. A higher lung-deposited dose afforded by the inhaled route demonstrates higher delivery efficiency to the lungs than the i.p. route. At 42 h post-inhaled dose, most of the mAb is cleared from the BAL (∼85% <30 min post-IH), which appears to be faster clearance rate than an expected lung half-life of ∼8 days reported in other studies.28
Table 2.
Estimated inhaled 1212C2 dose
| Nebulizer Consumption Rate | Aerosol Conc. | Aerosol mAb Conc. | Dose | Actual Dose (ave.) | |
|---|---|---|---|---|---|
| Description | mL/min | mL/L | mg/L | mg/kg | mg/kg |
| Nonspecific IgG Inhalation 25 mg/kg (cage 1) | 0.30 | 0.033 | 0.51 | 9.13 | 11.30 |
| Nonspecific IgG Inhalation 25 mg/kg (cage 2) | 0.25 | 0.049 | 0.75 | 13.48 | |
| No Virus, Saline Inhalation (cage 1) | 0.19 | Not measured | 0.00 | 0.00 | 0.00 |
| No Virus, Saline Inhalation (cage 2) | 0.17 | Not measured | 0.00 | 0.00 | |
| 1212C2 HLE-LALA Inhalation 25 mg/kg (cage 1) | 0.38 | 0.052 | 0.81 | 14.52 | 16.33 |
| 1212C2 HLE-LALA Inhalation 25 mg/kg (cage 2) | 0.38 | 0.065 | 1.01 | 18.15 |
Conc., concentration; ave., average.
Therapeutic activity of inhaled 1212C2 hmAb in hamsters
The crystallizable fragment (Fc) of 1212C2 was modified with the LALA mutation to reduce Fc receptor (FcR) binding and subsequent Fc-mediated effector functions,29 and further modified to increase half-life30 (referred to as 1212C2-HLE-LALA). To determine the efficacy of inhaled 1212C2 and evaluate the therapeutic dose-response, hamsters were infected i.n. with 2 × 105 plaque-forming units (PFU) SARS-CoV-2 and treated with a single dose of 1212C2 hmAb or isotype control hmAb 12 h later using IH or i.p. routes. As measures of efficacy, body weight was recorded, pulmonary lung lesions were measured, and viral titers quantitated from nasal turbinates and lungs on days 2 and 4 following infection.
At 2 dpi, the virus was detected in the nasal turbinates and lungs of all infected control hamsters (PBS and isotype control hmAb) (Figure 6A). Virus was not detectable in the lungs of any hamsters treated via IH with 1212C2-HLE-LALA at 16.3 mg/kg, and virus was lower in their nasal turbinates compared to controls. In contrast, virus was detectable at 2 dpi in the lungs of 50% of the hamsters treated i.p. with 1212C2-HLE-LALA at 25 mg/kg, and not significantly decreased in nasal turbinates compared to controls. At 2 dpi, virus was only detectable in the lungs of 25% and 50% of the hamsters treated via IH with lung-delivered doses of 3.2 mg/kg and 0.6 mg/kg 1212C2-HLE-LALA, respectively. At 2 dpi, 16.3 and 3.2 mg/kg inhaled 1212C2-HLE-LALA was superior in reducing viral titer in the lungs compared to i.p. administration.
Figure 6.
Therapeutic activity of inhaled 1212C2 hmAb in SARS-CoV-2-infected hamsters
(A and B) Golden Syrian hamsters were infected i.n. with 2 × 105 PFU SARS-CoV-2 and 12 h later treated with indicated hmAb by i.p. or IH administration. Virus present in nasal turbinates and lungs was determined by plaque assay at 2 dpi (A) and 4 dpi (B). Dotted line indicates limit of detection.
(C) Distribution of pathologic lesion were measured using ImageJ and represented as the percentage of the total lung surface area. ∗p < 0.0055 and ∗∗p < 0.00055 compared to PBS i.p.-treated group determined by 2-tailed t test.
At 4 dpi, virus was detected in the nasal turbinates and lungs of all infected control hamsters (saline and isotype hmAb), with a trend toward modest non-specific reduction in viral titer in nasal turbinates only with isotype control hmAb (Figure 6B). Viral titers were only sporadically detected in the nasal turbinates of the 1212C2 treated groups, with the exception of detectable virus in all of the 0.6 mg/kg inhaled 1212C2-HLE-LALA group. At 4 dpi, there was no detectable viral titer in the lungs of any 1212C2 treated hamsters, including the 0.6 mg/kg IH 1212C2-HLE-LALA group, representing at least a 2-log reduction compared to the control groups. Control PBS-treated hamsters exhibited an ∼10% maximum weight loss; the 1212C2 treated animals did not have appreciable weight loss (Figure S5). Lung pathology was evident at 4 dpi, particularly in the control groups; however, overall, relative to non-infected hamsters, no lung pathology was evident in 9/24 (37.5%) of the 1212C2 treated hamsters, compared with lung pathology evident in 11/12 (91.7%) of the control treated infected hamsters (Figure 6C). At 4 dpi, lung lesions were significantly decreased in hamsters that were treated with 3.2 mg/kg (p = 0.0028) of inhaled 1212C2 hmAb compared to PBS-treated hamsters.
With regard to effector function removal brought about by the addition of the LALA mutation, animals treated with 1212C2-HLE-LALA appeared to achieve comparable viral reduction as the unmodified 1212C2 mAb. Overall, these results confirm the therapeutic activity of 1212C2 hmAb against SARS-CoV-2 infection and suggest the increased efficacy of inhaled 1212C2 hmAb, with the elimination of viral lung burden with a single inhaled dose of only 0.6 mg/kg. As shown in Table S2, the lung-deposited dose in hamsters is ∼1.7% of the inhaled dose. This implies that the elimination of lung viral burden was achieved at day 4 post-infection with a lung dose of 0.01 mg/kg. Furthermore, if the relevant translation between species to achieve an efficacious dose is a normalization based on a lung-deposited dose per kilogram of lung weight, then, combined with the expectation that at least 50% of the inhaled dose from a similar commercial nebulizer would be deposited in the lung of a human, the human equivalent efficacious inhaled dose would be 0.03 mg/kg body weight (assumptions: 110 g hamster with 1 g lung weight; 70 kg human with 1,000-g lung weight).
Discussion
The SARS-CoV-2 global pandemic continues without an optimal targeted intervention to treat the infection. Our results clearly demonstrate that SARS-CoV-2 infection can result in memory B cells encoding high-affinity, high-potency NAbs specific for the RBD. The 1212C2 hmAb is able to significantly reduce the viral burden in SARS-CoV-2-infected hamsters when used either prophylactically or therapeutically. The in vitro affinity and the neutralization activity of 1212C2 compared favorably to hmAbs that are in late-stage clinical trials to treat SARS-CoV-2 (i.e., REGN10987, REGN10933, and CB6/JS016), suggesting that 1212C2 could have clinical activity. Of note is that 1212C2 bound consistently across RBD domain, S1 subunit, and full-length spike protein (S1S2), which is a property that is additionally different from that of the REGN10987, REGN10933, and CB6/JS016 mAbs (Figure 1B).
The resulting panel of SARS-CoV-2 neutralizing hmAbs indicate that RBD-specific IgG, IgA, and IgM memory B cells develop after infection; however, they exhibit only modest somatic hypermutation, which is consistent with an acute primary immune response. Several hmAbs, including 1212C2, 1212F5, 1213H7, and 1212F2, despite only modest somatic hypermutation (4.1%–8.2% VH), have remarkably high affinity for SARS-CoV-2 RBD (104–1,180 pM KD) and potent neutralization (<200 ng/mL NT50). The frequent usage of VH3-66 and Vk1-9 among the hmAbs may represent preferential RBD specificity among those germline genes. 1212C2 and 1212F5, which are members of the same clonal lineage, were isolated from IgM memory B cells, and use the VH1–2 heavy-chain gene. VH1–2 usage is a common feature of other antibodies targeting viral glycoproteins, including HIV envelope.31, 32, 33 VH1–2 usage is more pronounced among marginal zone B cells than naive B cells or switched memory B cells and increased among splenic marginal zone B cell lymphoma.34, 35, 36, 37 This may indicate that the 1212C2 lineage arose from a marginal zone B cell response.
1212C2 has higher affinity (KD = 104 pM) for SARS-CoV-2 RBD compared to 1212F5 (KD = 841 pM), which is consistent with its further somatic hypermutation. Additional lineage members were identified with greater somatic hypermutation, and may be used to guide rational improvement of 1212C2 affinity, neutralization, and ultimate efficacy against SARS-CoV-2 infection.
The resulting hmAbs neutralized SARS-CoV-2 at NT50 of ≤1 μg/mL, indicating their utility for investigating the mechanisms and epitopes mediating SARS-CoV-2 RBD-targeted antiviral activity. Although there was overall consistency that the hmAbs neutralized SARS-CoV-2 either when pre-incubated with the virus (pre-treatment) or after the cells were infected with the virus for 1 h (post-treatment), particularly 1212C2, 1212F5, and 1213H7 potently neutralized (NT50 <100 ng/mL) SARS-CoV-2 in both conditions, discordance was noted. 1206D1, which was tested in vivo, was ∼2-fold less effective neutralizing in post-treatment than in pre-treatment, which may contribute to its limited in vivo activity. In contrast, 1215D1, which had the highest affinity for RBD (KD = 71 pM), is ∼5-fold more effective in neutralizing in post-treatment than in pre-treatment. It is expected that the post-treatment neutralization assay may identify antibodies that are better able to mitigate cell-to-cell virus spreading, including antibodies targeting the fusiogenic activity of the S2 domain. Subsequently, 1215D1 should be evaluated for its in vivo efficacy and discern whether the post-treatment neutralization assay is a more sensitive indicator of in vivo efficacy. Epitope mapping suggests that 1212C2 has a large footprint on the RBD, blocking the binding of several other neutralizing hmAbs encoded by diverse heavy- and light-chain variable region genes. Efforts are ongoing to solve the 1212C2-RBD structure to adequately define the epitope.
The high-affinity binding of 1212C2 to SARS-CoV-2 RBD, mediating its ability to block RBD attachment to ACE2, and subsequent potent neutralization of SARS-CoV-2, even when added after virus has been added to culture, demonstrate its direct and substantial antiviral activity. These properties likely contribute to its prophylactic and therapeutic abilities to protect hamsters from SARS-CoV-2 challenge, reducing viral burden and the development of lung pathology. 1212C2 was similarly effective in treating SARS-CoV-2 infection in hamsters, reducing viral burden and lung pathology when administered via the parenteral route at clinically relevant doses. The emergence of SARS-CoV-2 variants, particularly those with mutations in RBD with clinical significance, necessitates that SARS-CoV-2-neutralizing hmAbs such as 1212C2 be thoroughly evaluated for their tolerance to RBD variations and breadth of activity against variants.
As the portal of entry for SARS-CoV-2 is the respiratory tract, with the lungs serving as the key target organ for pathogenesis, delivering mAbs directly to the lungs using IH is a logical approach. While aerosol delivery of drugs to the lungs is more inefficient in small animals as compared to humans (∼1% of the inhaled dose is deposited in the lungs in rodents as compared to ∼50% in humans), our data showed that inhaled delivery of mAbs to the lung BAL in hamsters is still substantially more efficient than that achieved using the i.p. route (Table S2). When comparing the inhaled route to the i.p. route in the hamster challenge study, therapeutic efficacy was achieved at a higher efficiency. Inhaled administration of 1212C2 hmAb resulted in sterilizing therapeutic protection at all tested doses, with the lowest inhaled dose at 0.6 mg/kg, which corresponds to 0.01 mg/kg of the lung-deposited dose (or a human equivalent inhaled dose of 0.03 mg/kg). In contrast, mAbs against SARS-CoV-2 that have been reported to date required at least ≥5 mg/kg to achieve therapeutic efficacy when administered parenterally.17,38, 39, 40 Therefore, inhaled 1212C2 has the potential to profoundly facilitate dose sparing and treatment coverage compared to conventional parenteral administration. There are several commercial inhaled protein therapeutic products and many more inhaled protein therapeutics that are in various stages of clinical evaluation, most with an attractive safety profile. Several mAbs have also been clinically evaluated as inhaled aerosols with demonstrated preliminary safety and tolerability,41, 42, 43 suggesting the practicality of formulating 1212C2 for self-administered IH. Over 90% of all symptomatic COVID-19 patients are not hospitalized, but they all still need treatment to minimize the potential for transmission and limit complications from viral infection. This is a large unmet COVID-19 population, and having a convenient self-administered dose that can be done on an outpatient basis or at home using commercially available nebulizer devices could materially affect treatment coverage, reducing transmissibility and, ultimately, the viral burden of the population.
Limitations of study
Epitope-mapping studies were performed by injecting multiple (n = 6) hmAbs sequentially over the same immobilized RBD surface. While this mapping format can obscure the details of specific hmAb epitopes, our goal was to rapidly identify distinct epitopes that could simultaneously bind to RBD. In aggregate, the mapping studies successfully identify four major Ab classes, with the 1212C2 overlapping all of the hmAbs, except 1215B11. More precise epitope resolution approaches such as X-ray crystallography are needed to accurately determine critical binding residues and epitope relation to other RBD-specific neutralizing mAbs reported. The recent emergence of SARS-CoV-2 variants precluded testing of the described hmAbs against these viruses, and such testing would inform the clinical utility of the hmAbs. Only a single animal model, the hamster, was used for this study, and testing in another animal model such as the lethal K18 hACE2 mouse model of SARS-CoV-2 infection would enable the determination of 1212C2 activity against more severe disease. These experiments are planned to further substantiate the results reported here.
STAR★Methods
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-ACE2 antibody | R&D System | Cat#AF933-SP |
| anti- CD19-APC-Cy7 (SJ25C1) | BD Biosciences | Cat# 557791; RRID: AB_396873 |
| HIV gp140 (F8)-AlexaFluor647 | N. Haigwood (OHSU)/Conjugated in Lab (Molecular Probes) | Cat# A20186 |
| CD3-BV510 (OKT3) | Biolegend | Cat# 317331; RRID: AB_2561376 |
| CD4-BV510 (HI30) | Biolegend | Cat# 304031 |
| IgD-FITC (IA6-2) | BD Biosciences | Cat# 555778; RRID: AB_396113 |
| CD27-PE (CLB-27/1) | Life Technologies | Cat# MHCD2704; RRID: AB_10392393 |
| Goat Anti-Human IgG Antibody (Peroxidase) | Jackson Immunoresearch | Cat#109-035-003; RRID:AB_2337577 |
| AffiniPure F(ab’)2 Fragment Goat Anti-Human IgG, F(ab’)2 fragment specific | Jackson Immunoresearch | Cat#109-006-097; RRID: AB_2337550 |
| human myeloma IgG1 | BioXcell | Cat#BE0297; RRID: AB_2687817 |
| REGN 10987 | IDT synthesized gBlock of variable region - M.Piepenbrink ligated into AbVec expression vectors | PMID: 32540901 |
| REGN 10933 | IDT synthesized gBlock of variable region - M.Piepenbrink ligated into AbVec expression vectors | PMID: 32540901 |
| CB6/JS016 | IDT synthesized gBlock of variable region - M.Piepenbrink ligated into AbVec expression vectors | MT470197.1/MT470196.1 |
| CR3022 | IDT synthesized gBlock of variable region - M.Piepenbrink ligated into AbVec expression vectors | DQ168569.1/DQ168570.1 |
| FITC-Anti-human IgG | Dako | Cat#F020202-2 |
| Fluorescein (FITC) AffiniPure Rabbit Anti-Human IgG (H+L) | Jackson Immunoresearch | Cat#309-095-003; RRID: AB_2339685 |
| Rabbit Anti-Mouse Immunoglobulins/FITC | Dako | Cat#F0261 |
| anti-NP monoclonal 1C7C7 antibody | Millipore Sigma | Cat#ZMS1075 |
| Bacterial and virus strains | ||
| DH5a | Thermo Fisher Scientific | Cat#18265017 |
| SARS-CoV-2 (isolate USA-WA1/2020) | BEI Resources | Cat#NR-52281 |
| Biological samples | ||
| Human Peripheral blood mononuclear cells | University of Alabama at Birmingham | N/A |
| Hamster lung | Texas Biomedical Research Institute | N/A |
| Hamster nasal turbinate | Texas Biomedical Research Institute | N/A |
| Hamster serum | Texas Biomedical Research Institute | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 Spike RBD (RBDhisavi) | M. Walter | uniprot P0DT2 |
| RBD-Fc | M. Walter | uniprot P0DT2 |
| SARS-CoV-2 S1+S2 | Sino Biological | Cat#40589-V08B1-B |
| SARS-CoV-2 D614G S1 | Sino Biological | Cat#40591-V08H3 |
| SARS-CoV-1 S | BEI Resources | Cat#NR-722 |
| SARS-CoV-2 Nucleocapsid | Sino Biological | Cat#40588-V08B |
| HepG2 whole cell lysate | Abcam | Cat#ab166833 |
| Vector pCAGGS Containing the SARS-Related Coronavirus 2, Wuhan-Hu-1 Spike Glycoprotein Receptor Binding Domain (RBD) | BEI Resources | Cat#NR-52309 |
| Urea | Invitrogen | Cat#ZU10001 |
| Phospate Buffered Saline (PBS) | Corning | Cat#21-040-CV |
| Fetal Bovine Serum (FBS) | Atlanta Biologicals | Cat#S12450H |
| Tween 20 | Biorad | Cat#1706531 |
| 2-Mercaptoethanol | Millipore Sigma | Cat# M3148 |
| DTT | Invitrogen | Cat#P2325 |
| RiboLock RNase Inhibitor | ThermoFisher | Cat#EO0381 |
| qScript cDNA synthesis kit | QuantaBio | Cat#95047-100 |
| DMEM | GIBCO | Cat#10313039 |
| Penicillin-Streptomycin L-glutamine 100X | Corning | Cat#30-009-CI |
| HyClone FetalClone II | Cytiva Lifesciences | Cat#SH30066.03 |
| Antibiotic-Antimycotic solution | Corning | Cat#30-004-CI |
| jetPRIME transfection reagent | PolyPlus | Cat#114-75 |
| Magna Protein A beads | Promega | Cat#G8782 |
| Bovine Serum Albumin (BSA) | Fisher Scientific | Cat#BP1600100 |
| Bovine Serum Albumin (BSA) | Millipore Sigma | Cat#A9647 |
| Trypsin-EDTA | Corning | Cat#25-051-CI |
| BirA biotin-protein ligase, bulk ligase kit | Avidity LLC | Cat#NA |
| Triton X-100 | Millipore Sigma | Cat#X100-500ML |
| HBS-P+ Buffer 10 × | Cytiva Lifesciences | Cat#BR100671 |
| Human Antibody Capture Kit | Cytiva Lifesciences | Cat#BR100839 |
| Mouse Antibody Capture Kit | Cytiva Lifesciences | Cat#BR100838 |
| 10 mM glycine pH 1.7 | Cytiva Lifesciences | Cat#BR100354 |
| autoMACs rinsing solution | Miltenyi Biotec | Cat#130-091-222 |
| MACS BSA Stock Solution | Miltenyi Biotec | Cat#130-091-376 |
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| Turbo DNA-free Kit | Invitrogen | Cat#AM1907 |
| Platinum Taq High Fidelity Polymerase | Invitrogen | Cat#11304011 |
| EconoTaq PLUS GREEN 2X Master Mix | Lucigen | Cat# 30033-2 |
| iProof High-Fidelity DNA Polymerase | Biorad | Cat# 1725302 |
| HotStarTaq Plus DNA Polymerase | QIAGEN | Cat# 203607 |
| E.Z.N.A. Gel Extraction Kit | Omega Bio-tek | Cat#D2501-02 |
| E.Z.N.A. Cycle Pure Kit | Omega Bio-tek | Cat#D6493-02 |
| E.Z.N.A. Plasmid Mini Kit I | Omega Bio-tek | Cat# D6942-02 |
| Amicon® Ultra-15 Centrifugal Filter Unit | Millipore Sigma | Cat# UFC910024 |
| ZymoPURE Plasmid Midi Prep Kit | Zymo Research Corporation | Cat#D4201 |
| 0.5M EDTA | Thermo Scientific | Cat#J15694-AE |
| 7-Aminoactinomycin D (7-AAD) | Molecular Probes | Cat#A1310 |
| Annexin V-PerCP-Cy5.5 | Biolegend | Cat# 640936 |
| Live/Dead aqua | Molecular Probes | Cat# L34957 |
| Streptavidin-Pacific Blue Conjugate | Invitrogen | Cat#s11222 |
| Opti-MEM | GIBCO | Cat#31985070 |
| Passive Lysis Buffer | Promega | Cat#E1941 |
| Luciferase substrate | Promega | Cat#E151A |
| Avicel PH-101 | Millipore Sigma | Cat#11365 |
| Formalin, neutral buffered, 10% | Millipore Sigma | Cat#HT501128 |
| Isoflurane | Covetrus | Cat#008957 |
| Critical commercial assays | ||
| Illumina MiSeq Reagent Kit v3 | Illumina Inc. | Cat#MS-102-3003 |
| VECTASTAIN® ABC-HRP Kit, Peroxidase, POD (Mouse IgG) | Vector Laboratory | Cat#P-4002 |
| DAB Substrate Kit, POD (HRP), with Nickel | Vector Laboratory | Cat#SK-4100 |
| Experimental models: cell lines | ||
| HEK293T | ATCC | Cat#CRL-3216; RRID:CVCL_0063 |
| HEK293T-hACE2 Cell Line | BEI Resources | Cat#NR-52511 |
| Vero E6 | ATCC | Cat#CRL-1586 |
| VeroE6/TMPRSS2 | JCRB Cell Bank | RRID:CVCL_YQ49 |
| Experimental models: organisms/strains | ||
| Golden Syrian hamsters (Mesocricetus auratus) | Charles River | https://www.criver.com/products-services/find-model/lvg-golden-syrian-hamster?region=3611 |
| Oligonucleotides | ||
| Primers for mAb generation and MiSeq | 31 | PMID:32577626 |
| Software and algorithms | ||
| CTL immunospot | Cellular Technology Limited | http://www.immunospot.com/ImmunoSpot-analyzers-software |
| ImageJ | NIH | https://imagej.nih.gov/ij/download.html |
| IMGT/V-QUEST | International | http://www.imgt.org/IMGT_vquest/vquest |
| Immunogenetics | ||
| Information System | ||
| GraphPad Prism v7.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Flowjo | BD | https://www.flowjo.com/ |
| HighV-QUEST | International | https://www.imgt.org/HighV-QUEST/home.action |
| Immunogenetics | ||
| Information System | ||
| Phylip’s protpars tool (version 3.695) | University of Washington | https://evolution.genetics.washington.edu/phylip.html |
| Cytoscape | Cytoscape Consortium | https://cytoscape.org/ |
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html |
| Other | ||
| Aerogen Aeroneb Solo nebulizer | Grayline Medical | Cat# 06-AG-AS3200 |
| Precellys tissue homogenizer | Bertin Instruments | Cat#P000062-PEVO0-A |
| Tissue Grinding CKMix50 - 7mL tubes 50 Prep | Thermo Fisher Scientific | Cat#P000939LYSK0A.0 |
| Rodent anesthesia machine | Veterinary Anesthesia Systems | http://vasinc.net/index.htm |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, James Kobie (jjkobie@uabmc.edu).
Materials availability
Limited quantities of newly generated materials associated with the paper are available under MTA.
Data and code availability
All relevant data are included within the manuscript and are available without restriction from the Lead Contact upon request.
Experimental model and subject details
Human subjects
Peripheral blood was collected at the University of Alabama at Birmingham from adult convalescent patients approximately 1 month following PCR confirmed infection with SARS-CoV-2. The patients were symptomatic for at least 7 days and were not hospitalized during SARS-CoV-2 infection. Patients were white, female and male, and > 45 years old. The subjects provided signed written informed consent. All procedures and methods were approved by the Institutional Review Board for Human Use at the University of Alabama at Birmingham, and all experiments were performed in accordance with relevant guidelines and regulations.
Golden Syrian hamster model
We have previously demonstrated that golden Syrian hamsters (Mesocricetus auratus) are susceptible to SARS-CoV2, showing loss of body weight and viral replication in the lungs and nasal turbinate.25 5 to 6-week old, female hamsters were purchased from Charles River and maintained in the animal facility at Texas Biomedical Research Institute (Texas Biomed) under specific pathogen-free conditions. with infectious SARS-CoV-2 were conducted under appropriate animal BSL3 (ABSL3) laboratories, respectively, at Texas Biomed. Experiments were approved by the Texas Biomed Institutional Biosafety (IBC) and Animal Care and Use (IACUC) committees.
Method details
Sample collection and B cell isolation
Peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation and cryopreserved. SARS-CoV-2 recombinant protein was generated, briefly SARS-CoV-2 Spike RBD domain consisted of residues Thr-333 to Thr-531 (uniprot P0DT2). The RBD sequence was cloned in frame with a his8 and avi-tag sequence, respectively (RBDhisavi). RBDhisavi was expressed in insect cells and purified by nickel affinity chromatography. Purified RBDhisavi was biotinylated in vitro using BIRA (https://www.avidity.com/) for subsequent formation of RBDhisavi-tetramers for B cell staining experiments. The same RBD sequence was cloned in frame with a murine IgG1 FC domain (RBD-FC). RBD-FC was expressed in insect cells and purified by protein A affinity chromatography. Cryopreserved cells were thawed and blocked with 0.5 μg anti-ACE2 antibody (AF933-SP, R&D System) for 5 min at room temperature and then stained for flow cytometry similar as previously described,44 using anti- CD19-APC-Cy7 (SJ25C1, BD Biosciences), HIV gp140-AlexaFluor647, SARS-CoV-2 RBD-BV421, CD3-BV510 (OKT3, Biolegend), CD4-BV510 (HI30, Biolegend), IgD-FITC (IA6-2, BD Biosciences), CD27-PE (CLB-27/1,Life Technologies), Annexin V-PerCP-Cy5.5 (Biolegend), and Live/Dead aqua (Molecular Probes).
Monoclonal antibody production
Single B cells were sorted using a FACSMelody (BD Biosciences) into 96-well PCR plates and immediately frozen at −80°C until thawed for reverse transcription and nested PCR performed for IgH, Igλ, and Igκ variable gene transcripts as previously described.31,44 Paired heavy and light chain genes cloned into IgG1 expression vectors and were transfected into HEK293T cells and culture supernatant was concentrated using 100,000 MWCO Amicon Ultra centrifugal filters (Millipore-Sigma, Cork, Ireland), and IgG captured and eluted from Magne Protein A beads (Promega, Madison, WI) as previously described.31,44 The hmAb 1069D6 or a human myeloma IgG1 (BioXcell, Labanon, NH) were used as an isotype control. 1212C2 was also generated with the incorporation of the LALA mutation to diminish Fc receptor binding45 and a mutation described to increase half-life.30 Immunoglobulin sequences were analyzed by IgBlast (www.ncbi.nlm.nih.gov/igblast) and IMGT/V-QUEST (http://www.imgt.org/IMGT_vquest/vquest) to determine which sequences should lead to productive immunoglobulin, to identify the germline V(D)J gene segments with the highest identity, and to scrutinize sequence properties. REGN 10987, REGN 10933, CB6/JS016, and CR3022 were previously described17,20,46 and heavy and light chain variable regions synthesized by IDT based on reported sequences (GenBank: MT470197.1, GenBank: MT470196.1, GenBank: DQ168569.1, GenBank: DQ168570.120) and cloned into IgG1 expression vector for production in HEK293T cell.
Binding characterization
ELISA plates (Nunc MaxiSorp; Thermo Fisher Scientific, Grand Island, NY) were coated with recombinant SARS-CoV-2 proteins at 1 μg/ml. Recombinant proteins used include SARS-CoV-2 S1+S2 (Sino Biological, Wayne, PA), SARS-CoV-2 D614G S1 (Sino Biological), SARS-CoV-1 S (BEI Resources), SARS-CoV-2 Nucleocapsid (Sino Biological), and HepG2 whole cell lysate (Abcam, Cambridge, MA). Purified hmAbs were diluted in PBS, and binding was detected with HRP-conjugated anti-human IgG (Jackson ImmunoResearch, West Grove, PA). In selected ELISAs, 8M urea were added to the ELISA plate and the plates incubated for 15 min at room temperature prior to washing with PBS plus 0.05% Tween20 and detection with anti-IgG-HRP to evaluate avidity. Immunofluorescence assay was used to determine hmAb binding to SARS-CoV-2 infected cells. Briefly, confluent monolayers of Vero E6 cells were mock infected or infected with the indicated SARS-CoV-2. At 24 hours post infection (hpi), cells were fixed with 4% paraformaldehyde (PFA) for 30 minutes and permeabilized with 0.5% Triton X-100–PBS for 15 min at room temperature, and blocked with 2.5% Bovine Serum Albumin at 37°C for 1 h. Cells were then incubated for 1 h at 37°C with 1 μg/ml of indicated hmAb. Then, cells were incubated with fluorescein isothiocyanate (FITC)-conjugated secondary anti-human Ab (Dako) for 1 h at 37°C. Images were captured using a fluorescence microscope and camera with a 10X objective.
Surface plasmon resonance (SPR)
SPR experiments were performed on a Biacore T200 (Cytiva) at 25°C using a running buffer consisting of 10mM HEPES, 150mM NaCl, 0.0075% P20, and 100 μg/mL BSA. For affinity measurements, antibodies were captured onto CM-5 sensor chips using the human IgG capture kit (Cytiva). All SPR affinity experiments were double referenced (e.g., sensorgram data was subtracted from a control surface and from a buffer blank injection). The control surface for all experiments consisted of the capture antibody. Approximately 100RU of each hmAb was captured onto the chip surface. RBD was injected over the NmAbs at four concentrations (80nM, 20nM, 5nM, and 1.25nM) for 90 s, followed by 300 s dissociation phase. After each injection, the surface was regenerated with a 30 s pulse of 3M MgCl2, followed by capture of fresh hmAb. The buffer flowrate for affinity analysis was 50 μL/min. Sensorgrams were globally fit to a 1:1 model, without a bulk index correction, using Biacore T-200 evaluation software version 1.0. Standard errors for the rate constants were obtained from the fitted sensorgram data. Replicate KD errors were estimated by repeating the experiments for four of NmAb-RBD interactions (1212C2, 1206D1, 1215D1 and CB6/JS016). Standard deviations from these experiments were used to define an average KD error of 11%.
Epitope mapping was performed by capturing RBD-FC to CM-5 sensor chips using an anti-murine FC capture kit (Cytiva). The running buffer was the same as used for RBD / hmAb affinity measurements. hmAbs were sequentially injected, up to 6 NmAbs in one series, over the RBD-FC surface. The first hmAb was injected at 100 nM concentration, and subsequent hmAbs were injected at 50nM concentrations. Each hmAb was injected for 90 s over the RBD-FC, followed by a 60 s dissociation phase. The flowrate for the epitope mapping studies was 30 μL/min. After the final injection, the surface was regenerated by a 3-minute injection of 10 mM glycine pH 1.7. hmAb epitopes were defined by measuring the reduction in hmAb binding (RU) to the RBD-FC surface in the presence of different hmAbs, relative to hmAb-RBD-FC alone.
ACE2 – RBD inhibition assay
All dilutions and cell suspensions were made in MACs buffer [autoMACs rinsing solution (Miltenyi Biotec, Auburn, CA) with 0.5% BSA]. For 20 μg of hmAb, 0.2 μg of biotinylated SARS-CoV-2 RBD was mixed in a total volume of 1 mL and incubated for 30 minutes on ice. HEK293T cells co-expressing human ACE2 and green fluorescent protein (BEI Resources) were grown in DMEM containing 4 mM L-glutamine, 4500 mg per L-glucose, 1 mM sodium pyruvate and 1500 mg per mL sodium bicarbonate, supplemented with 10% fetal bovine serum. On the day of experiment, cells were dissociated with 0.5 mM EDTA and a cell scraper. Cells were washed with MACs buffer and pelleted at 350 x g for 5 minutes. Cells were counted, distributed at 5 × 105 per tube, and pelleted. Cells pellets were suspended in the mAb – RBD mixture and allowed to interact for 60 m on ice. Unbound RBD was washed away by adding 1 mL MACs buffer and centrifuging for 5 minutes at 350 x g. Supernatant was carefully removed, and cells suspended in 100 μl MACs buffer. 7AAD and Streptavidin-Pacific Blue (Invitrogen) was added and the mixture incubated on ice for 30 minutes. To wash cells, 1 mL of MACs buffer was added and tubes centrifuged at 350 x g for 5 minutes. Supernatant was removed and the wash step was repeated with 2 mL of ice cold PBS. After removing the PBS wash, the cell pellet was suspended in 0.5 mL PBS and the cells passed through a cells strainer on a 5 mL tube (Falcon). Cells were analyzed on a FACSymphony (BD Biosciences) and gated on 7AAD negative GFP positive cells.
SARS-CoV-2 neutralizing activity
hmAbs were tested for neutralization of SARS-CoV2 (isolate USA-WA1/2020, BEI Resources) as previously described.24 For pre-treatment, 100-200 plaque forming units (PFU)/well of SARS-CoV-2 containing 2-folds dilutions (starting concentration 25 μg/ml) for hmAbs (or 1:100 dilution for human serum control) were mixed in 100 μL of media and incubated at 37°C for 1h. Vero E6 cells (96-well plate format, 4 × 104 cells/well, quadruplicate) were infected with virus-hmAb or virus-serum (control well) mixture for virus adsorption for 1h, followed by changing media with post-infection media containing 2% FBS, 1% Penicillin-Streptomycin L-glutamine (PSG), and 1% Avicel. For post-treatment, Vero E6 cells (96-well plate format, 4 × 104 cells/well, quadruplicate) were infected with 100-200 PFU/well of SARS-CoV-2. After 1 h of viral adsorption, the infection media was changed with the 100 μl of media containing 1% Avicel and 2-fold dilutions, starting at 25 μg/ml of hmAb (or 1:100 dilution for human serum control). At 24 hpi, infected cells were fixed with 10% neutral formalin for 24 h and were immune-stained using anti-NP monoclonal 1C7C7 antibody.24 Virus neutralization was evaluated and quantified using ELISPOT, and the percentage of infectivity calculated using sigmoidal dose response curves. The formula to calculate percent viral infection for each concentration is given as [(Average # of plaques from each treated wells – average # of plaques from “no virus” wells)/(average # of plaques from “virus only” wells - average # of plaques from “no virus” wells)] x 100. A non-linear regression curve fit analysis over the dilution curve can be performed using GraphPad Prism to calculate NT50. Mock-infected cells and viruses in the absence of hmAb were used as internal controls. hmAbs were also tested using a SARS-CoV-2 Spike protein pseudotyped virus (PsV) containing the gene for firefly luciferase. Virus neutralization can be measured by the reduction of luciferase expression. VeroE6/TMPRSS2 cells were seeded at 2 × 104 cells/well in opaque plates (Greiner 655083). The next day, PsV corresponding to 1-10 × 106 luciferase units was mixed in Opti-MEM with dilutions of hmAbs and incubated at RT for 1 h. Media was removed from the cells and 100 μl/well of the hmAb/PsV mix was added in triplicates. After 1 h incubation at 37C and 5% CO2, another 100 μL of media containing 2% FBS, was added, and cells were incubated for 24 more hr. After this time, luciferase activity was measured using Passive Lysis Buffer (Promega E1941) and Luciferase substrate (Promega E151A) following the manufacturer’s instructions. Neutralization was calculated as the percent reduction of luciferase readings as compared to no-antibody-controls.
Prophylactic and therapeutic protective activities of RBD specific hmAbs in hamsters
We have previously demonstrated that golden Syrian hamsters (Mesocricetus auratus) are susceptible to SARS-CoV2, showing loss of body weight and viral replication in the lungs and nasal turbinate.25 In a prophylactic experiment, 6-week old, female hamsters were IP injection of 10 mg/kg of hmAb 1212C2 (n = 6), 1206D1 (n = 3), human IgG1 Isotype control mAb (BioXcell, Labanon, NH; n = 6), or PBS (n = 5). Six hours later, hamsters were given 2x105 PFU of SARS-CoV-2, intranasally. Three animals were also mock infected with PBS at this time to serve as a negative control. For each day, lungs and nasal turbinates were collected removed and photographed on white paper towels to look for gross pathology using a macroscopic pathology scoring analysis measure the distributions of pathological lesions, including consolidation, congestion, and pneumonic lesions using ImageJ software (NIH). Resulting pathology were represented as the percent of the total lung surface area. Left side of the lungs were used for histopathology and the right side was used for viral titers.
The hmAb 1212C2 was also assessed in an in vivo therapeutic experiment. At the start of the experiment, 12, 6-week old, female hamsters were administered 2x105 PFU of SARS-CoV-2, intranasally. Four animals were mock infected with PBS at the same time. Six hours later, animals were given either 25 mg/kg hmAb 1212C2 (n = 4), 25 mg/kg isotype control (n = 4), or PBS (n = 4) IP. Animals were euthanized at 4 dpi and lungs and nasal turbinates collected for histopathology and viral titers as described for the prophylactic experiment.
Delivery of inhaled 1212C2
The therapeutic testing of 1212C2 was tested using 5-week old, female hamsters. Hamsters were challenged with 2x105 PFU of SARS-CoV2 IN and 12 hours later treated with hmAb IP or IH. The estimated inhaled dose of hmAb is measured by sampling the aerosolized atmosphere the hamsters are exposed at a known fixed flowrate during the entire course of their exposure. The gas drawn out of the exposure chamber by vacuum passes through a collection filter to capture the aerosolized droplets. The amount of mAb deposited in the filter is determined using A280 following extraction in formulation buffer, which determines the mAb aerosol concentration in the exposure chamber. The approximate inhaled dose over a 30 minutes whole body exposure is then derived from the aerosol concentration and minute volume of the hamsters. The target high dose level of nonspecific IgG or 1212C2-HLE-LALA was 25 mg/kg. The actual dose measured was 11.30 mg/kg for the non-specific IgG group and 16.3 mg/kg for the 1212C2-HLE-LALA group (Table 2).
The mid and low dose groups for 1212C2-HLE-LALA were 5-fold and 25-fold dilutions of the mAb solution used in the high dose, respectively, which correspond to 3.2 mg/kg and 0.6 mg/kg.
Deep-sequencing immunoglobulin repertoire analysis
10 million PBMCs were thawed and used for RNA isolation (QIAGEN, RNeasy Mini Kit). Removal of any residual genomic DNA was performed using the Turbo DNA-Free kit (Invitrogen) and cDNA was generated using the qScript cDNA synthesis kit (QuantaBio). Two rounds of PCRs were performed to generate the libraries with either global (VH1 – VH6) or individual VH targeted forward primers as previously described31,44 with the modifications adapted for Illumina Nextera approach. The final PCR products were gel extracted (QIAQuick, QIAGEN, Hilden, Germany) and further purified using the ProNex size-selective purification system (Promega) to select products between 500-700 bp range. Libraries were submitted to the Heflin Center for Genomic Sciences at the University of Alabama at Birmingham where DNA quantification was made by qPCR and sequenced on an Illumina MiSeq system (Illumina, Inc., CA, USA) using 2 × 300 bp paired-end kits (Illumina MiSeq Reagent Kit v3). Sequence analysis and assembly of lineage trees were performed using an in-house custom analysis pipeline as previously described.44,47 All sequences were aligned using IMGT.org/HighVquest.48 Lineage trees were generated by identifying the lineage (the cluster of sequences with identical VH, JH, and HCDR3 lengths and ≥ 85% HCDR3 similarity) containing the corresponding hmAb sequence.
Quantification and statistical analysis
Significance was determined using GraphPad Prism, v8.0. Two-tailed t tests were applied for evaluation of the results between treatments. Bonferroni correction for multi-outcomes was applied to the therapeutic treatment experiment of hamsters utilizing multiple hmAb doses and routes of administration (n = 9 outcomes), p < 0.0055 was considered significant. For statistical analysis viral titers were log transformed and undetectable virus was set to the limit of detection (200 PFU/ml).
Acknowledgments
We are grateful for the clinical research staff that enabled this project and for the technical assistance provided by Eric Carlin and Sarah Sterrett, the technical guidance provided by Justin Roth, and the assistance of the University of Alabama at Birmingham (UAB) Center for AIDS Research and its Flow Cytometry Core Facility under the direction of Olaf Kutsch and the UAB Heflin Genomics Core, UAB Multidisciplinary Molecular Interaction Core Facility. We thank Alexander Rosenberg and Christopher Fucile for providing immunoglobulin sequence analysis software. We are most grateful for the participation of the study volunteers. Funding for this work provided by institutional support from the University of Alabama at Birmingham (to J.J.K.) and the Texas Biomedical Research Institute (to L.M.-S.), and funding from Aridis Pharmaceuticals (to V.L.T.).
Author contributions
J.J.K., L.M.-S., M.R.W., P.A.G., and V.L.T. conceived the study. M.S.P., M.B., S.S., and J.J.K. isolated and screened the mAbs. C.W.B., R.E.B., M.S.P., J.J.K., M.R.W., and A.D. produced the mAbs. M.R.W. and A.D. designed and produced the recombinant RBD proteins and conducted the SPR analysis. M.S.P. and S.S. conducted the ACE2 binding inhibition. J.-G.P., F.S.O., C.Y., K.C., and L.M.-S. conducted the in vitro and in vivo antiviral assessments. M.S.P. and M.B. conducted the deep sequencing of the 1212C2 lineage. P.A.G. and N.B.E. acquired the clinical specimens. A.L. conducted the pseudovirus neutralization assay. J.W., P.L., D.S., and V.L.T. designed and conducted the experiments to determine the efficiency of the inhaled delivery of mAbs. J.J.K., L.M.-S., M.R.W., and V.L.T. supervised the work. J.J.K., L.M.-S., M.R.W., and V.L.T. wrote the manuscript. All of the authors reviewed the manuscript.
Declaration of interests
M.S.P., J.-G.P., A.D., F.S.O., M.B., S.S., N.B.E., P.A.G., M.R.W., L.M.-S., and J.J.K. are co-inventors on a patent that includes claims related to the hmAbs described. A.L., J.W., P.L., D.S., and V.L.T. are employees of Aridis Pharmaceuticals.
Published: February 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100218.
Contributor Information
Luis Martinez-Sobrido, Email: lmartinez@txbiomed.org.
James J. Kobie, Email: jjkobie@uabmc.edu.
Supplemental information
References
- 1.Cai Q., Chen F., Wang T., Luo F., Liu X., Wu Q., He Q., Wang Z., Liu Y., Liu L. Obesity and COVID-19 Severity in a Designated Hospital in Shenzhen, China. Diabetes Care. 2020;43:1392–1398. doi: 10.2337/dc20-0576. [DOI] [PubMed] [Google Scholar]
- 2.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC COVID-19 Response Team Preliminary Estimates of the Prevalence of Selected Underlying Health Conditions Among Patients with Coronavirus Disease 2019 - United States, February 12-March 28, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:382–386. doi: 10.15585/mmwr.mm6913e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worldometer Worldometer COVID-19 data. https://www.worldometers.info/coronavirus/about/
- 7.Lavery A.M., Preston L.E., Ko J.Y., Chevinsky J.R., DeSisto C.L., Pennington A.F., Kompaniyets L., Datta S.D., Click E.S., Golden T. Characteristics of Hospitalized COVID-19 Patients Discharged and Experiencing Same-Hospital Readmission - United States, March-August 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1695–1699. doi: 10.15585/mmwr.mm6945e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis N.M., Friedrichs M., Wagstaff S., Sage K., LaCross N., Bui D., McCaffrey K., Barbeau B., George A., Rose C. Disparities in COVID-19 Incidence, Hospitalizations, and Testing, by Area-Level Deprivation - Utah, March 3-July 9, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1369–1373. doi: 10.15585/mmwr.mm6938a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., Yu J., Chan C.N., Bondoc S., Starke C.E. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020;26:1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with high attack rate. J. Clin. Microbiol. 2020;58:e02107-20. doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Case J.B., Rothlauf P.W., Chen R.E., Kafai N.M., Fox J.M., Smith B.K., Shrihari S., McCune B.T., Harvey I.B., Keeler S.P. Replication-Competent Vesicular Stomatitis Virus Vaccine Vector Protects against SARS-CoV-2-Mediated Pathogenesis in Mice. Cell Host Microbe. 2020;28:465–474.e4. doi: 10.1016/j.chom.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv Z., Deng Y.Q., Ye Q., Cao L., Sun C.Y., Fan C., Huang W., Sun S., Sun Y., Zhu L. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 15.Ibrahim D., Dulipsingh L., Zapatka L., Eadie R., Crowell R., Williams K., Wakefield D.B., Cook L., Puff J., Hussain S.A. Factors Associated with Good Patient Outcomes Following Convalescent Plasma in COVID-19: A Prospective Phase II Clinical Trial. Infect. Dis. Ther. 2020;9:913–926. doi: 10.1007/s40121-020-00341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salazar E., Kuchipudi S.V., Christensen P.A., Eagar T., Yi X., Zhao P., Jin Z., Long S.W., Olsen R.J., Chen J. Convalescent plasma anti-SARS-CoV-2 spike protein ectodomain and receptor-binding domain IgG correlate with virus neutralization. J. Clin. Invest. 2020;130:6728–6738. doi: 10.1172/JCI141206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 18.Callaway E. 2020. What Pfizer’s landmark COVID vaccine results mean for the pandemic.https://www.nature.com/articles/d41586-020-03248-7 [DOI] [PubMed] [Google Scholar]
- 19.Callaway E. COVID vaccine excitement builds as Moderna reports third positive result. Nature. 2020;587:337–338. doi: 10.1038/d41586-020-03248-7. [DOI] [PubMed] [Google Scholar]
- 20.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J.-G., Oladduni F.S., Chiem K., Ye C., Pipenbrink M., Moran T., Walter M.R., Kobie J., Martinez-Sobrido L. Rapid in vitro assays for screening neutralizing antibodies and antivirals against SARS-CoV-2. J. Virol. Methods. 2021;287:113995. doi: 10.1016/j.jviromet.2020.113995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye C., Chiem K., Park J.G., Oladunni F., Platt R.N., 2nd, Anderson T., Almazan F., de la Torre J.C., Martinez-Sobrido L. Rescue of SARS-CoV-2 from a Single Bacterial Artificial Chromosome. MBio. 2020;11:e02168-20. doi: 10.1128/mBio.02168-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magyarics Z., Leslie F., Bartko J., Rouha H., Luperchio S., Schörgenhofer C., Schwameis M., Derhaschnig U., Lagler H., Stiebellehner L. Randomized, Double-Blind, Placebo-Controlled, Single-Ascending-Dose Study of the Penetration of a Monoclonal Antibody Combination (ASN100) Targeting Staphylococcus aureus Cytotoxins in the Lung Epithelial Lining Fluid of Healthy Volunteers. Antimicrob. Agents Chemother. 2019;63:e00350-19. doi: 10.1128/AAC.00350-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raabe O.G., Al-Bayati M.A., Teague S.V., Rasolt A. Regional Deposition of Inhaled Monodisperse Coarse and Fine Aerosol Particles in Small Laboratory Animals. Ann. Occup. Hyg. 1988;32:53–63. [Google Scholar]
- 28.Guilleminault L., Azzopardi N., Arnoult C., Sobilo J., Hervé V., Montharu J. Fate of inhaled monoclonal antibodies after the deposition of aerosolized particles in the respiratory system. J Control Release. 2014;196:344–354. doi: 10.1016/j.jconrel.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Tetro J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22:72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Booth B.J., Ramakrishnan B., Narayan K., Wollacott A.M., Babcock G.J., Shriver Z., Viswanathan K. Extending human IgG half-life using structure-guided design. MAbs. 2018;10:1098–1110. doi: 10.1080/19420862.2018.1490119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basu M., Piepenbrink M.S., Francois C., Roche F., Zheng B., Spencer D.A., Hessell A.J., Fucile C.F., Rosenberg A.F., Bunce C.A. Persistence of HIV-1 Env-Specific Plasmablast Lineages in Plasma Cells after Vaccination in Humans. Cell Rep. Med. 2020;1:100015. doi: 10.1016/j.xcrm.2020.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao H., Li S., Yu Y., Yue Y., Su K., Zheng Q., Jiang N., Zhang Z. Characteristics of Plasmablast Repertoire in Chronically HIV-Infected Individuals for Immunoglobulin H and L Chain Profiled by Single-Cell Analysis. Front. Immunol. 2020;10:3163. doi: 10.3389/fimmu.2019.03163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scharf L., West A.P., Jr., Gao H., Lee T., Scheid J.F., Nussenzweig M.C., Bjorkman P.J., Diskin R. Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc. Natl. Acad. Sci. USA. 2013;110:6049–6054. doi: 10.1073/pnas.1303682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michaeli M., Tabibian-Keissar H., Schiby G., Shahaf G., Pickman Y., Hazanov L., Rosenblatt K., Dunn-Walters D.K., Barshack I., Mehr R. Immunoglobulin gene repertoire diversification and selection in the stomach - from gastritis to gastric lymphomas. Front. Immunol. 2014;5:264. doi: 10.3389/fimmu.2014.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Julakyan U.L., Biderman B.V., Gemdzhian E.G., Sudarikov A.B., Savchenko V.G. [Molecular analysis of immunoglobulin genes in the tumor B cells in splenic marginal zone lymphoma] Ter. Arkh. 2015;87:58–63. doi: 10.17116/terarkh201587758-63. [DOI] [PubMed] [Google Scholar]
- 36.Bikos V., Darzentas N., Hadzidimitriou A., Davis Z., Hockley S., Traverse-Glehen A., Algara P., Santoro A., Gonzalez D., Mollejo M. Over 30% of patients with splenic marginal zone lymphoma express the same immunoglobulin heavy variable gene: ontogenetic implications. Leukemia. 2012;26:1638–1646. doi: 10.1038/leu.2012.3. [DOI] [PubMed] [Google Scholar]
- 37.Pujanauski L.M., Janoff E.N., McCarter M.D., Pelanda R., Torres R.M. Mouse marginal zone B cells harbor specificities similar to human broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA. 2013;110:1422–1427. doi: 10.1073/pnas.1213713110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreye J., Reincke S.M., Kornau H.C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.D. A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgess G., Boyce M., Jones M., Larsson L., Main M.J., Morgan F., Phillips P., Scrimgeour A., Strimenopoulou F., Vajjah P. Randomized study of the safety and pharmacodynamics of inhaled interleukin-13 monoclonal antibody fragment VR942. EBioMedicine. 2018;35:67–75. doi: 10.1016/j.ebiom.2018.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bodier-Montagutelli E., Mayor A., Vecellio L., Respaud R., Heuzé-Vourc’h N. Designing inhaled protein therapeutics for topical lung delivery: what are the next steps? Expert Opin. Drug Deliv. 2018;15:729–736. doi: 10.1080/17425247.2018.1503251. [DOI] [PubMed] [Google Scholar]
- 43.ClinicalTrials.gov. Dose Ranging Study of ALX-0171 in Infants Hospitalized for Respiratory Syncytial Virus Lower Respiratory Tract Infection (Respire). https://www.clinicaltrials.gov/ct2/show/NCT02979431.
- 44.Nogales A., Piepenbrink M.S., Wang J., Ortega S., Basu M., Fucile C.F., Treanor J.J., Rosenberg A.F., Zand M.S., Keefer M.C. A Highly Potent and Broadly Neutralizing H1 Influenza-Specific Human Monoclonal Antibody. Sci. Rep. 2018;8:4374. doi: 10.1038/s41598-018-22307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hezareh M., Hessell A.J., Jensen R.C., van de Winkel J.G., Parren P.W. Effector function activities of a panel of mutants of a broadly neutralizing antibody against human immunodeficiency virus type 1. J. Virol. 2001;75:12161–12168. doi: 10.1128/JVI.75.24.12161-12168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tipton C.M., Fucile C.F., Darce J., Chida A., Ichikawa T., Gregoretti I., Schieferl S., Hom J., Jenks S., Feldman R.J. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat. Immunol. 2015;16:755–765. doi: 10.1038/ni.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alamyar E., Duroux P., Lefranc M.P., Giudicelli V. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included within the manuscript and are available without restriction from the Lead Contact upon request.