Summary
Polymorphisms in MHC-I protein sequences across human populations significantly affect viral peptide binding capacity, and thus alter T cell immunity to infection. In the present study, we assess the relationship between observed SARS-CoV-2 population mortality and the predicted viral binding capacities of 52 common MHC-I alleles. Potential SARS-CoV-2 MHC-I peptides are identified using a consensus MHC-I binding and presentation prediction algorithm called EnsembleMHC. Starting with nearly 3.5 million candidates, we resolve a few hundred highly probable MHC-I peptides. By weighing individual MHC allele-specific SARS-CoV-2 binding capacity with population frequency in 23 countries, we discover a strong inverse correlation between predicted population SARS-CoV-2 peptide binding capacity and mortality rate. Our computations reveal that peptides derived from the structural proteins of the virus produce a stronger association with observed mortality rate, highlighting the importance of S, N, M, and E proteins in driving productive immune responses.
Keywords: MHC-I, CD8, SARS-CoV-2, EnsembleMHC, population dynamics, risk, epitope, immunoinformatics
Highlights
EnsembleMHC, a MHC-I presentation prediction algorithm, can predict SARS-CoV-2 epitopes
Countries show variation in the predicted MHC-I SARS-CoV-2 binding capacity
A population score combines the MHC-I binding capacity with MHC-I allele frequencies
The population score inversely correlates with observed deaths from SARS-CoV-2
Wilson et al. define a predicted MHC allele-specific hierarchy for the presentation of peptides derived from SARS-CoV-2 viral proteins. They find that a composite population-level metric combining predicted MHC allele SARS-CoV-2 binding capacity and endemic allele frequencies is inversely correlated with deaths per million.
Introduction
In December 2019, the novel coronavirus, severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) was identified from a cluster of cases of pneumonia in Wuhan, China.1,2 With >73.1 million cases and >1.6 million deaths, the viral spread was declared a global pandemic by the World Health Organization.3 Due to its high rate of transmission and unpredictable severity, there is an immediate need for information surrounding the adaptive immune response toward SARS-CoV-2.
A robust T cell response is integral for the clearance of coronaviruses and the generation of lasting immunity.4 The potential role of T cells for coronavirus clearance has been supported by the identification of immunogenic CD8+ T cell epitopes in the S (Spike), N (Nucleocapsid), M (Membrane), and E (Envelope) proteins.5 In addition, SARS-CoV-specific CD8+ T cells have been shown to provide long-lasting immunity, with memory CD8+ T cells being detected up to 17 years post-infection.4,6,7 The specifics of the T cell response to SARS-CoV-2 is still evolving. However, a recent screening of SARS-CoV-2 peptides revealed that a majority of the CD8+ T cell immune response is targeted toward viral structural proteins (N, M, S).8
A successful CD8+ T cell response is contingent on the efficient presentation of viral protein fragments by major histocompatibility complex I (MHC-I) proteins. MHC-I molecules bind and present peptides derived from endogenous proteins on the cell surface for CD8+ T cell interrogation. The MHC-I protein is highly polymorphic, with amino acid substitutions within the peptide binding groove drastically altering the composition of presented peptides. Consequently, the influence of MHC genotype to shape patient outcome has been well studied in the context of viral infections.9 For coronaviruses, there have been several studies of the association of MHC with disease susceptibility. A study of a Taiwanese and Hong Kong cohort of patients with SARS-CoV found that the MHC-I alleles HLA (histocompatibility leukocyte antigen)-B∗07:03 and HLA-B∗46:01 were linked to increased susceptibility, while HLA-Cw∗15:02 was linked to increased resistance.10, 11, 12 However, some of the reported associations did not remain after statistical correction, and it is still unclear whether MHC-outcome associations reported for SARS-CoV are applicable to SARS-CoV-2.13,14 Recently, a comprehensive prediction of SARS-CoV-2 MHC-I peptides indicated a relative depletion of high-affinity binding peptides for HLA-B∗46:01, hinting at a similar association profile in SARS-CoV-2.15 More important, it remains elusive whether such a depletion of putative high-affinity peptides will affect patient outcomes to SARS-CoV-2 infections.
The lack of large-scale genomic data linking individual MHC genotypes and outcomes from SARS-CoV-2 infections precludes a similar analysis as performed for SARS-CoV.10, 11, 12 Therefore, we endeavored to assess the relationship between the predicted SARS-CoV-2 binding capacity of a population and the observed SARS-CoV-2 mortality rate. Histroically, MHC-I prediction algorithms have been characterized by a high false positive rate, particularly when predicting peptides that are naturally presented.16,17 To minimize false positives and identify the highest-confidence SARS-CoV-2 MHC-I peptides, we developed a consensus algorithm, called EnsembleMHC, and predicted MHC-I peptides for a panel of 52 common MHC-I alleles.18 This prediction workflow integrates seven different algorithms that have been parameterized on high-quality mass spectrometry data and provides a confidence level for each identified peptide.17,19, 20, 21, 22, 23, 24 The distribution of the number of high-confidence peptides assigned to each allele was used to assess a country-specific SARS-CoV-2 binding capacity, called the EnsembleMHC population (EMP) score, for 23 countries (for selection criteria, please refer to the STAR Methods). This score was derived by weighing the individual binding capacities of the 52 MHC-I alleles by their endemic frequencies. We note a strong inverse correlation between the EMP score and observed population SARS-CoV-2 mortality. Furthermore, the correlation is demonstrated to become stronger when considering EMP scores based solely on SARS-CoV-2 structural proteins, underlining their potential importance in driving a robust immune response. Based on their predicted binding affinity, expression, and sequence conservation in viral isolates, we identified 108 peptides derived from SARS-CoV-2 structural proteins that are high-value targets for CD8+ T cell vaccine development.
Results
EnsembleMHC workflow offers more precise MHC-I presentation predictions than individual algorithms
The accurate assessment of differences in SARS-CoV-2 binding capacities across MHC-I allelic variants requires the isolation of MHC-I peptides with a high probability of being presented. EnsembleMHC provides the requisite precision through the use of allele- and algorithm-specific score thresholds and peptide confidence assignment.
MHC-I alleles substantially vary in both peptide binding repertoire size and median binding affinity.25 The EnsembleMHC workflow addresses this inter-allele variation by identifying peptides based on MHC allele- and algorithm-specific binding affinity thresholds. These thresholds were set by benchmarking each of the 7 component algorithms against 52 single MHC allele peptide datasets.17 Each dataset consists of mass spectrometry-confirmed MHC-I peptides that have been naturally presented by a model cell line expressing 1 of the 52 select MHC-I alleles. These experimentally validated peptides, denoted target peptides, were supplemented with a 100-fold excess of decoy peptides. Decoys were generated by randomly sampling peptides that were not detected by mass spectrometry, but were derived from the same protein sources as a detected target peptide. Algorithm- and allele-specific binding affinity thresholds were then identified through the independent application of each component algorithm to all of the MHC allele datasets. For every dataset and algorithm combination, the target and decoy peptides were ranked by predicted binding affinity to the MHC allele defined by that dataset. Then, an algorithm-specific binding affinity threshold was set to the minimum score needed to isolate the highest affinity peptides commensurate to 50% of the observed allele repertoire size (STAR Methods; Figure S1A). The observed allele repertoire size was defined as the total number of target peptides within a given single MHC allele dataset. Therefore, if a dataset had 1,000 target peptides, the top 500 highest affinity peptides would be selected, and the algorithm-specific threshold would be set to the predicted binding affinity of the 500th peptide. This parameterization method resulted in the generation of a customized set of allele- and algorithm-specific binding affinity thresholds in which an expected quantity of peptides can be recovered.
Consensus MHC-I prediction typically require a method for combining outputs from each individual component algorithm into a composite score. This composite score is then used for peptide selection. EnsembleMHC identifies high-confidence peptides based on filtering by a quantity called peptideFDR (STAR Methods, Equation 1). During the identification of allele- and algorithm-specific binding affinity thresholds, the empirical false detection rate (FDR) of each algorithm was calculated. This calculation was based on the proportion of target to decoy peptides isolated by the algorithm-specific binding affinity threshold. A peptideFDR is then assigned to each individual peptide by taking the product of the empirical FDRs of each algorithm that identified that peptide for the same MHC-I allele. Analysis of the parameterization process revealed that the overall performance of each included algorithm was comparable, and there was diversity in individual peptide calls by each algorithm, supporting an integrated approach to peptide confidence assessment (Figures S1B–S1D). Peptide identification by EnsembleMHC was performed by selecting all of the peptides with a peptideFDR ≤5%.26
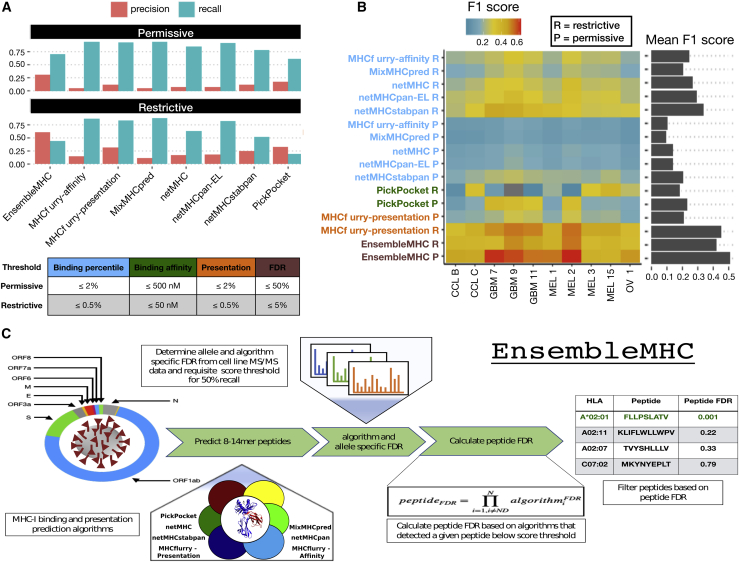
The efficacy of peptideFDR as a filtering metric was determined through the prediction of naturally presented MHC-I peptides derived from 10 tumor samples (Figure 1).17 Similar to the single MHC allele datasets, each tumor sample dataset consisted of mass spectrometry-detected target peptides and a 100-fold excess of decoy peptides. The performance of EnsembleMHC was assessed via comparison with individual component algorithms. Peptide identification by each algorithm was based on a restrictive or permissive binding affinity threshold (Figure 1A, table). For the component algorithms, the permissive and restrictive thresholds correspond to commonly used binding affinity cutoffs for the identification of weak and strong binders, respectively.27 The performance of each algorithm on the 10 datasets was evaluated through the calculation of the empirical precision, recall, and F1 score.
Figure 1.
Application of the EnsembleMHC prediction algorithm
The EnsembleMHC prediction algorithm was used to recover MHC-I peptides from 10 tumor sample datasets.
(A) The average precision and recall for EnsembleMHC and each component algorithm were calculated across all 10 tumor samples. Peptide identification by each algorithm was based on commonly used restrictive (strong) or permissive (strong and weak) binding affinity thresholds (see table below).
(B) The F1 score of each algorithm was calculated for all tumor samples. Each algorithm is grouped into 1 of 4 categories: binding affinity represented by percentile score (blue), binding affinity represented by predicted peptide half-maximal inhibitory concentration (IC50) value (green), MHC-I presentation prediction (orange), and EnsembleMHC (brown). The heatmap colors indicate the value of the observed F1 score (color bar) for a given algorithm (y axis) on a particular dataset (x axis). Warmer colors indicate higher F1 scores, and cooler colors indicate lower F1 scores. The average F1 score for each algorithm across all of the samples is shown in the marginal bar plot.
(C) The schematic for the application of the EnsembleMHC predication algorithm to identify SARS-CoV-2 MHC-I peptides.
The average precision and recall of each algorithm across all tumor samples demonstrated an inverse relationship (Figure 1A). In general, restrictive binding affinity thresholds produced higher precision at the cost of poorer recall. When comparing the precision of each algorithm at restrictive thresholds, EnsembleMHC demonstrated a 3.4-fold improvement over the median precision of individual component algorithms. EnsembleMHC also produced the highest F1 score, with an average of 0.51, followed by mhcflurry-presentation, with an F1 score of 0.45, both of which are 1.5- to 2-fold higher than the rest of the algorithms (Figure 1B). This result was shown to be robust across a range of peptideFDR cutoff thresholds (Figure S1E), alternative performance metrics (Figure S1F), and other consensus-based prediction algorithms (Figures S1G and S1H). Furthermore, EnsembleMHC demonstrated the ability to more efficiently prioritize peptides with experimentally established immunogenicity from the Hepatitis-C genome polyprotein, the Dengue virus genome polyprotein, and the HIV-1 POL-GAG protein (Figure S1I). These results demonstrate the enhanced precision of EnsembleMHC over individual component algorithms when using common binding affinity thresholds.
In summary, the EnsembleMHC workflow offers two desirable features. First, it determines allele-specific binding affinity thresholds for each algorithm at which a known quantity of peptides is expected to be successfully presented on the cell surface. Second, it assigns a confidence level to each peptide call made by each algorithm. These traits enhance the ability to identify MHC-I peptides with a high probability of successful cell surface presentation.
EnsembleMHC was used to identify MHC-I peptides for the SARS-CoV-2 virus (Figure 1C). The resulting identification of high-confidence SARS-CoV-2 peptides allows for the characterization of alleles that are enriched or depleted for predicted MHC-I peptides. The resulting distribution of allele-specific SARS-CoV-2 binding capacities will then be weighed by the normalized frequencies of the 52 alleles (Figure S2; STAR Methods, Equations 5 and 6) in 23 countries to determine the population-specific SARS-CoV-2 binding capacity or EMP score (STAR Methods, Equation 7). The potential impact of varying population SARS-CoV-2 binding capacities on disease outcomes can then be assessed by correlating population SARS-CoV-2 mortality rates with EMP scores.
The MHC-I peptide-allele distribution for SARS-CoV-2 structural proteins is especially disproportionate
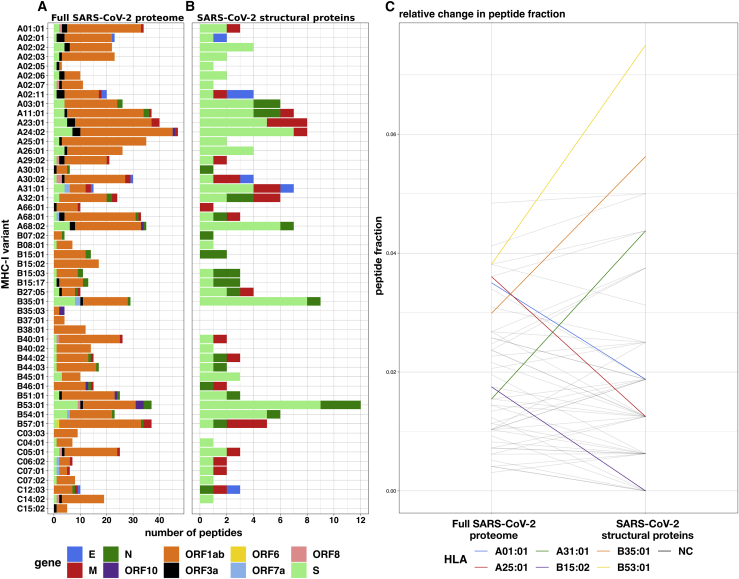
MHC-I peptides derived from the SARS-CoV-2 proteome were predicted and prioritized using EnsembleMHC. A total of 67,207 potential 8- to 14-mer viral peptides were evaluated for each of the considered MHC-I alleles. After filtering the pool of candidate peptides at the 5% peptideFDR threshold, the number of potential peptides was reduced from 3.49 million to 971 (658 unique peptides) (Figures S3A and S3B; Table S1). Illustrated in Figure 2A, the viral peptide-MHC allele (or peptide-allele) distribution for high-confidence SARS-CoV-2 peptides was determined by assigning the identified peptides to their predicted MHC-I alleles. There was a median of 16 peptides per allele, with a maximum of 47 peptides (HLA-A∗24:02), a minimum of 3 peptides (HLA-A∗02:05), and an interquartile range (IQR) of 16 peptides. Quality assurance of the predicted peptides was performed by computing the peptide length frequencies and binding motifs. The predicted peptides were found to adhere to expected MHC-I peptide lengths,28 with 78% of the peptides being 9 amino acids in length, 13% being 10 amino acids in length, and 8% of peptides accounting for the remaining lengths (Figures S3C and S3D). Similarly, logo plots generated from predicted peptides were found to closely reflect reference peptide binding motifs for considered alleles (Figure S3E).29 Overall, the EnsembleMHC prediction platform demonstrated the ability to isolate a short list of potential peptides that adhere to expected MHC-I peptide characteristics.
Figure 2.
Prediction of SARS-CoV-2 peptides across 52 common MHC-I alleles
(A and B) The EnsembleMHC workflow was used to predict MHC-I peptides for 52 alleles from the entire SARS-CoV-2 proteome or specifically SARS-CoV-2 structural proteins (E, S, N, and M).
(C) The peptide fractions for both protein sets were calculated by dividing the number of peptides assigned to a given allele by the total number of identified peptides for that protein set. Each line indicates the change in peptide fraction observed by a given allele when comparing the viral peptide-MHC allele distribution for the full SARS-CoV-2 proteome or structural proteins. Alleles showing a change of greater than the median peptide fraction, = 0.015, are highlighted in color.
The high expression, relative conservation, and reduced search space of SARS-CoV-2 structural proteins (S, E, M, and N) make MHC-I binding peptides derived from these proteins high-value targets for CD8+ T cell-based vaccine development. Figure 2B describes the peptide-allele distribution for predicted MHC-I peptides originating from the four structural proteins. This analysis markedly reduces the number of considered peptides from 658 to 108 (Table S1). The median number of predicted SARS-CoV-2 structural peptides assigned to each MHC-I allele was found to be 2, with a maximum of 12 peptides (HLAB∗53:01), a minimum of 0 peptides (HLA-B∗15:02, B∗35:03,B∗38:01,C∗03:03,C∗15:02), and an IQR of 3 peptides. Analysis of the molecular source of the identified SARS-CoV-2 structural protein peptides revealed that they originate from enriched regions that are highly conserved (Figure S4). This indicates that such peptides would be ideal candidates for targeted therapies as they are unlikely to be disrupted by mutation, and several peptides can be targeted using minimal stretches of the source protein. Consideration of the MHC-I peptides derived only from SARS-CoV-2 structural proteins reduces the number of potential peptides to a condensed set of high-value targets that is amenable to experimental validation.
Both the peptide-allele distributions, namely the ones derived from the full SARS-CoV-2 proteome, and those from the structural proteins were found to significantly deviate from an even distribution of predicted peptides as is apparent in Figures 2A and 2B and reflected in the Kolmogorov-Smirnov test p values (Figure S5; full proteome = 5.673e−7 and structural proteins = 1.45e−2). These results support a potential allele-specific hierarchy for SARS-CoV-2 peptide presentation.
To determine whether the MHC-I binding capacity hierarchy was consistent between the full SARS-CoV-2 proteome and SARS-CoV-2 structural proteins, the relative changes in the observed peptide fraction (number of peptides assigned to an allele/total number of peptides) between the two protein sets was visualized (Figure 2C). A total of 6 alleles demonstrated changes greater than the median peptide fraction ( = 0.015) when comparing the 2 protein sets. The greatest decrease in peptide fraction was observed for A∗25:01 (1.52 times the median peptide fraction), and the greatest increase was seen with B∗53:01 (2.38 times the median peptide fraction). Furthermore, the resulting SARS-CoV-2 structural protein peptide-allele distribution was found to be more variable than the distribution derived from the full SARS-CoV-2 proteome, with a quartile coefficient of dispersion of 0.6 compared to 0.44, respectively. This indicates that peptides derived from SARS-CoV-2 structural proteins experience larger relative inter-allele binding capacity discrepancies than peptides derived from the full SARS-CoV-2 proteome. These results indicate a potential MHC-I binding capacity hierarchy that is more pronounced for SARS-CoV-2 structural proteins.
Total population epitope load inversely correlates with reported death rates from SARS-CoV-2
The documented importance of MHC-I peptides derived from SARS-CoV-2 structural proteins,8 coupled with the observed MHC allele binding capacity hierarchy and the high immunogenicity rate of SARS-CoV-2 structural protein MHC-I peptides identified by EnsembleMHC (Figure S5D), prompts a potential relationship between MHC-I genotype and infection outcome. However, due to the absence of MHC genotype data for SARS-CoV-2 patients, we assessed this relationship at the population level by correlating predicted country-specific SARS-CoV-2 binding capacity (or EMP score) with observed SARS-CoV-2 mortality.
EMP scores were determined for 23 countries (Table S2) by weighing the individual binding capacities of 52 common MHC-I alleles by their normalized endemic expression (STAR Methods; Figure S2).18 Every country in the cohort is assigned two separate EMP scores—one calculated with respect to the 108 unique SARS-CoV-2 structural protein peptides (structural protein EMP) and the other with respect to the 658 unique peptides derived from the full SARS-CoV-2 proteome (full proteome EMP). The EMP score corresponds to the average predicted SARS-CoV-2 binding capacity of a select population. Therefore, individuals in a country with a high EMP score would be expected, on average, to present more SARS-CoV-2 peptides to CD8+ T cells than would individuals from a country with a low EMP score. The resulting EMP scores were then correlated with observed SARS-CoV-2 mortality (deaths per million) as a function of time (January–April 2020). Temporal variance in community spread within the cohort of countries was corrected by truncating the SARS-CoV-2 mortality dataset for each country to start after a certain minimum death threshold was met. For example, if the minimum death threshold was 50, then day 0 would be when each country reported at least 50 deaths. The number of countries included in each correlation decreases as the number of days increases due to discrepancies in the length of time that each country met a given minimum death threshold (Table S3). Therefore, the correlation between EMP score and SARS-CoV-2 mortality was only estimated at time points at which there were at least 8 countries. The 8-country threshold was chosen because it is the minimum sample size needed to maintain sufficient power when detecting large effect sizes (ρ > 0.85). The strength of the relationship between EMP score and SARS-CoV-2 mortality was determined using Spearman’s rank-order correlation (for details concerning the choice of statistical tests, please refer to STAR Methods). Accordingly, both EMP scores and SARS-CoV-2 mortality data were converted into ascending ranks, with the lowest rank indicating the minimum value and the highest rank indicating the maximum value. For instance, a country with an EMP score rank of 1 and death per million rank of 23 would have the lowest predicted SARS-CoV-2 binding capacity and the highest level of SARS-CoV-2-related mortality. Using the described paradigm, the structural protein EMP score and the full proteome EMP score were correlated with SARS-CoV-2-related deaths per million for 23 countries.
Total predicted population SARS-CoV-2 binding capacity exhibited a strong inverse correlation with observed deaths per million. This relationship was found to be true for correlations based on the structural protein EMP (Figure 3A) and full proteome EMP (Figure S5A) scores, with mean effect sizes of −0.66 and −0.60, respectively. Significance testing of the correlations produced by both EMP scores revealed that the majority of reported correlations are statistically significant, with 63% attaining a p ≤ 0.05. Correlations based on the structural protein EMP score demonstrated a 23% higher proportion of statistically significant correlations compared to the full proteome EMP score (74% versus 51%). Furthermore, correlations for EMP scores based on structural proteins produced narrower 95% confidence intervals (Figure S5B; Table S3). Due to relatively low statistical power of the obtained correlations (Figure S6), the positive predictive value (PPV) for each correlation (STAR Methods, Equation 8) was calculated. The resulting proportions of correlations with a PPV of ≥ 95% were similar to the observed significant p value proportions, with 62% of all measured correlations, 72% of structural protein EMP score correlations, and 52% of full proteome EMP score correlations (Figure S5B). The similar proportions of significant p values and PPVs supports that an overall true association is being captured. Furthermore, analysis of similar-size peptide sets sampled from the full SARS-CoV-2 proteome revealed that the observed distinction between the correlations produced by the two protein groups are unlikely to be due to differences in peptide set sizes (Figures S7A and S7B).
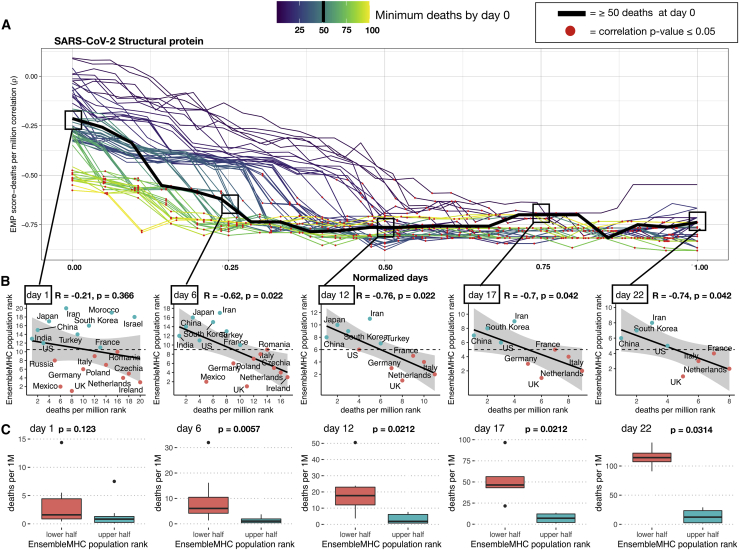
Figure 3.
Predicted total epitope load within a population inversely correlates with mortality
(A) SARS-CoV-2 structural protein-based EnsembleMHC population (EMP) scores were assigned to 23 countries (Table S2, A), and correlated with observed mortality rate (deaths per million). The correlation coefficient is presented as a function of time. Individual country mortality rate data were aligned by truncating each dataset to start after a minimum threshold of deaths was observed in a given country (line color). The Spearman’s rank correlation coefficient between structural protein EMP score and SARS-CoV-2 mortality rate was calculated every day following day 0 for each of the minimum death thresholds. Due to the differing lengths of time series analysis at each minimum death threshold, the number of days was normalized to improve visualization. Thus, normalized day 0 represents the day when qualifying countries recorded at least the number of deaths indicated by the minimum death threshold; normalized day 1 represents the final time point at which a correlation was measured. Correlations that were shown to be statistically significant (p ≤ 0.05) are indicated by a red point.
(B) The correlations between the structural protein EMP score (y axis) and deaths per million (x axis) were shown for countries meeting the 50 minimum deaths threshold at days 1, 6, 12, 17, and 22. Correlation coefficients and p values were assigned using Spearman’s rank correlation and the shaded region signifies the 95% confidence interval. Due to Spearman’s rank correlation only considering data rank, deaths per million and EMP score were converted to ascending rank values (low rank = low values, high rank = high values) to improve visualization of the measured relationship. Red points indicate a country that has an EMP rank that is less than the median EMP rank of all countries at that day, and blue points indicate a country with an EMP rank that is greater than the median EMP rank.
(C) The countries at each day were partitioned into an upper or a lower half based on the median observed EMP rank. Therefore, countries with an EMP rank greater than the median group EMP score were assigned to the upper half (red) and the remaining countries were assigned to the lower half (blue). p values were determined by the Mann-Whitney U test. The presented boxplots are in the style of Tukey (box defined by 25%, 50%, and 75% quantiles, and whiskers ±1.5 × IQR). The increasing gap between the red and the blue boxplots indicates a greater discrepancy in the number of deaths per million between the 2 groups.
The p values in (A)–(C) were corrected using the Benjamini-Hochberg procedure30 relative to the number of tests performed for each death threshold.
Finally, the reported correlations did not remain after randomizing the allele assignment of predicted peptides before peptideFDR filtering (Figures S7C and S7D) through the use of any individual algorithm (Figure S7E). This indicates that the observed relationship is contingent on the high-confidence peptide-allele distribution identified by EnsembleMHC. These data demonstrate that the MHC-I allele hierarchy characterized by EnsembleMHC is inversely associated with SARS-CoV-2 population mortality, and that the relationship becomes stronger when considering only the presentation of SARS-CoV-2 structural proteins.
The ability to use the structural protein EMP score to identify high- and low-risk populations was assessed using the median minimum death threshold (50 deaths) at evenly spaced time points (Figure 3A, squares). All of the correlations, with the exception of day 1, were found to be significant, with an average effect size of −0.71 (Figure 3B). Next, the countries at each day were partitioned into a high or low group based on whether their assigned EMP score was higher or lower than the median observed EMP score (Figure 3C). The resulting groups demonstrated a statistically significant difference in the median deaths per million between countries with low structural protein EMP scores and countries with high structural protein EMP scores. In addition, it was observed that deaths per million increased much more rapidly in countries with low structural protein EMP scores. These results indicate that the structural protein EMP score may be useful for assessing population risk from SARS-CoV-2 infections.
In summary, we make several important observations. First, there is a strong inverse correlation between predicted population SARS-CoV-2 binding capacity and observed deaths per million. This finding suggests that outcome to SARS-CoV-2 may be tied to total epitope load. Second, the correlation between predicted epitope load and population mortality is stronger for SARS-CoV-2 structural MHC-I peptides. This suggests that CD8+ T cell-mediated immune response may be driven primarily by the recognition of epitopes derived from these proteins, a finding supported by recent T cell epitope mapping of SARS-CoV-2.8 Finally, the EMP score can separate countries within the considered cohort into high- or low-risk populations.
Structural protein EMP score correlates better with population outcome than identified individual risk factors
Recent large-scale patient studies have identified several socioeconomic and health-related factors associated with the increased risk of death from SARS-CoV-2 infection.31,32 To delineate the relative importance of the structural protein EMP score as a SARS-CoV-2 severity descriptor, 12 additional risk factors were assessed for their ability to model population-level SARS-CoV-2 outcome in 21 countries (Table S2).
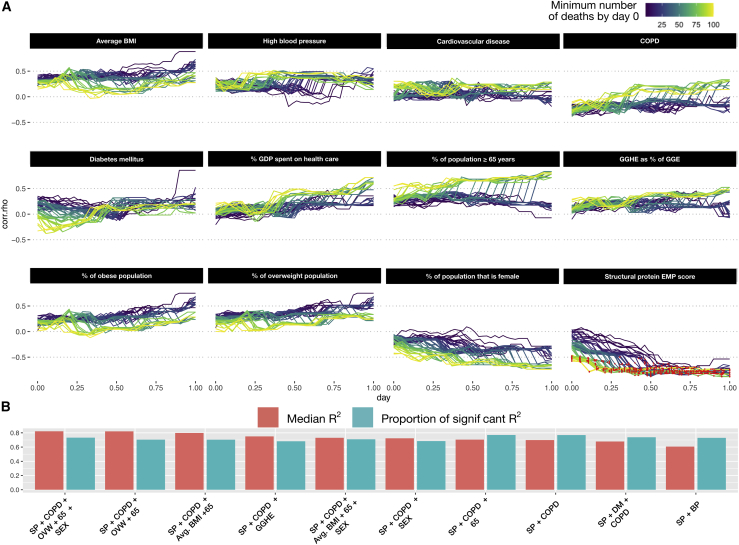
Overall, the structural protein EMP scores produced a significantly stronger association with population SARS-CoV-2 mortality compared to 12 other descriptors (Figure 4A). While various effect size trends were observed, all of the additional covariates failed to produce statistically significant correlations. To determine whether the modeling of the SARS-CoV-2 mortality rate could be improved by the combination of single socioeconomic or health-related risk factors with structural protein EMP scores, a set of linear models consisting of either a single risk factor (single-feature model) or that factor combined with structural protein EMP scores (combination model) were generated for every time point across each minimum death threshold (STAR Methods). Following model generation, the adjusted coefficient of determination (R2) and significance level of each individual model was extracted and aggregated by dependent variable (Figure S8). Single-feature models were characterized by low R2 ( = 0.0262), while combination models showed significant improvement ( = 0.496). Similarly, combination models demonstrated a substantially higher proportion of statistical significance (Figure S8B). To determine the set of features that produce the best-fitting model, all possible combinations of explanatory factors (risk factors and structural protein EMP score) were tested. Subsequently, the top 10 performing models, ranked by adjusted R2 value, were selected for analysis (Figure 4B). The identified models were found to be largely significant (average proportion of significant regressions = 72%) and produce strong fits to the data (average R2 = 0.7).
Figure 4.
Analysis of other SARS-CoV-2 covariates with observed SARS-CoV-2 population mortality and development of an integrative model
(A) A total of 12 covariates associated with SARS-CoV-2 mortality on the individual patient level were assessed for correlation with population-level mortality (Table S2, B). The correlation of each country-level covariate was determined at each time point after a minimum death threshold was met (line color). The x axis represents the number of days (normalized) following when a minimum death threshold was met, and the y axis indicates the observed effect size for that covariate at a given time point. Correlations achieving statistical significance are colored with a red dot.
(B) All possible combinations of covariates were used to fit a linear model. The top 10 models, ranked by median adjusted R2 (red bars), were identified. The proportion of regressions performed by that model that were found to be statistically significant (F-test p value ≤ 0.05) are represented by the blue bars.
Analysis of the dependent variables included in the top-performing models revealed that all models used structural protein EMP scores followed by deaths per million due to complications from chronic obstructive pulmonary disease (COPD) (90% of models). The median model size included 3 features, with a maximum of 5 features and a minimum of 2 features. The model producing the best fit (median R2 = 0.791) consisted of structural protein EMP scores, gender demographics, number of deaths due to COPD complications, the proportion of the population older than age 65 years, and proportion of the population that is overweight (Figure 4B). These results further indicate the robustness of the structural protein EMP score as a population-level risk descriptor.
Discussion
In the present study, we uncover evidence supporting an association between population SARS-CoV-2 infection outcome and MHC-I genotype. In line with related work highlighting the relationship between total epitope load and HIV viral control,33 we arrive at a working model that MHC-I alleles presenting more unique SARS-CoV-2 epitopes will be associated with lower mortality due to a higher number of potential T cell targets. The SARS-CoV-2 binding capacities of 52 common MHC-I alleles were assessed using the EnsembleMHC prediction platform. These predictions identified 971 high-confidence MHC-I peptides out of a candidate pool of nearly 3.5 million. In agreement with other in silico studies,15,34 the assignment of the predicted peptides to their respective MHC-I alleles revealed an uneven distribution in the number of peptides attributed to each allele. We discovered that the MHC-I peptide-allele distribution originating from the full SARS-CoV-2 proteome undergoes a notable rearrangement when considering only peptides derived from viral structural proteins. The structural protein-specific peptide-allele distribution produced a distinct hierarchy of allele-binding capacities. This finding has important clinical implications as a majority of SARS-CoV-2 specific CD8+ T cell response is directed toward SARS-CoV-2 structural proteins.8 Therefore, patients who express MHC-I alleles enriched with a large potential repertoire of SARS-CoV-2 structural proteins peptides may benefit from a broader CD8+ T cell immune response.
The variations in SARS-CoV-2 peptide-allele distributions were analyzed at epidemiological scale to track its impact on country-specific mortality. Each of the 23 countries were assigned a population SARS-CoV-2 binding capacity (or EMP score) based on the individual binding capacities of the selected 52 MHC-I alleles weighted by their endemic population frequencies. This hierarchization revealed a strong inverse correlation between EMP score and observed population mortality, indicating that populations enriched with high SARS-CoV-2 binding capacity MHC-I alleles may be better protected. The correlation was shown to be stronger when calculating the EMP scores with respect to only structural proteins, reinforcing their relevance to viral immunity. Finally, the molecular origin of the 108 predicted peptides specific to SARS-CoV-2 structural proteins revealed that they are derived from enriched regions with a minimal predicted impact from amino acid sequence polymorphisms.
The utility of structural protein EMP scores was further supported by a multivariate analysis of additional SARS-CoV-2 risk factors. These results emphasized the relative robustness of structural protein EMP scores as a population risk assessment tool. Furthermore, a linear model based on the combination of structural protein EMP scores and select population-level risk factors was identified as a potential candidate for a predictive model for pandemic population severity. As such, the incorporation of the structural protein EMP score in more sophisticated models will likely improve epidemiological modeling.
To achieve the highest level of accuracy in MHC-I predictions, the most up-to-date versions of each component algorithm were used. However, this meant that several of the algorithms (MHCflurry, netMHCpan-EL-4.0, and MixMHCpred) were benchmarked against subsets of mass spectrometry data that were used in the original training of these MHC-I prediction models. While this could result in an unfair weight applied to these algorithms in peptideFDR calculation, the individual FDRs of MHCflurry, netMHCpan-EL-4.0, and MixMHCpred were comparable to algorithms without this advantage (Figure S1C). Furthermore, the peptide selection of SARS-CoV-2 peptides was shown to be highly cooperative within EnsembleMHC (Figure S3A), and individual algorithms failed to replicate the strong observed correlations between population-binding capacity and observed SARS-CoV-2 mortality (Figure S7E).
In the future, the presented model could be applied to predict individual T cell capacity to mount a robust SARS-CoV-2 immune response. Evolutionary divergence of patient MHC-I genotypes has been shown to be predictive of the response to immune checkpoint therapy in cancer and HIV.35,36 However, confirmation will require large datasets associating individual patient MHC-I genotype and outcome. In addition, the future use of EnsembleMHC to design personalized T cell vaccines will require broad experimental validation of high-scoring peptides, since EnsembleMHC predicts MHC-I peptides with a high probability of antigen presentation as opposed to directly predicting peptide immunogenicity. While previous work has determined that a majority of successfully presented viral MHC-I peptides are immunogenic,37 there is an expectation that some presented SARS-CoV-2 MHC-I peptides will fail to produce an immune response.
The versatility of the proposed model will be improved by the consideration of additional MHC-I alleles. To reduce the presence of confounding factors, EnsembleMHC was parameterized on only a subset of common MHC-I alleles that had strong existing experimental validation. While the selected MHC-I alleles are among some of the most common, personalized risk assessment will require consideration of the full patient MHC-I genotype. The continued mass spectrometry-based characterization of MHC-I peptide-binding motifs will help in this regard. However, due to the large potential sequence space of the MHC-I protein, extension of this model will likely require the inference of binding motifs based on MHC variant clustering.
Limitations of study
This work demonstrated a strong association between a population-level metric, SARS-CoV-2 MHC-I peptide-binding capacity, and SARS-CoV-2 mortality rate by country. Other risk factors for SARS-CoV-2-specific mortality have been reported, including comorbidities, healthcare infrastructure, age, and gender. These risk factors are predicted to have a significant impact on individual patient outcome, which is not evaluated in this study. Other genetic determinants of severity, such as angiotensin-converting enzyme 2 (ACE2) polymorphism, were not considered.38 The impact of MHC-I genotype and SARS-CoV-2 antigen presentation capacity on outcomes will require the integration of individual patient genetic and clinical data. While this study evaluated EnsembleMHC for population SARS-CoV-2 binding capacity, its use has not yet been validated for other applications, such as other infectious diseases or individual MHC-I binding predictions.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| MHC-I peptide Mass spectrometry data | Sarkizova et al.17 | https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp |
| SARS-CoV-2 reference sequence | Wu et al.39 | https://www.ncbi.nlm.nih.gov/nuccore/MN908947.3/ |
| SARS-CoV-2 polymorphism data | Ahmed et al.40 | https://covidep.ust.hk/ |
| SARS-CoV-2 spike and Envelope protein crystal structure | RCSB (6VXX, 5X29) | http://www.rcsb.org/ |
| predicted structure of nucleocapsid and membrane proteins | Zhang et al.41 | https://zhanglab.ccmb.med.umich.edu/COVID-19/ |
| HLA frequency data | González-Galarza et al.18 | http://allelefrequencies.net/ |
| JHU CSSE COVID-19 Data | Dong et al.42 | https://github.com/CSSEGISandData/COVID-19 |
| population covariate data | Global Health Observatory data repository | https://apps.who.int/gho/data/node.main) |
| Viral peptides with known immunogenicity | IEDB | https://www.iedb.org/ |
| Software and algorithms | ||
| R 4.0 | R Core Team43 | https://www.r-project.org |
| EnsembleMHC | This paper | https://github.com/eawilson-CompBio/EnsembleMHC-Covid |
| VMD | Humphrey et al.44 | https://www.ks.uiuc.edu/Research/vmd/ |
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the Lead Contact, Karen Anderson (Karen.Anderson.1@asu.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data and code generated during this study are available at EnsembleMHC-Covid (https://github.com/eawilson-CompBio/EnsembleMHC-Covid).
Method details
EnsembleMHC component binding and processing prediction algorithms
EnsembleMHC incorporates MHC-I binding and processing predictions from 7 publicly available algorithms: MHCflurry-affinity-1.6.0,19 MHCflurry-presentation-1.6.0,19 netMHC-4.0,21 netMHCpan-4.0-EL,20netMHCstabpan-1.0,24 PickPocket-1.1,23 and, MixMHCpred-2.0.2.22 These algorithms were chosen based on the criteria of providing a free academic license, bash command line integration, and demonstrated accuracy for predicting SARS-CoV-2 MHC-I peptides with experimentally validated binding stability.45
Each of the selected algorithms cover components of MHC-I binding and antigen processing that roughly fall into two categories: ones based primarily on MHC-I binding affinity predictions and others that incorporate antigen presentation. To this end, MHCflurry-affinity, netMHC, PickPocket, and netMHCstabpan predict binding affinity based on quantitative peptide binding affinity measurements. netMHCstabpan also incorporates peptide-MHC stability measurements and PickPocket performs prediction based on binding pocket structural extrapolation. To model the effects of antigen presentation, MixMHCpred, netMHCpan-EL, and MHCflurry - presentation are trained on naturally eluted MHC-I ligands. Additionally, MHCflurry-presentation incorporates an antigen processing term.
Parameterization of EnsembleMHC using mass spectrometry data
EnsembleMHC is able to achieve high levels of precision in peptide selection through the use of allele and algorithm-specific binding affinity thresholds. These binding affinity thresholds were identified through the parameterization of each algorithm on high-quality mass spectrometry datasets.17 The mass spectrometry datasets used for algorithm parameterization were collected in the largest single laboratory MS-based characterization of MHC-I peptides presented by single MHC allele cell lines. These characteristics significantly reduces the number of artifacts introduced by differences in peptide isolation methods, mass spectrometry acquisition, and convolution of peptides in multiallelic cell lines. An overview of the EnsembleMHC parameterization is provided in supplemental figures (Figure S1A).
Fifty-two common MHC-I alleles were selected for parameterization based on the criteria that they were characterized in Sarkizova et al. datasets and that all 7 component algorithms could perform peptide binding affinity predictions for that allele. Each target peptide (observed in the MS dataset) was paired with 100 length-matched randomly sampled decoy peptides (not observed in the MS dataset) derived from the same source proteins. If a protein was less than 100 amino acids in length, then every potential peptide from that protein was extracted.
Each of the seven algorithms were independently applied to each of the 52 allele datasets. For each allele dataset, the minimum score threshold was determined for each algorithm that recovered 50% of the allele repertoire size (the total number of target peptides observed in the MS dataset for that allele). Additionally, the expected accuracy of each algorithm was assessed by calculating the observed false detection rate (the fraction of identified peptides that were decoy peptides) using the identified algorithm- and allele-specific scoring threshold. The parameterization process was repeated 1000 times for each allele through bootstrap sampling of half of the peptides in each single MHC allele dataset. The final FDR and score threshold for each algorithm at each allele was determined by taking the median value of both quantities reported during bootstrap sampling.
PeptideFDR calculation
Peptide confidence is assigned by calculating the peptideFDR. This quantity is defined as the product of the empirical FDRs of each individual algorithm that detected a given peptide. The peptideFDR is calculated using Equation 1,
| (1) |
, where N is the number of MHC-I binding and processing algorithms, ND represents an algorithm that did not detect a given peptide, and algorithmFDR represents the allele specific FDR of the Nth algorithm. The peptideFDR represents the joint probability that all MHC-I binding and processing algorithms that detected a particular peptide did so in error, and therefore returns a probability of false detection. Unless otherwise stated, EnsembleMHC selected peptides based on the criterion of a peptideFDR ≤ 5%.
Application of EnsembleMHC to tumor cell line data
Ten tumor samples were obtained from the Sarkizova et al. datasets. Tumor samples were selected for analysis if at least 50% of the expressed MHC-I alleles for that sample were included in the 52 MHC-I alleles supported by EnsembleMHC. For each dataset, decoy peptides were generated in a manner identical to the method used for algorithm parameterization on single MHC allele data.
Peptide identification by each algorithm was based on restrictive or permissive binding affinities thresholds. These thresholds correspond to commonly used score cutoffs for the identification of strong binders (restrictive) or all binders (permissive) (0.5% (percentile rank) or 50nM (IC50 value) for strong binders, and 2% (percentile rank) or 500nM (IC50 value) for all binders). Due to the lack of recommend score thresholds for MHCflurry-presentation-1.6.0, the raw presentation score was converted to a percentile score using presentation scores produced by 100,000 randomly generated peptides.
SARS-CoV-2 reference sequence
MHC-I peptide predictions for the SARS-CoV-2 proteome were performed using the Wuhan-Hu-1 (GenBank: MN908947.3) reference sequence.39 All potential 8-14-mer peptides (n = 67,207) were derived from the open reading frames in the reported proteome, and each peptide was evaluated by the EnsembleMHC workflow.
SARS-CoV-2 polymorphism analysis and protein structure visualizations
Polymorphism analysis of SARS-CoV-2 structural proteins were performed using 102,148 full length protein sequences obtained from the COVIDep database.40 Solved structures for the E (PDB: 5X29) and S (PDB: 6VXX) proteins (https://www.rcsb.org/)46 and predicted structures for the M and N proteins41 were visualized using VMD.44
Application of EnsembleMHC to determine population SARS-CoV-2 binding capacity
The peptides identified by the EnsembleMHC workflow were used to assess the SARS-CoV-2 population binding capacity by weighing individual MHC allele SARS-CoV-2 binding capacities by regional expression (for a schematic representation see Figure S2).
The selection of countries included in the EnsembleMHC population binding capacity assessment was based on several criteria regarding the underlying MHC-I allele data for that country (Figure S2). The MHC-I allele frequency data used in our model was obtained from the Allele Frequency Net Database (AFND),18 and frequencies were aggregated by country. However, the currently available population-based MHC-I frequency data has specific limitations and variances, which we have addressed as follows:
MHC allele data coverage within countries
We define MHC-typing breadth as the diversity of identified MHC-I alleles within a given country, and its depth as the ability to accurately achieve 4-digit MHC-I genotype resolution. High variability was observed in both the MHC-I genotyping breadth and depth (Figure S2 inset). Consequently, additional filter-measures were introduced to capture potential sources of variance within the analyzed cohort of countries. The thresholds for filtering the country-wide MHC-I allele data were set based on meeting two inclusion criteria: 1) MHC genotyping of at least 1000 individuals have been performed in that population, avoiding skewing of allele frequencies due to small sample size. 2) MHC-I allele frequency data for at least 51 of the 52 (95%) MHC-I alleles for which the EnsembleMHC was parameterized to predict, ensuring full power of the EnsembleMHC workflow.
Ethnic communities within countries
In instances where the MHC-I allele frequencies would pertain to more than one community, the reported frequencies were counted toward both contributing groups. For example, the MHC-I frequency data pertaining to the Chinese minority in Germany would be factored into the population MHC-I frequencies for both China and Germany. In doing so, this treatment resolves both ancestral and demographic MHC-I allele frequencies.
Normalization of MHC allele frequency data
The focus of this work was to uncover potential differences in SARS-CoV-2 MHC-I peptide presentation dynamics induced by the 52 selected alleles within a population. Accordingly, the MHC-I allele frequency data was carefully processed in order to maintain important differences in the expression of selected alleles, while minimizing the effect of confounding factors.
The MHC-I allele frequency data for a given population was first filtered to the 52 selected alleles. These allele frequencies were then converted to the theoretical total number of copies of that allele within the population (allele count) following
| (2) |
, where allelefreq is the observed allele frequency in a population and n is the population sample size for which that allele frequency was measured. The allele count is then normalized with respect to the total allele count of selected 52 alleles within that population using the following relationship
| (3) |
, where i is one of the 52 selected alleles. This normalization is required to overcome the potential bias toward hidden alleles (alleles that are either not well characterized or not supported by EnsembleMHC) as would be seen using alternative allele frequency accounting techniques (e.g., sample-weighted mean of selected allele frequencies or normalization with respect to all observed alleles within a population; Figure S6C). The SARS-CoV-2 binding capacity of these hidden alleles cannot be accurately determined using the EnsembleMHC workflow, and therefore important potential relationships would be obscured.
EnsembleMHC population score
The predicted ability of a given population to present SARS-CoV-2 derived peptides was assessed by calculating the EnsembleMHC Population (EMP) score. After the MHC-I allele frequency data filtering steps, 23 countries were included in the analysis. The calculation of the EnsembleMHC population score is as follows
| (4) |
, where norm allele count is the observed normalized allele count for a given allele in a population, Nnorm allele count ≠ 0 is the number of the 52 select alleles detected in a given population (range 51-52 alleles), and peptidefrac is the peptide fraction or the fraction of total predicted peptides expected to be presented by that allele within the total set of predicted peptides with a peptideFDR ≤ 5%.
Death rate-presentation correlation
The correlation between the EMP score and the observed deaths per million within the cohort of selected countries was calculated as a function of time. SARS-Cov-2 data covering the time dependent global evolution of the SARS-CoV-2 pandemic was obtained from Johns Hopkins University Center for Systems Science and Engineering42 covering the time frame of January 22nd to April 9th 2020. The temporal variations in occurrence of community spread observed in different countries were accounted for by rescaling the time series data relative to when a certain minimum death threshold was met in a country. This analysis was performed for minimum death thresholds of 1-100 total deaths by day 0, and correlations were calculated at each day sequentially following day 0 until there were fewer than 8 countries remaining at that time point. The upper-limit of 100-deaths was chosen due to a steep decline in average statistical power observed with day 1 death thresholds greater than 100 deaths (Figure S6E).
The time death correlation was computed using Spearman’s rank correlation coefficient (two-sided). This method was chosen due to the small sample size and non-normality of the underlying data (Figure S6D). The reported correlations of EMP score and deaths per million using other correlation methods can be seen in supplemental Figure S6A.
The low statistical power for some of the obtained correlations were addressed by calculating the Positive Predictive Value (PPV) of all correlations using the following equation47
| (5) |
,where 1 is the statistical power of a given correlation, R is the pre-study odds, and is the significance level. A PPV value of ≥ 95% is analogous to a p value of ≤ 0.05. Due to an unknown pre-study odd (probability that probed effect is truly non null), R was set to 1 in the reported correlations. The significance of partitioning high risk and low risk countries based on median EMP score was determined using Mann-Whitney U-test. Significance values were corrected for multiple tests using the Benjamini-Hochberg procedure.30
Sub-sampling of peptides from the Full SARS-CoV-2 proteome
108 unique peptides, derived from the Full SARS-CoV-2 proteome and passing the 5% peptideFDR filter, were randomly sampled. Then, the time series EMP score - death per million correlation analysis used to generate Figure 3 was applied to each sampled peptide set. The sub-sampling procedure was repeated for 1,000 iterations (Figure S7A). To quantitatively describe the similarity of the distributions, the Kullback-Leibler divergence (KLD), a measure of divergence between two probability distributions, was calculated for the correlation distribution of each sub-sample iteration relative to either the correlation distribution of the Full SARS-CoV-2 proteome or SARS-CoV-2 structural proteins (Figure S7B).
Additional SARS-CoV-2 risk factors
Twelve potential SARS-CoV-2 risk factors (Table S2) were selected for analysis. Country-specific data for each risk factor was obtained from the Global Health Observatory data repository provided by the World Health Organization (https://apps.who.int/gho/data/node.main). Countries were selected for analysis based on the criteria of having reported data in the WHO datasets and inclusion in the set of 23 countries for which EnsembleMHC population scores were assigned (Table S2A). Data regarding the total number of noncommunicable disease-related deaths (Cardiovascular disease, Chronic obstructive pulmonary disease, and Diabetes mellitus) were converted to deaths per million.
Correlation of additional risk factors with observed deaths per million
Correlation analysis of each additional factor was carried out in a similar manner to that of the EnsembleMHC population score. In short, Spearman’s correlation coefficient between each individual factor and observed deaths per million was estimated as a function of time from when a specified minimum death threshold was met (Figure 4). The significance level was set to p ≤ 0.05 and significant PPV was set to PPV ≥ 0.95 (Equation 8).
Linear models of SARS-CoV-2 mortality
For the single and combination models, individual linear models were constructed for each considered death threshold as a function of time (similar to the univariate correlation analysis). Each model consisted of 1 (a single socioeconomic or health-related risk factor) or 2 (a combination of 1 risk factor and structural protein EMP score) dependent variables and deaths per million as the independent variable. The adjusted R2 value and statistical significance of the model (F-test) were then extracted from each individual model and aggregated by dependent variable (Figure S8A).
The best performing models were determined by assessing all possible combinations of factors including structural protein EMP score. This resulted in the consideration of 4,083 different linear models. The top performing models were then selected by ranking each model by median adjusted R2.
Immunogenic viral peptide analysis
Individual algorithms were assessed for ability to prioritize viral peptides with known immunogenicity by calculating the precision (experimentally validated peptides / putative non-immunogenic peptides) when selecting n number of top scoring peptides as determined by a given algorithm. For example, if n = 25, then the precision of each algorithm would be calculated based on the top 25 highest scoring peptides according that algorithm. A Viral peptide dataset was generated by extracting all potential 8 – 14-mer peptides from the Hepatitis-C genome polyprotein (P26664), the Dengue virus genome polyprotein (P14340), and the HIV-1 POL-GAG protein (P03369). The resulting peptides were then checked against the Immune Epitope database48 (IEDB, https://www.iedb.org/) to identify peptides with experimentally validated immunogenicity. This resulted in the generation of a dataset comprised of 616 experimentally validated immunogenic peptides and 54,663 putative non-immunogenic peptides (this includes peptides experimentally determined to non-immunogenic or peptides with unknown immunogenicity). To benchmark EnsembleMHC against other Ensemble-based MHC-I peptide prediction algorithms, netCTLpan49 and MHCcons50 were included for comparison purposes.
Quantification and statistical analysis
Statistical tests were performed using R 4.0.343. All effect size estimations were performed using Spearman’s rank correlation. Mann-Whitney U test was used to test for significant testing of death rate stratification between countries with high and low EnsembleMHC score. The threshold for statistical significance was set to p values of ≤ 0.05 or positive predictive value of PPV ≥ 0.95. Where indicated, p value correction for multiple testing was accomplished using the Benjamini-Hochberg procedure.
Acknowledgments
We would like to thank Drs. Diego Chowell, Matthew Scotch, Sri Krishna, and Shay Ferdosi as well as Mr. John Vant, Mr. Ryan Boyd, and Ms. Mollie Peters for critical feedback and discussion. Finally, we would like to thank ASU Research computing for allocating the computational resources used in this study. We acknowledge start-up funds from the SMS and CASD at Arizona State University. A.S. acknowledges funding from the CAREER award from NSF (MCB-1942763) and the Gordon and Betty Moore foundation.
Author Contributions
E.A.W., A.S., and K.S.A. contributed to the study design and interpretation. E.A.W. performed the data collection and analysis. E.A.W., G.H., A.S., and K.S.A contributed to writing the manuscript. All of the authors reviewed and approved the final version of the manuscript.
Declaration of interests
E.A.W., A.S., and K.S.A. have a patent application on EnsembleMHC and T cell targeting using epitopes described in this article, licensed to SafeGen Therapeutics (K.S.A., co-founder). The authors declare no other competing interests.
Published: February 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100221.
Contributor Information
Abhishek Singharoy, Email: asinghar@asu.edu.
Karen S. Anderson, Email: karen.anderson.1@asu.edu.
Supplemental information
All predicted peptides with a peptideFDR ≤0.05.
All data pertaining to the correlations between EMP score and deaths per million. This includes rho estimates, 95% CI, non-normalized day values, and sample size for each correlation.
References
- 1.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296:E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 2014;59:118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janice Oh H.-L., Ken-En Gan S., Bertoletti A., Tan Y.J. Understanding the T cell immune response in SARS coronavirus infection. Emerg. Microbes Infect. 2012;1:e23. doi: 10.1038/emi.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng O.-W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 8.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matzaraki V., Kumar V., Wijmenga C., Zhernakova A. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 2017;18:76. doi: 10.1186/s13059-017-1207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin M., Tseng H.-K., Trejaut J.A., Lee H.-L., Loo J.-H., Chu C.-C., Chen P.-J., Su Y.-W., Lim K.H., Tsai Z.-U. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med. Genet. 2003;4:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S.-F., Chen K.H., Chen M., Li W.Y., Chen Y.J., Tsao C.H., Yen M.Y., Huang J.C., Chen Y.M. Human-leukocyte antigen class I Cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral Immunol. 2011;24:421–426. doi: 10.1089/vim.2011.0024. [DOI] [PubMed] [Google Scholar]
- 12.Ng M.H., Lau K.M., Li L., Cheng S.H., Chan W.Y., Hui P.K., Zee B., Leung C.B., Sung J.J. Association of human-leukocyte-antigen class I (B∗0703) and class II (DRB1∗0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J. Infect. Dis. 2004;190:515–518. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng M., Cheng S.H., Lau K.M., Leung G.M., Khoo U.S., Zee B.C.W., Sung J.J.Y. Immunogenetics in SARS: a case-control study. Hong Kong Med. J. 2010;16(5 Suppl 4):29–33. [PubMed] [Google Scholar]
- 14.Sanchez-Mazas A. HLA studies in the context of coronavirus outbreaks. Swiss Med. Wkly. 2020;150:w20248. doi: 10.4414/smw.2020.20248. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen A., David J.K., Maden S.K., Wood M.A., Weeder B.R., Nellore A., Thompson R.F. Human leukocyte antigen susceptibility map for SARS-CoV-2. J. Virol. 2020;94 doi: 10.1128/JVI.00510-20. e00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao W., Sher X. Systematically benchmarking peptide-MHC binding predictors: from synthetic to naturally processed epitopes. PLoS Comput. Biol. 2018;14:e1006457. doi: 10.1371/journal.pcbi.1006457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkizova S., Klaeger S., Le P.M., Li L.W., Oliveira G., Keshishian H., Hartigan C.R., Zhang W., Braun D.A., Ligon K.L. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 2020;38:199–209. doi: 10.1038/s41587-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González-Galarza F.F., Takeshita L.Y., Santos E.J., Kempson F., Maia M.H., da Silva A.L., Teles e Silva A.L., Ghattaoraya G.S., Alfirevic A., Jones A.R., Middleton D. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43(D1):D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Donnell T.J., Rubinsteyn A., Laserson U. MHCflurry 2.0: Improved Pan-Allele Prediction of MHC Class I-Presented Peptides by Incorporating Antigen Processing. Cell Syst. 2020;11:42–48.e7. doi: 10.1016/j.cels.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreatta M., Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32:511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassani-Sternberg M., Chong C., Guillaume P., Solleder M., Pak H., O Gannon P., Kandalaft L.E., Coukos G., Gfeller D. Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen predictions and identifies allostery regulating HLA specificity. PLoS Comput. Biol. 2017;13:e1005725. doi: 10.1371/journal.pcbi.1005725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H., Lund O., Nielsen M. The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: application to MHC-peptide binding. Bioinformatics. 2009;25:1293–1299. doi: 10.1093/bioinformatics/btp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmussen M., Fenoy E., Harndahl M., Kristensen A.B., Nielsen I.K., Nielsen M., Buus S. Pan-specific prediction of peptide–MHC class I complex stability, a correlate of T cell immunogenicity. J. Immunol. 2016;197:1517–1524. doi: 10.4049/jimmunol.1600582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul S., Weiskopf D., Angelo M.A., Sidney J., Peters B., Sette A. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J. Immunol. 2013;191:5831–5839. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols K. False discovery rate procedures. In: Penny W., Friston K., Ashburner J., Kiebel S., Nichols T., editors. Statistical Parametric Mapping. Elsevier; 2007. pp. 246–252. [Google Scholar]
- 27.Nielsen M., Andreatta M., Peters B., Buus S. Immunoinformatics: Predicting Peptide–MHC Binding. Annu. Rev. Biomed. Data Sci. 2020;3:191–215. doi: 10.1146/annurev-biodatasci-021920-100259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trolle T., McMurtrey C.P., Sidney J., Bardet W., Osborn S.C., Kaever T., Sette A., Hildebrand W.H., Nielsen M., Peters B. The length distribution of class I–restricted T cell epitopes is determined by both peptide supply and MHC allele–specific binding preference. J. Immunol. 2016;196:1480–1487. doi: 10.4049/jimmunol.1501721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rapin N., Hoof I., Lund O., Nielsen M. The MHC motif viewer: a visualization tool for MHC binding motifs. Curr. Protoc. Immunol. 2010;Chapter 18:17. doi: 10.1002/0471142735.im1817s88. [DOI] [PubMed] [Google Scholar]
- 30.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 31.Williamson E.J., Walker A.J., Bhaskara K., Bacon S., Bates C., Morton C.E., Curtis H.J., Mehrkar A., Evans D., Inglesby P. Factors associated with COVID-19 death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Lusignan S., Dorward J., Correa A., Jones N., Akinyemi O., Amirthalingam G., Andrews N., Byford R., Dabrera G., Elliot A. Risk factors for SARS-CoV-2 among patients in the Oxford Royal College of General Practitioners Research and Surveillance Centre primary care network: a cross-sectional study. Lancet Infect. Dis. 2020;20:1034–1042. doi: 10.1016/S1473-3099(20)30371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rolland M., Heckerman D., Deng W., Rousseau C.M., Coovadia H., Bishop K., Goulder P.J., Walker B.D., Brander C., Mullins J.I. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS ONE. 2008;3:e1424. doi: 10.1371/journal.pone.0001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell K.M., Steiner G., Wells D.K., Ribas A., Kalbasi A. Prediction of SARS-CoV-2 epitopes across 9360 HLA class I alleles. bioRxiv. 2020 doi: 10.1101/2020.03.30.016931. [DOI] [Google Scholar]
- 35.Chowell D., Krishna C., Pierini F., Makarov V., Rizvi N.A., Kuo F., Morris L.G.T., Riaz N., Lenz T.L., Chan T.A. Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat. Med. 2019;25:1715–1720. doi: 10.1038/s41591-019-0639-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arora J., Pierini F., McLaren P.J., Carrington M., Fellay J., Lenz T.L. HLA heterozygote advantage against HIV-1 is driven by quantitative and qualitative differences in HLA allele-specific peptide presentation. Mol. Biol. Evol. 2020;37:639–650. doi: 10.1093/molbev/msz249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croft N.P., Smith S.A., Pickering J., Sidney J., Peters B., Faridi P., Witney M.J., Sebastian P., Flesch I.E.A., Heading S.L. Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc. Natl. Acad. Sci. USA. 2019;116:3112–3117. doi: 10.1073/pnas.1815239116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., Wen F., Huang X., Ning G., Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmed S.F., Quadeer A.A., McKay M.R. COVIDep: a web-based platform for real-time reporting of vaccine target recommendations for SARS-CoV-2. Nat. Protoc. 2020;15:2141–2142. doi: 10.1038/s41596-020-0358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C., Zheng W., Huang X., Bell E.W., Zhou X., Zhang Y. Protein structure and sequence re-analysis of 2019-nCoV genome refutes snakes as its intermediate host or the unique similarity between its spike protein insertions and HIV-1. J. Proteome Res. 2020;19:1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2020 https://www.R-project.org/ [Google Scholar]
- 44.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 45.Prachar, M., Justesen, S., Bisgaard Steen-Jensen, D., Thorgrimsen, S., Jurgons, E.,Winther, O., and Bagger, F.O. (2020). COVID-19 Vaccine Candidates: Prediction and Validation of 174 SARS-CoV-2 Epitopes. bioRxiv 10.1101/2020.03.20.000794. [DOI] [PMC free article] [PubMed]
- 46.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Button K.S., Ioannidis J.P., Mokrysz C., Nosek B.A., Flint J., Robinson E.S., Munafò M.R. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013;14:365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- 48.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stranzl T., Larsen M.V., Lundegaard C., Nielsen M. NetCTLpan: pan-specific MHC class I pathway epitope predictions. Immunogenetics. 2010;62:357–368. doi: 10.1007/s00251-010-0441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karosiene E., Lundegaard C., Lund O., Nielsen M. NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics. 2012;64:177–186. doi: 10.1007/s00251-011-0579-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All predicted peptides with a peptideFDR ≤0.05.
All data pertaining to the correlations between EMP score and deaths per million. This includes rho estimates, 95% CI, non-normalized day values, and sample size for each correlation.
Data Availability Statement
All data and code generated during this study are available at EnsembleMHC-Covid (https://github.com/eawilson-CompBio/EnsembleMHC-Covid).