Abstract
Hyperinflammatory response caused by infections such as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) increases organ failure, intensive care unit admission, and mortality. Cytokine storm in patients with Coronavirus Disease 2019 (COVID-19) drives this pattern of poor clinical outcomes and is dependent upon the activity of the transcription factor complex nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kappaB) and its downstream target gene interleukin 6 (IL6) which interacts with IL6 receptor (IL6R) and the IL6 signal transduction protein (IL6ST or gp130) to regulate intracellular inflammatory pathways. In this study, we compare transcriptomic signatures from a variety of drug-treated or genetically suppressed (i.e. knockdown) cell lines in order to identify a mechanism by which antidepressants such as fluoxetine demonstrate non-serotonergic, anti-inflammatory effects. Our results demonstrate a critical role for IL6ST and NF-kappaB Subunit 1 (NFKB1) in fluoxetine’s ability to act as a potential therapy for hyperinflammatory states such as asthma, sepsis, and COVID-19.
Abbreviation: SSRI, selective serotonin reuptake inhibitor; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; COVID-19, Coronavirus Disease 2019; SHLH, secondary hemophagochytic lymphohistiocytosis; LINCS, library of integrated network-based cellular signatures; CGS, consensus gene knockdown signatures; IL6, Interleukin 6; IL6R, IL6 Receptor; sIL6R, Soluble IL6R; IL6ST, Interleukin 6 Signal Transducer; NF-kappaB, nuclear factor kappa-light-chain-enhancer of activated B cells; NFKB1, NF-kappaB Subunit 1; KD, knockdown
Keywords: Cytokine IL6, Transcription factor NF-κB, Inflammation, Cytokine storm, Antidepressants, Fluoxetine, Selective serotonin reuptake inhibitors, SSRIs, Severe acute respiratory syndrome coronavirus 2, SARS-CoV-2, Coronavirus disease 2019, COVID-19, Sepsis, Nuclear factor kappa B subunit 1
Graphical Abstract
1. Introduction
Evidence for the role of interleukin 6 (IL6) mediated cytokine storm in the development of Coronavirus Disease 2019 (COVID-19) associated morbidity and mortality suggests that therapeutic agents which decrease IL6 signal transduction may prevent serious COVID-19 disease outcomes [1]. Clinical consequences of COVID-19 infection such as multisystem organ failure, intensive care unit admission, and death result from an exaggerated immune response whereby increased IL6 drives a destructive cytokine storm [2], [3], [4], [5], [6]. This pattern of poor clinical outcomes parallels the Secondary Hemophagocytic Lymphohistiocytosis (SHLH) seen in another coronavirus-induced illness, Severe Acute Respiratory Syndrome (SARS) [7], [8]. Indeed, many infections result in an IL6-mediated cytokine storm that drives negative clinical outcomes. NF-kappaB-mediates increases in the transcriptional expression of the proinflammatory cytokine IL6 [9], [10]. Binding of IL6 to its receptor complex (IL6R) activates the IL6 signal transduction protein (IL6ST or gp130), a subunit of the IL6 receptor complex. IL6ST activation is integral to many intracellular cytokine-mediated inflammatory pathways, including the cytokine storm / cytokine release syndrome in COVID-19 patients [11]. Further, in vitro, in vivo, and clinical data demonstrate that many selective serotonin reuptake inhibitors (SSRIs) decrease IL6 signaling activity and reduce hyperinflammatory states [12], [13], [14], [15], [16], [17]. The SSRI fluoxetine causes pronounced inhibition of NF-kappaB signaling and IL6 expression correlating with decreased illness severity across a variety of animal disease models including cancer, pulmonary inflammation, and sepsis [17], [18], [19], [20], [21], [22].
Fluoxetine’s well-established safety profile and its inhibition of IL6 signal transduction suggests that this drug may be advantageous when used early in COVID-19 infection to prevent or reduce the cytokine storm. Evidence supporting this hypothesis can be found across multiple animal models of disease whereby fluoxetine not only reduced IL6 signaling cascades but also prevented end organ damage [16], [18], [23], [24], [25]. In rat models of chronic obstructive pulmonary disease, fluoxetine decreased lung injury by inhibiting the transcription factor NF-kappaB, thus reducing downstream IL6 production [18]. In a lipopolysaccharide (LPS)-induced sepsis model of hyperinflammatory illness, pretreatment with fluoxetine outperformed pretreatment with corticosteroids and resulted in diminished end organ damage that included decreased incidence of pulmonary arterial hypertension, decreased pulmonary arterial muscularization, and decreased extracellular matrix remodeling [23], [24]. Fluoxetine treatment in rats decreased bronchial asthma severity concurrent with reduced levels of inflammatory cytokines, including IL6 [24]. Fluoxetine has also demonstrated inhibition of NF-kappaB with corresponding decreases in inflammatory cytokines like IL6 in models of septic shock and allergic asthma, as well as in patients with depression [16], [25].
Recently, Zhou et al. performed phylogenetic analyses of multiple human coronavirus genomes to identify evolutionarily conserved targets for SARS-CoV-2 therapeutics. Their quantification of interactions between the human coronavirus-host interactome and subsequent gene set enrichment analyses comparing coronavirus-infected host cells and drug-treated human cells identified the SSRI paroxetine as a novel COVID-19 therapeutic candidate [26]. An observational study correlated antidepressant therapy with decreased risk of intubation or death for hospitalized patients with COVID-19 [27]. Preliminary reports also suggest that fluoxetine demonstrates direct inhibition of SARS-CoV-2 viral protein expression [28], although the present study is focused on fluoxetine’s anti-inflammatory activity in human pathophysiology.
To investigate the potential clinical utility of using fluoxetine to reduce IL6 mediated inflammation in COVID-19 patients and to elucidate the mechanism by which fluoxetine acts as an anti-inflammatory agent, we used transcriptomic signatures from the Library of Integrated Network-Based Cellular Signatures (LINCS) data repository to perform bioinformatic analyses comparing fluoxetine with 2 other antidepressants—bupropion and paroxetine—as well as an established anti-inflammatory therapeutic—the corticosteroid dexamethasone. The transcriptomic changes caused by targeted knockdown of 27 inflammatory genes potentially involved in fluoxetine’s anti-inflammatory mechanism of action were compared to the transcriptomic changes caused by antidepressant drug therapy. Our resultant data highlights the significance of IL6ST and NF-kappaB in fluoxetine’s anti-inflammatory mechanism of action and supports further exploration of fluoxetine as a candidate drug in the treatment of inflammatory pathologies such as those contributing to COVID-19.
2. Methods
2.1. Differential gene expression signatures
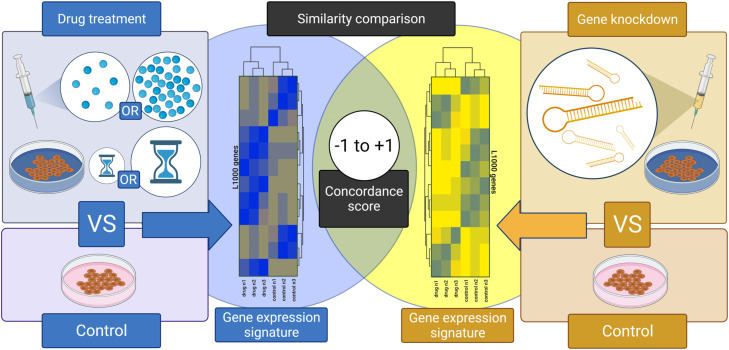
Fig. 1 summarizes our methodological workflow. We collected transcriptomic data from the Library of Integrated Network-Based Cellular Signatures (LINCS). LINCS is a program initiated by the National Institute of Health (NIH) to create a comprehensive repository of molecular interactions and responses to internal and external stressors. The LINCS project uses the L1000 assay which measures the expression of 978 “hub” genes which represent ~82% of the information present in the transcriptome. The iLINCS Portal (www.ilincs.org) provides a convenient gateway to access these signatures. We used the iLINCS application programing interface (API) to download differential gene expression signatures. We acquired all available signatures for fluoxetine (33), paroxetine (65), bupropion (29), or dexamethasone (309) treated cell lines. In total, 436 signatures were obtained for all 4 drug treatments, with data generated in 45 different cell lines, at different timepoints (6–24 h) and different doses (0.01uM - 80uM) (Table S1). To create high-confidence gene signatures, differential gene expression data were filtered such that only those genes with a log-fold change (LFC) ≥ 0.85 or ≤ −0.85 in drug-treated versus corresponding control cell lines were included in downstream gene signature analyses. This LFC threshold was found to effectively reduce excess noise from low-difference genes and reduce signatures to their most significant components O’Donovan, 2021, [30]. The resulting signatures were searched against the iLINCS Consensus Gene Knockdown (CGS) database to generate similarity indices between (a) the drug treatment signatures within a given cell line and (b) individual inflammatory gene knockdown signatures for 27 inflammatory genes with a potential role in fluoxetine’s anti-inflammatory mechanism of action (i.e. drug treatment gene signatures were compared to gene knockdown signatures generated only in the same cell line).
Fig. 1.
Experimental Design and Analytical Workflow. We generated differential gene expression signatures produced by fluoxetine, paroxetine, bupropion, or dexamethasone drug treatments across a variety of concentrations and treatment durations compared to control, untreated, wild-type cells. We also generated differential gene expression signatures produced by genetic knockdown of inflammatory genes related to the initiation and maintenance of IL6-mediated cytokine storm. Again, these signatures are in relation to control, untreated, wild-type cells. We then compared the drug treatment signatures with the genetic knockdown signatures to quantify similarity using concordance scores. When a drug treatment signature is highly concordant with a genetic knockdown signature, it generates a value approaching +1 and allows us to surmise that this drug and this genetic knockdown induce equivalent changes in gene expression. When no significant similarities exist, the concordance score approaches zero. When a drug treatment signature is highly discordant with a genetic knockdown signature, it generates a value approaching −1 and allows us to surmise that this drug and this genetic knockdown induce inverted changes in gene expression.
A concordance score is an adjusted correlation coefficient that can range from –1 to 1. iLINCS only reports a match if the concordance score is ≤ −0.2 or ≥ 0.2. Concordance scores outside of those parameters are considered statistically insignificant. In this way, we identified 779 total signatures (concordance ≤ −0.2 or ≥ 0.2) derived from comparisons of the 168 drug treatment gene signatures to CGS for 27 inflammatory genes in 11 different cell lines (Table S2). We again filtered our list to include only the highest absolute concordance score (range –0.7166 to 0.7982) for each drug treatment, knockdown, and cell line combination. This resulted in 395 concordance scores comparing 27 inflammatory genes and 4 drugs of interest across 11 cell lines (Table S3). Mean concordance scores are calculated by averaging the concordance scores for a given drug-gene comparison across cell lines. Statistically significant outliers (P < 0.5) within the list of drug-gene concordance scores among cell lines containing calculated concordance scores for less than 3 drugs of interest were identified using the extreme studentized deviate method (Grubbs’ test) and subsequently excluded.
2.2. Data and bioinformatic pipeline repository access
The entire process was performed using R version 3.6.3. The entire bioinformatic pipeline including raw data, analytical scripts, and results can be reproduced using our publicly available protocol at http://dx.doi.org/10.17504/protocols.io.bscjnaun and our publicly available repository at https://doi.org/10.5281/zenodo.4546412. Figures were created with BioRender.com, GraphPad Prism 9.0.2, Adobe Illustrator 25.1, and R 3.6.3.
3. Results
To assess the potential clinical utility of fluoxetine in the prevention or mitigation of cytokine storm syndromes, and to investigate the mechanism by which fluoxetine realizes its anti-inflammatory effect, we generated differential gene expression signatures produced by fluoxetine, paroxetine, bupropion, or dexamethasone drug treatments. We also generated differential gene expression signatures produced by genetic knockdown of inflammatory genes related to the initiation and maintenance of IL6-mediated cytokine storm. We then compared the drug treatment signatures with the genetic knockdown signatures to generate concordance scores which measure the degree to which a given drug treatment parallels genetic knockdown of a given inflammatory gene. When a drug treatment signature is highly concordant with a genetic knockdown signature, we surmise that this drug and this gene knockdown induce a similar pattern of gene expression changes [31].
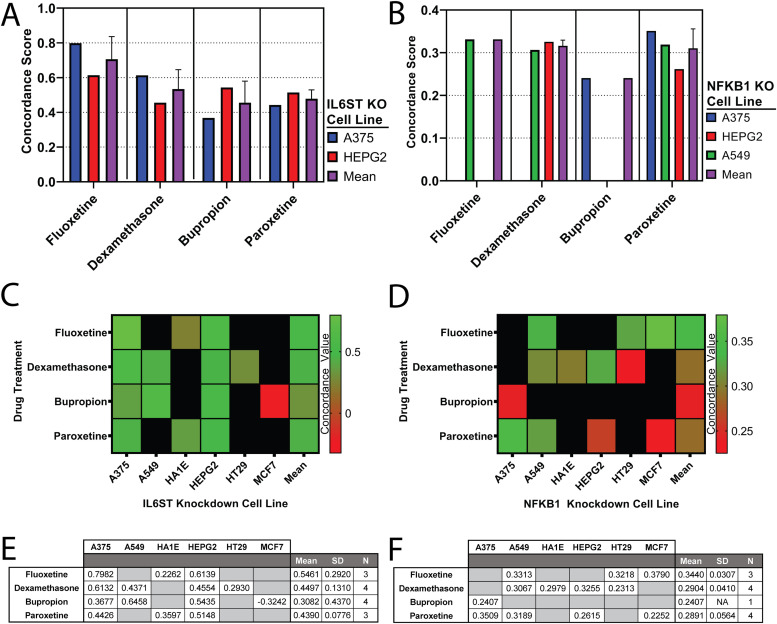
We compared gene signatures generated from cell lines that have been treated with fluoxetine, paroxetine, bupropion, and dexamethasone to matched cell lines in which IL6ST was genetically knocked down with siRNA ( Fig. 2A). In this way, the differential gene expression signatures for drug-treated A375 or HEPG2 cell lines were compared to IL6ST knockdown A375 or HEPG2 cell lines, respectively. This allowed us to quantify the degree of similarity between pharmacologically initiated mRNA changes and those resulting from targeted knockdown of a specific gene. In A375 cells, concordance scores quantifying the similarity between drug treated cells and IL6ST knockdown cells are as follows: 0.7982 for fluoxetine treated cells, 0.6132 for dexamethasone treated cells, 0.3677 for bupropion treated cells, and 0.4426 for paroxetine treated cells. In HEPG2 cells, concordance scores for the same comparisons are as follows: 0.6139 for fluoxetine treated cells, 0.4554 for dexamethasone treated cells, 0.5435 for bupropion treated cells, and 0.5148 for paroxetine treated cells. Mean concordance scores for these comparisons are as follows: 0.7061 (SD: 0.1303; N: 2) for comparisons of fluoxetine treated cells and IL6ST knockdown cells; 0.5343 (SD: 0.1116; N: 2) for comparisons of dexamethasone treated cells and IL6ST knockdown cells; 0.4556 (SD: 0.1243; N: 2) for comparisons of bupropion treated cells and IL6ST knockdown cells; and 0.4787 (SD: 0.0511; N: 2) for comparisons of paroxetine treated cells and IL6ST knockdown cells.
Fig. 2.
Quantification of similarity between fluoxetine, dexamethasone, bupropion, or paroxetine drug-treatment signatures and IL6ST or NFKB1 gene knockdown signatures. (A and B) Analyses of differential gene expression signatures elicited by fluoxetine, dexamethasone, bupropion, or paroxetine treatments compared to differential gene expression signatures elicited by (A) IL6ST knockdown or (B) NFKB1 knockdown. Similarity is quantified as concordance score. The maximum absolute concordance scores from all comparands are reported. Means derived from cell lines are also reported. (C−F) Heatmaps and corresponding data tables representing full concordance analyses comparing drug-treated cells and C, (E) matched IL6ST knockdown or D, (F) matched NFKB1 knockdown cell lines. For comprehensive coverage, the heatmaps and corresponding data tables represent additional concordance scores derived from cell lines lacking data set(s) within our inclusion parameters for one or more comparison (black boxes in heatmap; gray boxes in data table). Means and standard deviations (SD), derived from all available comparand cell lines (N) are also presented.
To further develop the relationship between our drugs of interest, IL6ST, and inflammatory pathways, we again analyzed differential gene expression signatures caused by drug treatments in A375 and HEPG2 cell lines. This time however, we compared drug treatment signatures to those produced by NFKB1 knockdown (Fig. 2B). As before, comparisons of these signatures generated concordance scores quantifying the degree of similarity between a drug treatment and NFKB1 knockdown. In A375 cells, these concordance scores are as follows: 0.2407 for bupropion treated cells, and 0.3509 for paroxetine treated cells. In HEPG2 cells, concordance scores for the same comparisons are as follows: 0.3255 for dexamethasone treated cells, and 0.2615 for paroxetine treated cells. Given our previously discussed statistical cut-off parameter for similarity, neither A375 nor HEPG2 cell lines were able to generate concordance scores for all four drugs of interest and NFKB1 knockdown. Thus, to increase the power of our NFKB1 analysis, we expanded our iLINCS query to include additional cell lines reporting transcriptomic signatures resulting from NFKB1 knockdown and at least three of our four drug treatments (Fig. 2B). In A549 cells, concordance scores quantifying the degree of similarity between drug-treated cells and NFBK1 knockdown cells are as follows: 0.3313 for fluoxetine treated cells, 0.3067 for dexamethasone treated cells, and 0.3189 for paroxetine treated cells. Mean concordance scores for these comparisons are as follows: 0.3313 (N: 1) for comparisons of fluoxetine treated cells and NFKB1 knockdown cells; 0.3161 (SD: 0.133; N: 2) for comparisons of dexamethasone treated cells and NFKB1 knockdown cells; 0.2407 (N: 1) for comparisons of bupropion treated cells and NFKB1 knockdown cells; and 0.3104 (SD: 0.453; N: 3) for comparisons of paroxetine treated cells and NFKB1 knockdown cells.
To contextualize IL6ST-drug concordance scores and NFKB1-drug concordance scores, and to increase the statistical power of our study, we further expanded our analyses to include all cell lines generating IL6ST-drug concordance scores (Fig. 2C) and NFKB1-drug concordance scores (Fig. 2D) for at least one drug of interest. In addition to the previously reported cell lines, these comparisons include the HA1E, HT29, and MCF7 cell lines. In A549 cells, concordance scores quantifying the similarity between drug-treated cells and IL6ST knockdown cells are as follows: 0.4371 for dexamethasone treated cells, and 0.6458 for bupropion treated cells, and 0.3067 for dexamethasone treated cells. In HA1E cells, the concordance scores quantifying the similarity between drug-treated cells and IL6ST knockdown cells are as follows: 0.2262 for fluoxetine treated cells, and 0.3597 for paroxetine treated cells. In HA1E cells, the concordance score quantifying the similarity between dexamethasone treated cells and NFKB1 knockdown cells is 0.2979. In HT29 cells, the concordance score quantifying the similarity between dexamethasone treated cells and IL6ST knockdown cells is 0.2930. In HT29 cells, the concordance scores quantifying the similarity between drug-treated cells and NFKB1 knockdown cells are as follows: 0.3218 for fluoxetine treated cells, and 0.2313 for dexamethasone treated cells. In MCF7 cells, the concordance scores quantifying the similarity between bupropion treated cells and IL6ST knockdown cells is −0.3242. In MCF7 cells, the concordance scores quantifying the similarity between drug-treated cells and NFBK1 knockdown cells are as follows: 0.379 for fluoxetine treated cells, and 0.2252 for paroxetine treated cells. We present summary data tables containing all calculated concordance scores, mean values, and standard deviations for IL6ST (Fig. 2E) and NFKB1 (Fig. 2F) drug-knockdown comparisons across A375, A549, HA1E, HEPG2, HT29, and MCF7 cell lines. In this way, we represent the concordance scores generated by comparisons of differential gene expression signatures elicited by drug treatments and differential gene expression signatures elicited by IL6ST or NFKB1 knockdown in each evaluated cell line, as well as the mean concordance scores calculated across all cell lines. The mean concordance scores quantifying the average similarity between drug-treated cells and IL6ST knockdown cells are as follows: 0.5461 (SD: 0.2920; N: 3) for fluoxetine treated cells, 0.4497 (SD: 0.1310; N: 4) for dexamethasone treated cells; 0.3082 (SD: 0.4370; N: 4) for bupropion treated cells; and 0.4390 (SD: 0.0776; N: 3) for paroxetine treated cells. The mean concordance scores quantifying the average similarity between drug-treated cells and NFKB1 knockdown cells are as follows: 0.3440 (SD: 0.0307;N: 3) for fluoxetine treated cells, 0.2904 (SD: 0.0410; N: 4) for dexamethasone treated cells; 0.2407 (N: 1) for bupropion treated cells; and 0.2891 (SD: 0.0564; N: 4) for paroxetine treated cells.
4. Discussion
When evaluating mean concordance scores across all IL6ST knockdown cell lines containing statistically significant differential gene expression signatures for all four drug treatments, fluoxetine treated cells generated transcriptional profiles highly similar to the transcriptional profiles of IL6ST knockdown cells (mean concordance score = 0.7061). Furthermore, fluoxetine treated cells also generate gene expression signatures similar to NFKB1 knockdown cells. When evaluating mean concordance scores across all NFKB1 knockdown cell lines containing statistically significant differential gene expression signatures for at least one drug treatment of interest, fluoxetine treated cells generate transcriptional profiles that parallel the transcriptional profiles of NFKB1 knockdown cells (mean concordance score = 0.3440). While bioinformaticians are still refining interpretations of concordance scores, it is generally accepted that concordance scores larger than 0.3210 are valuable in establishing significant relationships between comparands [32], [33].
These data are particularly interesting in contrast to dexamethasone-induced transcriptional changes. Surprisingly, treatments with fluoxetine approximate inflammatory IL6ST and NFBK1 gene knockdowns more significantly than dexamethasone—a steroid routinely used to treat cytokine induced hyperinflammation. Administration of dexamethasone in porcine respiratory coronavirus-infected pigs downregulated IL6 in the early disease stages suggesting that corticosteroids such as dexamethasone may be beneficial to COVID-19 patients if administered during the early acute phase of infection [34], [35]. A study comparing over 2000 patients being treated with dexamethasone to over 4000 control patients receiving usual care showed that hospitalized patients receiving respiratory support such as mechanical ventilation or oxygen demonstrate lower 28-day mortality when treated with dexamethasone [36]. Concordance scores generated by comparisons of fluoxetine treatment and IL6ST knockdown or NFKB1 knockdown are also distinctly higher than those comparing bupropion or paroxetine treatment with the same inflammatory gene knockdowns. This is striking given that both bupropion and paroxetine share monoamine neurotransmission profiles with fluoxetine. Previous reports describe the NF-kappaB family of transcription factors as regulating many pro-inflammatory target genes including IL6ST [9], [10], [37]. Taken together, our data suggest that fluoxetine induces a reduction in IL6ST-mediated signal transduction via NF-kappaB-mediated transcriptional changes. These data also suggest that the anti-inflammatory action of fluoxetine is independent of its action on monoaminergic pathways. Instead, fluoxetine may manifest its anti-inflammatory character through a mechanism of action related to the suppression of inflammatory genes such as IL6ST and NFKB1.
IL6 signaling is enormously complex and capable of eliciting both pro- and anti-inflammatory cellular behaviors [38], [39]. Classical IL6 pathways reliant upon membrane-bound IL6R tend to elicit anti-inflammatory responses. In contrast, trans-signaling-mediated responses are distinctly pro-inflammatory and proceed by way of IL6ST on cells that do not express membrane-bound IL6R [40]. These trans-signaling-mediated responses require soluble IL6R (sIL6R) to interact with IL6 and IL6ST proteins ( Fig. 3). A fluoxetine-mediated reduction in IL6ST signal transduction could therefore specifically decrease hyper-inflammatory IL6 pathways while maintaining the classic anti-inflammatory IL6 pathways critical to the re-establishment of homeostasis in hyperinflammatory disorders [41]. Indeed, our analysis shows insignificant similarity between fluoxetine treated cells and IL6R knockdown cells (concordance score = 0.2467; Tables S2 and S3).
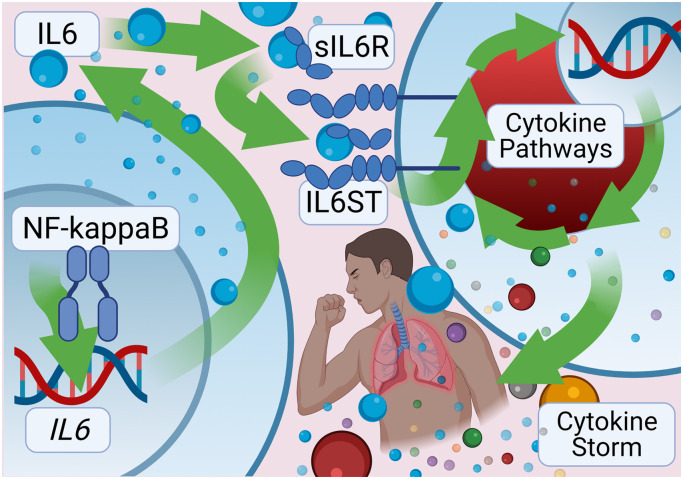
Fig. 3.
Pathway: NF-kappaB, IL6, sIL6R, IL6ST, and Cytokine Storm. The transcription factor NF-kappaB binds the IL6 gene to induce transcriptional expression of the proinflammatory cytokine. Pro-inflammatory trans-signaling-mediated responses involve IL6 proteins binding to soluble IL6 receptors (sIL6R) which activates the IL6 signal transduction protein (IL6ST or gp130). IL6ST activation is integral to many intracellular cytokine-mediated inflammatory pathways including the cytokine storm / cytokine release syndrome driving many of the negative clinic outcomes associated with COVID-19.
Recent work from our group demonstrates antidepressant-mediated inhibition of IL6 and NF-KappaB signaling pathways limit organ damage, decrease pro-inflammatory cytokine production, decrease intracellular migration of early-stage inflammatory response, and improve animal survival after an overwhelming systemic inflammatory response [42]. The present study further supports the potential of antidepressants like fluoxetine to prevent cytokine storm severity and reduce the risk of severe organ dysfunction and death for COVID-19 patients. Indeed, preliminary reports from a multi-site fluoxetine clinical trial in France [27], as well as our group’s ongoing fluoxetine clinical trial in America (NCT04377308) provide encouraging results suggesting fluoxetine treatment significantly reduces morbidity and mortality in COVID-19 patients.
5. Conclusion
Increased IL6 signal transduction causes cytokine storm which is associated with increased morbidity and mortality in COVID-19 patients. SARS-CoV-2 vaccine distribution is underway but immediate life-saving drug treatments are necessary to decrease case-fatality rates. NF-kappaB plays a pivotal role in cytokine storms through transcriptional activation of the pro-inflammatory cytokines IL6 and IL6ST. This study provides further evidence supporting the use of fluoxetine to decrease NF-kappaB signaling and thereby decrease the IL6ST signal transduction pathway driving cytokine storm in SARS-CoV-2 infection. Using fluoxetine to disrupt this NF-kappaB/IL6ST axis and mitigate the resultant cytokine storm may increase survival and decrease rates of hospitalization in COVID-19 patients.
Author Contributions
Conceptualization, R.E.M., J.F.C, A.S.I., C.B.M., S.M.O.; methodology, R.E.M., J.F.C, A.S.I., C.B.M., S.M.O., H.M.E.; software, R.E.M., J.F.C., A.S.I., H.M.E., S.M.O.; validation, A.S.I.; formal analysis, A.S.I.; investigation, R.E.M., J.F.C, A.S.I., C.B.M., S.M.O.; resources, R.E.M., J.F.C, A.S.I., C.B.M., S.M.O.; data curation, A.S.I.; writing - original draft preparation, J.F.C., A.S.I., S.M.O., C.B.M., E.A.; writing - review and editing, J.F.C, A.S.I., H.M.E. C.G., K.N.B., J.R., E.A., Z.K.P, S.M.D., R.E.M., C.B.M.; visualization, J.F.C., A.S.I., K.N.B; supervision, R.E.M., S.M.O., E.A., C.B.M.; project administration, R.E.M., C.B.M.; funding acquisition, R.E.M. and C.B.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by NIMH R01 MH107487 (R.E.M.), NIH R01 AG057598 (R.E.M.), and NIMH R01 MH121102 (R.E.M.), as well as NIA DA 049417 (C.B.M.), and SAMHSA H79-TI-081657 (C.B.M.).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2021.111437.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
References
- 1.Tanaka T., Narazaki M., Kishimoto T. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy. 2016;8(8):959–970. doi: 10.2217/imt-2016-0020. [DOI] [PubMed] [Google Scholar]
- 2.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., Koufargyris P., Karageorgos A., Katrini K., Lekakis V., Lupse M., Kotsaki A., Renieris G., Theodoulou D., Panou V., Koukaki E., Koulouris N., Gogos C., Koutsoukou A. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000. doi: 10.1016/j.chom.2020.04.009. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang N.L.S., Chan P.K.S., Wong C.K., To K.F., Wu A.K.L., Sung Y.M., Hui D.S.C., Sung J.J.Y., Lam C.W.K. Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin. Chem. 2005;51(12):2333–2340. doi: 10.1373/clinchem.2005.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beijing Group of National Research Project for S. Dynamic changes in blood cytokine levels as clinical indicators in severe acute respiratory syndrome. Chin. Med J. (Engl.) 2003;116(9):1283–1287. [PubMed] [Google Scholar]
- 9.Libermann T.A., Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol. Cell Biol. 1990;10(5):2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu T. NF-kappaB signaling in inflammation. Signal Transduct. Target Ther. 2017:2. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C., Wu Z., Li J.W., Zhao H., Wang G.Q. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taraz M., Khatami M.R., Dashti-Khavidaki S., Akhonzadeh S., Noorbala A.A., Ghaeli P., Taraz S. Sertraline decreases serum level of interleukin-6 (IL-6) in hemodialysis patients with depression: results of a randomized double-blind, placebo-controlled clinical trial. Int Immunopharmacol. 2013;17(3):917–923. doi: 10.1016/j.intimp.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 13.Gobin V., Van Steendam K., Denys D., Deforce D. Selective serotonin reuptake inhibitors as a novel class of immunosuppressants. Int Immunopharmacol. 2014;20(1):148–156. doi: 10.1016/j.intimp.2014.02.030. [DOI] [PubMed] [Google Scholar]
- 14.Basterzi A.D., Aydemir Ç., Kisa C., Aksaray S., Tuzer V., Yazici K., Göka E. IL-6 levels decrease with SSRI treatment in patients with major depression. Hum. Psychopharmacol. 2005;20(7):473–476. doi: 10.1002/hup.717. [DOI] [PubMed] [Google Scholar]
- 15.Durairaj H., Steury M.D., Parameswaran N. Paroxetine differentially modulates LPS-induced TNFalpha and IL-6 production in mouse macrophages. Int Immunopharmacol. 2015;25(2):485–492. doi: 10.1016/j.intimp.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L., Wang R., Liu L., Qiao D., Baldwin D.S., Hou R. Effects of SSRIs on peripheral inflammatory markers in patients with major depressive disorder: A systematic review and meta-analysis. Brain Behav. Immun. 2019;79:24–38. doi: 10.1016/j.bbi.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 17.Hashioka S., Klegeris A., Monji A., Kato T., Sawada M., McGeer P.L., Kanba S. Antidepressants inhibit interferon-gamma-induced microglial production of IL-6 and nitric oxide. Exp. Neurol. 2007;206(1):33–42. doi: 10.1016/j.expneurol.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 18.Cai Z., Liu J., Bian H., Cai J., Jin Q., Han J. Fluoxetine, an antidepressant drug, inhibited cigarette smoke-induced pulmonary inflammation and apoptosis in rats. Inflammation. 2017;40(4):1375–1381. doi: 10.1007/s10753-017-0580-y. [DOI] [PubMed] [Google Scholar]
- 19.Hu H.M., Li B., Wang X.D., Guo Y.S., Hui H., Zhang H.P., Wang B., Huang D.G., Hao D.J. Fluoxetine is neuroprotective in early brain injury via its anti-inflammatory and anti-apoptotic effects in a rat experimental subarachnoid hemorrhage model. Neurosci. Bull. 2018;34(6):951–962. doi: 10.1007/s12264-018-0232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D., Wang Z., Liu S., Wang F., Zhao S., Hao A. Anti-inflammatory effects of fluoxetine in lipopolysaccharide(LPS)-stimulated microglial cells. Neuropharmacology. 2011;61(4):592–599. doi: 10.1016/j.neuropharm.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 21.Guo Y., Song T., Liu J., Li H., Tian Z., Xu C. Disruption of NF-kappaB signaling by fluoxetine attenuates MGMT expression in glioma cells. Onco Targets Ther. 2015;8:2199–2208. doi: 10.2147/OTT.S85948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koh S.J., Kim J.M., Kim I.K., Kim N., Jung H.C., Song I.S., Kim J.S. Fluoxetine inhibits NF-kappaB signaling in intestinal epithelial cells and ameliorates experimental colitis and colitis-associated colon cancer in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;301(1):G9–G19. doi: 10.1152/ajpgi.00267.2010. [DOI] [PubMed] [Google Scholar]
- 23.Dong C., Zhang J., Yao W., Ren Q., Yang C., Ma M., Han M., Saito R., Hashimoto K. Effects of escitalopram, R-citalopram, and reboxetine on serum levels of tumor necrosis factor-alpha, interleukin-10, and depression-like behavior in mice after lipopolysaccharide administration. Pharm. Biochem Behav. 2016;144:7–12. doi: 10.1016/j.pbb.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Sherkawy M.M., Abo-youssef A.M., Salama A.A.A., Ismaiel I.E. Fluoxetine protects against OVA induced bronchial asthma and depression in rats. Eur. J. Pharm. 2018;837:25–32. doi: 10.1016/j.ejphar.2018.08.026. [DOI] [PubMed] [Google Scholar]
- 25.Roumestan C., Michel A., Bichon F., Portet K., Detoc M., Henriquet C., Jaffuel D., Mathieu M. Anti-inflammatory properties of desipramine and fluoxetine. Respir. Res. 2007;8:35. doi: 10.1186/1465-9921-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Disco. 2020;6(1):14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoertel N. Association between antidepressant use and reduced risk of intubation or death in hospitalized patients with COVID-19: results from an observational study. Mol. Psychiatr. 2021:1–14. doi: 10.1038/s41380-021-01021-4. [DOI] [PubMed] [Google Scholar]
- 28.Zimniak M., Kirschner L., Hilpert H., Seibel J., Bodem J. The serotonin reuptake inhibitor Fluoxetine inhibits SARS-CoV-2. Biorxiv. 2020 doi: 10.1038/s41598-021-85049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan S.M. Identification of candidate repurposable drugs to combat COVID-19 using a signature-based approach. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-84044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imami A.S., O’Donovan S.M., Creeden J.F., Wu X., Eby H., McCullumsmith C.B., Uvnäs-Moberg K., McCullumsmith R.E., Andari E. Oxytocin’s anti-inflammatory and proimmune functions in COVID-19: a transcriptomic signature-based approach. Physiol. Genom. 2020;52(9):401–407. doi: 10.1152/physiolgenomics.00095.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorman A.W. Accelerating drug discovery and repurposing by combining transcriptional signature connectivity with docking. bioRxiv. 2020 doi: 10.1126/sciadv.adj3010. 2020.11.25.399238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newton M.A., Quintana F.A., den Boon J.A., Sengupta S., Ahlquist P. Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Ann. Appl. Stat. 2007;1(1):85–106. [Google Scholar]
- 33.Pilarczyk M. Connecting omics signatures of diseases, drugs, and mechanisms of actions with iLINCS. bioRxiv. 2020 [Google Scholar]
- 34.Russell B., Moss C., Rigg A., Van Hemelrijck M. COVID-19 and treatment with NSAIDs and corticosteroids: should we be limiting their use in the clinical setting? Ecancermedicalscience. 2020;14:1023. doi: 10.3332/ecancer.2020.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X., Alekseev K., Jung K., Vlasova A., Hadya N., Saif L.J. Cytokine responses in porcine respiratory coronavirus-infected pigs treated with corticosteroids as a model for severe acute respiratory syndrome. J. Virol. 2008;82(9):4420–4428. doi: 10.1128/JVI.02190-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dexamethasone in hospitalized patients with Covid-19 − preliminary report. New Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schreiber J., Jenner R.G., Murray H.L., Gerber G.K., Gifford D.K., Young R.A. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc. Natl. Acad. Sci. U.S.A. 2006;103(15):5899–5904. doi: 10.1073/pnas.0510996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell I.L., Erta M., Lim S.L., Frausto R., May U., Rose-John S., Scheller J., Hidalgo J. Trans-signaling is a dominant mechanism for the pathogenic actions of interleukin-6 in the brain. J. Neurosci. 2014;34(7):2503–2513. doi: 10.1523/JNEUROSCI.2830-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baran P., Hansen S., Waetzig G.H., Akbarzadeh M., Lamertz L., Huber H.J., Ahmadian M.R., Moll J.M., Scheller J. The balance of interleukin (IL)-6, IL-6.soluble IL-6 receptor (sIL-6R), and IL-6.sIL-6R.sgp130 complexes allows simultaneous classic and trans-signaling. J. Biol. Chem. 2018;293(18):6762–6775. doi: 10.1074/jbc.RA117.001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zegeye M.M., Lindkvist M., Fälker K., Kumawat A.K., Paramel G., Grenegård M., Sirsjö A., Ljungberg L.U. Activation of the JAK/STAT3 and PI3K/AKT pathways are crucial for IL-6 trans-signaling-mediated pro-inflammatory response in human vascular endothelial cells. Cell Commun. Signal. 2018;16(1):55. doi: 10.1186/s12964-018-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schett G. Physiological effects of modulating the interleukin-6 axis. Rheumatology (Oxford) 2018;57(suppl_2):ii43–ii50. doi: 10.1093/rheumatology/kex513. [DOI] [PubMed] [Google Scholar]
- 42.Lu X., Wang J., Chen X., Jiang Y., Pan Z.K. Rolipram protects mice from gram-negative bacterium Escherichia coli-induced inflammation and septic shock. Sci. Rep. 2020;10(1):175. doi: 10.1038/s41598-019-56899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.
Supplementary material.