Abstract
Objective
Baricitinib seems a promising therapy for COVID-19. To fully-investigate its effects, we in-vitro evaluated the impact of baricitinib on the SARS-CoV-2-specific-response using the whole-blood platform.
Methods
We evaluated baricitinib effect on the IFN-γ-release and on a panel of soluble factors by multiplex-technology after stimulating whole-blood from 39 COVID-19 patients with SARS-CoV-2 antigens. Staphylococcal Enterotoxin B (SEB) antigen was used as a positive control.
Results
In-vitro exogenous addition of baricitinib significantly decreased IFN-γ response to spike- (median: 0.21, IQR: 0.01–1; spike+baricitinib 1000 nM median: 0.05, IQR: 0–0.18; p < 0.0001) and to the remainder-antigens (median: 0.08 IQR: 0–0.55; remainder-antigens+baricitinib 1000 nM median: 0.03, IQR: 0–0.14; p = 0.0013). Moreover, baricitinib significantly decreased SEB-induced response (median: 12.52, IQR: 9.7–15.2; SEB+baricitinib 1000 nM median: 8, IQR: 1.44–12.16; p < 0.0001). Baricitinib did modulate other soluble factors besides IFN-γ, significantly decreasing the spike-specific-response mediated by IL-17, IL-1β, IL-6, TNF-α, IL-4, IL-13, IL-1ra, IL-10, GM-CSF, FGF, IP-10, MCP-1, MIP-1β (p ≤ 0.0156). The baricitinib-decreased SARS-CoV-2-specific-response was observed mainly in mild/moderate COVID-19 and in those with lymphocyte count ≥1 × 103/µl.
Conclusions
Exogenous addition of baricitinib decreases the in-vitro SARS-CoV-2-specific response in COVID-19 patients using a whole-blood platform. These results are the first to show the effects of this therapy on the immune-specific viral response.
Keywords: Baricitinib, SARS-CoV-2, COVID-19, IGRA, Specific immune-response
Graphical abstract
Introduction
COronaVIrus Disease 2019 (COVID-19) is an emerging respiratory infection caused by SARS-CoV-2, reported for the first time in Wuhan (China) and now spread to almost all countries in the world.1
Clinical presentations of COVID-19 include mild/moderate disease, severe and critical disease.2, 3, 4 An over exuberant production of proinflammatory cytokines and chemokines5 mainly during critical disease, is a key aspect of SARS-CoV-2 pathogenesis. Therefore, the block of the cascade of proinflammatory immune factors was considered a promising therapeutic approach6. Beside the oxygen supply, several therapies were tested as antivirals, chloroquine and hydroxychloroquine, plasma from convalescent COVID-19 patients, corticosteroids or immunomodulators.6
Baricitinib, a Janus kinase (JAK) 1/2 inhibitor, is a drug approved for rheumatoid arthritis treatment.7 This drug was predicted through the BenevolentAI algorithm8 as candidate for COVID-19 treatment due to its dual effect of reducing cytokine release and potentially viral entry.9 , 10 Indeed, SARS-CoV-2 enters cells binding the Angiotensin-converting enzyme 2 (ACE2) receptor and the endocytosis is regulated by AP2-associated protein kinase-1 (AAK1) and cyclin G-associated kinase (GAK). Therefore, baricitinib-supposed mechanisms are the inhibition of kinases signalling, thus preventing viral-endocytosis, and inhibition of the cytokine release blocking JAK1/2. The anti-viral effects of baricitinib were demonstrated in primary human liver spheroids.8 Baricitinib, administered to COVID-19 patients showed a good safety profile,11 reduced mortality, intensive care unit (ICU)-admission and viral load detected from nasopharyngeal swabs.12 Recently, the Adaptive COVID-19 Treatment Trial (ACTT-2) showed that the combination of baricitinib plus remdesivir was superior to remdesivir alone in reducing recovery time and accelerating improvement in clinical status among patients with COVID-19, notably among those receiving high-flow oxygen or noninvasive ventilation. The combination was significantly associated with fewer serious adverse events in the combination group compared to those occurring in the control group.13 To note that in the SOLIDARITY trial supported by the World Health Organization14 , which was a large study conducted without placebo arm, the therapy with remdesivir was shown to have little or no effect on hospitalized patients with COVID-19, as indicated by overall mortality, initiation of ventilation, and duration of hospital stay.
T-cell immune response specific to SARS-CoV-2 antigens was extensively studied15, 16, 17, 18 and we and others also showed that SARS-CoV-2 specific T-cell responses may be measured in patients with COVID-19 using a whole-blood based platform.19 , 20 The whole-blood assay is a valid approach to measure antigen-specific responses that has been explored in the field of several infectious diseases.21, 22, 23, 24, 25 In a disease different from COVID-19 as tuberculosis, T-cell-specific response decreases overtime and this associates with cure.26, 27, 28, 29, 30
Therefore, since baricitinib has been suggested as an important therapy for COVID-19, we in-vitro evaluated the effect of baricitinib on the SARS-CoV-2-specific-response.
Material and method
Study population
The 39 COVID-19-patients were hospitalized inpatients and classified as mild, moderate, severe and critical according to WHO4 and scored with the highest severity score of the disease occurring during the hospitalization. Briefly, mild COVID-19 patients have symptoms but do not have viral pneumonia or hypoxia; moderate COVID-19 patients have pneumonia and SpO2 ≥9 0% on room air; severe COVID-19 patients have pneumonia and a respiratory rate > 30 breaths/min or severe respiratory distress or SpO2 < 90% on room air; critical COVID-19 patients have acute respiratory distress syndrome. Inclusion criteria for COVID-19 patients was a diagnosis based on a positive nasopharyngeal swab for SARS-CoV-2 and a disease with the clinical characteristics already described [Lazzaro Spallanzani National Institute of Infectious Diseases (INMI) Recommendations for COVID-19 management].3 Exclusion criteria were: HIV infection, inability to sign an informed consent and age younger than 18 years. For controls, 19 “NO COVID-19″-individuals were enrolled and were healthy donors (HD) (n = 8) volunteers from our laboratory or a convenience sample of consecutively patients hospitalized or followed as outpatients for other diseases as bacterial pneumonia (n = 2), active tuberculosis (n = 5) and latent tuberculosis infection (n = 4). Inclusion criteria for “NO COVID-19″ was a negative IgG SARS-CoV-2 serology, no symptoms of COVID-19 and in a portion of them, a negative swab for SARS-CoV-2. Demographic and clinical information were collected at enrollment.
Study approval
Ethical Committee of Lazzaro Spallanzani National Institute of Infectious Diseases (INMI) approved the study (60/2020) that was conducted in April 17th–July 20th. Informed, written consent was required to consecutively enroll patients and controls by physicians.
This study was conducted also evaluating samples from COVID-19 patients and “NO COVID-19″ individuals who were part of a previous cohort (study approval number 59/2020).20 All the enrolled individuals provided an informed written consent. All the participants’ data were anonymized and identified by codes.
Drugs and Stimuli
Baricitinib was kindly provided by Ely Lilly (Indianapolis, IN, USA); the powder was dissolved in dimethylsulfoxide (DMSO), aliquoted and frozen until use. Baricitinib was used at 10 nM and 1000 nM based on previous studies conducted in different settings not using the whole-blood platform from COVID-19 patients.9
SARS-CoV-2 MegaPool of peptides (MP), spike and remainder-antigens MPs, were already described.15 , 18 , 31 Briefly, the peptides prediction was carried out starting from the Wuhan-Hu-1 reference isolated (GenBank ID:MN908947); overlapping 15-mers by 10 spanning the entire spike protein (spike; n = 253) were derived, while the remainder of the SARS-CoV-2 proteome has been filtered by CD4-specific prediction algorithm using the 7-allele methodology as previously described (remainder-antigens; n = 221).31 All the peptides were synthesized as crude material (A&A, San Diego, CA), resuspended in DMSO, pooled separately followed by sequential lyophilization steps as previously described.
Whole-blood assay
Whole-blood (600 µl) was stimulated or not with the described MPs at 0.1 µg/mL and Staphylococcal Enterotoxin B (SEB) (Sigma-Aldrich, St. Loius, MO, USA) at 200 ng/mL as positive control for 24 h at 37 °C (5% CO2). Baricitinib was added to the culture 1 h prior antigen stimulation.
Interferon (IFN)-γ levels were evaluated on harvested plasma by an ELISA test routinely used in our lab (www.quantiferon.com). IFN-γ levels’ values were subtracted from the unstimulated control.
Multiplex analysis
A Bio-Plex Pro-Human Cytokine 27-plex Assay panel (Bio-Rad, Hercules, CA, USA) was carried out to measure cytokines, chemokines and growth factor [interleukin (IL)−1β, IL-1 receptor antagonist (ra), IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL-15, IL-17A, Eotaxin, basic fibroblast growth factor (FGF), granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), IFN-γ, -IFN-γ-induced protein 10 (IP-10), monocyte chemoattractant protein-1 (MCP-1), macrophage inflammatory protein (MIP)−1α, MIP-1β, platelet-derived growth factor (PDGF), regulated upon activation, normal t cell expressed and presumably secreted (RANTES), tumor necrosis factor (TNF)-α, vascular endothelial growth factor (VEGF)] in the leftover plasma from the whole-blood assay. The assay was performed as manufacturer instructions using the MagPix system and the Bio-Plex Manager software (all from Bio-Rad). Concentrations below the detection range were considered as 0, whereas concentrations above the detection range were converted to the highest value of the standard curve. Analyte levels were subtracted from the unstimulated control. Moreover, values generated from < 50 bead counts reading were excluded from the final analysis.
IgG serology
SARS-CoV-2 IgG ELISA was performed according to the manufacturer's instructions (DIESSE Diagnostica Senese S.p.a., Monteriggioni, Italy). The ratio between the optical density (OD) value of the sample and that of the cut-off reagent (index) has been calculated. The sample is considered positive if the index is >1.1, doubtful if the index is between 1.1 and 0.9, negative if the index is <0.9. In “NO COVID-19″-group, samples with a positive ELISA score, were tested with an indirect immunofluorescence (IF) as previously described.20 Although IF has been described as a test with high sensibility/specificity,32 here was used as confirmatory test as it is an home-made test.
Statistics
Data were analyzed using SPSS software (Version 19 for Windows, Italy SRL, Bologna, Italy), and GraphPad (GraphPad Prism 8 XML ProjecT). Kolmogorov–Smirnov test was performed and statistics was performed accordingly. For continuous measures, medians and IQR were calculated; the Friedman or Kruskal–Wallis tests were used for comparisons among several groups and the Wilcoxon or Mann–Whitney U tests with Bonferroni correction for pairwise comparisons. Chi square test was used for categorical variables. Spearman Rank Correlation was used for correlations with the following definitions: rs>0.7 was considered high correlation, 0.7<rs>0.5 moderate correlation and rs<0.5 low correlation.
Results
Demographic and clinical characteristics of study population
We enrolled 58 subjects: 39 COVID-19 patients and 19 “NO COVID-19″ individuals.
COVID-19 patients were classified based on disease severity4 in mild (n = 12, 30.8%), moderate (n = 16, 41.0%), severe (n = 3, 7.7%) and critical (n = 8, 20.5%).
Within the “NO COVID-19″-group, 7/19 had a negative swab result (swab performed within a week from the draw blood). Within the remaining 12 subjects that did not undergo swab testing, 8/12 were lab workers and have been evaluated as healthy up to the following 6 months after enrollment; the remaining 4/12 were subjects with LTBI (n = 3) or with active TB (n = 1) and they were all followed by our clinic for the appropriate therapy. None of these 4 individuals developed COVID-19 symptoms after more than 6 months from the study enrolment. One HD had a positive IgG-serology by ELISA; however, the IF showed an antibody pattern not specific for SARS-CoV-2. Therefore, all “NO COVID-19″-individuals were considered negative for SARS-CoV-2-serology.
The demographic and clinical characteristics of the enrolled individuals are shown in Table 1 . A flow chart of the subjects analysed is included as Supplementary Figure 1.
Table 1.
Demographical and clinical characteristics of the enrolled subjects.
| COVID-19 | NO COVID-19 | P value | |
|---|---|---|---|
| N (%) | 39 | 19 | |
| Age median (IQR) | 57 (42–76) | 49 (38–54) | 0.04 |
| Male N (%) | 18 (46.2) | 8 (42.1) | 0.77 |
| Origin N (%) | |||
| Western Europe | 22 (56.4) | 13 (68.4) | 0.54 |
| Eastern Europe | 1 (2.6) | 1 (5.3) | |
| Asia | 12 (30.8) | 2 (10.5) | |
| Africa | 3 (7.7) | 2 (10.5) | |
| North America | 0 (0) | 0 | |
| South America | 1 (2.6) | 1 (5.3) | |
| Swab positive results N (%)* | 39 (100) | 0 (0) | <0.0001 |
| Serology positive results N (%)§ | 21 (63.6) | 0 (0) | <0.0001 |
| Severity N (%) | |||
| mild | 12 (30.8) | – | |
| moderate | 16 (41.0) | – | |
| severe | 3 (7.7) | – | |
| critical | 8 (20.5) | – | |
Footnotes: COVID-19: COronaVIrus Disease 19; N: Number;* info available for 39 COVID-19 (100%) and 7 “NO COVID-19″ (36.8%) individuals. § info available from 33 COVID-19 (84.6%) and 19 “NO COVID-19″ (100%) individuals.
Exogenous addition of baricitinib decreases the SARS-CoV-2 specific response in COVID-19 patients
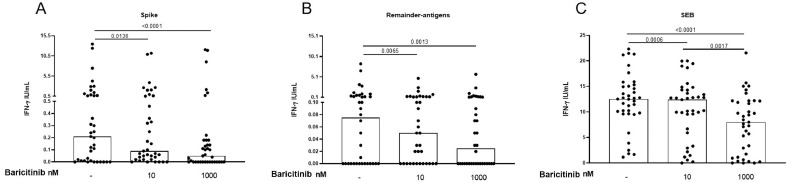
We evaluated the impact of the exogenous addition of baricitinib on the in vitro IFN-γ response to spike- and remainder-antigens MPs in 37/39 and 36/39 COVID-19 patients respectively and in 19 “NO COVID-19″ individuals. Three out of the 39 COVID-19 patients were not included in the following analysis because baricitinib conditions were either not available or available only at 1 concentration. In the COVID-19 patients, baricitinib 10 nM and baricitinib 1000 nM significantly decreased IFN-γ levels in response to spike-antigen [spike median: 0.21, interquartile range (IQR): 0.01–1; spike+baricitinib 10 nM median 0.09, IQR: 0.02–0.58; spike+baricitinib 1000 nM median 0.05, IQR: 0–0.18] (Fig. 1 A) compared to the untreated control (p = 0.0136 and p < 0.0001, respectively) (Fig. 1A). Similarly, baricitinib at both concentrations, significantly decreased the IFN-γ response to the remainder-antigens (remainder-antigens median: 0.08, IQR: 0–0.55; remainder-antigens+baricitinib 10 nM median 0.05, IQR: 0–0.23; remainder-antigens+baricitinib 1000 nM median 0.03, IQR: 0–0.14) compared to the untreated control (p = 0.0065 and p = 0.0013, respectively) (Fig. 1B).
Fig. 1.
The exogenous addition of baricitinib decreases the in vitro IFN-γ response to SARS-CoV-2 peptides in COVID-19 patients. Baricitnib 10 nM or 1000 nM decreased the IFN-γ levels after stimulating whole-blood with spike- (A) or remainder-antigens-MPs (B) or SEB (C). IFN-γ was measured by ELISA in stimulated plasma. Statistical analysis was performed using the Wilcoxon test with Bonferroni correction, and p value was considered significant if ≤0.016. Number of patients analysed: A) n = 37; B) n = 36; C) n = 37. Footnotes: IFN: Interferon; SEB: Staphylococcal Enterotoxin B.
Baricitinib 10 nM and baricitinib 1000 nM significantly reduced the IFN-γ response to SEB (p = 0.0006 and p < 0.0001 respectively) (SEB median: 12.52, IQR: 9.7–15.2; SEB+baricitinib 10 nM median 12.38, IQR: 6.3–14.3; SEB+baricitinib 1000 nM median 8, IQR: 1.44–12.16) (Fig. 1C).
Importantly, “NO COVID-19″ individuals showed a very low/absent IFN-γ response to both spike-antigen (spike median: 0.01, IQR: 0–0.1; spike+baricitinib 10 nM median 0.01, IQR: 0–0.04; spike+baricitinib 1000 nM median 0, IQR: 0–0.04) and remainder-antigens (remainder-antigens median: 0.01, IQR: 0–0.05; remainder-antigens+baricitinib 10 nM median 0.005, IQR: 0–0.03; remainder-antigens+baricitinib 1000 nM median 0, IQR: 0–0.02) (data not shown). However, baricitinib 1000 nM decreased the spike-specific response (p = 0.0073). Baricitinib did not reduce SEB-mediated immune response (SEB median: 12.94, IQR: 12.56–15.76; SEB+baricitinib 10 nM median 13.54, IQR: 11.76–14.58; SEB+baricitinib 1000 nM median 12.87, IQR: 11.92–15.32) (data not shown).
To better address the effect of baricitinib on the response to SARS-CoV-2-peptides, we analyzed the IFN-γ response considering only the IFN-γ values > 0 IU/mL (Supplementary Figure 1 and 2). In COVID-19, the decreasing effect of baricitinib was observed mainly in patients with an IFN-γ response higher than the cut-off already published for both spike- (0.16 IU/mL) (spike median: 0.31, IQR: 0.12–1.73; spike+baricitinib 10 nM median 0.17, IQR: 0.05–1.49; spike+baricitinib 1000 nM median 0.1, IQR: 0–0.18) and remainder-antigens-MPs (0.095 IU/mL) (remainder-antigens median: 0.26, IQR: 0.09–1.00; remainder-antigens+baricitinib 10 nM median 0.09, IQR: 0.03–0.3; remainder-antigens+baricitinib 1000 nM median 0.07, IQR: 0–0.27)20 (Supplementary Figure 2A-B, red lines indicate responses over the cut-off). Similar results were obtained in “NO COVID-19″ (spike median: 0.08, IQR: 0.02–0.4; spike+baricitinib 10 nM median 0.04, IQR: 0–0.33; spike+baricitinib 1000 nM median 0.02, IQR: 0–0.13; remainder-antigens median: 0.04, IQR: 0.01–0.16; remainder-antigens+baricitinib 10 nM median 0.03, IQR: 0.01–0.08; remainder-antigens+baricitinib 1000 nM median 0.01, IQR: 0–0.04) (Supplementary Figure 2C-D). In “NO COVID-19″ we observed that baricitinib decreased the IFN-γ response only if this response was present.
Exogenous addition of baricitinib reduces the SARS-CoV-2-specific response mainly in patients with mild/moderate COVID-19 and with a lymphocytes count ≥1 × 103/µl
We evaluated the impact of baricitinib on the IFN-γ response to SARS-CoV-2 peptides in COVID-19 patients stratifying for disease severity and lymphocyte counts. Only IFN-γ values>0 IU/mL were included in the analysis.
Firstly, we stratified the COVID-19 patients according to disease severity. Baricitinib 1000 nM significantly reduced the IFN-γ response to spike-MP in COVID-19 patients with mild and moderate disease (p = 0.002 and p = 0.001, respectively, Supplementary Figure 3A); similarly, baricitinib 10 nM significantly reduced the IFN-γ response to the remainder-antigens-MP (p = 0.0156 and p = 0.0117, respectively, Supplementary Figure 3B).
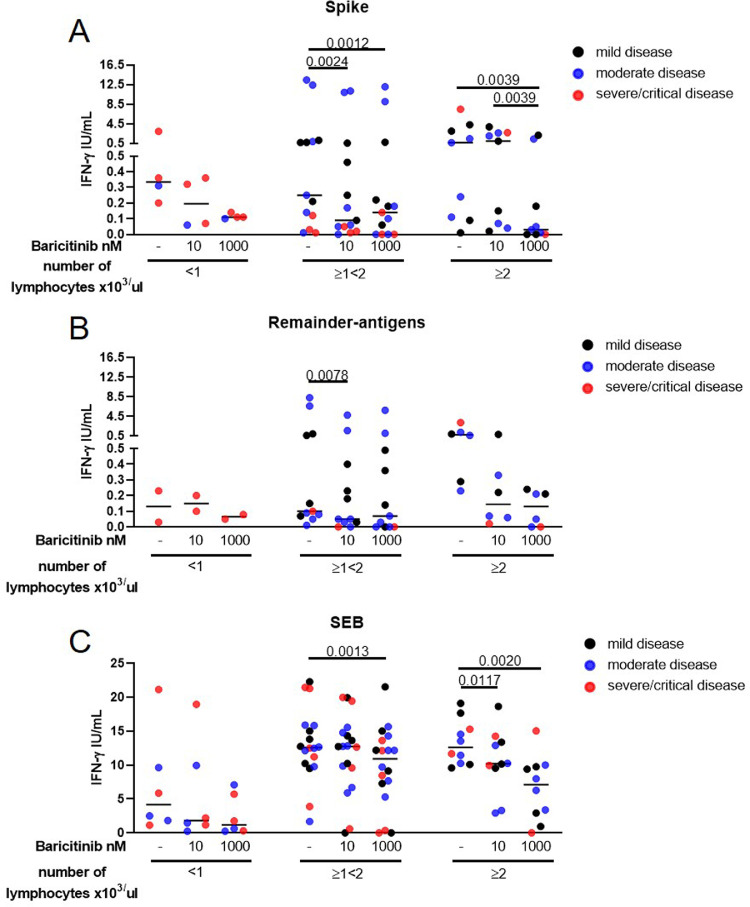
Moreover, we stratified COVID-19 patients based on the lymphocyte counts (Fig. 2 A-C) in relationship with disease severity (black dots: mild disease; blue dots: moderate disease; red dots: severe and critical disease). Lymphocytes counts were available in 26/37 patients for spike-antigen stimulation analysis and in 19/36 for remainder-antigens stimulation analysis. Three ranges were defined: <1 × 103/µl, ≥1 × 103/µl and <2 × 103/µl, ≥2 × 103/µl.
Fig. 2.
The exogenous addition of baricitinib modulates the IFN-γ in vitro response to SARS-CoV-2 peptides mainly in patients with a lymphocytes count higher than 1 × 103/µl.COVID-19 patients stratified based on the lymphocyte counts (A–C). Severity of each patient analysed is reported: black dots indicate mild disease, blue dots indicate moderate disease, red dots indicate severe and critical disease. Baricitinib at 10 nM and at 1000 nM significantly decreases the spike IFN-γ response in patients with ≥1<2 × 103/µl lymphocytes and in patients with more than 2 × 103/µl lymphocytes (A). Baricitinib 10 nM significantly decreases the IFN-γ response to the remainder-antigens in patients with ≥1<2 × 103/µl lymphocytes (B). Baricitinib 1000 nM decreases SEB-response in patients with ≥1<2 × 103/µl lymphocytes; baricitinib 10 nM and at 1000 nM decreases SEB-response in patients with ≥2 × 103/µl lymphocytes (C). IFN-γ was measured by ELISA in stimulated plasma. The horizontal lines represent the median; statistical analysis was performed using the Friedman or Kruskall-Wallis tests, Wilcoxon or Mann-Whitney tests with Bonferroni correction and p ≤ 0.016 was considered significant. Number of patients analysed for each subgroup: A) n = 4, 13, 9; B) n = 3, 11, 6; C) n = 6, 18. Footnotes: IFN: Interferon; SEB: Staphylococcal Enterotoxin B.
As expected, patients with mild disease were more likely to have lymphocyte count >1 × 103/µl.33
Exogenous addition of baricitinib at 10 nM and at 1000 nM significantly decreased the spike IFN-γ response in patients with ≥1<2 × 103/µl lymphocytes (p = 0.0024 and p = 0.0012, respectively) and in patients with more than 2 × 103/µl lymphocytes (p = 0.0039) (Fig. 2A). Baricitinib 10 nM significantly decreased the IFN-γ response to the remainder-antigens (p = 0.0078) in patients with ≥1<2 × 103/µl lymphocytes (Fig. 2B). Baricitinib 1000 nM significantly decreased SEB-induced IFN-γ response in patients with ≥1<2 × 103/µl lymphocytes (p = 0.0013). Moreover, baricitinib at both concentrations significantly reduced SEB-induced IFN-γ in patients with ≥2 × 103/µl lymphocytes (p = 0.0117 and p = 0.0028, respectively) (Fig. 2C). No significant correlations were found between lymphocyte counts and IFN-γ levels in response to these viral stimuli (data not shown).
Then, we evaluated the impact of the in vitro exogenous addition of baricitinib on the IFN-γ response to SARS-CoV-2 peptides according to other two clinical parameters as the symptoms onset and the therapy administered.
Info on symptoms onset were available in 19/37 COVID-19 patients for spike-analysis and 17/36 for remainder-antigens analysis (Supplementary Figure 4); the IFN-γ levels were stratified considering the symptoms onset within 15 days, within the following two weeks or more than a month in respect to the time of the experimental evaluation. Baricitinib at higher concentration significantly decreased the IFN-γ response to spike-MP in patients with symptoms onset started within 15 days (p = 0.0156) and in patients with symptoms onset started more than a month (p = 0.0039) (Supplementary Figure 4A).
Finally, the impact of cortisone or hydroxychloroquine (HCQ) therapy taken at the time of the blood sampling was considered to evaluate the in vitro effects of baricitinib on the viral-specific response. Information on therapy were available for 28/37 COVID-19 patients for spike-analysis and 21/36 for remainder-antigens analyisis. Baricitinib decreased the spike-response independently of cortisone or HCQ intake (p ≤ 0.0156) (Supplementary Figure 5A, C). Differently, baricitinib at both concentrations reduced the remainder-antigens-response only in patients not taking cortisone or HCQ (p ≤ 0.0037) (Supplementary Figure 5B, D).
Exogenous addition of baricitinib significantly decreased the in vitro SARS-CoV-2-specific response mediated by immune and growth factors
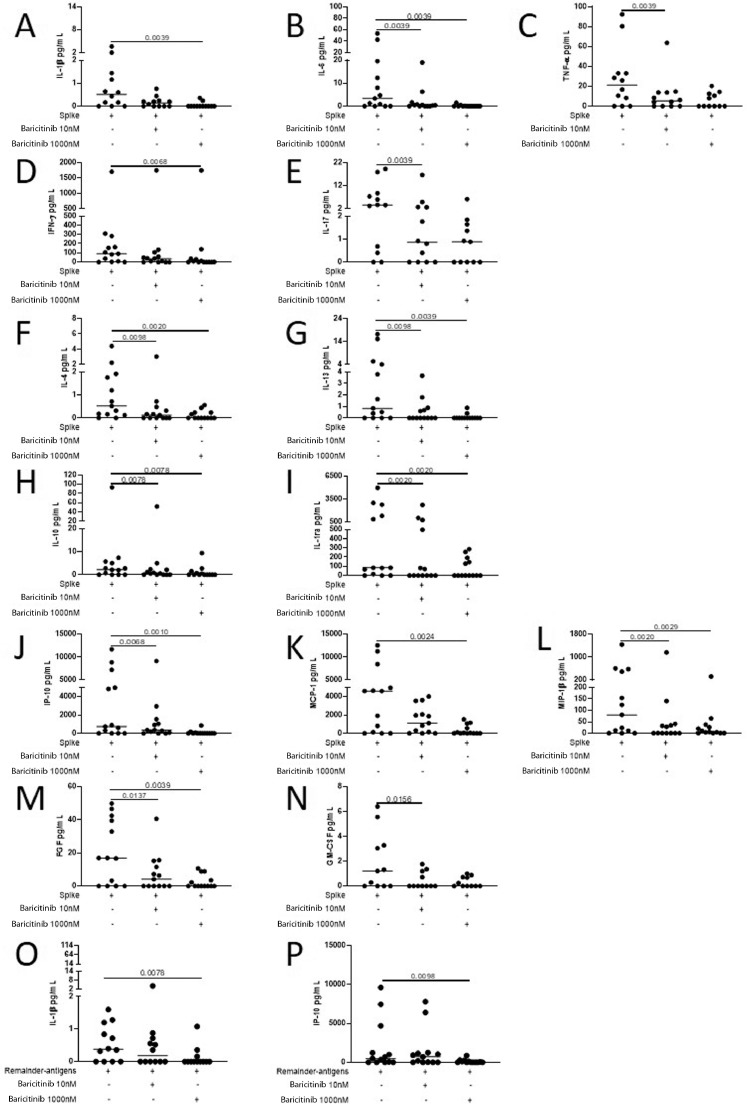
Baricitinib shuts down the signal of several cytokines by suppressing JAK1/2. Therefore, we evaluated the effect of baricitinib on the immune response elicited by SARS-CoV-2 peptides analyzing the production of several cytokines, chemokines and growth factors by multiplex technology (Fig. 3 and Supplementary Figures 6–8). Multiplex analysis was carried out in 13 COVID-19 patients from whom enough plasma-stimulated sample was available. Baricitinib at 1000 nM significantly decreased the in vitro spike-specific response mediated by the pro-inflammatory cytokines IL-1β, IL-6 and TNF-α (p = 0.0039, Fig. 3A-C respectively), the Th1- and Th17-cytokines IFN-γ and IL-17 (p = 0.0068 and p = 0.0039, Fig. 3D-E respectively), the Th2-cytokines IL-4 and IL-13 (p ≤ 0.0098 and p ≤ 0.0098, Fig. 3F-G respectively), the immunomodulatory factors IL-10 and IL-1ra (p = 0.0078 and p = 0.002, Fig. 3H-I, respectively), the chemokines IP-10, MCP-1 and MIP-1β (p ≤ 0.0068, p = 0.0024 and p ≤ 0.0029, Fig. 3J-L respectively) and the growth factors FGF and GM-CSF (p ≤ 0.0137 and p = 0.0156, Fig. 3M-N respectively). Baricitinib at 1000 nM decreased also IL-1β and IP-10 production in response to remainder-antigens (p = 0.0078 and p = 0.0098, Fig. 3O-P respectively). Moreover, although not significant, a trend of decreased levels of IL-2, IL-5, IL-12 in response to spike and of IFN-γ, MCP-1 and TNF-α in response to remainder-antigens was also observed (p ≤ 0.04) (Supplementary Figure 6). Neither significant differences in response to spike for the other factors tested (Supplementary Figure 7), nor significant modulations in response to remainder-antigens (Supplementary Figure 8) were observed.
Fig. 3.
In COVID-19 patients, the exogenous addition of baricitinib decreases the in vitro levels of pro-inflammatory, Th1, Th17, Th2 cytokines, immunomodulatory factors, chemokines and growth factor in response to SARS-Cov-2 peptides. Evaluation of 27 analytes in response to spike and to remainder-antigens by multiplex technology. In COVID-19 patients baricitinib at 1000 nM significantly decreases the in vitro cytokine spike-specific response mediated by the pro-inflammatory cytokines IL-1β, IL-6 and TNF-α (A-C), the Th1- and Th17-cytokines IFN-γ and IL-17 (D-E), the Th2-cytokines IL-4 and IL-13 (F-G), IL-10 and IL-1ra (H-I), the chemokines IP-10, MCP-1 and MIP-1β (J-L) and the growth factors FGF and GM-CSF (M-N). Baricitinib at 1000 nM decreased also IL-1β and IP-10 production in response to remainder-antigens (O-P). Analyte levels measured by luminex in stimulated plasma. The horizontal lines represent the median; statistical analysis was performed using the Wilcoxon test, and p value was considered significant if ≤0.016. Footnotes: IL: Interleukin; TNF: Tumor Necrosis Factor; IFN: Interferon; ra: receptor antagonist; IP: interferon-inducible protein; MCP: monocyte chemoattractant protein; MIP: macrophage inflammatory protein; FGF: fibroblast growth factor; GM-CSF: granulocyte-macrophage colony-stimulating factor.
Discussion
COVID-19 is a pandemic viral disease caused by the novel coronavirus SARS-CoV-2. Identification of effective therapy is crucial before large-scale vaccine administration would be available.6 Baricitinib combined with remdesivir has been recently shown in the ACTT-2 randomized controlled trial to be superior to remdesivir alone in reducing recovery time and accelerating improvement in clinical status among patients with COVID-19, notably among those receiving high-flow oxygen or noninvasive ventilation.13 This is likely due to the immune-modulatory and potential antiviral effects of the drug.8, 9, 10 , 12 However it is unknown the effect of this treatment on the modulation of the SARS-CoV-2-specific-response.
We showed for the first time in a whole-blood experimental setting, that exogenous in vitro addition of baricitinib decreased SARS-CoV-2-specific response to a broad set of immune factors including Th1, Th2 and chemokines. The effect of baricitinib in decreasing the viral-induced immune response was specific for COVID-19 mainly in patients with mild/moderate disease and with a lymphocytes count higher than 1 × 103/µl. These results highlight how this drug may be beneficial for COVID-19 treatment.
We recently showed that the immune response to spike peptides in COVID-19 patients is characterized by a high IFN-γ response20 and by the predominance of Th1 and low magnitude of Th2 cytokines.20 Here we confirm these findings and demonstrated that, in COVID-19 patients, baricitinib decreased the levels of a broad range of immune factors as IFN-γ, pro-inflammatory cytokines (IL-1β, IL-6 and TNF-α), IL-17, Th2 cytokines (IL-4, IL-13), IL-1ra, IL-10, growth factors (GM-CSF, FGF), chemokines (IP-10, MCP-1, MIP-1β). These factors are produced by both T and B cells as well as innate immunity cells and were already described as considerably increased in COVID-19 patients2 , 34 and associated with COVID-19 severity,5 although recent studies suggest that adaptive immune response is crucial to limit the severity of the disease.35
Our results indicate that exogenous addition of baricitinib in vitro shuts down the SARS-CoV-2-specific immune response, as shown in COVID-19-treated patients.36 Moreover, we observed an impact of baricitinib on FGF. Interestingly, FGF gene is up-regulated during the Middle East respiratory syndrome coronavirus (MERS‐CoV) infection.37 MERS-CoV is a virus close to SARS-CoV-2; therefore, this factor may have a role also in SARS-CoV-2 pathogenesis and its modulation may be involved in COVID-19 outcome.
Baricitinib does decrease the in vitro viral-specific response in patients with a high lymphocytes count and with mild/moderate COVID-19 and also in the severe and critical cases tested, although in these last the difference did not reach statistical significance. These data are in line with clinical studies in which baricitinib showed a good clinical efficacy in patients with mild-moderate disease.11 , 12 Moreover, baricitinib in vitro decreases the IFN-γ levels in response to SEB superantigen. Altogether, these in vitro results underline that baricitinib acts when a massive cytokine release takes place suggesting that clinically, this drug may be useful prior the manifestation of an exacerbated immune response. Consistently, baricitinib-treated COVID-19 patients have decreased pro-inflammatory cytokines (IL-6, TNF-α and IL-1β) compared to baseline levels.12 , 36 , 37 Moreover, the overtime evaluation of the whole-blood SARS-CoV-2-specific response may be useful for monitoring both, the biological effects of the drug clinically administered to patients and for disease outcome, as reported in other viral diseases as Cytomegalovirus (CMV). Indeed, this whole-blood assay has been already exploited to monitor congenital CMV.38
Cytokine responses after MPs stimulations were randomly found in “NO COVID-19″ patients as previous reported15 , 16 , 18 , 20 , 39 and baricitinib decreased the spike-specific response found mainly only in the 3 subjects in whom IFN-γ response was above the cut-off values.20
In addition to the important results of the ACTT-2 study,13 clinical administration of baricitinib was shown in smaller studies to improve the clinical conditions in moderate pneumonia of COVID-19 patients after 14 days of oral therapy and reducing ICU transfer and death. The clinical findings associated to a reduction of cytokines levels in plasmas as IL-612 and other immune factors as C-reactive proteins12 , 40 and of viral load as measured by decreased number of positive score to nasopharyngeal swabs. All these data indicated that this drugs had both anti-inflammatory and antiviral effects. In patients with rheumatoid arthritis, baricitinib therapy has been associated with a higher risk of venous thromboembolism. However in the ACTT-2, this adverse event was similar in the combination group and the control group and the difference was not significant. The absence of thromboembolism risk has been likely due to the prophylaxis with low-weight molecular heparin and to the fact that the baricitinib treatment was provided only for 2 weeks.
It is important to note that in other infectious diseases, i.e. tuberculosis, a decreased T-cell specific response, in this case to M. tuberculosis antigens, has been associated to cure.26, 27, 28, 29, 30 However, for the cure of the viral infection, evidence indicates that the cellular response, mainly Th1-mediated, remains essential.41
Limitations of the study
The limitations of this study include the small size of the samples evaluated and the lack of a prior sample size calculation. However, this is the first study analysing the effect of the baricitinib on the modulation of a specific response to SARS-CoV-2 in an easy-to-use system as whole-blood platform and deeply analysing this effect on a broad range of immune factors. The results obtained are robust because confirmed across the different COVID-19 stages and using several immune parameters beside IFN-γ. Another limit is the lack of the evaluation of the effect of clinically administered baricitinib on SARS-CoV-2-specific response in COVID-19 patients although it has been shown that baricitinib clinically administered decreases the pro-inflammatory plasma cytokines and increases the circulating number of B- and T-cells, in particular CD4 T-cells and within them, the effector memory cell subset.40 No overtime evaluation of the viral-specific-T cell response has been done yet.
In conclusion, we demonstrated for the first time to our knowledge that baricitinib down modulates the SARS-CoV-2-specific response in a whole-blood experimental assay. These results provide insights on the mechanisms potentially involved for the beneficial effects of baricitinib for COVID-19 treatment.
Declaration of Competing Interest
Dr Emanuele Nicastri reported consultancies from Gilead under $10,000 and outside the present work. Dr Alba Grifoni is listed as inventor on a provisional patent application on the diagnostic and therapeutic use of the MPs and peptides thereof filed on February 12, 2020. Dr Andrea Antinori reported consultancies, speaking fees and honoraria from Gilead Sciences, Janssen Cilag, Merk, VIIVHealthcare, Theratechnologies, all under $10,000 and outside the present work. Dr Alessandra Vergori received institutional grants from Gilead, travel grants and speaker's fees from Janssen and speaker's fee from MSD, all under $10,000 and outside the present work. Dr Delia Goletti reported consultancies from Quidel and Biomeriuex and speaking fees from Diasorin and Janssen, all under $10,000 and outside the present work.All the other authors have declared that no conflict of interest and/or financial disclosures exists.
Acknowledgments
Authors contributions
Study conception and design: DG.
Acquisition of data: LP, EP, TA, VV, GC, SNF, FP, GG, PV, EN, LL, AA, AV, DG.
Analysis and interpretation of data: LP, EP, TA, SNF, CC, AG, GI, FC, DG.
Drafting the article: LP, EP, TA, DG.
Revising the article critically for important intellectual content: LP, EP, TA, VV, GC, SNF, CC, FP, GG, PV, EN, LL, AG, AA, AV, GI, FC, DG.
Final approval of the version of the article to be published: LP, EP, TA, VV, GC, SNF, CC, FP, GG, PV, EN, LL, AG, AA, AV, GI, FC, DG.
Other study activities: AG, provided pool of peptides and expertise to carry out the T cell experiments.
Acknowledgements
This work was supported by Eli Lilly and Company (contract number ID: 2889530D-8A27–4FD8-A3CA-9384E65939F7), by Line one-Ricerca Corrente ‘Infezioni Emergenti e Riemergenti’, by Line four- Ricerca Corrente, by the projects COVID-2020–12371675, COVID-2020–12371817 and COVID-2020–12371735, all funded by Italian Ministry of Health, by the NIH NIAID contract Nr. 75N9301900065 to Alessandro Sette and Daniela Weiskopf and by European Virus Archive – GLOBAL (grants no. 653316 and no. 871029).
The authors are grateful to all the patients and nurses who helped to conduct this study. In particular we thank Sara Pantanella, Daniela Milordo, Luciana Dell'Arciprete and Lorella Tinari.
We wish to acknowledge Alessandro Sette for helpful discussions and providing peptide reagents.
The authors gratefully acknowledge the Collaborators Members of the National Institute for Infectious Diseases (INMI) COVID-19 study group: Maria Alessandra Abbonizio, Amina Abdeddaim, Chiara Agrati, Fabrizio Albarello, Gioia Amadei, Alessandra Amendola, Andrea Antinori, Mario Antonini, Tommaso Ascoli Bartoli, Francesco Baldini, Raffaella Barbaro, Barbara Bartolini, Rita Bellagamba, Martina Benigni, Nazario Bevilacqua, Gianlugi Biava, Michele Bibas, Licia Bordi, Veronica Bordoni, Evangelo Boumis, Marta Branca, Donatella Busso, Marta Camici, Paolo Campioni, Maria Rosaria Capobianchi, Alessandro Capone, Cinzia Caporale, Emanuela Caraffa, Ilaria Caravella, Fabrizio Carletti, Concetta Castilletti, Adriana Cataldo, Stefano Cerilli, Carlotta Cerva, Roberta Chiappini, Pierangelo Chinello, Carmine Ciaralli, Stefania Cicalini, Francesca Colavita, Angela Corpolongo, Massimo Cristofaro, Salvatore Curiale, Alessandra D'Abramo, Cristina Dantimi, Alessia De Angelis, Giada De Angelis, Maria Grazia De Palo, Federico De Zottis, Virginia Di Bari, Rachele Di Lorenzo, Federica Di Stefano, Gianpiero D'Offizi, Davide Donno, Francesca Faraglia, Federica Ferraro, Lorena Fiorentini, Andrea Frustaci, Matteo Fusetti, Vincenzo Galati, Roberta Gagliardini, Paola Gallì, Gabriele Garotto, Saba Gebremeskel Tekle, Maria Letizia Giancola, Filippo Giansante, Emanuela Giombini, Guido Granata, Maria Cristina Greci, Elisabetta Grilli, Susanna Grisetti, Gina Gualano, Fabio Iacomi, Giuseppina Iannicelli, Giuseppe Ippolito, Eleonora Lalle, Simone Lanini, Daniele Lapa, Luciana Lepore, Raffaella Libertone, Raffaella Lionetti, Giuseppina Liuzzi, Laura Loiacono, Andrea Lucia, Franco Lufrani, Manuela Macchione, Gaetano Maffongelli, Alessandra Marani, Luisa Marchioni, Andrea Mariano, Maria Cristina Marini, Micaela Maritti, Alessandra Mastrobattista, Giulia Matusali, Valentina Mazzotta, Paola Mencarini, Silvia Meschi, Francesco Messina, Annalisa Mondi, Marzia Montalbano, Chiara Montaldo, Silvia Mosti, Silvia Murachelli, Maria Musso, Emanuele Nicastri, Pasquale Noto, Roberto Noto, Alessandra Oliva, Sandrine Ottou, Claudia Palazzolo, Emanuele Pallini, Fabrizio Palmieri, Carlo Pareo, Virgilio Passeri, Federico Pelliccioni, Antonella Petrecchia, Ada Petrone, Nicola Petrosillo, Elisa Pianura, Carmela Pinnetti, Maria Pisciotta, Silvia Pittalis, Agostina Pontarelli, Costanza Proietti, Vincenzo Puro, Paolo Migliorisi Ramazzini, Alessia Rianda, Gabriele Rinonapoli, Silvia Rosati, Martina Rueca, Alessandra Sacchi, Alessandro Sampaolesi, Francesco Sanasi, Carmen Santagata, Alessandra Scarabello, Silvana Scarcia, Vincenzo Schininà, Paola Scognamiglio, Laura Scorzolini, Giulia Stazi, Fabrizio Taglietti, Chiara Taibi, Roberto Tonnarini, Simone Topino, Francesco Vaia, Francesco Vairo, Maria Beatrice Valli, Alessandra Vergori, Laura Vincenzi, Ubaldo Visco-Comandini, Serena Vita, Pietro Vittozzi, and Mauro Zaccarelli.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2021.02.023.
Appendix. Supplementary materials
References
- 1.ECDC. Report on COVID-19 pandemic. 2020:https://www.ecdc.europa.eu/en/covid-19-pandemic.
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;15;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicastri E., Petrosillo N., Ascoli Bartoli T., Lepore L., Mondi A., Palmieri F. National Institute for the Infectious Diseases "L. Spallanzani", IRCCS. Recommendations for COVID-19 clinical management. Infect Dis Rep. 2020;16;12(1):8543. doi: 10.4081/idr.2020.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO Clinical management of COVID-19. Interim Guid. 27 May 2020;2020 [Google Scholar]
- 5.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantini F., Goletti D., Petrone L., Najafi Fard S., Niccoli L., Foti R. Immune therapy, or antiviral therapy, or both for COVID-19: a systematic review. Drugs. 2020;Oct 17 doi: 10.1007/s40265-020-01421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantini F., Blandizzi C., Niccoli L., Petrone L., Goletti D. Systematic review on tuberculosis risk in patients with rheumatoid arthritis receiving inhibitors of Janus Kinases. Expert Opin Drug Saf. 2020;19(7):861–872. doi: 10.1080/14740338.2020.1774550. [DOI] [PubMed] [Google Scholar]
- 8.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;15;395(10223):e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stebbing J., Krishnan V., de Bono S., Ottaviani S., Casalini G., Richardson P.J. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol Med. 2020;7;12(8):e12697. doi: 10.15252/emmm.202012697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect Dis. 2020;20(4):400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantini F., Niccoli L., Matarrese D., Nicastri E., Stobbione P., Goletti D. Baricitinib therapy in COVID-19: a pilot study on safety and clinical impact. J Infect. 2020;81(2):318–356. doi: 10.1016/j.jinf.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantini F., Niccoli L., Nannini C., Matarrese D., Natale M.E.D., Lotti P. Beneficial impact of Baricitinib in COVID-19 moderate pneumonia; multicentre study. J Infect. 2020;81(4):647–679. doi: 10.1016/j.jinf.2020.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalil A.C., Patterson T.F., Mehta A.K., Tomashek K.M., Wolfe C.R., Ghazaryan V., Marconi V.C. Baricitinib plus Remdesivir for Hospitalized Adults with COVID-19. N Engl J Med. 2020;Dec 11 doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO Solidarity Trial Consortium. Pan H., Peto R., Henao-Restrepo A.M., Preziosi M.P., Sathiyamoorthy V. Repurposed antiviral drugs for COVID-19 - Interim WHO solidarity trial results. N Engl J Med. 2020 Dec 2 doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;25;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. .e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;Jul 15 doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 17.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Stralin K., Gorin J.B., Olsson A. Robust T cell immunity in convalescent individuals with Asymptomatic or Mild COVID-19. Cell. 2020;1;183(1):158–168. doi: 10.1016/j.cell.2020.08.017. .e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;26;5(48):eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murugesan K., Jagannathan P., Pham T.D., Pandey S., Bonilla H.F., Jacobson K. Interferon-gamma release assay for accurate detection of SARS-CoV-2 T cell response. Clin Infect Dis. 2020;Oct 9 doi: 10.1093/cid/ciaa1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrone L., Petruccioli E., Vanini V., Cuzzi G., Fard S.N., Alonzi T. A whole blood test to measure SARS-CoV-2 specific response in COVID-19 patients. Clin Microbiol Infect. 2020;Oct 10 doi: 10.1016/j.cmi.2020.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ong M.C.W., Migliori G.B., Raviglione M., MacGregor-Skinner G., Sotgiu G., Alffenaar J.W. Epidemic and pandemic viral infections: impact on tuberculosis and the lung. A consensus by the World Association for Infectious Diseases and Immunological Disorders (WAidid), Global Tuberculosis Network (GTN) and members(#) of ESCMID Study Group for Mycobacterial Infections (ESGMYC) Eur Respir J. 2020;Jul 2 doi: 10.1183/13993003.01727-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrone L., Vanini V., Amicosante M., Corpolongo A., Gomez Morales M.A., Ludovisi A. A T-cell diagnostic test for cystic echinococcosis based on Antigen B peptides. Parasite Immunol. 2017;39(12):e12499. doi: 10.1111/pim.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrone L., Vanini V., Petruccioli E., Ettorre G.M., Busi Rizzi E., Schininà V. IL-4 specific-response in whole blood associates with human Cystic Echinococcosis and cyst activity. J Infect. 2015;70(3):299–306. doi: 10.1016/j.jinf.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Dammermann W., Bentzien F., Stiel E.M., Kühne C., Ullrich S., Schulze Zur Wiesch J. Development of a novel IGRA assay to test T cell responsiveness to HBV antigens in whole blood of chronic Hepatitis B patients. J Transl Med. 2015;13;13 doi: 10.1186/s12967-015-0513-1. 157-015-0513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S.H. Interferon-γ Release Assay for Cytomegalovirus (IGRA-CMV) for risk stratification of posttransplant CMV infection: is it time to apply IGRA-CMV in routine clinical practice? Clin Infect Dis. 2020;Feb 20 doi: 10.1093/cid/ciz1211. [DOI] [PubMed] [Google Scholar]
- 26.Day C.L., Abrahams D.A., Lerumo L., Janse van Rensburg E., Stone L., O'rie T., Pienaar B. Functional capacity of Mycobacterium tuberculosis-specific T cell responses in humans is associated with mycobacterial load. J Immunol. 2011;Sep 1;187(5):2222–2232. doi: 10.4049/jimmunol.1101122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiacchio T., Petruccioli E., Vanini V., Cuzzi G., La Manna M.P., Orlando V., Pinnetti C., Sampaolesi A., Antinori A., Caccamo N., Goletti D. Impact of antiretroviral and tuberculosis therapies on CD4+ and CD8+ HIV/M. tuberculosis-specific T-cell in co-infected subjects. Immunol Lett. 2018;Jun;198:33–43. doi: 10.1016/j.imlet.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Petruccioli E., Petrone L., Vanini V., Sampaolesi A., Gualano G., Girardi E., Palmieri F., Goletti D. IFNγ/TNFα specific-cells and effector memory phenotype associate with active tuberculosis. J Infect. 2013;Jun;66(6):475–486. doi: 10.1016/j.jinf.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Carrara S., Vincenti D., Petrosillo N., Amicosante M., Girardi E., Goletti D. Use of a T cell-based assay for monitoring efficacy of antituberculosis therapy. Clin Infect Dis. 2004;Mar 1;38(5):754–756. doi: 10.1086/381754. [DOI] [PubMed] [Google Scholar]
- 30.Goletti D., Butera O., Bizzoni F., Casetti R., Girardi E., Poccia F. Region of difference 1 antigen-specific CD4+ memory T cells correlate with a favorable outcome of tuberculosis. J Infect Dis. 2006;Oct 1;194(7):984–992. doi: 10.1086/507427. [DOI] [PubMed] [Google Scholar]
- 31.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;8;27(4):671–680. doi: 10.1016/j.chom.2020.03.002. .e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurgel R.Q., de Sá L.C., Souza D.R.V., Martins A.F., Matos I.L.S., Lima A.G.A. SARS-CoV-2 has been circulating in northeastern Brazil since February 2020: evidence for antibody detection in asymptomatic patients. J Infect. 2020;Dec 1 doi: 10.1016/j.jinf.2020.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.Q. Correction: lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;29;5 doi: 10.1038/s41392-020-0148-4. 61-020-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;6;11(1) doi: 10.1038/s41467-020-17292-4. 3434-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;Sep 16 doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sims J.T., Krishnan V., Chang C.Y., Engle S.M., Casalini G., Rodgers G.H. Characterization of the cytokine storm reflects hyperinflammatory endothelial dysfunction in COVID-19. J Allergy Clin Immunol. 2020;Sep 10 doi: 10.1016/j.jaci.2020.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeung M.L., Yao Y., Jia L., Chan J.F., Chan K.H., Cheung K.F. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat Microbiol. 2016;22;1(3):16004. doi: 10.1038/nmicrobiol.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capretti M.G., Marsico C., Chiereghin A., Gabrielli L., Aceti A., Lazzarotto T. Immune Monitoring Using QuantiFERONⓇ-CMV Assay in Congenital Cytomegalovirus Infection: correlation With Clinical Presentation and CMV DNA load. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa704. [DOI] [PubMed] [Google Scholar]
- 39.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020 doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 40.Bronte V., Ugel S., Tinazzi E., Vella A., De Sanctis F., Canè S. Baricitinib restrains the immune dysregulation in severe COVID-19 patients. J Clin Invest. 2020;Aug 18 doi: 10.1172/JCI141772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Candia P., Prattichizzo F., Garavelli S., Matarese G. T Cells: warriors of SARS-CoV-2 Infection. Trends Immunol. 2021;42(1):18–30. doi: 10.1016/j.it.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.