Abstract
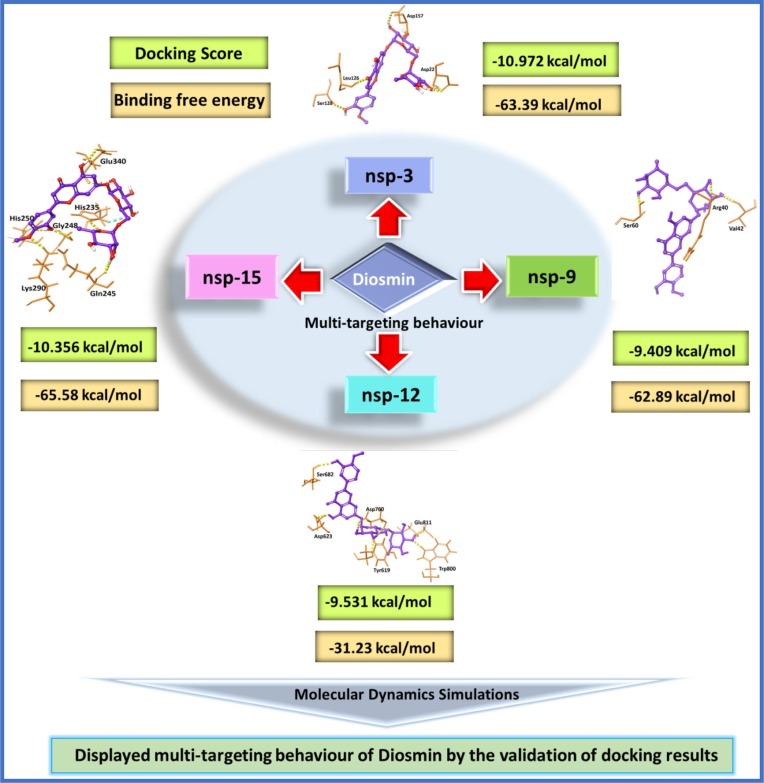
Novel coronavirus SARS-CoV-2 continues to spread rapidly worldwide and causing serious health and economic loss. In the absence of any effective treatment, various in-silico approaches are being explored towards the therapeutic discovery against COVID-19. Targeting multiple key enzymes of SARS-CoV-2 with a single potential drug could be an important in-silico strategy to tackle the therapeutic emergency. A number of Food and Drug Administration (FDA) approved drugs entered into clinical stages were originated from multi-target approaches with an increased rate, 16–21% between 2015 and 2017. In this study, we selected an FDA-approved library (Prestwick Chemical Library of 1520 compounds) and implemented in-silico virtual screening against multiple protein targets of SARS-CoV-2 on the Glide module of Schrödinger software (release 2020-1). Compounds were analyzed for their docking scores and the top-ranked against each targeted protein were further subjected to Molecular Dynamics (MD) simulations to assess the binding stability of ligand–protein complexes. A multi-targeting approach was optimized that enabled the analysis of several compounds’ binding efficiency with more than one protein targets. It was demonstrated that Diosmin (6) showed the highest binding affinity towards multiple targets with binding free energy (kcal/mol) values of −63.39 (nsp3); −62.89 (nsp9); −31.23 (nsp12); and −65.58 (nsp15). Therefore, our results suggests that Diosmin (6) possesses multi-targeting capability, a potent inhibitor of various non-structural proteins of SARS-CoV-2, and thus it deserves further validation experiments before using as a therapeutic against COVID-19 disease.
Keywords: SARS-CoV-2, Multi-targeting, Molecular docking, MD simulation, FDA-approved drugs
1. Introduction
Novel coronavirus (SARS-CoV-2) has emerged in late 2019 from a cluster of pneumonia cases in people associated with sea-food market in Wuhan, China [1], [2]. SARS-CoV-2 caused a viral infection in human and the disease was named as COVID-19 [3], [4]. As of February 2, 2021, the virus spread has infected over 102.3 million with a fatality of over 2.21 million people in the world [5]. Of particular note, United States of America, India, Brazil and Russia have officially reported the highest number of COVID-19 cases [6]. In contrast to the previously reported SARS (Severe Acute Respiratory Syndrome) in 2003, the SARS-CoV-2 has much lower estimated mortality ratio (~2.96%) [7]. However, the number of cases is high and has an immense growth rate in case of the SARS-CoV-2. The majority of the infection cases are asymptomatic, and thus raising serious health concerns as the virus continues infecting others [8]. Currently, researchers from academia and industries are in the dire need for finding treatments against COVID-19 including vaccines to limit the spread of the virus [9].
Discovering therapeutics [10], [11] based on targeting multiple enzymes represents an important approach towards the anti-COVID drug discovery. As such, applications of in-silico methods are easily explorable and time saving to accomplish search of potential molecules against multiple protein targets [12], [13], [14]. Interestingly, multiple target strategy has been successfully demonstrated to develop therapeutics for various diseases including Cancer, Alzheimer’s and Parkinson’s with complex causative factors [15] and few drugs have entered into the clinical trials [15], [16].
FDA approved Prestwick Chemical Library (PCL) of 1520 compounds [17] was tested for antiviral activity against two previously reported coronaviruses, SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) [18]. Frieman et al. [18] demonstrated 27 compounds active against both SARS-CoV and MERS-CoV. Lately, Frieman and his co-workers [19] analyzed the top 20 FDA approved drug out of 27 compounds against SARS-CoV-2, which showed in-vitro antiviral activity against SARS and MERS coronaviruses. Out of 20 compounds, 17 were successfully investigated against SARS-CoV-2 with IC50 values of 2.36–43.02 µM. Likewise, another study presented the potency of the PCL compounds in inhibiting the replication of SARS-CoV-2 [20]. Encouraged with this, we selected PCL compounds and decided to perform multi-targeting virtual screening against non-structural proteins (nsp3, nsp9, nsp12 and nsp15) of SARS-CoV-2. The non-structural proteins are responsible for the survival of the virus and its protection in host environment. Molecular docking results were further substantiated with MD simulations.
2. Methods
2.1. Computational methods and databases employed
The crystal structures of selected target proteins i.e. the ADP ribose phosphatase (nsp3, PDB ID: 6W02), replicase (nsp9, PDB ID: 6W4B), RNA dependent RNA polymerase (nsp12, PDB ID: 6M71), and endonuclease (nsp15, PDB ID: 6W01) protein structures were downloaded down from the RCSB website (https://www.rcsb.org). The computational work was performed using Schrödinger software (release 2020-1).
2.1.1. Preparation of protein structure
The SARS-CoV-2 virus protein structures (https://www.rcsb.org) were prepared in the protein preparation wizard and prime module of Schrödinger suite to remove any associated defects prior to docking [21], [22]. The structural errors assosciated with the protein structures were removed as per the reported literature [8].
2.1.2. Ligand preparation
The ligand structures of PCL containing 1520 compounds were prepared using Schrödinger’s Ligprep tool [23]. The Epike module of the software was used to determine the ionized state of ligands at pH 7 ± 2 [24].
2.1.3. Molecular docking of PCL compounds
Molecular docking of 1520 compounds was carried out in extra precision (XP) mode to get their respective binding pose and docking score [26]. It was performed in three successive modes provided by the Glide module [27]. The first screening was performed in high throughput virtual screening (HTVS) mode where all compounds were screened and only top 10% of structures were further screened by standard precision (SP) mode. Then again only top 10% structures from SP mode were selected into the next stage of screening by XP mode. The Van der Waals radii scaling factor and partial charge cutoff of 0.8 and 0.15, respectively, were used for docking. The binding free energy for these complexes were also calculated by prime MMGBSA [28].
2.1.4. Molecular Dynamics (MD) simulation
MD simulation was carried out to validate docking observations for the top ranked ligand–protein complexes. The simulation was performed for a 100 nano seconds (ns) time period on the Desmond software (D. E. Shaw Research, New York, NY, 2015) to visualize the binding and conformational stability of compound within binding pocket of the respective protein [31]. The complex was solvated by TIP3P water model prior to simulation, where 0.15 M NaCl added to mimic a physiological ionic concentration.
2.1.5. Workflow of the study
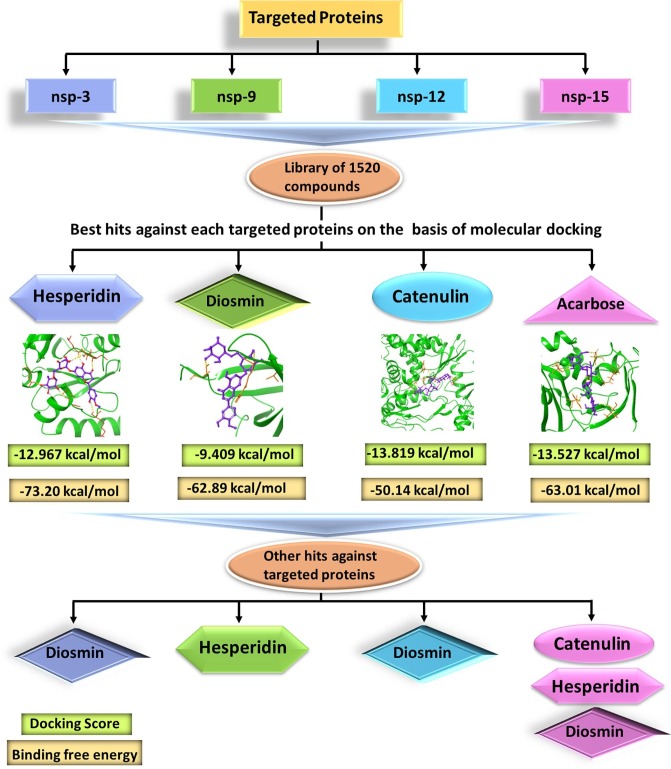
The main motive of this study is to find a multi-targeting inhibitor against several non-structural proteins of SARS-CoV-2. The observation from the study was that four compounds out of 1520 screened compounds showed best results against four different proteins on the basis of molecular docking experiments. MD simulation was also carried out on these four compounds with respective proteins, which revealed the stability of ligand–protein complexes. Then, other hit compounds were also analyzed against each targeted protein and it was found that only one compound (Diosmin, 6) successfully targeted all the proteins. A flow chart representing overall study is shown in Fig. 1 .
Fig. 1.
Flow chart of all the experiments carried out during the study. nsp3, nsp9, nsp12 and nsp15 protein of SARS-CoV-2 were screened with an approved library of 1520 compounds. The observation from the study revealed top compound against each targeted protein were: Hesperidin (1) (nsp3), Diosmin (6) (nsp9); Catenulin (13) (nsp12), and Acarbose (16) (nsp15) on the basis of molecular docking experiments. MD simulation was also carried out for these four compounds with respective protein, which revealed the stability of ligand–protein complex. Next, other hits compounds were also analyzed against each targeted protein and it was found that Diosmin was the one targeting all the proteins and hence considered for further studies.
3. Result and discussion
The objective of our study was centered at the discovery of multi-targeting inhibitors against key protein targets of SARS-CoV-2 i.e. nsp3, nsp9, nsp12 and nsp15, anticipating their applications as therapeutics against COVID-19. The number of amino acid (aa) present in these selected proteins for SARS-CoV-2 are found to be 170 aa for nsp3, 117 aa for nsp9, 942 aa for nsp12 and 370 aa for nsp15. The FDA-approved chemical library of 1520 compounds was downloaded from Prestwick database [17]. The docking results and MD simulations against individual protein targets are described separately.
3.1. nsp3 ADP ribose phosphatase
The non-structural protein, nsp3 is a papain-like proteinase protein with a long sequence, it possesses several conserved domains: ssRNA binding, ADP ribose binding, G-quadruplex binding, protease (papain-like protease, and nsp4 binding), and transmembrane domain. It also exhibits deubiquitinating (DUB) and deISGylating (deISG) activities and interacts with nsp4 and nsp6 [32], [33]. Among 16 non-structural proteins known in SARS-CoV-2, only nsp3, nsp4, and nsp6 have transmembrane domains [34]. ADP ribose phosphatase domain (Fig. S1A; supporting information) of nsp3 is known to interfere with the immune response, possibly due to its ability to remove ADP-ribose from ADP-ribosylated proteins and RNA, which is part of the host’s immune response’s intricacies [35]. Considering this important protease activity to release the essential proteins for viral activity, the inhibition of nsp3 protease activity represents an important strategy to find antiviral drugs [32].
3.1.1. nsp3-Molecular docking studies of PCL compounds
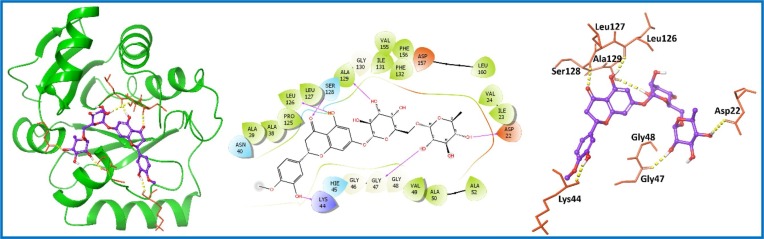
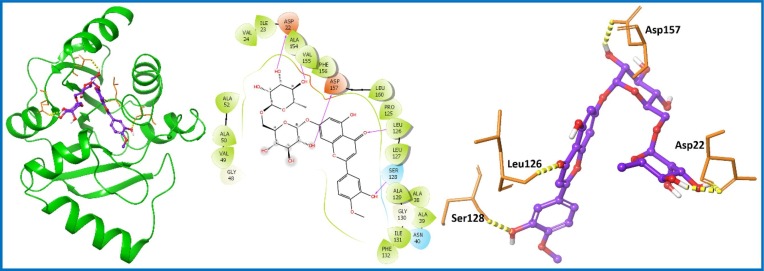
An extensive, in-silico study of the PCL compounds was carried out targeted nsp3 protein and hit compounds were selected on the basis of docking score, XP Gscore and binding free energy (Table S1, entry 1–8; supporting information). The top compound found among screened library was Hesperidin (PubChem ID: 10621; 1) with a docking score of −12.967 kcal/mol and the binding free energy of −73.20 kcal/mol (Table S1, entry 1; supporting information). Hesperidin (1) is a flavanone glycoside isolated in the year 1828 from citrus peels [36]. It is known to treat hemorrhoids (blood vessel condition), alone or in combination with other citrus bioflavonoids [37]. Recently published articles suggested lopinavir as a known inhibitor of the nsp3 protein [38], [39]. Thus, our molecular docking result were compared with known inhibitor, which displayed docking score of −7.478 kcal/mol and binding free energy of −65.24 kcal/mol (Table S1, entry 9; supporting information). The result was lower than the top-ranked Hesperidin (1) (Table S1, entry 1; supporting information) as well other shortlisted compounds (Table S1, entry 2–8; supporting information). Hesperidin (1) showed six H-bond interactions with the aa residues Asp22, Lys44, Gly47, Leu126, Ser128 and Ala129 of targeted nsp3 protein (Fig. 2 ).
Fig. 2.
Molecular interaction of the ligand, Hesperidin (1) with nsp3 protein is shown. Hydrogen bond interactions with amino acid (aa) residues (Asp22, Lys44, Gly47, Leu126, Ser128 and Ala129) of nsp3 protein, are shown in right panel with yellow dotted line, whereas the docked pose of ligand within crystal structure of nsp3 protein is shown in left panel. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.1.2. MD simulation study of Hesperidin (1)-nsp3 complex
To validate the docking results of Hesperidin (1), MD simulation was performed for a 100 ns. The root mean square deviation (RMSD) of Cα of nsp3 protein in complex with Hesperidin (1) remained stable within the range from 0.75 to 1.2 Å throughout the simulation period (Fig. S2A; supporting information). There was fluctuation noticed in ligand RMSD (up to 7 Å) for the first 60 ns after which it became stable and fluctuated within an acceptable limit (i.e. 3 Å). This change in RMSD is mainly due to both pyran rings of Hesperidin (1). The ligand root mean square fluctuation (RMSF) was also noticed due to the presence of pyran rings while less fluctuation for those ligand atoms, which are in contact with protein (Fig. S2B; supporting information). Overall, the compound remained in the binding pocket of the targeted protein. The RMSF plot of Cα of nsp3 protein showed fluctuations in Cα residues as well as with other residues interacting with compound, indicated by green color line (Fig. S2C; supporting information). Hesperidin (1) displayed several interaction with nsp3 protein residues such as: H-bond interaction (Asp22, Ala38, Asn40, Lys44, Gly46, Gly47, Gly48, Val49, Leu126, Ser128, Ala129, Ile131, Phe156, Asp157), hydrophobic interaction (Ala38, Val49, Ala50, Ala52, Ala129, Ile131, Phe132, Phe156), water bridge (Asp22, Ile23, Val24, Asn37, Ala39, Asn40, Lys44, His45, Gly46, Gly47, Gly48, Val49, Lys55, Val95, Gly97, Leu126, Ser128, Ala129, Ile131, Ala154, Phe156, Asp157), and salt bridge (Asp22, Gly48, Asp157) (Fig. S2D; supporting information). There were several residues identified with more than one type of interactions. The MD study strongly supported the molecular docking result of protein nsp3 with Hesperidin.
3.2. nsp9 replicase
Another targeted protein, nsp9 is a non-structural protein, encoded by ORF1a and related to the viral RNA synthesis. It contains a single 5-folded β-barrel, which is unique, unlike the other single protein domain proteins. The crystal structure of nsp9 replicase is a dimeric protein (Fig. S1B; supporting information). The protein replicase specifically binds to the RNA, which further interacts with the nsp8 protein and activates the mechanism of action for its functioning [40], [41]. It plays a crucial role in viral replication, but the mechanism of the RNA binding within nsp9 protein is still unidentified [42]. The induction of disturbance to the dimerization of nsp9 can be a way to overcome SARS-CoV-2 [43], [44], representing a potential drug target against SARS-CoV-2, providing a hope for new drugs inhibiting the viral progression.
3.2.1. nsp9-Molecular docking studies of PCL compounds
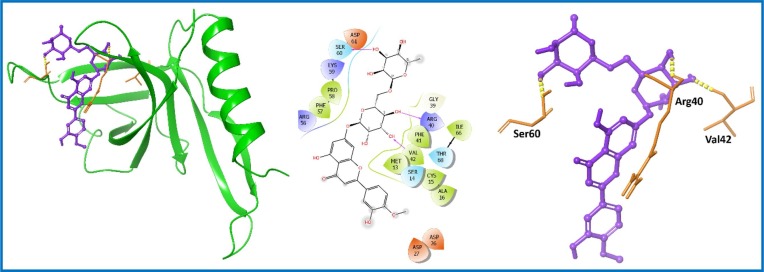
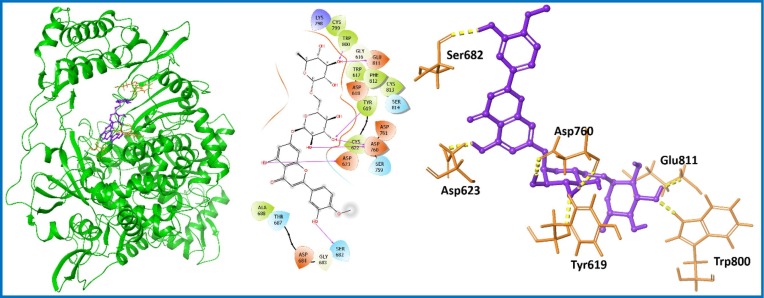
Diosmin (PubChem ID: 5281613; 6) [45] was found to possess better docking score (−9.409 kcal/mol) and binding free energy (−62.89 kcal/mol) among the screened PCL compounds (Table S1, entry 10; supporting information). Diosmin (6) is approved for the treatment of hemorrhoids or chronic venous diseases. It is a flavone extracted from natural occurring plant Teucrium gnaphalodes [46]. There are some scaffolds reported to bind with nsp9 protein such as Withanone (PubChem ID: 21679027) [47]. Here, we have analyzed docking score (−4.673 kcal/mol) and binding free energy (−44.43 kcal/mol) of Withanone (Table S1, entry 19; supporting information). Interestingly, Diosmin (6) showed improved docking results in comparison to Withanone (Table S1, entry 10; supporting information). Diosmin (6) displayed three important H-bond interactions with residues Arg40, Val42 and Ser60 of nsp9 protein as represented in Fig. 3 .
Fig. 3.
Molecular interaction of the ligand, Diosmin (6) with nsp9 protein is shown. Hydrogen bond interactions with aa residues (Arg40, Val42 and Ser60) of nsp9 protein, are shown in right panel with yellow dotted line, whereas the docked pose of ligand within crystal structure of nsp9 protein is shown in left panel. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2.2. MD simulation study of Diosmin (6)-nsp9 complex
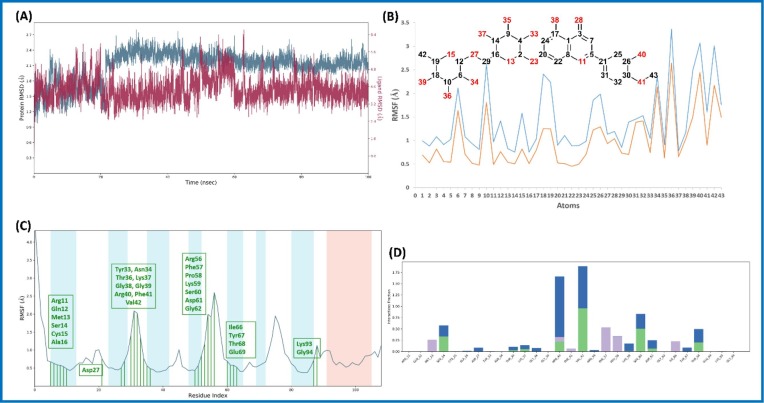
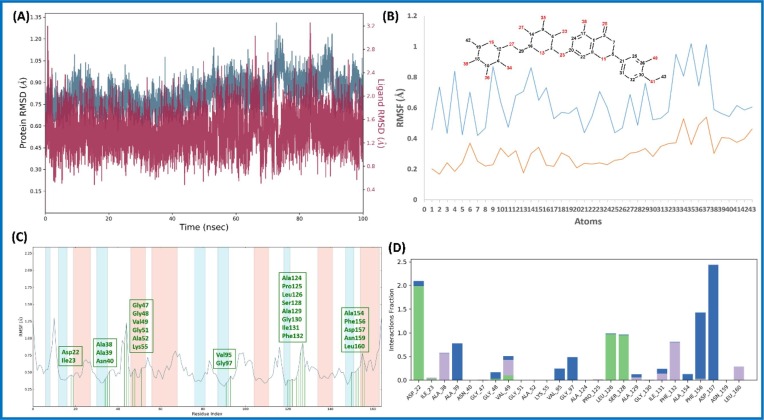
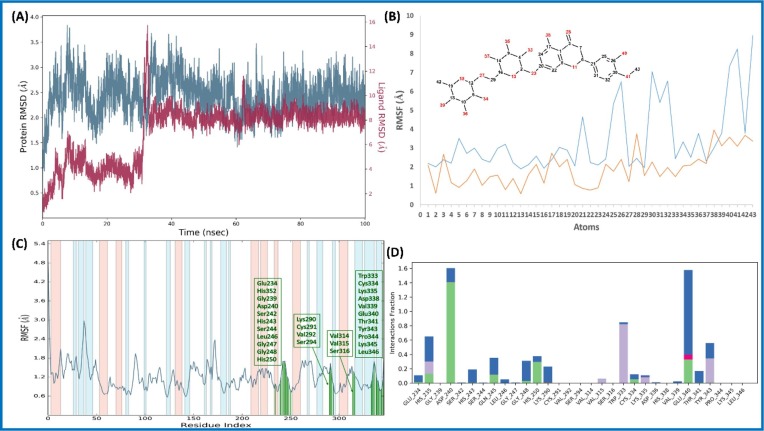
The RMSD value of Cα within the complex of Diosmin (6)-nsp9 increased for the initial 20 ns, and remained stable during the simulation period (100 ns) (Fig. 4 A). Likewise, ligand (6) RMSD was also found to be stable throughout the simulations, indicating ligand remains within the proximity of the binding site (Fig. 4 B ). The RMSF plot of Cα residues lie in the loop region of nsp9 protein and showed more fluctuations than those residues, which lie in the secondary structure (Fig. 4 C). Diosmin (6) showed three types of interactions with the aa residues. Three types of interaction include 1) H-bond (Ser14, Thr36, Lys37, Arg40, Val42, Ser60, Asp61, Thr68); 2) hydrophobic interaction (Met13, Arg40, Phe41, Phe57, Pro58, Ile66); and 3) water bridge (Ser14, Ala16, Asp27, Thr36, Lys37, Gly38, Arg40, Val42, Arg56, Lys59, Ser60, Asp61, Tyr67, Thr68) as shown in Fig. 4D. It can be observed that water is playing important role in ligand stability within the binding pocket.
Fig. 4.
Molecular dynamic simulation analysis of Diosmin (6)-nsp9 complex: (A) RMSD plot; (B) ligand RMSF plot, where light blue line indicates the ligand fluctuations with respect to the binding site residues present on target protein and orange line shows the fluctuations where the ligand in each frame is aligned on the ligand in the first reference frame; (C) protein Cα RMSF plot, residues are shown in three letter code with green color belong to binding site interacting to Diosmin (6); and (D) ligand contacts histogram showing Diosmin (6)-nsp9 complex forming H-bond interaction (green), water interaction (blue), hydrophobic interaction (purple), and salt bridge interaction (pink) during a 100 ns simulation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. nsp12 RNA dependent RNA polymerase
The other target protein nsp12, also known as RNA dependent RNA polymerase (RdRp), catalyzes the synthesis of viral RNA and hence plays an important role in the replication process of SARS-CoV-2 virus proliferation (Fig. S1C; supporting information). The replication process is achieved possibly with the assistance of nsp7 and nsp8 as co-factors [48]. Because of the crucial role of RdRp in ssRNA replication, it has been targeted for drug design and development against various viral infections such as hepatitis C virus (HCV) [49] zika virus (ZIKV) [50] and coronaviruses (CoVs) [51]. The nucleoside triphosphate (NTP) entry channel for RdRp polymerase was known to be formed by aa residues Lys545, Arg553 and Arg555 whereas aa stretch from 759 to 761 responsible for formation of catalytic site [52]. Hence, this active site stretches of aa of RdRp poses a potential inhibitory drug target for SARS-CoV-2.
3.3.1. nsp12-Molecular docking studies of PCL compounds
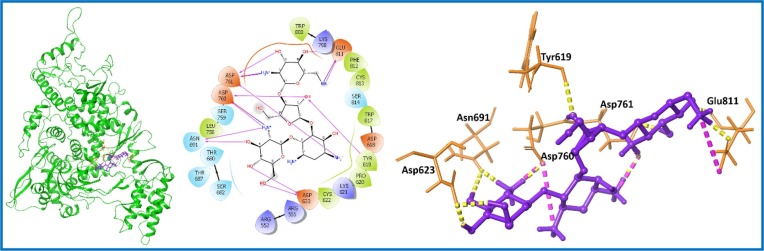
Docking score, XP Gscore and binding free energy are given in Table S1, entry 20–23; supporting information. Catenulin (PubChem ID: 4689; 13) [53] was identified as best among four hit compounds with high docking score, XP Gscore, and binding free energy of −13.819, −14.449, and −50.14 kcal/mol respectively (Table S1, entry 20; supporting information). Catenulin (13) is generic name for Paromomycin, which is an antiprotozoal and antibacterial agent [54]. Next, the observed docking results were compared with known inhibitor of RdRp i.e. Remdesivir triphosphate (remdesivirTP) [55], [56]. RemdesivirTP displayed docking score, XP Gscore, and binding free energy values, −8.199, −9.494, and −25.26 kcal/mol, respectively (Table S1, entry 24; supporting information); which is not up to scratch as compared to top-ranked Catenulin (13) (Table S1, entry 20; supporting information) and other hit compounds of nsp12 (Table S1, entry 21–23; supporting information). Catenulin (13) was later analyzed for its interaction with aa residues of nsp12 protein and found to form twelve crucial H-bond interactions (Tyr619, Asp623, Asn691, Asp760, Asp761, and Glu811) and three salt bridge interaction (Asp760, Asp761, and Glu811) needed for the stability of the ligand with the protein (Fig. 5 ).
Fig. 5.
Molecular interaction of the ligand, Catenulin (13) with nsp12 protein is shown. Hydrogen bond interactions with aa residues (Tyr619, Asp623, Asn691, Asp760, Asp761, and Glu811) of nsp12 protein, are shown in right panel with yellow dotted line and salt bridge interactions with residues Asp760, Asp761, and Glu811 are shown with violet dotted line, whereas the docked pose of ligand within crystal structure of nsp12 protein is shown in left panel. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3.2. MD simulation study of Catenulin (13)-nsp12 complex
MD simulation study revealed that the RMSD of Cα of nsp12 protein on complex with Catenulin (13) increased initially for initial 40 ns after that it became stable and remained stable at an average RMSD value of 3.5 Å till the complete simulation (Fig. S3A; supporting information). A similar trend was observed for ligand RMSD which increased initially from 0.9 Å and became stable at an average RMSD value of 4.8 Å. The ligand (13) remained within the proximity of the binding site (Fig. S3B; supporting information). The RMSF plot of Cα of nsp12 protein shown high fluctuations for the residues present in the loop region as compared to other residues in secondary structure. The aa residues showing interaction with the ligand are represented in green color (Fig. S3C; supporting information). The residues interacting with Catenulin (13) were involved either in loop formation or sheet formation. Catenulin (13) displayed several type of interaction with the aa residues of targeted protein viz. H-bond (Asp618, Lys621, Asp623, Asp760, Asp761, Lys798, Trp800, His810, Glu811, Ser814), water bridge (Lys551, Arg553, Arg555, Trp617, Asp618, Tyr619, Lys621, Cys622, Asp623, Thr687, Ser759, Asp760, Asp761, Ala762, Lys798, Trp800, His810, Glu811, Cys813, Ser814), and salt bridge interaction (Lys545, Lys551, Arg553, Arg555, Asp618, Lys621, Asp623, Ser759, Asp760, Asp761, Lys798, His810, Glu811, Cys813, Ser814) as depicted in Fig. S3D; supporting information. There were several residues exhibiting more than one type of interaction, but without hydrophobic interaction between the targeted protein and the ligand. The residues common in molecular docking and MD simulation study included Asp623, Asp760, Asp761, and Glu811 displaying H-bond; and Asp760, Asp761 and Glu811 displaying salt bridge interaction.
3.4. nsp15 endonuclease
The nsp15 is non-structured protein of SARS-CoV-2, uridylate-specific endoribonuclease structure. This protein nsp15 of SARS-CoV-2 represents a drug target probably due to its high sequence similarity with nsp15 protein of SARS and MERS [57]. The catalytic function of nsp15 resides in the C-terminal NendoU domain (Fig. S1D; supporting information). The active site is located in a shallow groove between two β-sheets which carries six key aa residues conserved among SARS-CoV-2, SARS-CoV and MERS-CoV proteins: His235, His250, Lys290, Thr341, Tyr343, and Ser294 [58]. The protein is reported to be involved in the RNA replication and processing of sub-genomic RNAs but the exact function is still not clearly understood [57].
3.4.1. nsp15-Molecular docking studies of PCL compounds
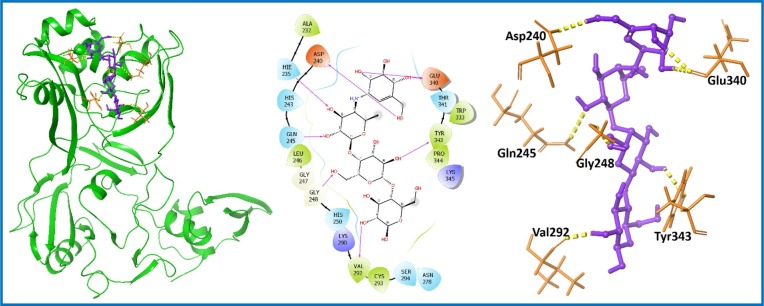
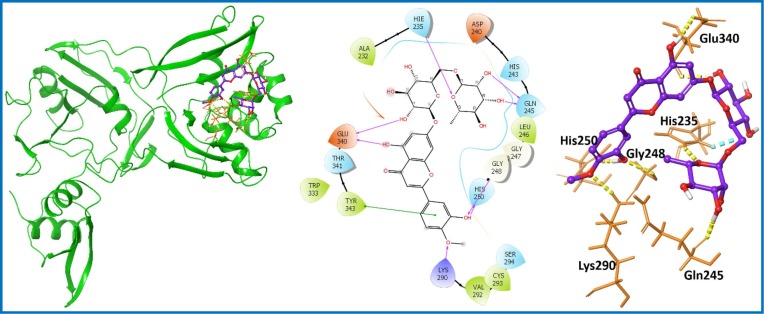
Saikosaponins were reported to show potency against nsp15 of SARS-CoV-2. Of particular note, Saikosaponin V (PubChem ID: 100958093) was the top hit with a docking score of −8.538 kcal/mol [59]. Thus, Saikosaponin V was selected as a positive control for comparison with the PCL compounds. PCL compounds displaying better results in comparison to Saikosaponin V were shortlisted as hit analogs. A total of seven compounds showed improved results (Table S1, entry 25–31; supporting information). Compound 16 (Acarbose; PubChem ID: 41774) [60] was identified as the top hit with docking score and binding free energy values of −13.527 and −63.01 kcal/mol, respectively (Table S1, entry 25; supporting information). Acarbose (16) is a pseudotetrasaccharide displaying antihyperglycemic activity, which also inhibits alpha-glucosidase, preventing the degradation of essential larger carbohydrates [60]. Acarbose displayed seven H-bond interactions with aa residues (His235, Asp240, Gln245, Gly248, Val292, Glu340, and Tyr343) as represented in Fig. 6 . The compound was further studied for MD simulation to evaluate the stability of Acarbose (16)-nsp15 complex.
Fig. 6.
Molecular interaction of the ligand, Acarbose (16) with nsp15 protein is shown. Hydrogen bond interactions with aa residues (Asp240, His235, Gln245, Gly248, Val292, Glu340, and Tyr343) of nsp15 protein, are shown in right panel with yellow dotted line, whereas the docked pose of ligand within crystal structure of nsp15 protein is shown in left panel. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4.2. MD simulation study of Acarbose (16)-nsp15 complex
MD simulation study suggested that the RMSD of Cα of nsp15 protein within complex with Acarbose (16) increased initially from 0.8 Å and stabilized at 2.6 Å and then remained stable as displayed in Fig. S4A; supporting information. Similarly, ligand RMSD was also increased initially from 0.8 Å and became stable at 50 ns. The ligand (16) remained within the proximity of the binding site of nsp15 protein (Fig. S4B; supporting information) during the entire simulation process. The RMSF plot of Cα of nsp15 protein showed high fluctuations for the aa residues, which lie in the loop region as compared to the residues lying in secondary structure. The aa residues showing interaction with Acarbose (16) represented as green line (Fig. S4C; supporting information). Acarbose (16) showed four types of interaction with different aa residues of nsp15 protein namely; 1) H-bond interaction (His235, Asp240, Gln245, Gly248, His250, Lys290, Val292, Ser294, Glu340, Thr341, Tyr343); 2) hydrophobic interaction (His243, Tyr343); 3) water bridge (His235, Asp240, Ser242, His243, Gln245, Leu246, Gly248, His250, Asn278, Lys290, Val292, Ser294, Lys335, Glu340, Thr341, Tyr343, Pro344, Lys345, Leu346); and 4) salt bridge interaction (Asp240, Tyr343, Lys345, Leu346) (Fig. S4D; supporting information). On other hand, many residues showed more than one type of interaction.
3.5. Multi-targeting PCL compounds
The main objective of our study is to find out multi-targeting inhibitors against large number of proteins of SARS-CoV-2. The four hit compounds: 1) Hesperidin (1) (nsp3; Table S1, entry 1; supporting information); 2) Diosmin (6) (nsp9; Table S1, entry 10); 3) Catenulin (13) (nsp12; Table S1, entry 20); and 4) Acarbose (16) (nsp15; Table S1, entry 25), were selected to analyse their interactions with other proteins (nsp3, nsp9, nsp12 and nsp15) individually to obtain an insight about their multi-targeting capability. Interestingly, Hesperidin (1) found to show high binding affinity towards two more proteins, nsp9 (Table S1, entry 14; supporting information) and nsp15 (Table S1, entry 28) other than nsp3. Likewise, Diosmin (6) targeted three more proteins, nsp3 (Table S1, entry 6); nsp12 (Table S1, entry 23) and nsp15 (Table S1, entry 27) other than nsp9. Catenulin (13) also targeted one additional protein, nsp15 (Table S1, entry 31; supporting information) other than nsp12. On other hand, Acarbose (16) was found to be selective against nsp15 only. Overall, three compounds Hesperidin (1), Diosmin (6) and Catenulin (13) showed binding affinity towards multiple protein targets. The best multi-targeting capacity was displayed by Diosmin (6), which showed interaction with four proteins (Fig. 7), whereas Hesperidin (1) showed interactions with three targeted proteins. A literature report by Meiyanto et al. [61], found Hesperidin (1) and Diosmin (6) as best binding molecules analyzed against different proteins of SARSCoV-2 (TMPRSS, 3CL-pro, S2-RBD, and PD-ACE2). These two also possess chemopreventive properties against cancer, viral infection and inflammatory symptoms that could further help in combating the comorbidities of COVID-19 [61]. Notably, these two compounds are under clinical trial phase 1 and explored as treatment option against COVID-19 [62]. Interestingly, nsp15 protein was found to be a common target for four compounds and displayed remarkable interactions with key residues, Gln245 and Glu340 present on the active site of the protein (Table S2; supporting information). Therefore, nsp15 represents an attractive target for the development of therapeutics against SARS-CoV-2.
Fig. 7.
The multi-targeting behavior of Diosmin (6) against nsp3, nsp9, nsp12 and nsp15 proteins of SARS-CoV-2.
3.5.1. Molecular docking and MD simulation study of Diosmin (6) -nsp3 complex
Diosmin (6) showed a docking score, XP Gscore and binding free energy of −10.972, −10.972 and −63.39 kcal/mol respectively; (Table S1, entry 6; supporting information) against nsp3 protein. It was analyzed for its interactions with nsp3 protein and displayed five H-bond with residues Asp22, Leu126, Ser128, Val155 and Asp157 (Fig. 8). Further, complex was subjected for MD simulations evalute the stability, and RMSD of Cα of nsp3 protein in complex with ligand remained stable within the range from 0.6 to 1.2 Å during the simulation period (Fig. 9 A). Ligand RMSD fluctuated in the range between 0.8 and 2.8 Å, showing that the compound was in the proximity of the binding site (Fig. 9 B). The RMSF plot of Cα of targeted nsp3 protein showed fluctuations for the residues which are present in loop region as well as in secondary structure. The plot also displayed fluctuation for the residues interacting with Diosmin (6) indicated by green color line (Fig. 9 C). Diosmin (16) showed essential interaction with the residues needed for the stability of ligand-protein complex: 1) H-bond interaction (Asp22, Ile23, Gly48, Val49, Leu126, Ser128); 2) hydrophobic interaction (Ala38, Val49, Ala129, Ile131, Phe132, Leu160); and 3) water bridge (Asp22, Ala39, Gly48, Val49, Val95, Gly97, Ala129, Ile131, Ala154, Phe156, Asp157) as presented in Fig. 9 D.
Fig. 8.
Molecular interaction of the ligand, Diosmin (6) with nsp3 protein is shown. Hydrogen bond interactions with aa residues (Asp22, Leu126, Ser128, Val155 and Asp157) of nsp3 protein, are shown in right panel with yellow dotted line, whereas the docked pose of ligand within crystal structure of nsp3 protein is shown in left panel. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 9.
Molecular dynamic simulation analysis of Diosmin (6)-nsp3 complex: (A) RMSD plot; (B) ligand RMSF plot, where light blue line indicates the ligand fluctuations with respect to the binding site residues present on target protein and orange line shows the fluctuations where the ligand in each frame is aligned on the ligand in the first reference frame; (C) protein Cα RMSF plot, residues are shown in three letter code with green color belong to binding site interacting to Diosmin (6); and (D) ligand contacts histogram showing Diosmin (6)-nsp3 complex forming H-bond interaction (green), water interaction (blue), hydrophobic interaction (purple), and salt bridge interaction (pink) during a 100 ns simulation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5.2. Molecular docking and MD simulation study of Diosmin (6)-nsp12 complex
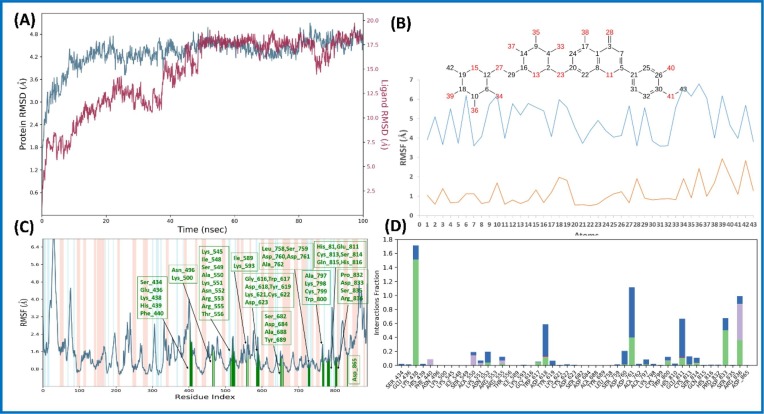
Diosmin (6) within nsp12 complex exhibited docking score, XP Gscore, and binding free energy of −9.531, −9.531, and −31.23 kcal/mol, respectively (Table S1, entry 23; supporting information). Seven important H-bond interactions were shown by Diosmin (6) with residues Asp623, Asp760, Glu811, Ser682, Tyr619, and Trp800 of nsp12 protein (Fig. 10). Next, Diosmin (6)-nsp12 complex was further demonstrated for 100 ns MD simulations. The RMSD of Cα of nsp12 protein complexed with Diosmin (6) increased for first 20 ns and afterwards remained at an average value of 4.0 Å till the simulation completed (Fig. 11 A). Similarly, ligand RMSD increased initially for first 50 ns and attained conformational stability at 17.5 Å with fluctuation in the acceptable range. The trajectory analysis suggested that the Diosmin (6) was shifted from its initial docking site to achieve energetically favorable conformational state where it interacted with the residue Lys438 (binding domain for nsp7) as well as with Asp760, which is a catalytic site for the polymerase activity. These interactions caused shifting in the binding domain residue (Asn416-Gly427) which may further affect binding stability of nsp2 to its cofactor nsp7. After 50 ns, the ligand remained within the proximity of new binding site (Fig. 11B). The residues interacting with the Diosmin (6) in RMSF plot are shown in green line (Fig. 11C). Diosmin (6) showed three types of important interactions with the residues of targeted nsp12: 1) H-bond interactions (Lys438, Trp617, Asp618, Asp761, Trp800, Glu811, Asp833, Arg836); 2) hydrophobic interactions (Phe440, Ala550, Lys551, Arg555, Arg836); and 3) water bridge (Lys438, Ala550, Lys551, Asn552, Arg555, Asp618, Tyr619, Asp760, Asp761, Ala797, Trp800, His810, Glu811, Cys813, Arg814, Asp833, Arg836) as depicted in Fig. 11D. Finally, MD study indicated the stability of the Diosmin (6)-nsp12 protein complex.
Fig. 10.
Molecular interaction of the ligand, Diosmin (6) with nsp12 protein is shown. Hydrogen bond interactions with aa residues (Asp623, Asp760, Glu811, Ser682, Tyr619, and Trp800) of nsp12 protein, are shown in right panel with yellow dotted line, whereas the docked pose of ligand within crystal structure of nsp12 protein is shown in left panel. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 11.
Molecular dynamic simulation analysis of Diosmin (6)-nsp12 complex: (A) RMSD plot; (B) ligand RMSF plot, where light blue line indicates the ligand fluctuations with respect to the binding site residues present on target protein and orange line shows the fluctuations where the ligand in each frame is aligned on the ligand in the first reference frame; (C) protein Cα RMSF plot, residues are shown in three letter code with green color belong to binding site interacting to Diosmin (6); and (D) ligand contacts histogram showing Diosmin (6)-nsp12 complex forming H-bond interaction (green), water interaction (blue), hydrophobic interaction (purple), and salt bridge interaction (pink) during a 100 ns simulation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5.3. Molecular docking and MD simulation study of Diosmin (6)-nsp15 complex
Diosmin (6) in complex with nsp15 protein showed docking score, XP Gscore, and binding free energy of −10.356, −10.356, and −65.58 kcal/mol, respectively (Table S1, entry 27; supporting information). It showed eight H-bond interactions with the aa residues (His235, Gln245, Gly248, His250, Lys290, and Glu340) and also displayed pi-pi interaction with Tyr343 residue (Fig. 12 ). The RMSD plot of Cα of nsp15 protein complexed with Diosmin (6) increased initially from 1.0 Å and attained stability at 2.5 Å (Fig. 13 A). Likewise, the ligand RMSD initially increased upto 4.0 Å and became stable at 36 ns and maintained its stable conformation. The major fluctuation observed in atoms 21, 25, 26, 30–32, 40–41, and 43, which belongs to 2-methoxyphenyl moiety of the compound. Ligand remained within the proximity of the binding site as shown in Fig. 13 B. The residues in RMSF plot, showing interaction with compound are represented as green line (Fig. 13 C). Diosmin (6) showed interaction with the residues by H-bond (His235, Asp240, Gln245, Gly248, His250, Cys334, Glu340); hydrophobic interaction (His235, Val315, Trp333, Lys335, Tyr343); water bridge (Glu234, His235, Asp240, His243, Gln245, Leu246, Gly248, His250, Lys290, Trp333, Cys334, Lys335, Val339, Glu340, Thr341, Tyr343); and salt bridge interaction (Glu340) as presented in Fig. 13D. MD simulation study indicated that the Diosmin (6) was stable in the binding proximity of the targeted nsp15 protein.
Fig. 12.
Molecular interaction of the ligand, Diosmin (6) with nsp15 protein is shown. Hydrogen bond interactions with aa residues (His235, Gln245, Gly248, His250, Lys290, and Glu340) are shown in right panel with yellow dotted line and pi-pi interaction with residue Tyr343 (blue dotted line), whereas the docked pose of ligand within crystal structure of nsp15 protein is shown in left panel. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 13.
Molecular dynamic simulation analysis of Diosmin (6)-nsp15 complex: (A) RMSD plot; (B) ligand RMSF plot, where light blue line indicates the ligand fluctuations with respect to the binding site residues present on target protein and orange line shows the fluctuations where the ligand in each frame is aligned on the ligand in the first reference frame; (C) protein Cα RMSF plot, residues are shown in three letter code with green color belong to binding site interacting to Diosmin (6); and (D) ligand contacts histogram showing Diosmin (6)-nsp15 complex forming H-bond interaction (green), water interaction (blue), hydrophobic interaction (purple), and salt bridge interaction (pink) during a 100 ns simulation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.6. Stereochemical geometry analysis of protein after MD simulation
The stereochemical geometry for all the aa residues of all 4 targeted proteins complexed with respective ligands were analyzed. All protein complexes have outlier residues either one or zero (Table S3; supporting information). The large percentage of the aa residues were found to be in favored, additional allowed and generously allowed regions for all the complexes, indicating stability of the complexes (Fig. S5; supporting information).
3.7. Off-target prediction
The NCBI BLAST results showed that the sequence alignment for the nsp3 binding site has sequence similarity with the residues SAGIF in protein 3Q6Z, 3VFQ, and 2X47 found in humans. It is possible that the compound (6) may interfere with the functioning of these proteins (Fig. S6; supporting information). However, nsp9 protein showed very less query cover (21%), indicating the percentage of sequence aligned to the sequence in gene bank, particularly related to humans. This suggests that the absence of the proteins in human with binding site similar to nsp9 protein (Fig. S7; supporting information). We could not find similar sequence for the proteins, nsp12 and nsp15 binding site residues in the BLAST search (Figs. S8 and S9; supporting information).
4. Conclusions and future directions
In the current work, we virtually validated the PCL compounds against multiple targets, non-structural proteins of SARS-CoV-2. In-silico molecular docking aided with MD simulations suggested that four top-ranked compounds against each targeted protein include Hesperidin (1) (nsp3), Diosmin (6) (nsp9), Catenulin (13) (nsp12), and Acarbose (16) (nsp15). Among all, only Diosmin (6) showed notable interactions with all four proteins. Molecular docking results indicated important H-bond interactions for Diosmin (6) with all the proteins with binding free energy (kcal/mol) of −63.39 (nsp3); −62.89 (nsp9); −31.23 (nsp12); and −65.58 (nsp15). Next, MD simulation study revealed the conformational flexibility of the Diosmin (6)-protein complex, including other characteristics like RMSD, RMSF, H-bond interaction, and hydrophobic interactions suggested the stability of the complex. MD simulation studies provide the validation of the docking experiments supporting multi-targeting ability of Diosmin (6) with non-structural proteins of SARS-CoV-2. Next approach could be extending observed simulation results for biochemical validations to obtain new insights that could enhance the understanding towards the drug discovery against COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Poonam is thankful to Department of Science and Technology, Government of India for financial support (DST/TDT/AGRO-54/2019). BR acknowledges Science and Engineering Research Board for financial support under CRG scheme (CRG/2020/005800). SK and CU are highly grateful to CSIR for senior research fellowship, PPS acknowledges DBT, Govt. of India for junior research fellowship.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymeth.2021.02.017.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sumit K., Poonam R.B. Coronavirus Disease COVID-19: a new threat to public health. Curr. Top. Med. Chem. 2020;20(8):599–600. doi: 10.2174/1568026620999200305144319. [DOI] [PubMed] [Google Scholar]
- 4.Chhikara B.S., Rathi B., Singh J., Fnu P. Corona virus SARS-CoV-2 disease COVID-19: Infection, prevention and clinical advances of the prospective chemical drug therapeutics: a review on Corona Virus Disease COVID-19, epidemiology, prevention, and anticipated therapeutic advances. Chem. Biol. Lett. 2020;7(1):63–72. [Google Scholar]
- 5.World Health Organization: Weekly operational update on COVID-19 - 1 February 2021 (https://www.who.int/publications/m/item/weekly-operational-update-on-covid-19-1-february-2021), 20201. (Accessed Feb. 2, 2021).
- 6.Worldometer: COVID-19 CORONAVIRUS PANDEMIC (https://www.worldometers.info/coronavirus/), 2020. (Accessed Oct. 19, 2020).
- 7.World Health Organisation: Coronavirus disease (COVID-2019) Weekly Epidemiological and Operational updates October 5, 2020 (https://www.who.int/docs/default-source/coronaviruse/situation-reports/20201005-weekly-epi-update-8.pdf), 2020. (Accessed Oct. 7, 2020).
- 8.Kumar S., Sharma P.P., Shankar U., Kumar D., Joshi S.K., Pena L., Durvasula R., Kumar A., Kempaiah P., Poonam B.R. Discovery of new hydroxyethylamine analogs against 3CLpro Protein Target of SARS-CoV-2: molecular docking, molecular dynamics simulation, and structure-activity relationship studies. J. Chem. Inf. Model. 2020;60(12):5754–5770. doi: 10.1021/acs.jcim.0c00326. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention: Vaccines & Immunizations; Vaccine Testing and the Approval Process (https://www.cdc.gov/vaccines/basics/test-approve.html), 2020. (Accessed Oct. 10, 2020).
- 10.Ojha P.K., Kar S., Krishna J.G., Roy K., Leszczynski J. Therapeutics for COVID-19: from computation to practices—where we are, where we are heading to. Mol. Divers. 2020 doi: 10.1007/s11030-020-10134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khatri M., Mago P. Nitazoxanide/Camostat combination for COVID-19: an unexplored potential therapy. Chem. Biol. Lett. 2020;7(3):192–196. [Google Scholar]
- 12.Vincetti P., Caporuscio F., Kaptein S., Gioiello A., Mancino V., Suzuki Y., Yamamoto N., Crespan E., Lossani A., Maga G., Rastelli G., Castagnolo D., Neyts J., Leyssen P., Costantino G., Radi M. Discovery of multitarget antivirals acting on both the dengue virus NS5-NS3 interaction and the Host Src/Fyn Kinases. J. Med. Chem. 2015;58(12):4964–4975. doi: 10.1021/acs.jmedchem.5b00108. [DOI] [PubMed] [Google Scholar]
- 13.Raghavendra N.M., Pingili D., Kadasi S., Mettu A., Prasad S.V.U.M. Dual or multi-targeting inhibitors: the next generation anticancer agents. Eur. J. Med. Chem. 2018;143:1277–1300. doi: 10.1016/j.ejmech.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 14.Chandel V., Raj S., Rathi B., Kumar D. In silico identification of potent FDA approved drugs against Coronavirus COVID-19 main protease: a drug repurposing approach. Chem. Biol. Lett. 2020;7(3):166–175. [Google Scholar]
- 15.Ramsay R.R., Popovic-Nikolic M.R., Nikolic K., Uliassi E., Bolognesi M.L. A perspective on multi-target drug discovery and design for complex diseases. Clin. Transl. Med. 2018;7(1):3. doi: 10.1186/s40169-017-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin H.-H., Zhang L.-L., Yan R., Lu J.-J., Hu Y. Network analysis of drug–target interactions: a study on FDA-approved new molecular entities between 2000 to 2015. Sci. Rep. 2017;7(1):12230. doi: 10.1038/s41598-017-12061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.PRESTWICK CHEMICAL LIBRARY: NEW LIBRARY OF 1520 APPROVED DRUGS. (http://prestwickchemical.fr/libraries-screening-lib-pcl.html#.html), 2020. (Accessed May 5, 2020).
- 18.Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Repurposing of clinically developed drugs for treatment of middle east respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58(8):4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weston S., Coleman C.M., Haupt R., Logue J., Matthews K., Li Y., Reyes H.M., Weiss S.R., Frieman M.B. Broad anti-coronavirus activity of food and drug administration-approved drugs against SARS-CoV-2 in vitro and SARS-CoV in vivo. J. Virol. 2020;94(21):e01218–20. doi: 10.1128/JVI.01218-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Touret F., Gilles M., Barral K., Nougairède A., van Helden J., Decroly E., de Lamballerie X., Coutard B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 2020;10(1):13093. doi: 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrödinger Release 2020-1: Protein Preparation Wizard; Epik, Schrödinger, LLC, New York, 2020.
- 22.Schrödinger Release 2020-1: Protein Preparation Wizard; Prime, Schrödinger, LLC, New York, NY, 2020.
- 23.Schrödinger Release 2020-1: LigPrep, Schrödinger, LLC, New York, NY, 2020.
- 24.Schrödinger Release 2020-1: Epik, Schrödinger, LLC, New York, NY, 2020.
- 26.Kumar A., Dhar S.K., Subbarao N. In silico identification of inhibitors against Plasmodium falciparum histone deacetylase 1 (PfHDAC-1) J. Mol. Model. 2018;24(9):232. doi: 10.1007/s00894-018-3761-1. [DOI] [PubMed] [Google Scholar]
- 27.Schrödinger Release 2020-1: Glide, Schrödinger, LLC, New York, NY, 2020.
- 28.Schrödinger Release 2020-1: Prime, Schrödinger, LLC, New York, NY, 2020.
- 31.Schrödinger Release 2020-1: Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2020. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2020.
- 32.Báez-Santos Y.M., St S.E., John A.D.M. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.A Review of the SARS-CoV-2 (COVID-19) Genome and Proteome (https://www.genetex.com/MarketingMaterial/Index/SARS-CoV-2_Genome_and_Proteome), 2020. (Accessed 14 Sept., 2020).
- 34.Sakai Y., Kawachi K., Terada Y., Omori H., Matsuura Y., Kamitani W. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Y.M.O. Alhammad, M.M. Kashipathy, A. Roy, J.-P. Gagné, L. Nonfoux, P. McDonald, P. Gao, K.P. Battaile, D.K. Johnson, G.G. Poirier, S. Lovell, A.R. Fehr, The SARS-CoV-2 conserved macrodomain is a highly efficient ADP-ribosylhydrolase (http://biorxiv.org/content/early/2020/06/01/2020.05.11.089375), bioRxiv (2020) 2020.05.11.089375.
- 36.Inderjit K.M.M.D. Hesperetin 7-rutinoside (hesperidin) and taxifolin 3-arabinoside as germination and growth inhibitors in soils associated with the weed, Pluchea lanceolata (DC) C.B. Clarke (Asteraceae) J. Chem. Ecol. 1991;17(8):1585–1591. doi: 10.1007/BF00984690. [DOI] [PubMed] [Google Scholar]
- 37.Mott T., Latimer K., Edwards C. Hemorrhoids: diagnosis and treatment options. Am. Fam. Phys. 2018;97(3):172–179. [PubMed] [Google Scholar]
- 38.Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J. Biomol. Struct. Dyn. 2020;1–6 doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 39.Deshpande R.R., Tiwari A.P., Nyayanit N., Modak M. In silico molecular docking analysis for repurposing therapeutics against multiple proteins from SARS-CoV-2. Eur. J. Pharmacol. 2020;886 doi: 10.1016/j.ejphar.2020.173430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chandel V., Sharma P.P., Raj S., Choudhari R., Rathi B., Kumar D. Structure-based drug repurposing for targeting Nsp9 replicase and spike proteins of severe acute respiratory syndrome coronavirus 2. J. Biomol. Struct. Dyn. 2020;1–14 doi: 10.1080/07391102.2020.1811773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L.M., Diprose J., Alderton D., Walsh M., Grimes J.M., Stuart D.I. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12(2):341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Littler D.R., Gully B.S., Colson R.N., Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. iScience. 2020;23(7) doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu T., Chen C., Li H., Dou Y., Zhou M., Lu D., Zong Q., Li Y., Yang C., Zhong Z., Singh N., Hu H., Zhang R., Yang H., Su D. Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9. Protein Sci. 2017;26(5):1037–1048. doi: 10.1002/pro.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egloff M.-P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. PNAS. 2004;101(11):3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Center for Biotechnology Information. PubChem Compound Summary for CID 5353588, Diosmetin-7-O-rutinoside (https://pubchem.ncbi.nlm.nih.gov/compound/Diosmetin-7-O-rutinoside), 2020. (Accessed Sept. 3, 2020).
- 46.Prabhu V.V., Sathyamurthy D., Ramasamy A., Das S., Anuradha M., Pachiappan S. Evaluation of protective effects of diosmin (a citrus flavonoid) in chemical-induced urolithiasis in experimental rats. Pharm. Bio. 2016;54(9):1513–1521. doi: 10.3109/13880209.2015.1107105. [DOI] [PubMed] [Google Scholar]
- 47.D.K.S. Maurya, D., Evaluation of Traditional Ayurvedic Preparation for Prevention and Management of the Novel Coronavirus (sars-cov-2) Using Molecular Docking Approach. DOI:10.26434/chemrxiv.12110214.v1, ChemRxiv. (2020).
- 48.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. PNAS. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shirota Y., Luo H., Qin W., Kaneko S., Yamashita T., Kobayashi K., Murakami S. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 2002;277(13):11149–11155. doi: 10.1074/jbc.M111392200. [DOI] [PubMed] [Google Scholar]
- 50.Godoy A.S., Lima G.M.A., Oliveira K.I.Z., Torres N.U., Maluf F.V., Guido R.V.C., Oliva G. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat. Commun. 2017;8(1):14764. doi: 10.1038/ncomms14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.National Center for Biotechnology Information (2020). PubChem Compound Summary for CID 4689, Catenulin. (https://pubchem.ncbi.nlm.nih.gov/compound/Catenulin). (Accessed Sept. 3, 2020).
- 54.Davidson R.N., den Boer M., Ritmeijer K. Paromomycin. Trans. R. Soc. Trop. Med. Hyg. 2009;103(7):653–660. doi: 10.1016/j.trstmh.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 55.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J. Med. Chem. 2017;60(5):1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 56.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295(20):6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishnan D.A., Sangeetha G., Vajravijayan S., Nandhagopal N., Gunasekaran K. Structure-based drug designing towards the identification of potential anti-viral for COVID-19 by targeting endoribonuclease NSP15. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29(7):1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinha S.K., Shakya A., Prasad S.K., Singh S., Gurav N.S., Prasad R.S., Gurav S.S. An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J. Biomol. Struct. Dyn. 2020;1–12 doi: 10.1080/07391102.2020.1762741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.National Center for Biotechnology Information PubChem Compound Summary for CID 41774, Acarbose (https://pubchem.ncbi.nlm.nih.gov/compound/Acarbose.) 2020 Accessed Sept. 3, 2020.
- 61.Utomo R.Y., Putri D.D.P., Salsabila I.A., Meiyanto E. The chemopreventive potential of diosmin and hesperidin for COVID-19 and its comorbid diseases. Indonesian J. Cancer Chemopreven. 2020;11(3):154–167. [Google Scholar]
- 62.Hesperidin and Diosmin for Treatment of COVID-19 (https://clinicaltrials.gov/ct2/show/NCT04452799), 2020. (Accessed Jan. 31, 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.