Abstract
Leucine-rich alpha-2-glycoprotein-1 (LRG1) has been shown to compete with apoptosis activating factor-1 (Apaf-1) for binding cytochrome c (Cyt c) and could play a role in inhibition of apoptosis. Employing MCF-7 breast cancer cells, we report that intracellular LRG1 does protect against apoptosis. Thus, cells transfected with the lrg1 gene and expressing higher levels of LRG1 were more resistant to hydrogen peroxide-induced apoptosis than parental cells, while cells in which LRG mRNA was knocked down by short hairpin (sh) RNA-induced degradation were more sensitive. The amount of Cyt c co-immunoprecipitated with Apaf-1 from the cytosol of apoptotic cells was inversely related to the level of LRG1 expression. In lrg1-transfected cells partially-glycosylated LRG1 was found in the cytosol and there was an increase in cytosolic Cyt c in live lrg1-transfected cells relative to parental cells. However, apoptosis was not spontaneously induced because Cyt c was bound to LRG1 and not to Apaf-1. Cyt c was the only detectable protein co-immunoprecipitated with LRG1. Following hydrogen peroxide treatment degradation of LRG1 allowed for induction of apoptosis. We propose that intracellular LRG1 raises the threshold of cytoplasmic Cyt c required to induce apoptosis and, thus, prevents onset of the intrinsic pathway in cells where Cyt c release from mitochondria does not result from committed apoptotic signaling. This mechanism of survival afforded by LRG1 is likely to be distinct from its extracellular survival function that has been reported by several research groups.
Keywords: Cyt c, LRG1, Apaf-1, Survival factor, MCF-7 cells
Introduction
Cytochrome c (Cyt c) plays a critical role in the induction of the intrinsic pathway of apoptosis and is important in the amplification of the extrinsic pathway [1,2]. In response to a variety of stress stimuli Cyt c is released from mitochondria and binds to cytoplasmic apoptotic protease activating factor-1 (Apaf-1). Apaf-1 undergoes a conformational change allowing for binding of dATP/ATP and cleavage of pro-caspase-9 that then activates downstream caspases, ultimately leading to cell death [2]. The initial release of Cyt c from mitochondria into the cytoplasm can be blocked in stressed cells by Bcl-2 [3,4]. Furthermore, after Cyt c release, apoptosis may still be inhibited by binding of the heat shock protein, Hsp27, or nucleotides to Cyt c blocking formation of the apoptosome [5,6].
A decade ago we identified leucine-rich alpha-2-glycoprotein-1 (LRG1) as a ligand of Cyt c and provided evidence that it acts as a survival factor protecting lymphocytes from apoptosis induced by extracellular Cyt c [7]. The mechanism for protection was not defined but it appeared to involve active signaling rather than steric blockade of Cyt c. More recently, several other research groups have also shown that LRG1 acts as a survival signal for other normal cells and cancer cell lines [8–11]. However, the mechanism(s) for protection by LRG1 has (have) not been clearly defined. Paradoxically, in other studies extracellular LRG1 has been shown to promote apoptosis or growth suppression by activating the TGF-β1 signaling pathway [12,13].
LRG1 has partial amino acid sequence homology to Apaf-1 and binds to Cyt c, as Apaf-1 does, in the region around residue 72. The binding of LRG1 inhibits the interaction between Cyt c and Apaf-1 in vitro [7]. Formation of the apoptosome and activation of the intrinsic pathway of apoptosis could, therefore, be compromised in cells that express cytoplasmic LRG.
Gene expression analyses reveal LRG1 to be maximally up-regulated 24 to 72 hours in certain tissues following toxin-induced stress consistent with a possible survival function [14,15]. While LRG1 is produced in abundance by the liver and secreted into the blood, it is also expressed at lower levels in most tissues. The level of RNA expression for LRG1 is greater than that for Apaf-1 in appendix, bone marrow, and gall bladder, for example, while its expression is less than that for Apaf-1 in lymph nodes and brain tissue [16].
In this study MCF-7 breast adenocarcinoma cells were employed to evaluate the influence of LRG1 on the survival of cells exposed to hydrogen peroxide, a common apoptosis inducing agent [17]. Increased LRG1 expression was accomplished by transfecting the cells with the lrg1 gene and decreased expression was obtained by targeting LRG1 mRNA for degradation using short hairpin RNA (shRNA). Cell survival in response to hydrogen peroxide was shown to correlate with the level of LRG1 expression. Increased Cyt c binding to Apaf-1 was not observed in lrg-transfected MCF-7 cells despite increased levels of Cyt c in the cytosol indicating that cell survival in the absence of committed apoptotic signaling is affected by the ability of cytoplasmic LRG1 to block Cyt c binding to Apaf-1.
Materials and methods
Antibodies and other reagents
A mAb (3C9.D5) specific for human LRG1 peptide 274–283 with the amino acid sequence DLYRWLQAQK was obtained by fusing splenocytes of an immunized BALB/c mouse with P3X63 myeloma cells using polyethylene glycol [18]. The mouse was injected twice, 3 weeks apart, with the peptide coupled to hemocyanin using glutaraldehyde. Antibody-secreting hybridomas were selected by ELISA and subcloned in limiting dilution using normal mouse splenocytes as feeder cells. All animal procedures were approved by the University of Minnesota Institutional Animal Use and Care Committee. Anti-LRG1 mAb 2F5.A2 specific for the native glycoprotein was previously described [19]. The mAb 7H8.2C12 used for Western blotting of Cyt c has been reported [1]. Antibodies for Western blotting of LRG1 were obtained from Abnova (Taipei, Taiwan). A mouse mAb specific for actin was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit antibodies against GRASP65 and Cyt c were purchased from BioLegend (San Diego, CA) and Santa Cruz Biotechnology, respectively. A mouse mAb specific for the FLAG tag was obtained from Sigma-Aldrich (St. Louis, MO). The COX IV specific antibody was obtained from Abcam (Cambridge, MA). Bortezomib was purchased from Calbiochem (San Diego, CA). Horse Cyt c was obtained from Sigma-Aldrich and further purified by ion exchange chromatography on carboxymethyl-Sephadex in 65 mM sodium phosphate, pH 7.5.
Cells and transfections
MCF-7 cells were maintained in a 5% CO2 incubator at 37° C in Advanced medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum, glutamine, and antibiotics (penicillin/streptomycin). A recombinant form of the lrg1 gene was obtained by PCR amplification of commercial cDNA encoding human LRG1 (Open Biosystems, Huntsville, AL) using the 5’ primer GAG CTC GGA TCC AAC CCT AGG ACC ATG TCC TCT TGG AGC AGA and the 3’ primer CAG TAG CTC GAG TCA CTT GTC ATC GTC GTC TTT GTA GTC CTG GGA CTT GGC CAC TGC. The 5’ primer encodes a BamHI site and the 3’ primer encodes a XhoI site for insertion of the amplified DNA into the pcDNA3.0 vector for transfection. In addition, the 3’ primer encodes a FLAG tag sequence allowing for detection and immunoprecipitation of recombinant LRG1 using anti-FLAG tag antibodies.
MCF-7 cells were transfected with the pcDNA3.0 vector (Invitrogen, Carlsbad, CA) that contains a neomycin resistance gene (neo cells) or the same vector containing a cDNA encoding human lrg1. Transfection was accomplished using FUGENE6 Transfection Reagent (Roche, Madison, WI) according to the manufacturer’s instructions. Transfectants were selected for growth in the antibiotic G418 (1 mg/ml). Cells were passaged following trypsinization by diluting 1:20 in fresh medium. Individual clones were removed from tissue culture plates by trypsinization and, following expansion, were analyzed for LRG1 expression by RT-PCR and for LRG1 protein by Western blotting.
Expression of LRG1 was downregulated using a shRNA construct. The duplex insert had the following sequences: 5’- GAT CCA ACC CGC TTA ACA AAT AAT CCG AAG CTT GGG ATT ATT TGT TAA GCG GGT TTT TTT TGG AAG- 3’ and antisense 5’- AAT TCT TCC AAA AAA AAC CCG CTT AAC AAA TAA TCC CAA GCT TCG GAT TAT TTG TTA AGC GGG TTG- 3’. The vector, pGSH1 (Genlantis, San Diego, CA), contains BamH1 and EcoR1 insertion sites. FUGENE6 (Roche) was used for transfection and G418 (1mg/ml) was the selecting antibiotic. The particular oligonucleotide sequence used for the insert was shortened from the sequence of a commercial siRNA tested in a preliminary experiment (GCAACCCGCUUAACAAAUAAUCCUG duplexed with the reverse complement having a 5’ GA overhang; Integrated DNA Technologies, Coralville, IA). Thus, the DNA homolog of the sequence CCCGCUUAACAAAUAAUCC within the siRNA was duplexed and inserted into the shRNA vector. The negative control vector expressed an irrelevant siRNA sequence with limited homology to any known sequence and was shown in other experiments not to affect expression of specific mRNAs (A. Kelekar, unpublished). For lentiviral infection four commercial shRNAs inserted into the pLKO.1 vector were obtained from Open Biosystems, Inc. (TRCN series: 146912, 148302, 148225, and 148199). Cells (293T) plated on polylysine-coated plates were transfected individually with each vector (4 μg). Also included in the transfection were pLP/VSVG (Invitrogen) which encodes the envelope protein from VSV-G and pCMVΔR8.91 which encodes the structural and regulatory proteins required for virus production [20]. The following day fresh media was added to cells. After a further 48 hr incubation culture supernates were harvested, filtered through a 0.45 μm syringe filter, aliquoted and either used immediately to transduce cells or stored at −80° C. Most consistent LRG knock down in MCF-7 cells was observed with the vectors TRCN146912 and TRCN148199. Cells infected with lentivirus expressing these two vectors showed similar sensitivities to peroxide-induced death.
SDS-PAGE and Western blotting
Total cell lysates were prepared in radioimmunoprecipitation assay buffer (150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecylsulfate, PBS pH 7.4) containing a protease inhibitor cocktail (Sigma-Aldrich). Cells were fractionated by nitrogen cavitation and differential centrifugation [21]. Protein concentration was measured using BCA™ Protein Assay Protein Quantification Kit (Pierce Biotechnology, Rockford, IL). For immunoprecipitation anti-Apaf-1-Ab-agarose beads (Santa Cruz Biotechnology), anti-FLAG tag-Ab agarose beads (Pierce Biotechnology), and anti-LRG1-Sepharose beads were employed. The latter were obtained by coupling mAb 2F5.A2 to CNBr-activated Sepharose 4B. The immunoprecipitation reactions were incubated overnight and washed 3 times in 1 ml PBS. For deglycosylation, PNGase F (New England BioLabs, Ipswich, MA) was used according to the manufacturer’s instructions. Proteins (30μl/lane) were subjected to electrophoresis in a 4–20% polyacrylamide gel in SDS after boiling (Pierce Biotechnology) and transferred to a nitrocellulose membrane (Hybond-ECL, GE Healthcare Biosciences, Piscataway, NJ) and Western blot analysis was performed using standard techniques with appropriate primary antibodies and secondary antibodies coupled to HRP (Sigma-Aldrich), the chemiluminescent substrates SuperSignal West Femto or Dura (Pierce Biotechnology), and Hyperfilm-ECL (GE Healthcare Biosciences).
RT-PCR
Expression of LRG1 mRNA in cells was analyzed using the Cells-to-cDNA™ II Kit (Invitrogen). An aliquot of 2.5 × 105 cells was lysed and treated with DNase to degrade genomic DNA. The DNase was then inactivated and two-step RT-PCR was carried out. Amplification of lrg1 was performed with primers 5’-CCATCTCCTGTCAACCACCT-3’ and 5’-GGTTAGAT CCAGCACCCTCA-3’ to produce a 215 base pair product. Amplification of the actin gene yielding a product containing 294 base pairs served as the internal control. Samples were subjected to a total of 33 or 35 cycles of the polymerase chain reaction: 1 × 94°C for 2 minutes; 33 x (94°C for 1 min; 60 °C for 1 min; 72 °C for 1.5 min), 1 × 72 °C for 10 min. PCR products were separated on 1% or 1.5% aragose gels and visualized by ethidium bromide staining under UV light. Data were acquired using the AlphaImager v5.5 Imaging system (Alpha Innotech Corporation, San Leandro, CA).
Viability assays
Viability assays were carried out as previously described [22]. Data were collected using a FLUOstar plate reader (BMG Labtech, Durham, NC). Cell survival was quantified by the fluorescence of fluorescein (absorption at 492 nm, emission at 517 nm) derived from cleavage of fluorescein diacetate (FDA) by cellular esterases. Control, lrg-transfected, and LRG mRNA knockdown MCF-7 cells were cultured in 96-well plates in triplicate. After 50 to 75% confluent cell growth was observed, wells were treated with dilutions of hydrogen peroxide for 8 hrs. at 37° C.
After incubation with hydrogen peroxide, wells were washed with 150μl PBS and 100 μl FDA working solution (10μg/ml in PBS) was then added to each well and incubated for 1 hour at room temperature in the dark. Viable cells were able to incorporate the compound and hydrolyze it to fluorescein. Percent viability was calculated as: V= 100 x (A-Y) / (B-Y) where A is the average fluorescence of cells treated with hydrogen peroxide, B is the average fluorescence of untreated cells, and Y is the average background fluorescence (no cells in the well).
Mass spectrometry (MS)
MS was performed on trypsin-digested protein slices from SDS-PAGE gels as previously described [23]. Data were acquired on a 4800 MALDI TOF/TOF mass spectrometer (AB Sciex, Foster City, CA) as previously described [24]. Data dependent tandem MS settings included acquisition of the top 20 most intense ion signals per spot. Tandem mass spectra were analyzed with Protein Pilot 2.0.1 software (AB Sciex) which uses Paragon scoring algorithm [25]. The search parameters included the NCBI reference sequence human protein database (June, 2008), iodoacetamide cysteine alkylation reagent, trypsin enzyme, 4800 instrument, biological modifications, and thorough search effort.
ELISA
LRG1 secreted into the culture fluid by MCF-7 cells was quantified by an ELISA in which horse Cyt c was employed to capture LRG1 and then bound LRG1 was detected indirectly using mAb 2F5.A2, followed by goat anti-mouse IgG coupled to horseradish peroxidase (HRP; Sigma-Aldrich), and analysis of the enzymatic activity of the HRP [19].
Immunofluorescent Confocal Microscopy
Cells were grown to 50–75% confluency on 8 chamber slides (Nalge Lab-Tek II, ThermoFisher, Rochester, NY), washed in PBS, and fixed for 2 hrs. in Streck tissue fixative (Streck Laboratories, Omaha, NE). Cells were permeabilized with 0.5% Triton X-100 in 2% fetal bovine serum, 2% horse serum, phosphate-buffered saline, pH 7.4. Antigens were visualized indirectly using the secondary antibodies Alexa 488-conjugated goat anti-mouse IgG (Molecular Probes, Eugene, OR) or Cy3-conjugated goat anti-rabbit IgG (Jackson Immunoresearch Labs, West Grove, PA). Nuclei were stained with DAPI. Immunofluorescent micrographs were viewed employing an Olympus BX61 Fluoview confocal microscope with the 60X objective. Images were acquired using Olympus Fluoview software (version 1.7a). Isotype-matched IgG control antibodies yielded negative staining results in all cases.
Results
Manipulation of LRG1 expression in MCF-7 cells
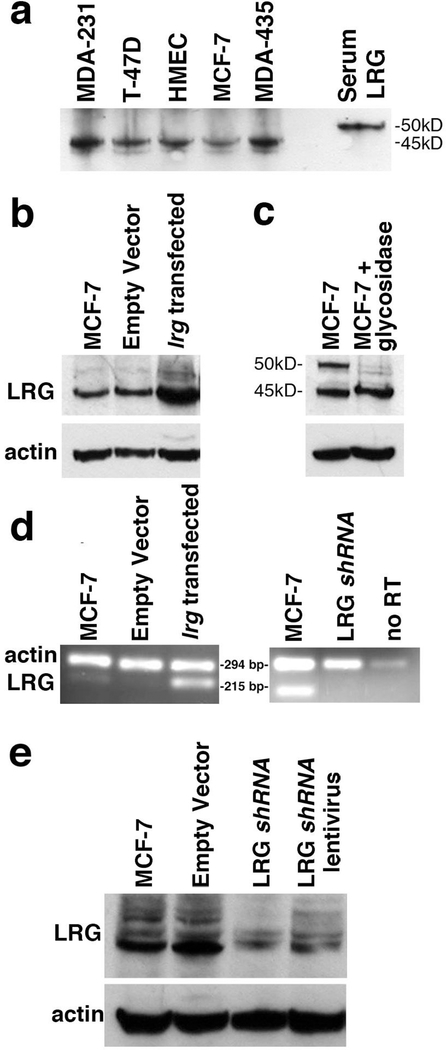
LRG1 mRNA is expressed in mammary tissue and the LRG1 protein has been identified by immunohistochemistry [26,27]. Therefore, we examined several human breast cancer cell lines for levels of LRG1 protein expression in comparison with primary human, mammary epithelial cells (HMEC), to identify a cell line that could be used to analyze LRG1 function. In Western blotting a band at 45 kD reactive with a commercial anti-LRG1 antibody was observed in all of the cell lines examined (Fig. 1a). The smaller size of LRG1 in the cell lines relative to the serum glycoprotein (50 kD) results from differential glycosylation (see below). Because MCF-7 cells have been frequently used as models for apoptosis research and are amenable to transfection, they were chosen for further study [21,28].
Figure 1. Manipulation of LRG1 expression in MCF-7 cells.
(a) Western blot analysis of LRG1 expression in primary human mammary epithelial cells (HMEC) and in several breast cancer cell lines shows lower molecular weights compared to that of serum LRG1.
(b) Transfection of MCF-7 cells with lrg1 resulted in increased protein expression relative to actin shown by Western blotting.
(c) N-glycosidase (PNGase F) treatment of proteins extracted from MCF-7 cells resulted in a decrease in intensity of the 50 kDa band indicating that it is a fully glycosylated form of LRG1.
(d) RT-PCR analysis confirmed increased LRG1 mRNA expression in transfected cells as detected by ethidium bromide staining and decreased expression in a clone transfected with LRG1 shRNA. In the absence of reverse transcriptase (RT) there was little amplification indicating near absence of genomic DNA. Samples in the right panel were amplified through two additional cycles compared to the samples in the left panel.
(e) Western blotting confirmed decreased expression of LRG1 in the clone transfected with LRG1 shRNA and partial knock down in MCF-7 cells infected with a recombinant lentivirus encoding an LRG1 shRNA distinct from that used for transfection.
Transfection of cells with an empty vector or one containing a nonsense shRNA insert (not shown) had no effect on LRG1 expression.
A representative clone of cells transfected with the lrg1 gene showed increased protein expression with a prominent band at 45 kD in Western blotting relative to parental MCF-7 cells or cells transfected with the vector without the lrg1 insert (Empty Vector, Fig. 1b). Actin was employed as a gel loading control in SDS-PAGE prior to the blotting procedure. A weaker band at 50 kD likely represents a fully-glycosylated LRG1 polypeptide as its size was reduced by treatment with peptide: N-glycosidase F (PNGase F; Fig. 1c; ref. 29). The difference in intensity of the 50 kD band between Fig. 1b and Fig. 1c is likely due to differences between the culture conditions at the time of cell harvest such as pH, cell density, and nutrient levels as variability in glycosylation is known to be affected by these factors (30).
Consistent with increased protein expression, RT-PCR showed that mRNA expression was increased in lrg1-transfected cells (the band at 215 base pairs in Fig. 1d) compared with both parental cells and vector controls. Amplification of actin cDNA (the band at 294 base pairs in Fig. 1d) served as an internal control. A clone transfected with a LRG1-specific shRNA showed decreased levels of LRG1 mRNA by RT-PCR compared to parental MCF-7 cells in two additional rounds of amplification (Fig. 1d).
LRG1 knockdown was confirmed by Western blotting (Fig. 1e). The multiple bands detected in this particular blot likely represent various glycosylated forms of LRG1. We also infected cells with lentiviral constructs encoding four different commercial LRG1-specific shRNAs. Partial knock down of LRG1 protein expression was observed for two of the shRNAs (see Materials and methods). A representative Western blot result for cells infected with one of these lentiviral shRNA constructs is shown in Fig. 1e.
Protection from cell death correlates with the level of expression of LRG1 expression
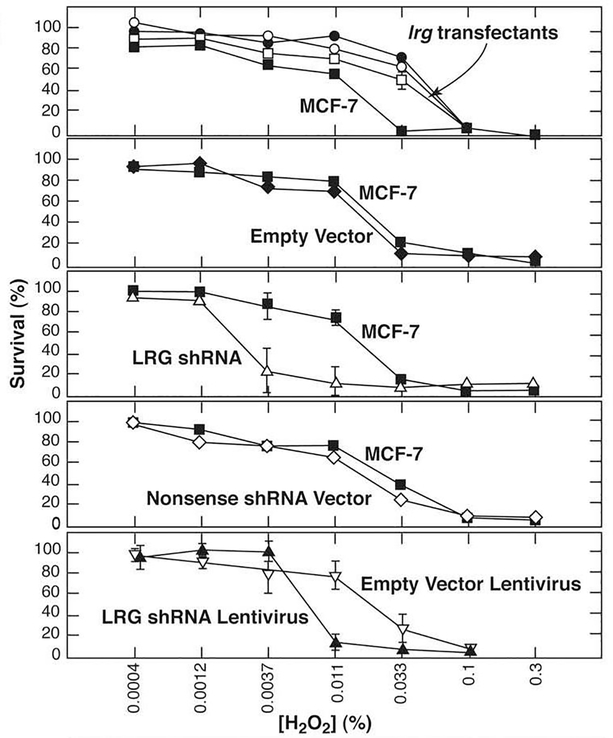
Apoptosis was induced in the cells by treatment with the oxidant, hydrogen peroxide. Viability was monitored by uptake of fluorescein diacetate (FDA) into live cells and subsequent cleavage by cellular esterases to produce fluorescein [22]. Induction of apoptosis was demonstrated in Western blots detecting cleavage of pro-caspases 7 and 9 (results not shown). Furthermore, upstream of caspase activation, Cyt c was found to co-immunoprecipiate with Apaf-1 from the cytosolic fraction of MCF-7 cells treated with hydrogen peroxide (see below, Fig. 3c). As shown in Fig. 2, after 8 hour treatment with hydrogen peroxide three independent lrg1 gene-transfected clones were approximately 3-fold more resistant to peroxide-induced apoptosis than control cells, including both parental MCF-7 cells and cells transfected with the empty vector (Fig. 2, top two panels). In contrast, cells transfected with the LRG1-specific shRNA plasmid were about 10-fold more sensitive than control cells (Fig. 2, middle panel) and cells infected with a recombinant lentivirus encoding a different LRG1-knockdown were about 3-fold more sensitive (Fig. 2, bottom panel). Taken together, the results indicate that the level of LRG1 expression directly correlates with the extent of cell survival during hydrogen peroxide-induced apoptosis.
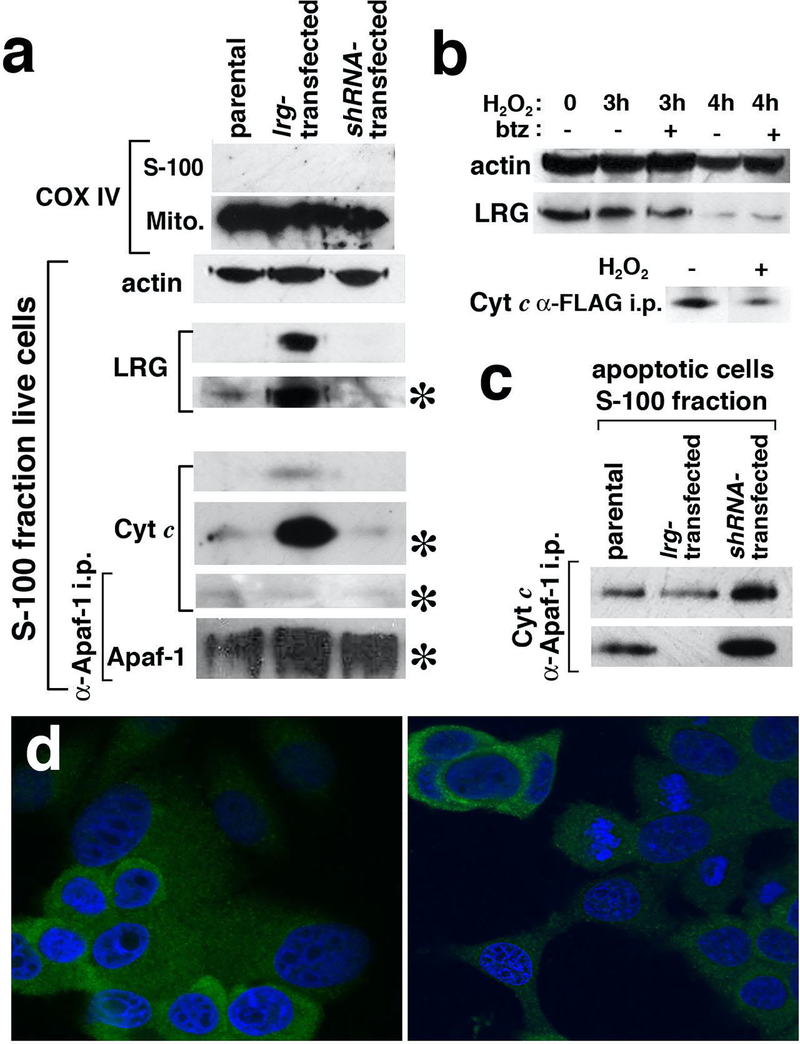
Figure 3. Western blotting in conjunction with immunoprecipitation identifies increased cytoplasmic Cyt c in lrg1-transfected cells and, following degradation of LRG1 in apoptotic cells, binding of Cyt c to Apaf-1.
(a) Antibodies specific for COX IV, a mitochondrial marker, confirmed by Western blotting the purity of the subcellular fractions of MCF-7 cells expressing different amounts of LRG1. LRG1 was detected in the cytosol (S-100 fraction) of lrg-transfected MCF-7 cells and to a lesser extent in the cytosol of parental cells using a more sensitive detection reagent. Cyt c was also found in the cytosol of live lrg1-transfected cells, although the amount of Cyt c-co-immunoprecipitated with Apaf-1 was weakly detected. Asterisks indicate the use of a more sensitive detecting reagent.
(b) LRG1 was degraded in peroxide-treated cells, however, degradation was not blocked by the proteasome inhibitor bortezomib (btz). Cyt c in the cytosol of lrg1-transfected cells was co-immunoprecipitated with recombinant LRG1. Less Cyt c was present in the pull down from cells treated with hydrogen peroxide, consistent with degradation of LRG1.
(c) In the cytosol of apoptotic cells, the amount of Cyt c co-immunoprecipitated with Apaf-1 was inversely related to the amount of LRG1 expressed. Results for two independent trials are shown.
(d) Degradation of LRG1 in hydrogen-peroxide-treated parental MCF-7 cells was confirmed by confocal microscopy. Cells were labeled using mAb 3C9.D5 specific for LRG1 and nuclei were stained using DAPI. Live cells are on the left and cells treated for 3 hrs. with 0.1% hydrogen peroxide are on the right.
Figure 2. LRG1 expression levels correlate with the ability of MCF-7 cells to survive apoptotic stress.
Cells transfected with the lrg1 gene were more resistant to cell death in response to hydrogen peroxide than were control cells, while cells transfected with LRG1-specific shRNA or infected with a lentivirus encoding a different LRG1 shRNA were more sensitive. Data represent averages of triplicate determinations ± standard deviations.
Increased Cyt c in the cytosol of lrg1-transfected MCF-7 cells without induction of apoptosis
Live and apoptotic cells were subjected to nitrogen cavitation and differential centrifugation to isolate the cytosolic (S-100) and mitochondrial fractions. A Western blot (Fig. 3a) showing COX IV, a mitochondrial protein, in the mitochondrial fractions but not in the S-100 fractions confirms effective separation of the two fractions. Actin served as a gel loading control for the cytosolic fractions.
LRG (45 kD) was readily detected in the cytosol of live lrg1-transfected cells by Western blotting using the Super Signal West Dura reagent (Fig. 3a). Further exposure of the blot to the Super Signal West Femto reagent (indicated by the asterisk) revealed a lesser amount of the 45 kD band in the cytosol of parental cells and even less in the cytosol of LRG1 knockdown cells. Similar results were observed in additional trials.
There was an unexpected increase of Cyt c in the cytosolic fraction from lrg1-transfected cells relative to the parental cells that was reproduced in other experiments. Cytosolic Cyt c was visible in Western blotting when the West Dura reagent was employed and was amplified to visibility in the parental and shRNA-transfected cells only with the West Femto reagent (Fig. 3a). The small amount of Cyt c that co-immunoprecipitated with Apaf-1 from all S-100 fractions (Fig. 3a) is likely attributable to the small number of dead cells in the cultures. Thus, apoptosis was not spontaneously induced in the lrg1-transfected cells despite the increase in cytosolic Cyt c. Survival of MCF-7 cells and other cells despite increased Cyt c in the cytoplasm has been observed previously and explained by regulatory mechanisms blocking apoptosis induction [31,32].
LRG1 degradation promotes binding of cytosolic Cyt c to Apaf-1 in apoptotic cells
Cyt c in live lrg-transfected cells was found associated with LRG1 as shown by Western blotting of the immunoprecipitates from the S-100 fraction (Fig. 3b). Following apoptosis induction with 0.1% hydrogen peroxide, Cyt c that immunoprecipitated with recombinant LRG1 decreased and this was consistent with degradation of LRG1 (Fig. 3b). To rule out relocation of LRG1 to subcellular sites other than the cytosol confocal microscopy was employed (Fig. 3d). A decrease in intensity of LRG1 in hydrogen peroxide-treated parental MCF-7 cells visualized with a specific mAb confirms that LRG1 is degraded. This is most evident within cells with apoptotic nuclei. LRG1 harbors a KEN box at residues 113–115 that is known to lead to ubiquitination in other proteins and subsequent degradation [33, 34]. The observed degradation of LRG1 did not appear to involve the proteasome as it was not inhibited in the presence of the proteasome inhibitor, bortezomib (40 nM, Fig. 3b) [35].
Note that Cyt c is not degraded in apoptosis of cultured cells and, several hours after apoptosis activation, is released into the extracellular space as an intact protein [36]. In the early phase of apoptosis as apoptosome formation is occurring some Cyt c molecules move to the endoplasmic reticulum causing calcium release and amplification of Cyt c translocation from mitochondria and Cyt c has also been shown to translocate to the nucleus [37].
In apoptotic cells at 3 hours during treatment with 0.03% hydrogen peroxide, the amount of Cyt c bound to Apaf-1 was observed to be inversely related to the level of LRG1 expression (Fig. 3c). Thus, in the S-100 fraction of cells in which LRG1 was knocked down by shRNA transfection there was more Cyt c associated with Apaf-1 than in the S-100 fraction of parental cells (Fig. 3c). Also, lesser amounts of Cyt c were associated with Apaf-1 in the S-100 fraction of lrg-transfected cells relative to that in parental cells. Results from two apoptosis trials are shown in Fig. 3c to demonstrate some variability in capturing cells at the same timepoint during apoptosis progression.
The secreted and intracellular forms of LRG1 in lrg1-transfected MCF-7 cells are differentially glycosylated
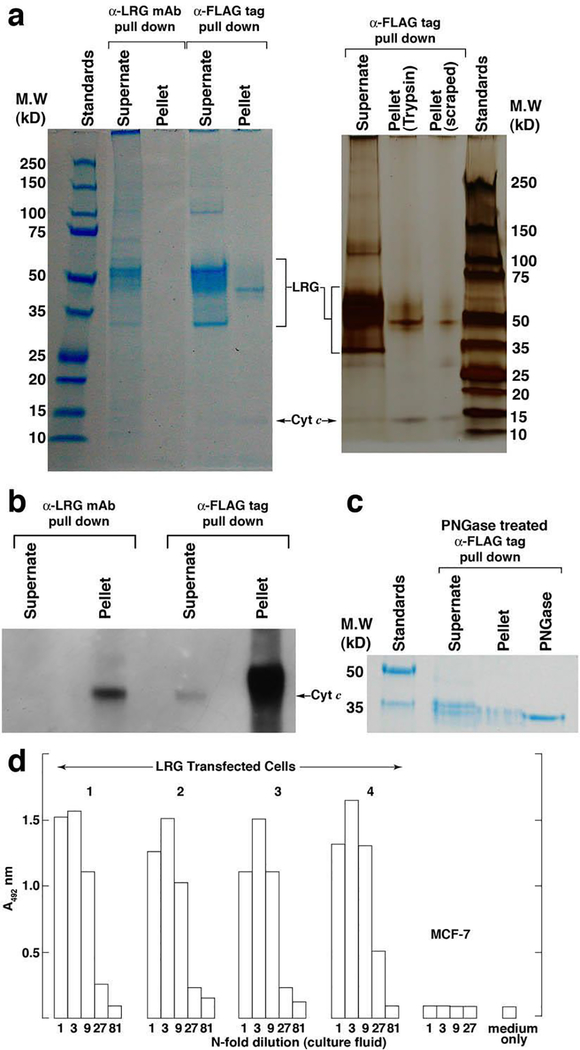
We compared LRG1 present in the supernate of lrg1-transfected cells to the intracellularly-retained protein with respect to glycosylation (Fig. 1a). Both the supernate and detergent-solubilized pellet of lrg1-transfected cells were incubated with anti-FLAG agarose beads or anti-LRG1 mAb-coated Sepharose 4B beads to immunoprecipitate recombinant LRG1, FLAG-tagged at the carboxyl terminus. Immunoprecipitated proteins were detected in SDS-PAGE gels with either the GelCode Blue stain or, the more sensitive, silver staining. In anti-FLAG tag immunoprecipitation, a prominent band at 45 kD was present in the pellet of the transfected cells, whether they were harvested by trypsinization or scraping, along with a band at 12.5 kD that was identified by Western blotting as Cyt c (Fig. 4b). The 45 kD band was identified as human LRG1 by mass spectrometry (MS) analysis (Table I). Although the LRG1-specific mAb 2F5.A2 was less effective in immunoprecipitating the cell-associated LRG (likely due to the decreased carbohydrate content of intracellular LRG1 that affected antibody recognition), Cyt c was still detectable in the pulldown by Western blotting. Importantly, Cyt c appeared to be the only protein detectable that co-immunoprecipitated with LRG1 in the cell pellets.
Figure 4. Lrg1-transfected MCF-7 cells express variably glycosylated forms of LRG1 most of which are secreted, while a 45 kD form capable of binding Cyt c is retained in the cells.
(a) A heterogeneous population of LRG1 polypeptides visualized by SDS-PAGE was immunoprecipitated from the supernate of lrg1-transfected cells employing antibody-coupled beads specific for LRG (2F5.A2) and the FLAG tag present in recombinant LRG1. On the left the gel was stained with GelCode Blue and on the right a separate gel was silver stained. Major bands were identified as LRG1 by MS (see Table I). A uniquely-sized polypeptide (45 kD) was retained in the cell pellet. Co-immunoprecipitation of Cyt c was also apparent.
(b) Western blotting confirmed the presence of Cyt c in the immunoprecipitates in “a.”
(c) PNGase F reduced the molecular weight of LRG1 present in both the supernate and cell pellet to a size approximating the non-glycosylated polypeptide (34 kD).
(d) Indirect ELISA demonstrated the presence of LRG1 in the supernates of four clones of lrg1-transfected MCF-7 cells but not in the supernate of non-transfected cells.
Table 1.
Confidence levels (%) for MS identification of individual peptides derived from recombinant polypeptides of lrg-transfected MCF-7 cells*
| Tryptic Peptides | Supernate: 33 kD | Supernate: 50 kD | Cell Pellet: 45 kD |
|---|---|---|---|
| 1–6 (GalN 2) | - | - | - |
| 7–12 | 89 | 95 | 61 |
| 13–58 (GlcN 44) | - | - | - |
| 59–84 | 99 | 99 | 99 |
| 85–90 | 43 | - | 84 |
| 91–113 | - | - | - |
| 114–129 | - | - | 99 |
| 130–140 | 99 | 99 | 99 |
| 141–142 | - | - | - |
| 143–156 (GlcN 151) | 99 | - | - |
| 144–155 (GlcN 151) | 99 | - | - |
| 157–174 | 99 | 99 | 99 |
| 175–181 | 98 | 75 | 97 |
| 182–204 | 99 | 99 | - |
| 182–188 | - | - | - |
| 189–204 | 99 | - | - |
| 189–194 | - | - | - |
| 195–204 | 99 | 99 | 99 |
| 205–212 | 99 | 99 | 99 |
| 213–215 | - | - | - |
| 216–225 | 99 | 99 | 99 |
| 213–225 | - | - | - |
| 226–256 (GlcN 234) | - | - | - |
| 257–277 | 99 | - | 99 |
| 278–283 | - | - | - |
| 284–285 | - | - | - |
| 286–293 (GlcN 290) | - | - | - |
| 294–301 | - | - | - |
| 302–310 | - | - | - |
| 294–310 | 99 | - | - |
| 311–312 | - | - | - |
| Total Coverage | 74% | 55% | 57% |
Protein confidence levels as determined by Protein Pilot 2.0.1 software; peptide mass error range was 3–48 ppm
The small amount of Cyt c in the supernates compared to that in the immunoprecipitated cell pellets (Fig. 4b) likely represents protein released from a few dying cells in the culture [36]. A heterogeneous mixture of polypeptides was present in the immunoprecipitated supernates resulting from cells over-expressing LRG1. This mixture contained differentially glycoslyated forms of LRG1 that were reduced in size to around 35 kD by PNGase F treatment (Fig. 4c). The cell-associated polypeptide was reduced to similar size. The more extensive deglycosidation observed in Fig. 4c compared to Fig. 1c is probably due to the greater purity of the material that was subjected to PNGase treatment (immunoprecipitated protein versus total cell extract). Both the dominant 50 kD band and a minor 34 kD band in the supernate were identified as LRG1 by MS (Table I). The 34 kD band is the size expected for the non-glycosylated protein [33]. Consistent with this interpretation this band contained two overlapping peptides comprising residues 144–155 not found at the expected mass in the MS of the 45 kD and 50 kD polypeptides likely due to N-glycosylation at residue 151. The 34 kD polypeptide is likely to be full-length given that it contains peptides corresponding to residues 7–12 and residues 294–310, where the carboxyl terminus is residue 312. Other non-glycosylated forms of peptides that are glycosylated in the 50 kD protein were not observed by MS in the 34 kD polypeptide, perhaps due to poor recovery or because their masses were outside the range screened in the analysis.
Parental MCF-7 cells do not secrete detectable levels of LRG1
To determine whether LRG1 is secreted by parental MCF-7 cells, the supernate was screened using an indirect ELISA for LRG1 [19]. Prior to culture the fresh medium was adsorbed on Cyt c-Sepharose beads to remove bovine LRG1 present in fetal bovine serum that would have interfered with the assay which involves capture of LRG1 by plate-adsorbed Cyt c in the first step. As shown in Fig. 4d, LRG1 was not detected in the supernate of MCF-7 cells but was readily detected in the supernates of four different lrg1-transfected clones in amounts approximating 1 μg/ml at the time of culture fluid testing based on a standard curve from an assay of serum LRG. Furthermore, in several cell lines, including MCF-7 cells, 45 kD LRG1 was the dominant species (Fig. 1a). This suggests that LRG1 is not a substantive secretion product of these cell lines.
Subcellular localization of LRG1 in parental and lrg-transfected MCF-7 cells visualized by confocal microscopy
LRG1 is expected to localize, at least in part, to the Golgi complex in cells because the serum protein is N-glycosylated at four sites and O-glycosylated at one site (Table I). The LRG1-specific mAb employed for ELISA reacted poorly with incompletely glycosylated forms of LRG1 (Fig. 4a) so we generated a mouse mAb (mAb 3C9.D5) specific for amino acid residues 274–283 of human LRG1, a segment not targeted for glycosylation. By Western blot this mAb reacted poorly with more fully glycosylated forms of LRG1 (results not shown). We used mAb 3C9.D5 along with rabbit polyclonal antibodies specific for the Golgi marker, GRASP 65, to visualize the localization of LRG1 in MCF-7 cells relative to the Golgi by indirect immunofluorescent confocal microscopy. Merging of the red (GRASP 65) and green (LRG1) fluorescent channels yielded yellow showing substantial co-localization of the two markers in non-transfected MCF-7 cells (Fig. 5a, right panel).
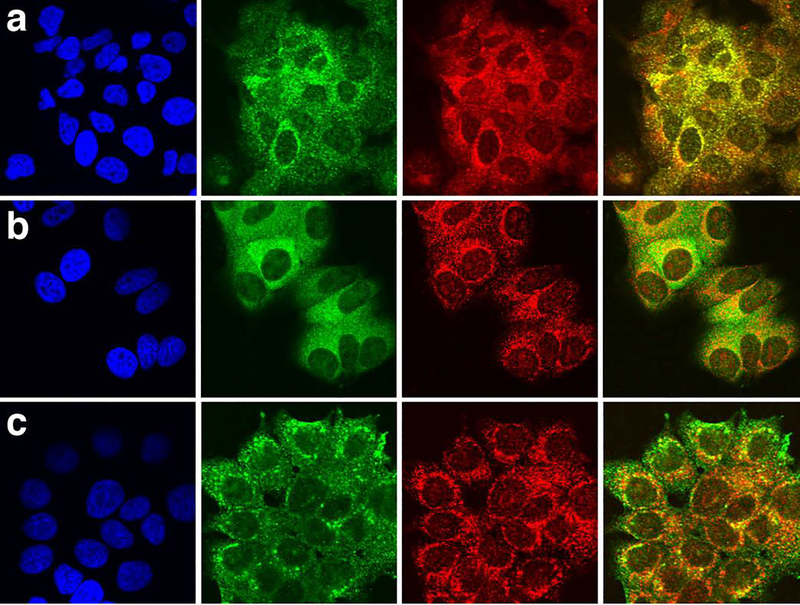
Figure 5. Intracellular localization of LRG1 in parental MCF-7 cells and in cells transfected with lrg1 by confocal immunofluorescent microscopy.
(a) In MCF-7 cells, LRG1 (green) co-localized with the Golgi marker, GRASP 65 (red). LRG was detected with LRG1 peptide-specific mAb 3C9.D5. Nuclei (blue) were stained with DAPI (left panel). In the far right panel the green and red fluorescent images are merged.
(b) In lrg1-transfected MCF-7 cells, LRG1 detected with mAb 3C9.D5 (green) co-localized with GRASP 65 (red) and also appeared in a diffuse pattern outside the Golgi.
(c) Recombinant LRG1 detected with an anti-FLAG tag mAb (green) partially co-localized with GRASP 65 (red) in lrg1-transfected MCF-7 cells and was also prominent just below the cell surface consistent with its secretion from lrg1-transfected cells (see Fig. 4d).
LRG1 in lrg1-transfected cells partially localized to the Golgi complex using the same mAb (Fig. 5b) but was more prominent outside the Golgi in the cytoplasm as shown by the diffuse green staining in the merged image (Fig. 5b, last panel). This is consistent with identification of LRG1 in the S-100 fraction of lrg1-transfected cells (Fig. 3a).
An antibody specific for the FLAG tag of recombinant LRG1 (Fig. 5c, green label) detected LRG1 in lrg-transfected MCF-7 cells partially co-localized with GRASP 65 but more prominently in a punctate pattern below the plasma membrane. This likely represents staining in secretory vesicles, consistent with the secretion of recombinant LRG1 (Fig. 4d).
Discussion
The evolutionarily conserved region around residue 72 on Cyt c is known to be involved in the interaction between Cyt c and Apaf-1 [38,39]. We previously reported that LRG1 also binds in this region and inhibits the interaction between Apaf-1 and Cyt c in vitro [7]. Therefore, we posited that LRG1 may act as a survival factor delaying the onset of apoptosis in cells that may express it in the cytoplasm by blocking the binding of Apaf-1 to Cyt c and preventing formation of the apoptosome. In this study resistance to hydrogen peroxide-induced apoptosis directly correlated with the level of LRG1 expression. The protection afforded by lrg1 transfection is comparable to the protection provided against peroxide-induced apoptosis by bcl-2 overexpression in mouse epidermal cells [40].
We show that although more Cyt c could be detected in the cytosol of live lrg1-transfected cells than in the cytosol of either parental cells or LRG1 knockdown cells, it was not bound to Apaf-1 and, therefore, unable to induce apoptosis. Overexpression of LRG1 appears to have allowed trapping Cyt c molecules that are released from mitochondria into the cytosol at low levels. The affinity of LRG1 for Cyt c has been reported to be among the highest for any interaction between proteins (KD=1.58 × 10−13 M, ref. 41). Following committed apoptotic signaling as a consequence of hydrogen peroxide treatment, cytosolic LRG1 was degraded allowing Cyt c to bind Apaf-1 and activate the intrinsic apoptotic pathway.
LRG1 was secreted by lrg1-transfected cells but not by parental MCF-7 cells, at least in amounts that could be detected. LRG1 was found in the cytosol of lrg1-transfected cells, detected at a lower level in the cytosol of parental cells, and at an even lower level in lrg-knockdown cells. Furthermore, the amount of Cyt c in the cytosol of apoptotic cells in association with Apaf-1 was inversely related to the expression of LRG1. Thus, protection of MCF-7 cells from apoptosis in this study appears to be due to cytoplasmic LRG1 and not to extracellular LRG1, a survival mechanism we previously described which protects lymphocytes from the toxicity of extracellular Cyt c [7].
Secreted LRG1 in lrg1-transfected MCF-7 cells was varied in size owing to different extents of glycosylation, although the dominant form was approximately 50 kD which is the size of serum LRG1 [33]. This form was not prominent in parental MCF-7 cells or in other cells of mammary tissue origin.
A partially glycosylated form of LRG1 (45 kD) that had the capacity to bind Cyt c was retained in the lrg1-transfected cells. A similarly-sized LRG1 polypeptide was the dominant form of LRG1 in non-transfected MCF-7 cells, other breast cancer cell lines, and primary, mammary epithelial cells by Western blotting. There is considerable evidence for the presence of partially glycosylated proteins in cellular compartments outside the ER and Golgi [42]. For example, clusterin (apolipoprotein J) which plays an anti-apoptotic role by preventing BAX-mediated Cyt c release from mitochondria, has been reported to translocate from the Golgi to the cytosol under stress conditions [43,44].
Our findings contribute to an understanding of the function of LRG1 and present a role for intracellular LRG1 in cell survival. In the cytoplasm LRG1 binds molecules of Cyt c that may be released at low levels from mitochondria in the absence of committed apoptotic signaling. This may explain why LRG1 mRNA is transiently upregulated in cells recovering from chemical trauma [14,15]. There are now several ligands of Cyt c including Hsp27, nucleotides, and LRG1 that have been shown to block formation of the apoptosome [5,6]. While their apoptosis-inhibiting functions appear to be redundant, these ligands have other roles and they are independently regulated. Thus, each may serve to interfere with activation of the intrinsic pathway of apoptosis under the specialized conditions in which they are optimally expressed.
The mechanism we describe here may not explain the survival function of extracellular LRG1 that we reported previously for lymphocytes and that other researchers have since observed for other cell types [8–11]. Cyt c is well documented as a damage-associated molecular pattern (DAMP) when released from dying cells [45,46]. Previously we detected Cyt c in cultures of live lymphocytes presumably derived from dying cells and, in combination with extracellular LRG1, enhanced cell viability was observed [7]. We proposed that the protection afforded by LRG1 involved cell signaling based on two findings. First, a low molar ratio of LRG1 to Cyt c promoted cell survival indicating that blockade of Cyt c was not the mechanism. Second, while human LRG1 binds both human and mouse Cyt c, it did not promote survival of mouse lymphocytes suggesting interaction of LRG1 with a species-specific component in the mouse cells.
Paradoxically, in some studies extracellular LRG1 was reported to have an opposite effect, inducing apoptosis or growth suppression, via a TGF-β1 signaling pathway [12,13]. In addition, LRG1 has been shown to promote angiogenesis via TGF-β1 signaling [47]. We have found that TGF-β1 and Cyt c bind LRG1 independently of one another (unpublished). Thus, it is possible that signaling pathways impacted by extracellular LRG1 could be directed not only by its association with TGF-β1 but also by binding Cyt c, either in combination, or in conjunction with some other molecule(s). A recent study implicates EGF receptor signaling in the role of LRG1 as promoter of cell survival [10]. This raises the possibility that TGF-β1 receptor signaling via LRG1 induces apoptosis, while LRG1 signaling through the EGF receptor promotes cell viability. It remains to be determined if Cyt c plays a role in EGF receptor-mediated cell-survival signaling.
Acknowledgments
The authors thank Tim Leonard for preparation of the figures, Celia Choh for help with the viability assays, and Samantha Van Hove for assisting with the lentivirus infections. This work was supported by grants from the Minnesota Medical Foundation and University of Minnesota Graduate School (to R.J.). The authors recognize the Center for Mass Spectrometry and Proteomics at the University of Minnesota and various supporting agencies, including the National Science Foundation for Major Research Instrumentation grants 9871237 and NSF-DBI-0215759 used to purchase the instruments employed in this study and National Institutes of Health grant R01 CA157971 (to A.K.).
Footnotes
Conflicts and Competing Interests
The University of Minnesota and/or R.J. have commercially licensed two of the monoclonal antibodies employed in this study. There are no other potentially conflicting interests.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Contributor Information
Ronald Jemmerson, Department of Microbiology and Immunology, University of Minnesota Medical School, Minneapolis, MN 55455 USA.
Katherine Staskus, Department of Microbiology and Immunology.
LeeAnn Higgins, Department of Biochemistry, Molecular Biology, and Biophysics.
Kathleen Conklin, Department of Genetics, Cell Biology, and Biophysics, Masonic Cancer Center.
Ameeta Kelekar, Department of Laboratory Medicine and Pathology, Masonic Cancer Center.
References
- 1.Liu X, Kim CN, Yang J, Jemmerson R, Wang X (1996) Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 2.Jiang X, Wang X (2004) Cytochrome c-mediated apoptosis. Annu. Rev. Biochem. 73, 87–106 [DOI] [PubMed] [Google Scholar]
- 3.Yang et al. (1997) Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science 275, 1129–1132 [DOI] [PubMed] [Google Scholar]
- 4.Kluck RM et al. (1997) The release of cytochrome c from mitochondria: a primary Site for Bcl-2 regulation of apoptosis. Science 275, 1132–1136 [DOI] [PubMed] [Google Scholar]
- 5.Bruey JM et al. (2000) Hsp27 negatively regulates cell death by interacting with cytochrome c. Nature Cell Biol. 2, 645–652 [DOI] [PubMed] [Google Scholar]
- 6.Chandra D et al. (2006) Intracellular nucleotides act as critical prosurvival factors by binding to cytochrome c and inhibiting apoptosome. Cell 125, 1333–1346 [DOI] [PubMed] [Google Scholar]
- 7.Codina R, Vanasse A., Kelekar A, Vezys V, Jemmerson R (2010) Cytochrome c-induced lymphocyte death from the outside in: inhibition by serum leucine-rich alpha-2-glycoprotein-1. Apoptosis 15, 139–152 [DOI] [PubMed] [Google Scholar]
- 8.Zhong D, Zhao S, He G, Li J, Lang Y, Ye W, Li Y, Jiang C, Li X (2015) Stable knockdown of LRG1 by RNA interference inhibits growth and promotes apoptosis of glioblastoma cells in vitro and in vivo. Tumour Biol. 36, 4271–4278 [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, Zhong X, Zhang J, Fang J, Ge Z, Li X (2017) Lrg1 promotes proliferation and inhibits apoptosis in colorectal cancer cells via RUNX1 activation. PLOS One 12(4): e175122. doi. 10.1371/journal.pone0175122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie ZB, Zhang YF, Jin C, Mao YS, Fu DL (2019) LRG1 promotes pancreatic cancer growth and metastasis via modulation of the EGFR/p38 signaling. J. Exper. Clin. Cancer Res. 38, Article 75 doi: 10.1186/s13046-019-1088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Luo R, Cheng Y, Liu T, Dai W, Li Y, Ge S, Xu G (2020) Leucine rich α2-glycoprotein-1 upregulation in plasma and kidney of patients with lupus nephritis. BMC Nephrology 21, Article 122 doi: 10.1186/s12882-020-01782-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takumoto N et al. , (2015) Leucine rich α−2-glycoprotein promotes TGF-β1 mediated growth suppression in the Lewis lung carcinoma cell line. Oncotarget 6, 11009–11022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin J et al. (2019) LRG1 promotes apoptosis and autophagy through TGF-β-Smad 1/5 signaling. Neuroscience 413, 123–134. [DOI] [PubMed] [Google Scholar]
- 14.Sciuto AM et al. (2005) Genomic analysis of murine pulmonary tissue following carbonyl chloride inhalation. Chem. Res. Toxicol. 18, 1654–1660 [DOI] [PubMed] [Google Scholar]
- 15.Zhao P et al. (2002) Slug is a novel downstream target of MyoD. Temporal profiling in muscle regeneration. J. Biol. Chem. 277, 30091–10101 [DOI] [PubMed] [Google Scholar]
- 16. ncbi.nlm.nih.gov (gene resources)
- 17.Xiang J, Wan C, Guo R, Guo D (2016) Is hydrogen peroxide a suitable apoptosis inducer for all cell types? BioMed Research International doi/ 10.1155/2016/7343965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kearney JF, Radbruch A, Liesegang B, Rajewsky K (1979) A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J. Immunol. 123, 1548–1550 [PubMed] [Google Scholar]
- 19.Weivoda S et al. (2008) ELISA for human serum leucine-rich alpha-2-glycoprotein-1 employing cytochrome c as the capturing ligand. J. Immunol. Methods 336, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D (1997) Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15, 871–875 [DOI] [PubMed] [Google Scholar]
- 21.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A (2007) Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death & Differ. 14, 500–510 [DOI] [PubMed] [Google Scholar]
- 22.Farah IO (2005) Assessment of cellular responses to oxidative stress using MCF-7 breast cancer cells, black seed (N. Sativa L.) extracts and H2O2. Int. J. Environ. Res. & Pub. Health 2, 411–419 [DOI] [PubMed] [Google Scholar]
- 23.Cummings C, Walder J, Treeful A, Jemmerson R (2006) Serum leucine-rich alpha-2-glycoprotein-1 binds cytochrome c and inhibits antibody detection of this apoptotic marker in enzyme-linked immunosorbent assay. Apoptosis 11, 1121–1129 [DOI] [PubMed] [Google Scholar]
- 24.Akkina SK, Zhang Y, Nelsestuen GL, Oetting WS, Ibrahim H N (2009) Temporal stability of the urinary proteome after kidney transplant: more sensitive than protein composition? J. Proteome Res. 8, 94–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shilov IV et al. (2007) The Paragon Algorithim, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 26.Bini L et al. (1997) Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis 18, 2832–2841 [DOI] [PubMed] [Google Scholar]
- 27.Uhlen M et al. (2005) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell Proteomics 4, 1920–1932 [DOI] [PubMed] [Google Scholar]
- 28.Kottke TJ et al. (2002) Lack of correlation between caspase activation and caspase activity assays in paclitaxel-treated MCF-7 breast cancer cells. J. Biol. Chem. 277, 804–815 [DOI] [PubMed] [Google Scholar]
- 29.Maley F, Trimble RB, Tarentino AL, Plummer TH (1989) Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180, 195–204 [DOI] [PubMed] [Google Scholar]
- 30.Hossler P, Khattak SF, Zheng J (2009) Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology 19, 936–949 [DOI] [PubMed] [Google Scholar]
- 31.Li F et al. (1997) Cell-specific induction of apoptosis by microinjection of cytochrome c. J. Biol. Chem. 272, 30299–30305 [DOI] [PubMed] [Google Scholar]
- 32.Oliver L et al. (2005) Constitutive presence of cytochrome c in the cytosol of a chemoresistant leukemic cell line. Apoptosis 10, 277–287 [DOI] [PubMed] [Google Scholar]
- 33.Takahashi N, Takahashi Y, Putnam FW (1985) Periodicity of leucine and tandem repetition of a 24-amino acid segment in the primary structure of leucine-rich α2-glycoprotein of human serum. Proc. Natl. Acad. Sci. U.S.A. 82, 1906–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfleger CM, Kirschner MW (2000) The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes & Develop. 14, 655–665 [PMC free article] [PubMed] [Google Scholar]
- 35.Roelofs J (2015) Proteosome inhibition by bortezomib: A left hook and a right punch. E Bio Medicine 2, 619–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jemmerson R, LaPlante B, Treeful A (2002) Release of intact, monomeric cytochrome c from apoptotic and necrotic cells. Cell Death and Differentiation 9, 538–548 [DOI] [PubMed] [Google Scholar]
- 37.Katiuska G-A et al. (2019) New moonlighting functions of mitochondrial cytochrome c in the cytoplasm and nucleus. FEBS Letters 593, 3101–3119 [DOI] [PubMed] [Google Scholar]
- 38.Kluck RM et al. (2000) Determinants of cytochrome c pro-apoptotic activity: The role of lysine 72 trimethylation. J. Biol. Chem. 275, 16127–16133 [DOI] [PubMed] [Google Scholar]
- 39.Yu T, Wang X, Purring-Koch C, Wei Y, McLendon G (2001) A mutational epitope for cytochrome c binding to the apoptosis protease activation factor (Apaf-1). J. Biol. Chem. 276, 13034–13038 [DOI] [PubMed] [Google Scholar]
- 40.Amstad PA et al. (2001) BCL-2 is involved in preventing oxidant-induced cell death and in decreasing oxygen radical production. Redox Report 6, 351–362 [DOI] [PubMed] [Google Scholar]
- 41.Shirai R, Gotou R, Hirano F, Ikeda K, Inoue S (2010) Autologous extracellular cytochrome c is an endogenous ligand for leucine-rich alpha-2-glycoprotein and beta-type phospholipase A2 inhibitor. J. Biol. Chem. 285, 21607–21614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Funakoshi Y, Suzuki T (2009) Glycobiology in the cytosol: The bitter side of a sweet world. Biochim. Biophys. Acta 1790, 81–94 [DOI] [PubMed] [Google Scholar]
- 43.Zhang H et al. (2005) Clusterin inhibits apoptosis by interacting with activated BAX. Nat. Cell Biol. 7, 909–915 [DOI] [PubMed] [Google Scholar]
- 44.Nizard P et al. (2007) Stress-induced retrotranslocation of clusterin/ApoJ into the cytosol. Traffic 8, 554–565 [DOI] [PubMed] [Google Scholar]
- 45.Eleftheriadis T, Pissas G, Liakopoulos V, Stefanidis I (2016) Cytochrome c as a potentially clinical useful marker of mitochondrial and cellular damage. Front. Immunol. doi. 10.3389/fimmu.2016.00279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Comish PB, Carlson D, Kang R, Tang D (2020) Damage-associated molecular patterns and the systemic consequences of severe thermal injury. J. Immunol. 205, 1189–1197 [DOI] [PubMed] [Google Scholar]
- 47.Wang X et al. (2013) LRG1 promotes angiogenesis by modulating endothelial TGF-β signaling. Nature 499, 306–311 [DOI] [PMC free article] [PubMed] [Google Scholar]