Abstract
Multiciliated cells (MCCs) are extremely highly-differentiated, presenting >100 cilia and basal bodies. Therefore, MCC fate is thought to be terminal and irreversible. We analyzed how MCCs are removed from the airway-like mucociliary Xenopus epidermis during developmental tissue remodeling. We found that a subset of MCCs undergoes lateral line-induced apoptosis, but that the majority coordinately trans-differentiate into Goblet secretory cells. Both processes are dependent on Notch signaling, while the cellular response to Notch is modulated by Jak/STAT, Thyroid hormone and mTOR signaling. At the cellular level, trans-differentiation is executed through loss of ciliary gene expression including foxj1 and pcm1, altered proteostasis, cilia retraction, basal body elimination as well as initiation of mucus production and secretion. Our work describes two modes for MCC loss during vertebrate development, the signaling regulation of these processes, and demonstrates that even cells with extreme differentiation features can undergo direct fate conversion.
Keywords: cilia, basal body, mucus, autophagy, proteasome, apoptosis, transdifferentiation, pericentriolar material, Xenopus, goblet cell
eTOC
Tasca et al. describe how multiciliated cells are removed from airway-like mucociliary epithelia during developmental tissue remodeling. In addition to cell death, these highly specialized cells can also change their identity towards another cell type, which was considered impossible, challenging the concept of terminal differentiation.
Graphical Abstract
Introduction:
The fate of terminally-differentiated cells appears fixed, but direct fate change through reprogramming techniques or trans-differentiation in regeneration and pathogenesis demonstrate that cell identity is more flexible than previously anticipated (Merrell and Stanger, 2016; Wells and Watt, 2018). This suggests that maintaining cell identity is not an intrinsic feature of differentiation but relies on extrinsic cues from the environment. While it is accepted that specialized cells can be induced from more generic types, e.g. fibroblasts or stem cells (Soldner and Jaenisch, 2018), it is debated to which degree conversion is possible in cells that have generated highly specialized morphological features.
Multiciliated cells (MCCs) are the only cells that form >100 motile cilia and basal bodies, i.e. modified centrioles generated through deuterosome-mediated amplification (Klos Dehring et al., 2013). MCCs are also subject to a permanent cell cycle block due to the deployment of centrioles to ciliogenesis, mutually exclusive with cell division (Izawa et al., 2015). Hence, it is widely considered impossible for MCCs to undergo fate change during normal development and in regeneration.
In mucociliary epithelia, such as the airway epithelium or the embryonic epidermis of Xenopus tadpoles, correct balance between MCCs and secretory cells provides the functional basis for removal of particles and pathogens to prevent infections and to maintain organismal oxygenation (Walentek and Quigley, 2017). Mucociliary epithelial remodeling and MCC loss are observed in human chronic lung disease as well as during metamorphosis in Xenopus, however, it remains unresolved how and why MCCs are lost in various conditions (Hogan et al., 2014).
We studied the process of MCC loss in the developing epidermis of Xenopus and found that MCCs are first locally removed by lateral line-dependent apoptosis, but later globally removed through trans-differentiation into a mucus-secretory cell type. Both processes are induced by a changing Notch signaling environment and modulated by Jak/STAT, Thyroid hormone and mTOR signals. We show that MCC trans-differentiation occurs during normal vertebrate development, involves coordinated cilia retraction, cytoskeletal rearrangement, elimination of ciliary and basal body material as well as the initiation of mucus production and secretion. Our data indicate that trans-differentiation is controlled by changing the balance between autophagy and proteasomal degradation, similar to primary cilia retraction during the cell cycle, thus, suggesting a broadly employed molecular mechanism for cilia maintenance in various cell types.
Results:
Local loss of MCCs correlates with lateral line organ emergence
Like the mammalian airway epithelium, the Xenopus embryonic epidermis is composed of MCCs, ionocytes (ISCs), mucus-secreting cells (Goblet cells and small secretory cells; SSCs), and sub-epithelial basal stem cells (Haas et al., 2019; Walentek and Quigley, 2017). Once established by stage (st.) 32, cell type composition is generally invariable between individuals (Haas et al., 2019), but MCCs were previously reported to be lost at the onset of metamorphosis in amphibians (Nishikawa et al., 1992; Smith et al., 1988). Using scanning electron microscopy (SEM), we found that MCCs were strongly reduced at st. 45 and largely lost by st. 47, while ISCs, SSCs and Goblet cells persisted (Figure 1A).
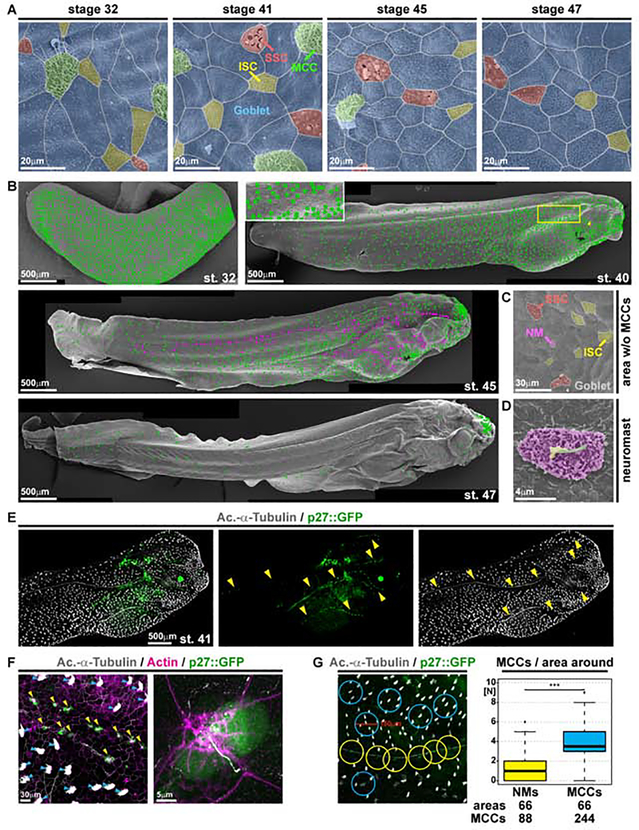
Figure 1. Characterization of epidermal MCC loss during Xenopus development.
(A-D) Pseudo-colored scanning electron micrographs from developmental stages (st.) 32 through 47. Cf. Figure S1 for non-colored images. (A) Analysis of the changing composition of mucociliary cell types in the epidermis shows progressive loss of MCCs (green), while ionocytes (ISCs, yellow), small secretory cells (SSCs, red) and mucus-secreting Goblet cells (blue) remain present. (B) Analysis of MCC-loss patterns on whole tadpoles. MCCs are marked by green dots. Neuromasts are marked by magenta dots. Box indicates magnified area. Arrowhead indicates MCC loss ventral to the eye. (C) Magnification of skin area devoid of MCCs at st. 45 reveals presence of lateral line neuromast (NM, purple). (D) Magnified image of a neuromast. Kinocilium (green), stereocilia (yellow), support cells (purple). (A-D) st. 32 N = 7; st. 40/41 N = 6; st. 45 N = 3; st. 47 N = 3 embryos. (E) Confocal micrograph of NMs (p27::GFP, green; yellow arrowheads) and MCCs (Ac.-α-Tubulin, grey). The picture was reconstructed from multiple images. (F) Confocal micrograph of p27::GFP (green; NMs marked by yellow arrowheads) transgenic tadpole stained for cilia (Ac.-α-Tubulin, grey; MCCs marked by blue arrowheads) and F-actin (Actin, magenta) shows lack of MCCs around NMs and GFP expression in a subset of ciliated neuromast cells. (E, F) N = 3 embryos. (G) Quantification of MCCs in areas with 150μm diameter around NMs (yellow) or MCCs (blue) shows reduced density of MCCs around NMs in the specimen depicted in (E). Mann Whitney test, *** P < 0.001. Images in B, E and F were reconstructed from multiple individual micrographs.
We first detected local loss of MCCs at st. 40/41 in areas where lateral line neuromasts (NMs) emerge by st. 45 (Figure 1B–D and S1A–F). The lateral line develops from placodes around the otic vesicles that migrate in multiple streams to populate the head, trunk and tail with numerous NMs (Winklbauer, 1989). NMs sense external fluid flows for orientation, and MCCs produce fluid flow by coordinated beating of motile cilia. Thus, we wondered if MCCs are removed to allow for lateral line function. Using correlative light and electron microscopy, immunofluorescent staining for Acetylated-α-tubulin (Ac.-α-tubulin), and a transgenic NM marker (p27::GFP) (Carruthers et al., 2003; Rubbini et al., 2015), we confirmed that p27::GFP is expressed in Xenopus NMs and that MCC loss is initially restricted to GFP(+) areas (Figure 1E–G and S1E). In areas with decreased MCC density, we also detected cells with altered morphology that indicated shedding (Figure S1H,I).
We transplanted the lateral line primordium by transfer of the otic vesicle and adjacent tissue at st. 18–22 from membrane RFP injected donors into hosts in which MCCs were marked by GFP expression using a MCC-specific promoter (αtub::GFP) (Stubbs et al., 2006). This revealed MCCs above the lateral line leading edge, but lack of MCCs at positions where NMs have emerged, indicating MCC removal during NM intercalation (Figure S2A–C).
Lateral line-derived Notch signaling induces MCC apoptosis
The lateral line primordium and NMs are signaling centers expressing ligands from the Notch, Wnt and FGF signaling pathways that regulate morphogenesis and proliferation (Dalle Nogare and Chitnis, 2017). Notch signaling inhibits MCC specification (Deblandre et al., 1999), and mature mammalian MCCs keep expressing Notch receptors, although at lower levels than secretory cells (Pardo-Saganta et al., 2015). We therefore tested if Notch could also be employed to induce MCC removal in Xenopus.
Analysis of all Notch ligands revealed high levels of jagged1 (jag1) expression in the lateral line primordium as well as in mature NMs (Figure 2B,D and S2D–G). Expression of the Notch target gene hes1 further demonstrated transient Notch signaling activity in NMs and epidermal cells in contact with the lateral line, and hes1 was downregulated after Notch inhibition using the γ-Secretase inhibitor DAPT (Figure 2E and S2E–J). TUNEL staining showed apoptosis in the epidermis along the paths of lateral line migration (Winklbauer, 1989) (Fig. 2A,C and S2D–F), and MCCs close to the lateral line specifically stained for activated Caspase 3 protein (Figure 2F). These data suggested that Notch ligand expression by the lateral line could induce local Notch signaling in MCCs and cell type-specific apoptosis.
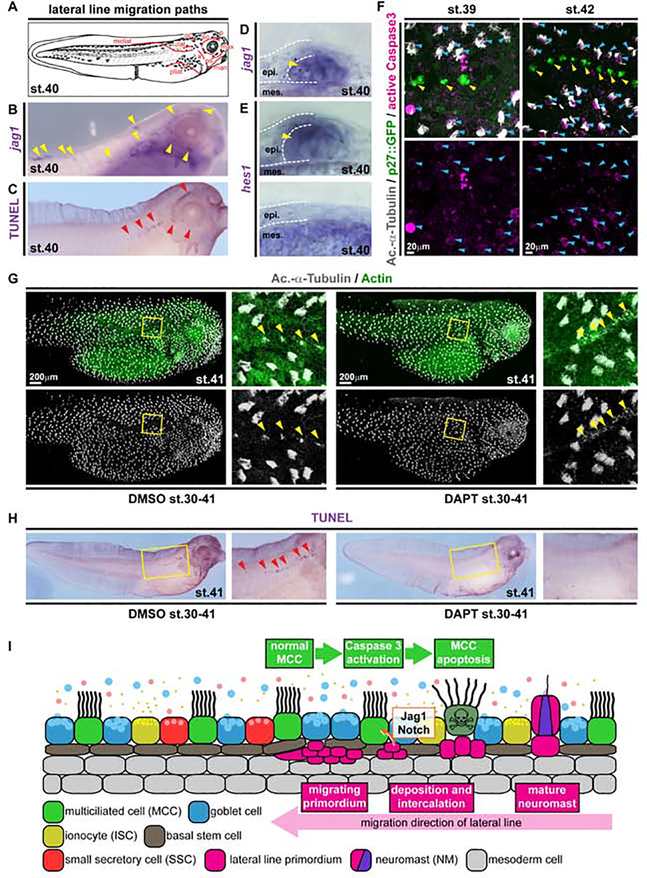
Figure 2. Local loss of MCCs is induced by Notch signaling from the emerging lateral line.
(A) Schematic representation of lateral line migration patterns in st. 40 Xenopus tadpoles. (B) In situ hybridization shows jagged1 (jag1, purple) expression in the lateral line primordium and in NMs (yellow arrowheads). N = 3 embryos. (C) TUNEL staining (purple) reveals apoptotic cells clustering along the lateral line migration paths (red arrowheads). N = 13 embryos. (D-E) Sections of NMs and epidermal area after in situ hybridization staining for jag1 (D) and hes1 (E) at stage 40. (D) N = 4 embryos. (E) N = 4 embryos. (F) Active Caspase 3 (magenta) staining in MCCs (Ac.-α-Tubulin, grey; blue arrowheads) located in proximity to NMs (p27::GFP, green; yellow arrowheads) at stages 39 (N = 6 embryos) and 42 (N = 5 embryos). (G) Notch signaling inhibition by DAPT prevents lateral line induced MCC (Ac.-α-Tubulin, grey) loss in the presence of neuromasts (yellow arrowheads). F-actin (Actin, green) was used as counterstain in confocal images. (DMSO N = 11; DAPT N = 13 embryos) (H) Notch signaling inhibition by DAPT application prevents induction of apoptosis (TUNEL staining, purple; red arrowheads) by the lateral line. Magnified areas in G,H are indicated by yellow boxes. Cf. Figure S3 for quantification of results. (DMSO N = 24; DAPT = 25 embryos) Images in F,G were reconstructed from multiple individual micrographs. (I) Schematic representation of lateral line induced MCC removal.
Next, we tested the functional relationship between lateral line, Notch and MCC loss. Ablation of the pre-migratory lateral line primordium in transgenic p27::GFP embryos at st. 28–30 resulted in loss of NMs and prevented MCC removal (Figure S3A).
Notch inhibition by DAPT treatment also prevented local loss of MCCs, reduced TUNEL and Caspase 3 signals along the lateral line, but without interfering with lateral line migration or jag1 expression (Figure 2G,H and S3B–K). Interestingly, NMs failed to properly intercalate in DAPT treated specimens, indicating that MCCs have to be removed by Notch signaling to make space in the epidermis for NM emergence (Fig. S3C,H).
In summary, these experiments revealed that high Notch signaling derived from the migrating lateral line primordium and maturing NMs induces local cell type-specific apoptosis in MCCs (Figure 2I).
Notch and Jak/STAT signaling regulate MCC to Goblet cell trans-differentiation
Extensive epidermal TUNEL staining was missing from areas farther away from the lateral line (Figure S2D–F) indicating an alternative mode of MCC removal there. We stained tadpoles between st. 32 and 46 for MCC cilia (Ac.-α-tubulin) and secretory cell mucus (PNA) to identify cell types, and for F-actin (Actin) to outline cell borders and to assess cell morphology. Confocal microscopy revealed altered apical F-actin morphology in a subset of MCCs, which also stained positive for mucus (Figure 3A). While the overall number of identifiable MCCs decreased over time, the proportions of PNA(+) MCCs and Goblet cells increased in areas at distance to the lateral line (Figure 3A,B, S4A and S5A). This suggested MCC to Goblet cell trans-differentiation as an additional mechanism for MCC removal in Xenopus development.
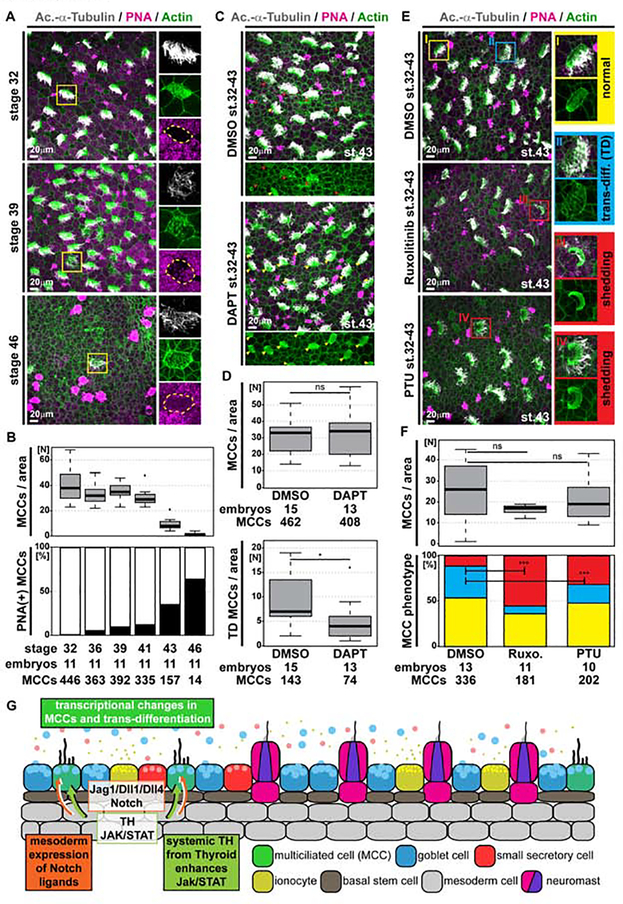
Figure 3. Global loss of MCCs through trans-differentiation into Goblet cells is regulated by Notch, Jak/STAT and Thyroid hormone signaling.
(A,C,E) Confocal micrographs of the epidermis stained for MCCs (Ac.-α-Tubulin, grey), F-actin (Actin, green) and mucus (PNA staining, magenta). (A) Analysis of samples from stage 32 to 46 reveals MCCs with altered apical F-actin morphology, which stain positive for mucus, indicating MCC trans-differentiation into Goblet cells. Magnified MCCs are indicated by yellow boxes. N = 11 embryos per stage. (B) Quantification of total MCC numbers and proportion of PNA(+) MCCs between stage 32 and 46. (C) Notch inhibition by DAPT treatment reduces the number of MCCs showing signs of trans-differentiation (red arrowheads). Normal MCCs indicated by yellow arrowheads. DMSO N = 15; DAPT N = 13 embryos. (D) Quantification of total MCC numbers and number of MCCs showing trans-differentiation (TD) morphology. Mann Whitney test, ns P > 0.05 = not significant, * P < 0.05. (E) Inhibition of Jak1/2 signaling by Ruxolitinib (Ruxo.) or inhibition of Thyroid hormone signaling by Propylthiouracil (PTU) decreases the number of MCCs showing signs of trans-differentiation (TD, blue box) and increases the number of MCCs with apoptotic/shedding morphology (red boxes). Normal MCC indicated by yellow box. DMSO N = 13; Ruxolitinib N = 11; PTU N = 10 embryos. (F) Quantification of total MCC numbers (upper panel; Mann Whitney test, ns P > 0.05 = not significant) and proportions of normal (yellow), trans-differentiating (blue), apoptotic/shedding (red) MCC phenotypes (lower panel; χ2 test, *** P < 0.001). (G) Schematic representation of mesoderm induced MCC trans-differentiation.
MCC to Goblet cell trans-differentiation was proposed in conditions associated with persistent inflammation such as Asthma or COPD (Chronic obstructive pulmonary disease) (Tyner et al., 2006), but this idea remains controversial as lineage tracing experiments failed to detect labeled cells after fate conversion in the mouse airway (Pardo-Saganta et al., 2013) and the opposite conversion was proposed as well (Ruiz García et al., 2019). In the mammalian airways, Interleukin-13 (IL-13) activates Janus kinase 1 (Jak1) that phosphorylates Signal transducer and activator of transcription 6 (STAT6) (Tyner et al., 2006). Active STAT6 (STAT6-p) promotes expression of the transcription factor Spdef, which in turn activates Mucin5AC transcription (Park et al., 2007; Parker et al., 2013). STAT6 and Notch were further shown to synergistically promote Goblet cells in the mouse (Danahay et al., 2015; Guseh et al., 2009). STAT6-p also directly represses Foxj1, a transcription factor required for motile cilia (Gomperts et al., 2007). The IL-13 induced effects are modulated by Epidermal growth factor (EGF) signaling, which enhances Jak1 activity and exhibits an anti-apoptotic effect, a prerequisite for the proposed trans-differentiation (Tyner et al., 2006) (Figure S5B).
To determine if Notch and Jak/STAT signaling could play a similar role to induce MCC to Goblet cell trans-differentiation in Xenopus, we first analyzed the expression of the Notch ligands jag1, delta-like1 (dll1) and delta-like4 (dll4) on histological sections. This showed transient upregulation of Notch ligand expression in the mesoderm underlying the epidermis in stages of MCC trans-differentiation (Figure S4B–D). Additional analysis of hes1 expression indicated increased Notch signaling activation in the epidermis, but at lower levels as compared to cells directly overlaying the lateral line primordium (Figure 2E and S4E).
We then tested treatment of tadpoles with DAPT throughout later stages of development (st. 32–43) and analyzed MCCs at st. 43. We found that DAPT significantly reduced the number of MCCs with trans-differentiation morphology (Figure 3C,D). We also tested the effects of Jak1/2 inhibition by Ruxolitinib treatment (st. 32–43). Ruxolitinib reduced the number of trans-differentiating MCCs and instead induced shedding morphology and MCC loss at st. 43 (Figure 3E,F). This suggested possible anti-apoptotic effects for Jak1/2 in Xenopus similar to reports from mammalian airway MCCs.
We wondered how Jak/STAT signaling could be elevated systemically to account for the observed effects. Thyroid hormone (TH) was shown to potentiate STAT as well as EFGR signaling via the MAPK/ERK cascade (Cheng et al., 2010). The Xenopus thyroid primordium forms at st. 33 and matures into the thyroid gland until st. 43 (Fini et al., 2012), thus, coinciding with MCC trans-differentiation. We blocked TH production by application of Propylthiouracil (PTU; st. 32–43). PTU treatment also induced shedding morphology and a reduction in MCCs equivalent to Ruxolitinib at st. 43 (Figure 3E,F). Interestingly, we found an additional area with locally restricted MCCs displaying shedding morphology and which were Caspase 3 positive, where the frog pronephros develops at st. 44, i.e. after development of the thyroid gland is completed (Movie 1). The pronephros also expresses high levels of jag1 (Figure S4B), suggesting that TH cannot block shedding and apoptosis in MCCs that are exposed to high Notch ligand levels. Within close proximity, we also detected MCCs with trans-differentiation morphology, which were Caspase 3 negative (Movie 1), further supporting the need for direct cell-cell interactions and high levels of Notch signaling for the induction of shedding and apoptosis in MCCs.
Although in part correlative, these data collectively indicated that mesoderm-derived intermediate Notch signaling levels cause MCC to Goblet cell trans-differentiation in the presence of TH-induced elevated Jak/STAT signaling, which likely has an anti-apoptotic effect (Figure 3G and S5C).
Ectopic Notch activation in MCCs induces apoptosis and cell fate change
To support our findings on the dual role of Notch signaling in MCC apoptosis and cell fate change based on loss-of-function experiments, we turned to a Notch gain-of-function approach using overexpression of constitutively-active Notch intracellular domain (NICD) (Deblandre et al., 1999; Walentek, 2018). To this end, we used the MCC-specific α-tubulin (αtub) promoter (Stubbs et al., 2006) to drive GFP-fused NICD expression in MCCs after cell fate specification.
We confirmed broad GFP-NICD expression shortly after zygotic genome activation (st. 10) under the control of a human cytomegalovirus promoter (CMV::GFP-NICD), while GFP fluorescence from αtub promoter constructs (αtub::GFP and αtub::GFP-NICD) was observed in MCCs after specification (st. 14) (Figure 4A,B). TUNEL assays on control and GFP-NICD overexpressing specimens confirmed that GFP-NICD can induce ectopic apoptosis when expressed ubiquitously (CMV::GFP-NICD) as well as specifically in MCCs (αtub::GFP-NICD) (Figure 4C,D).
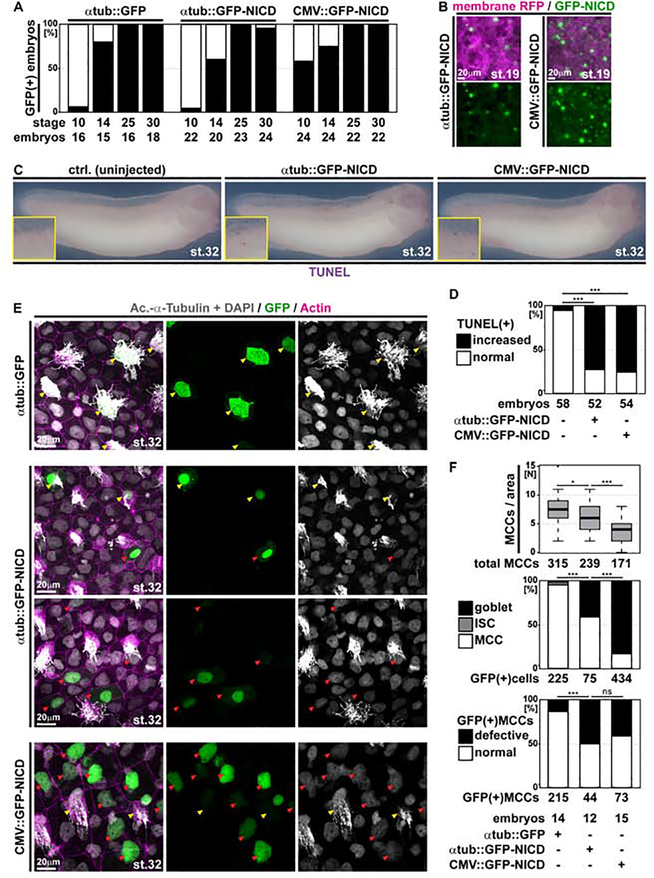
Figure 4. Ectopic Notch signaling induces apoptosis and cell fate change in MCCs.
(A) Analysis of GFP-expression in embryos injected with a control construct (αtub::GFP) or GFP-NICD constructs under control of a MCC-specific promoter (αtub::GFP-NICD) or a ubiquitous promoter (CMV::GFP-NICD) show different onset of promoter activation. Graph depicts GFP fluorescence in % of analyzed embryos. N embryos per stage and construct analyzed are indicted below the graph. (B) Epifluorescent micrographs of epidermal areas at stage 19 in αtub::GFP-NICD and CMV::GFP-NICD injected embryos show broader GFP expression in the CMV-driven construct. N = 3 embryos per construct. (C) GFP-NICD overexpression induces ectopic apoptosis (TUNEL staining, purple) in embryos at stage 32. Ctrl. N = 58; αtub::GFP-NICD N = 52; CMV::GFP-NICD N = 54 embryos. (D) Quantification of TUNEL assay results. χ2 test, *** P < 0.001. (E) Confocal images of control or GFP-NICD construct-injected specimens show cytoplasmic GFP (green) localization in normal looking MCCs (Ac.-α-Tubulin, grey; yellow arrowheads) in controls (αtub::GFP-NICD), but nuclear (DAPI, grey) GFP localization in defective MCCs as well as in Goblet cells (red arrowheads) upon GFP-NICD expression from both promoter constructs (αtub::GFP-NICD and CMV::GFP-NICD). αtub::GFP N = 14; αtub::GFP-NICD N = 12; CMV::GFP-NICD N = 15 embryos. (F) Quantification of MCCs per area (Mann Whitney test, * P < 0.05, *** P < 0.001), identity of GFP(+) cells (χ2 test, *** P < 0.001), and defective MCCs (χ2 test, ns P > 0.05 = not significant, *** P < 0.001).
Next, we analyzed the effects of elevated Notch signaling on MCC cell fate, ciliation and morphology by immunofluorescent staining and confocal microscopy at st. 32. Injection of the control construct (αtub::GFP) revealed cytoplasmic GFP expression specifically in MCCs, which displayed normal ciliation and morphology (Figure 4E,F). In contrast, expression of GFP-NICD lead to nuclear GFP localization and reduced the total number of MCCs, which was more apparent with the CMV::GFP-NICD construct than with αtub::GFP-NICD (Figure 4E,F). The strong reduction in MCC number after CMV::GFP-NICD injection mimicked previous results using nicd mRNA injections that inhibit MCC specification (Quigley and Kintner, 2017). Thus, these data confirmed the earlier onset of expression from the CMV promoter as well as the functionality of the GFP-NICD fusion protein. Consequently, the vast majority of GFP(+) cells in CMV::GFP-NICD injected embryos showed characteristic Goblet cell morphology, including large cell size and specific apical actin patterns, which both strikingly differ from MCC morphology (Figure 4E,F).
Importantly, a significant proportion of GFP(+) cells also showed Goblet cell morphology upon injection of αtub::GFP-NICD, demonstrating that Notch signaling activation in MCCs can trigger fate change towards Goblet cell identity after MCC fate specification (Figure 4E,F). In addition to altered cell fates in GFP-NICD injected MCCs, we also observed reduced ciliation and abnormal apical morphology, similar to our observations in naturally trans-differentiating MCCs at later stages (Figure 4E,F and 3A).
Together, these experiments provide evidence that ectopic Notch signaling can induce apoptosis as well as cell fate conversion in MCCs.
MCCs retract cilia, loose basal body components and change their expression profile
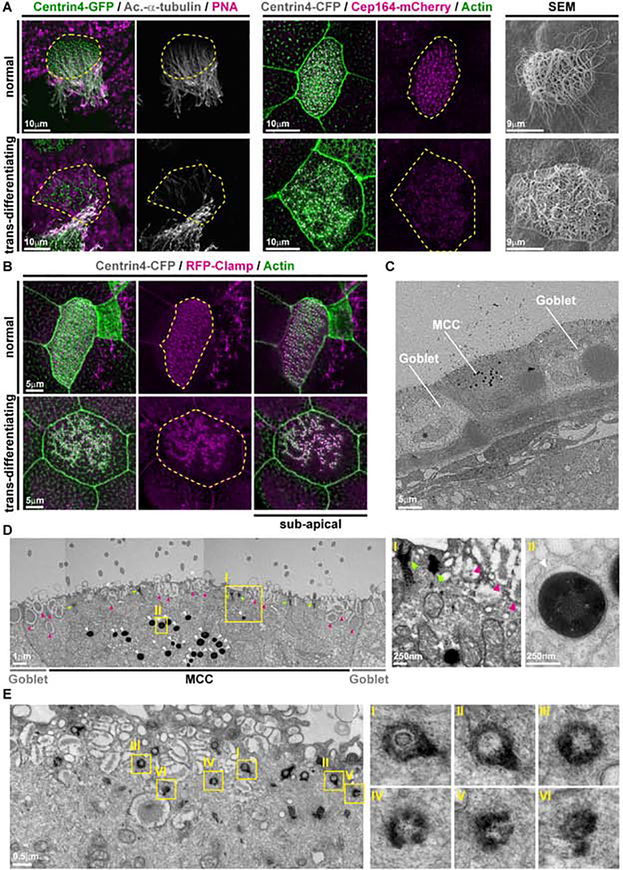
To study how MCC to Goblet cell trans-differentiation is executed at the cellular level, we investigated ciliation, basal bodies and F-actin organization during this process through direct comparison of trans-differentiating and normal MCCs within the same specimens. This revealed a decrease in acetylation of MCC cilia (Ac.-α-tubulin), loss of normal basal body spacing (Centrin4-GFP/CFP), and reduced Cep164-mCherry localization to basal bodies (Figure 5A and S5D,E). Additionally, we observed remodeling of the apical and sub-apical F-actin network in trans-differentiating MCCs, which resulted in an apical cell morphology similar to Goblet cells and to the loss of polarized basal body alignment visualized by RFP-Clamp (Figure 5B and S5E). In contrast to apoptotic MCCs over the lateral line, we did not detect increased levels of active Caspase 3 staining in trans-differentiating MCCs at st. 41–45, in which Caspase 3 levels were comparable to the levels found in newly matured MCCs at st. 32 (Figure S5F).
Fig. 5. Cellular changes in trans-differentiating MCCs.
(A) Confocal and scanning electron microscopy (SEM) micrographs of normal and trans-differentiating MCCs reveal disorganized basal bodies (Centrin4-GFP, green / Centrin4-CFP, grey), cilia de-acetylation (Ac.-α-Tubulin, grey), F-actin remodeling (Actin, green) and loss of basal body distal appendages (Cep164-mCherry) during MCC cilia retraction. Cf. Fig. S5E for quantifications. Centrin4-GFP/Ac.-α-Tubulin/PNA N = 1 embryo; N = 8 normal MCCs; N = 2 trans-differentiating MCCs. Centrin4-CFP/Cep164-mCherry/Actin N = 3 embryos; N = 15 normal MCCs; N = 15 trans-differentiating MCCs. SEM N = 2 embryos; N = 15 normal MCCs; N = 11 trans-differentiating MCCs. (B) Confocal micrographs of a normal and a trans-differentiating MCC stained for basal bodies (Centrin4-CFP, gray), rootlets (RFP-Clamp, magenta) and F-actin (Actin, green) reveals loss of basal body alignment as well as apical and sub-apical F-actin organization. N = 8 embryos. (C,D) Transversal sectioning and transmission electron microscopy (TEM) shows normal and abnormal basal bodies (green arrowheads), mucus granules (magenta arrowheads) and an enrichment of lysosomes (white arrowheads) in a trans-differentiating MCC. Magnified areas are indicated by yellow boxes. Image in D was reconstructed from multiple individual micrographs of cell depicted in C. N = 1 embryo. (E) Parallel section and TEM shows normal and abnormal basal bodies in a trans-differentiating MCC. Magnified areas are indicated by yellow boxes. N = 1 embryo.
While Ac.-α-tubulin staining was strongly reduced or absent in trans-differentiating MCCs, SEM analysis showed persisting cilia with various degree of length reduction in MCCs with an altered apical surface morphology (Figure 5A). The cilia length reduction appeared similar to reports on primary cilia retraction upon cell cycle reentry (Izawa et al., 2015). Trans-differentiating MCCs also lost expression of the ciliary transcription factor foxj1 and of the cell cycle inhibitor p27 indicating inhibition of the MCC transcriptional program (Figure S6A–C). Transmission electron microscopy (TEM) further confirmed the presence of large mucus granules as well as release of secretory material from trans-differentiating MCCs (Figure 5C,D).
Together these data indicated a switch in the transcriptional program in trans-differentiating MCCs, which leads to removal of cilia and initiates mucus production and secretion.
Autophagy stimulation or proteasomal inhibition prevent cilia loss and MCC trans-differentiation
We wondered what happens to the >100 basal bodies, which are a specific feature of MCCs. We used membrane GFP, which labels also the ciliary membrane, together with Centrin4-CFP to label basal bodies, and analyzed tadpoles at st. 44/45. This revealed late stage trans-differentiating MCCs with few cilia and basal bodies, which localized to the cytoplasm (Figure S6D). In rare cases, cytoplasmic Centrin4-CFP signals were associated with vesicle-like membranes, suggesting potential basal body packaging for degradation (Figure S6D inset).
TEM and analysis of MCCs at various stages of trans-differentiation (as judged by the amount of mucus granules) detected undocked cytoplasmic basal bodies, but none enclosed by a membrane or within vesicles (Figure S6E). Instead, trans-differentiating MCCs were enriched for electron-dense structures, reminiscent of lysosomes (Hurbain et al., 2017) (Figure 5C,D and S6E). We confirmed this by Lysotracker-green staining in membrane RFP-injected specimens (Figure S7A,B). Furthermore, we frequently detected structurally incomplete basal bodies showing various degrees of dissociation by TEM (Figure 5D,E).
To visualize a possible uptake of basal body material into early lysosomes, we co-injected mRNAs encoding Centrin4-CFP and Lamp1-mCherry, and analyzed MCCs at st. 43–46. We found that MCCs were enriched for Lamp1-mCherry-vesicles, but we were not able to detect Lamp1-mCherry(+) trans-differentiating MCCs at st. 43/44 although trans-differentiating Lamp1-mCherry(−) MCCs were readily detectable within the same areas (Figure S7C). Furthermore, ciliation persisted until st. 46 in Lamp1-mCherry injected specimens (Figure S7D). These findings indicated that Lamp1 overexpression interfered with trans-differentiation.
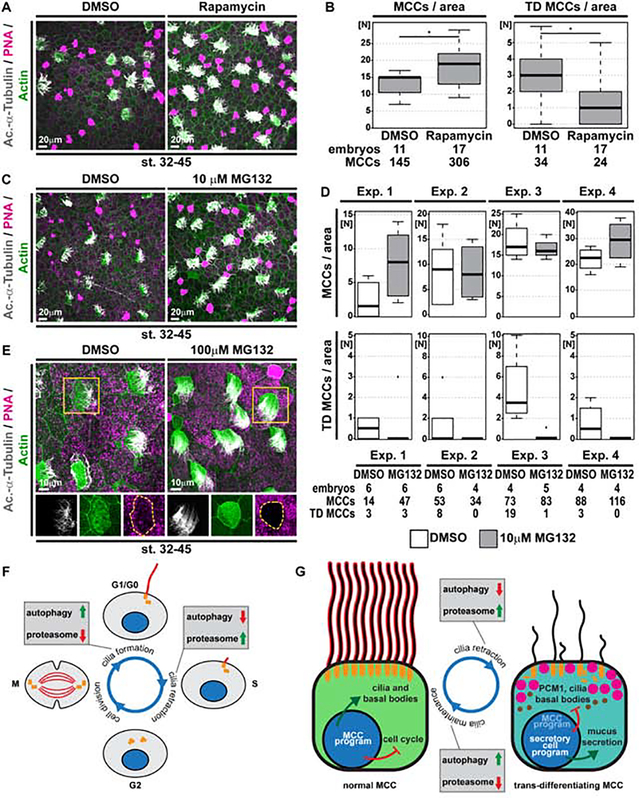
Impaired lysosome function upon Lamp1 overexpression was demonstrated in human pancreatic cancer cells (Chen et al., 2019). Additionally, lysosomal dysfunction and the resulting stress was reported to trigger autophagy, similar to nutrient deprivation (Huber and Teis, 2016; Pan et al., 2019). This suggested that upregulated autophagy could prevent MCC trans-differentiation. We treated tadpoles with Rapamycin to stimulate autophagy through specific inhibition of the mTOR complex 1 (mTORC1). Rapamycin treatment (st. 32–45) and analysis of MCCs at st. 45 revealed significantly increased MCC numbers and a reduction of MCCs showing features of trans-differentiation (Figure 6A,B). This supported our hypothesis that MCC and cilia loss can be inhibited by upregulation of autophagy.
Figure 6. Activation of autophagy or inhibition of proteasomal degradation prevents cilia retraction and MCC loss.
(A,C,E) Confocal micrographs of the epidermis stained for MCCs (Ac.-α-Tubulin, grey), F-actin (Actin, green) and mucus (PNA staining, magenta) demonstrate an increase in MCCs and reduced cilia loss in Rapamycin (A) and MG132 (C,E) treated tadpoles. (A) DMSO N = 11; Rapamycin N = 17 embryos. (C) DMSO N = 20; MG132 N = 19 embryos. (E) Altered F-actin and cell morphology was observed even in fully ciliated MCCs after MG132 treatment. DMSO N = 20; MG132 N = 5 embryos. (B,D) Quantification of total MCC numbers and number of MCCs showing trans-differentiation (TD) morphology. Mann Whitney test, * P < 0.05. Due to greater variability between individual experiments with 10μM MG132, individual replicates are depicted separately in (D) for transparency. (F,G) Schematic summary of protein degradation pathways in primary cilia (E) and MCC cilia (F) maintenance and retraction.
A shift in the balance between autophagy and proteasomal degradation was demonstrated to regulate ciliogenesis versus cilia retraction during the cell cycle in mono-ciliated cells (Boukhalfa et al., 2019; Tang et al., 2013). Primary cilia formation can be induced by nutrient deprivation, which leads to mTORC1 inhibition and upregulation of autophagy. In contrast, cilia retraction is associated with increased proteolysis of basal body-associated proteins by the proteasome. We wondered if a similar shift could be employed during MCC cilia retraction and basal body elimination in Xenopus. We inhibited the proteasomal machinery by application of MG132 (st. 32–45) and analyzed MCC presence and ciliation at st. 44–45. MG132 prevented MCC trans-differentiation and cilia loss, similar to Rapamycin, although we encountered greater variability in those experiments (Figure 6C,D). Additionally, exposure of MCCs to high doses of MG132 lead to elongated MCC cilia (Figure 6E).
These data indicate that the regulation of proteostasis through autophagy versus the proteasome could play a similar role in MCC ciliation as implicated for primary cilia, arguing for a universal protein control mechanism in the maintenance and removal of cilia across ciliated cell types (Figure 6F,G).
PCM1 is required for MCC cilia maintenance
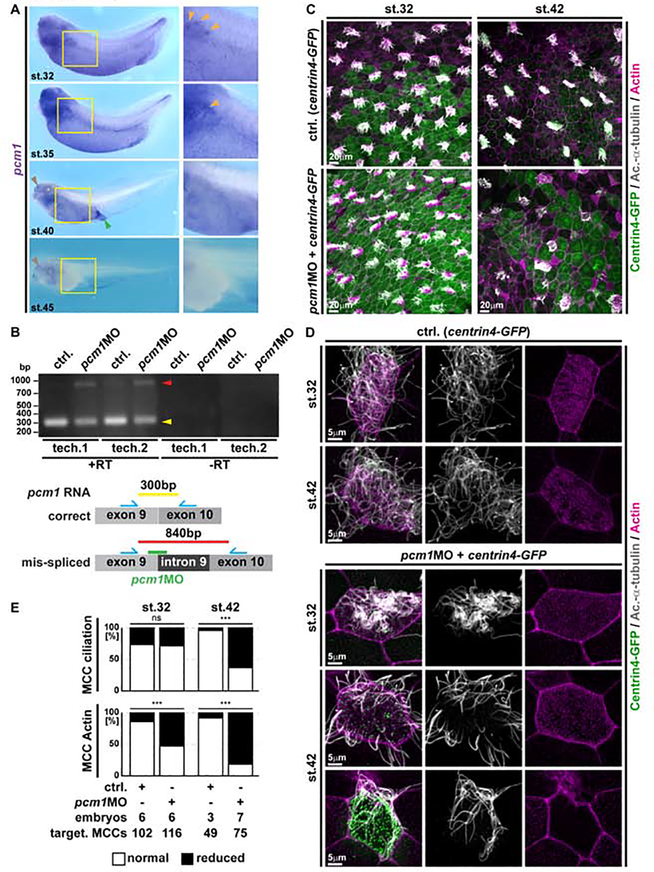
Loss of Pericentriolar material 1 (PCM1) from basal bodies is a key event during primary cilia retraction and leads to exposure of basal body proteins to proteasomal degradation when autophagy is downregulated (Joachim et al., 2017; Wang et al., 2016). Transcriptionally, pcm1 expression is regulated downstream of Notch through the multi-ciliogenesis cascade (including Multicilin and Foxj1) in Xenopus, and pcm1 was designated a core-MCC transcript (Quigley and Kintner, 2017). We analyzed pcm1 expression in Xenopus development and confirmed strong signals in the epidermis as well as other multi-ciliated tissues, e.g. nephrostomes, nasal pits and the cloaca (Figure 7A). As seen for foxj1, pcm1 expression was lost from the epidermis during stages of MCC trans-differentiation (Figure 7A and S6A). We therefore wondered if PCM1 might play a role in regulating cilia loss in trans-differentiating MCCs.
Figure 7. Loss of PCM1 leads to premature MCC de-ciliation during trans-differentiation.
(A) In situ hybridization shows pcm1 (purple) expression in the epidermis and other multiciliated areas (nephrostomes, orange arrowheads; nasal pit, brown arrowheads; cloaca, green arrowhead) and progressive loss of expression in the epidermis during stages (32 to 45) of MCC trans-differentiation. Yellow boxes indicate location of magnified images. St. 32 N = 13; st. 35 N = 7; st. 40 N = 7; st. 45 N = 12 embryos. (B) RT-PCR in controls and pcm1MO injected specimens confirms pcm1 intron 9 retention after injection of the splice-blocking MO. Schematic representation of correct and mis-spliced transcripts, expected amplicons and localization of primers (blue) as well as MO-targeting (green). N = 3 biological and 2 technical replicates each. (C-D) Confocal micrographs of centrin4-GFP injected (basal bodies, green) control (ctrl.) and pcm1MO treated MCCs reveal normal ciliation (Ac.-α-Tubulin, grey) and only mild effects on apical F-actin organization (Actin, magenta) at stage 32, but reduced ciliation and severe apical F-actin defects in targeted MCCs already shortly after onset of trans-differentiation at stage 42. Ctrl. st. 32 N = 6; ctrl. st. 42 N = 3; pcm1MO st. 32 N = 6; pcm1MO st. 42 N = 7 embryos. (E) Quantification of ciliation and F-actin defects in targeted MCCs at stages 32 and 42. χ2 test, ns P > 0.05 = not significant, *** P < 0.001.
To reduce PCM1 levels, we designed a splice-inhibiting morpholino oligonucleotide (MO) targeting the splice-donor site of pcm1 protein-coding exon 9, which leads to intron 9 retention (Figure 7B), frameshift, and the generation of a shortened protein (492 instead of 2034 amino acids) that lacks the conserved C-terminal PCM1-C superfamily domain. Injection of pcm1MO did not lead to defective MCC cilia formation at st. 32, but premature loss of cilia and apical F-actin structures in targeted MCCs at st. 42, i.e. a stage when trans-differentiation is ongoing (Figure 7C–E). Additionally, we observed uncoupling from the apical membrane and cytoplasmic localization of basal bodies in PCM1-deficient MCCs (Movie 2).
Together with the observed shift from autophagy towards proteasomal degradation, these data suggest that loss of PCM1 production during MCC trans-differentiation leads to the initiation of cilia retraction and basal body protein degradation, similar to the proposed mechanism employed for primary cilia retraction in cells re-entering the cell cycle.
Discussion:
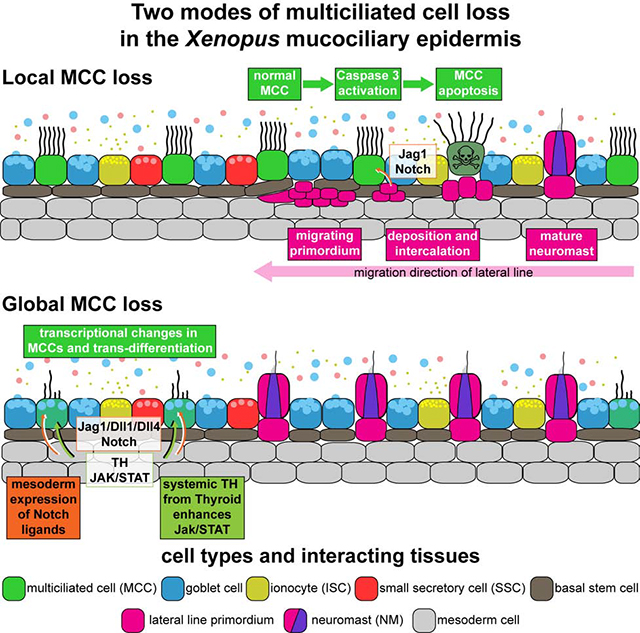
We demonstrate that Xenopus epidermal MCCs can be lost by apoptosis or by trans-differentiation during developmental tissue remodeling. Both processes are induced by Notch signaling, which becomes elevated in the epidermis through tissue interactions with the lateral line or the mesoderm. Trans-differentiation occurs in MCCs that are protected from apoptosis by Thyroid-dependent elevation of Jak/STAT signaling, which occurs after initial onset of lateral line organ development. Additionally, the two different modes of MCC removal could also reflect the perceived increase in Notch signaling over time, that could be faster and stronger in the case of lateral line induced apoptosis as judged by the stronger expression of hes1 in those areas. This notion is supported by occurrence of localized MCC apoptosis also during later stages of development, in areas adjacent to the developing pronephros, which also expresses jag1 at high levels, similar to the lateral line.
The specific reactions of MCCs to elevated Notch signaling as compared to secretory cell types (Goblet cells, SSCs and ISCs) could be achieved through a combination of cell type-specific expression levels of Notch receptors, as previously demonstrated in mammalian airway cells (Pardo-Saganta et al., 2015), with distinct requirements for Notch signaling for cell type specification and maintenance, i.e. secretory cells are induced and maintained at higher Notch signals than MCCs (Lafkas et al., 2015; Quigley and Kintner, 2017).
MCC to Goblet secretory cell trans-differentiation involves cilia de-acetylation and shortening, similar to primary cilia retraction, as well as F-actin remodeling and loss of basal body proteins (e.g. Cep164). These processes can be inhibited by Rapamycin or MG132. While further work is necessary to elucidate the precise molecular mechanism of this process, our MCC data suggest a similar positive role for autophagy in MCC ciliation as described for primary cilia (Wang et al., 2015). Primary cilia formation is induced upon serum starvation, which inhibits mTORC1 and promotes autophagy (Boukhalfa et al., 2019). How precisely autophagy modulates cilia formation, length and maintenance remains incompletely resolved. Nevertheless, emerging evidence suggests that the balance between autophagy and proteasomal degradation influences cilia formation and resorption as well as maintenance of centrioles and basal bodies (Boukhalfa et al., 2019; Holdgaard et al., 2020; Izawa et al., 2015; Liang et al., 2016; Paridaen et al., 2013).
Our data support the hypothesis that de-ciliation and basal body loss could be controlled by cellular levels of PCM1. PCM1 is involved in the protection of basal body associated proteins from ubiquitination and proteasomal degradation via interactions with Mib1 (Wang et al., 2016). Pericentriolar material loss was also implicated as step in centriolar elimination during oocyte maturation in various species (Schoborg and Rusan, 2016), and PCM1 was shown to recruit Plk1 to the pericentriolar cloud (Wang et al., 2013), while loss of Pkl1 induced centriolar degradation in Drosophila (Pimenta-Marques et al., 2016). Together with our MCC data, these observations raise the possibility of a potentially common mechanism for cilia, basal body and centriole maintenance across cell types and species.
Interestingly, we also observed an increase in lysosomes in trans-differentiating MCCs, but a clear role for lysosomal degradation in the regulation of cilia has not been established to date. Therefore, our findings encourage exploration of additional connections between regulated proteostasis and ciliation, especially in light of emerging data on the crosstalk between the autophagy and the proteasomal systems (Kocaturk and Gozuacik, 2018).
Our results further reveal surprising parallels between mucociliary epithelial remodeling in the Xenopus epidermis and the diseased mammalian lung. Changes in the MCC transcriptional program, loss of ciliation, initiation of mucus production and secretion were all reported in mammalian MCCs stimulated with IL-13 directly or by agents that are known to induce IL-13 release (Gomperts et al., 2007; Tyner et al., 2006). Nevertheless, the question whether MCC to Goblet cell trans-differentiation really occurs in the mammalian airways remains the topic of controversial debate in the field. An important consideration is that genetic lineage tracing data from the mouse failed to detect secretory cells tracing back to trans-differentiated MCCs (Pardo-Saganta et al., 2013), and due to technical limitations in the Xenopus system, we cannot provide genetic tracing data for MCCs in the mucociliary epidermis at this point.
Nevertheless, our work contributes valuable insights and establishes the Xenopus tadpole as an accessible model to study the molecular mechanisms underlying trans-differentiation, proteostasis, and the role of pericentriolar material in vertebrate MCCs. Additionally, our findings challenge the idea that there is a fundamental limit for cell fate change in differentiated cell types.
STAR Methods:
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peter Walentek (peter.walentek@medizin.uni-freiburg.de).
Materials Availability
Plasmids generated in this study are available from the Lead Contact.
p27::GFP [Xla.Tg(Xtr.cdknx:GFP)Papal, RRID:EXRC_0043] frozen spern was obtained and is available from the European Xenopus Resource Centre (EXRC) at University of Portsmouth, School of Biological Sciences, UK.
Data and Code Availability
The datasets (imaging files) supporting the current study have not been deposited in a public repository due to large size but are available from the corresponding author on request. This study did not generate code.
Experimental models and subject details
Xenopus laevis
Wild-type Xenopus laevis were obtained from the European Xenopus Resource Centre (EXRC) at University of Portsmouth, School of Biological Sciences, UK. Frog maintenance and care was conducted according to standard procedures and based on recommendations provided by the international Xenopus community resource centers NXR and EXRC as well as by Xenbase (http://xenbase.org).
Ethics statements on animal experiments
This work was done in compliance with German animal protection laws and was approved under Registrier-Nr. G-18/76 by the state of Baden-Württemberg.
Method details
Fertilization and microinjection of Xenopus embryos
X. laevis eggs were collected and in vitro-fertilized with wild-type or transgenic sperm, then cultured and/or microinjected by standard procedures (Sive et al., 2000). In vitro fertilization with p27::GFP [Xla.Tg(Xtr.cdknx:GFP)Papal, RRID:EXRC_0043] (Carruthers et al., 2003) frozen sperm was performed by NXR protocol. Frozen sperm was removed from liquid nitrogen and immediately swirled in a 37°C water bath for 30sec, then resuspended in 500μl room-temperature 1/3x Modified Frog Ringer’s solution (MR) by using a micropestle. Sperm mixture was then added to the clutch of eggs and mixed into the monolayer for 3–5min, then flooded with 1/3x MR. After fertilization, embryos were injected with mRNAs and DNAs at the four-cell stage using a PicoSpritzer setup in 1/3x MR with 2.5% Ficoll PM 400 (GE Healthcare, #17–0300-50), and were transferred after injection into 1/3x MR containing 50μg/ml Gentamycin. Drop size was calibrated to about 7–8nl per injection.
mRNAs and DNA constructs and MO used in this study
mRNAs encoding membrane-RFP/GFP (gift from the Harland lab), Centrin4-GFP/CFP (Antoniades et al., 2014; Park et al., 2008), Clamp-RFP (Park et al., 2008), Cep164-mCherry (Tu et al., 2018), hLamp1-mCherry (this study) were prepared using the Ambion mMessage Machine kit using Sp6 (#AM1340) supplemented with RNAse Inhibitor (Promega #N251B), and diluted to 30–80ng/μl for injection into embryos. GFP-NICD constructs were generated by subcloning of NICD from pCS2+ (Chitnis et al., 1995) to pCS107 (CMV promoter), or to α-tub::GFP (Chung et al., 2014) after removal of the GFP-Utrophin coding sequence. Then, the 5’-end of the NICD sequence was modified to add a Cla1 restriction site using the Q5 site directed mutagenesis kit (NEB #E0554S), and GFP was fused N-terminally to NICD in both constructs. Additionally, a hormone-inducible GR-domain (Kolm and Sive, 1995) was fused C-terminally to the GFP-NICD sequences in an attempt to generate inducible constructs, but due to the strong nuclear localization signal present within the NICD sequence, the construct shows nuclear localization even without hormonal induction. hLamp1-mCherry was generated by subcloning hLamp1-mCherry from pcDNA3.1 (Addgene #45147) to pCS107. The α-tub::GFP, α-tub::GFP-NICD (this study) and CMV::GFP-NICD (this study) plasmid were injected at 10–30ng/μl after purification using the PureYield Midiprep kit (Promega, #A2492). pcm1MO (Gene tools) was used at 5pmol concentration. For validation of pcm1MO, embryos were injected in four of four blastomeres at 4-cell stage, and processed for total RNA extraction using a standard Trizol (Invitrogen #15596026) protocol, cDNA synthesis conducted according to the manufacturer’s protocol using M-MLV reverse transcriptase (Promega #M3681). RT-PCR reactions were carried out on a BioRad S1000 thermocycler, and resulting amplicons were analyzed by standard gel-electrophoresis and imaging using gel documentation setup (PeqLab E-Box-VX2). These experiments were carried out on samples from three experiments, with two technical replicates each. –RT controls were processed the same way as +RT samples, but no RT enzyme was added to the cDNA synthesis reaction. For antisense in situ hybridization probes, jag1, hes1, dll1, dll4, and pcm1 fragments were cloned from whole-embryo cDNAs derived from stages between 10 and 30 using primers listed below (ISH-primers). All sequences were verified by Sanger sequencing.
Morpholino nucleotide and cloning primer sequences:
| Name | Sequence |
|---|---|
| jag1-ISH-F | 3‘-ATGCTCAAAGGTGTGGTGTG-5‘ |
| jag1-ISH-R | 3‘-CTCTGTGCTGTTTCTAAGTCTC-5‘ |
| hes1-ISH-F | 3‘-AAAGGATCCATGCCGGCTGATGTGATGGAG-5‘ |
| hes1-ISH-R | 3‘-AAAGTCGACTTACCAGGGCCTCCAAACAG-5‘ |
| dll1-ISH-F | 3‘-GATGACTGTGCTTCCTTCC-5‘ |
| dll1-ISH-R | 3‘-TTGACGTTGAGTAGGCAGAG-5‘ |
| dll4-ISH-F | 3‘-CACAAGAAATCCCTGCCTG-5‘ |
| dll4-ISH-R | 3‘-TATACCTCTGTAGCTATCACAC-5‘ |
| NICD-BstB1-F | 3’-AAATTCGAAAATAAGAAGCGTCGCCGTGAACACG-5’ |
| NICD-Sal1-R | 3’-AAAGTCGACCTTGAAAGCTTCAGGTATGTGGGTGCG-5’ |
| Q5Cla1ATG-F | 3’-TATGAATAAGAAGCGTCGCCGT-5’ |
| Q5Cla1ATG-R | 3’-TCGATTCGATGGGATCCTGCAAAAA-5’ |
| Cla1-GFP-F | 3’-AAAAAAATCGATATGGTGAGCAAGGGCGAGGAG-5’ |
| GFP-Cla1-R | 3’-AAAAAAATCGATCTTGTACAGCTCGTCCATGCC-5’ |
| BamH1-hLamp1mCherry-F | 3‘-AAAAAA GGATCCATGGCGGCCCCCGGCAGCGC-5‘ |
| hLamp1mCherry-EcoR1-R | 3‘-AAAAAAGAATTCTTACTTGTACAGCTCGTCCATG-5‘ |
| pcm1-ISH-F | 3’-ATACCATCTACTCTGAAGTTGCCA-5‘ |
| pcm1-ISH-R | 3’-TTTCATCTTCACCACTGCCA-5‘ |
| pcm1MO | 5’-CTTTGTCTCCACAACTTACATGCAG-3‘ |
| pcm1Spl9-F | 3’-CACCAGATAACAGGAGGCA-5’ |
| pcm1Spl9-R | 3’-ATCAGAAGTCAAGGCGGA-5’ |
Quantification of MCCs around neuromasts and other MCCs
Multiple individual micrographs used for reconstruction of embryo depicted in Figure 1E that contained p27::GFP signals from NMS were analyzed in ImageJ. For that a circular area of 150μm diameter with either a NM (blue circles) or a MCC (yellow circles) in the center was overlaid, and MCC numbers were quantified.
Lateral line placode transplantation
The otic vesicle and adjacent lateral line placode tissue were transplanted at stage 18–22 from membrane-RFP injected donor embryos into hosts that were injected with the α-tub::GFP construct to label MCCs. Grafting was conducted in 1x MBS (88mM NaCl, 1mM CaCl2, 1mM MgSO4, 5mM HEPES at pH 7.8, and 2.5mM NaHCO3) (Sive et al., 2000) containing 50μg/ml Gentamycin. For recovery, embryos were kept in 0.1x MBS with Gentamycin. After recovery, embryos were maintained in 1/3MR with Gentamycin until fixation in 4%PFA.
Lateral line ablation
Lateral line ablation was conducted on st. 28–30 embryos after anesthesia using buffered MS222 (Sigma-Aldrich #E10521) in 1x MBS medium containing 50μg/ml Gentamycin. An incision was made using a dissection knife (F.S.T. #10055–12) from dorsal-posterior of the otic vesicle to ventral-anterior of the heart-anlage. After wound healing for approximately 15min in 1xMBS, the embryos were transferred to 1/3MR with Gentamycin until fixation in 4%PFA.
Drug treatments of Xenopus embryos
Drug treatment of embryos started/was terminated at the indicated stages. Embryos were incubated in 24-well plates containing 2ml of 1/3MR + Gentamycin. DMSO (Roth #A994.2) was used as vehicle control at the same volume as specific drugs where indicated. Media were replaced every day. All treatments were carried out at room temperature (RT) and protected from light. DAPT (N-[(3,5-Difluorophenyl)acetyl]-L-al-anyl-2-phenyl]glycine-1,1-dimethylethyl ester; Sigma-Aldrich #05942) was dissolved in DMSO and used at 100–200μM (Myers et al., 2014). PTU (Propylthiouracil; Sigma-Aldrich #1578000; 10mM stock stored at −80°C for long term and −20°C for short term) was dissolved in Ultrapure water (ThermoFisher #10977035) and used at 1mM (Veenendaal et al., 2013). Rapamycin (Sigma-Aldrich #37094) was dissolved in DMSO and used at 100nM. Ruxolitinib (Selleckchem #S1378) was dissolved in DMSO and used at 10μM. MG132 (Calbiochem #474790) was dissolved in DMSO and used at 10μM and 100μM.
Characterization of onset of GFP expression
Embryos injected with α-tub::GFP, α-tub::GFP-NICD or CMV::GFP-NICD plasmids and co-injected with membrane RFP were imaged live at the indicated developmental stages. GFP(+) embryos were defined as showing GFP fluorescent expression above background in membrane RFP injected areas.
LysoTracker use in Xenopus embryos
For LysoTracker (Invitrogen #L7526 Green DND-26) treatment, embryos were incubated in 12-well plates containing 4ml of 1/3x MR and 50μg/mL of Gentamycin. 50μM of LysoTracker was added to the medium for 5min in the dark at RT. Embryos were washed 3× 3min in 1/3x MR and finally anesthetized in buffered MS222 (Sigma-Aldrich #E10521) solution during mounting and live cell imaging. Non-LysoTracker controls were not incubated with the compound and anesthetized in buffered MS222 solution during mounting and live cell imaging using the same microscope settings.
Immunofluorescence staining and sample preparation
Whole Xenopus embryos, were fixed at indicated stages in 4% Paraformaldehyde (PFA) in PBS at 4°C overnight or 2h at RT, then washed 3× 15min with PBS, 2× 30min in PBST (0.1% Triton X-100 in PBS), and were blocked in PBST-CAS (90% PBS containing 0.1% Triton X-100, 10% CAS Blocking; ThermoFischer #00–8120) for 1h at RT. All antibodies were applied in 100% CAS Blocking over night at 4°C or 2h at RT (for secondary antibodies). Primary antibodies used: mouse monoclonal anti-Acetylated-α−tubulin (1:1000; Sigma/Merck #T6793), anti-α-tubulin (1:1000; Abcam #ab7291), rabbit anti-CleavedCaspase3 (1:400; Cell Signaling #D176). Secondary antibodies used: AlexaFluor 405-labeled goat anti-mouse antibody (1:500; MolecularProbes #A31553), AlexaFluor 555-labeled goat anti-rabbit antibody (1:500; MolecularProbes #A21428). Actin was stained by incubation (30–120min at RT) with AlexaFluor 488- or 647-labeled Phalloidin (1:40 in PBST; Molecular Probes #A12379 and #A22287), mucus-like compounds in Xenopus were stained by incubation (overnight at 4°C) with AlexaFluor 647-labeled PNA (1:1000 in PBST; Molecular Probes #L32460), and nuclei were stained with DAPI (1:500; Invitrogen D1306). Detailed protocol was published in (Walentek, 2018).
Whole mount in situ hybridization
Embryos were fixed in MEMFA (100mM MOPS pH7.4, 2mM EGTA, 1mM MgSO4, 3.7% (v/v) Formaldehyde) overnight at 4°C and stored in 100% Ethanol at −20°C until used. DNAs were purified using the PureYield Midiprep kit and were linearized before in vitro synthesis of anti-sense RNA probes using T7 or Sp6 polymerase (Promega, #P2077 and #P108G), RNAse inhibitor and dig-labeled rNTPs (Roche, #3359247910 and 11277057001). Embryos were in situ hybridized according to (Harland and Biology, 1991), bleached (Sive et al., 2000) after staining with BM Purple (Roche #11442074001) and imaged. Sections were made after embedding in gelatin-albumin with Glutaraldehyde at 50–70μm as described in (Walentek et al., 2012).
TUNEL assay
Embryos were fixed in MEMFA overnight at 4°C and stored in 100% Ethanol at −20°C until use. Embryos were bleached before staining. TUNEL staining was performed according to respective manufacturer protocol using Terminal Deoxynucleotidyl Transferase kits (Invitrogen #10533065 or Roche #03333574001), dig-UTP (Roche, #3359247910), anti-Digoxigenin AP antibody (1:3000; Roche #11093274910), and NBT/BCIP (Roche #11681451001). Samples were stopped in PBS, fixed briefly with 4% PFA in PBS and imaged.
Sample preparation for electron microscopy
Sample primary fixation was done by 4% PFA, 2% Glutaraldehyde (Carl Roth #4157) in 0.1M Cacodylate Buffer (Science Services #11650). For transmission electron microscopy (TEM), the tissue was then post-fixed in 0.5% Osmium tetroxide (Science Services #E19150) in ddH2O for 60min on ice and then washed 6x in ddH2O. The tissue was incubated in 1% aqueous Uranyl acetate solution (Science Services #E22400–1) for 2h in the dark and washed 2x in ddH2O. Dehydration was performed by 15min incubation steps in 30%, 50%, 70%, 90% and 2× 100% Ethanol (Fisher Scientific #32205) and 2× 100% Aceton (Sigma-Aldrich #179124). After embedding in Durcupan resin (Sigma-Aldrich #44611 and #44612), ultrathin sections (55nm) were performed using a UC7 Ultramicrotome (Leica), collected on Formvar-coated (Science Services #E15830–25) copper grids (Plano #G2500C). Post-staining was done for 1min with 3% Lead Citrate (Delta Microscopies #11300). For scanning electron microscopy (SEM), the fixated samples were dehydrated in 70%, 80%, 90% and 100% Ethanol (each step for 1h at RT) and incubated in a 1:1 solution of Ethanol and Hexamethyldisilazan (HMDS; Carl Roth #3840.2) for 30min. After incubation in 100% HMDS, the solvent was allowed to evaporate. The dehydrated tissue was mounted onto sample holders and sputtered with gold using a Polaron Cool Sputter Coater E 5100.
Light imaging, electron microscopy and image processing
All confocal imaging was performed using a Zeiss LSM880 (Zeiss objectives: 10x Plan apochromat 10x/0.45; 25x LD LC/Plan apochromat 25x/0.8 DIC; 63x Plan apochromat 63x/1.40) and Zeiss Zen Black software. Whole embryo bright-field or fluorescent images were done on a Zeiss AxioZoom setup and Zeiss Zen Pro Blue software. Sections were imaged on a AxioZoom or AxioImager.Z1 microscope and Zeiss Zen Pro Blue software. Fluorescence images were processed in ImageJ/Fiji (maximum intensity projections, reconstruction of tile scans, selection of indicated planes, brightness/contrast adjustment, merging of channels) (Schindelin et al., 2012). Brightfield images were adjusted for color balance, brightness and contrast using Adobe Photoshop. TEM imaging was done using a Zeiss Leo 912 transmission electron microscope. SEM imaging was done using a scanning electron microscope (Leo 1450 VP scanning). Electron microscopy including TEM (transmission electron microscopy) and SEM (scanning electron microscopy) images were adjusted for brightness and contrast, and pseudo-colored using Adobe Photoshop.
Analysis of trans-differentiated MCCs (ciliation, Actin network, basal body integrity)
Imaging was performed using the same settings within individual experiments on embryos which were injected with equal amounts of mRNAs. Images were processed using ImageJ to adjust brightness/contrast and to generate maximum intensity projections. For quantification of ciliation rates, images from similar areas were acquired (cf. Figure S5 A). Trans-differentiated MCCs were designated by presence of mucins and/or impaired apical actin network and impaired ciliation. For quantification of cilia acetylation, basal body polarization and presence of Cep164-mCherry, normal and trans-differentiated MCCs from the same embryos were analyzed. For presentation of some representative examples, deep optical planes from the F-actin channel containing muscle fibers were manipulated to remove muscle signals.
Quantification and Statistical Analysis
Statistical evaluation
Stacked bar graphs were generated in Microsoft Excel, boxplots were generated in R (the line represents the median; 50% of values are represented by the box; 95% of values are represented within whiskers; values beyond 95% are depicted as outliers). Statistical evaluation of experimental data was performed using Wilcoxon sum of ranks (Mann-Whitney) test (https://astatsa.com/WilcoxonTest/), or χ2 test (http://www.physics.csbsju.edu/stats/t-test.html) as indicated in figure legends. Sample sizes for all experiments were chosen based on previous experience and used embryos derived from at least two different females. No randomization or blinding was applied. All experiments were conducted in embryos derived from at least two different females and independent in vitro fertilizations unless specified.
Supplementary Material
Supplemental Movie 1: MCC loss over the nephrostome area at st. 44 (Related to Figure 3)
Two times image series (unannotated first, annotated second) through the z-planes (from apical to deep tissue layers) of a confocal micrograph (left panels) and stills (right panels) of a normal MCC, several apoptotic MCCs (apopt. MCC 1–3) with shedding morphology, a post apoptotic site (after apoptosis), and a trans-differentiating MCC (TD MCC) in a specimen at stage 44 stained for Ac.-α-Tubulin (grey), active Caspase 3 (magenta) and F-actin (Actin, green). Please note the presence of apoptotic cells overlaying the developing kidney tubules (yellow arrowheads). The MCCs depicted in stills are indicated by labeled yellow boxes.
Supplemental Movie 2: PCM1 knockdown leads to basal body uncoupling from the apical membrane (Related to Figure 7)
Image series through the z-planes (from apical to basal) of confocal micrographs of stage 42 control-injected (ctrl.) and pcm1MO-injected MCCs depicted in Figure 7D. Ac.-α-Tubulin (grey), F-actin (magenta) and basal bodies (Centrin4-GFP, green).
Supplemental Table 1: Experiment log and use of data in figures and graphs (Related to Figure 1–7)
List of experiments, species, assay, condition, number of specimens (N), and their use in figure are indicated.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-Acetylated-α-tubulin | Sigma/Merck | T6793 |
| mouse anti-α-tubulin | Abcam | ab7291 |
| rabbit anti-active-Caspase3 | Cell Signaling | D176 |
| anti-Digoxigenin AP antibody | Roche | 11093274910 |
| AlexaFlour 405-labeled goat anti-mouse | Molecular Probes | A31553 |
| AlexaFlour 555-labeled goat anti-rabbit | Molecular Probes | A21428 |
| Bacterial and Virus Strains | ||
| NEB® 5-alpha Competent E. coli (High Efficiency) | New England Biolabs | C2987H |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trizol | Invitrogen | 15596026 |
| RNase Inhibitor | Promega | N251B |
| Dig-labeled rNTPs | Roche | 3359247910, 11277057001 |
| BM Purple | Roche | 11442074001 |
| NBT/BCIP | Roche | 11681451001 |
| DMSO | Roth | A994.2 |
| DAPT (N-[(3,5-Difluorophenyl)acetyl]-L-al-anyl-2-phenyl]glycine-1,1-dimethylethyl ester | Sigma-Aldrich | 05942 |
| PTU (Propylthiouracil) | Sigma-Aldrich | 1578000 |
| Ultrapure water | ThermoFisher | 10977035 |
| Rapamycin | Sigma-Aldrich | 37094 |
| Ruxolitinib | Selleckchem | S1378 |
| MG132 | Calbiochem | 474790 |
| MS222 | Sigma-Aldrich | E10521 |
| Glutaraldehyde | Carl Roth | 4157 |
| Cacodylate Buffer | Science Services | 11650 |
| Osmium tetroxide | Science Services | E19150 |
| Uranyl acetate solution | Science Services | E22400-1 |
| Ethanol | Fisher Scientific | 32205 |
| Aceton | Sigma-Aldrich | 179124 |
| Durcupan resin | Sigma-Aldrich | 44611 and 44612 |
| Formvar | Science Services | E15830-25 |
| Lead Citrate | Delta Microscopies | 11300 |
| Hexamethyldisilazan | Carl Roth | 3840.2 |
| CAS Blocking | ThermoFischer | 00-8120 |
| DAPI | Invitrogen | D1306 |
| AlexaFluor 488-labeled Phalloidin | Molecular Probes | A12379 |
| AlexaFluor 647-labeled Phalloidin | Molecular Probes | A22287 |
| AlexaFluor 647-labeled PNA | Molecular Probes | L32460 |
| Critical Commercial Assays | ||
| M-MLV reverse transcriptase | Promega | M3681 |
| PureYield Midiprep kit | Promega | A2495 |
| Ambion mMessage Machine kit | Thermo Fisher | AM1340 |
| Q5 Site-directed mutagenesis kit | NEB | E0554S |
| LysoTracker | Invitrogen | L7526 Green DND-26 |
| Terminal Deoxynucleotidyl Transferase | Invitrogen | 10533065 |
| Terminal Deoxynucleotidyl Transferase | Roche | 03333574001 |
| T7 RNA polymerase | Promega | P2077 |
| SP6 RNA polymerase | Promega | P108G |
| Experimental Models: Organisms/Strains | ||
| Xla.Tg(Xtr.cdknx:GFP)Papal | EXRC | RRID:EXRC_0043 |
| wt Xenopus laevis | EXRC | N/A |
| Oligonucleotides | ||
| jag1-ISH-F 3‘-ATGCTCAAAGGTGTGGTGTG-5‘ | Sigma-Aldrich | N/A |
| jag1-ISH-R 3‘-CTCTGTGCTGTTTCTAAGTCTC-5‘ | Sigma-Aldrich | N/A |
| hes1-ISH-F 3‘-AAAGGATCCATGCCGGCTGATGTGATGGAG-5‘ | Sigma-Aldrich | N/A |
| hes1-ISH-R 3‘-AAAGTCGACTTACCAGGGCCTCCAAACAG-5‘ | Sigma-Aldrich | N/A |
| dll1-ISH-F 3‘-GATGACTGTGCTTCCTTCC-5‘ | Sigma-Aldrich | N/A |
| dll1-ISH-R 3‘-TTGACGTTGAGTAGGCAGAG-5‘ | Sigma-Aldrich | N/A |
| dll4-ISH-F 3‘-CACAAGAAATCCCTGCCTG-5‘ | Sigma-Aldrich | N/A |
| dll4-ISH-R 3‘-TATACCTCTGTAGCTATCACAC-5‘ | Sigma-Aldrich | N/A |
| NICD-BstB1-F 3’-AAATTCGAAAATAAGAAGCGTCGCCGTGAACACG-5’ | Sigma-Aldrich | N/A |
| NICD-Sal1-R 3’-AAAGTCGACCTTGAAAGCTTCAGGTATGTGGGTGCG-5’ | Sigma-Aldrich | N/A |
| Q5Cla1ATG-F 3’-TATGAATAAGAAGCGTCGCCGT-5’ | Sigma-Aldrich | N/A |
| Q5Cla1ATG-R 3’-TCGATTCGATGGGATCCTGCAAAAA-5’ | Sigma-Aldrich | N/A |
| Cla1-GFP-F 3’-AAAAAAATCGATATGGTGAGCAAGGGCGAGGAG-5’ | Sigma-Aldrich | N/A |
| GFP-Cla1-R 3’-AAAAAAATCGATCTTGTACAGCTCGTCCATGCC-5’ | Sigma-Aldrich | N/A |
| BamH1-hLamp1mCherry-F 3‘-AAAAAAGGATCCATGGCGGCCCCCGGCAGCGC-5‘ | Sigma-Aldrich | N/A |
| hLamp1mCherry-EcoR1-R 3‘-AAAAAAGAATTCTTACTTGTACAGCTCGTCCATG-5‘ | Sigma-Aldrich | N/A |
| pcm1-ISH-F 3’-ATACCATCTACTCTGAAGTTGCCA-5‘ | Sigma-Aldrich | N/A |
| pcm1-ISH-R 3’-TTTCATCTTCACCACTGCCA-5‘ | Sigma-Aldrich | N/A |
| pcm1MO 5’-CTTTGTCTCCACAACTTACATGCAG-3‘ | Gene Tools | N/A |
| pcm1Spl9-F 3’-CACCAGATAACAGGAGGCA-5’ | Sigma-Aldrich | N/A |
| pcm1Spl9-R 3’-ATCAGAAGTCAAGGCGGA-5’ | Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| hLamp1-mCherry pcDNA3.1 | Addgene | 45147 |
| Software and Algorithms | ||
| Zeiss Zen Pro Blue | Zeiss | N/A |
| Zeiss Zen Black | Zeiss | N/A |
| ImageJ/Fiji | imagej.nih.gov/ij/download.html | N/A |
| Adobe Photoshop | Adobe | N/A |
Highlights.
Xenopus multiciliated cells are lost during developmental epidermal remodeling
Multiciliated cell loss is induced by elevated Notch signaling
Notch can induce apoptosis or secretory cell fate change in multiciliated cells
Multiciliated cell cilia loss resembles key aspects of primary cilia retraction
Acknowledgments:
We thank: S. Schefold, J. Groth and S. Kayser for expert technical help; Walz and Driever labs for sharing resources; S. Arnold, A. Classen, L. Davidson, W. Driever, R. Harland, G. Pyrowolakis, A. Schambony, J. Wallingford for discussions and/or critical reading; Xenbase (RRID:SCR_004337), NXR (RRID:SCR_013731) and EXRC for Xenopus resources; Light Imaging Center Freiburg and EM Core Facility for microscope use. This study was supported by the Deutsche Forschungsgemeinschaft (DFG) under the Emmy Noether Programme (grant WA3365/2-1) and under Germany’s Excellence Strategy (CIBSS – EXC-2189 – Project ID 390939984) to PW. BM is supported from NIH-NIGMS (GM089970).
Footnotes
Declaration of Interests:
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Antoniades I, Stylianou P, and Skourides PA (2014). Making the Connection: Ciliary Adhesion Complexes Anchor Basal Bodies to the Actin Cytoskeleton. Dev. Cell 28, 70–80. [DOI] [PubMed] [Google Scholar]
- Boukhalfa A, Miceli C, Ávalos Y, Morel E, and Dupont N (2019). Interplay between primary cilia, ubiquitin-proteasome system and autophagy. Biochimie 166, 286–292. [DOI] [PubMed] [Google Scholar]
- Carruthers S, Mason J, and Papalopulu N (2003). Depletion of the cell-cycle inhibitor p27Xic1 impairs neuronal differentiation and increases the number of ElrC+ progenitor cells in Xenopus tropicalis. Mech. Dev 120, 607–616. [DOI] [PubMed] [Google Scholar]
- Chen H, Li L, Hu J, Zhao Z, Ji L, Cheng C, Zhang G, Zhang T, Li Y, Chen H, et al. (2019). UBL4A inhibits autophagy-mediated proliferation and metastasis of pancreatic ductal adenocarcinoma via targeting LAMP1. J. Exp. Clin. Cancer Res 38, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SY, Leonard JL, and Davis PJ (2010). Molecular aspects of thyroid hormone actions. Endocr. Rev 31, 139–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, and Kintner C (1995). Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature 375, 761–766. [DOI] [PubMed] [Google Scholar]
- Chung M-I, Kwon T, Tu F, Brooks ER, Gupta R, Meyer M, Baker JC, Marcotte EM, and Wallingford JB (2014). Coordinated genomic control of ciliogenesis and cell movement by RFX2. Elife 3, e01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle Nogare D, and Chitnis AB (2017). A framework for understanding morphogenesis and migration of the zebrafish posterior Lateral Line primordium. Mech. Dev 148, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danahay H, Pessotti ADD, Coote J, Montgomery BEE, Xia D, Wilson A, Yang H, Wang Z, Bevan L, Thomas C, et al. (2015). Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Rep 10, 239–252. [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Wettstein DA, Koyano-nakagawa N, and Kintner C (1999). A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development 126, 4715–4728. [DOI] [PubMed] [Google Scholar]
- Fini JB, Le Mével S, Palmier K, Darras VM, Punzon I, Richardson SJ, Clerget-Froidevaux MS, and Demeneix BA (2012). Thyroid hormone signaling in the Xenopus laevis embryo is functional and susceptible to endocrine disruption. Endocrinology 153, 5068–5081. [DOI] [PubMed] [Google Scholar]
- Gomperts BN, Kim LJ, Flaherty SA, and Hackett BP (2007). IL-13 regulates cilia loss and foxj1 expression in human airway epithelium. Am. J. Respir. Cell Mol. Biol 37, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, and Rajagopal J (2009). Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 136, 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M, Gómez Vázquez JL, Sun DI, Tran HT, Brislinger M, Tasca A, Shomroni O, Vleminckx K, and Walentek P (2019). ΔN-Tp63 Mediates Wnt/β-Catenin-Induced Inhibition of Differentiation in Basal Stem Cells of Mucociliary Epithelia. Cell Rep 28, 3338–3352.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM, and Biology C (1991). In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol 36, 685–695. [DOI] [PubMed] [Google Scholar]
- Hogan BLM, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CCW, Niklason L, Calle E, Le A, Randell SH, et al. (2014). Repair and Regeneration of the Respiratory System: Complexity, Plasticity, and Mechanisms of Lung Stem Cell Function. Cell Stem Cell 15, 123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdgaard SG, Cianfanelli V, and Cecconi F (2020). Cloud hunting: doryphagy, a form of selective autophagy that degrades centriolar satellites. Autophagy 16, 379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber LA, and Teis D (2016). Lysosomal signaling in control of degradation pathways. Curr. Opin. Cell Biol 39, 8–14. [DOI] [PubMed] [Google Scholar]
- Hurbain I, Romao M, Bergam P, Heiligenstein X, and Raposo G (2017). Analyzing Lysosome-Related Organelles by Electron Microscopy In Lysosomes: Methods and Protocols, Methods in Molecular Biology, Öllinger K, and Appelqvist H, eds. (New York, NY: Springer New York; ), pp. 43–71. [DOI] [PubMed] [Google Scholar]
- Izawa I, Goto H, Kasahara K, and Inagaki M (2015). Current topics of functional links between primary cilia and cell cycle. Cilia 4, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachim J, Razi M, Judith D, Wirth M, Calamita E, Encheva V, Dynlacht BD, Snijders AP, O’Reilly N, Jefferies HBJ, et al. (2017). Centriolar Satellites Control GABARAP Ubiquitination and GABARAP-Mediated Autophagy. Curr. Biol. 27, 2123–2136.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klos Dehring DA, Vladar EK, Werner ME, Mitchell JW, Hwang P, and Mitchell BJ (2013). Deuterosome-mediated centriole biogenesis. Dev. Cell 27, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocaturk NM, and Gozuacik D (2018). Crosstalk between mammalian autophagy and the ubiquitin-proteasome system. Front. Cell Dev. Biol 6, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolm PJ, and Sive HL (1995). Efficient hormone-inducible protein function in Xenopus laevis. Dev. Biol 171, 267–272. [DOI] [PubMed] [Google Scholar]
- Lafkas D, Shelton A, Chiu C, De Leon Boenig G, Chen Y, Stawicki SS, Siltanen C, Reichelt M, Zhou M, Wu X, et al. (2015). Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature 528, 127–131. [DOI] [PubMed] [Google Scholar]
- Liang Y, Meng D, Zhu B, and Pan J (2016). Mechanism of ciliary disassembly. Cell. Mol. Life Sci 73, 1787–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell AJ, and Stanger BZ (2016). Adult cell plasticity in vivo: De-differentiation and transdifferentiation are back in style. Nat. Rev. Mol. Cell Biol 17, 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CT, Appleby SC, and Krieg PA (2014). Use of small molecule inhibitors of the Wnt and Notch signaling pathways during Xenopus development. Methods 66, 380–389. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Hirata J, and Sasaki F (1992). Fate of ciliated epidermal cells during early development of Xenopus laevis using whole-mount immunostaining with an antibody against chondroitin 6-sulfate proteoglycan and anti-tubulin: transdifferentiation or metaplasia of amphibian epidermis. Histochemistry 98, 355–358. [DOI] [PubMed] [Google Scholar]
- Pan HY, Alamri AH, and Valapala M (2019). Nutrient deprivation and lysosomal stress induce activation of TFEB in retinal pigment epithelial cells. Cell. Mol. Biol. Lett 24, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Saganta A, Law BM, Gonzalez-Celeiro M, Vinarsky V, and Rajagopal J (2013). Ciliated cells of pseudostratified airway epithelium do not become mucous cells after ovalbumin challenge. Am. J. Respir. Cell Mol. Biol 48, 364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Saganta A, Tata PR, Law BM, Saez B, Chow RDW, Prabhu M, Gridley T, and Rajagopal J (2015). Parent stem cells can serve as niches for their daughter cells. Nature 523, 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen JTML, Wilsch-Bräuninger M, and Huttner WB (2013). Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell 155, 333. [DOI] [PubMed] [Google Scholar]
- Park K-S, Korfhagen TR, Bruno MD, Kitzmiller JA, Wan H, Wert SE, Khurana Hershey GK, Chen G, and Whitsett J. a. (2007). SPDEF regulates goblet cell hyperplasia in the airway epithelium. J. Clin. Invest 117, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Mitchell BJ, Abitua PB, Kintner C, and Wallingford JB (2008). Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat. Genet 40, 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JC, Thavagnanam S, Skibinski G, Lyons J, Bell J, Heaney LG, and Shields MD (2013). Chronic IL9 and IL-13 Exposure Leads to an Altered Differentiation of Ciliated Cells in a Well-Differentiated Paediatric Bronchial Epithelial Cell Model. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta-Marques A, Bento I, Lopes CAM, Duarte P, Jana SC, and Bettencourt-Dias M (2016). A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science (80-. ). 353, 1–16. [DOI] [PubMed] [Google Scholar]
- Quigley IK, and Kintner C (2017). Rfx2 Stabilizes Foxj1 Binding at Chromatin Loops to Enable Multiciliated Cell Gene Expression. PLoS Genet 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbini D, Robert-Moreno À, Hoijman E, and Alsina B (2015). Retinoic acid signaling mediates hair cell regeneration by repressing p27kip and sox2 in supporting cells. J. Neurosci 35, 15752–15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz García S, Deprez M, Lebrigand K, Cavard A, Paquet A, Arguel M-J, Magnone V, Truchi M, Caballero I, Leroy S, et al. (2019). Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development 146, dev.177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg TA, and Rusan NM (2016). Taking Centrioles to the Elimination Round. Dev. Cell 38, 10–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, and Harland RM (2000). Early Development of Xenopus laevis (Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Smith SC, Lannoo MJ, and Armstrong JB (1988). Lateral‐line neuromast development in Ambystoma mexicanum and a comparison with Rana pipiens. J. Morphol 198, 367–379. [DOI] [PubMed] [Google Scholar]
- Soldner F, and Jaenisch R (2018). Stem Cells, Genome Editing, and the Path to Translational Medicine. Cell 175, 615–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Davidson L, Keller R, and Kintner C (2006). Radial intercalation of ciliated cells during Xenopus skin development. Development 133, 2507–2515. [DOI] [PubMed] [Google Scholar]
- Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, Franco B, and Zhong Q (2013). Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature 502, 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu F, Sedzinski J, Ma Y, Marcotte EM, and Wallingford JB (2018). Protein localization screening in vivo reveals novel regulators of multiciliated cell development and function. J. Cell Sci 131, jcs206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner JW, Kim EY, Ide K, Pelletier MR, Roswit WT, Morton JD, Battaile JT, Patel AC, Patterson GA, Castro M, et al. (2006). Blocking airway mucous cell metaplasia by inhibiting EGFR antiapoptosis and IL-13 transdifferentiation signals. J. Clin. Invest 116, 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenendaal NR, Ulmer B, Boskovski MT, Fang X, Khokha MK, Wendler CC, Blum M, and Rivkees SA (2013). Embryonic exposure to propylthiouracil disrupts left‐right patterning in Xenopus embryos. FASEB J 27, 684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walentek P (2018). Manipulating and Analyzing Cell Type Composition of the Xenopus Mucociliary Epidermis. In Methods in Molecular Biology, pp. 251–263. [DOI] [PubMed] [Google Scholar]
- Walentek P, and Quigley IK (2017). What we can learn from a tadpole about ciliopathies and airway diseases: Using systems biology in Xenopus to study cilia and mucociliary epithelia. Genesis 55, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walentek P, Beyer T, Thumberger T, Schweickert A, and Blum M (2012). ATP4a is required for Wnt-dependent Foxj1 expression and leftward flow in Xenopus left-right development. Cell Rep 1, 516–527. [DOI] [PubMed] [Google Scholar]
- Wang G, Chen Q, Zhang X, Zhang B, Zhuo X, Liu J, Jiang Q, and Zhang C (2013). PCM1 recruits Plk1 to the pericentriolar matrix to promote primary cilia disassembly before mitotic entry. J. Cell Sci 126, 1355–1365. [DOI] [PubMed] [Google Scholar]
- Wang L, Lee K, Malonis R, Sanchez I, and Dynlacht BD (2016). Tethering of an E3 ligase by PCM1 regulates the abundance of centrosomal KIAA0586/Talpid3 and promotes ciliogenesis. Elife 5, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Livingston MJ, Su Y, and Dong Z (2015). Reciprocal regulation of cilia and autophagy via the MTOR and proteasome pathways. Autophagy 11, 607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JM, and Watt FM (2018). Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature 557, 322–328. [DOI] [PubMed] [Google Scholar]
- Winklbauer R (1989). Development of the Lateral Line System. Prog. Neurobiol 32, 181–206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Movie 1: MCC loss over the nephrostome area at st. 44 (Related to Figure 3)
Two times image series (unannotated first, annotated second) through the z-planes (from apical to deep tissue layers) of a confocal micrograph (left panels) and stills (right panels) of a normal MCC, several apoptotic MCCs (apopt. MCC 1–3) with shedding morphology, a post apoptotic site (after apoptosis), and a trans-differentiating MCC (TD MCC) in a specimen at stage 44 stained for Ac.-α-Tubulin (grey), active Caspase 3 (magenta) and F-actin (Actin, green). Please note the presence of apoptotic cells overlaying the developing kidney tubules (yellow arrowheads). The MCCs depicted in stills are indicated by labeled yellow boxes.
Supplemental Movie 2: PCM1 knockdown leads to basal body uncoupling from the apical membrane (Related to Figure 7)
Image series through the z-planes (from apical to basal) of confocal micrographs of stage 42 control-injected (ctrl.) and pcm1MO-injected MCCs depicted in Figure 7D. Ac.-α-Tubulin (grey), F-actin (magenta) and basal bodies (Centrin4-GFP, green).
Supplemental Table 1: Experiment log and use of data in figures and graphs (Related to Figure 1–7)
List of experiments, species, assay, condition, number of specimens (N), and their use in figure are indicated.
Data Availability Statement
The datasets (imaging files) supporting the current study have not been deposited in a public repository due to large size but are available from the corresponding author on request. This study did not generate code.