Abstract
Nanoparticles must recognize, adhere to, and/or traverse multiple barriers in sequence to achieve cytosolic drug delivery. New nanoparticles often exhibit a unique ability to cross a single barrier (i.e. the vasculature, cell membrane, or endosomal compartment), but fail to deliver an adequate dose to intracellular sites of action because they cannot traverse other biological barriers for which they were not optimized. Here, we developed poly(acrylamide-co-methacrylic acid) nanogels that were modified in a modular manner with bioactive peptides. This nanogel does not recognize target cells or disrupt endosomal vesicles in its unmodified state, but can incorporate peptides with molecular recognition or environmentally responsive properties. Nanogels were modified with up to 15 wt% peptide without significantly altering their size, surface charge, or stability in aqueous buffer. Nanogels modified with a colon cancer-targeting oligopeptide exhibited up to a 324% enhancement in co-localization with SW-48 colon cancer cells in vitro, while influencing nanogel uptake by fibroblasts and macrophages to a lesser extent. Nanogels modified with an endosome disrupting peptide failed to retain its native endosomolytic activity, when coupled either individually or in combination with the targeting peptide. Our results offer a proof-of-concept for modifying synthetic nanogels with a combination of peptides that address barriers to cytosolic delivery individually and in tandem. Our data further motivate the need to identify endosome disrupting moieties which retain their activity within poly(acidic) networks.
Keywords: Nanogel, Peptide, Bioconjugation, Cell Targeting, Endosomal Escape
Graphical Abstract
Bioactive peptides were covalently coupled to poly(acidic) nanoscale hydrogels (nanogels). Target cell co-localization was conserved and proportional to the amount of peptide coupled. Endosomolytic activity was lost upon peptide conjugation.
1. Introduction:
Nanogels have been developed to deliver a variety of payloads (i.e. small molecules, peptides, proteins, nucleic acids) across barriers at the organ (i.e. skin1, gastrointestinal tract2) or cellular level (i.e. cell membrane3,4, endosome5,6). Nanogels (i.e. water-swollen, crosslinked, polymeric networks) are particularly attractive for these drug delivery applications because they can be designed for stability in physiological fluid, active/passive targeting capability, and responsiveness to their environment7–9.
Previous studies have explored ligand conjugation to nanoparticles and nanogels as a means of targeting tumors for drug delivery. Generally speaking, research groups identify a receptor that is overexpressed within a particular tumor population, and conjugate a peptide, protein, or nucleic acid ligand for that receptor to drug-loaded nanogels10. An ideal targeting ligand will identify and adhere to target cells without promoting nanogel co-localization with off-target cells (i.e. epithelial cells, fibroblasts, immune cells etc.). In vitro, peptide conjugation has been shown to increase nanogel uptake by target cells for a number of applications11,12. There is greater debate over the extent to which the conjugation of peptides or other targeting moieties influences the biodistribution of nanoparticles in vivo.13
Once a nanogel enters the cell by endocytosis, if it is to deliver its encapsulated payload to an intracellular site of action, it must disrupt the endosome14,15. Membrane disrupting moieties can facilitate endosomal escape. Validation of reliable mechanisms through which drug delivery vehicles can escape endosomes is an active area of research16,17. One proposed mechanism, consistent with the mechanism of intracellular delivery using lipoplexes18, is that cationic cell penetrating peptides deposit within and destabilize the endosomal membrane19. A second mechanism underlying the endosome disrupting ability of cationic networks and polyplexes is termed the proton sponge effect20,21. In the proton sponge effect, polybases with a pKa between the extracellular and lysosyomal pH (4.5 to 7.4)22 buffer the endosomal pH, alter the osmotic pressure of the endosome, and rupture it5,23.
Many studies on new nanogels or nanoparticles neglect to address the need for nano-scale drug delivery systems to overcome sequential biological barriers24–26. Researchers have explored the conjugation of multiple ligands to some extent27–31, for sequential targeting or for enhancing the ability of encapsulated payloads to act on intracellular targets. Further work is needed to fully understand the extent to which multiple encapsulated, adsorbed, or covalently incorporated ligands can (i) retain their bioactivity within the nanocarrier and (ii) overcome sequential barriers to drug delivery. Our understanding of the extent to which ligand density and position within the nanoparticle architecture contribute to their retention of biological activity is also incomplete.
Here, we developed a nanogel platform technology, which was by itself minimally bioactive, that could be modified to include multiple ligands, that are optimized individually to recognize, respond to, or traverse biological barriers. We developed a poly(acrylamide-co-methacrylic acid) nanogel, which has a unique modular design component32. The unmodified nanogel is responsive to the pH environment, able to complex hydrophilic small molecule payloads, and non-toxic to multiple cell lines at a high dose (0.5 wt% in media)33. The acid groups within the nanogel are amenable to carbodiimide-mediated bioconjugation, which has been applied previously to couple active biomolecules to numerous poly(acid) or poly(amine) biomaterials34,35. We initially modified the acid groups to possess pendant hydrophobic (tyramine) or cationic (n,n-dimethylethylenediamine) small molecules, peptides (multiple) and proteins (peroxidase, wheat germ agglutinin). The extent of nanogel modification was tunable, and in each tested case the ligand activity (i.e. payload complexation, enzymatic activity, molecular recognition) was conserved32.
For the purpose of organizing this proof-of-concept study, we selected colorectal cancer as a disease focus. Colorectal cancer is the third most common cancer in the US, affecting more than 140,000 new patients annually 36–38. Colorectal cancer incidence in young adults is increasing, rising by 22% from 2000 to 2013 39. In isolation, or in combination with targeted therapies, cocktails of cytotoxic chemotherapeutics are administered to treat colorectal cancer (i.e. 5-fluorouracil 40,41, irinotecan 42, trifluridine 41, tipiracil 41, and oxaliplatin 43). All of these chemotherapeutics, with the exception of irinotecan, are hydrophilic (octanol/water partition coefficient (logP) values of less than 0.07), resulting in rapid clearance from the bloodstream. Therefore, nanogels which extend the circulation time, tumor targeting, cell uptake, and cytosolic delivery of hydrophilic chemotherapeutics would be useful for treating colorectal cancer.
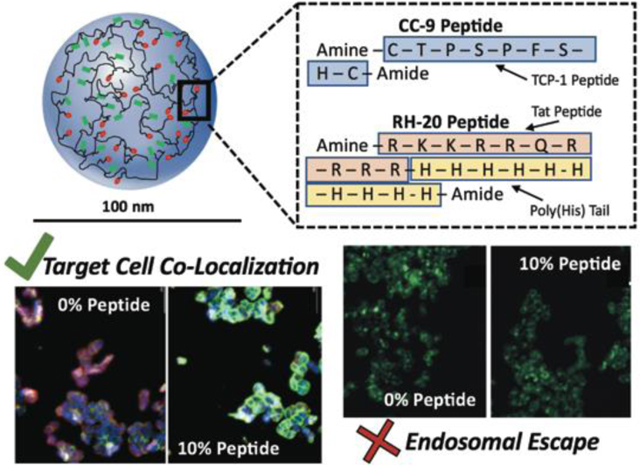
In this study, we selected peptides which have published experimental evidence for their colon cancer targeting and endosome escape ability. We selected a peptide (here called CC-9 after its sequence (CTPSPFSHC)) to target colorectal cancer. This peptide was identified by Li et al.44 by in vivo phage display, where it partitioned in colorectal tumors 11- to 94-fold more than in other organs (i.e. distal colon, brain, heart). We selected a histidine-modified Tat peptide for endosomolysis. Lo et al. 23 identified that complexes of the Tat peptide (RKKRRQRRRR) with a polyhistidine (5–20 H subunits) tail act as a potent facilitator of endosomal escape (up to a 700-fold increase in gene transfection).
The purpose of this study was to investigate the impact of peptide conjugation to a nanogel backbone on its bioactivity, specifically for overcoming the cell membrane and endosomal barriers. Our design purpose was to determine the influence of the peptide weight percentage, individually or when multiple unique peptides are used in tandem, on target cell co-localization and endosomal escape. We hypothesized that the nanogel chemistry will dictate the spatial arrangement of peptide ligands, which could influence its targeting capabilities or ability to interact with endosomal membranes, particularly if it is burried within the nanogel bulk. Similarly, we hyothesized that the nanogels‟ buffering capacility could influence the proton sponge effect, altering the extent of endosomolysis.
2. Materials and Methods
2.1. Nanogel Synthesis and Purification
Poly(acrylamide-co-methacrylic acid) nanogels were synthesized by inverse emulsion polymerization as described previously 33. The monomer feed was comprised of 75 mol% acrylamide, 22.5 mol% MAA, and 2.5 mol% methylenebisacrylamide, dissolved in ultrapure water at 42 wt%. This aquous monomer soution, after addition of 50 μL of TEMED was added to a stirring 50mL solution of Brij 30 (2.952 g) and AOT (0.724 g) in hexanes. The inverse emulsion was purged with nitrogen (20 min). Polymerization was initiated by adding 10 mg of APS (100 mg/mL in ultrapure water, nitrogen purged), and was allowed to proceed under constant mixing, at room temperature, for two hours. Nanogels were purified by precipitation in ethanol (three times) and dialysis against a water/ethanol gradient, as described previously 33. Purified nanogels were lyophilized and stored at room temperature.
2.2. Nanogel Modification with CC-9 and RH-20 Peptides
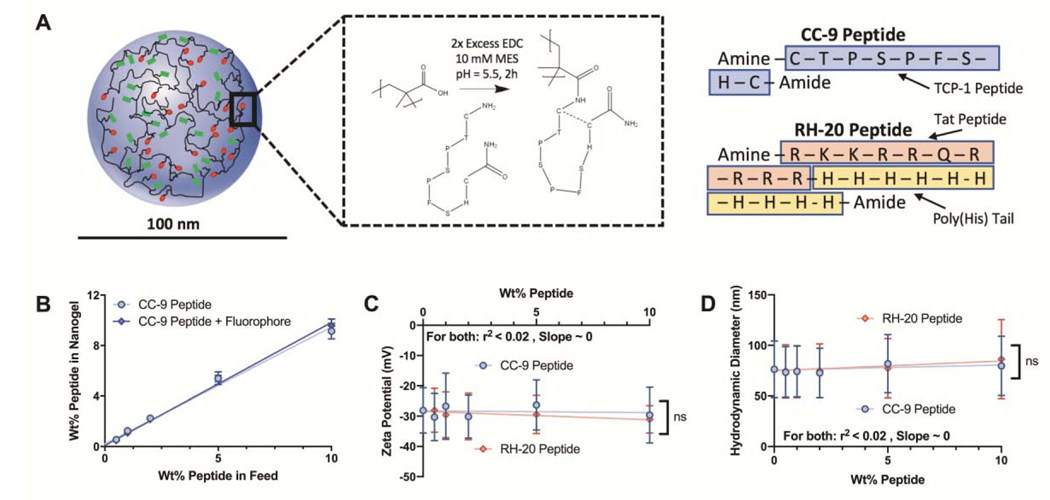
Dried nanogels were suspended in 10 mM MES buffer at 10 mg/mL, vortexted/sonicated to disperse uniformly, and adjusted to pH = 5.5 with 1N hydrochloric acid. They were transferred (3 mL per reaction) into 15 mL conical tubes. The CC-9 (sequence = CTPSPFSHC) and RH-20 (sequence = RKKRRQRRRRHHHHHHHHHH) peptides were dissolved at 5 mg/mL in 10 mM MES, and adjusted to pH = 5.5. An amine-terminated fluorophore (5-(amino acetamido) fluorescein) was dissolved at 1.5 mg/mL in 10 mM MES (pH = 5.5). Immediately prior to the coupling reaction, EDC was dissolved at 56 mg/mL in 10 mM MES (pH = 5.5). We added 600 μL of EDC solution (33.6 mg EDC per reaction, a 2-fold molar excess EDC to MAA groups in the nanogels) to each conical tube, immediately followed by the peptide and/or fluorphpore solutions. Peptides were added at of 0–10 wt%, relative to the nanogel weight. Fluorophore mass added was 0.5 wt%, relative to the nanogel weight. After mixing, the conical tube was placed on an orbital spinner, and was mixed end-over-end for two hours. A schematic of the peptide modification reaction is given in Figure 1.
Figure 1: Validation of peptide conjugation to modular P(AAm-co-MAA) nanogels.
(A) Schematic depiction of peptide modification by carbodiimide mediated coupling. CC-9 peptide shown, in its conjugated, loop conformation. Sequences for the CC-9 and RH20 peptide. (B) CC-9 peptide conjugation, as measured by a MicroBCA assay, was linearly related to the amount of peptide in the conjugation reaction. The conjugated CC-9 or RH-20 peptide, up to 10 wt%, did not significantly impact the (C) zeta potential, or (D) hydrodynamic diameter of the nanogels. Further there was no significant relationship between either parameter and peptide weight fraction. Shown are (B) mean ± sd, (C) zeta potential ± zeta deviation, (D) z-average diameter ± PDI width. (n = 3).
All peptide-nanogel conjugates were prepared in the manner described above. To test the peptide conjugation efficiency, we synthesized nanogels with 0–10 wt% CC-9, 0–10 wt% RH-20, and combinations of CC-9 and RH-20 (2–10 wt% each). We fabricated nanogels with 0.5 wt% fluoropore and peptide (CC or RH, different wt%) for cell co-localization studies. Peptide-modified nanogels without fluorphore were prepared for endosomal escape studies. All peptide-nanogel conjugates were purified by dialysis against ultrapure water (72 hours, twice daily water changes). They were lyophilized and stored at −20ºC.
2.3. Dynamic Light Scattering and Zeta Potential Measurement
The nanogels were characterized by dynamic light scattering (DLS) in 1x PBS and zeta potential measurement in 5mM sodium phosphate buffer. Nanogels were suspended in their respective buffers at 2 mg/mL, sonicated to suspend uniformly, and adjusted to pH = 7.4 with 1N sodium hydroxide or hydrochloric acid. Measurements were obtained using a ZetaSizer NanoZS system (Malvern). Presented results are the average of at least three measurements, where each measurement included a minimum of ten runs. DLS data are presented as the z-average diameter, plus or minus the PDI width. Zeta potential measurements are presented as the average zeta potential, plus or minus the zeta deviation.
2.4. MicroBCA Assay
The total CC-9 peptide content was quantified using a MicroBCA colorimetric assay (Thermo). MicroBCA quanitifies the total protein/peptide content of a solution through the reduction of copper, and is influenced strongly by cysteine, tyrosine, and tryptophan residues 45,46. Therefore, it was a sensitive assay for determining the amount of CC-9 peptide within nanogel conjugates. The MicroBCA assay exhibited poor sensitivity to RH-20 peptide content (ΔA562nm for a 200 μg/mL stock of RH-20 peptide = 0.09) Therefore, it was not suitable for quanitifying RH-20 peptide incorporation. Nanogels (50–200 μg/mL) were incubated in equal volume ratio with MicroBCA working reagent under orbital mixing for 2 hours. The total CC-9 peptide content within nanogels was quantified by solution absorbance at λ =562 nm using a microplate reader (BioTek), relative to a standard curve generated from unconjugated CC-9 peptide.
2.5. High-Throughput Assay for Nanogel Co-localization
Fluorescently tagged nanogels (50–400 μg/mL) were suspended in 2% FBS phenol-red free DMEM and incubated in the presence of each cell line for 2 hr. After incubation, the nanogels were aspirated, and the microwell plate was washed three times with cold DPBS (100 μL/well). Cells were fixed with 50 μL of cold IC Fixation Buffer (Invitrogen), and stained with AlexaFluor 594 Wheat Germ Agglutinin (membrane) and DAPI (nucleus). In between each fixation or staining step, the cells were washed three times with cold Hanks Balanecd Salt Solution (HBSS) to ensure removal of unbound or unreacted species. The microplates, containing fixed and stained cells, were stored in the dark at 4°C until imaged.
Images were aquired using the Cytation 3 Cell Imaging Multi-Mode Reader (BioTek) and processed usig Gen5 software (BioTek). Four wells were imaged for each treatment (cell, nanogel identity, concentration) and four images were aquired per well. Images were acquired using DAPI, GFP, and Texas Red filters (DAPI: excitation = 377 nm, emission = 447 nm, Texas Red: excitation = 586 nm, emission = 647 nm, GFP: excitation = 469 nm, emission = 525 nm) and an Olympus 20× objective. Each well was autofocused to in the DAPI and Texas Red channels. GFP images were acquired at the DAPI focal length. Imaging parameters (i.e. LED intensity, gain, and integration time) were optimized for the most fluorescent sample (RAW 264.7 cells, all formulations) to prevent saturation. These parameters were held constant for the purpose of quantitative image analysis (DAPI: Intensity = 5, Integration time = 53, Gain = 0, Texas Red: LED intensity = 10, Integration time = 100, Gain = 12.3, GFP: LED intensity = 10, Integration time = 271, Gain = 15.6).
Images were quantified using the cellular analysis tool in Gen5. From each image, we subtracted the background fluorescence (DAPI, Texas Red, and GFP channels) and performed a cell count with the DAPI channel. The nanogel fluorescence (GFP) was normalized to the cell count. Further, we adjusted this normalized fluorescence by a correction factor, corresponding to the slope of a fluorescence calibration curve for each nanogel formulation. This ensured that observed trends in nanogel co-localization (measured by normalized fluorescence) are in fact due to nanogel co-localization and not differences in fluorescent labeling.
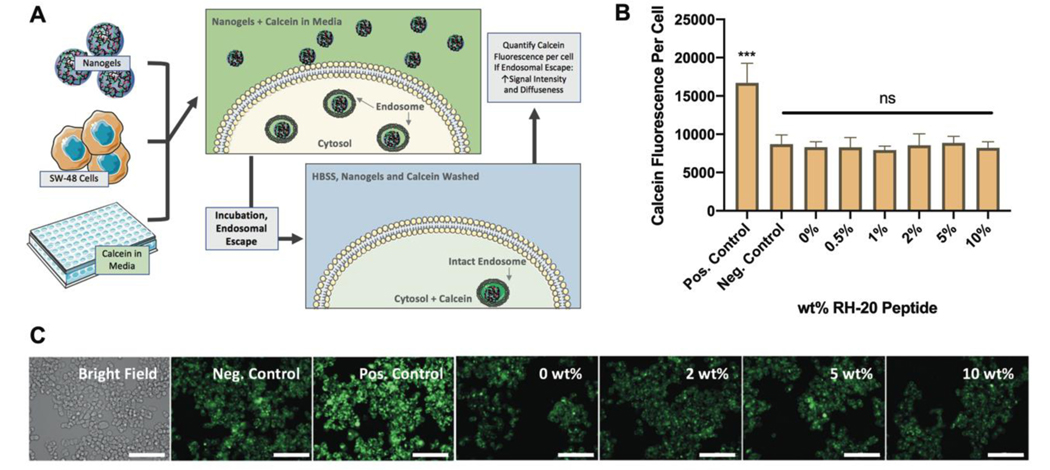
2.6. Calcein Assay for Endosmal Escape
We probed peptide-enhanced endosomal escape in SW-48 cells with a calcein assay developed by Kongkatigumjorn et al. 47 and modified by Deshpande et al 48. Nanogels (100–400 μg/mL) and calcein (100 μg/mL) suspended in 2% FBS phenol redfree DMEM were incubated with SW-48 cells for 4 hr. Cells incubated with calcein (100 μg/mL) only were used as a negative control. Cells incubated with calcein (100 μg/mL) and free RH-20 peptide (200 μg/mL) were used as a positive control. After incubation, the nanogels and calcein were aspirated, and the cells were washed three times with cold DPBS.
The cells were imaged immediately using the Cytation 3, with a 20x Olympus objective, using the bright field and GFP filters. Each sample was foccused to the bright field channel, and the GFP image was acquired at the bright field focal height. The bright field image was used to obtain a cell count, and the GFP channel enabled visualization of vessicular/free calcein within the cytosol.
Endosomal escape images were quantified using the cellular analysis tool in Gen5. Disruption of the endosome will lead to diffuse calcein in the cytosol, accompanied by an increase in green fluorescence 48,49. Therefore, the green fluorescence per cell, relative to positive and negative controls, was used as a surrogate measure of endosomal escape.
3. Results and Discussion:
3.1. Feasibility of Modular Approach:
Prior to modification, the poly(acrylamide-co-methacrylic acid) (P(AAm-co-MAA) nanogels had a z-average hydrodynamic diameter of 76.3 ± 28.0 nm. To make estimations, regarding peptide modification of the nanogels, we made a few simplifying assumptions. We assumed that the nanogel swelling ratio for P(AAm-co-MAA) nanogles is similar to the thin films of the same formulation. We fabricated P(AAm-co-MAA) films of the same composition, and measured the film weight in a completely dried and equilibrium-swollen state. We calculated that:
where Q is the weight swelling ratio, mp is the polymer mass in the nanogels, and mw is the water mass in the nanogels. We assumed that the polymer density is approximately equal to the density of water.
Assuming that, consistent with our previous study, all monomers were incorporated at feed ratio, we next computed the number of MAA moieties within each nanogel:
where Rh is the nanogel hydrodynamic diameter, is the weight fraction of MAA in the nanogel, 86.06 g/mol is the molecular weight of MAA, and NA is Avogadro‟s number. There were an average of 2953 molecules of MAA polymerized into each P(AAm-co-MAA) nanogel.
We determined the feasibility of modifying the nanogel to 10 wt% of each peptide (as a weight fraction of the polymer dry weight). A 10 wt% CC-9 and 10 wt% RH-20 peptide nanogel requires modification of 114 and 39 MAA molecules, respectively. This requires conversion of only 3.9% and 1.3% of the total MAA. For fluorescently tagged nanogels, 0.5 wt% fluorphore requires further modification of another 13 (0.4% of total) MAA molecules per nanogel. As the highest extent of modification (10 wt% CC-9 and 10 wt% RH-20, fluorescent) requires modifying only 4.6% of the MAA groups, we determined that all extents of modification pursued in this study were feasible.
3.2. Nanogel Structure and Surface Coverage
P(AAm-co-MAA) nanogels possess pendant carboxylic acid moieties throughout their bulk that interact with the aqueous environment. In comparison to similarly-sized polymeric nanoparticles, metal nanoparticles, or liposomes, P(AAm-co-MAA) nanogels have more water-accessible acid groups, which can be modified with peptides in a modular manner. As a result, P(AAm-co-MAA) nanogels have the potential to be modified covalently, and with a substantial mass fraction, of bioactive peptides. In this study, we leverage this unique property of P(AAm-co-MAA) nanogels to achieve significant (up to 20 wt%) peptide modification.
While we have shown previously that proteins coupled to P(AAm-co-MAA) retain their bioactivity32, it was previously unclear the extent to which peptides immobilized within nanogels retain bioactivity. Our modular bioconjugation schema, connecting N-terminal amines on peptides to nanogel carboxylic acid moieties, couples indiscriminately to the nanogel surface and bulk. If the CC-9 peptide is sufficiently burried within the nanogel bulk, it may be unable to engage with cell surface receptors to facilitate co-localization. Conversely, RH-20 peptides within the nanogel bulk are intended to facilitate endosomal escape through a proton sponge effect. Because the RH-20 peptide is net-cationic, it could also act as a cell penetrating peptide if concentrated at the nanogel surface. Here, we acknowledge that the spatial distribution of peptides within the nanogel could influence their ability to exert a useful biological function. Our modular system does not enable spatial selectivity, which is a noted limitation.
A second aspect to peptide modification of nanogels is minimizing its impact on nanogel stability in aqueous solution. In a recent paper, we noted that P(AAm-co-MAA) modification to high extents with a hydrophobic ligand (modifying >50% of the carboxylic acid groups) resulted in nanogel aggregation32. Our P(AAm-co-MAA) nanogels rely on hydrophilicity and a highly negative zeta potential to prevent aggregation. The CC-9 peptide containes multiple hydrophobic amino acids and the RH-20 peptide is cationic. If cationic or hydrophobic moieties are present at too high of a density on the nanogel surface, they could promote nanogel aggregation and reduce efficacy for intracellular delivery.
To aid our analysis, we estimated the peptide surface coverage of each nanogel. We made a simplifying assumption that the peptide groups are bound only to the nanogel surface, and not throughout the bulk. The purpose of this assumption was to determine the upper limit for surface coverage, given a specific cocktail and quantity of peptides. Estimated from their respective molecular weights and the Stokes-Einstein equation, the CC-9 peptide is 1.39 nm and the RH-20 peptide is 1.98 nm in diameter. Modeling the nanogels and each peptide as a uniform sphere, 10 wt% modification corresponds to 3.8% and 2.6% surface coverage the CC-9 and RH-20 peptide, respectively. Therefore, it is reasonable to assume that the nanogel with the highest extent of modification retains at least 93.6% of its native surface.
3.3. Nanogel Characterization
The umodified P(AAm-co-MAA) nanogels possessed a z-average hydrodynamic diameter of 76.3 ± 28.0 nm and a zeta potential of −28.1 ± 7.5 mV. As the RH-20 peptide does not contain cysteine, tyrosine, or tryptophan, microBCA analysis did not enable direct quantification of immobilized RH-20. The CC-9 peptide, when immobilized within the nanogels was detected sensitively by a microBCA, applied as documented previously.50 Nanogel modification with the CC-9 peptide trended linearly with the concentration (r2 = 0.99) and the peptide coupling/recovery efficiency was greater than than 90% efficient (Fig 1b). There was no significant difference in peptide modification efficiency as a result of simultaneous fluorphore conjugation (Fig S1a). Further, through direct quantification of free thiols by Ellman‟s assay, we determined that more than 99% of the conjugated CC-9 peptide was in a loop confirmation, with a loss of free thiols and adoption of a disulfide linkage (Fig S1b). For in vitro studies, we generated three libraries of peptide-modified nanogels; those modified with 0.5 wt% to 10 wt% CC-9, 0.5% to 10 wt% RH-20, and combinationt of 2 wt% to 10 wt% CC-9 and RH-20. In each case a fluorescently tagged formulation was synthesized for cell co-localization studies, while a non-fluorescent analogue was synthesized for endosomal escape studies.
Consistent with the expectation that peptide and/or fluorphore modification will react with less than 5% of the total number of methacrylic acid moieties in the nanogel, peptide modification did not significantly alter the nanogels‟ zeta potential (Figure 1c). Peptide modification increased the hydrodynamic diameter of the nanogels, although the increase in diameter was not statistically significant (Figure 1d). The peptide identity (CC-9 or RH-20) and amount (0–10 wt%) of peptide did not significantly influence the nanogel hydrodynamic diameter, as compared to unmodified nanogels. The average polydispersity index for the modified and unmodified nanogels was 0.185. Full grouped (i.e. extent of modification, peptide, fluorophore) size and zeta potential data, with statistical analyses, are given in Fig. S2. These results support our hypothesis that peptide modification is indiscriminate to the nanogel surface and bulk. Further, it was encouraging that peptide modification up to 10 wt% did not lead to nanogel aggregation nor a loss of nanogel stability in aqueous solution.
3.4. CC-9 Peptide Conjugation Enhances Nanogel Co-localization by Colorectal Carcinoma Cells, without Altering Co-localization by Fibroblasts or Macrophages:
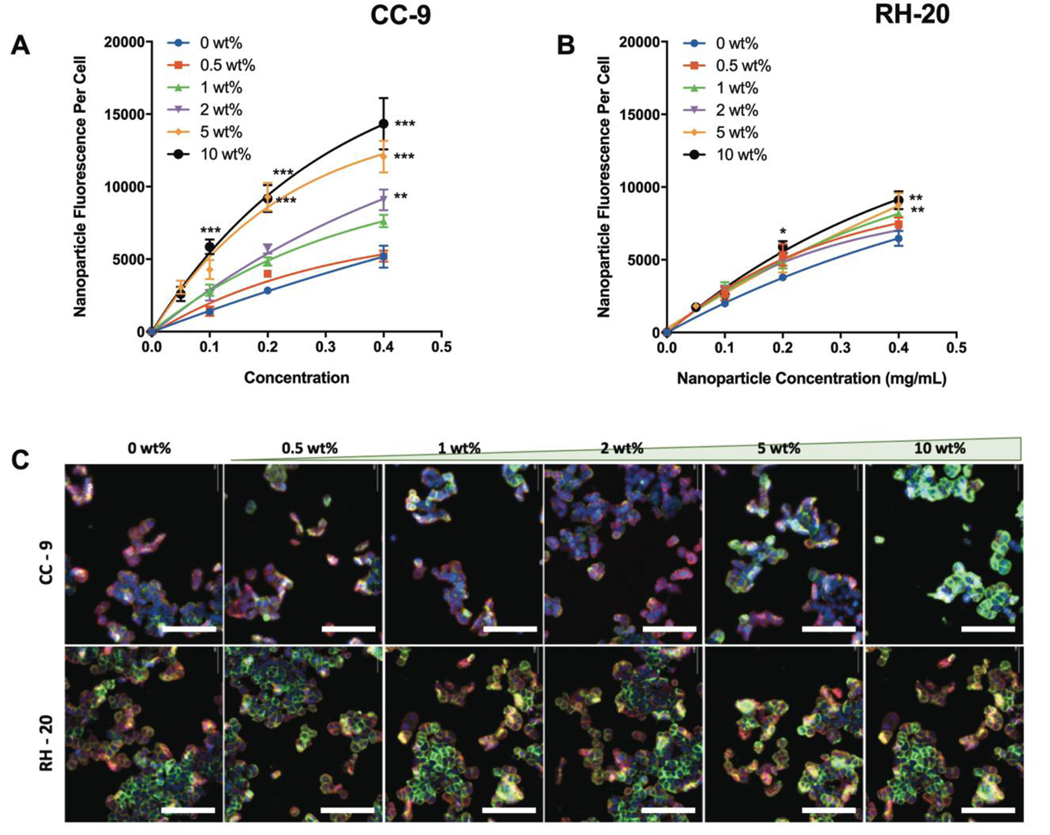
The CC-9 peptide significantly increased nanogel co-localization with SW-48 cells (Fig 2a). A minimum of 2 wt% CC-9 was necessary to observe a significant enhancement in co-localization, and the extent of enhancement increased as the CC-9 weight percent increased. The 10 wt% CC-9 nanogel enhanced co-localization with SW-48 cells by 324 ± 72% at a 100 μg/mL dose, and 177 ± 69% at a 400 μg/mL dose (both p < 0.001). This demonstrated that the improvement in SW-48 cell co-localization was (i) dependent on the CC-9 concentration within the nanogel and (ii) particularly pronounced at low nanoparticle to cell ratios. Both findings are consistent with a molecular recognition event between the SW-48 cells and CC-9 ligand.
Figure 2: Influence of peptide conjugation on nanogel co-localization with SW-48 colon carcinoma cells.
Nanoparticle fluorescence per cell, indicative of the relative-co-localization, was measured as a function of nanogel concentration in the medium (2 h incubation) for (A) nanogels modified to different extents with CC-9 or (B) nanogels modified to different extents with RH-20. As more CC-9 was incorporated, the extent of nanogel co-localization increased. (C) Representative images for nanogel co-localization (2 h, 400 μg/mL) by fluorescence imaging, as a function of peptide identity and peptide weight percentage in the dried nanogel (blue = nucleus, red = membrane, green = nanogels, scale bar = 100 μm). (n = 4, *p < 0.05, **p < 0.01, ***p < 0.001, relative to unmodified nanogels at the same concentration, 2-way ANOVA with tukey post-test).
The RH-20 conjugated nanogels also co-localizatized with SW-48 colorectal carcinoma cells to a greater extent than unmodified controls, but the extent of enhancement was less pronounced than was observed with CC-9 (Figure 2b). The SW-48 cells exhibited enhanced co-localization of nanogels modified with 5 wt% and 10 wt% RH-20 (36 ± 25% and 41 ± 19% increase at a 400 μg/mL dose, relative to unmodified controls, respectively). Unlike the significant increase in co-localization observed by CC-9 modified nanogels at low concentrations, there were no signifiant differences in SW-48 co-localization of RH-20 modified nanogels at a 100 μg/mL dose. This nanogel dose-dependent enhancement in co-localization, where effects are seen only when significant doses are achieved, is consistent with the hypothesis that surface immobilized RH-20 acts as a non-specific cell penetrating peptide.
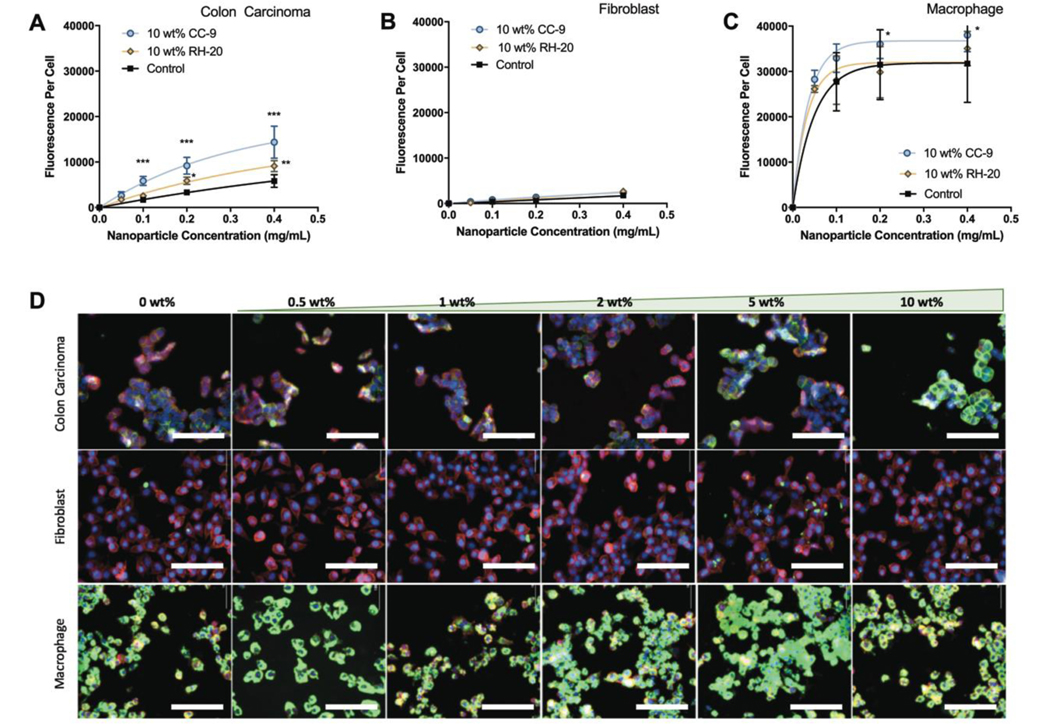
To investigate, in a general sense, the extent to which CC-9 and RH-20 influence nanogel interactions with cells, we compared our co-localization results for SW-48 cells (Fig 3a) to fibroblasts (L929, Fig 3b) and macrophages (RAW 264.7, Fig 3c) under the same conditions. If CC-9 modified nanogels exhibited enhanced co-localization with SW-48 cells due to a specific molecular recognition event, we expected that CC-9 would not enhance nanogel co-localization with L929 or RAW 264.7 cells. Each „nanoparticle fluorescence per cell‟ value was the green fluorescence intensity, normalized to cell count and calibrated for nanogel fluorescence intensity (i.e. standardized to calibration curves generated for each nanogel formulation). Figure 3 is plotted on a common scale, so that differences in nanogel co-localization across cell lines is more visually apparent. Nanogel co-localization data are presented for each cell line-formulation pair in Figure S3 on variable y-axes to facilitate comparisons between formulations.
Figure 3: Peptide conjugation influences cellular co-localization in a cell-dependent manner.
Nanoparticle fluoresence per cell is given as a function of cell line, nanoparticle concentration in the medium, and peptide identity (10 wt% peptide, 2 h incubation). (A) Modification with the CC-9 peptide increased nanogel co-localization with colon carcinoma cells to a greater extent than modification with RH-20 peptide. (B) Nanogels did not significantly co-localize with fibroblasts irrespective of concentration or peptide modification. (C) Nanogels co-localized in substantial quantity with macrophages irrespective of peptide modificaiton. (D) Representative fluorescence images of nanogels, modified to different extents with CC-9 peptide and incubated individually with each cell line (2 h, 400 μg/mL). Colon carcinoma images are reproduced from Figure 2 for visual comparison with fibroblast and macrophage cell lines. (blue = nucleus, red = membrane, green = nanogels, scale bar = 100 μm). (n = 4, *p < 0.05, **p < 0.05, ***p < 0.001, 2 way ANOVA with tukey post-test).
Fibroblasts did not co-localize with substantial quantities of any formulation. Nanogel co-localization with fibroblasts trended linearly with nanogel concentration, and CC-9 or RH-20 modification at 10 wt% did not significantly increase co-localization. The extent and identity of peptide modification did not predict the extent of co-localization. Only the 1 wt% and 2 wt% CC-9 nanogels, dosed at 400 μg/mL, co-localized to a greater extent than unmodified controls, and this increase was small (59 ± 62% and 62 ± 47% increase, respectively, Fig. S3). Both of these results indicated that the fibroblast-nanogel interaction is non-specific irrespective of the extent of CC-9 or RH-20 modification, and that both the modified and unmodified nanogels largely evade interaction with and uptake by fibroblasts.
The macrophages interacted with substantial quantities of all nanogel formulations, irrespective of the dose or extent of peptide modification (Figures 3c and 3d). Co-localization of the 1 wt%, 5 wt% and 10 wt% CC-9 peptide nanogels was greater than unmodified controls (p < 0.001 for 1 wt%, p < 0.05 for 5 wt% and 10 wt%) (Figure S3). There was no relationship between the extent of CC-9 modification and macrophage co-localization. Peptide (CC-9) modification increased nanogel co-localization with macrophages by an average of 35% (range = −18% to 64%) and this enhancement did not trend with nanoparticle dose (0.1 to 0.4 μg/mL) or the extent to which CC-9 was incorporated. Therefore, in comparison to the influence of CC-9 on co-localization with SW-48 cells, CC-9 appears to enhance macrophage co-localization to a lesser extent. No significant differences were observed in the co-localization of RH-20 modified nanogels by murine macrophages, dosed at up to 400 μg/mL.
We compared the relative extent of nanogel association with each cell line. The nanogels associated to a significantly greater extent with macrophages than colon carcinoma cells (p<0.001). Their co-localization was also greater with colon carcinoma cells than fibroblasts (p<0.05) (Fig 3d, Fig S3). This relative co-localization (macrophages > colon carcinoma > fibroblasts) indicates that, while the nanogels do not evade uptake by phagocytotic cells, they are preferentially co-localize with SW-48 colon cancer cells over model cells of the connective tissue (L929 fibroblasts).
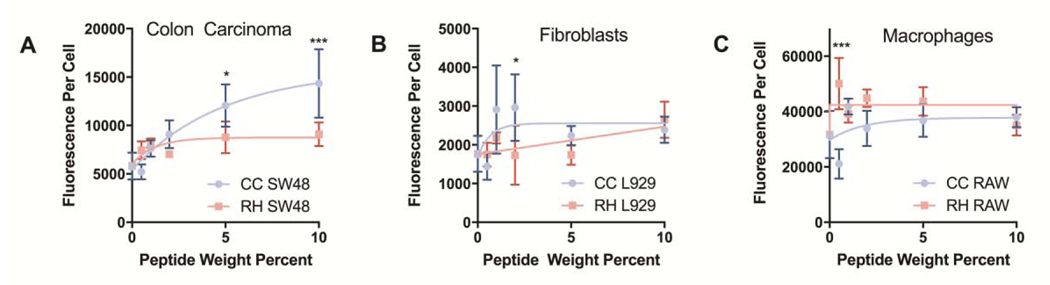
In addition, we compared the extent to which CC-9 and RH-20 enhanced nanogel co-localization between cell lines. As shown in Figure 4, peptide modification had a greater impact on nanogel co-localization with colon cancer cells than macrophages or fibroblasts. A non-linear one phase association model was fit to each data set for visualization. Only with the colon carcinoma cell line and CC-9 peptide modified nanogels was the extent of peptide modification (in wt%) predicive of co-localization (r2 = 0.77). For the RH-20 nanogels with colon carcinoma cells, as well as both peptide conjugates with fibroblasts and macrophages, the variation in peptide weight fraction explained less than 30% of the variation in nanogel-cell co-localization.
Figure 4: Influence of peptide weight fraction on cellular co-localization.
Fluorescence intensity per cell was measured to quantify the relative extent of nanogel co-localization with each cell line. Only in the case of (A) colon carcinoma cells, and not with (B) fibroblasts or (C) macrophages did the extent of cellular co-localization trend significantly with CC-9 wt% in the dry nanogel (r2 for goodness of fit = 0.77 for CC-9 wt% and co-localization with SW-48 cells). Higher weight fractions of CC-9 (5–10 wt%) significantly increased nanogel co-localization with SW-48 cells, relative to inclusion of a similar weight fraction RH-20. When the same nanogels were incubated with fibroblasts or macrophages, no significant difference in co-localization were observed. (mean fluorescence ± sd, n = 4–8, *p < 0.05, ***p < 0.001, 2-way ANOVA with tukey post-test).
Taken together, our results indicate that peptide modification is an effective method for imparting cell recognition properties in P(AAm-co-MAA) nanogels. The extent of cell recognition was dependent on the suitability of the peptide ligand (in this case CC-9) for a given cell type and its expressed receptors. The distribution of CC-9 or RH-20 within the nanogel surface and bulk did not deter cell recognition, although we are unable to determine if similar recognition properties could have been achieved with a lower degree of modification if those ligands were concentrated on the nanocarrier surface. It is worth noting that, throughout all of our studies, we measured cell co-localization rather than uptake. This focus was due to the resolution limitation of our high-throughput fluorescence imaging assay. By focusing on co-localization, we were able to look at the extent to which nanogel decoration with peptides influences the extent of nanogel-cell interaction in a general sense. Future experiments and measurements, to clarify the extent to which nanogel decoration influences cellular trafficking of the carrier and encapsulated cargoes, could involve confocal microscopy or flow cytometry. In such a study, it would be necessary to consider how physical properties of the nanogel, such as stiffness51,52, influence the endocytosis mechanism and extent of uptake5.
3.5. Free RH-20 Peptide, but not Peptide-Nanogel Conjugates, Facilitates Endosomal Escape
The extent to which peptide modification enhanced nanogel endosomal escape in SW-48 colon carcinoma cells was determined using a Calcein assay with fluorescence microscopy (Fig 5a). For these experiments, non-fluorescent nanogels modified to different extents with the RH-20 peptide were used, as not to provide background green fluorescence. In isoloation (i.e. not conjugated to a nanogel) RH-20 significantly enhanced calcein delivery to the cytosol, indicative of endosomal escape (p < 0.001). Therefore for all further experiments, free RH-20 at 200 μg/mL was used as a positive control. Calcein in media (no peptide or nanogels) was used as a negative control. Nanogels and nanogel-peptide conjugates did not alter the amount of calcein delivered to the cytosol, when compared to the negative control (Fig 5b). Therefore, neither the unmodified nanogels, nor nanogels modified to any extent with RH-20 peptide, facilitated endosomal escape. Representative images, illustrating the spatial distribution and intensity of calcein fluorescence are given in Figure 5c.
Figure 5: Endosome disruption by RH-20.
(A) Schematic depiction of endosomal escape assay by calcein imaging. (B) While free RH-20 peptide (200 μg / mL, positive control) significantly disrupted endosomes, resulting in siginficant and diffuse calcein fluorescence in the cytosol of SW-48 cells, nanogels modified with up to 10 wt% RH-20 failed to disrupt endosomes. The calcein fluorescence per cell for RH-20 modified nanogels was indistiguishable from that of the negative control (media with calcein, no nanogels). (C) Representative images for SW-48 cells in each incubation condition (green = calcein, scale bar = 100 μm). Images are enlarged and reprinted in Figure S4. (mean fluorescence ± sd, n = 3, ***p < 0.001, 1 way ANOVA).
It was particularly interesting that RH-20 did not retain its endosomolytic activity when conjugated to a polyacidic network. Based on the results published by Lo et al. 23 the proton sponge effect is critical to the endosomolytic activity of the Tat peptide with a polyhistidine tail. It appears that the polyacidic network, which accounts for the majority of the nanogel by weight, may in fact impair the RH-20 peptide‟s ability to disrupt endosomes. An alternate explanation is that, in the previous work, the RH-20 peptide was co-administered as a DNA complex, whereas the peptide was conjugated in a network for this study. Mobility of the RH-20 peptide, for membrane insertion and disruption purposes, may be critical to its function.
From our cell co-localization experiments, we knew that CC-9 modification can significantly enhance nanogel interaction with SW-48 cells. Therefore, we further sought to determine if dual peptide-modified nanogels could recognize SW-48 cells through their CC-9 ligand and facilitate endosomal escape with RH-20.
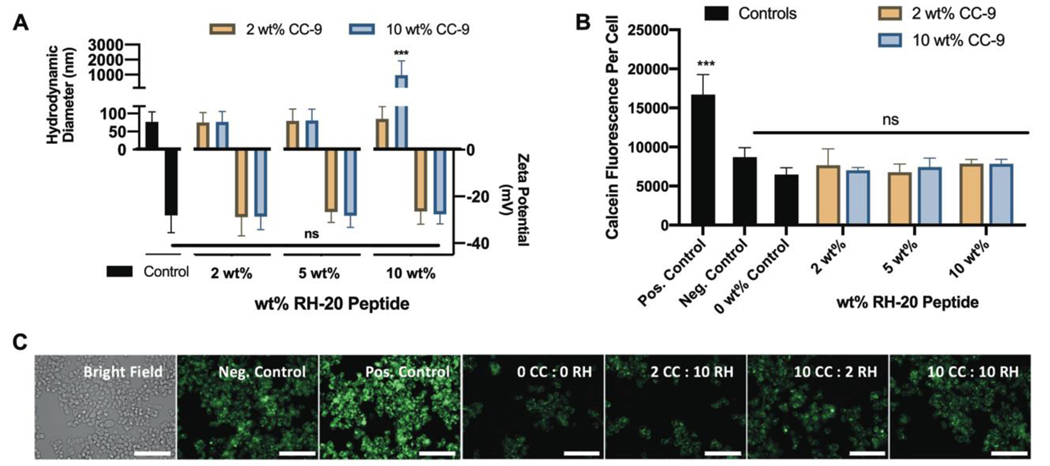
We first determined the extent to which nanogels could be modified with both the CC-9 and RH-20 peptide without loss of stability in aqueous solution. Nanogels modified with less than 15 wt% peptide (various combinations of CC-9 and RH-20) were stable in 1x PBS. Peptide modification up to 15 wt% increased the hydrodynamic diameter of the nanogels slightly, although this increase was not statistically different from unmodified control nanogels. Modification with increasing amounts of RH-20 peptide was associated with an increase in zeta potential, although this increase was also not statistically significant (Figure 6a). Nanogels modified with 20 wt% peptide (10 wt% CC-9 and 10 wt% RH-20) aggregated significantly (z-average diameter = 962 nm, PDI = 1) (Figure 6a).
Figure 6: Dual peptide-modified nanogels fail to disrupt endosomes.
(A) Nanogel modification with both CC-9 and RH-20 to different extents (2 – 10 wt%) did not significantly influence the nanogel zeta potential or hydrodynamic diameter, and led to aggregation only in the case of the 20 wt% (equal weight CC-9 and RH-20) conjugate (control = unmodified nanogels, z-average diameter ± PDI width, zeta potential ± zeta deviation, n = 3). (B) Dual peptide-modified nanogels fail to facilitate endosomal escape in SW-48 cells, suggesting that enhanced nanogel co-localization with 10 wt% CC-9 is insufficient to enable endosome disruption by RH-20 (positive control = 200 μg / mL RH-20, negative control = calcein and media only, 0 wt% control = unmodified nanogels in calcein solution). (C) Representative images for SW-48 cells in each incubation condition, labeled by peptide modification cocktail (green = calcein, scale bar = 100 μm). Images are enlarged and reprinted in Figure S4 (mean fluorescence ± sd, n = 3, ***p < 0.001, 2-way ANOVA).
We also validated that the CC-9 peptide retains its active conformation when conjugated to nanogels in combination with RH-20. CC-9 conjugation efficiency, as measured by MicroBCA, was greater than 83% for all dual-peptide modification reactions (Fig S1c). Similar to the nanogels modified with CC-9 peptide alone, more than 99% of the CC-9 peptide within dual-modified nanogels was in a loop conformation (Fig S1d).
Similar to what was observed with P(AAm-co-MAA) nanogels coupled with RH-20 alone (up to 10 wt%) dual peptide conjugates containing CC-9 and RH-20 also failed to enhance calcein release to the SW-48 cell cytosol, indicating an inability to disrupt endosomes (Fig 6 b,c). The extent of CC-9 conjugation (2 wt%, 10 wt%) did not affect endosomolytic activity, indicating that the extent of nanogel co-localization with SW-48 cells was not predictive of endosome escape. This further supports that the observed loss of RH-20’s endosomolytic activity results from its coupling to the polyacidic P(AAm-co-MAA) network.
4. Conclusion:
In the present study, we developed a modular method for modifying nanoscale P(AAm-co-MAA) networks (i.e. nanogels) with bioactive peptides, with a design goal of (i) co-localizing with target cells and (ii) disrupting endosomes upon uptake. Two peptides, CC-9 and RH-20, were identified from literature reports23,44 for their ability to target colon cancer cells and disrupt endosomes. Both peptides were efficiently integrated (>83% efficiency in all cases) within the nanogels via carbodiimide mediated coupling. The high density of bulk, modifyable acid groups in P(AAm-co-MAA) made it particularly amenable to a high degree of peptide incorporation (up to 10 wt% CC-9 or RH-20, and up to 15 wt% when the two peptides were combined) without a significant change to the network hydrodynamic diameter, zeta potential, or stability in aqueous buffer.
The CC-9 peptide promoted nanogel co-localization by SW-48 colorectal carcinoma cells, exhibiting an increase in total co-localization of up to 324%. The extent of CC-9 modification (in weight percent) was largely predictive of the nanogel co-localization with for the SW-48 cells. While peptide modification also effected nanogel co-localization with the other cell lines (RAW 264.7 macrophages, L929 fibroblasts), the extent to which modification increased co-localization was at least five times less than what was observed with colon cancer epithelial cells. Further, the extent of nanogel modification with CC-9 (i.e. from 0.5 to 10 wt%) was predictive of the extent of nanogel co-localization only for SW-48 colon cancer cells. The endosomolytic activity of RH-20 was lost when conjugated to P(AAm-co-MAA), alone or in combination with CC-9. This is a limitation for P(AAm-co-MAA) to be applied as a vehicle to deliver drugs to the cell cytosol, but could be useful for delivery to the lysosome or cellular transport by transcytosis.
In summary, we developed and validated a method for incorporating peptides into synthetic nanogels, with particular insight into how retention of the peptides‟ molecular recognition capability can be applied to tune the cell-nanogel interaction. The physical stability of these networks, with up to 15 wt% peptide, throughout synthesis, lyophilization, storage, and reconstitution in biological media could be particularly attractive for specific drug delivery applications. In the future, further studies must determine the extent to which presenting a high quantity of bioactive peptides (i.e. individually or in combination) to the surrounding environment by route of a synthetic nanogel can usefully alter the nanogel‟s biodistribution or other drug delivery properties.
Supplementary Material
6. Acknowledgements:
The authors gratefully recognize financial support from the National Institutes of Health (NIH) (EB022025 to NAP), Cockrell Regents Family Chair in Engineering (UT Austin) and UT-Portugal Collaborative Research Program. JRC was supported by an NSF Graduate Research Fellowship (DGE-1610403). JS, JG, and AKV were supported by undergraduate research fellowships from the UT Austin Office of Undergraduate Research. The schematic in Figure 5a was generated using freely available graphics from Servier Medical Art.
This contribution is dedicated to Professor Jindrich Kopecek of the University of Utah on his 80th birthday. Dr. Kopecek has been a leading authority in biomaterials and drug delivery and we have all benefited from his lucid, detailed publications and his imaginative synthetic chemistry. The senior author (NAP) met Prof. Kopecek at an IUPAC meeting in 1979 and has been the recipient of much sage advice from him over most of his career. The very imaginative and detailed chemistry he has devised plus the superb understanding of biological process have helped all of us in our understanding of disease treatment.
Footnotes
Declaration of Interest:
None.
CRediT Author Statement:
John R. Clegg: Conceptualization, Data curation, Analysis, Investigation, Methodology, Writing - Original Draft, Writing - review & editing. Jessie A. Sun: Data curation, Investigation, Methodology. Joann Gu: Data curation, Investigation, Methodology. Abhijeet K. Venkataraman: Data curation, Investigation, Methodology. Nicholas A. Peppas: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. References:
- 1.Zabihi F et al. Intradermal drug delivery by nanogel-peptide conjugates; specific and efficient transport of temoporfin. Journal of Controlled Release 242, 35–41 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Shimoda A, Sawada S. i. & Akiyoshi K Cell specific peptide conjugated polysaccharide nanogels for protein delivery. Macromolecular bioscience 11, 882–888 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Ding L, Jiang Y, Zhang J, Klok H-A & Zhong Z pH-sensitive coiled-coil peptide-cross-linked hyaluronic acid nanogels: synthesis and targeted intracellular protein delivery to CD44 positive cancer cells. Biomacromolecules 19, 555–562 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Li D, van Nostrum CF, Mastrobattista E, Vermonden T & Hennink WE Nanogels for intracellular delivery of biotherapeutics. Journal of Controlled Release 259, 16–28 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Liechty WB, Scheuerle RL, Vela Ramirez JE & Peppas NA Uptake and function of membrane destabilizing cationic nanogels for intracellular drug delivery. Bioengineering & translational medicine 4, 17–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson CE et al. Balancing cationic and hydrophobic content of PEGylated siRNA polyplexes enhances endosome escape, stability, blood circulation time, and bioactivity in vivo. ACS nano 7, 8870–8880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckmann D, Composto R, Tsourkas A & Muzykantov V Nanogel carrier design for targeted drug delivery. Journal of Materials Chemistry B 2, 8085–8097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajebi S et al. Stimulus-responsive polymeric nanogels as smart drug delivery systems. Acta biomaterialia 92, 1–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh JK, Drumright R, Siegwart DJ & Matyjaszewski K The development of microgels/nanogels for drug delivery applications. Progress in Polymer Science 33, 448–477 (2008). [Google Scholar]
- 10.Steichen SD, Caldorera-Moore M & Peppas NA A review of current nanoparticle and targeting moieties for the delivery of cancer therapeutics. European journal of pharmaceutical sciences 48, 416–427 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Zou Y, Zhong Z & Haag R Cyclo (RGD) decorated reduction-responsive nanogels mediate targeted chemotherapy of integrin overexpressing human glioblastoma in vivo. Small 13, 1601997 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Nukolova NV et al. LHRH-targeted nanogels as a delivery system for cisplatin to ovarian cancer. Molecular pharmaceutics 10, 3913–3921 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilhelm S et al. Analysis of nanoparticle delivery to tumours. Nature reviews materials 1, 1–12 (2016). [Google Scholar]
- 14.Liu S et al. Virus Spike and Membrane-Lytic Mimicking Nanoparticles for High Cell Binding and Superior Endosomal Escape. ACS applied materials & interfaces 10, 23630–23637 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Smith SA, Selby LI, Johnston AP & Such GK The endosomal escape of nanoparticles: Toward more efficient cellular delivery. Bioconjugate chemistry 30, 263–272 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM & Andresen TL The possible “proton sponge” effect of polyethylenimine (PEI) does not include change in lysosomal pH. Molecular Therapy 21, 149–157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vermeulen LM, De Smedt SC, Remaut K & Braeckmans K The proton sponge hypothesis: Fable or fact? European Journal of Pharmaceutics and Biopharmaceutics 129, 184–190 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Degors IMS, Wang C, Rehman ZU & Zuhorn IS Carriers Break Barriers in Drug Delivery: Endocytosis and Endosomal Escape of Gene Delivery Vectors. Acc Chem Res 52, 1750–1760, doi: 10.1021/acs.accounts.9b00177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans BC et al. An anionic, endosome-escaping polymer to potentiate intracellular delivery of cationic peptides, biomacromolecules, and nanoparticles. Nat Commun 10, 5012, doi: 10.1038/s41467-019-12906-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J et al. Metal-Phenolic Coatings as a Platform to Trigger Endosomal Escape of Nanoparticles. ACS Nano 13, 11653–11664, doi: 10.1021/acsnano.9b05521 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Wojnilowicz M, Glab A, Bertucci A, Caruso F & Cavalieri F Super-resolution imaging of proton sponge-triggered rupture of endosomes and cytosolic release of small interfering RNA. ACS nano 13, 187–202 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Tai W & Gao X Functional peptides for siRNA delivery. Advanced drug delivery reviews 110, 157–168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo SL & Wang S An endosomolytic Tat peptide produced by incorporation of histidine and cysteine residues as a nonviral vector for DNA transfection. Biomaterials 29, 2408–2414 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Cao J, Huang D & Peppas NA Advanced engineered nanoparticulate platforms to address key biological barriers for delivering chemotherapeutic agents to target sites. Advanced Drug Delivery Reviews, 154 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Zhao Z, Ukidve A, Kim J & Mitragotri S Targeting Strategies for Tissue-Specific Drug Delivery. Cell 181, 151–167 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Patel S, Kim J, Herrera M, Mukherjee A, Kavanov AV, Sahay G Brief update on endocytosis of nanomedicines. Advanced Drug Delivery Reviews 144, 90–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quan C-Y et al. Dual targeting of a thermosensitive nanogel conjugated with transferrin and RGD-containing peptide for effective cell uptake and drug release. Nanotechnology 20, 335101 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Zhou Z, Yan Y, Wang L, Zhang Q & Cheng Y Melanin-like nanoparticles decorated with an autophagy-inducing peptide for efficient targeted photothermal therapy. Biomaterials 203, 63–72, doi: 10.1016/j.biomaterials.2019.02.023 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Chen J et al. Receptor and microenvironment dual-recognizable nanogel for targeted chemotherapy of highly metastatic malignancy. Nano letters 17, 4526–4533 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Chen J, He H, Deng C, Yin L & Zhong Z Saporin-loaded CD44 and EGFR dual-targeted nanogels for potent inhibition of metastatic breast cancer in vivo. International Journal of Pharmaceutics 560, 57–64 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Jiang Z et al. Nanogels of dual inhibitor-modified hyaluronic acid function as a potent inhibitor of amyloid β-protein aggregation and cytotoxicity. Scientific reports 8, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clegg JR et al. Synthetic networks with tunable responsiveness, biodegradation, and molecular recognition for precision medicine applications. Science advances 5, eaax7946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong JX, Clegg JR, Ander EW & Peppas NA Tunable poly (methacrylic acid co acrylamide) nanoparticles through inverse emulsion polymerization. Journal of Biomedical Materials Research Part A 106, 1677–1686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Psarra E et al. In situ monitoring of linear RGD-peptide bioconjugation with nanoscale polymer brushes. ACS omega 2, 946–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Culver HR, Sharma I, Wechsler ME, Anslyn EV & Peppas NA Charged poly (N-isopropylacrylamide) nanogels for use as differential protein receptors in a turbidimetric sensor array. Analyst 142, 3183–3193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller KD et al. Cancer treatment and survivorship statistics, 2016. CA: a cancer journal for clinicians 66, 271–289 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Siegel R, DeSantis C & Jemal A Colorectal cancer statistics, 2014. CA: a cancer journal for clinicians 64, 104–117 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Torre LA et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians 65, 87–108 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Siegel RL et al. Colorectal cancer statistics, 2017. CA: a cancer journal for clinicians 67, 177–193 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Hurwitz H et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. New England journal of medicine 350, 2335–2342 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Longley DB, Harkin DP & Johnston PG 5-fluorouracil: mechanisms of action and clinical strategies. Nature Reviews Cancer 3, 330 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Xu Y & Villalona-Calero M Irinotecan: mechanisms of tumor resistance and novel strategies for modulating its activity. Annals of oncology 13, 1841–1851 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Raymond E, Faivre S, Woynarowski JM & Chaney SG in Seminars in oncology. 4–12. [PubMed] [Google Scholar]
- 44.Li ZJ et al. A novel peptide specifically targeting the vasculature of orthotopic colorectal cancer for imaging detection and drug delivery. Journal of controlled release 148, 292–302 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Smith PK et al. Measurement of protein using bicinchoninic acid. Analytical biochemistry 150, 76–85 (1985). [DOI] [PubMed] [Google Scholar]
- 46.Wiechelman KJ, Braun RD & Fitzpatrick JD Investigation of the bicinchoninic acid protein assay: identification of the groups responsible for color formation. Analytical biochemistry 175, 231–237 (1988). [DOI] [PubMed] [Google Scholar]
- 47.Kongkatigumjorn N et al. Probing endosomal escape using phlexi nanoparticles. Macromolecular bioscience 17, 1600248 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Deshpande S, Patil S & Singh N Enhancing Gene-Knockdown Efficiency of Poly (N-isopropylacrylamide) Nanogels. ACS omega 3, 8042–8049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallach DH & Steck T Fluorescence Techniques in the Microdetermination of Metals in Biological Materials. Utility of 2, 4-Bis-[N, Ń-di-(carboxymethyl) aminomethyl] Fluorescein in the Fluorometric Estimation of Al+ 3, Alkaline Earths, Co+ 2, Cu+ 2, Ni+ 2, and Zn+ 2 in Micromolar Concentrations. Analytical Chemistry 35, 1035–1044 (1963). [Google Scholar]
- 50.Clegg JR et al. Characterization of protein interactions with molecularly imprinted hydrogels that possess engineered affinity for high isoelectric point biomarkers. Journal of Biomedical Materials Research-Part A 105, 1565–1574 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hui Y et al. Nanoparticle elasticity regulates phagocytosis and cancer cell uptake. Science Advances 6, eaaz4316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu W et al. Uptake of hydrogel particles with different stiffness and its influence on HepG2 cell functions. Soft Matter 8, doi: 10.1039/c2sm26001h (2012). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.