Summary
The niche controls stem cell self-renewal and progenitor differentiation for maintaining adult tissue homeostasis in various organisms. However, it remains unclear if the niche is compartmentalized to control stem cell self-renewal and stepwise progeny differentiation. In the Drosophila ovary, inner germarial sheath (IGS) cells form a niche for controlling germline stem cell (GSC) progeny differentiation. In this study, we have identified four IGS subpopulations, which form linearly arranged niche compartments for controlling GSC maintenance and multi-step progeny differentiation. Single-cell analysis of the adult ovary has identified four IGS subpopulations (IGS1-4), which identities and cellular locations have been further confirmed by fluorescent in situ hybridization. IGS1 and IGS2 physically interact with GSCs and mitotic cysts to control GSC maintenance and cyst formation, respectively, whereas IGS3 and IGS4 physically interact with 16-cell cysts to regulate meiosis, oocyte development and cyst morphological change. Finally, one follicle cell progenitor population has also been transcriptionally defined for facilitating future studies on follicle stem cell regulation. Therefore, this study has structurally revealed that the niche is organized into multiple compartments for orchestrating stepwise adult stem cell development, and has also provided useful resources and tools for further functional characterization of the niche in the future.
Graphical Abstract
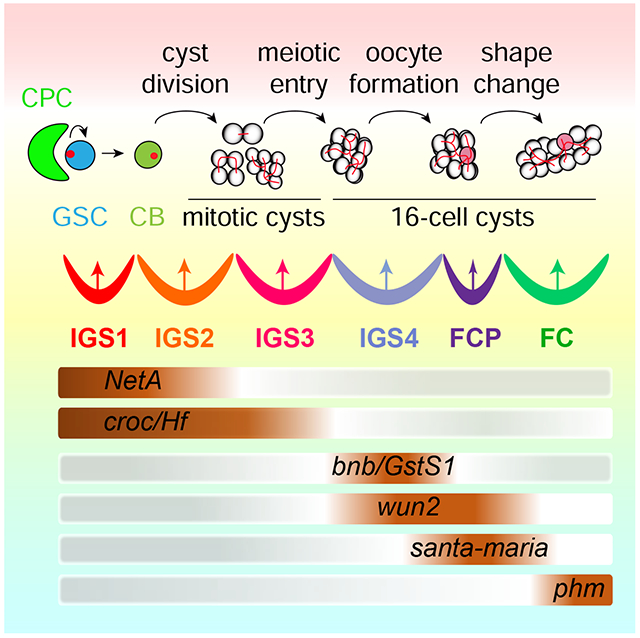
Tu et al. show that the niche forms linearly arranged compartments (IGS1-4) for controlling multi-step GSC development. IGS1 controls GSC self-renewal, whereas IGS2-4 sequentially regulate germline cyst formation, meiosis, timely oocyte determination and cyst shape change. One follicle cell progenitor population is also transcriptionally defined.
Introduction
Stem cells maintain adult tissue homeostasis through continuous self-renewal and generation of differentiated cells. Their self-renewal is shown to be controlled by the niche in the organisms ranging from Drosophila to mammals 1–3. Recently, stem cell progeny differentiation has also been proposed to be regulated by the niche in the Drosophila ovary 4. The differentiation process often consists of multiple developmental steps for generating one or several functional cell types. However, it remains largely unclear how the niche controls these differentiation steps at the cellular level.
The Drosophila ovary is an effective model for studying niche functions in regulating GSC self-renewal and differentiation 5, 6. At the tip of the germarium, two or three GSCs contact cap cells anteriorly and inner germarial sheath cells (IGS; previously known as escort cells) laterally in the region 1 (Figure 1A). Immediate GSC progeny, cystoblasts (CBs), divide four times synchronously with incomplete cytokinesis to form interconnected mitotic cysts (MCs; 2-cell, 4-cell, 8-cell cysts) and 16-cell cysts 7. IGS cells wrap around CBs and MCs in the region 1 as well as 16-cell cysts in the region 2a (Figure 1A). Follicle cells begin to surround 16-cell cysts in the region 2b and then form stage-1 egg chambers in the region 3 (Figure 1A). Cap cells and anterior IGS cells form a niche for controlling GSC self-renewal through Dpp/BMP-mediated signaling and E-cadherin-mediated cell adhesion 8–12, whereas IGS cells form a niche for promoting differentiation partly by preventing BMP signaling 4, 12. IGS cells utilize Hh, Wnt, EGFR and Jak-Stat signaling to prevent BMP signaling in GSC progeny 11, 13–21. Long IGS cellular processes are regulated by Hh and Rho-CDC42 small GTPase signaling, and cellular process-mediated direct interactions are important for CB differentiation and cyst formation 4, 14, 22. Ecdysone signaling prevents IGS transformation into cap cells during development, and is also required in IGS cells for cyst formation, meiosis and egg chamber formation 23–25. Therefore, two niches coordinately control GSC development in the Drosophila ovary.
Figure 1. scRNA-seq reveals five IGS subpopulations in the Drosophila germarium.
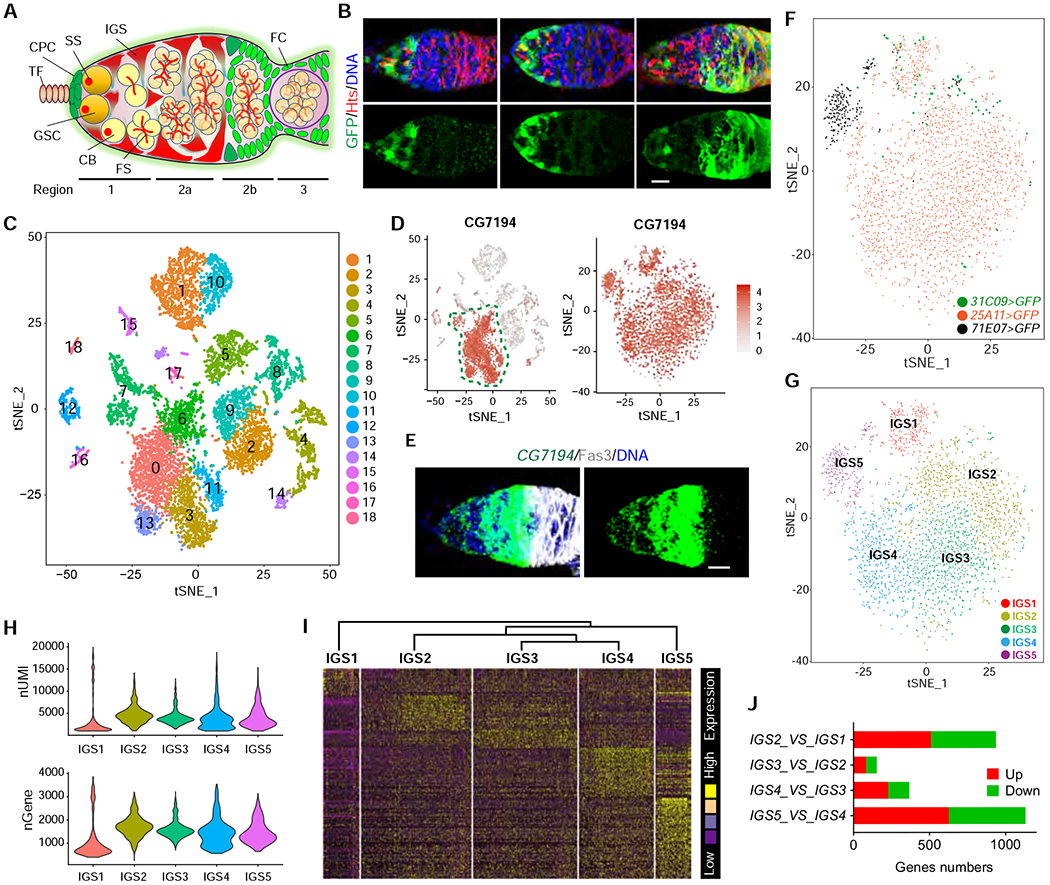
(A) Schematic diagram of the germarium (TF: terminal filament; CPC: cap cell; GSC: germline stem cell, IGS: inner germarial sheath cell, SS: spectrosome; FS: fusome; FC: follicle cell).
(B) Confocal images of UAS-GFP driven by 31C09-Gal4, 25A11-Gal4 and 71E07-Gal4 showing higher GFP expression in anterior, anterior/ middle, and posterior IGS cells, respectively. Scale bars, 10 μm.
(C) Based on unsupervised gene clustering, t-SNE plot shows 18 cell clusters from combined purified GFP-positive single IGS cells from 31C09-Gal4>UAS-GFP, 25A11-Gal4>UAS-GFP and 71E07-Gal4>UAS-GFP ovaries.
(D) t-SNE plots of CG7194 expression patterns in 18 cell clusters and IGS cell clusters. CG7194-expressed IGS cell clusters are identified based on high expression levels and close IGS relationships.
(E) CG7194 is expressed specifically in IGS cells based on FISH. Scale bars, 10 μm.
(F and G) t-SNE plot of single GFP-positive IGS cells. Colors in t-SNE plot images indicate the cells labeled by three Gal4 lines (F) and five cell clusters (G).
(H) nUMI and nGene per IGS cluster.
(I) Hierarchical clustering dendrogram and gene expression heatmap of five IGS clusters.
(J) Gene expression comparisons among IGS1, IGS2, IGS3, IGS4 and IGS5 clusters showing IGS2, IGS3 and IGS4 exhibit fewer up/down-regulated genes than IGS1 and IGS5.
See also Figures S1 and S2.
While interacting with IGS cells, newly formed 16-cell cysts in the region 2a undergo three important cellular events. First, those 16-cell cysts undergo two important meiotic events, chromosomal pairing and meiotic recombination 26, 27. Second, both pro-oocytes form synaptonemal complexes initially and only one of them become the oocyte. Third, 16-cell cysts change their round shape into the lens-like shape for ensuring exactly one cyst to be packaged into an egg chamber by follicle cells. Thus, it is tantalizing to speculate that IGS cells function as a niche for regulating these three important germ cell developmental events. Consistently, ecdysone signaling functions in IGS cells to promote meiotic entry and egg chamber formation possibly by maintaining IGS identity 23. In this study, we use 10xgenomics single-cell RNA-sequencing (scRNA-seq) to identify IGS subpopulations, which form linearly arranged niche compartments for orchestrating GSC self-renewal, CB differentiation, meiotic recombination, timely oocyte development and cyst shape change. Therefore, we propose that the niche forms distinct subcompartments to control GSC maintenance and stepwise progeny differentiation in the Drosophila ovary.
Results
scRNA-seq analysis identifies IGS subpopulations interacting with GSCs and early progeny in the adult ovary
To investigate if distinct IGS subpopulations physically interact with different developmental stages of GSC progeny, we screened the publicly available Gal4 lines by crossing with UAS-GFP to identify three Gal4 lines with distinct expression patterns in IGS subpopulations, 31C09, 25A11 and 71E07 (Figure 1B). To visualize GSCs and their early progeny, we immunostained the ovaries for Hu-li tai shao (Hts) protein, which labels spherical spectrosomes in GSCs and CBs and branched fusomes in mitotic and 16-cell cysts, as well as membrane cytoskeletons in follicle cells 28 (Figure 1B). Three reporters, UAS-GFP, UAS-mGFP (membrane-tethered GFP) and UAS-nLacZ (nuclear LacZ) were used to independently confirm that 31C09, 25A11 and 71E07-expressing IGS cells cover the region 1, regions 1-2a and posterior region 2a, respectively (Figures 1B and S1A). In addition, 71E07 and 31C09 are also expressed in other somatic cells, including follicle cell progenitors (FCPs) and stalk cells (Figure S1A). 31C09, 25A11 and 71E07 express Gal4 under the control of the regulatory sequences of string, wnt4 and dally, respectively 29. These results support the existence of IGS subpopulations in the adult germarium.
To further define IGS sub-populations, we used 31C09-, 25A11- and 71E07-driven GFP expression to purify IGS cells for 10xgenomics scRNA-seq (Figure S1B). The machine learning algorithm, t-distributed stochastic neighbor embedding (tSNE), was used to identify 18 different populations among the 31C09-, 25A11- and 71E07-expressing ovarian somatic cells (Figure 1C). Because of 71E07 and 31C09 expression in other ovarian somatic cells, we have used our previously identified IGS-specific marker CG7194, which is confirmed by mRNA fluorescent in situ hybridization (FISH), to take out CG7194-expressing clusters for further detailed analysis (Figure 1D and 1E). By unsupervised gene clustering, those CG7194-expressing clusters are composed of five subpopulations, IGS1-5, suggesting the existence of IGS subpopulations (Figures 1F–1G). Those purified 31C09-expressing IGS cells in the region 1 are almost exclusively restricted to the IGS1 and IGS2 subpopulations, whereas those purified 25A11-expressing IGS cells in the regions 1 and 2a are distributed to IGS1-4 (Figures 1B and 1G). 71E07-expressing IGS cells in the region 1 and somatic cells in 2b and 3 are allocated into IGS1-2 and IGS5 subpopulations (Figures 1B and 1G). These results suggest that IGS1-2, IGS3-4 and IGS5 are located along the anterior-to-posterior germarial axis.
Our final scRNA-seq dataset has an average of ~5,000 unique molecular identifiers (UMIs) and ~1,500 genes per cell, with each cell type having variable levels of mRNA content and gene expression (Figure 1H). The hierarchical clustering dendrogram and gene expression heatmap also show that IGS2-4 are much closer to one another, while IGS1 and IGS5 are more distant (Figure 1I). By comparing gene expression profiles between different cell populations, IGS2 shows 510 upregulated genes and 427 downregulated genes compared to IGS1, whereas IGS3 exhibits 83 upregulated genes and 70 downregulated genes compared to IGS2; IGS4 shows 229 upregulated genes and 138 downregulated genes compared to IGS3, whereas IGS5 exhibits 627 upregulated genes and 504 downregulated genes compared to IGS4 (Figure 1J). Based on GO term overrepresentation, IGS1 and IGS5 are also more distinct from the IGS2-4 subpopulations, which are closer to one another (Figures S2A–S2E). The top two GO term categories for the differentially enriched genes in IGS2-5 subpopulations are metabolic processes and biological regulation, which are likely important for IGS cells to support dynamic GSC development. Thus, our results suggest that multiple IGS subpopulations exist to perform distinct biological functions in the regulation of GSC development.
mRNA FISH identifies four IGS subpopulations and one follicle cell progenitor (FCP) population
To further confirm the cellular locations of IGS subpopulations, we performed mRNA FISH experiments for the IGS subpopulation-enriched genes in adult ovaries. phantom (phm) mRNA, which encodes a cytochrome P450 involved in ecdysone biosynthesis 30, highlights follicle cells based on its co-expression with known follicle cell marker Fas3 and its complete absence from CG7194-expressing IGS cells 31, 32 (Figures S3A–S3D). Netrin-A (NetA), which encodes a secreted Netrin protein 33, is expressed in IGS1 and IGS2 interacting with GSCs and CBs/MCs in the region 1 (Figure 2A). Based on our observation that IGS1 is more distant from IGS2-5 in gene expression profiles (Figure 1I), the GSC-contacting IGS subpopulation should be IGS1 because IGS2-5 interact with early GSC progeny. Logically, IGS2 should interact with CBs and MCs. Based on its overlapped expression with NetA, crocodile (croc) mRNA, which encodes a forkhead transcription factor required in IGS cells for promoting CB differentiation 34, must be expressed in IGS1 and IGS2. In addition, croc should also be expressed in IGS3, which posteriorly contact bangles and beads (bnb)- and Glutathione S transferase S1 (GstS1)-expressing IGS cells (Figures 2B and 2C). Bnb is a protein with unknown functions, whereas GstS1 is a glutathione transferase essential for Drosophila development 35, 36. Supporting the idea, there is a regional gap between NetA-expressing and bnb-expressing IGS cells, which should be covered by IGS3 (Figure 2A). GstS1- and bnb-expressing IGS cells anteriorly contact the croc-expressing IGS cells, suggesting that GstS1 and bnb are expressed in IGS4 (Figures 2B–2E). santa-maria, which encodes a scavenger receptor important for vitamin A synthesis 37, is expressed in IGS5 since its expressing region is anteriorly adjacent to GstS1- and bnb-expressing IGS4 (Figures 2B–2E). wunen2 (wun2), which encodes a lipid phosphatase required for germ cell migration 38, is expressed in the IGS cells that posteriorly contact phm-expressing cells and anteriorly overlap with bnb-expressing cells, indicating that wun2 is expressed in IGS4 and IGS5 (Figure 2F). Helical factor (Hf), which encodes a secreted immune-regulated cytokine 39, shares a similar expression pattern in IGS1-3 with croc (Figure 2G). Therefore, the five IGS subpopulations are successfully mapped onto different regions of the germarium with overlapped marker gene expression in adjacent IGS subpopulations (Figures 2H and 2I). It is worth noting that scRNA-seq and mRNA FISH results on NetA, croc and wun2 are very consistent, but the results on bnb and GstS1 do not quite match, suggesting that scRNA-seq results require validation by RNA FISH (Figure S4).
Figure 2. Mapping IGS subpopulation-enriched genes onto the germarium by FISH.
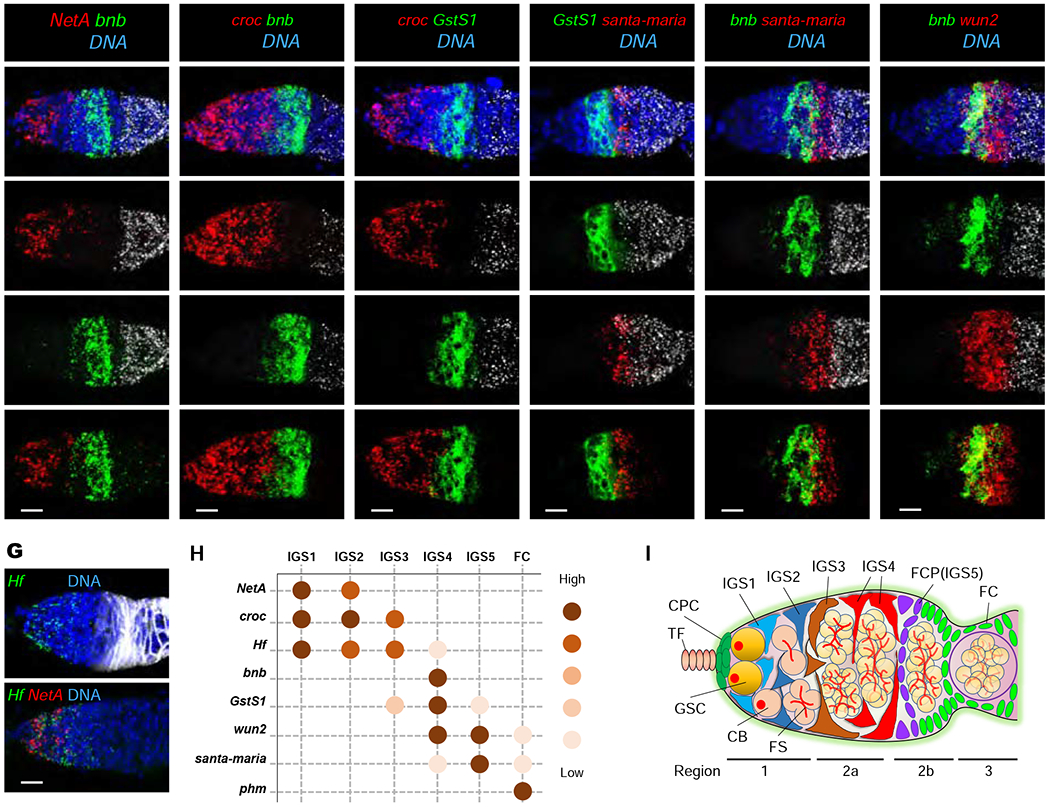
(A-G) Multi-channel mRNA FISH confocal images of NetA/bnb/phm (A), croc/bnb/phm (B), croc/GstS1/phm (C), GstS1/santa-maria/phm (D), bnb/santa-maria/phm (E), bnb/wun2/phm (F) and Hf/Fas3, Hf/NetA (G). DNA is labeled by DAPI (Blue). Scale bars, 10 μm.
(H) Summary of NetA, croc, Hf, bnb, GstS1, wun2, santa-maria and phm expression patterns in germarium.
(I) Diagram showing the arrangement of IGS1-4 and FCP (IGS5) in the Drosophila germarium.
See also Figures S3, S4 and S5.
Previous studies have identified a follicle cell progenitor (FCP) population located between IGS cells and Fas3-expressing follicle cells 31, 32, 40. Based on its germarial location, the santa-maria-positive IGS5 could be the FCP population (Figures 2D, 2E and S5A). Since PZ1444 is a widely used IGS marker 41, the most posterior PZ1444-positive IGS cells also express IGS4 marker bnb, indicating that IGS4 is the most posterior IGS subpopulation (Figure S5B). As predicted, the highly santa-maria-expressing IGS5 anteriorly contacts the most posterior PZ1444-positive IGS cells anteriorly and Fas3-positive follicle cells posteriorly (Figure S5B–S5D). Based on these results, IGS5 is indeed the same as the FCP population, which specifically expresses santa-maria (Figures 2D and 2E). Therefore, this study has identified four IGS subpopulations and the FCP (Figures 2H and 2I).
Four IGS subpopulations dynamically express dpr17
Based on the scRNA-seq result, dpr17 is expressed in IGS4 at higher levels than other IGS subpopulations (Figure 3A). Surprisingly, mRNA FISH results show that dpr17 exhibits four different expression patterns in IGS subpopulations in addition to terminal filament cells: 1) no expression in any IGS cells; 2) IGS1 and IGS2 cells; 3) IGS3 and IGS4 cells; 4) IGS1-4 cells (Figure 3B). Then, we used Cas9/CRISPR to knock the yeast Gal4 gene into dpr17 to generate dpr17-Gal4 for confirming its dynamic IGS expression patterns. By using the UAS-nLacZ reporter, we show that dpr17-Gal4 exhibits similar dynamic IGS expression patterns to its mRNA, and the percentages of the germaria with the same IGS expression pattern for dpr17-Gal4 and dpr17 mRNA are comparable (Figures 3C and 3D). dpr17 encodes an immunoglobin-domain containing protein involved in synapse recognition and specificity determination by engaging with another Ig-domain containing protein DIP-ε or DIP-γ 42. These results suggest that IGS subpopulations are molecularly dynamic in nature. However, it remains unclear about the biological significance, prevalence and regulation of this dynamic gene expression.
Figure 3. dpr17 is dynamically expressed in different IGS subpopulations.
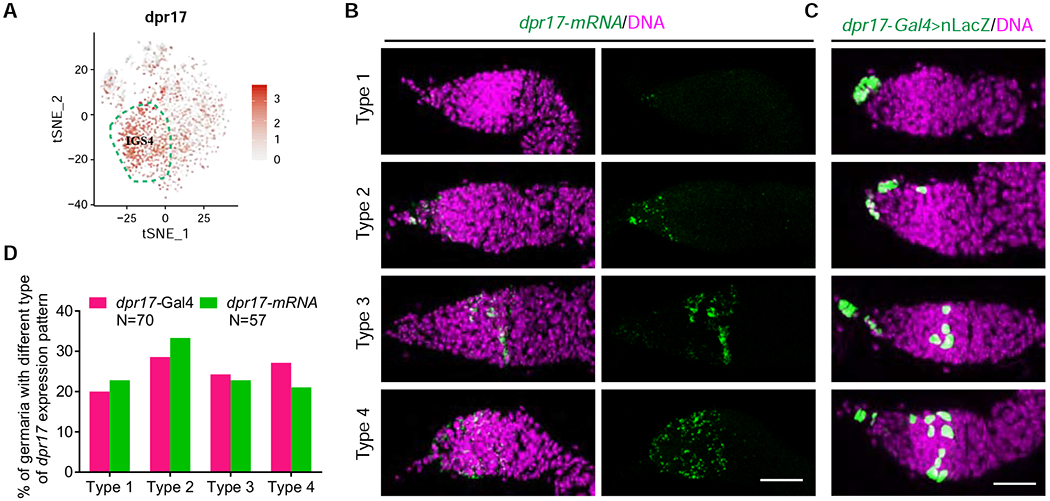
(A) t-SNE plot showing dpr17 is enriched in the IGS4 (highlighted by broken lines) and is also expressed in other IGS subpopulations at low levels.
(B-C) dpr17 mRNA FISH (B) images and dpr17-Gal4>UAS-nLacZ expression patterns (C) showing variable expression patterns of dpr17 in Drosophila germaria. Scale bars, 20 μm.
(D) Quantification results of dpr17-Gal4 and dpr17 mRNA expression patterns.
scRNA-seq helps define new IGS-specific gene markers
Based on scRNA-seq results, biniou (bin), vein (vn), Neurotactin (Nrt), mirror (mirr), CG42458 and dunce (dnc) exhibit similar expression patterns in the IGS and the FCP as CG7194 (Figure 4A). To visualize follicle cells, we performed mRNA FISH in combination with Fas3 immunostaining to confirm that bin, vn, Nrt, mirr and CG42458 are expressed in IGS1-4 and FCP (Figure 4B). In addition, they are also lowly expressed in the Fas3-positive follicle cells (Figure 4B). Further, we used the publicly available GFP protein tagged lines, mirr-GFP and dnc-GFP, to confirm that mirr and dnc are highly enriched in IGS1-4 and FCP based on GFP immunostaining (Figure 4C). bin encodes a forkhead transcription factor important for visceral muscle development, whereas CG42458 encodes a RNA-binding protein with unknown function (Zaffran et al., 2001). Interestingly, vn, Nrt, mirr and dnc encode neuregulin-like EGFR ligand, transmembrane adhesion protein, homeobox transcription factor and cAMP-specific phosphodiesterase, respectively, which are known to regulate neuronal development and functions 43–47. These observations suggest that IGS cells use various pathways to control their interaction with GSCs and their progeny.
Figure 4. FISH validation of new IGS-enriched genes identified by scRNA-seq.
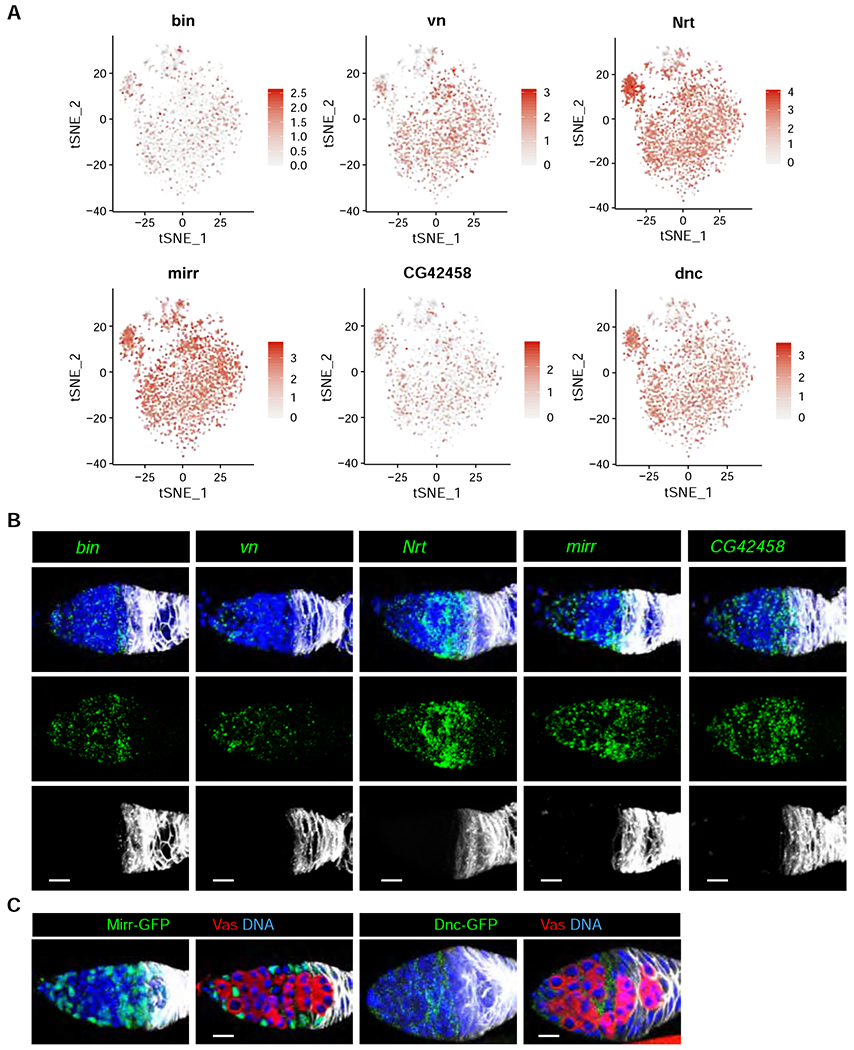
(A) t-SNE plots of bin, vn, Nrt, mirr, CG42458 and dnc showing their expression in all IGS cells.
(B) mRNA FISH (Green channel) and immunostaining (Grey channel, anti-Fas3) confocal images of bin/Fas3, vn/Fas3, Nrt/Fas3, mirr/Fas3 and CG42458/Fas3 showing their expression in IGS1-4 and FCP. Scale bars, 10 μm.
(B) Confocal images of mirr-GFP and dnc-GFP germaria labeled for GFP, Fas3, Vas and DAPI (DNA) showing Mirr-GFP and Dnc-GFP expression in IGS cells. Scale bars, 10 μm.
IGS-enriched genes, bin and vn, are required extrinsically for controlling CB differentiation
Then, we used c587-Gal4;tub-Gal80ts (c587ts) to knock down the IGS-enriched genes, bin and vn, to delineate their functions in adult IGS cells for regulating GSC progeny differentiation. c587-Gal4 is highly expressed in all the IGS cells 48. tub-Gal8ts ubiquitously expresses a temperature-sensitive mutant GAL80 protein in all cells, including IGS cells; the mutant GAL80 protein is functional at 18 °C to repress c587-driven UAS-shRNA expression in IGS cells, but 29 °C inactivates its function to allow shRNA expression in IGS cells 49. When the newly eclosed adult c587ts; UAS-shRNA females at 18 °C are shifted to 29 °C, the expression of a specific shRNA in adult IGS cells can be achieved as we have done previously 34, 50. PZ1444 is expressed in both IGS cells and cap cells, which morphology and location easily distinguish each other 41. Spectrosome-containing GSCs and CBs can be distinguished from each other based on the fact that cap cells physically contact GSCs, but not CBs 28. IGS-specific bin knockdown (binKD) for 7 days does not affect GSC maintenance since the binKD germaria and the luciferase knockdown (lucKD) control germaria (firefly luc, a non-Drosophila gene, is used as the control for non-specific RNAi effect) have normal 2 or 3 GSCs (Figures 5A, 5B, S6A and S6B). However, those binKD germaria accumulate excess CB-like spectrosome-containing single germ cells (SGCs) compared to the control germaria with an average of one CB, indicating that bin functions in IGS cells to promote CB differentiation (Figures 5A and 5B). Since our previous studies have shown that a severe IGS cell loss also causes the germ cell differentiation defect 4, 34, 50, we then examined and quantified PZ1444-positive IGS cells in the control and binKD germaria. In contrast with the control germaria carrying 30–35 PZ1444-positive IGS cells, most of the binKD germaria completely lose their IGS cells (Figures 5A and 5B). These results demonstrate that homeobox transcription factor Bin promotes IGS maintenance and consequently GSC progeny differentiation.
Figure 5. IGS-enriched bin and vn are required for GSC progeny differentiation.
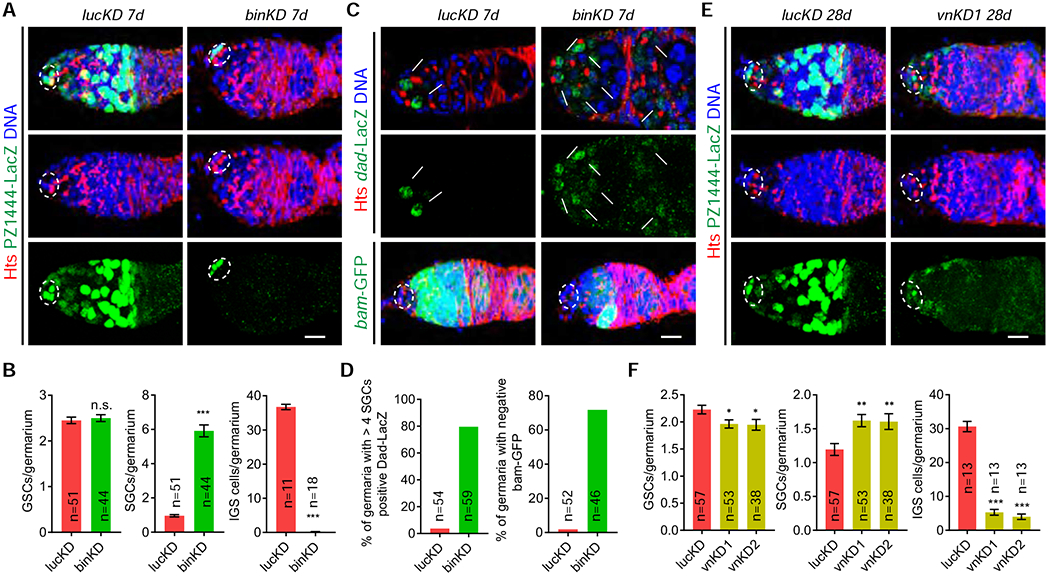
The germaria (A and E) are labeled for PZ1444-LacZ (IGS cells and cap cells) and DAPI (nuclei). Cap cells and GSCs are highlighted by broken ovals.
(A-B) Merged confocal images showing that c587ts-driven bin knockdown causes the accumulation of significantly more spectrosome-containing SGCs (one in control and six in binKD indicated by arrowheads) and the loss of IGS cells (B: quantification results; n = No. of germaria). Scale bars, 10 μm. (Student’s t-test: ***, P ≤ 0.001; n.s., no significance).
(C-D) c587ts-driven binKD germaria accumulate more Dad-LacZ-positive (single section confocal images) and bam-GFP-negative SGCs (some by arrows) (merged confocal images) compared to the control with Dad-lacZ-positive and bam-GFP-negative GSCs (two arrows, C). (D: quantification results; n = No. of germaria). Scale bars, 10 μm.
(E-F) c587ts-driven vn knockdown causes the accumulation of slightly increased SGCs (one in control and two in vnKD indicated by arrowheads) and a significant loss of IGS cells. (F: quantification results; n = No. of germaria). (Student’s t-test: ***, P ≤ 0.001; **, P ≤ 0.01, *, P ≤ 0.05). Scale bars, 10 μm.
See also Figure S6.
Since the IGS loss disrupts GSC progeny differentiation by elevating BMP signaling 4, 12, 51, we then used two BMP signaling reporters, Dad-lacZ and bam-GFP, to verify BMP signaling in the control and binKD germaria. In the control germaria, Dad-lacZ is expressed in GSCs, whereas bam-GFP expression is repressed in GSCs and upregulated in differentiated GSC progeny (Figures 5C and 5D). By contrast, the binKD germaria show Dad-lacZ upregulation and bam-GFP repression in the accumulated SGCs located a few cells away from cap cells (Figures 5C and 5D). These results further confirm that IGS cells are required for preventing BMP signaling in GSC progeny.
EGFR signaling is required in IGS cells for their maintenance and for promoting GSC progeny differentiation 50, whereas Vn is a neuregulin-like ligand activating EGFR in Drosophila 43. Consistently, 28 days after knockdown in IGS cells, the vn knockdown (vnKD) germaria gradually lose most of IGS cells, still leaving a few IGS cells in the anterior germarial region (Figures 5E, 5F, S6C and S6D). The vnKD germaria show slightly reduced GSCs, increased SGCs and excess 16-cells cyst (Figures 5E and 5F). Since EGFR signaling has previously been shown to be important for FSC regulation 52, the accumulated 16-cell cysts are likely caused by facultative follicle cell development due to the loss of IGS-expressed Vn in the regulation of FSCs and FCPs. Since EGFR signaling in IGS cells is known to be activated by ligands in underneath germ cells 53, this study suggests that IGS-expressed Vn can also contributes to EGFR signaling for promoting IGS maintenance and GSC progeny differentiation.
Anterior IGS-expressed genes, NetA and Hf, are required extrinsically for maintaining GSCs
To further validate NetA protein expression in the germarium, we analyzed the Drosophila strain carrying a recombination-mediated cassette exchange of a Mi[MIC] insertion, which results in the expression of NetA protein tagged with EGFP-FlAsH-StrepII-TEV-3xFlag. Consistently, NetA-GFP is expressed in IGS1 and IGS2, but at lower levels in IGS2 (Figure 6A). NetA-GFP is absent in germ cells, which are marked by Vasa expression (Figure 6A). c587ts-driven expression of two UAS-shRNA lines against NetA can knock down NetA mRNA expression effectively and specifically in adult IGS cells (Figures S6E and S6F). Compared to the control germaria, the NetAKD germaria carry significantly fewer GSCs and consequently fewer SGCs since SGCs are produced by GSCs (Figures 6B and 6C). In addition, the NetAKD germaria have the normal number of IGS cells compared to the control, indicating that NetA is dispensable for IGS maintenance (Figures 6D and 6E). These results indicate that NetA is required in IGS1 and possibly IGS2 for maintaining GSCs. Since NetA is a secreted signaling molecule, it remains to be determined if NetA maintains GSCs by direct signaling in the future.
Figure 6. Anterior IGS-enriched NetA and Hf are required for maintaining GSCs.
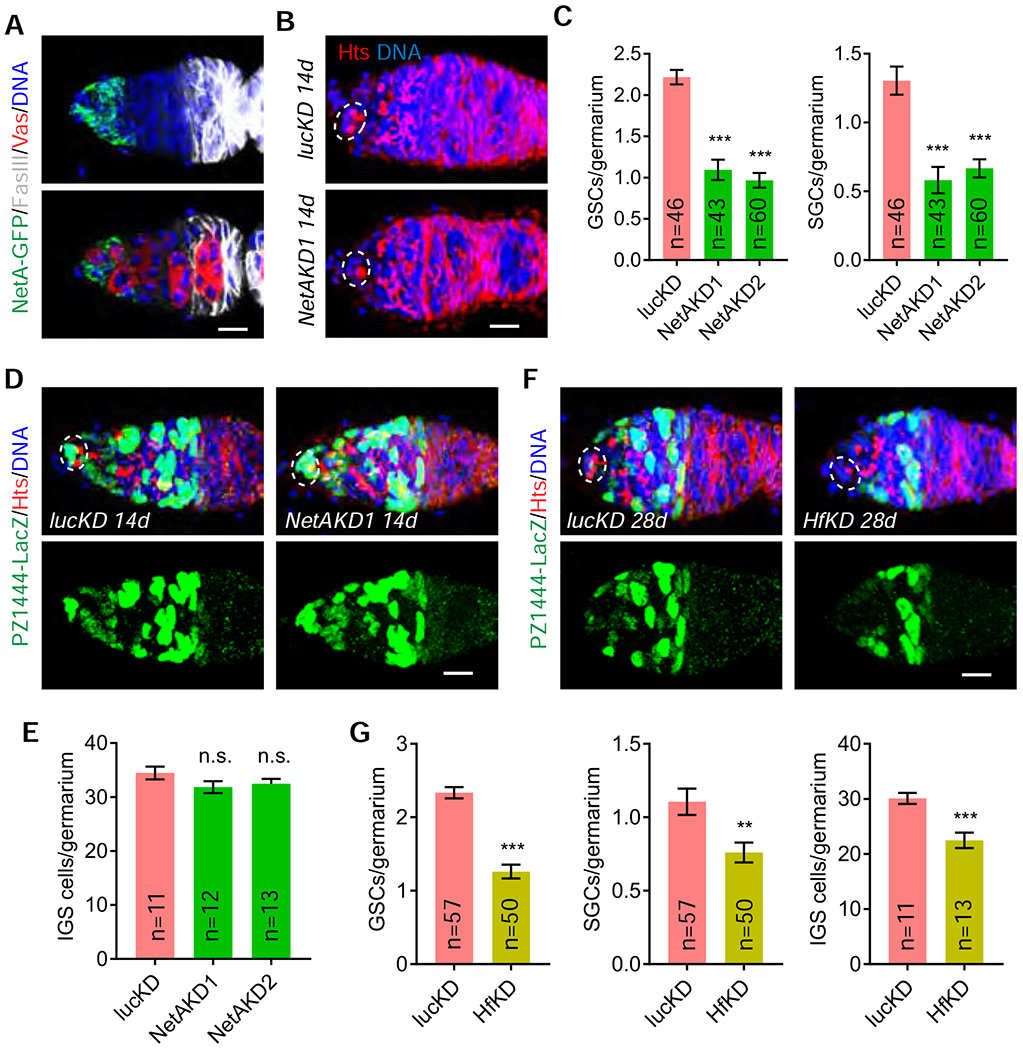
Cap cells and GSCs are highlighted by broken ovals.
(A) Confocal images showing high expression of NetA-GFP in IGS1 and its weak expression in IGS2. Scale bars, 10 μm.
(B-C) c587ts-driven NetA knockdown decreases GSCs and SGCs (C: quantification results; n = No. of germaria). Scale bars, 10 μm. (Student’s t-test: ***, P ≤ 0.001).
(D-E) c587ts-driven NetA knockdown does not significantly affect IGS cells (E: quantification results; n = No. of germaria). Scale bars, 10 μm. (Student’s t-test: n.s., no significance)
(F-G) c587ts-driven Hf knockdown decreases GSCs, SGCs and IGS cells (E: quantification results; n = No. of germaria). Scale bars, 10 μm. (Student’s t-test: ***, P ≤ 0.001; **, P ≤ 0.01). Scale bars, 10 μm.
See also Figure S6.
Although Hf functions like a cytokine to induce anti-bacteria immune response in S2 cells, the in vivo biological function remains unclear 39. c587ts-driven expression of a UAS-Hf shRNA can knock down Hf expression effectively in adult IGS cells (Figures S6G and S6H). Compared to the control germaria, the HfKD germaria show significantly fewer GSCs and SGCs (Figures 6F and 6G). Interestingly, IGS cells are also significantly reduced in the knockdown germaria, indicating that Hf also maintains IGS cells. Along with the NetA findings, these results have further supported the idea that IGS1 is also a part of the niche for maintaining GSCs.
Posterior IGS-enriched genes, wun2 and GstS1, are required for meiotic recombination, timely oocyte specification and cyst shape change
Early16-cell cysts in the region 2a produce double-stranded DNA breaks (DSBs) for meiotic recombination, which can be identified by H2AvD, a phosphorylated form of histone variant H2A; these DSBs are successfully repaired in 16-cell cysts in the regions 2b and 3 54 (Figure 7A). Then, we used c587ts-driven expression of two independent UAS-shRNAs against wun2 and GstS1 to knock down their expression in IGS4 cells efficiently to investigate if they regulate early meiotic entry (Figures S6I–S6L). Interestingly, wun2 or GstS1 knockdown in IGS cells decreases H2AvD expression in early 16-cell cysts compared to the control, indicating that Wun2 and GstS1 are required in IGS4 cells for meiotic recombination in 16-cell cysts (Figures 7A and 7B). In addition, IGS-specific wun2 or GstS1 knockdown also results in the accumulation of round-shaped 16-cell cysts moving pairwisely to the region 2b, where normally only one lens-shaped 16-cell cyst waits to be covered by follicle cells, suggesting that Wun2 and GstS1 are required in IGS4 cells for promoting cyst shape changes (Figures 7C and 7D). However, because of wun2 expression in FCPs, we could not rule out the possibility that Wun2 also regulates FCPs to change cyst shape. These results suggest that Wun2 and GstS1 are required in IGS4 and possibly IGS3 (due to their low expression in IGS3) for regulating meiosis and cyst shape changes.
Figure 7. Posterior IGS3 and IGS4 are required for meiotic recombination, oocyte specification and cyst shape change.
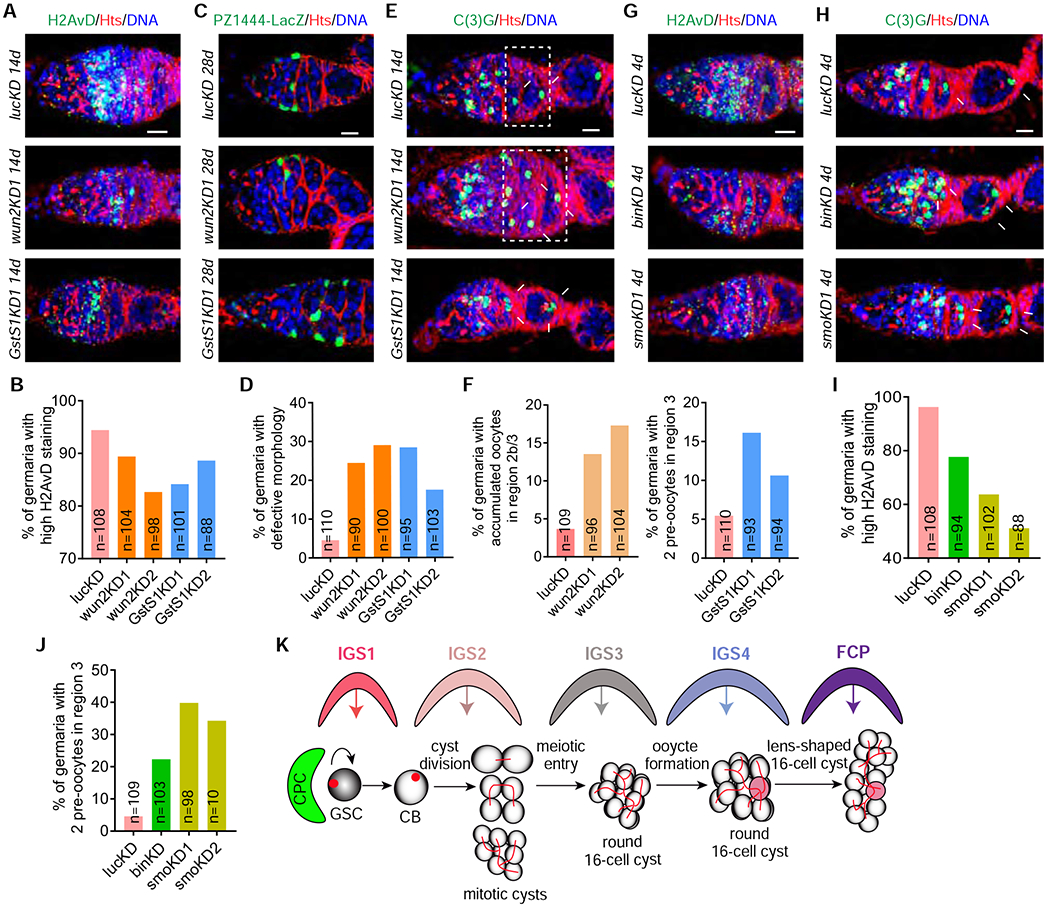
(A-B) Adult IGS-specific c587ts-driven wun2 or GstS1 knockdown decreases the H2AvD expression in meiotic 16-cell cysts compared to the control, in which meiotic 16-cell cysts in the region 2a express high H2AvD (B: quantification results; n = No. of germaria). Scale bars, 10 μm.
(C-D) Single section confocal images showing adult IGS-specific c587ts-driven wun2 or GstS1 knockdown causes the accumulation of cysts with abnormal shapes in the regions 2b and region 3 (highlighted by asterisk, D: quantification results; n = No. of germaria). Scale bars, 10 μm.
(E-F) In the regions 2b and 3, control germarium contains 16-cell cysts with one C(3)G-positive oocyte, c587ts-driven wun2KD germarium accumulates more 16-cell cysts with one C(3)G-positive oocyte (highlighted by broken rectangle), but c587ts-driven GstS1KD germarium carries 16-cell cysts with two C(3)G pro-oocytes (pointed by arrows) (F: quantification results; n = No. of germaria). Scale bars, 10 μm.
(G-J) Adult stage H2126-specific bin and smo knockdown germaria contain more meiotic 16-cell cysts expressing low H2AvD in the region 2a (G) and more 16-cell cysts with two C(3)G-positive pro-oocytes in the region 3 (pointed by arrows) (H). (I and J: quantification results; n = No. of germaria). Scale bars, 10 μm.
(K) A working model explaining how IGS subpopulations regulate stepwise GSC progeny differentiation through direct signaling, which needs further experiments for validation.
See also Figures S6 and S7.
Normally, 16-cell cysts in the region 2a contain two pro-oocytes, but only one oocyte is retained in stage-1 egg chambers in the region 3. Both pro-oocytes and oocytes can be reliably labeled by an antibody against the synaptonemal complex protein C(3)G 55, 56. To test if IGS cells influence oocyte formation, we examined the number of C(3)G-positive oocytes in the control and wun2KD and GstS1KD germaria. The wun2KD germaria still carry the 16-cell cysts with one C(3)G-positive oocyte in the regions 2b and 3 as the control ones (Figures 7E and 7F). However, the 16-cell cysts in the regions 2b and 3 of the GstS1KD germaria frequently contain two C(3)G-positive pro-oocytes (Figures 7E and 7F). Therefore, our findings suggest that GstS1 is required in the IGS4 for timely oocyte formation (Figures 7E and 7F). It will be of great interest to investigate how IGS cells influence the timely elimination of the extra pro-oocyte in the future.
Bin and Smoothened (Smo) are required for posterior IGS4 for meiosis and oocyte specification
A recent study has identified H2126-switchGal4 as a posterior IGS-specific Gal4 57. switchGal4 is an inducible Gal4 system, in which the yeast Gal4 gene is fused with human progesterone receptor (GR) to produce a Gal4-GR fusion protein capable of binding to the UAS promoter upon progesterone analog RU486 administration 58. We first used UAS-nLacZ to confirm that H2126-switchGal4 is expressed in posterior IGS cells and at lower levels in FCPs upon RU486 administration (Figure S7A). Then, we applied mRNA FISH in combination with Fas3 antibody immunostaining to show that H2126-switchGal4 is expressed in IGS4 cells and also a few IGS3 cells (Figures S7B and S7C). Bin is described earlier shown to, while Smo, a Hh receptor, is previously shown to, maintain IGS cells and promote CB differentiation 14, 34 To further define the function of IGS3 and IGS4 in the regulation of meiosis and oocyte formation, we used H2126-switchGal4 and previously validated UAS-shRNA lines 14, 34 to knock down bin and smo in adult IGS3 and IGS4 cells for 4 days, which did not cause severe IGS loss based on the morphologies of the knockdown germaria (Figures 7G–7J). Consistently, knocking down bin or smo in adult IGS3 and IGS4 also decreases H2AvD expression in early 16-cell cysts in the region 2a, and increases the presence of two oocytes in the 16-cell cysts of stage-1 egg chambers (Figures 7G–7J). Therefore, these results further confirm that IGS3 and IGS4 control meiotic recombination and timely oocyte determination in 16-cell cysts.
Discussion
Although IGS cells and their cellular processes form a niche for controlling GSC progeny differentiation, including CB differentiation, cyst formation and the meiotic entry 4, 12, 23, it remains unclear if IGS cells form distinct niche compartments that control different differentiation steps. In this study, we used scRNA-seq to identify four IGS subpopulations, IGS1-4, which are organized linearly along the germarium to interact with GSCs and their early progeny. We have further used genetic manipulations to show that IGS1-4 subpopulations form functionally separate compartments for controlling GSC maintenance, CB differentiation, meiotic recombination, timely oocyte specification and cyst shape change (Figure 7K). In addition, we have molecularly defined the FCP population, which remain poorly studied due to the lack of suitable molecular markers. Therefore, this study has identified four IGS subpopulations that form subsequential niche compartments for controlling different steps of GSC progeny differentiation and has also molecularly defined the poorly studied FCP population, which opens the door for in-depth studies of IGS and FCP populations in the future.
Four IGS subpopulations form linearly arranged niche compartments
Two recent scRNA-seq studies on the whole Drosophila ovary have identified different somatic cell types important for oogenesis 59, 60. However, those two studies failed to identify the IGS subpopulations due to high complexities of somatic cell types and similarities of IGS cells. In this study, we have used GFP-based cell sorting and scRNA-seq to successfully identify four IGS subpopulations, IGS1-4, and have further used gene-specific Gal4 lines and mRNA FISH to show their linear arrangement in the anterior germarium. IGS1 and IGS2 reside in the region 1 to interact with GSCs and CBs/MCs, while IGS3 and IGS4 are located in the region 2a to contact 16-cell cysts. IGS4 is the most posterior population directly contacting FSCs and FCPs, and should be the niche for FSCs based on the previous studies 31, 40, 61. Unfortunately, we have not identified unique markers for IGS1-3 subpopulations except IGS4, which specifically expresses bnb and GstS1. However, the combinatory gene expression patterns can still reliably separate IGS1-3 subpopulations. For example, NetA is specifically expressed in IGS1 and IGS2, while croc and Hf are expressed in IGS1-3. In the future, we will use the split-Gal4 strategy to generate IGS subpopulation-specific Gal4 lines for further defining their molecular signatures and functions 62.
Previous studies have identified three general IGS markers, c587-Gal4, 13C06-Gal4 and PZ1444 41, 48, 61. This study has identified bin, vn, Nrt, mirr, dnc, CG7194 and CG42458 as new molecular markers for IGS and FCP cells. In addition, our RNAi knockdown results have demonstrated that bin and vn maintain IGS cells and promote GSC progeny differentiation. In addition to germ cell-mediated EGFR activation in IGS cells 53, 63, this study has shown that IGS-expressing neuregulin-like EGFR Vn also contributes to EGFR signaling and thus IGS maintenance. Since Wnt, Hh and EGFR signaling are known to maintain IGS cells 14, 17, 34, it will be important to determine if forkhead transcription factor Bin functions downstream of these pathways to maintain IGS cells. Transmembrane adhesion molecule Dpr17 is dynamically expressed in different IGS sub-populations in different germaria, suggesting that IGS cells could change their adhesive property dynamically. This could be a potentially exciting finding since CBs, MCs and 16-cell cysts have to move along the germarium by disengaging one IGS subpopulation and then engaging a new IGS subpopulation. This speculation needs future experimental confirmation. Therefore, this study has provided important insight into IGS subpopulations by uncovering new markers and functions, and has also improved our ability to probe new IGS functions in regulating GSC development in the future.
Different IGS subpopulations have distinct functions in the regulation of GSC lineage development
Although recent studies have shown that IGS cells maintain GSCs and promote CB differentiation 4, 12, 14, 16, 17, 19–22, 34, 50, none of these studies have attributed the functions to any specific IGS subpopulations. This study shows that secreted signaling molecules, NetA and Hf, are expressed in IGS1-2 to maintain GSCs. However, we could not definitively demonstrate that only IGS1-expressing NetA and Hf contribute to GSC maintenance due to the lack of IGS1-specific Gal4 lines. Although we show that IGS cells are required for CB differentiation and IGS2 directly contacts CBs/MCs, we could not directly demonstrate that IGS2 directly controls the differentiation of CBs and MCs into 16-cell cysts due to the lack of IGS2-specific Gal4 lines.
This study also shows that IGS3 and IGS4 control meiosis, oocyte specification and cyst shape. IGS3 and IGS4 in the region 2a extend their long cellular processes to wrap around newly formed H2AvD-positive pre-meiotic and meiotic 16-cell cysts. These 16-cell cysts still have two pro-oocytes, but they only retain one oocyte and also undergo the round-to-lens shape change when surrounded by follicle cells in regions 2b. Knocking down IGS4-expressing wun2 and GstS1 decreases the H2AvD-positive 16-cell cysts in the region 2a and increases the presence of round16-cell cysts in the regions 2b, which fail to become lens-shaped, suggesting that IGS4 cells regulate meiotic recombination and cyst shape change. In addition, IGS-specific knockdown of GstS1, but not wun2, causes the presence of 2 pro-oocytes in stage 1 egg chambers. Consistently, H2126-meidated bin and smo knockdown in IGS4 can also decrease the H2AvD-positive 16-cell cysts and increase the frequency of stage 1 egg chambers with 2 pro-oocytes, further supporting that IGS4 regulates meiosis and oocyte specification. However, we could not completely rule out the possibility that IGS3 cells might also regulate meiotic recombination and timely oocyte specification due to the lack of IGS3-specific Gal4 lines since IGS3 expresses low levels of wun2 and GstS1 and some of them also express H2126. In addition to one previous study showing that Ecdysone signaling in IGS cells regulates meiotic entry, this study has, for the first time, shown that IGS cells regulate meiotic recombination in 16-cell cysts. Since the oocyte was previously thought to be determined entirely by the intrinsic mechanism, the differential RNA and protein transport caused by asymmetrically localized microtubules and fusomes 64–68, this is also the first time to show that IGS cells influence timely oocyte determination in Drosophila. Therefore, this study has provided important insight into IGS subpopulations in the regulation of GSC maintenance, cyst formation, meiotic recombination, timely oocyte determination and cyst shape change, but the underlying signaling mechanisms await future investigation through generation of new genetic tools (Fig. 7K).
STAR ★ Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ting Xie (tgx@stowers.org).
Materials Availability
New reagents generated in this study are available via the lead contact.
Data Availability
Original scRNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE143817). Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1575.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila culture
Flies were maintained and crossed at room temperature on standard cornmeal/molasses/agar media unless specified. For maximizing the effect of RNAi-mediated knockdown or gene overexpression, newly eclosed flies were shifted to 29°C for the specified time before analyzing ovarian phenotypes. For the GeneSwitch Gal4-mediated knockdown, RU486 administration was performed at adult stage according to the previous publication 57.
METHOD DETAILS
Generation of Gal4 lines using Cas9/CRISPR
dpr17-Gal4 were generated according to previous publication 69. Briefly, homology arms of approximately 800-1000 bp were amplified by PCR from the Drosophila genomic DNA. The sgRNA sequence for dpr17 is gaaattatgctgatctgtgccgg. ggtttatcccgttaaggaagc and gatcagcataatttcatttgcat primers were used to amplify the left arm, while acaagcgaaggcagaatcag and caggtgaactttggcactca primers were used to amplify the right arm.
Immunostaining and confocal imaging
Immunostaining was performed according to our previous published procedures (Song et al.,2002; Xie and Spradling, 1998). The following antibodies were used in this study: mouse monoclonal anti-Hts antibody (1:50, 1B1, DSHB), rabbit polyclonal anti-β-galactosidase antibody (1:500, #08559761, MP Biomedical), rabbit polyclonal anti-pS137 H2AvD antibody (1:2000, #600-401-914S, Rockland), chicken polyclonal anti-GFP antibody (1:500, Invitrogen, #A10262), mouse monoclonal anti-C3G (1:500, gift from Dr. Hawley, Stowers Institute), rabbit polyclonal anti-C3G (1:10000, gift from Dr. Lilly, NICHD/DIR).
Fluorescence-activating cell sorting (FACS) of GFP-positive IGS cells
31C09-Gal4, 25A11-Gal4, 71E07-Gal4 were used to drive UAS-GFP expression in different IGS cell populations. After being cultured for 1 week at 25 °C, Drosophila ovaries were dissected and placed in the Grace’s medium (Sigma-Aldrich; G9771), washed twice by 1×PBS and centrifuged at 700×g for 1 minute. The ovaries were incubated with a prewarmed Collagenase solution (50D11833; Worthington) in a 15 ml conical tube at a 37 °C water bath for 3 minutes with gentle shaking. Enzyme reaction was stopped after 3 minutes of incubation following the addition of the cold 1×DPBS+2% FBS. Dissociated samples were washed by 1×DPBS and then centrifuged at 700×g and 4 °C for 5 minutes. The cell pellet was resuspended in 1×DPBS and filtered with the 70 μm Filcon (BD; 340605). Cells were centrifuged and then resuspended in 200 μl of 1×DPBS for sorting the GFP-positive cells at 45 psi with 70 μm tip (BD; InFlux) immediately. The samples were processed with the PrimeFlow RNA Assay kit (ThermoFisher) following the manufacturers protocol.
Fluorescent RNA in situ hybridization (FISH)
Hybridization chain reaction (HCR) v3.0 method was used to achieve mRNA FISH at high sensitivity and specificity. Probe sets against mRNA were ordered from Molecular Instruments, Inc. For the combined immunostaining and FISH staining experiments, the immunostaining procedures, including primary antibody incubation, secondary antibody incubation, and ovary postfixation and dehydration, were performed according to the previous publications 70, 71. Then the HCR v3.0 protocol for whole-mount fruit fly embryos were applied from the detection stage to the end. At the end of HCR in situ hybridization, DAPI was stained at 0.2 μg/μl for 10 min in the 5×SSCT buffer, and then washed four times in the 5×SSCT buffer for 15 min each. Finally, specimen mounting and image capturing were done as we previously described.
10x Chromium single-cell RNA-seq library construction (v2)
After the dissociated cells were sorted into the Schneider’s media, they were further assessed for their concentration and viability via a Nexcelom Cellometer Auto T4. Only when the sorted cells were over 40% viable, they were loaded on a Chromium Single Cell Controller (10x Genomics, Pleasanton, CA), based on live cell concentration, with a target of ~3,000-5,000 cells per sample. Libraries were prepared using the Chromium Single Cell 3’ Library & Gel Bead Kit v2 (10x Genomics) according to manufacturer’s directions. Resulting short fragment libraries were checked for quality and quantity using an Agilent 2100 Bioanalyzer and Thermo Fisher Qubit Fluorometer. Libraries were pooled at equal molar concentrations and sequenced to a depth necessary to achieve at least 50,000 mean reads per cell - ~130M reads each - on an Illumina HiSeq 2500 instrument using Rapid SBS v2 chemistry with the following paired read lengths: 26 bp Read1, 8 bp I7 Index and 98 bp Read2. Approximately 700-2500 cells were captured and used for analysis.
Single cell RNA-seq 10X data preprocessing
cDNA libraries were sequenced as paired-end reads on the Illumina HiSeq 2500 machine. Raw sequencing data were processed using 10x Genomics Cell Ranger pipeline v2.1., and the reads were demultiplexed into Fastq file format using cellranger mkfastq. Genome index was built by cellranger mkref using Drosophila genome dm6, ensembl 84 gene model. Data were aligned by STAR aligner and cell counts tables were generated using cellranger count function with default parameters.
Single cell RNA-seq data analysis
Cellranger’s raw gene count matrices were further analyzed using the Seurat (v2.3.3) R package in standard protocols. The cells that have less than 1000 UMIs were excluded from downstream analysis. Gene expression results were log-normalized, and then regressed on the number of UMIs. Principle component analysis (PCA) was done using the highly variable genes, and the first 25 principle components (PCs) were used for clustering analysis to identify distinct cell clusters based on PCElbowPlot. tSNE plots were used to visualize the clustering results. Known and de novo markers were used to classify the cells into different IGS subpopulations.
QUANTIFICATION AND STATISTICAL ANALYSIS
GSCs, SGCs and IGS cells were quantified according to our previous studies under the fluorescent microscope 50, 72. Briefly, the spectrosome-containing single germ cells attached to the cap cells are defined as GSCs, whereas those single germ cells away from cap cells are identified as differentiating SGCs; PZ1444 is used to label IGS cells and cap cells, which can be easily distinguished based on size and location. The statistical analysis was done using GraphPad Prism 7 with the Student’s t-test method. P values are indicated in figure legends, and the results are presented as mean or mean ± s.e.m. (***: P ≤ 0.001; **: P ≤ 0.01; *: P ≤ 0.05; n.s., no significance).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Hts | Developmental Studies Hybridoma Bank | Cat# hts RC; RRID: AB_528289 |
| Rabbit polyclonal anti-β-galactosidase antibody | MP BIOMEDICALS | Cat# 08559761 |
| Rabbit polyclonal anti-pS137 H2AvD antibody | Rockland | Cat# 600-401-914 |
| Rat polyclonal anti-Vasa antibody | Developmental Studies Hybridoma Bank | Cat# anti-vasa; RRID: AB_760351 |
| Chicken polyclonal anti-GFP antibody | Invitrogen | Cat# A10262; RRID: AB_2534023 |
| Mouse monoclonal anti-C3G | gift from Dr. Scott Hawley | PMID: 15767569 |
| Rabbit polyclonal anti-C3G | gift from Dr. Mary Lilly | PMID: 12588841 |
| Deposited Data | ||
| scRNA-sequencing dataset | GEO: GSE143817 | This study |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: C587-Gal4 | The Xie lab | FlyBase: FBti0037960 |
| D. melanogaster: tubP-Gal80ts (ChrIII) | Bloomington Drosophila Stock Center | BDSC: 7017 |
| D. melanogaster: tubP-Gal80ts (ChrII) | Bloomington Drosophila Stock Center | BDSC: 7019 |
| D. melanogaster: PZ1444 | The Xie lab | FlyBase: FBti0003776 |
| D. melanogaster: UAS-mGFP | Bloomington Drosophila Stock Center | BDSC: 5137 |
| D. melanogaster: UAS-GFP | Bloomington Drosophila Stock Center | BDSC: 5431 |
| D. melanogaster: NetA-GFP | Bloomington Drosophila Stock Center | BDSC: 59409 |
| D. melanogaster: mirr-GFP | Bloomington Drosophila Stock Center | BDSC: 68183 |
| D. melanogaster: bam-GFP | The Xie lab | FlyBase: FBtp0016968 |
| D. melanogaster: dnc-GFP | Bloomington Drosophila Stock Center | BDSC: 60535 |
| D. melanogaster: GMR31C09-Gal4 | Bloomington Drosophila Stock Center | BDSC: 49670 |
| D. melanogaster: GMR25A11-Gal4 | Bloomington Drosophila Stock Center | BDSC: 49106 |
| D. melanogaster: GMR71E07-Gal4 | Bloomington Drosophila Stock Center | BDSC: 39590 |
| D. melanogaster: UAS-nLacZ | Bloomington Drosophila Stock Center | BDSC: 3955 |
| D. melanogaster: RNAi of luc | Bloomington Drosophila Stock Center | BDSC: 31603 |
| D. melanogaster: RNAi of NetA | Bloomington Drosophila Stock Center | BDSC: 31288 |
| D. melanogaster: RNAi of NetA | Bloomington Drosophila Stock Center | BDSC: 31665 |
| D. melanogaster: RNAi of Hf | Bloomington Drosophila Stock Center | BDSC: 77335 |
| D. melanogaster: RNAi of bin | Bloomington Drosophila Stock Center | BDSC: 34718 |
| D. melanogaster: RNAi of vn | Bloomington Drosophila Stock Center | BDSC: 67844 |
| D. melanogaster: RNAi of vn | Bloomington Drosophila Stock Center | BDSC: 56950 |
| D. melanogaster: RNAi of GstS1 | Bloomington Drosophila Stock Center | BDSC: 28885 |
| D. melanogaster: RNAi of GstS1 | Bloomington Drosophila Stock Center | BDSC: 53238 |
| D. melanogaster: RNAi of wun2 | Bloomington Drosophila Stock Center | BDSC: 32381 |
| D. melanogaster: RNAi of wun2 | Bloomington Drosophila Stock Center | BDSC: 32423 |
| D. melanogaster: dpr17-Gal4 | This study | This study |
| D. melanogaster: H2126-SwitchGal4 | gift from Dr. Hwei-Jan Hsu | PMID: 31018943 |
| D. melanogaster: RNAi of smo | Bloomington Drosophila Stock Center | BDSC: 27037 |
| D. melanogaster: RNAi of smo | Bloomington Drosophila Stock Center | BDSC: 62987 |
| Software and Algorithms | ||
| GraphPad Prism 7 | GraphPad Software | RRID: SCR 002798 |
| Adobe Illustrator | Adobe Inc. | Adobe Illustrator 2020 24.0.1 |
| ImageJ | NIH | N/A |
| Other | ||
| HCR probe of NetA | Molecular Instruments, Inc. | LOT: PRE373 |
| HCR probe of croc | Molecular Instruments, Inc. | LOT: PRE366 |
| HCR probe of bnb | Molecular Instruments, Inc. | LOT: PRE390 |
| HCR probe of GstS1 | Molecular Instruments, Inc. | LOT: PRE388 |
| HCR probe of santa-maria | Molecular Instruments, Inc. | LOT: PRE433 |
| HCR probe of wun2 | Molecular Instruments, Inc. | LOT: PRE381 |
| HCR probe of phm | Molecular Instruments, Inc. | LOT: PRE441 |
| HCR probe of bin | Molecular Instruments, Inc. | LOT: PRE408 |
| HCR probe of vn | Molecular Instruments, Inc. | LOT: PRE403 |
| HCR probe of mirr | Molecular Instruments, Inc. | LOT: PRE369 |
| HCR probe of Nrt | Molecular Instruments, Inc. | LOT: PRE392 |
| HCR probe of Hf | Molecular Instruments, Inc. | LOT: PRE411 |
| HCR probe of CG7194 | Molecular Instruments, Inc. | LOT: PRD613 |
| HCR probe of CG42458 | Molecular Instruments, Inc. | LOT: PRD614 |
| HCR probe of dpr17 | Molecular Instruments, Inc. | LOT: PRE379 |
Highlights.
scRNA-seq identifies four linearly arranged GSC niche compartments, IGS1-4
IGS1 and IGS2 control GSC maintenance and cyst formation, respectively
IGS3 and IGS4 regulate meiosis, oocyte development and shape change of 16-cell cysts
A population of follicle cell progenitors is transcriptionally defined
ACKNOWLEDGEMENTS
We would like to thank the members of the Xie laboratory for advice and discussion, Flybase, Bloomington Drosophila Stock Center, the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947), Developmental Studies Hybridoma Bank, HS Hawley, S. Heidmann, HJ Hsu and M Lilly for reagents. This work was supported by Stowers Institute for Medical Research (T. X.) and a grant from National Institutes of Health (R01HD097664 to T. X.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no conflicts of interest.
REFERENCES
- 1.Li L, and Xie T (2005). Stem cell niche: structure and function. Annu Rev Cell Dev Biol 21, 605–631. [DOI] [PubMed] [Google Scholar]
- 2.Losick VP, Morris LX, Fox DT, and Spradling A (2011). Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev Cell 21, 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison SJ, and Spradling AC (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirilly D, Wang S, and Xie T (2011). Self-maintained escort cells form a germline stem cell differentiation niche. Development 138, 5087–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spradling A, Fuller MT, Braun RE, and Yoshida S (2011). Germline Stem Cells. Cold Spring Harbor perspectives in biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie T (2013). Control of germline stem cell self-renewal and differentiation in the Drosophila ovary: concerted actions of niche signals and intrinsic factors. WIREs Dev Biol 2, 261–273. [DOI] [PubMed] [Google Scholar]
- 7.de Cuevas M, Lilly MA, and Spradling AC (1997). Germline cyst formation in Drosophila. Annu Rev Genet 31, 405–428. [DOI] [PubMed] [Google Scholar]
- 8.Song X, Wong MD, Kawase E, Xi R, Ding BC, McCarthy JJ, and Xie T (2004). Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development 131, 1353–1364. [DOI] [PubMed] [Google Scholar]
- 9.Xie T, and Spradling AC (1998). decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 94, 251–260. [DOI] [PubMed] [Google Scholar]
- 10.Song X, Zhu CH, Doan C, and Xie T (2002). Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science 296, 1855–1857. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, and Page-McCaw A (2018). Wnt6 maintains anterior escort cells as an integral component of the germline stem cell niche. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Pan L, Wang S, Zhou J, McDowell W, Park J, Haug J, Staehling K, Tang H, and Xie T (2011). Histone H3K9 trimethylase Eggless controls germline stem cell maintenance and differentiation. PLoS genetics 7, e1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng CY, Su YH, Yang SM, Lin KY, Lai CM, Rastegari E, Amartuvshin O, Cho Y, Cai Y, and Hsu HJ (2018). Smad-Independent BMP Signaling in Somatic Cells Limits the Size of the Germline Stem Cell Pool. Stem cell reports 11, 811–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu T, Wang S, Gao Y, Mao Y, Yang Z, Liu L, Song X, Ni J, and Xie T (2015). COP9-Hedgehog axis regulates the function of the germline stem cell progeny differentiation niche in the Drosophila ovary. Development 142, 4242–4252. [DOI] [PubMed] [Google Scholar]
- 15.Huang J, Reilein A, and Kalderon D (2017). Yorkie and Hedgehog independently restrict BMP production in escort cells to permit germline differentiation in the Drosophila ovary. Development 144, 2584–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mottier-Pavie VI, Palacios V, Eliazer S, Scoggin S, and Buszczak M (2016). The Wnt pathway limits BMP signaling outside of the germline stem cell niche in Drosophila ovaries. Dev Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Gao Y, Song X, Ma X, Zhu X, Mao Y, Yang Z, Ni J, Li H, Malanowski KE, et al. (2015). Wnt signaling-mediated redox regulation maintains the germ line stem cell differentiation niche. Elife 4, e08174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo L, Wang H, Fan C, Liu S, and Cai Y (2015). Wnt ligands regulate Tkv expression to constrain Dpp activity in the Drosophila ovarian stem cell niche. J Cell Biol 209, 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamada-Kawaguchi N, Nore BF, Kuwada Y, Smith CI, and Yamamoto D (2014). Btk29A promotes Wnt4 signaling in the niche to terminate germ cell proliferation in Drosophila. Science 343, 294–297. [DOI] [PubMed] [Google Scholar]
- 20.Upadhyay M, Martino Cortez Y, Wong-Deyrup S, Tavares L, Schowalter S, Flora P, Hill C, Nasrallah MA, Chittur S, and Rangan P (2016). Transposon Dysregulation Modulates dWnt4 Signaling to Control Germline Stem Cell Differentiation in Drosophila. PLoS genetics 12, e1005918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maimon I, Popliker M, and Gilboa L (2014). Without children is required for Stat-mediated zfh1 transcription and for germline stem cell differentiation. Development 141, 2602–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banisch TU, Maimon I, Dadosh T, and Gilboa L (2017). Escort cells generate a dynamic compartment for germline stem cell differentiation via combined Stat and Erk signalling. Development 144, 1937–1947. [DOI] [PubMed] [Google Scholar]
- 23.Morris LX, and Spradling AC (2012). Steroid signaling within Drosophila ovarian epithelial cells sex-specifically modulates early germ cell development and meiotic entry. PloS one 7, e46109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konig A, Yatsenko AS, Weiss M, and Shcherbata HR (2011). Ecdysteroids affect Drosophila ovarian stem cell niche formation and early germline differentiation. EMBO J 30, 1549–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gancz D, Lengil T, and Gilboa L (2011). Coordinated regulation of niche and stem cell precursors by hormonal signaling. PLoS Biol 9, e1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes SE, Miller DE, Miller AL, and Hawley RS (2018). Female Meiosis: Synapsis, Recombination, and Segregation in Drosophila melanogaster. Genetics 208, 875–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lake CM, and Hawley RS (2012). The molecular control of meiotic chromosomal behavior: events in early meiotic prophase in Drosophila oocytes. Annu Rev Physiol 74, 425–451. [DOI] [PubMed] [Google Scholar]
- 28.Lin H, Yue L, and Spradling AC (1994). The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 120, 947–956. [DOI] [PubMed] [Google Scholar]
- 29.Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A 105, 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domanitskaya E, Anllo L, and Schupbach T (2014). Phantom, a cytochrome P450 enzyme essential for ecdysone biosynthesis, plays a critical role in the control of border cell migration in Drosophila. Dev Biol 386, 408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song X, and Xie T (2002). DE-cadherin-mediated cell adhesion is essential for maintaining somatic stem cells in the Drosophila ovary. Proc Natl Acad Sci U S A 99, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, and Kalderon D (2001). Hedgehog acts as a somatic stem cell factor in the Drosophila ovary. Nature 410, 599–604. [DOI] [PubMed] [Google Scholar]
- 33.Timofeev K, Joly W, Hadjieconomou D, and Salecker I (2012). Localized netrins act as positional cues to control layer-specific targeting of photoreceptor axons in Drosophila. Neuron 75, 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tu R, Duan B, Song X, and Xie T (2020). Dlp-mediated Hh and Wnt signaling interdependence is critical in the niche for germline stem cell progeny differentiation. Sci Adv 6, eaaz0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eberl DF, Perkins LA, Engelstein M, Hilliker AJ, and Perrimon N (1992). Genetic and developmental analysis of polytene section 17 of the X chromosome of Drosophila melanogaster. Genetics 130, 569–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, and Rubin GM (1999). The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153, 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang T, Jiao Y, and Montell C (2007). Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J Cell Biol 177, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renault AD, Sigal YJ, Morris AJ, and Lehmann R (2004). Soma-germ line competition for lipid phosphate uptake regulates germ cell migration and survival. Science 305, 1963–1966. [DOI] [PubMed] [Google Scholar]
- 39.Malagoli D, Accorsi A, Sacchi S, Basile V, Mandrioli M, Pinti M, Conklin D, and Ottaviani E (2012). Drosophila Helical factor is an inducible protein acting as an immune-regulated cytokine in S2 cells. Cytokine 58, 280–286. [DOI] [PubMed] [Google Scholar]
- 40.Nystul T, and Spradling A (2007). An Epithelial Niche in the Drosophila Ovary Undergoes Long-Range Stem Cell Replacement. Cell Stem Cell 1, 277–285 [DOI] [PubMed] [Google Scholar]
- 41.Margolis J, and Spradling A (1995). Identification and behavior of epithelial stem cells in the Drosophila ovary. Development 121, 3797–3807. [DOI] [PubMed] [Google Scholar]
- 42.Tan L, Zhang KX, Pecot MY, Nagarkar-Jaiswal S, Lee PT, Takemura SY, McEwen JM, Nern A, Xu S, Tadros W, et al. (2015). Ig Superfamily Ligand and Receptor Pairs Expressed in Synaptic Partners in Drosophila. Cell 163, 1756–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnepp B, Grumbling G, Donaldson T, and Simcox A (1996). Vein is a novel component in the Drosophila epidermal growth factor receptor pathway with similarity to the neuregulins. Genes Dev 10, 2302–2313. [DOI] [PubMed] [Google Scholar]
- 44.Nighorn A, Healy MJ, and Davis RL (1991). The cyclic AMP phosphodiesterase encoded by the Drosophila dunce gene is concentrated in the mushroom body neuropil. Neuron 6, 455–467. [DOI] [PubMed] [Google Scholar]
- 45.Speicher S, Garcia-Alonso L, Carmena A, Martin-Bermudo MD, de la Escalera S, and Jimenez F (1998). Neurotactin functions in concert with other identified CAMs in growth cone guidance in Drosophila. Neuron 20, 221–233. [DOI] [PubMed] [Google Scholar]
- 46.Kehl BT, Cho KO, and Choi KW (1998). mirror, a Drosophila homeobox gene in the Iroquois complex, is required for sensory organ and alula formation. Development 125, 1217–1227. [DOI] [PubMed] [Google Scholar]
- 47.Zaffran S, Kuchler A, Lee HH, and Frasch M (2001). biniou (FoxF), a central component in a regulatory network controlling visceral mesoderm development and midgut morphogenesis in Drosophila. Genes Dev 15, 2900–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu CH, and Xie T (2003). Clonal expansion of ovarian germline stem cells during niche formation in Drosophila. Development 130, 2579–2588. [DOI] [PubMed] [Google Scholar]
- 49.Lee T, and Luo L (1999). Mosaic analysis with a repressible neurotechnique cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461. [DOI] [PubMed] [Google Scholar]
- 50.Mao Y, Tu R, Huang Y, Mao D, Yang Z, Lau PK, Wang J, Ni J, Guo Y, and Xie T (2019). The exocyst functions in niche cells to promote germline stem cell differentiation by directly controlling EGFR membrane trafficking. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma X, Wang S, Do T, Song X, Inaba M, Nishimoto Y, Liu LP, Gao Y, Mao Y, Li H, et al. (2014). Piwi is required in multiple cell types to control germline stem cell lineage development in the Drosophila ovary. PloS one 9, e90267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim-Yip RP, and Nystul TG (2018). Wingless promotes EGFR signaling in follicle stem cells to maintain self-renewal. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schultz C, Wood CG, Jones DL, Tazuke SI, and Fuller MT (2002). Signaling from germ cells mediated by the rhomboid homolog stet organizes encapsulation by somatic support cells. Development 129, 4523–4534. [DOI] [PubMed] [Google Scholar]
- 54.Jang JK, Sherizen DE, Bhagat R, Manheim EA, and McKim KS (2003). Relationship of DNA double-strand breaks to synapsis in Drosophila. J Cell Sci 116, 3069–3077. [DOI] [PubMed] [Google Scholar]
- 55.Page SL, and Hawley RS (2001). c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev 15, 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hong A, Lee-Kong S, Iida T, Sugimura I, and Lilly MA (2003). The p27cip/kip ortholog dacapo maintains the Drosophila oocyte in prophase of meiosis I. Development 130, 1235–1242. [DOI] [PubMed] [Google Scholar]
- 57.Ke YT, and Hsu HJ (2019). Generation of Inducible Gene-Switched GAL4 Expressed in the Drosophila Female Germline Stem Cell Niche. G3 (Bethesda) 9, 2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osterwalder T, Yoon KS, White BH, and Keshishian H (2001). A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A 98, 12596–12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Slaidina M, Banisch TU, Gupta S, and Lehmann R (2020). A single-cell atlas of the developing Drosophila ovary identifies follicle stem cell progenitors. Genes Dev. 34, 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jevitt A, Chatterjee D, Xie G, Wang XF, Otwell T, Huang YC, and Deng WM (2020). A single-cell atlas of adult Drosophila ovary identifies transcriptional programs and somatic cell lineage regulating oogenesis. PLoS Biol. 18, e3000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sahai-Hernandez P, and Nystul TG (2013). A dynamic population of stromal cells contributes to the follicle stem cell niche in the Drosophila ovary. Development 140, 4490–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dionne H, Hibbard KL, Cavallaro A, Kao JC, and Rubin GM (2018). Genetic Reagents for Making Split-GAL4 Lines in Drosophila. Genetics 209, 31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu M, Lim TM, and Cai Y (2010). The Drosophila female germline stem cell lineage acts to spatially restrict DPP function within the niche. Science signaling 3, ra57. [DOI] [PubMed] [Google Scholar]
- 64.de Cuevas M, and Spradling AC (1998). Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development 125, 2781–2789. [DOI] [PubMed] [Google Scholar]
- 65.Koch EA, and Spitzer RH (1983). Multiple effects of colchicine on oogenesis in Drosophila: induced sterility and switch of potential oocyte to nurse-cell developmental pathway. Cell Tissue Res 228, 21–32. [DOI] [PubMed] [Google Scholar]
- 66.Theurkauf WE, Alberts BM, Jan YN, and Jongens TA (1993). A central role for microtubules in the differentiation of Drosophila oocytes. Development 118, 1169–1180. [DOI] [PubMed] [Google Scholar]
- 67.Grieder NC, de Cuevas M, and Spradling AC (2000). The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development 127, 4253–4264. [DOI] [PubMed] [Google Scholar]
- 68.Deng W, and Lin H (1997). Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev Biol 189, 79–94. [DOI] [PubMed] [Google Scholar]
- 69.Diao F, Ironfield H, Luan H, Diao F, Shropshire WC, Ewer J, Marr E, Potter CJ, Landgraf M, and White BH (2015). Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell Rep. 10, 1410–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zimmerman SG, Peters NC, Altaras AE, and Berg CA (2013). Optimized RNA ISH, RNA FISH and protein-RNA double labeling (IF/FISH) in Drosophila ovaries. Nat. Protoc 8, 2158–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zou F, Tu R, Duan B, Yang Z, Ping Z, Song X, Chen S, Price A, Li H, Scott A, et al. (2020). Drosophila YBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs. Proc Natl Acad Sci U S A 117, 3603–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma X, Zhu X, Han Y, Story B, Do T, Song X, Wang S, Zhang Y, Blanchette M, Gogol M, et al. (2017). Aubergine Controls Germline Stem Cell Self-Renewal and Progeny Differentiation via Distinct Mechanisms. Dev Cell 41, 157–169 e155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original scRNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE143817). Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1575.


