Abstract
The yeast Saccharomyces boulardii is well known for its probiotic effects such as treating or preventing gastrointestinal diseases. Due to its ability to survive in stomach and intestine, S. boulardii could be applied as a vehicle for producing and delivering bioactive substances of interest to human gut. In this study, we cloned the gene lecC encoding the antilisterial peptide leucocin C from lactic acid bacterium Leuconostoc carnosum in S. boulardii. The constructed S. boulardii strain secreted a peptide, which had molecular weight corresponding to leucocin C in SDS-PAGE. The peptide band inhibited Listeria monocytogenes in gel overlay assay. Likewise, concentrated S. boulardii culture supernatant inhibited the growth of L. monocytogenes. The growth profile and acid tolerance of the leucocin C secreting S. boulardii were similar as those of the strain carrying the empty vector. We further demonstrated that the cells of the leucocin C producing S. boulardii efficiently killed L. monocytogenes, also without antibiotic selection pressure. These results showed that antilisterial activity could be added to the arsenal of probiotic activities of S. boulardii, demonstrating its potential as a carrier for therapeutics delivery.
Electronic supplementary material
The online version of this article (10.1007/s12602-020-09676-1) contains supplementary material, which is available to authorized users.
Keywords: Probiotic yeast, Saccharomyces boulardii, Bacteriocin, Leucocin C, Heterologous expression
Introduction
The probiotic yeast Saccharomyces boulardii, originally isolated from litchi fruit by Henri Boulard in the 1920s, is a subtype of Saccharomyces cerevisiae [1]. S. boulardii is the only approved probiotic yeast in human nutrition [2] and has been granted with the GRAS (generally regarded as safe) status by the Food and Drug Administration (FDA) of USA [3]. The benefits of S. boulardii have been widely assessed. Controlled clinical trials have indicated that oral administration of S. boulardii can treat or prevent gastrointestinal diseases such as antibiotic-associated diarrhea [4], acute diarrhea in children [5, 6], and AIDS-associated diarrhea [7]. Additionally, some studies have revealed that S. boulardii can control inflammation in the inflammatory bowel disease (IBD) like Crohn’s disease and ulcerative colitis [8].
Despite the probiotic effects of S. boulardii are well understood, only a few studies have focused on expanding its probiotic properties by production of heterologous proteins, such as cytokines or antimicrobial proteins. In the previous work, Douradinha et al. [9] investigated factors concerning the genetic manipulation of S. boulardii, such as plasmids transformation and screening of positive strains. Regarding heterologous protein expression, researchers have produced mouse interleukin-10 (IL-10) [10, 11] and Microneme-2 proteins of chicken parasite Eimeria tenella [12] in S. boulardii using antibiotics as selection markers. In addition, green fluorescent protein (GFP) [13], human lysozyme [14], and ovalbumin (OVA) [15] have been secreted in auxotrophic mutant of S. boulardii. Thus, regardless of the limited number of genetic studies, it is possible to produce heterologous proteins in S. boulardii. As a carrier to deliver drugs or antimicrobial agents to gastrointestinal tracts for treating disorders or killing pathogens, S. boulardii is more suitable than S. cerevisiae, due to the better tolerance of S. boulardii to high temperature and low pH [1]. To the best of our knowledge, so far there are no studies reported about the production of antimicrobial proteins from bacteria in S. boulardii.
Bacteriocins are ribosomally synthesized antimicrobial proteins produced by bacteria. Studies have shown that a large amount of lactic acid bacteria (LAB) strains including Lactobacillus, Lactococcus, and Leuconostoc can secrete bacteriocins, of which many have been well characterized [16]. Bacteriocins generally comprise three classes [17]. Among the class II group, the subgroup class IIa (pediocin-like) bacteriocins have high killing activity against Listeria monocytogenes [18] by holing its cell membrane and causing internals release [19]. L. monocytogenes is a foodborne pathogen and widely distributed in the environment, including water [20], foods [21], and human feces [22]. In human, L. monocytogenes causes infection known as listeriosis, one of the most severe foodborne diseases [23]. In 2017, there were 786 listeriosis cases reported in the USA [24] and 2480 cases in EU [25].
In our previous work, we have successfully produced the class IIa bacteriocin leucocin C from Leuconostoc carnosum 4010 [26] in Lactococcus lactis NZ9000 [27]. Therefore, in this present study, we chose leucocin C as the target antimicrobial peptide. The main aim was to design a recombinant S. boulardii strain, which could produce antilisterial leucocin C besides its probiotic characteristics. Based on the results of antimicrobial tests, SDS-PAGE, gel overlay assay, and L. monocytogenes killing assay, active leucocin C was shown to be successfully secreted by S. boulardii. The benefits of S. boulardii could thus be enhanced with the combination of antimicrobial and probiotic effects.
Materials and Methods
Plasmids, Strains, and Media
All the plasmids and strains used in this study are listed in Table 1. Escherichia coli DH5α was grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl) at 37 °C, supplemented with kanamycin (50 μg/ml) for selecting the transformants. Saccharomyces boulardii CNCM I-745 was grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose) at 37 °C, and 20 μg/ml blasticidin S (Thermo Fisher Scientific, Waltham, MA, USA) was used to maintain plasmids in yeasts [28]. L. monocytogenes was grown in BHI (brain heart infusion, Lab M, Lancashire, UK) agar or in broth at 37 °C.
Table 1.
Plasmids and strains
| Plasmids and strains | Description | Source |
|---|---|---|
| Plasmids | ||
| pSF-Blast | pSF-TEF1-TPI1-Blast (OG539), E. coli-yeast shuttle vector | Oxford Genetics, Oxford, UK |
| pSF-Blast-lecC | pSF-TEF1-TPI1-Blast-lecC, plasmid inserted with leucocin C expression cassette | GenScript, Piscataway, NJ, USA |
| Strains | ||
| Escherichia coli DH5α | Library Efficiency™ DH5α™ Competent Cells; intermediate host for preserving plasmids | Invitrogen, Carlsbad, CA, USA |
| Sb-wild type | Saccharomyces boulardii CNCM I-745, wild-type strain | Capsule PRECOSA, Biocodex, Espoo, Finland |
| Sb-vector | S. boulardii CNCM I-745 carrying pSF-TEF1-TPI1-Blast (OG539) | This study |
| Sb-LecC | S. boulardii CNCM I-745 carrying pSF-TEF1-TPI1-Blast-lecC for leucocin C expression | This study |
| Listeria monocytogenes WSLC 1018 | Indicator strain, sensitive to leucocin C, ATCC 19118 | Prof. Martin Loessner, ETH Zürich, Switzerland |
Plasmid and Transformants Construction
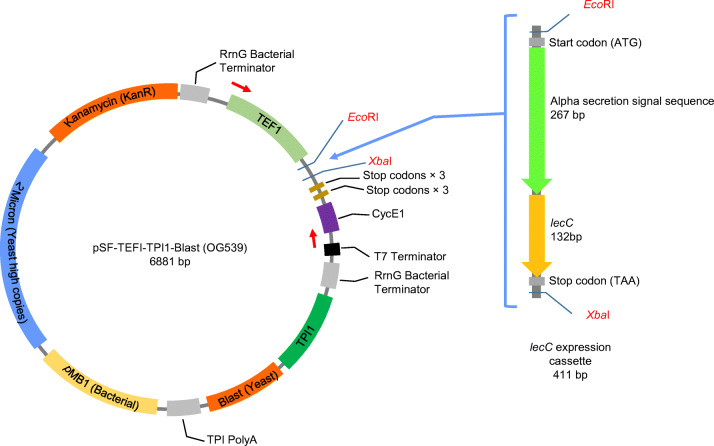
For heterologous expression of leucocin C in S. boulardii, the plasmid pSF-TEF1-TPI1-Blast (pSF-Blast) was used as the expression vector (Fig. 1). This vector carries a blasticidin S resistance cassette (Blast), which is driven from the strong yeast constitutive triosephosphate isomerase (TPI1) gene promoter. Blast is the yeast selection marker, while the kanamycin-resistant gene (KanR) is to select bacterial transformants. The plasmid also contains a constitutive promoter of yeast translation elongation factor 1 (TEF1), which is the strongest promoter known for the protein expression in S. cerevisiae [29, 30]. EcoRI and XbaI were selected as the restriction sites for the gene insertion.
Fig. 1.
Construction of the plasmid pSF-Blast-lecC. The lecC expression cassette was cloned into EcoRI and XbaI sites in the vector pSF-TEF1-TPI1-Blast (OG539), resulting in the leucocin C secretion plasmid pSF-TEF1-TPI1-Blast-lecC. The red arrows on the plasmid map indicate the location where the plasmid specific primers would bind
The DNA fragment for the leucocin C secretion (Fig. 1) was synthesized by GenScript (Piscataway, NJ, USA). This fragment consists of the S. cerevisiae α-mating factor signal sequence to guide the extracellular secretion [31] and the lecC gene sequence [27]. Codon optimization was achieved by GenScript using OptimumGene™ algorithm to enhance the efficiency of gene expression. After optimization, the synthesized fragment was cloned into pSF-Blast plasmid at the EcoRI/XbaI sites. To increase the amount of plasmid for yeast transformation, the resulting plasmid pSF-Blast-lecC (pSF-TEF1-TPI1-Blast-lecC) was introduced into E. coli DH5α by electroporation [32]. Afterwards, the enriched pSF-Blast-lecC plasmid was isolated from E. coli with EZ-10 Spin Column Plasmid Mini-Preps Kit (BBI Life Sciences, Shanghai, China).
After that, 2.7 μg of recombinant plasmid was transferred into S. boulardii via electroporation [33]. Correct transformants were identified by PCR using the plasmid specific primers (F, 5′-CATATCACATAGGAAGCAACAG-3′, hybridizing before the TEF1 promoter; R, 5′-CTACGATACCGATAGAGATGG-3′, hybridizing before T7 terminator), which resulted in PCR products of 1400 bp from pSF-Blast-lecC and 1050 bp from the plain vector.
Secretion of Leucocin C
For the production of leucocin C, S. boulardii carrying pSF-Blast-lecC (Sb-LecC) was grown in 250 ml of broth at 37 °C for 36 h with shaking. S. boulardii wild type (Sb-wild type) and S. boulardii carrying empty vector pSF-TEF1-TPI1-Blast (Sb-vector) were used as negative controls. Cell-free supernatants were collected by centrifugation at 5000×g for 10 min at 4 °C and filtered through the 0.22-μm sterilized Millipore filter (Merck Millipore, Bedford, MA, USA). The leucocin C in the cell-free supernatant of Sb-LecC was precipitated with 40% ammonium sulfate and collected by centrifugation at 10,000×g for 30 min as previously described [26, 34]. The protein precipitation was dissolved in 500 μl of 20 mM PBS buffer (pH 7.2). The final concentrated samples were kept at − 20 °C for further tests. The concentrated supernatant from the leucocin C producing L. lactis NZ9000 constructed in our previous study [27] was used as a positive control.
Antimicrobial Tests
An agar inhibition assay [35] was performed to roughly estimate the antimicrobial activity of recombinant S. boulardii cells. Yeast cells were grown in the YPD liquid culture overnight and washed with Ringer’s solution (Sigma-Aldrich, St. Louis, MO, USA) for two times. Ten microliters of the washed cells were added and let dry on the Listeria indicator plate. After overnight incubation, the inhibition halos were observed to determine the antimicrobial activity of yeast cells.
The bacteriocin activity of the concentrated supernatant was then tested by using agar well diffusion assay [36]. The indicator strain L. monocytogenes was grown for overnight and spread onto the BHI agar plate. Wells were made on the plate with sterile cork borer (7 mm in diameter, MRS Scientific Ltd., Wickford, UK), and the bottom of well was sealed with 20 μl of agar. After the concentrated supernatant was loaded, plates were incubated overnight at 37 °C.
Identification of Leucocin C Using SDS-PAGE and Gel Overlay
Tricine-SDS-PAGE (16.5% resolving gel, 10% spacer gel) [37] and gel overlay assay [38] were performed to verify the presence of leucocin C in the supernatant. For SDS-PAGE, proteins from 100 μl of concentrated supernatant were collected with 10 μl of StrataClean resin (Agilent Technologies, Santa Clara, CA, USA). The resin beads were resuspended with 40 μl of Tricine-SDS-PAGE loading buffer (Bio-Rad, Hercules, CA, USA) to release the proteins. The mixture was boiled for 3 min and insoluble fraction was removed by centrifugation (10,000×g, 1 min). The soluble proteins were then analyzed by Tricine-SDS-PAGE. Two identical gels were prepared under the same condition. Ten microliters of concentrated Sb-LecC supernatant was loaded into the gel, and the electrophoresis was performed at 4 °C under 30 V for 30 min and 200 V for the rest of 2 h. After electrophoresis, SDS-PAGE gels were fixed for 30 min in the fixing solution (50% methanol, 10% acetic acid, 100 mM ammonium acetate). Then, one gel was stained with Coomassie Brilliant Blue R-250 (Sigma-Aldrich). The other gel was rinsed in distilled water for 2 h, then placed on BHI agar plate, and covered with 15 ml BHI soft agar containing 200 μl of the overnight culture of L. monocytogenes. The plate with gel was incubated overnight at 37 °C.
Growth Profile
Bioscreen C™ Automated Microbiology Growth Curve Analysis System (Growth Curves, Helsinki, Finland) was used to determine the growth profile of yeasts. A single colony was inoculated into YPD medium and grown for overnight. On the next day, the fresh overnight culture was transferred to pre-warmed YPD medium with 2% inoculum. Two hundred microliters of the mixture were loaded into the honeycomb plate. The Bioscreen C™ system collected the optical density at 600 nm (OD600) every 1 h at 37 °C with continuous shaking. To measure the growth profile of yeasts under various pH conditions, YPD media were adjusted to pH 2 or 4, respectively.
L. monocytogenes Killing Assay Using Recombinant S. boulardii Cells
In order to further quantify the capacity of Sb-LecC cells against L. monocytogenes, we tested the antimicrobial activity of the cell cultures without antibiotic selection pressure. First, Sb-LecC and L. monocytogenes were separately grown overnight. Colony forming unit (CFU) was calculated to obtain the initial cell numbers of L. monocytogenes. Then, L. monocytogenes was serially diluted (up to 1:1000) with Ringer’s solution. Sb-LecC cells were centrifuged and washed two times with Ringer’s solution to remove the antibiotic blasticidin S. After final resuspension in YPD medium, 900 μl of Sb-LecC cells were mixed with 100 μl of different concentrations of L. monocytogenes. We used two experimental approaches: (i) yeasts and Listeria cells were incubated in YPD medium with shaking, and (ii) mixed cells were centrifuged (5000×g, 3 min) to form a pellet, thereby reducing the physical distance between the cells. After incubation at 37 °C for 4 h, the viable cells of L. monocytogenes were determined by colony counting after serial dilutions. Nystatin of 50 μg/ml (Sigma-Aldrich) [39] was added into BHI plates to kill yeast cells for counting Listeria colonies. Listeria incubated without yeast and Listeria incubated with the yeast Sb-vector were used as controls. Then, the survival ratio was calculated based on the ratio between viable cell numbers and initial cell numbers of Listeria. If the survival ratio was less than one, we would consider that Sb-LecC killed Listeria. Survival ratio higher than one indicates that Listeria had grown.
Results
Cloning of the Leucocin C Gene lecC in S. boulardii
The aim of this work was to improve the probiotic potential of S. boulardii CNCM I-745 by enabling it to secrete the antimicrobial peptide leucocin C from Leuconostoc carnosum. Our strategy for this was to clone the leucocin C gene lecC with a yeast secretion signal in a plasmid vector. The lecC expression plasmid and the empty vector were then introduced into S. boulardii, resulting in the strains Sb-LecC and Sb-vector. The presence of the plasmids in S. boulardii was confirmed by PCR using plasmid specific primers. Correct bands of 1400 and 1050 bp were obtained from Sb-LecC and Sb-vector, respectively (Online Source 1). No band was obtained from the S. boulardii wild-type strain.
Antimicrobial Activity of S. boulardii Sb-LecC
Antimicrobial activity of the constructed strain S. boulardii Sb-LecC was tested against L. monocytogenes. In the agar inhibition assay, Sb-LecC cells caused a clear inhibition zone on Listeria lawn, whereas the Sb-wild type and Sb-vector did not inhibit Listeria (Online Source 2a). To examine the secretion of the antimicrobial substance, the strains were cultured in liquid media. Concentrated supernatant of Sb-LecC and leucocin C from L. lactis NZ9000 as a control showed inhibition against L. monocytogenes, whereas the supernatants from Sb-wild type and Sb-vector were not antimicrobial (Online Source 2b). These results demonstrated the secretion of antilisterial compound by the Sb-LecC strain.
Identifying Leucocin C in Sb-LecC Growth Supernatant
To verify that the inhibition effect of the strain Sb-LecC was caused by secreted leucocin C, concentrated cell-free supernatant of the Sb-LecC was analyzed in Coomassie-stained SDS-PAGE and in gel overlay assay. In stained SDS-gel, concentrated supernatant of Sb-LecC gave a band of the same size as leucocin C from L. lactis, between 4.6 and 10 kDa (Online Source 3a). According to the amino acid sequence, the calculated molecular weight of leucocin C is 4.6 kDa. To confirm that the obtained band was the active antimicrobial leucocin C, the supernatants were loaded into another SDS-gel, which was subjected to overlay assay with L. monocytogenes. Sb-LecC supernatant and leucocin C from L. lactis formed inhibition zones corresponding to the bands in the stained SDS-gel (Online Source 3b). Neither the supernatant of Sb-wild type nor the supernatant of Sb-vector showed any inhibition. Therefore, it could be concluded that the constructed Sb-LecC secretes leucocin C.
Growth of S. boulardii Strains
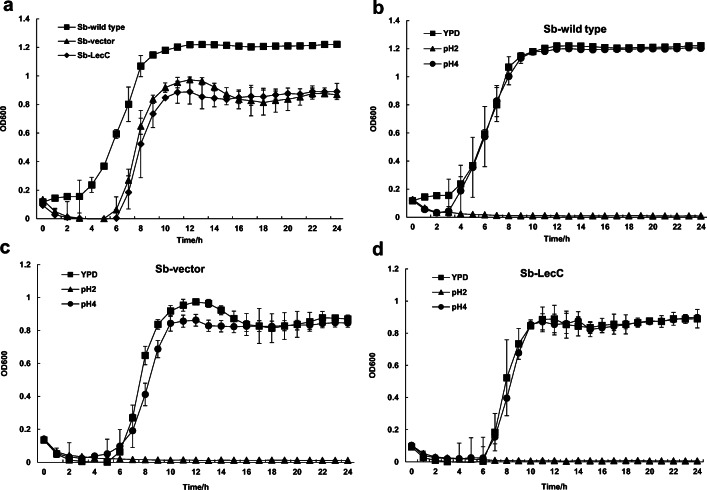
The effect of the introduced plasmids on growth was investigated. Carrying heterologous plasmid made the lag phase longer and decreased the cell concentration in the stationary growth phase of both Sb-LecC and Sb-vector strains, compared with the wild-type strain (Fig. 2a). However, as there was no significant difference between the growth of Sb-LecC and Sb-vector strain, it can be deduced that the production of the bacteriocin did not disturb the growth.
Fig. 2.
Growth of S. boulardii strains. a Growth curve of Sb-wild type, Sb-vector, and Sb-LecC in YPD medium. b–d Growth curves of Sb-wild type, Sb-vector, and Sb-LecC in YPD medium of pH 2 and pH 4. All experiments were performed in triplicate. Data is presented as mean value with standard deviation
In order to determine whether the transformants could tolerate acidity similar to that in the human stomach, the growth of S. boulardii strains was tested at pH 2 and pH 4. All three S. boulardii strains grew in the same way at pH 4 as in YPD medium without pH adjustment, but they could not grow at pH 2 (Fig. 2b–d). Nevertheless, all the strains were still viable after 24-h incubation at pH 2 (data not shown). These results indicated that even though the growth of Sb-LecC and Sb-vector was disturbed by introduced plasmids, their resistance to low pH was maintained.
Killing of L. monocytogenes Without Selection Pressure
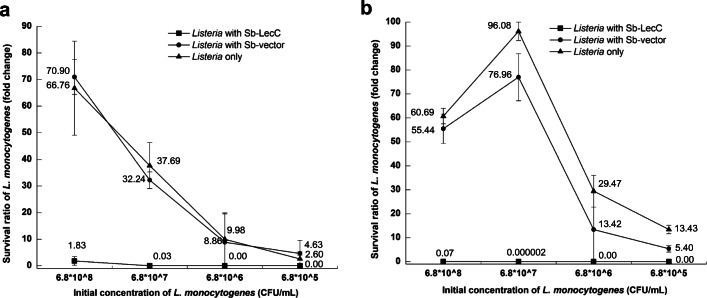
The last aim in this study was to assess the killing capacity of Sb-LecC, as well as to determine whether the strain can retain its antimicrobial activity without selection pressure. To answer these questions, S. boulardii strains were co-cultured with serially diluted L. monocytogenes in media without blasticidin S. As shown in Fig. 3a, S. boulardii Sb-LecC killed Listeria effectively. When L. monocytogenes was incubated with Sb-LecC at the initial Listeria concentration of 6.8 × 107 CFU/ml, only 3% of Listeria cells were viable. With lower initial Listeria concentrations, no live cells were found. Although Listeria count in liquid culture with Sb-LecC almost doubled (1.83) at the initial cell concentration of 6.8 × 108 CFU/ml, its survival ratio was much lower than when incubated with Sb-vector (70.90) or without yeast (66.76).
Fig. 3.
Killing of Listeria by S. boulardii. a S. boulardii and L. monocytogenes were co-cultured in the YPD medium with shaking. b S. boulardii and L. monocytogenes were incubated in the form of cell pellet after centrifugation to make a closer contact. The survival ratio of L. monocytogenes after incubation is shown in charts. The cell density of original L. monocytogenes culture was 6.8 × 109 CFU/ml, and then it was diluted to 6.8 × 108–6.8 × 105 CFU/ml for incubation. The cell density of S. boulardii added was 3.4 × 107 CFU/ml. All experiments were performed in triplicate. All data is presented as mean value with standard deviation
In addition, we wanted to examine if the killing efficiency could be increased when the bacteriocin producing yeast and Listeria were closer to each other. To test that, S. boulardii and Listeria cultures were mixed, the mixtures were centrifuged, supernatants were removed, and the cell pellets were incubated per se to keep the cells in close contact. Compared with the results from liquid co-cultures, Sb-LecC killed L. monocytogenes more effectively when the two strains were incubated in pellet, also at the highest initial listerial cell concentration (Fig. 3b). The Sb-LecC strain killed 93% of Listeria cells at the highest initial concentration of 6.8 × 108 CFU/ml. At the initial concentration of 6.8 × 107 CFU/ml, the amount of viable Listeria dropped after incubating with S. boulardii Sb-LecC, with only a marginal survival ratio of 2 × 10−6. With lower initial concentrations, no live L. monocytogenes cells were found (Fig. 3b). Quite the opposite, when Listeria was incubated without yeast cells, or with the S. boulardii vector strain, Listeria counts increased after the incubation. These results indicated that S. boulardii producing leucocin C killed the pathogen L. monocytogenes without antibiotic selection pressure and that the killing efficiency was higher when the cells were in close proximity.
Discussion
In this study, the probiotic yeast S. boulardii CNCM I-745 was used as the host to secrete antilisterial peptide leucocin C. Some of the unique features of S. boulardii make it a superior drug delivery vehicle compared with other microorganisms. For example, S. boulardii exhibits better heat and acid tolerance than S. cerevisiae [1], making it a stronger survivor under the harsh conditions in the gastrointestinal tract. In addition, since S. boulardii is not a natural colonizer in the intestine [1, 40], it can easily be removed after treatment [41]. Moreover, nowadays yeast cultures can be produced in large scale at low cost, which makes the genetically modified S. boulardii an economical vehicle for drug delivery.
To realize the purposes mentioned above, it is essential to examine or develop tools for genetic manipulation for S. boulardii. To date, the genome sequences of a few S. boulardii strains, such as ATCC MYA-796 [42], Unique 28, and CNCM I-745 [43], have been published, but the information about promoters or functional genes of S. boulardii is still scarce. One of the few studies about S. boulardii genetics is the work by Douradinha et al. [9], who confirmed the existence of the constitutive promoters PGK1, PYK1, and ENO1 in the genome of the strain ATCC MYA-796. Therefore, genetic manipulation of S. boulardii mainly relies on the genetic tools for S. cerevisiae. For example, S. cerevisiae promoters like TEF1 and PGK1 [12], TDH3 [14], and Gal1 [15] were successfully applied for the heterologous protein expression in S. boulardii. In our study, expression of lecC was mediated by the constitutive promoter TEF1, which has been shown to be constantly active during cultivation [29, 30]. To facilitate the secretion of leucocin C, the commonly used α-mating factor signal sequence [31] was fused to the lecC gene. As for the selection marker, we have previously tested the sensitivity of wild-type S. boulardii to blasticidin S and determined its minimum lethal dose (20 μg/ml) [28]. The successful secretion of leucocin C in our study demonstrated that the TEF1 promoter and α-mating factor signal sequence of S. cerevisiae can be used for S. boulardii CNCM I-745. Yet, the vector used here is a high copy number plasmid, which decreased the growth of the host [44]. Indeed, the transformed strains Sb-vector and Sb-LecC exhibited a prolonged lag phase and yielded a lower cell density at the stationary phase (Fig. 2a) compared with the Sb-wild type strain, even without the selective agent blasticidin S in the medium (data not shown).
In SDS-PAGE and gel overlay assay, the leucocin C secreted by Sb-LecC had a corresponding molecular weight and inhibition band between 4.6 and 10 kDa. These results are in consistence with the results of Fu et al. [45] and Wan et al. [27], which both used L. lactis as the host. SDS-PAGE gives only a rough estimation of the molecular weight [46], explaining inaccurate results when molecular weight of leucocin C was determined with SDS-PAGE. However, our aim here was only to demonstrate the presence and activity of leucocin C in the Sb-LecC supernatant, as when we tested its antimicrobial activity against L. monocytogenes, it indeed gave an inhibition halo (Online Source 2). These results revealed that S. boulardii is able to act as a host to secrete heterologous and active leucocin C.
Undoubtedly, it is important that the yeast to be used as a probiotic survives and, if possible, is metabolically active in the gastrointestinal tract. In previous studies, S. boulardii has shown an enhanced growth compared with S. cerevisiae in media adjusted to pH 4 [1, 47]. Thus, we investigated whether the transformants we constructed could retain tolerance to low pH, resembling the condition in stomach. The Sb-LecC constructed in this study grew well at pH 4 (Fig. 2d), and it could recover after 24-h incubation at pH 2, indicating that it survives the acidic conditions in human stomach and can possibly regain its activity in human intestinal environment. Thus, it seems possible that the leucocin C-secreting S. boulardii we constructed can function as a vehicle to deliver leucocin C into the small intestine. Correspondingly, S. boulardii may be modified to secrete other recombinant therapeutics into human intestine. Michael et al. [10] constructed an IL-10 producing S. boulardii and tested its anti-inflammatory function in the colitis mice model. No significant differences were observed between the treated and untreated mice groups, suggesting that IL-10 was not secreted in the intestine in sufficient quantities to decrease the inflammatory response. A later research by Hudson et al. [13] demonstrated that their GFP-producing recombinant S. boulardii strain could be recovered from Peyer’s patches of mice fed with the strain and that the isolated yeast cells had maintained their GFP production capacity. Most recently, Bagherpour et al. [15] successfully applied an OVA-producing S. boulardii to bring antigenic peptide ovalbumin into intestinal lumen via mice oral administration and observed a significant increase of antibody response in treated group compared with control groups. This work indicated the potential of this new delivery platform.
The use of probiotic microorganisms expressing antimicrobial peptides has been studied as a dual therapy to control bacterial infectious diseases [48]. Probiotic bacteria like Lactococcus lactis [49], Lactobacillus plantarum [50], and Propionibacterium freudenreichii [51] have been successfully applied as hosts to produce bacteriocin. As probiotic yeast, S. boulardii has no direct antimicrobial effects, except binding to some enteric pathogens like Salmonella [52], E. coli [53], and Shigella [54]. In our case, we focused on improving the probiotic effects of S. boulardii by adding the ability to kill Listeria. Thus, the leucocin C expressed in this study enabled S. boulardii to directly kill Listeria. As a foodborne pathogen, L. monocytogenes can get into the human gastrointestinal tract, interacting with intestinal epithelium [55, 56], and cause gastroenteritis [57]. It is important to eliminate or prevent L. monocytogenes in the human intestine. In this study, we demonstrated that the Sb-LecC could efficiently kill L. monocytogenes in vitro. When we incubated L. monocytogenes and Sb-LecC cells in a pellet formed by centrifugation, L. monocytogenes was killed more efficiently compared with incubating free cells together in liquid culture. Clearly, incubating the cells in the centrifugation pellet provides a better chance for cell contact and interactions, and leucocin C could thereby easily reach the target. The concentration of leucocin C in the cell pellet is likely to be higher than when leucocin C is freely secreted to the growth medium, which is the case when the two strains are co-cultured in a liquid culture. The results also demonstrated that the leucocin C secretion was relatively stable, as Sb-LecC was still secreting active leucocin C even without selection pressure, and the yield was enough to kill Listeria. Washing Sb-LecC cells directly after they were separated from the growth medium did not release leucocin C into the supernatant (data not shown), suggesting that produced leucocin C does not stick to the outer surface of Sb-LecC cells.
Taken together, our study showed that it is possible to add antibacterial capacity to the probiotic yeast S. boulardii by making it to secrete bacteriocin. In addition, we gave an insight about the possibility of applying genetically engineered S. boulardii as a carrier for therapeutics delivery. Further studies will focus on integrating the expression cassette into the chromosome of the S. boulardii and future testing of their improved probiotic effects in vivo in animal model.
Electronic Supplementary Material
(DOCX 487 kb)
(DOCX 24111 kb)
(DOCX 2398 kb)
Acknowledgments
We would like to thank Dr. Leena Räsänen for her advice and revision for our manuscript.
Funding Information
Open access funding provided by University of Helsinki including Helsinki University Central Hospital. This work was supported by the China Scholarship Council (Grant 201706910094 to Ran Li) and Magnus Ehrnrooth Foundation of Finland to Per E.J. Saris.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Edwards-Ingram L, Gitsham P, Burton N, Warhurst G, Clarke I, Hoyle D, Oliver SG, Stateva L. Genotypic and physiological characterization of Saccharomyces boulardii, the probiotic strain of Saccharomyces cerevisiae. Appl Environ Microbiol. 2007;73(8):2458–2467. doi: 10.1128/AEM.02201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markowiak P, Slizewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients. 2017;9(9):1021. doi: 10.3390/nu9091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czerucka D, Piche T, Rampal P. Yeast as probiotics-Saccharomyces boulardii. Aliment Pharmacol Ther. 2007;26(6):767–778. doi: 10.1111/j.1365-2036.2007.03442.x. [DOI] [PubMed] [Google Scholar]
- 4.Ehrhardt S, Guo N, Hinz R, Schoppen S, May J, Reiser M, Schroeder MP, Schmiedel S, Keuchel M, Reisinger EC (2016) Saccharomyces boulardii to prevent antibiotic-associated diarrhea: a randomized, double-masked, placebo-controlled trial. In: Open Forum Infect Dis 3(1) Oxford University Press [DOI] [PMC free article] [PubMed]
- 5.Feizizadeh S, Salehi-Abargouei A, Akbari V. Efficacy and safety of Saccharomyces boulardii for acute diarrhea. Pediatrics. 2014;134(1):e176–e191. doi: 10.1542/peds.2013-3950. [DOI] [PubMed] [Google Scholar]
- 6.Dinleyici E, Kara A, Dalgic N, Kurugol Z, Arica V, Metin O, Temur E, Turel O, Guven S, Yasa O. Saccharomyces boulardii CNCM I-745 reduces the duration of diarrhoea, length of emergency care and hospital stay in children with acute diarrhoea. Benef Microbes. 2015;6(4):415–421. doi: 10.3920/BM2014.0086. [DOI] [PubMed] [Google Scholar]
- 7.Saint-Marc T, Rossello-Prats L, Touraine J. Efficacy of Saccharomyces boulardii in the treatment of diarrhea in AIDS. Ann Med Interne Paris. 1991;142(1):64–65. [PubMed] [Google Scholar]
- 8.Thomas S, Metzke D, Schmitz J, Dörffel Y, Baumgart DC. Anti-inflammatory effects of Saccharomyces boulardii mediated by myeloid dendritic cells from patients with Crohn's disease and ulcerative colitis. Am J Physiol Gastrointest Liver Physiol. 2011;301(6):G1083–G1092. doi: 10.1152/ajpgi.00217.2011. [DOI] [PubMed] [Google Scholar]
- 9.Douradinha B, Reis VC, Rogers MB, Torres FA, Evans JD, Marques ET., Jr Novel insights in genetic transformation of the probiotic yeast Saccharomyces boulardii. Bioengineered. 2014;5(1):21–29. doi: 10.4161/bioe.26271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michael S, Keubler LM, Smoczek A, Meier M, Gunzer F, Pöhlmann C, Krause-Buchholz U, Hedrich H-J, Bleich A. Quantitative phenotyping of inflammatory bowel disease in the IL-10-deficient mouse by use of noninvasive magnetic resonance imaging. Inflamm Bowel Dis. 2012;19(1):185–193. doi: 10.1002/ibd.23006. [DOI] [PubMed] [Google Scholar]
- 11.Pöhlmann C, Thomas M, Förster S, Brandt M, Hartmann M, Bleich A, Gunzer F. Improving health from the inside: use of engineered intestinal microorganisms as in situ cytokine delivery system. Bioengineered. 2013;4(3):172–179. doi: 10.4161/bioe.22646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T, Sun H, Zhang J, Liu Q, Wang L, Chen P, Wang F, Li H, Xiao Y, Zhao X. The establishment of Saccharomyces boulardii surface display system using a single expression vector. Fungal Genet Biol. 2014;64:1–10. doi: 10.1016/j.fgb.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Hudson LE, Fasken MB, McDermott CD, McBride SM, Kuiper EG, Guiliano DB, Corbett AH, Lamb TJ. Functional heterologous protein expression by genetically engineered probiotic yeast Saccharomyces boulardii. PLoS One. 2014;9(11):e112660. doi: 10.1371/journal.pone.0112660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu JJ, Kong II, Zhang GC, Jayakody LN, Kim H, Xia PF, Kwak S, Sung BH, Sohn JH, Walukiewicz HE, Rao CV, Jin YS. Metabolic engineering of a probiotic Saccharomyces boulardii. Appl Environ Microbiol. 2016;82(8):2280–2287. doi: 10.1128/AEM.00057-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bagherpour G, Ghasemi H, Zand B, Zarei N, Roohvand F, Ardakani EM, Azizi M, Khalaj V. Oral administration of recombinant Saccharomyces boulardii expressing ovalbumin-CPE fusion protein induces antibody response in mice. Front Microbiol. 2018;9:723. doi: 10.3389/fmicb.2018.00723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zacharof M, Lovitt R. Bacteriocins produced by lactic acid bacteria a review article. APCBEE Procedia. 2012;2:50–56. [Google Scholar]
- 17.Mokoena MP. Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uropathogens: a mini-review. Molecules. 2017;22(8):1255. doi: 10.3390/molecules22081255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colombo NSR, Chalón MC, Navarro SA, Bellomio A. Pediocin-like bacteriocins: new perspectives on mechanism of action and immunity. Curr Genet. 2018;64(2):345–351. doi: 10.1007/s00294-017-0757-9. [DOI] [PubMed] [Google Scholar]
- 19.Tiwari SK, Noll KS, Cavera VL, Chikindas ML. Improved antimicrobial activities of synthetic-hybrid bacteriocins designed from enterocin E50-52 and pediocin PA-1. Appl Environ Microbiol. 2015;81(5):1661–1667. doi: 10.1128/AEM.03477-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renterghem BV, Huysman F, Rygole R, Verstraete W. Detection and prevalence of Listeria monocytogenes in the agricultural ecosystem. J Appl Bacteriol. 1991;71(3):211–217. doi: 10.1111/j.1365-2672.1991.tb04450.x. [DOI] [PubMed] [Google Scholar]
- 21.Kozak J, Balmer T, Byrne R, Fisher K. Prevalence of Listeria monocytogenes in foods: incidence in dairy products. Food Control. 1996;7(4–5):215–221. [Google Scholar]
- 22.McLauchlin J (1987) Listeria monocytogenes, recent advances in the taxonomy and epidemiology of listeriosis in humans. J Appl Bacteriol 63(1):1–11 [DOI] [PubMed]
- 23.Low J, Donachie W. A review of Listeria monocytogenes and listeriosis. Vet J. 1997;153(1):9–29. doi: 10.1016/s1090-0233(97)80005-6. [DOI] [PubMed] [Google Scholar]
- 24.Centers for Disease Control and Prevention (CDC) (2018) 2017 Annual Tables of Infectious Disease Data. From: National Notifiable Diseases Surveillance System. https://wonder.cdc.gov/nndss/static/2017/annual/2017-396 table2i.html
- 25.European Centre for Disease Prevention and Control (ECDC) (2017) Listeriosis, reported cases in 2017. From: Surveillance ATLAS of infectious diseases. https://atlas.ecdc.europa.eu/public/index.aspx
- 26.Budde BB, Hornbæk T, Jacobsen T, Barkholt V, Koch AG. Leuconostoc carnosum 4010 has the potential for use as a protective culture for vacuum-packed meats: culture isolation, bacteriocin identification, and meat application experiments. Int J Food Microbiol. 2003;83(2):171–184. doi: 10.1016/s0168-1605(02)00364-1. [DOI] [PubMed] [Google Scholar]
- 27.Wan X, Li R, Saris PE, Takala TM. Genetic characterisation and heterologous expression of leucocin C, a class IIa bacteriocin from Leuconostoc carnosum 4010. Appl Microbiol Biotechnol. 2013;97(8):3509–3518. doi: 10.1007/s00253-012-4406-4. [DOI] [PubMed] [Google Scholar]
- 28.Wu R (2017) Heterologous human intrinsic factor expression from probiotic yeast Saccharomyces boulardii. Dissertation, University of Helsinki
- 29.Partow S, Siewers V, Bjorn S, Nielsen J, Maury J. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010;27(11):955–964. doi: 10.1002/yea.1806. [DOI] [PubMed] [Google Scholar]
- 30.Sun J, Shao Z, Zhao H, Nair N, Wen F, Xu JH, Zhao H. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol Bioeng. 2012;109(8):2082–2092. doi: 10.1002/bit.24481. [DOI] [PubMed] [Google Scholar]
- 31.Brake AJ, Merryweather JP, Coit DG, Heberlein UA, Masiarz FR, Mullenbach GT, Urdea MS, Valenzuela P, Barr PJ. Alpha-factor-directed synthesis and secretion of mature foreign proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1984;81(15):4642–4646. doi: 10.1073/pnas.81.15.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zabarovsky ER, Winberg G. High efficiency electroporation of ligated DNA into bacteria. Nucleic Acids Res. 1990;18(19):5912. doi: 10.1093/nar/18.19.5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai S, Hashimoto W, Murata K. Transformation of Saccharomyces cerevisiae and other fungi: methods and possible underlying mechanism. Bioeng Bugs. 2010;1(6):395–403. doi: 10.4161/bbug.1.6.13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan X, Saris PE, Takala TM. Genetic characterization and expression of leucocin B, a class IId bacteriocin from Leuconostoc carnosum 4010. Res Microbiol. 2015;166(6):494–503. doi: 10.1016/j.resmic.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Köhler GA, Assefa S, Reid G. Probiotic interference of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 with the opportunistic fungal pathogen Candida albicans. Infect Dis Obstet Gynecol. 2012;2012:1–14. doi: 10.1155/2012/636474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehrer RI, Rosenman M, Harwig SS, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137(2):167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 37.Schägger HJ. Tricine-SDS-PAGE. Nat Protoc. 2006;1(1):16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 38.Bhunia AK, Johnson M, Ray B. Direct detection of an antimicrobial peptide of Pediococcus acidilactici in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J Ind Microbiol Biotechnol. 1987;2(5):319–322. [Google Scholar]
- 39.Rajkowska K, Kunicka-Styczyńska A. Probiotic activity of Saccharomyces cerevisiae var. boulardii against human pathogens. Food Technol Biotechnol. 2012;50(2):230–236. [Google Scholar]
- 40.Berg R, Bernasconi P, Fowler D, Gautreaux M. Inhibition of Candida albicans translocation from the gastrointestinal tract of mice by oral administration of Saccharomyces boulardii. J Infect Dis. 1993;168(5):1314–1318. doi: 10.1093/infdis/168.5.1314. [DOI] [PubMed] [Google Scholar]
- 41.Klein SM, Elmer GW, McFarland LV, Surawicz CM, Levy RH. Recovery and elimination of the biotherapeutic agent, Saccharomyces boulardii, in healthy human volunteers. Pharm Res. 1993;10(11):1615–1619. doi: 10.1023/a:1018924820333. [DOI] [PubMed] [Google Scholar]
- 42.Batista T, Marques E, Franco G, Douradinha B. Draft genome sequence of the probiotic yeast Saccharomyces cerevisiae var. boulardii strain ATCC MYA-796. Genome Announc. 2014;2(6):e01345–e01314. doi: 10.1128/genomeA.01345-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khatri I, Tomar R, Ganesan K, Prasad G, Subramanian S. Complete genome sequence and comparative genomics of the probiotic yeast Saccharomyces boulardii. Sci Rep. 2017;7(1):371. doi: 10.1038/s41598-017-00414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karim AS, Curran KA, Alper HS. Characterization of plasmid burden and copy number in Saccharomyces cerevisiae for optimization of metabolic engineering applications. FEMS Yeast Res. 2013;13(1):107–116. doi: 10.1111/1567-1364.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu Y, Mu D, Qiao W, Zhu D, Wang X, Liu F, Xu H, Saris P, Kuipers OP, Qiao M. Co-expression of nisin z and leucocin c as a basis for effective protection against Listeria monocytogenes in pasteurized milk. Front Microbiol. 2018;9:547. doi: 10.3389/fmicb.2018.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969;244(16):4406–4412. [PubMed] [Google Scholar]
- 47.Fietto JL, Araújo RS, Valadão FN, Fietto LG, Brandão RL, Neves MJ, Gomes FC, Nicoli JR, Castro IM. Molecular and physiological comparisons between Saccharomyces cerevisiae and Saccharomyces boulardii. Can J Microbiol. 2004;50(8):615–621. doi: 10.1139/w04-050. [DOI] [PubMed] [Google Scholar]
- 48.Mandal SM, Silva ON, Franco OL. Recombinant probiotics with antimicrobial peptides: a dual strategy to improve immune response in immunocompromised patients. Drug Discov Today. 2014;19(8):1045–1050. doi: 10.1016/j.drudis.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 49.Sánchez J, Borrero J, Gómez-Sala B, Basanta A, Herranz C, Cintas LM, Hernández PE. Cloning and heterologous production of hiracin JM79, a sec-dependent bacteriocin produced by Enterococcus hirae DCH5, in lactic acid bacteria and Pichia pastoris. Appl Environ Microbiol. 2008;74(8):2471–2479. doi: 10.1128/AEM.02559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flynn S, van Sinderen D, Thornton GM, Holo H, Nes IF, Collins JK. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology. 2002;148(4):973–984. doi: 10.1099/00221287-148-4-973. [DOI] [PubMed] [Google Scholar]
- 51.Chikindas M, Venema K, Ledeboer A, Venema G, Kok J. Expression of lactococcin A and pediocin PA-1 in heterologous hosts. Lett Appl Microbiol. 1995;21(3):183–189. doi: 10.1111/j.1472-765x.1995.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 52.Martins FS, Vieira AT, Elian SD, Arantes RM, Tiago FC, Sousa LP, Araújo HR, Pimenta PF, Bonjardim CA, Nicoli JR, Teixeira MM. Inhibition of tissue inflammation and bacterial translocation as one of the protective mechanisms of Saccharomyces boulardii against Salmonella infection in mice. Microbes Infect. 2013;15(4):270–279. doi: 10.1016/j.micinf.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 53.Dahan S, Dalmasso G, Imbert V, Peyron JF, Rampal P, Czerucka D. Saccharomyces boulardii interferes with enterohemorrhagic Escherichia coli-induced signaling pathways in T84 cells. Infect Immun. 2003;71(2):766–773. doi: 10.1128/IAI.71.2.766-773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mumy KL, Chen X, Kelly CP, McCormick BA. Saccharomyces boulardii interferes with Shigella pathogenesis by postinvasion signaling events. Am J Physiol Gastrointest Liver Physiol. 2008;294(3):G599–G609. doi: 10.1152/ajpgi.00391.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG (2007) MyD88-mediated signals induce the bactericidal lectin RegIIIγ and protect mice against intestinal Listeria monocytogenes infection. J Exp Med 204(8):1891–1900 [DOI] [PMC free article] [PubMed]
- 56.Daniels JJ, Autenrieth IB, Goebel W. Interaction of Listeria monocytogenes with the intestinal epithelium. FEMS Microbiol Lett. 2000;190(2):323–328. doi: 10.1111/j.1574-6968.2000.tb09306.x. [DOI] [PubMed] [Google Scholar]
- 57.Barbuddhe SB, Chakraborty T. Molecular mechanisms of bacterial infection via the gut. Berlin, Heidelberg: Springer; 2009. Listeria as an enteroinvasive gastrointestinal pathogen; pp. 173–195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 487 kb)
(DOCX 24111 kb)
(DOCX 2398 kb)