Abstract
Long runs of homozygosity (ROH) are contiguous stretches of homozygous genotypes, which are a footprint of inbreeding and recessive inheritance. The presence of recessive loci is suggested for Alzheimer’s disease (AD); however, their search has been poorly assessed to date. To investigate homozygosity in AD, here we performed a fine-scale ROH analysis using 10 independent cohorts of European ancestry (11,919 AD cases and 9181 controls.) We detected an increase of homozygosity in AD cases compared to controls [βAVROH (CI 95%) = 0.070 (0.037–0.104); P = 3.91 × 10−5; βFROH (CI95%) = 0.043 (0.009–0.076); P = 0.013]. ROHs increasing the risk of AD (OR > 1) were significantly overrepresented compared to ROHs increasing protection (p < 2.20 × 10−16). A significant ROH association with AD risk was detected upstream the HS3ST1 locus (chr4:11,189,482‒11,305,456), (β (CI 95%) = 1.09 (0.48 ‒ 1.48), p value = 9.03 × 10−4), previously related to AD. Next, to search for recessive candidate variants in ROHs, we constructed a homozygosity map of inbred AD cases extracted from an outbred population and explored ROH regions in whole-exome sequencing data (N = 1449). We detected a candidate marker, rs117458494, mapped in the SPON1 locus, which has been previously associated with amyloid metabolism. Here, we provide a research framework to look for recessive variants in AD using outbred populations. Our results showed that AD cases have enriched homozygosity, suggesting that recessive effects may explain a proportion of AD heritability.
Subject terms: Genomics, Psychiatric disorders
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder that is the leading cause of dementia worldwide1. AD presents a strong genetic component. Autosomal dominant mutations have been linked to familial early onset AD (EOAD) (<65 years): mutations in presenilin 1 (PSEN1)2, presenilin 2 (PSEN2)3, and amyloid precursor protein (APP)4. These findings lead to the role of amyloid metabolism as disease-causing mechanism5. Despite that, dominant causes account for a minority of both familial and sporadic EOAD cases, suggesting that autosomal recessive loci might cause most EOAD cases (∼90%)6. However, only two recessive mutations in the APP gene (A673V and E693Δ) have been described to date7,8, and this mode of inheritance for AD remains controversial.
The sporadic form of late-onset AD (LOAD) (>65 years) has a polygenic background. Heritability estimation for LOAD is, roughly, 70%6. Although, near to 40 loci has been associated with LOAD risk9–12, genetic variance captured by genome-wide strategies fall in a range of 7 –31%9,13, explaining a limited part of disease heritability. Current genetic findings were made using an additive mode of inheritance, which overlooks the relevance of non-additive genetic components, i.e., the recessive model. Despite the fact these components could explain a fraction of disease heritability.
It is well known that inbreeding increases the incidence of recessive diseases. The probability of detecting a recessive locus increases in offspring of consanguineous unions14, because the partners share alleles identical-by-descent. This recent parental relatedness points to genuine regions of autozygosity. Long runs of homozygosity (ROHs)—long stretches of consecutive homozygous genotypes (>1 Mb)—are a recognized signature of recessive inheritance and provide a measure of inbreeding in studied populations. Thus far, they have been used for homozygosity mapping15. Population history, e.g., historical bottlenecks or geographical isolation, also influences homozygosity levels in individual genomes16,17.
An excess of homozygosity has been associated with the risk of AD in individuals of Caribbean-Hispanic and African-American ancestries18–20, suggesting the presence of inbreeding and potentially autosomal recessive AD (arAD) cases nested in these populations. Conversely, this association was not replicated for individuals of European ancestry21,22. Several factors might explain these inconsistencies, among them it has been estimated that large sample sizes (12,000‒65,000) are required to detect an excess of homozygosity in outbred populations23. Thus, previous studies might be underpowered.
The limited number of deeply characterized consanguineous families, the difficulties in finding familial information for sporadic AD individuals (mainly due to the late onset of the disease) and the reduced size of intragenerational pedigrees in western countries make the search for recessive patterns of inheritance in AD complex. Furthermore, follow-up of candidate ROHs in sequencing data might be a necessary step in the definitive mapping of an arAD locus, but it has been poorly assessed to date.
Assessing the impact of homozygosity in the genetic architecture of AD, and subsequent follow-up of homozygous regions remains a challenge. To the best of our knowledge, this is the largest genomic data set exploring the influence of homozygosity in AD (n = 21,100). First, we investigated whether AD individuals from a European outbred population presented an excess of homozygosity relative to controls. Next, we measured the degree of inbreeding in AD cases. To prioritize regions with potential recessive loci, we constructed a homozygosity map of genomic regions overrepresented in detected inbred AD cases. Finally, we performed further exploration of several promising candidate ROHs using whole-exome sequencing (WES) data.
Patients and methods
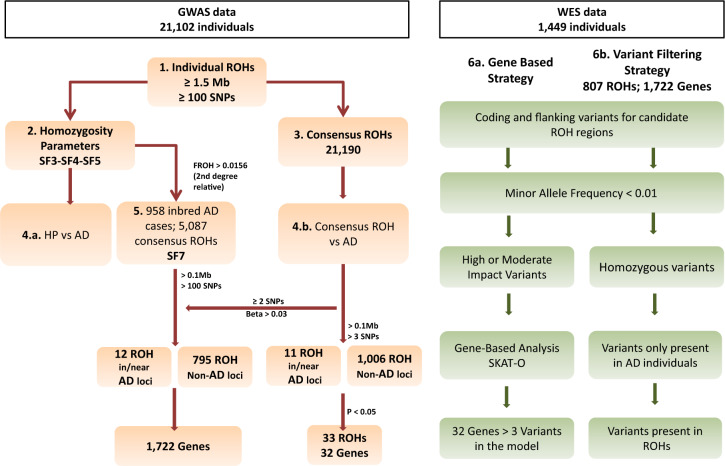
The overview of the proposed strategy for ROH detection and subsequent prioritization is depicted in Fig. 1.
Fig. 1. Schematic of the stepwise for ROH prioritization.
1. Identification of ROH segments per individual; 2. Estimation of: homozygosity parameters, and 3. Consensus ROHs; 4. Association analysis between: a) Homozygosity parameters and AD status, and b) Consensus ROH and AD status; 5. Identification of inbred AD cases and ROH prioritization; 6. Exploration of selected ROH segments in WES data applying: a) Gene-based strategy, and b) Variant filtering strategy.
Genotyping data
This study includes 10 independent genome-wide data sets comprising a total sample of 21,100 unrelated individuals (11,921 AD cases and 9181 individual controls) of European ancestry (Supplementary Table 1). The recruitment and phenotyping, has been described previously12.
Genotype-level data for each cohort was processed by applying identical quality control and imputation procedures, as previously reported12. Next, we generated a merged data set combining imputed genotypes (MAF > 0.05; imputation quality R2 > 0.90) from available data sets. We calculated identity-by-descendent (IBD) with PLINK 1.9 to generate a cohort of unrelated individuals of European ancestry (Supplementary Fig. 1). All possible pairs had Pi-hat < 0.1875, a Z0 ≥ 0.75 and a Z1 ≤ 0.25. Imputed markers with call rates >0.95 and MAF > 0.05 in the merged data set were selected for ROH calling (NSNPs = 2,678,325).
Runs of homozygosity (ROHs) exploration
1-Identification of individual ROHs
Individual ROH calling was conducted using the observational genotype-counting approach implemented in PLINK (v1.09) (https://www.cog-genomics.org/plink/1.9/), as it outperforms additional methods in ROH detection and it is applicable to outbred populations24. ROH detection was performed for each individual study and for the merged data set using imputed genotypes. We used a sliding window of 50 SNPs of 5000 Kb in length to scan the genome. In order to manage genomic regions with a small number of genotyping errors and discrete missingness, one heterozygote and five missing calls per window were tolerated. These parameters were similar to those described previously25. The minimal number of SNPs in a ROH was set to 100 SNPs26,27. We empirically explored two minimal length cut-offs to consider a ROH, 1 Mb and 1.5 Mb. ROHs < 1.5 Mb might reflect LD patterns of ancient origin rather than the consanguineous cultural practices and genetic isolation captured with ROHs > 1.5 Mb28. SNPs were included in a ROH if >5% of the sliding window was homozygous. The maximum distance between two consecutive SNPs was set to 1000 Kb apart, and SNP density to at least 1 SNP in 50 Kb.
2-Exploration of homozygosity parameters
To assess the data quality and genetic architecture of detected ROHs (>1 Mb and >1.5 Mb) in each individual study and in the whole dataset, we calculated: (a) the mean of the total length of ROH or sum of ROH (SROH); (b) the average ROH length (AVROH); (c) the number of ROHs (NROH); and (d) ROH-based estimates of the inbreeding coefficient, F, (FROH) per individual. AVROH is the SROH divided by NROH per subject. FROH represents the proportion of homozygous segments in the autosomal genome per individual (Eq. 1). For individuals, this would be the SROH detected divided by a factor of 3,020,190 Kb, the total autosomal genome length according to the GRCh37.p13 assembly. We further explored whether the effect of homozygosity parameters was similar when: (1) ROH length was set to 1 or 1.5 Mb; and (2) the analysis was performed per data set or in the final merged database. Results emerging from these exploratory analyses are shown in Supplementary Figs. 2–3, Supplementary Tables 2–3, and Supplementary Methods. According to them, we decided to conduct downstream analyses with ROH calling at 1.5 Mb in the merged data.
| 1 |
Copy number variants (CNV), particularly hemizygous deletions, are known to cause spurious ROHs. However, prior studies have demonstrated that the impact of performing ROH calling with or without CNVs is only 0.3% of the total ROH length28. To assess the impact of CNVs deletions, we also conducted ROH calling after removing common CNV deletions extracted from the Database of Genomic Variants (DGV) (http://dgv.tcag.ca/)29. The same exercise was conducted after removing CNVs detected in GR@ACE dataset. Further description of CNV calling is provided in Supplementary material.
3-Identification of consensus ROHs
Consensus ROHs were defined as overlapping segments between individual ROHs observed in different individuals, with DNA segment match of at least 95% for non-missing SNP markers. Consensus ROH calling was performed using PLINK 1.9. To prevent the detection of false-positive ROHs, we extracted those consensus ROHs with a length >100 Kb and >3 SNPs.
4-Analyses
4a-Association analysis between homozygosity parameters and AD risk
To assess the quality of the data in each individual study, we explored sample distribution for each of four homozygosity parameters: NROH, SROH, AVROH, and FROH. An exploratory analysis was depicted with violin plots, which combine a box plot with a kernel density plot, using the ggplot2 package from R (Supplementary Figs. 4 and 5). The inverse rank normal transformation was performed to generalize homozygosity parameters using “rankNorm” option in the RNOmni package in R. Transformed distributions are shown in Supplementary Fig. 6. To test the association of homozygosity parameters with AD status, we developed a generalized linear model for a binominal outcome, using R for individual-level data. We tested three models, adjusting per: (1) cohort and the first four principal components (PCs) resulting from ancestry analysis. See Eq. 2; (2) cohort, PCs and age; (3) cohort, PCs, age and gender. We also conducted a sensitivity analysis excluding control individuals <60 years old (See the “Results” section),
| 2 |
4b-Association analysis between consensus ROHs and AD
The association between the phenotype and consensus ROHs was explored using a logistic model, for ROHs in or near to previously identified AD loci extracted from de Rojas et al.30 and non-AD ROHs. The model was adjusted per cohort, and the first four PCs as covariates for downstream analysis. Covariate models adjusted for age and gender, in addition to cohort and PCs, were also calculated. Regression-based results were corrected for multiple testing using a Bonferroni correction.
Next, we sought to estimate whether there was an overrepresentation of risk (β > 0) or protective (β < 0) consensus ROHs in our association results at different levels of length and SNP number per consensus ROH. We applied a binominal test using R.
5-The homozygosity map of inbred AD individuals
5a-Identificationn of inbred individuals
We used FROH to detect the subset of inbred individuals within our dataset. FROH has been previously shown to better correlate with the unobserved pedigree inbreeding23,31. The cut-off between inbred and non-inbred individuals was set to FROH > 0.015632, which corresponds to a second-degree relation. It was assumed that there are no different biological effects below 0.0156 than in the general population33. The efficient capture of inbred individuals is shown in Supplementary Fig. 7. Next, to explore whether the frequency of consanguinity was higher in AD cases than in controls, we calculated the odds ratio and chi square p values using the epitools package in R.
5b-ROHs prioritization based on inbred AD cases
ROH detection was conducted in the subset of inbred AD cases, applying similar criteria to those previously described. Briefly, considering the long size of homozygous tracts for inbred individuals, there is a higher probability of finding a consensus ROH by chance within consanguineous AD cases than in the general population. Hence, we applied stringent criteria to define consensus ROHs. Consensus ROHs from inbred AD cases with ROH lengths >100 Kb and ROH > 100 SNPs were given priority for further analysis. Shared overlapping regions between inbred AD cases and the whole data set were also identified (See bash code in Supplementary Code Material) and selected based on their overrepresentation in AD cases relative to controls (β > 0.03). Prioritized regions were then explored in sequencing data. We also explored the overlapping of these regions with previously identified AD loci30.
WES data
To meet the objective of exploring most promising ROH candidates in the sequencing data, we used the Knight-ADRC-NIA-LOAD (KANL) cohort34. We excluded autosomal dominant familial cases and sporadic AD cases harboring well-known disease-causing mutations, as they could explain disease status. Thus, this study comprised 986 AD cases and 463 control individuals of European ancestry (See Supplementary Table 1 and Supplementary Fig. 1). Of these, 488 subjects presented both GWAS and WES data available for this study. Detailed descriptions of cohort characteristics and quality control for WES data have been provided previously34.
6-Candidate gene prioritization strategies using WES
6a-Gene-based analysis
To prioritize genes in consensus ROH regions, we performed gene-based analysis (986 cases vs 463 controls) (Fig. 1). To generate variant sets, variants were filtered out according to minor allele frequency (MAF < 0.01) and functional impact. The allele frequency cut-off was established according to the Exome Aggregation Consortium (ExAC), non-Finnish European Exome Sequencing project (ESP), and 1000 G. Only those variants predicted to have a high or moderate effect according to SnpEff were included35. To compute p-values per gene set, SKAT-O model was applied using R. The models were adjusted to consider the impact of the first two PCs and sex. Genes were filtered out from results if the number of variants included in the model was ≤3.
6b-Variant filtering strategy for inbred AD cases with WES and GWAS data available
ROH segments emerging from inbred AD cases are the most promising candidates to harbor autosomal recessive variants. Therefore, we deeply explored ROHs by applying a variant filtering strategy. We explored 488 AD cases with complementary GWAS and WES data. Because there is a low likelihood to identify any novel or causative mutation in available databases, variants with MAF > 0.01 were excluded. All heterozygous variants were removed. Finally, only the variants mapped in individual ROHs were selected.
To map genes within ROHs, we first extracted all the variants located in ROH regions. Next, we individually annotated each one.
Results
ROH parameters are associated with AD risk
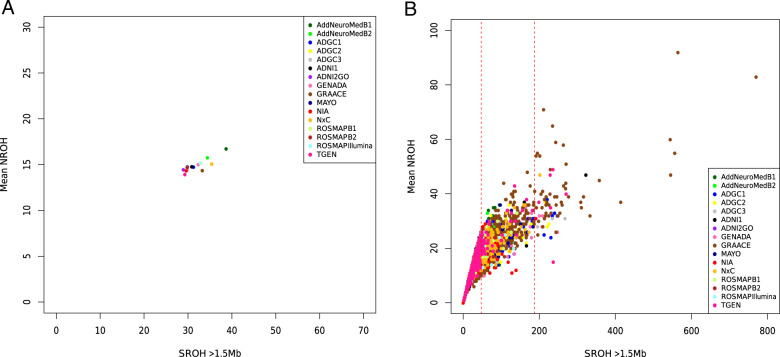
We examined the typical characteristics of the four ROH parameters (SROH, NROH, AVROH, FROH) in 21,100 unrelated European individuals from 10 independent cohorts (Supplementary Tables 1–2 and Supplementary Fig. 4). Relationships between the mean NROH and SROH are shown in Fig. 2. The mean NROH was 14.6 ± 4.6, the AVROH was 2.11 ± 0.61 Mb, and the SROH was 31.9 ± 22.2 Mb. These estimations are in accordance with those observed in European individuals32, except for the NROH parameter, which was higher than in the previous studies32.
Fig. 2. Runs of homozygosity per cohort and per individual.
A Mean number of ROHs versus mean total sum of ROHs in Mb for the 10 cohorts explored. B Mean number of ROHs versus mean total sum of ROHs in Mb per individual explored. Red dashed lines represent the threshold for the inbreeding coefficient of 0.0156 (second cousins’ offspring) and 0.0625 (first cousins’ offspring).
Next, we tested the association of the four homozygosity parameters with AD risk. We found that (i) higher inbreeding coefficient (FROH) increased the risk of suffering AD [βFROH (CI95%) = 0.043 (0.009–0.076); p value = 0.013] (Table 1); (ii) AD patients presented higher average lengths of ROHs compared to controls [βAVROH (CI95%) = 0.07 (0.037–0.104); p value = 3.91 × 10−5]; (iii) ROH number was not associated with AD risk after adjusting for age [βNROH (CI 95%) = 0.010 (−0.024–0.044); p value = 0.571] (Table 1). Results per cohort are shown in Supplementary Table 4. Notably, a sensitivity analysis conducted excluding: (1) known deletions, i.e., hemizygous segments29; and, (2) deletions identified in GR@ACE CNV study; provided comparable results (Supplementary Table 5). After excluding control individuals <60yo, a stable and significant effect remains for AVROH [βAVROH (CI 95%) = 0.07 (0.031–0.103); p value = 3.51 × 10−5] (Supplementary Table 5).
Table 1.
Effect of genome-wide homozygosity measures in Alzheimer’s disease for the joint analysis.
| Dataset | Model 1 | Model 2 | Model 3 | |||
|---|---|---|---|---|---|---|
| Beta (CI 95%) | P value | Beta (CI 95%) | P value | Beta (CI 95%) | P value | |
| FROH | 0.051 (0.023–0.078) | 3.25 × 10−4 | 0.044 (0.010–0.077) | 0.011 | 0.043 (0.009–0.076) | 0.013 |
| AVROH | 0.027 (0.000–0.055) | 0.051 | 0.074 (0.040–0.106) | 2.16 × 10−5 | 0.070 (0.037–0.104) | 3.91 × 10−5 |
| NROH | 0.043 (0.015–0.071) | 2.48 × 10−3 | 0.010 (−0.024–0.044) | 0.559 | 0.010 (−0.024–0.044) | 0.571 |
Model 1: adjusted per Cohort and PCs; Model 2: Adjusted by cohort, PCs, and age; Model 3: Adjusted by cohort, PCs, age and gender.
Results for the association of excess of homozygosity (FROH), average ROH lenght (AVROH), and number of ROH (NROH) with Alzheimer disease status.
OR, Odds ratio; with 95% confidence interval (CI 95%) and level of statistical significance (P value).
Association between homozygosity parameters and AD status, adjusted per Cohort, PCs, Age and Sex, was conducted in individuals with all available data; N = 19,253.
ROH analysis of AD risk using the whole data set
We identified 21,190 consensus ROHs in the merged data set (N = 21,100). We observed a significant over-representation of ROHs increasing the risk of suffering AD (p value < 2.20 × 10−16) (Table 2). The same over-representation of risk associations was detected after filtering at several levels based on the length and number of SNPs per consensus ROH (Table 2). When the test was conducted with results adjusted for cohort, PCs, age, and gender, the over-representation of risk associations remained highly significant (p value < 2.20 × 10−16).
Table 2.
Frequency of consensus ROHs with a potential risk or protective effect in Alzheimer’s disease.
| N ROH | Risk associations | Protective associations | P value | Probability of success | |
|---|---|---|---|---|---|
| Whole dataset | 21190 | 11974 | 9216 | < 2.2 × 10−16 | 0.56 |
| Category A | 1017 | 593 | 424 | < 2.2 × 10−16 | 0.58 |
| Category B | 926 | 537 | 389 | 1.30 × 10−6 | 0.57 |
| Category C | 858 | 499 | 359 | 1.98 × 10−6 | 0.58 |
| Category D | 42 | 33 | 9 | 2.7 × 10−4 | 0.79 |
| Whole dataset/map of inbreed AD cases | 6636 | 3969 | 2667 | < 2.2 × 10−16 | 0.60 |
Strategy A, ROHs > 100 kb; >3 SNPs.
Strategy B, ROHs > 100 kb; >25 SNPs.
Strategy C, ROHs > 100 kb; >50 SNPs.
Strategy D, ROHs > 100 kb; >3 SNPs, P < 0.05.
We then tested the association of 11 consensus ROH (≥100 Kb and ≥3 SNPs) located in or near to previously identified AD loci (N = 38)30, with AD status (Supplementary Table 6). For these analyses, Bonferroni corrected significance threshold of p = 1.32 × 10−3 was pre-specified. We detected a strong association near to HS3ST1 locus (consensus ROH length = 115.9 Kb; chr4:11,189,482‒11,305,456), (45 AD cases vs 12 controls, β (CI 95%) = 1.09 (0.48–1.48), p value = 9.03 × 10−4). This region survived age and gender adjustments (Supplementary Table 6), and was detected across 12 out of 16 datasets (Supplementary Table 7). The replication of this specific locus with AD, using ROH methodology, provides new insights of a potential recessive mechanism for this dementia locus. Among other ROH regions in or near to known AD loci (Supplementary Tables 6 and 10), we highlighted a 237 Kb ROH upstream the APP gene (chr21: 26,903,551–27,141,292), by its known role in AD7,8,36; detected in 38 AD cases vs 26 controls (26 vs 12 inbred individuals, respectively). For non-previously associated AD regions, none ROH (N = 1006) reached the significance threshold (Bonferroni correction of p = 4.97 × 10−5). Previous significant consensus ROH (chr8: 37835460–38143780) associated with AD in Europeans21 was not detected in this study, which is in line of results from Sims et al.22, failing replication.
We then explored the genes located in significant risk consensus ROHs (p value < 0.05) in gene-based analysis from WES data as well (Fig. 1). A total of 33 ROHs comprising 32 genes were analyzed (included > 3 SNPs in the model; Bonferroni correction p value = 0.0015). The NECAB1 locus (chr8:91,803,921-91,971,630) presented the most significant signal (p = 0.01) (Supplementary Table 8), but none loci reached the multiple test correction threshold.
Homozygosity mapping of AD using DNA segments identified in inbred cases
We detected 1621 individuals (958 Cases and 663 Controls) presenting a FROH ≥ 0.0156 among the total sample (N = 21,100) (Fig. 2) (Supplementary Table 9). Interestingly, inbreeding over the second degree of consanguinity was associated with a higher risk of suffering AD [OR (95%, CI) = 1.12 (1.01–1.25); p value = 0.027), which is in line with our previous results. This supports the idea that an excess of consanguineous individuals is present in the AD population. Accordingly, the search for recessive loci that play a role in AD can first be assessed in consanguineous cases.
After ROH calling in inbred AD cases, we detected 5087 ROHs, and extracted those with ≥100 Kb and ≥100 SNPs. We then selected only over-represented regions in AD cases relative to controls in the general analysis (Fig. 1). We prioritized 807 consensus homozygous segments from inbred cases (8.6% of the total autosomal genome) (Fig. 3 and Supplementary Table 10). Among them, 12 ROHs were in or near to a previously identified AD loci (Supplementary Table 10).
Fig. 3. Circos plot for the prioritized regions.
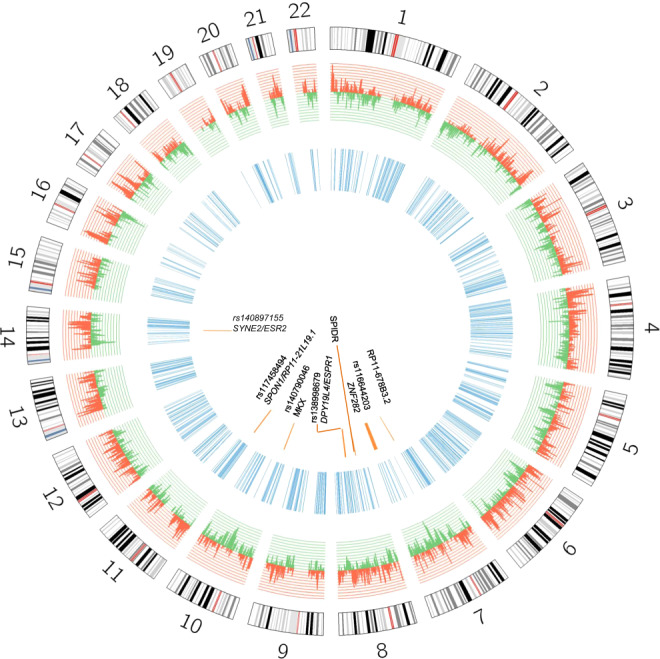
Histogram for the effect of the 21,190 consensus ROHs identified in the whole sample is shown. Risk ROH associations are shown in red; protective ROH associations are shown in green. Blue regions represent prioritized ROHs from consanguineous AD cases. Orange segments represent prioritized regions harboring potential recessive variants.
After exploring genes in identified ROHs by gene-based analysis from WES data, none of them remained associated after multiple corrections (Ngenes tested = 1136; p value = 3.47 × 10−5) (Supplementary Table 11). Our top signal was detected in the FRY locus (p value = 0.001) (Supplementary Table 11).
Considering that recessive variants are expected at very low frequencies, even gene-based analysis would be underpowered to detect significant associations. Therefore, we decided to further prioritize loci by searching homozygous mutations within selected consensus ROHs from inbred AD subjects (Fig. 1). We identified seven AD cases that had eight new (or extremely rare) homozygous variants within long ROH segments (Table 3). All ROH segments with homozygote variants were detected in more than 6 cohorts. Two of these individuals were consanguineous (FROH > 0.156). One had a missense variant (rs140790046, c.926A > G) that encodes p.Asn309Ser change within the MKX locus. Another carried a rare variant (rs116644203) in the ZNF282 locus, which was in an extremely large region of homozygosity (14.9 Mb) (Table 3). Furthermore, three additional homozygous variants were detected: (i) a variant (rs117458494) in the SPON1 locus, previously related with amyloid metabolism37, and (ii) two potential causative variants, carried only by this individual, within a previously identified AD region (TP53INP/NDUFAF6)12. One (rs73263258-ESRP1; in TP53INP/NDUFAF6 region) is a missense variant (c.475G > A) that encodes p.Ala159Thr change (Table 3). Further notes and functional effect predictions for these variants are provided in Supplementary Table 12.
Table 3.
Candidate recessive variants after ROH prioritization focused on inbred AD cases.
| Individual | Froh ROH>1500 kb | SROH (Kb) | CHR | ROH start | ROH end | ROH lenght | ROH SNPs | Rs | Variant | Near locus | Ref Allele | Alt Allele | MAF | Impact | Variant Effect |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.0058 | 17421.6 | 7 | 53232690 | 55262627 | 2029.938 | 4213 | NA | 7:53930748 | RP11-678B3.2 | C | A | <0.0000 | MODIFIER | non_coding_exon |
| 2 | 0.0185 | 55861.4 | 7 | 135617715 | 150518393 | 14900.679 | 12426 | rs116644203 | 7:148903805 | ZNF282 | C | T | <0.0000 |
LOW MODIFIER |
synonymous sequence_feature non_coding_exon |
| 3 | 0.0099 | 29868.8 | 8 | 47018484 | 49544784 | 2526.301 | 1265 | NA | 8:48352954 | SPIDR | T | A | 0.004 |
MODERATE LOW MODIFIER |
missense sequence_feature 3_prime_UTR upstream_gene non_coding_exon |
| 4 | 0.0153 | 46194.5 | 8 | 91282507 | 96735572 | 5453.066 | 4734 | rs73263258 | 8:95658495 | ESPR1* | G | A | 0.002 |
MODERATE MODIFIER |
missense 5_prime_UTR non_coding_exon |
| rs138998679 | 8:95780656 | DPY19L4* | A | G | 0.003 | LOW | synonymous | ||||||||
| 5 | 0.0179 | 54137.7 | 10 | 24516308 | 28414933 | 3898.626 | 6248 | rs140790046 | 10:27964291 | MKX | T | C | 0.0005 | MODERATE | missense |
| 6 | 0.0137 | 41392.6 | 11 | 12032272 | 16358722 | 4326.451 | 4473 | rs117458494 | 11:14282085 |
SPON1 RP11-21L19.1 |
G | A | <0.0000 | MODIFIER |
upstream_gene downstream_gene intronic |
| 7 | 0.0115 | 34603.4 | 14 | 63906601 | 66333670 | 2427.07 | 2633 | rs140897155 | 14:64688393 |
SYNE2 ESR2 |
G | A | 0.001 |
MODERATE LOW MODIFIER |
missense sequence_feature 3_prime_UTR downstream_gene intronic non_coding_exon_ |
MAF < 0.0000; Variants with non reported allele frequency in ExAC, ESP, 1000G reference database.
Discussion
This study represents the largest analysis of homozygosity conducted for AD. Our estimates of homozygosity provide a robust evidence supporting that recessive allelic architecture might define a portion of AD heritability.
Previous AD ROH studies in European populations have shown negative results for the association of ROH parameters with AD21,22. First studies had very modest sample sizes (N < 3000, vs Npresent study = 21,100)21,22, and likely were underpowered. Then, these studies used a ROH calling lengths set to 1 Mb21,22. This generates substantial inflation in the inbreeding coefficient (FROH) and makes undetectable the enrichment in consanguinity due to unspecific noise (Supplementary material). These reasons might explain initial failures. We encourage other groups to conduct ROH analysis in new unrelated populations, but with large enough sample sizes and redefining the ROH lengths at least to 1.5 Mb, to better capture the recessive component of AD.
At the present study we identified a study-wise significant ROH association close to the HS3ST1 gene (~200 Kb). Genetic markers near to this ROH (~300 kb) have been previously associated with AD using additive models38,39, and HS3ST1 locus was differentially expressed in the brain of AD cases versus controls38. Our finding reinforces the association of this region with AD, and further suggests the role of recessiveness in explaining underlying associations. High-resolution mapping across this ROH could help to identify the causative mutation.
This study failed replication of previously detected ROH at chr8:37835460–3814378021. Although, both studies include TGEN cohort, overlapping to some extent, the default technical parameters for ROH definition were completely different (ROH calling: 1 Mb vs 1.5 Mb). We assume that technical differences of the present study respect to prior ones, might be critical points impacting replication of ROH findings, in addition to other causes, e.g. population-specific genetic patterns, or, even, random chance.
A strength of the present study comes from our effort to prioritize consensus ROHs according to the homozygosity map of inbred AD individuals, performed by the first time in AD, and our capacity to explore them in sequencing data. This strategy lets us to find interesting candidate recessive variants in: MKX and ZNF282 genes, identified in two independent inbred AD cases; TP53INP1/NDUFAF6 genomic region, previously associated with AD12,40; and SPON1 locus. The SPON1 locus deserves a further explanation as it is directly related with APP metabolism, a key player in AD physiopathology. APP cleavage through β-secretases produces amyloid-beta (Aβ), which later accumulates in AD brains5. SPON1 has been found to bind to APP, inhibiting its α/β cleavage37, and to APOE family of receptors41. Markers in this gene have been related to dementia severity42 and with the rate of cognitive decline43. Considering prior findings and the present result, it would be biologically plausible that the presence of recessive variants in APP7,8, or its biological partners directly influences the amyloid cascade. Thus, we believe that SPON1 could be considered an interesting candidate, which deserves future resequencing efforts.
Our observations are subject to limitations that need to be considered. Data sets used in this study were genotyped using different platforms and shared a small proportion of directly genotyped markers. Given that lower SNP density could impact the accuracy of the study32, high quality imputed markers were used (r2 > 0.90, MAF > 0.05). Second, to reduce dataset heterogeneity we use a set of European individuals; applied the same GWAS quality control per study; generated a merged dataset including common variants across datasets; and controlled all our analyses by cohort, to account with potential confounding.
We assumed that differences in the ROH parameters between the cases and controls are modest. In that sense, we are not expecting a very large percentage of recessive AD cases, but we expect a fraction, in the same way, that it occurs for autosomal dominant forms (<1%). Considering that, the reported findings are supporting the hypothesis of this work, a group of recessive mutation may explain a portion of AD cases. However, we suspect that the existence of a large non-allelic heterogeneity is preventing its identification.
Our gene-based analysis strategy did not show significant associations. With a decreasing allele frequency and high locus heterogeneity, the power to detect genes of interest also decreases. Despite our effort to include WES data in the present study, the available sample size could be underpowered.
The potential impact of CNV deletions on ROH analysis must be taken into consideration. Thus, we assessed its effect on our analyses, but no differences were found before and after CNV exclusion (Supplementary Table 5), which is in agreement with the previous studies25. Clonal mosaicism, due to aging44, could also generate spurious ROHs. At the present study an age-dependent increase in the NROH was detected in the control group (Supplementary Table 13), which partially disappeared after excluding consensus ROHs associated with age (p < 0.05) (See Supplementary Material, and Supplementary Table 13 and 14). We assumed that these DNA segments might contain somatic alterations, confounding ROH associations. Among age-related ROH regions, we identified some loci previously associated with AD, e.g., RORA, CD2AP, HS3ST1, and amyloid-beta burden, e.g., GLIS345; suggesting that some known AD regions could be affected by this phenomenon. These findings deserve future investigations. Despite the existence of ROH segments associated to age and somatic mosaicism phenomena, our most significant findings largely supported adjustments by age. Therefore, we feel that the major observations of this study are not affected by age-related instability of the human genome.
In summary, we demonstrated the existence of an inbreeding effect in AD and efficiently captured a fraction of inbred individuals from outbred populations, providing an improved strategy to look for recessive alleles, and to conduct future large-scale homozygosity mapping studies in AD. Furthermore, the exploration of complementary sequencing data gave an added value to this research, providing a subset of potential candidates harboring recessive variants. In any case, the proposed candidates would need confirmation in larger series. Greater efforts and larger collections of individuals with GWAS and sequencing data are needed to confirm the present findings.
Our understanding of the dynamics of population genomics in AD is far from complete, but ROH analyses provide us with a means to go further and might be an alternative strategy to uncover the genetic loci underlying AD.
Supplementary information
Acknowledgements
We would like to thank patients and controls who participated in this project. The Genome Research at Fundació ACE project (GR@ACE) is supported by Fundación bancaria “La Caixa,” Grifols SA, Fundació ACE, and ISCIII (Ministry of Health, Spain). We also want to thank the private sponsors who support the basic and clinical projects of our institution (Piramal AG, Laboratorios Echevarne, Araclon Biotech S.A., and Fundació ACE). We are indebted to the Trinitat Port-Carbó legacy and her family for their support of Fundació ACE research programs. Fundació ACE is a participating center in the Dementia Genetics Spanish Consortium (DEGESCO). A.R. and M.B. receive support from the European Union/EFPIA Innovative Medicines Initiative Joint Undertaking ADAPTED and MOPEAD projects (Grants No. 115975 and 115985, respectively). M.B. and A.R. are also supported by national grants PI13/02434, PI16/01861, PI17/01474, and PI19/01301. Acción Estratégica en Salud is integrated into the Spanish National R + D + I Plan and funded by ISCIII (Instituto de Salud Carlos III)-Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER- “Una manera de Hacer Europa”). L.M.R. is supported by Consejería de Salud de la Junta de Andalucía (Grant PI-0001/2017). Control samples and data from patients included in this study were provided in part by the National DNA Bank Carlos III (www.bancoadn.org, University of Salamanca, Spain) and Hospital Universitario Virgen de Valme (Sevilla, Spain); they were processed following standard operating procedures with the appropriate approval of the ethical and scientific committees of these institutions. The present work was performed as part of the Biochemistry, Molecular Biology and Biomedicine doctoral program of S. Moreno-Grau at Universitat Autònoma de Barcelona (Barcelona, Spain). This work was supported by grants from the National Institutes of Health (R01AG044546, P01AG003991, RF1AG053303, R01AG058501, U01AG058922, RF1AG058501, and R01AG057777) and the Alzheimer’s Association (NIRG-11-200110, BAND-14-338165, AARG-16-441560, and BFG-15-362540). The recruitment and clinical characterization of research participants at Washington University were supported by NIH P50 AG05681, P01 AG03991, and P01 AG026276. This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders, and the Departments of Neurology and Psychiatry at Washington University School of Medicine. For additional information about GR@ACE & DEGESCO consortia, please contact with aruiz@fundacioace.com The GR@ACE study group is composed by: Abdelnour C, Aguilera N, Alarcon E, Alegret M, Benaque A, Boada M, Buendia M, Cañabate P, Carracedo A, Corbatón A, de Rojas I, Diego S, Espinosa A, Gailhajenet A, García González P, Gil S, Guitart M, González Pérez A, Hernández I, Ibarria, M, Lafuente A, Macias J, Maroñas O, Martín E, Martínez MT, Marquié M, Mauleón A1, Monté-Rubio G, Montrreal L, Moreno-Grau S, Moreno M, Orellana A, Ortega G, Pancho A, Pelejà E, Pérez-Cordon A, Pineda JA, Preckler S, Quintela I, Real LM3,8, Rodríguez-Gómez O, Rosende-Roca M, Ruiz A, Ruiz S, Sáez ME, Sanabria A, Santos-Santos MA, Serrano-Rios M, Sotolongo-Grau O, Tárraga L, Valero S, Vargas L. The DEGESCO consortium is composed by: Adarmes-Gómez A.D, Alarcón-Martín E, Álvarez I, Álvarez V, Amer-Ferrer G, Antequera M, Antúnez C, Baquero M, Bernal M, Blesa R, Boada M, Buiza-Rueda D, Bullido M.J, Burguera J.A, Calero M, Carrillo F, Carrión-Claro M, Casajeros MJ, Clarimón J, Cruz-Gamero J.M, de Pancorbo MM, de Rojas I, del Ser T Diez-Fairen M, Fortea J, Franco E, Frank-García A, García-Alberca JM, Garcia Madrona S, Garcia-Ribas G, Gómez-Garre P, Hernández I, Hevilla S, Jesús S, Labrador Espinosa MA, Lage C, Legaz A, Lleó A, López de Munáin A, López-García S, Macias D, Manzanares S, Marín M, Marín-Muñoz J, Marín T, Marquié M, Martín Montes A, Martínez B, Martínez C, Martínez V, Martínez-Lage Álvarez P, Medina M, Mendioroz Iriarte M, Menéndez-González M, Mir P, Molinuevo J.L, Monté-Rubio G, Montrreal L, Moreno-Grau S, Orellana A, Pastor A.B, Pastor P, Pérez Tur J, Periñán-Tocino T Piñol Ripoll G, Rábano A, Real de Asúa D, Rodrigo S, Rodríguez-Rodríguez E, Royo J.L, Ruiz A, Sanchez del Valle Díaz R, Sánchez-Juan P, Sastre I, Sotolongo-Grau O, Tárraga L, Valero S, Vicente M.P, Vivancos L. Data collection and sharing for this project was partially funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging and the National Institute of Biomedical Imaging and Bioengineering, as well as through generous contributions from the following: AbbVie; the Alzheimer’s Association; the Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institute of Health Research provides funds to support ADNI clinical sites in Canada. Private-sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study was coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. The AddNeuroMed data are from a public-private partnership supported by EFPIA companies and SMEs as part of InnoMed (Innovative Medicines in Europe), an integrated project funded by the European Union under the Sixth Framework program priority FP6-2004-LIFESCIHEALTH-5. Clinical leads responsible for data collection are Iwona Kłoszewska (Lodz), Simon Lovestone (London), Patrizia Mecocci (Perugia), Hilkka Soininen (Kuopio), Magda Tsolaki (Thessaloniki) and Bruno Vellas (Toulouse). Imaging leads are Andy Simmons (London), Lars-Olad Wahlund (Stockholm) and Christian Spenger (Zurich). Bioinformatics leads are Richard Dobson (London) and Stephen Newhouse (London). Funding support for the Alzheimer’s Disease Genetics Consortium (ADGC) was provided through the NIA Division of Neuroscience (U01-AG032984). The genotypic and associated phenotypic data used in the study “Multi-Site Collaborative Study for Genotype-Phenotype Associations in Alzheimer’s Disease (GenADA)” were provided by GlaxoSmithKline, R&D Limited. The data sets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000219. The Mayo Clinic Alzheimer’s Disease Genetic Studies, led by Dr. Nilüfer Ertekin-Taner and Dr. Steven G. Younkin at the Mayo Clinic in Jacksonville, FL, used samples from the Mayo Clinic Study of Aging, the Mayo Clinic Alzheimer’s Disease Research Center, and the Mayo Clinic Brain Bank. Data collection was supported through funding from NIA grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216 and R01 AG003949, NINDS grant R01 NS080820, the CurePSP Foundation, and the Mayo Foundation. The Neocodex-Murcia study was funded by the Fundación Alzheimur (Murcia), the Ministerio de Educación y Ciencia (Gobierno de España), Corporación Tecnológica de Andalucía, Agencia IDEA (Consejería de Innovación, Junta de Andalucía), the Diabetes Research Laboratory, and the Biomedical Research Foundation. University Hospital Clínico San Carlos has been supported by CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM); CIBERDEM is an ISCIII Project. The ROS/MAP study data were provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago. Data collection was supported through funding from NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG30146, R01AG36836, U01AG32984 and U01AG46152, the Illinois Department of Public Health, and the Translational Genomics Research Institute. The TGEN study was supported by Kronos Life Sciences Laboratories, the National Institute on Aging (Arizona Alzheimer’s Disease Center grants P30 AG19610 and RO1 AG023193, the Mayo Clinic Alzheimer’s Disease Center grant P50 AG16574, and the Intramural Research Program), the National Alzheimer’s Coordinating Center (U01 AG016976) and the state of Arizona. The results published here are in part based on data obtained from the AMP-AD Knowledge Portal accessed at 10.7303/syn2580853. We thank the International Genomics of Alzheimer’s Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i–Select chip was funded by the French National Foundation on Alzheimer’s Disease and Related Disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2, and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant no 503480), Alzheimer’s Research UK (Grant no 503176), the Wellcome Trust (Grant no 082604/2/07/Z), and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grants 01GI0102, 01GI0711, and 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC–10–196728.
Author contributions
S.M.G. and A.R. contributed to the study design. S.M.G. analyzed GWAS data. M.V.F., F.F., and J.P.B. analyzed WES data. P.G.G. performed CNV calling in GR@ACE dataset. I.H., M.A., E.A.M., S.V., M.M., and J.A.P., J.M., and L.M.R. contributed to phenotyping, database curation and patient selection from GR@ACE dataset. S.M.G., I.R., L.M., and A.O. carried out DNA extraction and quality control for GR@ACE dataset. I.Q., O.M., and A.C. carried out genotyping procedures for GR@ACE dataset. J.C., M.J.B., G.G.R., P.P., P.S.J., V.A., G.P.R., J.M.A., J.L.R., E.F.M., P.M., M.C., M.M., A.A., J.A., C.A., and A.R. are members of DEGESCO consortium. S.M.G., A.G., L.M., and M.E.S. contributed to data processing methods. A.B., L.T., M.B., C.C., and A.R. participated in funds procurement. S.M.G. prepared the first draft of the manuscript. S.M.G., M.V.F., C.C., and A.R. were responsible for the critic revision of the manuscript. All authors read and approved the final manuscript.
Code availability
Script used to identify shared overlapping regions between inbred AD cases and the whole data set is available at Supplementary Code Material. Additional scripts used to conduct ROH calling are available from the authors upon request.
Data availability
GR@ACE datased analysed during the current study is available into the European Genome-phenome Archive (EGA) (https://ega-archive.org), which is hosted by the EBI and the CRG, under accession number EGAS00001003424 (https://ega-archive.org/studies/EGAS00001003424). Additional GWAS datasets used in these study are available upon request in dbGaP. WES dataset used in the study can be downloaded from https://dss.niagads.org/studies/sa000004/. Data used in preparing this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators can be found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lists of authors and their affiliations appear at the end of the paper.
Contributor Information
Agustín Ruiz, Email: aruiz@fundacioace.org.
The GR@ACE study group:
C. Abdelnour, N. Aguilera, E. Alarcón-Martín, M. Alegret, A. Benaque, M. Boada, M. Buendía, P. Cañabate, A. Carracedo, A. Corbatón, I. de Rojas, S. Diego, A. Espinosa, A. Gailhajenet, P. García González, S. Gil, M. Guitart, A. González Pérez, I. Hernández, M. Ibarria, A. Lafuente, J. Macías, O. Maroñas, E. Martín, M. T. Martínez, M. Marquié, A. Mauleón, G. Monté-Rubio, L. Montrreal, S. Moreno-Grau, M. Moreno, A. Orellana, G. Ortega, A. Pancho, E. Pelejà, A. Pérez-Cordon, J. A. Pineda, S. Preckler, I. Quintela, L. M. Real, O. Rodríguez-Gómez, M. Rosende-Roca, A. Ruiz, S. Ruiz, M. E. Sáez, A. Sanabria, M. A. Santos-Santos, M. Serrano-Ríos, O. Sotolongo-Grau, L. Tárraga, S. Valero, and L. Vargas
DEGESCO consortium:
A. D. Adarmes-Gómez, E. Alarcón-Martín, I. Álvarez, V. Álvarez, G. Amer-Ferrer, M. Antequera, C. Antúnez, M. Baquero, M. Bernal, R. Blesa, M. Boada, D. Buiza-Rueda, M. J. Bullido, J. A. Burguera, M. Calero, F. Carrillo, M. Carrión-Claro, M. J. Casajeros, J. Clarimón, J. M. Cruz-Gamero, M. M. de Pancorbo, I. de Rojas, T. del Ser, M. Diez-Fairen, J. Fortea, E. Franco, A. Frank-García, J. M. García-Alberca, S. García Madrona, G. Garcia-Ribas, P. Gómez-Garre, I. Hernández, S. Hevilla, S. Jesús, M. A. Labrador Espinosa, C. Lage, A. Legaz, A. Lleó, A. López de Munáin, S. López-García, D. Macías, S. Manzanares, M. Marín, J. Marín-Muñoz, T. Marín, M. Marquié, A. Martín-Montes, B. Martínez, C. Martínez, V. Martínez, P. Martínez-Lage Álvarez, M. Medina, M. Mendioroz Iriarte, M. Menéndez-González, P. Mir, J. L. Molinuevo, L. Montrreal, S. Moreno-Grau, A. Orellana, A. B. Pastor, P. Pastor, J. Pérez-Tur, T. Periñán-Tocino, G. Piñol-Ripoll, A. Rábano, D. Real de Asúa, S. Rodrigo, E. Rodríguez-Rodríguez, J. L. Royo, A. Ruiz, R. Sanchez del Valle Díaz, P. Sánchez-Juan, I. Sastre, O. Sotolongo-Grau, L. Tárraga, S. Valero, M. P. Vicente, and L. Vivancos
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-020-01145-1.
References
- 1.2019 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 15, 321–387 (2019)
- 2.Sherrington R, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 3.Sherrington R, et al. Alzheimer’s disease associated with mutations in presenilin 2 is rare and variably penetrant. Hum. Mol. Genet. 1996;5:985–988. doi: 10.1093/hmg/5.7.985. [DOI] [PubMed] [Google Scholar]
- 4.Goate A, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 5.Hardy J, Selkoe DJ. The Amyloid Hypothesis of Alzheimer’s Disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 6.Wingo TS, Lah JJ, Levey AI, Cutler DJ. Autosomal recessive causes likely in early-onset Alzheimer disease. Arch. Neurol. 2012;69:59–64. doi: 10.1001/archneurol.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Fede G, et al. A recessive mutation in the APP gene with dominant-negative effect on Amyloidogenesis. Science. 2009;323:1473–1477. doi: 10.1126/science.1168979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomiyama T, et al. A new amyloid β variant favoring oligomerization in Alzheimer’s-type dementia. Ann. Neurol. 2008;63:377–387. doi: 10.1002/ana.21321. [DOI] [PubMed] [Google Scholar]
- 9.Kunkle BW, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019;51:414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansen IE, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sims R,J, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 2017;49:1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreno-Grau S, et al. Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer’s disease and three causality networks: the GR@ACE project. Alzheimer’s Dement. 2019;15:1333–1347. doi: 10.1016/j.jalz.2019.06.4950. [DOI] [PubMed] [Google Scholar]
- 13.Ridge PG, et al. Assessment of the genetic variance of late-onset Alzheimer’s disease. Neurobiol. Aging. 2016;41:200.e13–200.e20. doi: 10.1016/j.neurobiolaging.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrod AE. The incidence of alkaptonuria: a study in chemical individuality. Lancet. 1902;1616:1902–1911. [Google Scholar]
- 15.Lander E, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- 16.Jakkula E, et al. The Genome-wide patterns of variation expose significant substructure in a founder population. Am. J. Hum. Genet. 2008;83:787–794. doi: 10.1016/j.ajhg.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross A, et al. Population-genetic comparison of the Sorbian isolate population in Germany with the German KORA population using genome-wide SNP arrays. BMC Genet. 2011;12:67. doi: 10.1186/1471-2156-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vardarajan BN, et al. Inbreeding among Caribbean Hispanics from the Dominican Republic and its effects on risk of Alzheimer disease. Genet. Med. 2015;17:639–643. doi: 10.1038/gim.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghani M, et al. Evidence of recessive Alzheimer disease loci in a Caribbean Hispanic data set: genome-wide survey of runs of homozygosity. JAMA Neurol. 2013;70:1261–1267. doi: 10.1001/jamaneurol.2013.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghani M, et al. Association of long runs of homozygosity with Alzheimer Disease among African American individuals. JAMA Neurol. 2015;72:1313–1323. doi: 10.1001/jamaneurol.2015.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nalls MA, et al. Extended tracts of homozygosity identify novel candidate genes associated with late-onset Alzheimer’s disease. Neurogenetics. 2009;10:183–190. doi: 10.1007/s10048-009-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sims R, et al. No evidence that extended tracts of homozygosity are associated with Alzheimer’s disease. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2011;156:764–771. doi: 10.1002/ajmg.b.31216. [DOI] [PubMed] [Google Scholar]
- 23.Keller MC, Visscher PM, Goddard ME. Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics. 2011;189:237–249. doi: 10.1534/genetics.111.130922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howrigan DP, Simonson MA, Keller MC. Detecting autozygosity through runs of homozygosity: a comparison of three autozygosity detection algorithms. BMC Genomics. 2011;12:460. doi: 10.1186/1471-2164-12-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi PK, et al. Directional dominance on stature and cognition in diverse human populations. Nature. 2015;523:459–462. doi: 10.1038/nature14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lencz T, et al. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc. Natl Acad. Sci. USA. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christofidou P, et al. Runs of homozygosity: association with coronary artery disease and gene expression in monocytes and macrophages. Am. J. Hum. Genet. 2015;97:228–237. doi: 10.1016/j.ajhg.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McQuillan R, et al. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rojas, I de. et al. Common variants in Alzheimer’s disease: novel association of six genetic variants with AD and risk stratification by polygenic risk scores. medRxiv. 19012021 (2020)
- 31.Gazal S, et al. Inbreeding coefficient estimation with dense SNP data: comparison of strategies and application to HapMap III. Hum. Hered. 2014;77:49–62. doi: 10.1159/000358224. [DOI] [PubMed] [Google Scholar]
- 32.Ceballos FC, Hazelhurst S, Ramsay M. Runs of Homozygosity in sub-Saharan African populations provide insights into a complex demographic and health history. Hum. Genet. 2019;138:1123–1142. doi: 10.1007/s00439-019-02045-1. [DOI] [PubMed] [Google Scholar]
- 33.Bittles AH, Black ML. Consanguinity, human evolution, and complex diseases. Proc. Natl Acad. Sci. USA. 2010;107:1779–1786. doi: 10.1073/pnas.0906079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernández MV, et al. Analysis of neurodegenerative Mendelian genes in clinically diagnosed Alzheimer Disease. PLoS Genet. 2017;13:e1007045. doi: 10.1371/journal.pgen.1007045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shulman JM, et al. Genetic susceptibility for Alzheimer disease neuritic plaque pathology. JAMA Neurol. 2013;70:1150–1157. doi: 10.1001/jamaneurol.2013.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho A, Südhof TC. Binding of F-spondin to amyloid-beta precursor protein: a candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage. Proc. Natl Acad. Sci. USA. 2004;101:2548–2553. doi: 10.1073/pnas.0308655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broce IJ, et al. Dissecting the genetic relationship between cardiovascular risk factors and Alzheimer’s disease. Acta Neuropathol. 2018;137:209–226. doi: 10.1007/s00401-018-1928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witoelar A, et al. Meta-analysis of Alzheimer’s disease on 9,751 samples from Norway and IGAP study identifies four risk loci. Sci. Rep. 2018;8:18088. doi: 10.1038/s41598-018-36429-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Escott-Price V, et al. Gene-wide analysis detects two new susceptibility genes for Alzheimer’s disease. PLoS ONE. 2014;9:e94661. doi: 10.1371/journal.pone.0094661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoe H-S, et al. F-Spondin Interaction with the Apolipoprotein E receptor ApoEr2 affects processing of amyloid precursor protein. Mol. Cell Biol. 2005;25:9259–9268. doi: 10.1128/MCB.25.21.9259-9268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jahanshad N, et al. Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. Proc. Natl Acad. Sci. USA. 2013;110:4768–4773. doi: 10.1073/pnas.1216206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherva R, et al. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimer’s Dement. 2014;10:45–52. doi: 10.1016/j.jalz.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laurie CC, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deming Y, et al. Genome-wide association study identifies four novel loci associated with Alzheimer’s endophenotypes and disease modifiers. Acta Neuropathol. 2017;133:839–856. doi: 10.1007/s00401-017-1685-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Script used to identify shared overlapping regions between inbred AD cases and the whole data set is available at Supplementary Code Material. Additional scripts used to conduct ROH calling are available from the authors upon request.
GR@ACE datased analysed during the current study is available into the European Genome-phenome Archive (EGA) (https://ega-archive.org), which is hosted by the EBI and the CRG, under accession number EGAS00001003424 (https://ega-archive.org/studies/EGAS00001003424). Additional GWAS datasets used in these study are available upon request in dbGaP. WES dataset used in the study can be downloaded from https://dss.niagads.org/studies/sa000004/. Data used in preparing this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators can be found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.