Abstract
Dental caries is a biofilm-mediated, dynamic disease with early onset. A balanced salivary microbiota is a foundation of oral health, while dysbiosis causes tooth decay. We compared the saliva microbiota profiles in children with and without caries. The study consisted of 617 children aged 9–12 years from the Finnish Health in Teens (Fin-HIT) study with available register data on oral health. Caries status was summarised based on Decayed, Missing, and Filled Teeth (DMFT) index in permanent dentition. The children were then classified into the following two groups: DMFT value ≥ 1 was considered as cavitated caries lesions (hereafter called ‘caries’) (n = 208) and DMFT = 0 as ‘cavity free’ (n = 409). Bacterial 16S rRNA gene (V3–V4 regions) was amplified using PCR and sequenced by Illumina HiSeq. The mean age (SD) of the children was 11.7 (0.4) years and 56% were girls. The children had relatively good dental health with mean DMFT of 0.86 (1.97). Since sex was the key determinant of microbiota composition (p = 0.014), we focused on sex-stratified analysis. Alpha diversity indexes did not differ between caries and cavity free groups in either sexes (Shannon: p = 0.40 and 0.58; Inverse Simpson: p = 0.51 and 0.60, in boys and girls, respectively); neither did the composition differ between the groups (p = 0.070 for boys and p = 0.230 for girls). At the genus level, Paludibacter and Labrenzia had higher abundances in the caries group compared to cavity free group in both sexes (p < 0.001). Taken together, there were minor differences in saliva microbiota between children with and without caries. Potential biomarkers of caries were the sugar metabolisers Paludibacter and Labrenzia. These bacteria presumably enhance salivary acidification, which contributes to progression of dental caries. The clinical relevance of our findings warrants further studies.
Subject terms: Microbial communities, Pathogens, Sequencing, Diseases, Medical research, Risk factors
Introduction
Human saliva consists of diverse and numerous microorganisms including bacteria, fungi, viruses, archaea, and protozoa, commonly called the saliva microbiota1. The saliva microbiota is an important part of the human microbiome. Dysbiosis in the saliva microbiota is associated with various human diseases, including obesity2, inflammatory bowel diseases3, celiac disease4, Sjögren's syndrome5, and Kostmann syndrome6.
The oral cavity is a major route into the human body, and, therefore, the saliva microbiota is vital in maintaining both systemic and oral health7. Recent evidence suggests that the normal saliva microbiota protects against colonization of pathogenic bacteria, plays a fundamental role in maintaining oral homeostasis, and prevents development of various oral diseases, including dental caries8–10. In general, the saliva microbiota is considered a good indicator of oral health11.
Dental caries is a biofilm-mediated, dynamic disease with early onset and a significant oral health problem in humans worldwide 12,13. The prevalence of caries varies significantly between various parts of the world. A high prevalence is reported in many developing countries in Asia and Africa and also in some Central and Eastern European countries14. The aetiology of caries is complex and multifactorial and includes lifestyle factors (such as dietary habits, especially frequent consumption of dietary sugars), oral hygiene, use of antibiotics, fluorides, susceptible tooth surface, and biofilms12. In addition, recent evidence suggests that caries is a polymicrobial disease with some non-infectious species. However, increases in cariogenic microbial species result in tooth demineralization9. Moreover, caries is enhanced due to an ecological imbalance in the commensal microbes of the oral cavity, including sugar-fermenting and acidogenic bacteria, especially Streptococcus mutans15.
Several studies have reported that individual bacteria in saliva are positively associated with dental caries, including Streptococcus16, Lactobacilli17, and Bifidobacteria18. Recently, the association of saliva microbiota with dental caries has been examined in adults and in the elderly in Japan, Sweden, Germany, and China19–22. However, these results were not consistent and implied that the association is complex and likely depends on the population. Some pilot studies have suggested that alterations in the saliva microbiota correlate with caries development in children23,24. However, neither age nor sex was considered in the analysis; we, and others, have demonstrated that the composition of saliva microbiota is sex-specific in children2,25,26.
To the best of our knowledge, there have been no large-scale studies that compared saliva microbiota profiles in children with and without caries. Therefore, the present study sought to investigate the composition, diversity, and abundance of saliva microbial communities in Finnish children with and without caries history in permanent dentition. The study will provide a basis for a better understanding of the microbial aetiology of caries in children and serve as the foundation for novel therapeutic strategies for caries prevention.
Results
Characteristics of the study population
The present study included 617 participants. Of these, 33.7% had a history of caries; 36.8% of girls and 29.8% of boys had experienced caries based on the register data. Participants with a history of cavitated caries lesions (hereafter called ‘caries’) and no history of cavitated caries (hereafter called ‘cavity free’) were compared with respect to sex, age, body mass index (BMI) z-score, native language, Decayed, Missing, and Filled Teeth (DMFT) index, and oral hygiene risk (Table 1). Among these, native language (Finnish, Swedish or other) differed between caries and cavity free groups (p = 0.017). The proportion of girls appeared higher in the caries group than in the cavity free group, however, the difference was not statistically significant (61.1% [127/208] vs 53.3% [218/409]; p = 0.072). The average (standard deviation, SD) age of participants was 11.7 (0.4) years and ranged from 10.6 to 14.1 years. Median age was similar in the caries and cavity free group [caries 11.7 (0.3) vs cavity free 11.8 (0.4); p = 0.051]. In the entire group, the mean DMFT score was 0.86 (1.97) with minimum 0 and maximum 24. The maximum Community Periodontal Index for Treatment Needs (CPITN) value varied from 0 to 2, and 34.8% of the children had healthy gingival tissue (hereafter called ‘no oral hygiene risk’) with a maximum CPITN value of 0.
Table 1.
Comparisons performed between caries and cavity free groups for whole sample and separately for boys and girls.
| Whole sample (N = 617) | Boys (n = 272) | Girls (n = 345) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| caries (n = 208) | cavity free (n = 409) | p-value | caries (n = 81) | cavity free (n = 191) | p-value | caries (n = 127) | cavity free (n = 218) | p-value | |
| Age in years, median (SD) | 11.7 (0.3) | 11.8 (0.4) | 0.051£ | 11.7 (0.4) | 11.7 (0.4) | 0.302£ | 11.7 (0.4) | 11.7 (0.3) | 0.079£ |
| BMI z-score, mean (SD)a | 0.1 (1.0) | 0.1 (1.0) | 0.730£ | -0.1 (1.0) | 0.1 (1.1) | 0.311£ | 0.2 (0.9) | 0.1 (0.9) | 0.269£ |
| Native language, n (%) | |||||||||
| Finnish | 174 (83.7) | 354 (86.6) | 0.017# | 71 (87.6) | 163 (85.3) | 0.328# | 103 (81.1) | 191 (87.6) | 0.007# |
| Swedish | 20 (9.6) | 46 (11.2) | 5 (6.2) | 21 (11) | 15 (11.8) | 25 (11.5) | |||
| Other | 14 (6.7) | 9 (2.2) | 5 (6.2) | 7 (3.67) | 9 (7.1) | 2 (0.9) | |||
| Gingival health, n (%) | |||||||||
| No risk | 67 (32.2) | 148 (36.2) | 0.372# | 24 (29.6) | 53 (27.7) | 0.430# | 43 (33.9) | 95 (43.6) | 0.048# |
| Elevated risk | 141 (67.8) | 261 (63.8) | 57 (70.4) | 138 (72.3) | 84 (66.1) | 123 (56.4) | |||
| Alpha diversity indexes, mean (SD) | |||||||||
| Shannon | 2.92 (0.29) | 2.92 (0.28) | 0.613£ | 2.97 (0.27) | 2.92 (0.27) | 0.396£ | 2.91 (0.29) | 2.91 (0.30) | 0.581£ |
| Inverse Simpson | 10.32 (3.25) | 10.01 (3.01) | 0.831£ | 10.62 (2.88) | 10.12 (3.05) | 0.509£ | 10.13 (3.47) | 9.91 (2.95) | 0.595£ |
#Fisher’s exact test, £Independent samples t-test.
aData available for n = 603.
BMI = body mass index, SD = standard deviation.
Alpha and beta diversity
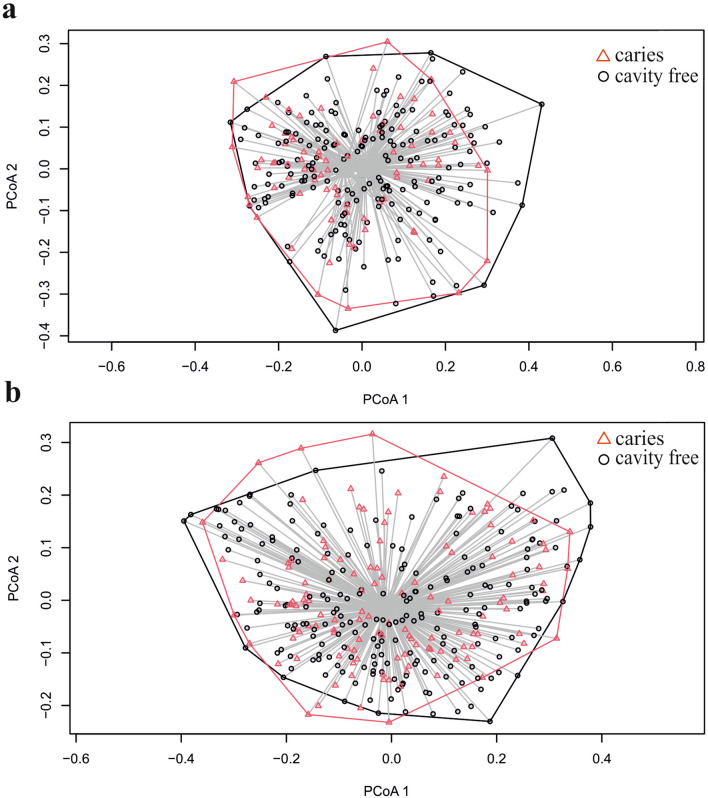
Alpha diversity (the diversity of microbes within a sample) indexes did not differ between caries and cavity free groups when both sexes were combined (Shannon: p = 0.613; Inverse Simpson [Invsim]: p = 0.831) (Table 1), indicating that diversity and the richness of the microbial communities were similar between groups (S1 Fig). Microbiota composition in terms of beta diversity (differences in microbial communities between two samples) was determined (Table 2) firstly by sex (p = 0.014), secondly by gingival health (p = 0.026), and finally by caries (p = 0.044) but not by native language (p = 0.162). Based on these findings, we performed sex-stratified analyses in order to extract the association of caries status with saliva microbiota in homogenous subsets.
Table 2.
Determinants of beta diversity in whole group, and separately in boys and girls.
| R2 | F-value | p-value | |
|---|---|---|---|
| Whole group | |||
| Sex | 0.004 | 2.551 | 0.014 |
| Gingival health | 0.003 | 2.106 | 0.026 |
| Caries status | 0.003 | 2.047 | 0.044 |
| Native language | 0.004 | 1.315 | 0.164 |
| Sex-stratified | |||
| Boys | |||
| Caries status | 0.007 | 1.787 | 0.070 |
| Gingival health | 0.006 | 1.489 | 0.142 |
| Girls | |||
| Caries status | 0.004 | 1.240 | 0.230 |
| Gingival health | 0.005 | 1.658 | 0.105 |
PERMANOVA analysis for the beta diversity using Bray Curtis dissimilarity index among the study participants adjusted for age, gender, BMI z-score, native language, and gingival health.
Analyses stratified by sex
Alpha and beta diversity
We compared alpha diversity indexes by caries status separately in boys and girls. In boys, Shannon and Invsim indexes did not differ between the groups (p = 0.396 and p = 0.509, respectively) (Table 1). Similarly, in girls, no significant differences in alpha diversity (Shannon: p = 0.581; Invsim: p = 0.595) were observed between caries and cavity free groups (Table 1). There were no differences in beta diversity between caries and cavity free groups among boys (p = 0.070) or among girls (p = 0.230) (Fig. 1; Table 2).
Figure 1.
Principal coordinates analysis (PCoA) plot of abundance based on Bray–Curtis dissimilarity (beta diversity) of the saliva microbiota composition in caries and cavity free groups in boys (a) and girls (b).
Differential abundance
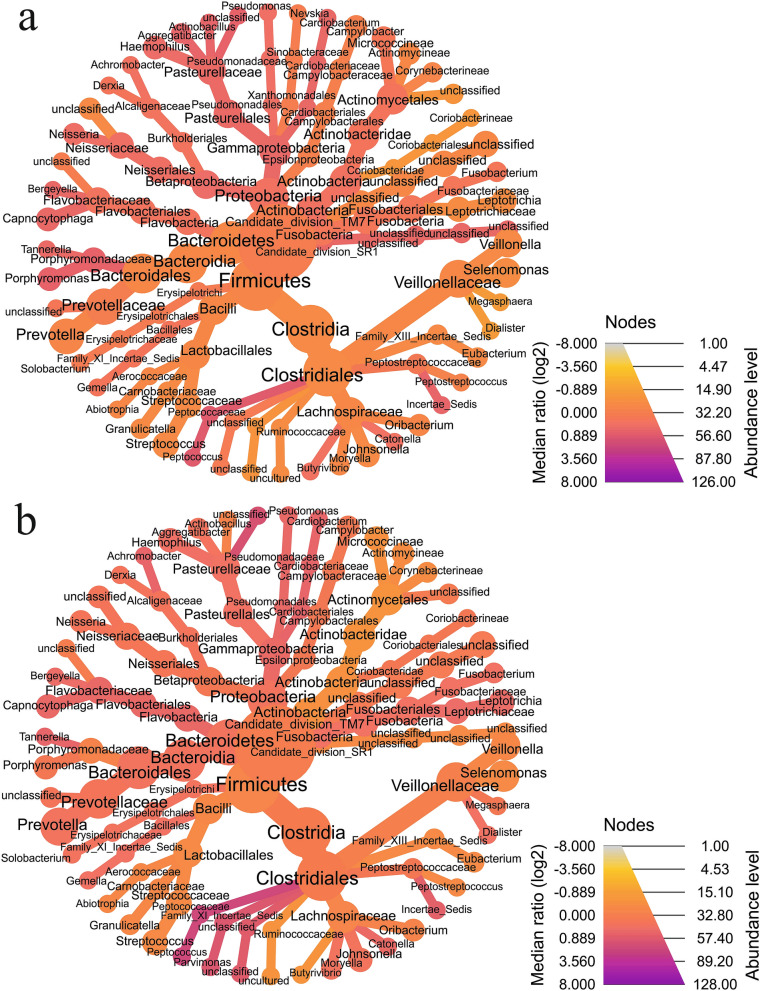
In boys, the five top genera Rothia, Neisseria, Haemophilus, Paludibacter, and Labrenzia were more abundant in the caries group (adjusted p-value < 0.001), while Anaerococcus (p = 0.005), Caulobacter (p = 0.004), Macrococcus (p = 0.004), Phenylobacterium (p = 0.004), and Acinetobacter (p = 0.004) were less abundant with caries than cavity free group (Table 3; Fig. 2a).
Table 3.
List of differentially abundant genera in boys in caries group compared with cavity free group.
| OTUs | Genus | log2FoldChange | p-value | Adjusted p-value |
|---|---|---|---|---|
| Otu000007 | Rothia | + 11.094 | 0.001 | < 0.001 |
| Otu000006 | Neisseria | + 10.735 | 0.001 | < 0.001 |
| Otu000008 | Haemophilus | + 10.563 | < 0.001 | < 0.001 |
| Otu000130 | Paludibacter | + 1.556 | < 0.001 | < 0.001 |
| Otu000176 | Labrenzia | + 1.189 | < 0.001 | < 0.001 |
| Otu000222 | Shuttleworthia | − 1.379 | 0.010 | 0.012 |
| Otu000492 | Mogibacterium | − 3.329 | < 0.001 | 0.003 |
| Otu000349 | Brachymonas | − 3.964 | 0.012 | 0.013 |
| Otu000748 | Selenomonas | − 4.228 | 0.005 | 0.006 |
| Otu004001 | Stenotrophomonas | − 5.577 | 0.002 | 0.004 |
| Otu000804 | Pelomonas | − 6.145 | 0.002 | 0.004 |
| Otu001818 | Acinetobacter | − 6.173 | 0.002 | 0.004 |
| Otu002378 | Phenylobacterium | − 6.247 | 0.002 | 0.004 |
| Otu001497 | Macrococcus | − 6.353 | 0.002 | 0.004 |
| Otu005048 | Caulobacter | − 6.577 | 0.002 | 0.004 |
| Otu005348 | Anaerococcus | − 7.577 | 0.004 | 0.005 |
log2FoldChange value (+) means higher and (–) means lower abundancy in caries group compared with cavity free group.
Figure 2.
Heat tree analysis leverages the hierarchical structure of taxonomic classifications to quantitatively (using median abundance) and statistically (using non-parametric Wilcoxon Rank Sum test) depict taxonomic differences between microbial communities in boys (a) and girls (b). The image is drawn using web-based platform MicrobiomeAnalyst27 (https://www.microbiomeanalyst.ca/) and trees are automatically created and arranged when there are multiple roots to the taxonomy28.
Similarly, in girls, the five top genera Prevotella, Selenomonas, Actinomyces, Paludibacter and Labrenzia were highly abundant in the caries group (adjusted p-value < 0.001), while Hyphomicrobium, Bdellovibrio, Weissella, Alistipes, and Xylanibacter were less abundant with caries than cavity free group (adjusted p-value < 0.001) (Table 4; Fig. 2b).
Table 4.
List of differentially abundant genera in girls in caries group compared with cavity free group.
| OTUs | Genus | log2FoldChange | p-value | Adjusted p-value |
|---|---|---|---|---|
| Otu000003 | Prevotella | + 11.482 | < 0.001 | < 0.001 |
| Otu000009 | Selenomonas | + 10.168 | < 0.001 | < 0.001 |
| Otu000033 | Actinomyces | + 7.182 | < 0.001 | < 0.001 |
| Otu000130 | Paludibacter | + 1.719 | < 0.001 | < 0.001 |
| Otu000176 | Labrenzia | + 0.853 | < 0.001 | < 0.001 |
| Otu001039 | Rhodanobacter | − 3.719 | < 0.001 | < 0.001 |
| Otu001497 | Macrococcus | − 4.219 | 0.006 | 0.007 |
| Otu000648 | Dolosigranulum | − 4.249 | 0.001 | 0.002 |
| Otu000804 | Pelomonas | − 4.912 | 0.002 | 0.003 |
| Otu002174 | Leuconostoc | − 6.804 | < 0.001 | 0.001 |
| Otu002378 | Phenylobacterium | − 6.804 | < 0.001 | 0.001 |
| Otu003167 | Xylanibacter | − 6.804 | < 0.001 | 0.001 |
| Otu004423 | Alistipes | − 6.804 | < 0.001 | 0.001 |
| Otu005632 | Weissella | − 7.554 | < 0.001 | 0.001 |
| Otu005084 | Bdellovibrio | − 7.564 | < 0.001 | 0.001 |
| Otu004975 | Hyphomicrobium | − 7.627 | < 0.001 | 0.001 |
log2FoldChange value (+) means higher and (–) means lower abundancy in caries group compared with cavity free group.
Taken together, the shared saliva microbes found with greater abundancy in both sexes were Paludibacter and Labrenzia in the caries group than in the cavity free group, while, Phenylobacterium, Macrococcus and Pelomonas were less abundant. On the other hand, Selenomonas showed an inconsistent abundancy in boys and girls.
Discussion
This study employed 16S rRNA amplicon sequencing to characterize the saliva microbial communities in preadolescent children with and without history of caries in permanent dentition. In total, the history of cavitated caries lesions was recorded in 34% of our participants. Girls were somewhat overrepresented in the caries group. Sex was a stronger determinant of the saliva microbiota than caries. Caries was associated with differences in the abundance of several taxa but not with overall microbial composition or diversity. Paludibacter and Labrenzia were the key caries-related taxa inhabitants both in preadolescent boys and girls, suggesting a possible pathogenic role of these genera in caries development.
Paludibacter is an anaerobic, chemoorganotrophic bacterium in the phylum Bacteroidetes. Recently, this genus was found in the subgingival plaque of periodontally healthy individuals29. Similar to other common saliva microbiota (such as Fusobacterium and Gemella), Paludibacter also helps in energy metabolism pathways and cell motility30. Labrenzia is a genus of bacteria from the phylum Proteobacteria and only a few species have been identified in this genus. Labrenzia is mainly found in soil and marine environments31. We believe the genus Labrenzia has not been linked with caries before. Along with other acid producers, Labrenzia are assumed to enhance salivary acidification, which contributes to the progression of dental caries. Paludibacter and Labrenzia are the two shared saliva microbes positively associated with caries in our study both in boys and girls.
The aetiology of caries is complex and alterations in the saliva microbiota are associated with the development of caries in children9,23,24. We observed minor differences in the abundance of several taxa between caries and cavity free groups in sex-stratified analysis. Sex was the major determinant of saliva microbiota composition; we are unaware if previous studies have considered sex in their analysis. Similar to our findings, Jiang et al.32 also found minor differences in the abundance of several species in caries and cavity free groups in children (3–4 years old) without formal statistical significance. However, they did not observe any difference in salivary microbiota diversity or composition between boys and girls32. In our study, beta diversity was similar between caries and cavity free groups. In subjects 60 years and older, Jiang et al.22 observed differences in beta diversity between caries and cavity free groups which is inconsistent with our study22. However, most of previous studies included very small sample sizes and were performed at different geographic locations9,22,24,32. Evidently, we have conducted the largest study on saliva microbiota and the history of cavitated caries lesions in children.
The sex-stratified analysis revealed potential pathogens, suggesting similar pathogenesis of caries in girls and boys. The timing of dental development differs slightly between boys and girls33. However, our results are consistent with other studies that demonstrate that caries is more common in girls than in boys34,35. Ortiz et al.25 examined the sex-specific differences in the saliva microbiota in children with active caries and identified several species (including Rothia aeria, Neisseria flavescens, and Haemophilus pittmaniae) that were higher in boys with active caries. Our findings are in line with these observations. While the genus Rothia is a key member of the saliva microbiota in persons with caries, its role is largely unknown36,37. The majority of the Neisseria species are assumed to be acid producers and therefore enhance the development of a cariogenic environment25. We found that Haemophilus was higher in boys with caries; however, the genus Haemophilus was also reported in the saliva microbiota in those with low levels of caries38. We found that the genera Phenylobacterium, Macrococcus and Pelomonas were less abundant in the caries group than in the cavity free group in both sexes. Phenylobacterium are strictly aerobic non-motile bacteria that belong to Caulobacteraceae family, and the role of Phenylobacterium species in saliva microbiota has not been elucidated yet39. Macrococcus genus belongs to the family Staphylococcaceae, and is closely related to the genus Staphylococcus, and this genus is considered nonpathogenic40. We have previously reported that Macrococcus were less abundant in the saliva of overweight children compared with normal weight children2. Pelomonas is a genus from the family Comamonadaceae and the abundance of Pelomonas has shown to decrease with age in the microbiota of gingival crevicular fluid and tongue back in healthy individuals41. Our finding on Selenomonas is discordant between sexes. Ortiz et al.25 showed that Selenomonas sps were associated with caries in girls, and a notably greater abundance of Selenomonas Genus probe 1 was observed with less frequent teeth brushing in adolescents with caries20, which is in line with our finding in girls. Taken together, we found that most of the caries-associated taxa were acid producers that cause demineralisation of tooth enamel. Accordingly, children with a greater abundance of acid producers may be more prone to caries development.
In addition to oral hygiene, frequent consumption of dietary sugars is the leading cause of caries; bacteria in the oral cavity break down dietary sugars and produce acids that destroy tooth enamel, slowly leading to tooth decay42. Our results highlight dissimilarly of saliva microbiota between sexes, while support similar taxa related to cavitated caries lesions. Tooth-brushing and eating habits were not considered here, although they may differ between girls and boys, as reported before 20,43. Moreover, variation in the quality of tooth enamel, saliva flow and composition may also contribute to gender differences in saliva microbiota as well as in caries44,45. Thus, further studies are needed to address their effects on caries.
The strengths of the present study include a large sample size, the inclusion of a homogeneous age group of both sexes, and availability of primary healthcare visit data collected in dental appointments. Caries was diagnosed by a dental professional and was not self-reported as in many other studies46,47. However, our study has several limitations. First, the nature of the study is descriptive due to a cross-sectional study design. From the randomly selected 1000 saliva samples, we limited our material to 617 based on the available records on dental examinations within 12 months. The mean time difference between the saliva sampling and dental appointment was 4.4 months, which we consider a fairly representative timeframe. Our study compared groups with or without history of cavitated caries lesions, not caries activity, in relatively low caries-risk paediatric population, thus a wider timeframe was considered justified. Secondly, participants in the present study were children with late mixed or fully permanent dentition. The eruption schedule of permanent teeth is thoroughly studied and well-known48; deciduous teeth are typically exfoliated at 6 to 12 years of age and during this time children have a mixture of permanent and deciduous teeth. Since records on deciduous teeth were limited, the analyses were based on permanent dentition. Hence, early caries experience in deciduous teeth was not included, which may affect our results. Moreover, information on tooth brushing, smoking and other dental hygiene habits is not known.
Dental health measured with the DMFT index has improved among Finnish children since the 1990s49. The mean DMFT index values were 0.7 in 12-year-olds in 2009 and our observation is in agreement with this49. For our study, the DMFT index was available as a composite variable, which is an indicator of individual’s history of cavitated caries lesions rather than current caries activity. This discrepancy and the overall usability of the DMFT index has been widely discussed50, and more elaborate indexes have been suggested, such as the T-Health index51. For periodontal or gingival health status, information was obtainable solely as CPITN index scores. Since actual diseases of the periodontium are rare in this age group, we dichotomized the variable and considered it as a reflection of the level of dental hygiene. Finally, our microbiota analysis was limited to the genus level. However, species-level identification would provide a more precise identity in terms of potential biomarkers related to caries. We demonstrated that Paludibacter and Labrenzia are the ‘keystone pathogens’ that influence the caries process by altering the ‘healthy’ microbiota to a disease state. Such observation have also been reported in periodontal disease52.
To conclude, cavitated caries lesions in permanent dentition is associated with relatively minor changes in the saliva microbiota in preadolescents with relatively good dental health. We identified Paludibacter and Labrenzia as potential biomarkers of caries; both are sugar metabolisers. Paludibacter and Labrenzia likely enhance salivary acidification, which contributes to the progression of dental caries. However, the clinical relevance of our findings warrants further studies.
Materials and methods
Study design
This study utilized saliva samples from the Finnish Health in Teens (Fin-HIT) study, which includes approximately 11,400 children aged mainly 9–12 years. Data were collected between 2011 and 2014 mostly in schools across Finland. The study protocol has been described in detail elsewhere53. Altogether 1000 samples were randomly selected from this cohort to obtain an unbiased representation of the population. Age and native language (Finnish, Swedish or other) were reported by the participants or their parents. BMI z-score was calculated based on measured weight and height. Participants with antibiotic use during the 3 months prior to sampling54 (n = 31) or without sufficient information on oral health (n = 255) were excluded. After saliva processing, 25 participants withdrew their consent. Furthermore, after processing the microbiota we found that 72 samples had low sequence depth, which may influence the diversity (< 10,000 sequences). These samples were also excluded. The Fin-HIT study protocol was approved by the Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa in Finland (169/13/03/00/10). Written informed consent was obtained from the children and one of their parents. All study procedures were performed in accordance with the Helsinki Declaration.
Oral health
Data on oral health variables were collected from Avohilmo maintained by the Finnish Institute for Health and Welfare (THL) (https://thl.fi/en/web/thlfi-en). Variables regarding the prevalence and history of dental caries were scores of decayed (D) permanent teeth, missing (M) permanent teeth due to caries, and filled (F) permanent teeth. The information on caries status was collected from dental appointments closest to the saliva sampling within 12 months. The mean time difference between the saliva sample collection and dentist appointment was 4.4 (SD 2.25) months. The DMFT index was summarised based on caries status scores in permanent dentition50,55, and the composite DMFT index was used as the main outcome. Children were classified into the following two groups: DMFT value ≥ 1 was considered as ‘caries’ and DMFT = 0 as ‘cavity free’. The final study population consisted of 617 participants (n = 208 for caries and n = 409 for cavity free).
Gingival health status was registered in Avohilmo records with the CPITN as reported by Ainamo et al.56; this was used as an indicator of dental hygiene status. We categorized the study participants in two groups by their highest CPITN value; CPITN value = 0 was considered as ‘no oral hygiene risk’ and CPITN value 1–2 indicated ‘elevated risk regarding oral hygiene’.
16S rRNA gene sequencing and processing of sequencing data
We have previously reported associations of saliva microbiota composition and diversity with weight status2, meal regularity57, breast feeding58, and lifelong antimicrobial purchases54. Saliva samples (unstimulated, up to 2 ml) were collected from each participant in Oragene-DNA (OG-500) self-collection kits (DNA Genotek Inc., Canada) containing a stabilizing reagent and transported to the laboratory for DNA extraction. The samples were lysed using 50 ml lysozyme (10 mg/ml, Sigma-Aldrich), 6 ml mutanolysin (25 KU/ml, Sigma-Aldrich), and 3 ml lysostaphin (4000 U/ml, Sigma-Aldrich), after which a 500-ml aliquot of cell suspension was added and further incubated for 1 h at 37 °C. After lysis, genomic DNA was extracted using a CMG-1035 saliva kit and Chemagic MSM1 nucleic acid extraction robot (PerkinElmer)2,54. 16S rRNA amplification was performed with an in-house protocol as described previously59. The V3—V4 variable regions were amplified using primers reported previously60. DNA quantity was assessed using an Agilent 2100 Bioanalyzer and PCR products were processed for paired-end sequencing (2 × 270 bp) on a HiSeq1500 platform (Illumina, CA, USA).
The paired-end reads were merged together to reconstruct full-length sequences using mothur pipeline (Version v.1.35.1). Sequencing quality was performed and further processed using the MiSeq SOP61. The sequence reads containing ambiguous bases (N), homopolymer stretches (> 8 bases), and small reads (< 330 bases) were removed using mothur pipeline62. UCHIME algorithm incorporated in the mothur was used to remove chimeric sequences63. The high-quality sequence reads were then aligned to the Silva 16S rRNA reference database64 (Ver V119) and clustered at > 98% homology to identify Operational Taxonomic Units (OTUs). The most abundant bacterial taxa were recognized at the genus level. Alpha diversity indexes (Shannon index and Invsim index) were calculated to illustrate the microbiota diversity and richness in each sample. Beta diversity was calculated using Bray Curtis dissimilarity index. These were performed using the R-package ‘vegan’ (version 2.5-6)65.
Statistical analysis
The normality of the distributions was visually examined, and appropriate tests were used for the analysis. Independent samples t-test (for continuous variables) and Fisher’s exact test (for categorical variables) were performed to examine differences between the caries and cavity free groups. Permutational analysis of variance (PERMANOVA) test was used to test the differences in microbial community composition between individuals with caries vs cavity free using the adonis and betadisper function in Phyloseq R-package (version 1.32.0). Differentially abundant OTUs were identified at the genus taxonomic level using DESeq2 incorporated in Phyloseq R-package (version 1.32.0). The results are presented with mean (SD) unless indicated otherwise. All statistical analyses were conducted using IBM SPSS for Windows, version 20 (IBM, Chicago, IL, USA). The statistical significance level was set at 5%. p-values were calculated and adjusted by the false discovery rate (FDR).
Supplementary information
Acknowledgements
We acknowledge all the children and parents who participated in the study and all fieldworkers for data collection and technical assistance.
Author contributions
Conceptualization: M.M., J.F., H.V.; Data curation: M.M., S.R., C.S., H.V.; Formal analysis: M.M., S.L., S.R., H.V.; Funding acquisition: M.M., C.S., H.V.; Investigation: M.M., S.L., S.R., H.V.; Methodology: M.M., J.F., S.L., E.E., H.V.; Project administration: C.S., E.E.; Supervision: H.V.; Visualization: M.M., S.L., H.V.; Writing—original draft: M.M., H.V.;Writing—review & editing: M.M., S.L., J.F., C.S., E.E., S.R., H.V.
Funding
This project received financial support from the Academy of Finland (Grant No. 332459), the Swedish Cultural Foundation in Finland, the Päivikki and Sakari Sohlberg Foundation, the Minerva Foundation / Selma and Maja-Lisa Selander Fund, and the Folkhälsan Research Foundation. The funders had no role in study design, data collection and analysis, preparation of the manuscript or decision to publish.
Data availability
All relevant data are within the manuscript and its Supporting Information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-83846-1.
References
- 1.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raju SC, et al. Gender-Specific Associations Between Saliva Microbiota and Body Size. Front. Microbiol. 2019;10:767. doi: 10.3389/fmicb.2019.00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xun Z, Zhang Q, Xu T, Chen N, Chen F. Dysbiosis and ecotypes of the salivary microbiome associated with inflammatory bowel diseases and the assistance in diagnosis of diseases using oral bacterial profiles. Front. Microbiol. 2018;9:1136. doi: 10.3389/fmicb.2018.01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francavilla R, et al. Salivary microbiota and metabolome associated with celiac disease. Appl. Environ. Microbiol. 2014;80:3416–3425. doi: 10.1128/AEM.00362-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rusthen S, et al. Dysbiotic salivary microbiota in dry mouth and primary Sjögren’s syndrome patients. PLoS ONE. 2019;14:e0218319. doi: 10.1371/journal.pone.0218319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topcuoglu N, Erdem AP, Karacan I, Kulekci G. Oral microbial dysbiosis in patients with Kostmann syndrome. J. Med. Microbiol. 2019;68:609–615. doi: 10.1099/jmm.0.000964. [DOI] [PubMed] [Google Scholar]
- 7.Llena-Puy C. The rôle of saliva in maintaining oral health and as an aid to diagnosis. Med. Oral. Patol. Oral. Cir. Bucal. 2006;11:449–455. [PubMed] [Google Scholar]
- 8.Hemadi AS, Huang R, Zhou Y, Zou J. Salivary proteins and microbiota as biomarkers for early childhood caries risk assessment. Int. J. Oral Sci. 2017 doi: 10.1038/ijos.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L, et al. Dynamic alterations in salivary microbiota related to dental caries and age in preschool children with deciduous dentition: a 2-year follow-up study. Front. Physiol. 2018 doi: 10.3389/fphys.2018.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedersen AML, Belstrøm D. The role of natural salivary defences in maintaining a healthy oral microbiota. J. Dent. 2019;80:S3–12. doi: 10.1016/j.jdent.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Sultan AS, Kong EF, Rizk AM, Jabra-Rizk MA. The oral microbiome: a Lesson in coexistence. PLOS Pathog. 2018;14:e1006719. doi: 10.1371/journal.ppat.1006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selwitz RH, Ismail AI, Pitts NB. Dental caries. Lancet. 2007;369:51–59. doi: 10.1016/S0140-6736(07)60031-2. [DOI] [PubMed] [Google Scholar]
- 13.Machiulskiene V, et al. Terminology of dental caries and dental caries management: consensus report of a workshop organized by ORCA and cariology research group of IADR. Caries Res. 2020;54:7–14. doi: 10.1159/000503309. [DOI] [PubMed] [Google Scholar]
- 14.Petersen PE. Challenges to improvement of oral health in the 21st century—the approach of the WHO Global Oral Health Programme. Int Dent J. 2004;54:329–343. doi: 10.1111/j.1875-595X.2004.tb00009.x. [DOI] [PubMed] [Google Scholar]
- 15.Metwalli KH, Khan SA, Krom BP, Jabra-Rizk MA. Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog. 2013;9:e1003616. doi: 10.1371/journal.ppat.1003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forssten SD, Björklund M, Ouwehand AC. Streptococcus mutans, caries and simulation models. Nutrients. 2010;2:290–298. doi: 10.3390/nu2030290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nase L, et al. Effect of long-term consumption of a probiotic bacterium, Lactobacillus rhamnosus GG, in milk on dental caries and caries risk in children. Caries Res. 2001;35:412–420. doi: 10.1159/000047484. [DOI] [PubMed] [Google Scholar]
- 18.Beighton D, et al. Oral bifidobacteria: caries-associated bacteria in older adults. J. Dent Res. 2010;89:970–974. doi: 10.1177/0022034510369319. [DOI] [PubMed] [Google Scholar]
- 19.Takeshita T, et al. Bacterial diversity in saliva and oral health-related conditions: the Hisayama Study. Sci. Rep. 2016;6:22164. doi: 10.1038/srep22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eriksson L, Lif Holgerson P, Johansson I. Saliva and tooth biofilm bacterial microbiota in adolescents in a low caries community. Sci. Rep. 2017;7:5861. doi: 10.1038/s41598-017-06221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupf S, et al. Comparison of initial oral microbiomes of young adults with and without cavitated dentin caries lesions using an in situ biofilm model. Sci. Rep. 2018;8:14010. doi: 10.1038/s41598-018-32361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Q, Liu J, Chen L, Gan N, Yang D. The oral microbiome in the elderly with dental caries and health. Front. Cell Infect. Microbiol. 2019 doi: 10.3389/fcimb.2018.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vieira AR, et al. Profiling microorganisms in whole saliva of children with and without dental caries. Clin Exp Dent Res. 2019;5:438–446. doi: 10.1002/cre2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurley E, et al. Comparison of the salivary and dentinal microbiome of children with severe-early childhood caries to the salivary microbiome of caries-free children. BMC Oral Health. 2019 doi: 10.1186/s12903-018-0693-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortiz S, et al. Sex-specific differences in the salivary microbiome of caries-active children. J. Oral Microbiol. 2019 doi: 10.1080/20002297.2019.1653124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minty M, et al. Gender-associated differences in oral microbiota and salivary biochemical parameters in response to feeding. J. Physiol Biochem. 2020 doi: 10.1007/s13105-020-00757-x. [DOI] [PubMed] [Google Scholar]
- 27.Chong J, Liu P, Zhou G, Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020;15:799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 28.Foster ZS, Sharpton TJ, Grünwald NJ. Metacoder: an R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 2017;13(2):e1005404. doi: 10.1371/journal.pcbi.1005404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, et al. Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J. 2018;12:1210–1224. doi: 10.1038/s41396-017-0037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi W, Tian J, Xu H, Zhou Q, Qin M. Distinctions and associations between the microbiota of saliva and supragingival plaque of permanent and deciduous teeth. PLoS ONE. 2018;13:e0200337. doi: 10.1371/journal.pone.0200337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghani NA, Norizan SNM, Chan XY, Yin WF, Chan KG. Labrenzia sp. BM1: a quorum quenching bacterium that degrades N-acyl homoserine lactones via lactonase activity. Sensors. 2014;14:11760–11769. doi: 10.3390/s140711760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang S, Gao X, Jin L, Lo ECM. Salivary microbiome diversity in caries-free and caries-affected children. Int. J. Mol. Sci. 2016 doi: 10.3390/ijms17121978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demirjian A, Levesque GY. Sexual differences in dental development and prediction of emergence. J. Dent Res. 1980;59:1110–1122. doi: 10.1177/00220345800590070301. [DOI] [PubMed] [Google Scholar]
- 34.Cheng YH, Liao Y, Chen DY, Wang Y, Wu Y. Prevalence of dental caries and its association with body mass index among school-age children in Shenzhen, China. BMC Oral Health. 2019;19:270. doi: 10.1186/s12903-019-0950-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferraro M, Vieira AR. Explaining gender differences in caries: a multifactorial approach to a multifactorial disease. Int. J. Dent. 2010 doi: 10.1155/2010/649643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jagathrakshakan SN, Sethumadhava RJ, Mehta DT, Ramanathan A. 16S rRNA gene-based metagenomic analysis identifies a novel bacterial co-prevalence pattern in dental caries. Eur. J. Dent. 2015;9:127–132. doi: 10.4103/1305-7456.149661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao C, Ran S, Huang Z, Liang J. Bacterial diversity and community structure of supragingival plaques in adults with dental health or caries revealed by 16S pyrosequencing. Front. Microbiol. 2016 doi: 10.3389/fmicb.2016.01145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belstrøm D, et al. Salivary microbiota in individuals with different levels of caries experience. J. Oral Microbiol. 2017 doi: 10.1080/20002297.2016.1270614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberspächer, J., Lingens, F. The genus Phenylobacterium. The prokaryotes. New York. 10.1007/0-387-30745-1_13 (2006).
- 40.Götz F, Bannerman T, Schleifer KH. The genera staphylococcus and macrococcus. The prokaryotes. New York: Springer; 2006. [Google Scholar]
- 41.Liu S, Wang Y, Zhao L, Sun X, Feng Q. Microbiome succession with increasing age in three oral sites. Aging. 2020;12:7874. doi: 10.18632/aging.103108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moynihan P. Sugars and dental caries: Evidence for setting a recommended threshold for intake. Adv. Nutr. 2016;7:149–156. doi: 10.3945/an.115.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuusela S, Honkala E, Kannas L, Tynjala J, Wold B. Oral hygiene habits of 11-year-old schoolchildren in 22 European countries and Canada in 1993/1994. J. Dent Res. 1997;76:1602–1609. doi: 10.1177/00220345970760091301. [DOI] [PubMed] [Google Scholar]
- 44.Percival RS, Challacombe SJ, Marsh PD. Flow rates of resting whole and stimulated parotid saliva in relation to age and gender. J. Dent. Res. 1994;73:1416–1420. doi: 10.1177/00220345940730080401. [DOI] [PubMed] [Google Scholar]
- 45.Lukacs JR. Sex differences in dental caries experience: clinical evidence, complex etiology. Clin. Oral Invest. 2011;15:649–656. doi: 10.1007/s00784-010-0445-3. [DOI] [PubMed] [Google Scholar]
- 46.Lintula T, et al. Self-reported oral health and associated factors in the North Finland 1966 birth cohort at the age of 31. BMC Oral Health. 2014 doi: 10.1186/1472-6831-14-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mattila A, et al. Self-reported oral health and use of dental services among asylum seekers and immigrants in Finland—a pilot study. Eur. J. Public Health. 2016;26:1006–1010. doi: 10.1093/eurpub/ckw116. [DOI] [PubMed] [Google Scholar]
- 48.Wise GE, King GJ. Mechanisms of tooth eruption and orthodontic tooth movement. J. Dent Res. 2008;87:414–434. doi: 10.1177/154405910808700509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Widström E, Järvinen S. Caries prevalence and use of dental services in Finnish children and adolescents in 2009. Oral. Health Dent. Manag. 2011;10:185–192. [Google Scholar]
- 50.Larmas M. Has dental caries prevalence some connection with caries index values in adults? Caries Res. 2010;44:81–84. doi: 10.1159/000279327. [DOI] [PubMed] [Google Scholar]
- 51.Bernabe E, Suominen-Taipale AL, Vehkalahti M, Nordblad A, Sheiham A. The T-Health index: a composite indicator of dental health. Eur. J. Oral Sci. 2009;117:385–389. doi: 10.1111/j.1600-0722.2009.00649.x. [DOI] [PubMed] [Google Scholar]
- 52.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Figueiredo RADO, et al. Cohort profile: the Finnish health in teens (Fin-HIT) study: a population-based study. Int. J. Epidemiol. 2019;48:22–24H. doi: 10.1093/ije/dyy189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raju SC, et al. Antimicrobial drug use in the first decade of life influences saliva microbiota diversity and composition. Microbiome. 2020;8:121. doi: 10.1186/s40168-020-00893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klein H, Palmer CE, Knutson JW. Studies on dental caries: I. Dental status and dental needs of elementary school children. Public Heal Rep. 1938;53:751–765. doi: 10.2307/4582532. [DOI] [Google Scholar]
- 56.Ainamo J, Barmes D, Beagrie G, Cutress T, Martin J, Sardo-Infirri J. Development of the World Health Organization (WHO) community periodontal index of treatment needs (CPITN) Int. Dent J. 1982;32:281–291. [PubMed] [Google Scholar]
- 57.Viljakainen J, et al. Meal regularity plays a role in shaping the saliva microbiota. Front. Microbiol. 2020;11:757. doi: 10.3389/fmicb.2020.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eshriqui I, Viljakainen HT, Ferreira SRG, Raju SC, Weiderpass E, Figueiredo RAO. Breastfeeding may have a long-term effect on oral microbiota: results from the Fin-HIT cohort. Int. Breastfeed. J. 2020;15:42. doi: 10.1186/s13006-020-00285-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raju SC, et al. Reproducibility and repeatability of six high-throughput 16S rDNA sequencing protocols for microbiota profiling. J. Microbiol. Methods. 2018;147:76–86. doi: 10.1016/j.mimet.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 60.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013 doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010 doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 63.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.