Abstract
Objective
Antimicrobial peptides (AMPs) play essential roles in maintaining gut health and are associated with IBD. This study is to elucidate the effect of angiogenin (ANG), an intestine-secreted AMP, on gut microbiota and its relevance with IBD.
Design
The effect of ANG on microbiota and its contribution to colitis were evaluated in different colitis models with co-housing and faecal microbiota transplantation. ANG-regulated bacteria were determined by 16S rDNA sequencing and their functions in colitis were analysed by bacterial colonisation. The species-specific antimicrobial activity of ANG and its underlying mechanism were further investigated with microbiological and biochemical methods. ANG level and the key bacteria were characterised in IBD faecal samples.
Results
ANG regulated microbiota composition and inhibited intestinal inflammation. Specifically, Ang1 deficiency in mice led to a decrease in the protective gut commensal strains of Lachnospiraceae but an increase in the colitogenic strains of α-Proteobacteria. Direct binding of ANG to α-Proteobacteria resulted in lethal disruption of bacterial membrane integrity, and consequently promoted the growth of Lachnospiraceae, which otherwise was antagonised by α-Proteobacteria. Oral administration of ANG1 reversed the dysbiosis and attenuated the severity of colitis in Ang1-deficient mice. The correlation among ANG, the identified bacteria and IBD status was established in patients.
Conclusion
These findings demonstrate a novel role of ANG in shaping gut microbe composition and thus maintaining gut health, suggesting that the ANG-microbiota axis could be developed as a potential preventive and/or therapeutic approach for dysbiosis-related gut diseases.
INTRODUCTION
A healthy intestine is inhabited by trillions of bacteria and has evolved a fine-tuned communication between host and microbe.1,2 In contrast, microbial imbalance, referred to as ‘dysbiosis’, is associated with IBD, including UC and Crohn’s disease (CD), yet the treatment of IBD is limited to immunotherapy. Hence, understanding the host–microbe interactions that contribute to colitis holds promise for developing novel microbiome-based therapeutic and even preventive options.
The immune system participates in the organisation of a healthy host–microbe interface, through mechanisms such as production of IgA, secretion of mucus and induction of antimicrobial peptides (AMPs).3 AMPs are produced by the host during symbiotic interaction with microbiota. An important function of AMPs is to establish the host–microbe homeostasis rather than to eliminate the microbial symbiont population.4 For example, α-defensin 5 and cathelin-related antimicrobial peptide (CRAMP), two major AMPs in mammals, are known to shape the composition of the intestinal bacterial community and maintain mucosal homeostasis.5,6 Given the crucial contribution of AMPs to microorganism homeostasis, dysregulation of AMP production or function is associated with intestinal diseases such as IBD.7 Currently, it is recognised that AMPs display multiple biological activities and influence the host–microbe interplay through various mechanisms, such as direct killing of bacterium by disrupting its cell membrane, prevention of bacterial invasion by suppressing motility of flagellated bacterium or modulation of innate immune response.8,9
Angiogenin (ANG), originally isolated as a tumour angiogenic factor,10 is also recognised as an AMP.11 To date, one ANG gene has been identified in human but five paralogs (Ang1, Ang2, Ang4, Ang5 and Ang6) have been documented in mouse, as the result of tandem duplication of ancient Ang gene (the human ANG ortholog).12 Among the proteins expressed, human ANG, mouse ANG1 and ANG4 exhibit broad-spectrum antimicrobial activities against bacteria and fungi by disrupting cell membranes or by degrading cellular RNAs as demonstrated in vitro.13 However, the in vivo role of ANG in intestinal microbial ecology and its contribution to gut health are largely unknown.
To explore the antimicrobial activity of ANG in vivo, we employed mice knockout of Ang1, which shares the same gene structure and expression pattern with human ANG (online supplementary figure S1A,B),14,15 and found that Ang1-deficient mice exhibited gut dysbiosis and an increased susceptibility to colitis induction. Further experiments revealed that ANG maintained a proper ratio of α-Proteobacteria to Lachnospiraceae by directly inhibiting the α-Proteobacteria strains, which in turn favoured the growth of Lachnospiraceae strains. Oral ANG1 gavage was able to restore gut microbe composition and mitigate the severity of colitis, reinforcing the role of ANG in maintaining healthy gut microbial ecology.
MATERIALS AND METHODS
Mice
Ang1−/− mice and their wild-type (WT) control were all generated from the same heterozygous Ang1+/− parents. Littermates were randomly assigned to experimental groups. All mice were maintained in pathogen-free conditions at the Laboratory Animal Centre of Zhejiang University (ZJU), and provided with water and a standard laboratory diet ad libitum except noted otherwise. All animal studies were performed in compliance with the guide for the care and use of laboratory animals and adopted the protocol that has been approved by the Medical Experimental Animal Care Commission of ZJU.
Human subjects
The faecal samples of IBD patients and healthy subjects were collected from the Inflammatory Bowel Disease Centre, Sir Run Run Shaw Hospital affiliated to ZJU School of Medicine. Fresh stool samples were frozen at −20°C within 2 hour of collection, transported on dry ice to laboratory and stored at −80°C freezer for further analysis within 1 week. CD or LTC diagnosis was based on a standard combination of clinical, endoscopic, histological and radiological criteria. The Partial Mayo Score (PMS) for UC and the Harvey-Bradshaw Index (HBI) for CD were used to assess disease status. Active disease was defined as a PMS >2 in UC and HBI >5 in CD. Colitis severity was assessed with pathological evidence of mucosal inflammation. Basic information about patients and healthy subjects was summarised in online supplementary table S1.
Statistical analysis
All analyses were performed with GraphPad Prism 8 (GraphPad Software). Data were presented as mean with SEM or violin plot. The difference between two groups was assessed using paired or unpaired Student’s t-test. The difference between variables was assessed by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test. The significant separation of the microbiome composition was assessed by analysis of similarities test. The correlation between two bacterial abundance was assessed by linear regression analysis. P value <0.05 was considered statistically significant; p value results were denoted by asterisks in the figures (*p<0.05; **p<0.01; ***p<0.001 and ****p<0.0001).
RESULTS
ANG inhibits colitis through maintaining healthy gut microbiota
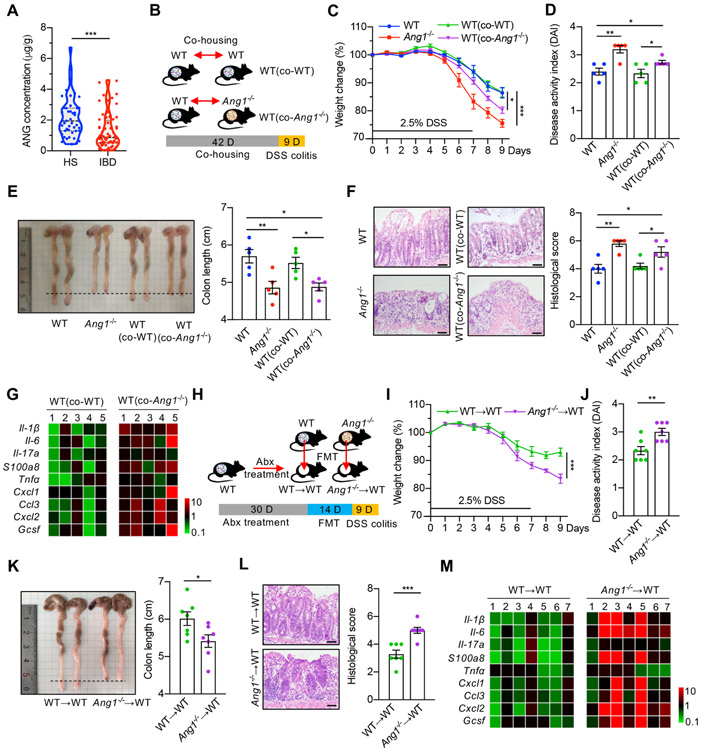
To determine if ANG is clinically relevant to human colitis, we compared faecal ANG level between 45 healthy subjects and 64 IBD patients (online supplementary table S1), and found that the level was 1.31±0.15 μg/g in disease group, which was 1.6-fold lower (p<0.001) than the amount of 2.17±0.20 μg/g in the control group (figure 1A). Consistent with deceased ANG expression in IBD patients, faecal ANG1 level progressively declined in WT mice on administration with dextran sodium sulphate (DSS) (online supplementary figure S1C). Moreover, Ang1−/− mice displayed more severe colitis than their WT counterparts in response to DSS treatment (figure 1C-F). These findings, together with the knowledge that ANG is antimicrobial11 and is highly expressed in Paneth cells (online supplementary figure S1D,E), prompted us to hypothesise that ANG suppresses intestinal inflammation by regulating certain gut microbes. Therefore, we conducted microbiota-intervention experiments. WT mice were co-housed with age-and-gender-matched Ang1−/− mice (WT (co-Ang1−/−)) or with the WT littermates (WT (co-WT)) derived from Ang1+/− parents for 6 weeks before treatment with 2.5% DSS (figure 1B). WT (co-Ang1−/−) mice developed more severe inflammation than non-co-housed WT or WT (co-WT) ones as judged by weight loss, Disease Activity Index (DAI), colon length, histological score and cytokine expression (figure 1C-G), suggesting that the enhanced colitis in WT (co-Ang1−/−) mice might be caused by changes in gut microbiota through consumption of faeces from Ang1−/− mice. To confirm this possibility, we transferred faeces from age-and-gender-matched WT or Ang1−/− mice into the gastrointestinal tract of antibiotic-treated WT mice via oral gavage every other day for 2 weeks (figure 1H). WT mice receiving faeces from Ang1−/− mice developed more severe colitis as compared with those receiving faeces from WT littermates (figure 1I-M).
Figure 1.
Ang1 deficiency exacerbates colitis. (A) ANG concentrations in the faeces from healthy subjects (HS, n=45) or IBD patients (n=64). (B) The design of co-housing experiment. (C) Body weight of WT, Ang1−/−, WT (co-WT) or WT (co-Ang1−/−) mice during DSS-induced colitis, n=5. (D–F) Disease Activity Index (DAI) (D), colon length (E) and histological score (F) of WT, Ang1−/−, WT (co-WT) or WT (co-Ang1−/−) mice on day 9 after DSS induction, n=5. Scale bar, 50 μm. (G) Expression levels of cytokines in the colons of WT (co-WT) or WT (co-Ang1−/−) mice. The heatmap represents the expression of indicated genes in each mouse after normalised to the average of WT (co-WT) group, n=5. (H) Diagram of gut microbiota transplantation experiment. Abx: antibiotic cocktail. (I) Body weight of WT→WT or Ang1−/−→WT mice during DSS-induced colitis, n=7. (J–L) DAI (J), colon length (K) and histological score (L) of WT→WVT or Ang1−/−→WT mice on day 9 after DSS induction, n=7. Scale bar, 50 μm. (M) Expressions of indicated cytokines in the colons of WT→WT or Ang1−/−→WT mice. The heatmap represents the expression of indicated genes in each mouse after normalised to the average of WT→WT group, n=7. Data are presented as mean±SEM; *p<0.05; **p<0.01; ***p<0.001 by unpaired Student’s t-test (A, I–L) or one-way ANOVA test (C–F). ANG, angiogenin; ANOVA, analysis of variance; DSS, dextran sodium sulphate; FMT, faecal microbiota transplantation; WT, wild-type.
To further test our hypothesis, we examined the effect of ANG1 in additional colitis models, that is, acute 2,4,6-trinitrobenzene sulfonic acid solution (TNBS)-induced colitis model and interleukin-10 knockout (Il-10−/−) spontaneous colitis model. On TNBS treatment, faecal ANG1 level dramatically declined in WT mice (online supplementary figure S2A), and Ang1−/− mice showed more severe colitis compared with WT ones (online supplementary figure S2B-F). In Il-10 mice, the faecal ANG1 level started to decrease around the beginning of colonic inflammation (online supplementary figure S3A). To test the direct contribution of ANG1 in this model, we further deleted Ang1 in Il-10 mice (Ang1−/−; Il-10−/−), and found that the double knockout mice progressed to colitis faster than the Il-10−/− ones (online supplementary figure S3B-F). We also observed faeces from Ang1−/− mice exacerbated colitic symptoms in these two models (online supplementary figures S2B-F and S3G-L). These results indicate that ANG1 plays similar roles in different colitis models.
Together, our data reveal an association among ANG level, gut microbiota and colitis severity, suggesting that the enhanced susceptibility of Ang1−/− mice to experimental colitis is related to the changes of gut microbiota.
Ang1 deficiency results in a dysbiotic microbiome
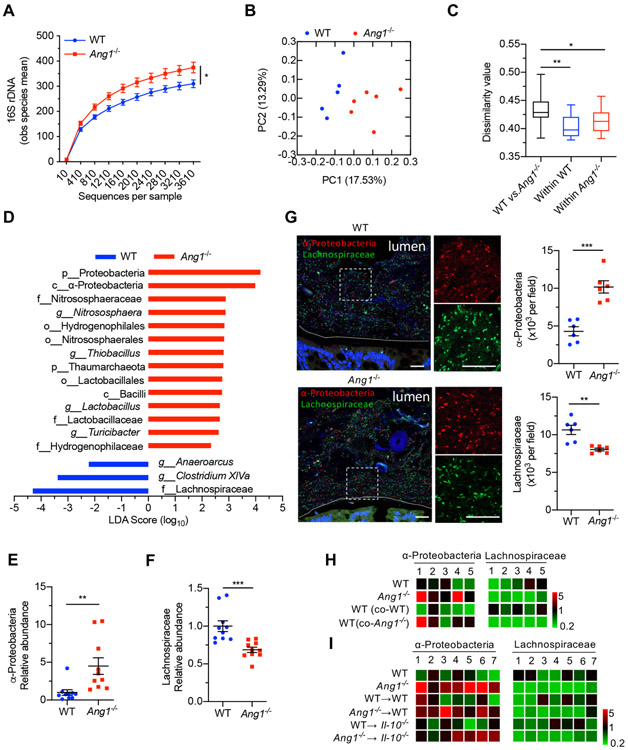
To identify microbes regulated by ANG1, we performed high-throughput sequencing analysis of 16S rDNA in faecal bacterial DNAs isolated from WT and Ang1−/− littermates that were originally generated from the same Ang1+/− parents housed in our animal facility for more than five generations. Rarefaction analysis16 was used to compare bacterial diversity among individual mouse within a group, and the results showed that Ang1−/− mice harboured more diverse microbiota (figure 2A, p<0.05) with a distinct community composition as compared with WT mice (figure 2B). Dissimilarity analysis within and between groups indicated that the microbiome difference between WT and Ang1−/− mice was significantly greater than that within each genotype (figure 2C, calculated from figure 2B). Linear discriminant analysis effect size17 showed several differences in the baseline faecal microbiota composition between WT and Ang1−/− mice (figure 2D). The Proteobacteria phylum, in particular the α-Proteobacteria class, was enriched, while the Lachnospiraceae family was reduced in Ang1−/− mice (figure 2D). This finding was confirmed by quantitative PCR (qPCR) analysis (figure 2E,F) and fluorescence in situ hybridisation (FISH) (figure 2G). The increase of α-Proteobacteria and decrease of Lachnospiraceae were also observed in Ang1−/−; Il-10−/− mice (online supplementary figure S3M,N). Notably, the levels of these two microbes in WT and Il-10−/− mice receiving Ang1−/− faeces transfer were similar to those in Ang1−/− mice (figure 2H,I), further supporting the role of ANG1 in shaping the composition of gut microbiota.
Figure 2.
Ang1 deficiency results in a dysbiotic microbiome. (A) High-throughput sequencing of 16S rDNA in faecal bacterial DNAs from WT (n=5) or Ang1−/− (n=6) mice. The Y-axis is the observed (obs) species mean, indicative of bacterial diversity. (B) Unweighted UniFrac principal-coordinate analysis of the β-diversity of microbiota composition in WT (n=5) or Ang1−/− (n=6) mice. (C) Quantification of unweighted UniFrac distance in (B). The five lines of boxplot from bottom to top represent the minimum value, the first quartile, median, the third quartile and the maximum value, respectively. (D) The most differentially abundant taxa of gut microbiota between WT and Ang1−/− mice. LDA Score: linear discriminant analysis score. (E and F) The abundance of α-Proteobacteria or Lachnospiraceae in the gut microbiota of WT or Ang1−/− mice, n=10. (G) FISH on colonic lumen sections from WT or Ang1−/− mice: α-Proteobacteria (red), Lachnospiraceae (green) and DNA (blue). Scale bar, 25 μm. At least five sections per group were analysed. (H and I) The relative bacterial abundance of α-Proteobacteria or Lachnospiraceae in mice from co-housing experiment (H) or FMT experiment (I). The heatmap represents the relative abundance of indicated bacterium in each mouse after normalised to the average of WT group. Data are presented as mean±SEM; *p<0.05; **p<0.01; ***p<0.001 by unpaired Student’s t-test (A, E–G) or analysis of similarities test (C). ANG, angiogenin; FISH, fluorescence in situ hybridisation; FMT, faecal microbiota transplantation; WT, wild-type.
ANG1-regulated bacteria are associated with colitis
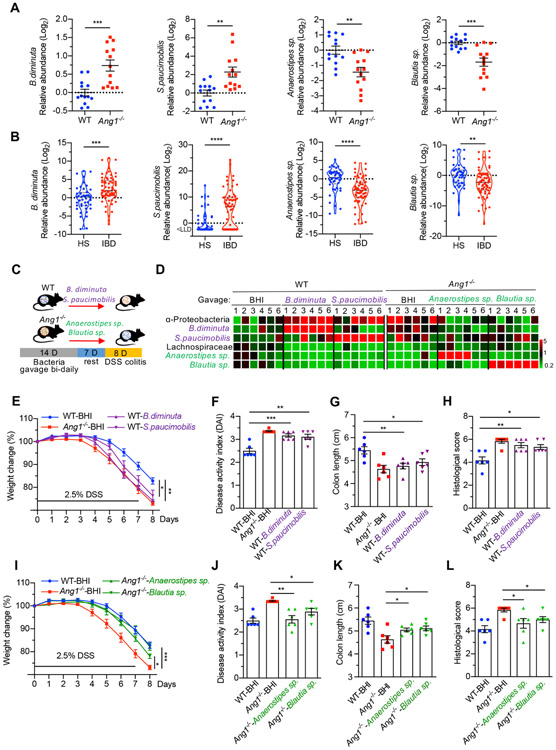
The correlation between enhanced vulnerability to inflammation and increased α-Proteobacteria but decreased Lachnospiraceae levels in Ang1−/− mice suggests that ANG may prevent colitis by regulating the ratio of these two gut bacteria. Therefore, we examined the faecal levels of both bacteria in DSS-induced WT mice and IBD patients. As expected, α-Proteobacteria was increased while Lachnospiraceae was decreased in DSS-treated mice (online supplementary figure S4A,B) and in IBD patients (online supplementary figure S4C,D). Moreover, mice with high α-Proteobacteria level lost more weight as compared with those with low α-Proteobacteria level (online supplementary figure S4E), whereas mice with high Lachnospiraceae level had less weight loss (online supplementary figure S4F). These data suggest that the ANG1-regulated bacteria are associated with the severity of colitis.
We further identified two α-Proteobacteria strains, Brevundimonas diminuta (B. diminuta) and Sphingomonas paucimobilis (S. paucimobilis), as well as two Lachnospiraceae strains, Anaerostipes species (Anaerostipes sp.) and Blautia species (Blautia sp.), as ANG1-regulated bacteria from mouse faeces (online supplementary table S2). To establish their correlations with colitis, we first examined the levels of these strains in mouse and human faecal samples. As shown in figure 3A,B, the abundances of B. diminuta and S. paucimobilis were increased by fold and 4.9-fold in Ang1−/− mice and by 5.2-fold and fold in IBD patients, while Anaerostipes sp. and Blautia sp. were decreased by 2.6-fold and 3.2-fold in Ang1−/− mice and by 9.8-fold and 5.3-fold in IBD patients as compared with WT mice and to healthy human subjects, respectively. To determine whether the bacteria can regulate colitis, we inoculated WT and Ang1−/− mice with these strains via oral gavage for 14 days, followed by 1 week of ‘rest’ before treatment with DSS (figure 3C). High bacterial colonisation efficiency in mice was confirmed by 16S rDNA qPCR (figure 3D), and minimal effect of feeding with bacteria or vehicle control (brain heart infusion, used for growing bacteria) on body weight before DSS treatment was observed (online supplementary figure S5A). Interestingly, on DSS administration, WT mice gavaged with the two α-Proteobacteria strains suffered from more severe colitis, including more significant weight loss, higher DAI Score, shorter colon length and more serious colon histopathology (figure 3E-H, online supplemental figure S5B,C). In contrast, gavage with the two Lachnospiraceae strains suppressed colitis in Ang1−/− mice (figure 3I-L, online supplementary figure S5D,E). It is worth noting that WT mice receiving B. diminuta and S. paucimobilis strains phenocopied Ang1−/− mice (figure 3E), while Ang1−/− mice receiving Anaerostipes sp. and Blautia sp. strains mirrored WT mice (figure 3I) in terms of colitis severity. Together, these results indicate that ANG1 maintains intestinal microbiota community composition and stability by balancing the levels of α-Proteobacteria and Lachnospiraceae, leading to protection against colitis.
Figure 3.
ANG1-regulated bacteria are associated with colitis. (A) The relative abundance of B. diminuta, S. paucimobilis, Anaerostipes sp. or Blautia sp. in gut microbiota of WT or Ang1−/− mice, n=13. (B) The relative abundance of B. diminuta, S. paucimobilis, Anaerostipes sp. or Blautia sp. in faeces from healthy subjects (HS, n=45) or IBD patients (n=64). LLD, lower limit of detection. (C) Diagram of bacterial gavage and DSS induction for analysing the relationship between identified bacteria and colitis progression. (D) The relative abundance of indicated bacterium in gavaged mice. The heatmap represents the relative bacterial abundance in each mouse after normalised to the average of WT-BHI group, n=6. (E–H) The body weight (E), DAI (F), colon length (G) and histological score (H) of WT colitis mice pre-treated with B. diminuta, S. paucimobilis or BHI, n=6. (I–L) The body weight (I), DAI (J), colon length (K) and histological score (L) of Ang1−/− colitis mice pre-treated with Anaerostipes sp., Blautia sp. or BHI, n=6. Data are presented as mean±SEM; *p<0.05; **p<0.01; ***p<0.001 by unpaired Student’s t-test (A and B) or one-way ANOVA (E–L). ANG, angiogenin; ANOVA, analysis of variance; BHI, brain heart infusion; DAI, Disease Activity Index; DSS, dextran sodium sulphate; WT, wild-type.
ANG promotes Lachnospiraceae outgrowth at the expense of α-Proteobacteria
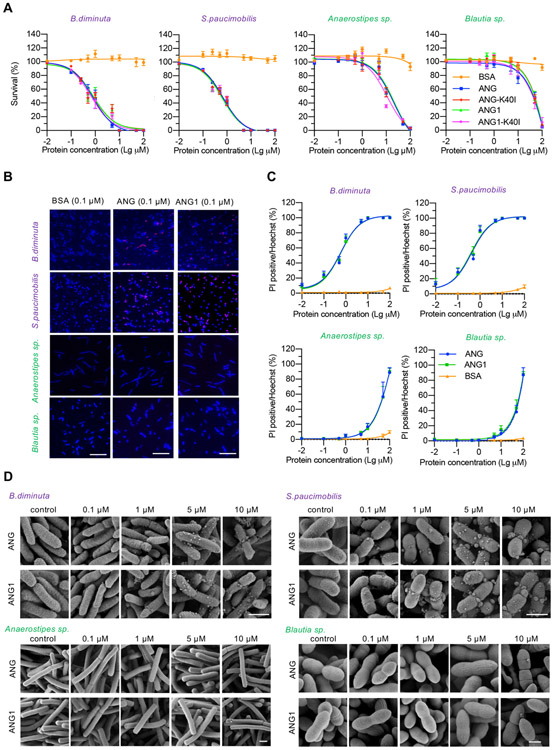
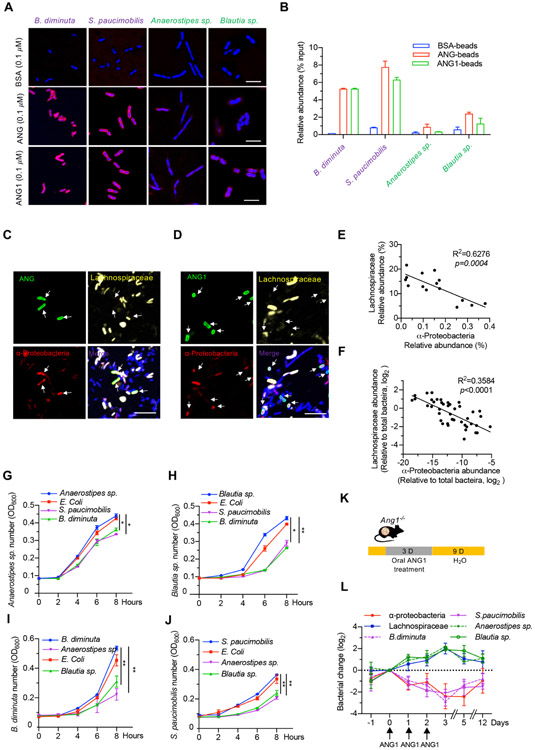
To understand how ANG regulates the balance between α-Proteobacteria and Lachnospiraceae, we examined the bactericidal activity of recombinant human ANG and mouse ANG1 against the four identified bacterial strains. The biological activity of recombinant ANG and ANG1 proteins was confirmed by angiogenic and ribonucleolytic assay (online supplementary figure S6A,B). The median lethal dose (LD50) of ANG and ANG1 against Lachnospiraceae (212.9 and 242.9 μM for Blautia sp., 20.75 and 22.50 μM for Anaerostipes sp.) was 25–250-fold higher than those against α-Proteobacteria (0.81 and 0.85 μM for B. diminuta, 0.69 and 0.63 μM for S. paucimobilis) (figure 4A, online supplementary table S3), indicating a selective antibacterial activity of ANG toward α-Proteobacteria. To understand the mode of action of ANG and the reason of differential bactericidal activities, we examined the effect of ANG on cellular RNA degradation and on cell membrane disruption, two known mechanisms by which ANG inhibits bacteria growth.13 Degradation of cellular RNA requires its ribonucleolytic activity; we therefore compared the antibacterial capacity of WT and ribonuclease-inactive variant (K40I) of ANG and found that the ribonucleolytic activity was not required (figure 4A, online supplementary table S3), ruling out degrading bacteria RNA as the bactericidal mechanism. Next, we examined the impact of ANG on the integrity of cell membrane by propidium iodide (PI) staining. Incubation with ANG or ANG1 at a physiological concentration (about 0.1 μM) resulted in PI-positive staining in 25% of B. diminuta and S. paucimobilis but only 2% of Blautia sp. and Anaerostipes sp. (figure 4B), suggesting that ANG treatment increased membrane permeabilisation. Further study indicated that ANG was able to disrupt bacterial membrane in a dose-dependent manner (figure 4C). We then used scanning electron microscope to examine the bacterial membrane morphology and found that treatment with low concentrations of ANG or ANG1 changed the membrane structure from a smooth surface to a bubble-like layer in B. diminuta and to a larger bleb-like structure in S. paucimobilis (figure 4D). In contrast, detrimental structure changes could not be observed on the two Lachnospiraceae strains until reaching much higher concentrations (10 μM ANG or ANG1) (figure 4D). These data suggest that ANG preferentially permeabilises the membrane of B. diminuta and S. paucimobilis strains, leading to cytotoxicity.
Figure 4.
The antibacterial activity of ANG. (A) Antibacterial efficacy of recombinant ANG, ANG1, ANG-K40I, ANG1-K40I or bovine serum albumin (BSA) against B. diminuta, S. paucimobilis, Anaerostipes sp. or Blautia sp. Each experiment was performed at least three times independently. (B) PI staining of B. diminuta, S. paucimobilis, Anaerostipes sp. or Blautia sp. after treatment with 0.1 μM of ANG, ANG1 or BSA. Scale bar, 20 μm. (C) Dose–response curve of the indicated bacterium treated with ANG, ANG1 or BSA. each experiment was performed at least three times independently. (D) Scanning electron microscopy micrograph of B. diminuta, S. paucimobilis, Anaerostipes sp. or Blautia sp. after treatment with different concentrations of ANG or ANG1 for 2 hours at 37°C. Scale bar, 1 μm. ANG, angiogenin; PI, propidium iodide.
We also found that ANG and ANG1 directly bound to B. diminuta and S. paucimobilis as shown by immunofluorescence analysis (figure 5A) and pull-down experiment (figure 5B). Faecal smear slide staining revealed that ANG was selectively co-localised with and enriched on the membrane of α-Proteobacteria in the faeces from both IBD patients (figure 5C) and colitic mice (figure 5D). These data indicate that ANG prefers binding the membrane of B. diminuta and S. paucimobilis to induce their permeabilisation.
Figure 5.
ANG promotes Lachnospiraceae outgrowth at the expense of α-Proteobacteria. (A) Immunofluorescence staining of ANG, ANG1 or BSA in bacteria (B. diminuta, S. paucimobilis, Anaerostipes sp. or Blautia sp.) after treatment with 0.1 μM respective recombinant protein. Scale bar, 10 μm. (B) The relative binding ability of recombinant ANG, ANG1 or BSA with bacteria (B. diminuta, S. paucimobilis, Anaerostipes sp. or Blautia sp.) detected by pull-down assay. Results represent the relative enrichment of indicated bacterium after normalised to the input control. (C and D) Combined FISH with ANG (C) or ANG1 (D) immunostaining on human or mouse faecal smear slide. α-Proteobacteria (red), Lachnospiraceae (yellow), ANG or ANG1 (green) and DNA (blue). Scale bar, 25 μm. (E and F) Relationship between α-Proteobacteria and Lachnospiraceae in human (E, n=15) or mouse (F, n=45) gut microbiota. (G and H) Bacterial growth curve of Anaerostipes sp. (G) or Blautia sp. (H) after adding spent medium of E. coli (unrelated control), B. diminuta or S. paucimobilis. (I and J) Bacterial growth curve of B. diminuta (I) or S. paucimobilis (J) with spent medium of E. coli (unrelated control), Anaerostipes sp. or Blautia sp. (K) Scheme of oral ANG1 treatment in Ang1−/− mice for gut microbe analysis. (L) The relative abundance of indicated bacterium in ANG1-treated Ang1−/− mice at different time points, n=5. Data are normalised to the mice at Day 0. Data are presented as mean±SEM; *p<0.05; **p<0.01 by one-way ANOVA test (G–J) or linear regression analysis (E and F). ANG, angiogenin; ANOVA, analysis of variance; FISH, fluorescence in situ hybridisation.
Accumulating evidence suggests that commensal bacteria–bacteria interactions contribute to the maintenance of a homogeneous microbiome.18 To understand whether ANG-mediated suppression of α-Proteobacteria is a reason for Lachnospiraceae outgrowth, we first examined the potential antagonistic effect of α-Proteobacteria on Lachnospiraceae growth. In both mouse (figure 5E) and human (figure 5F) faeces, the abundance of Lachnospiraceae was inversely correlated with α-Proteobacteria. Further, the spent media of B. diminuta and S. paucimobilis inhibited the growth of Anaerostipes sp. and Blautia sp. (figure 5G,H), and vice versa (figure 5I,J).
To confirm our in vitro results, we first detected whether orally administered recombinant ANG1 could influence these bacteria in Ang1−/− mice (figure 5K). The results showed that the abundance of endogenous α-Proteobacteria including B. diminuta and S. paucimobilis was continuously declined on ANG1 treatment but recovered after withdrawal of this protein. At the same time, the level of Lachnospiraceae including Anaerostipes sp. and Blautia sp. displayed the opposite trend (figure 5L). Second, we found that replenishing WT mice with the two α-Proteobacteria strains significantly inhibited the growth of endogenous Lachnospiraceae in the gut, while inoculating the two Lachnospiraceae strains into Ang1−/− mice antagonised endogenous intestinal α-Proteobacteria (figure 3D). These data indicate that ANG1 suppresses the growth of α-Proteobacteria which in turn promotes the growth of Lachnospiraceae in the intestine.
Taken together, these results suggest that ANG directly binds to and lethally permeabilises the membrane of α-Proteobacteria, which consequently favours the growth of the antagonistic Lachnospiraceae.
ANG1 treatment prevents dysbiosis and suppresses DSS-induced colitis in Ang1−/− mice
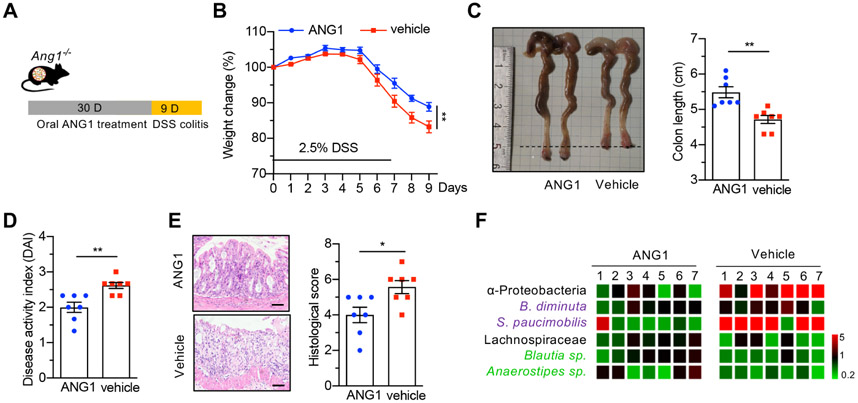
As shown in figure 5L, oral administration of recombinant ANG1 could change the ratio of α-Proteobacteria to Lachnospiraceae in Ang1−/− mice, suggesting a potential therapeutic benefit of ANG in colitis. To explore this possibility, we orally treated Ang1−/− mice with ANG1 for 30 days, followed by DSS administration for 7 days to induce colitis (figure 6A). Mice pretreated with ANG1 exhibited attenuated clinical signs of colitis, including increased body weight (figure 6B), longer colon length (figure 6C), decreased DAI (figure 6D) and lower histological score (figure 6E). Amelioration of the colitic symptoms was accompanied by a significant decrease in α-Proteobacteria and an increase in Lachnospiraceae (figure 6F), suggesting that reversion of dysbiosis underlies, at least partially, the therapeutic efficacy of ANG1 in experimental colitis.
Figure 6.
ANG1 treatment reverses dysbiosis and suppresses DSS-induced colitis in Ang1−/− mice. (A) Scheme of oral ANG1 treatment in Ang1−/− mice for analysing its preventive potential. (B) Body weight of vehicle-treated or ANG1-treated Ang1−/− mice during DSS-induced colitis, n=7. (C–E) Colon length (C), DAI (D) and histological score (E) of vehicle-treated or ANG1-treated Ang1−/− mice on day 9 after DSS induction, n=7. Scale bar, 50 μm. (F) The relative abundance of indicated bacterium in vehicle-treated or ANG1-treated Ang1−/− mice. The heatmap represents the relative bacterial abundance in each mouse after normalised to the average of ANG1 group, n=7. Data are presented as mean±SEM; *p<0.05; **p<0.01 by unpaired Student’s t-test (B–E). ANG, angiogenin; DSS, dextran sodium sulphate.
DISCUSSION
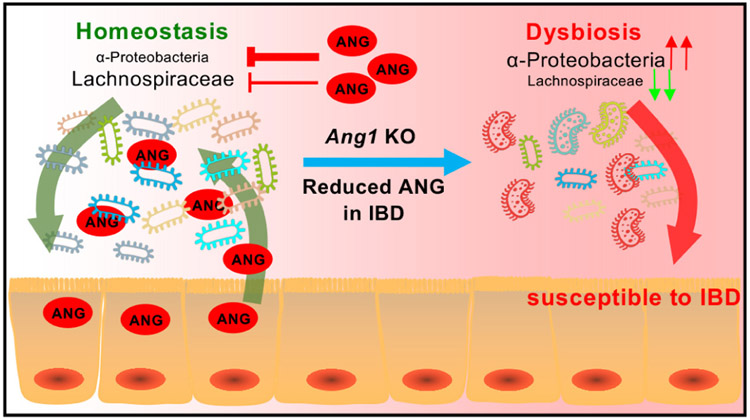
IBD-associated microbe dysbiosis generally displays an outgrowth of certain colitogenic bacteria at the expense of protective commensal bacterial species. Gut AMPs play a crucial role in maintaining a delicate and beneficial composition of the microbiota. In this study, we found that the absence of ANG1, a known AMP, results in gut dysbiosis via increasing α-Proteobacteria level to antagonise the growth of Lachnospiraceae. Such alteration in gut microenvironment instigated by ANG deficiency facilitates the progression of colitis (figure 7).
Figure 7.
Graphical abstract. Working model of ANG in maintaining gut microbe homeostasis to prevent colitis. ANG, angiogenin.
As a key player in mammalian physiology and pathology, the gut microbiota is influenced by both host genetics and the microenvironment. Any changes in these factors can predispose the host to inflammatory disorders such as IBD.19 Our results showed that Ang1 deletion had differential effects on particular members of microbiota (ie, α-Proteobacteria and Lachnospiraceae) and thus likely changed the host immune responses. Consistent with previous findings that the abundance of α-Proteobacteria, such as Novosphingobium sp., Sphingomonas sp., Rhizobiaceae and Bradyrhizobiaceae, was increased in IBD patients,20-22 we found that two other α-Proteobacteria strains, B. diminuta and S. pauciinobilis, were enriched in colitis mice and IBD patients. These bacteria appear benign when they are maintained in proper proportion, but become colitogenic under particular gut status such as Ang1 deficiency.23 Although the molecular mechanism underlying α-Proteobacteria-promoted colitis is unclear, it is commonly accepted that a bloom of α-Proteobacteria in the gut reflects dysbiosis or an unstable structure of gut microbial community.24,25 On the other hand, a decrease in the abundance of Lachnospiraceae has been observed in IBD patients.26,27 Our results showed that certain Lachnospiraceae strains such as Anaerostipes sp. and Blautia sp. were reduced in colitis, and their restoration protected mice from intestinal inflammation. Accumulating evidence indicates that gut Lachnospiraceae is the primary producer of butyrate which enhances epithelial barrier integrity and inhibit inflammation.28,29 Consistent with these reports, we found that loss of Lachnospiraceae species in Ang1−/− mice resulted in a lower butyrate level (data not shown).
Cooperative and competitive interactions happen between different gut microbes. Although advances have been made with the help of high-throughput sequencing techniques in identifying the species composition of host-associated microbe community, our understanding of ecological interaction within the microbiota is very limited.30 While a cooperative microbial network is indeed found to be very productive in the short term, competition between different microbes drives long-term microbiome stability which is beneficial for the host.30,31 By analysing pair-wise interactions between the culturable microbes in vitro, it was observed that at least 86% of interactions between strains or species are competitive.32,33 Here we presented both in vitro and in vivo evidence to establish a mutually inhibitory interaction between α-Proteobacteria and Lachnospiraceae, supporting the notion that microbe-microbe competition is essential for maintaining a healthy status of the gut microbiota.
ANG has been identified as an AMP for nearly 20 years; however, the understanding of its antimicrobial function was solely from in vitro studies. Furthermore, whether or not it contributes to the health of gut was completely unknown. In this study, we employed Ang1 knockout mice as a model to evaluate its effect on gut microbiota and found two types of ANG-regulated bacteria. To our knowledge, this is the first report that ANG directly controls bacterial growth under pathophysiological conditions. Mechanistically, we found that ANG kills α-Proteobacteria through binding to the membrane and causing membrane permeabilisation. Our data clearly demonstrated that ANG displays a selective bactericidal activity against Gram-negative bacteria (ie, B. diminuta and S. paucimobilis), but not Gram-positive bacteria (ie, Anaerostipes sp. and Blautia sp.). A similar observation has been reported in other Gram-negative (ie, Escherichia coli and Bacteroides thetaiotaomiron)13 and Gram-positive bacteria (ie, Enterococcus faecalis, Listeria monocytogenes and Streptococcus pneumoniae), but again merely with in vitro experimental systems.11 The antibacterial selectivity may be due to the difference in bacterial cell wall composition. The Gram-positive bacteria possess a thick (20–80 nm) cell wall as outer shell. In contrast, Gram-negative bacteria have a relatively thin (<10nm) layer of cell wall, but harbour an additional outer membrane with several pores and appendices.34 These differences in bacterial envelope confer various properties to the germs, in particular their responses to external stresses, such as heat, ultraviolet radiation and AMPs. Our data showed that ANG has a significant preference for binding and killing B. diminuta and S. paucimobilis. Further affinity studies of ANG toward different bacterial wall as well as membrane components would be helpful to illuminate the detailed mechanism of action.
As a colitis-related AMP, ANG was observed to be reduced in IBD patients and three different colitic mice. It is known that Paneth cells are the main ANG-secreting cell type in intestine, and clinical data have shown that the Paneth cell dysfunction and AMPs deficiency are the typical features of IBD patients, especially in CD patients.35,36 Meanwhile, it has been reported that the Paneth cells in colitic mouse display alterations in morphology, differentiation and AMPs production.37,38 Therefore, we reason that the diminished ANG level is likely a consequence of Paneth cell abnormality caused by the inflammation. Further study is certainly warranted to reveal the mechanism regulating ANG production in Paneth cells under physiological and pathological conditions.
Although the gut microbiota of mouse and human harbour many host species-specific bacterial signatures, the patterns of the abundance of these clades may be similar to each other.39,40 Moreover, the mouse gut microbiome is functionally similar to its human counterpart, with 95.2% of its Kyoto Encyclopaedia of Genes and Genomes orthologous groups in common by metagenome analysis.41 In line with these findings, the bacteria we identified in mice were also observed in human faeces, and were correlated to IBD progression. Although mouse genome possesses the largest ANG family, including five ANG genes and three ANG pseudogenes (Ang-ps1-3),42 only Ang1 shows the same gene structure and expression profile as the human ANG gene.14,15 More importantly, the antimicrobial activity assay showed equivalent function between human ANG and mouse ANG1. Therefore, the results from Ang1 knockout mice and disease models provide useful references for how ANG regulates human intestinal microbes. Given that endogenous AMPs, of which ANG is a kind, offer promising opportunities to explore into novel therapeutic antibiotics as they do not readily induce bacterial resistance,43 fine-tuning ANG level might be a new avenue for regulating microbiota composition of the chronically inflamed gut.
In conclusion, our data indicate that ANG in the intestine is critically vital for the control of bacterial community structure, particularly the balance of α-Proteobacteria and Lachnospiraceae. Our work provides a valuable example of the intricate interplay between the host and microbiota and among bacteria themselves during intestinal inflammation, highlighting the importance of ANG in keeping dysbiosis and intestinal inflammation at bay.
Supplementary Material
Significance of this study.
What is already known on this subject?
Dysregulation of antimicrobial peptide (AMP) production or function is associated with IBD.
Angiogenin (ANG) is an intestine-secreted AMP with potent species-specific antimicrobial activity.
ANG inhibits bacterial growth through degrading cellular RNA or disrupting cell membrane.
AMPs are promising candidates for developing novel therapeutic antibiotics.
What are the new findings?
ANG is significantly decreased in faeces of IBD patients and colitic mice.
Ang1 deficiency alters gut microbiota contributing to increased susceptibility to experimental colitis.
ANG disrupts the membrane of α-Proteobacteria to suppress its growth and consequently promotes the flourishing of Lachnospiraceae.
Oral ANG1 treatment prevents dysbiosis and suppresses colitis in Ang1-deficient mice.
How might it impact on clinical practice in the foreseeable future?
The study provides a novel preventive and/or therapeutic strategy for IBD basing on the regulatory activity of ANG on gut bacterial community.
Acknowledgements
We thank Prof. Wei Liu, Prof. Di Wang and Prof. Stijn van der Veen (Zhejiang University School of Medicine) for critical suggestions to the project conceptualisation and experimental design, Yanwei Li and Shuangshuang Liu of the Core Facilities (of Zhejiang University School of Medicine) for technical assistance in protein purification and microscopy analysis, Dandan Song of the Centre of Cryo-Electron Microscopy (of Zhejiang University) for technical assistance on scanning electron microscope analysis and Chunyan Wu (Realbio technology) for 16S rDNA high-throughput sequencing analysis.
Funding This study was supported by the following grants: National Natural Foundation of China (No. 81790631, No. 31770867, No. 31570786, No. 81602557, No. 81800474 and No. 31741026); Zhejiang Provincial Natural Science Foundation of China (No. LY18H030006 and No. LY17H160021); Fundamental Research Funds for the Central Universities (No. 2017FZA7006).
Footnotes
Competing interests None declared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication Not required.
Ethics approval Ethics approval for these studies involving human samples was obtained from the Ethics Committee of Zhejiang University School of Medicine (#2018-020, 2018 updated). The animal research was performed under the protocol that has been approved by the Medical Experimental Animal Care Commission of Zhejiang University (#12364, 2019 updated).
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability
The bacterial 16S rDNA sequences were deposited to the sequence read archive. The accession number for the 16S sequencing data reported in this paper is PRJNA559351.
See online supplementary materials and methods for additional information.
Data availability statement
Data are available in a public, open access repository. The bacterial 16S rDNA sequences were deposited to the sequence read archive. The accession number for the 16S sequencing data reported in this paper is PRJNA559351.
REFERENCES
- 1.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell 2016;164:337–40. [DOI] [PubMed] [Google Scholar]
- 2.Robertson SJ, Goethel A, Girardin SE, et al. Innate immune influences on the gut microbiome: lessons from mouse models. Trends Immunol 2018;39:992–1004. [DOI] [PubMed] [Google Scholar]
- 3.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012;336:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mergaert P Role of antimicrobial peptides in controlling symbiotic bacterial populations. Nat Prod Rep 2018;35:336–56. [DOI] [PubMed] [Google Scholar]
- 5.Salzman NH, Hung K, Haribhai D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 2010;11:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshimura T, McLean MH, Dzutsev AK, et al. The antimicrobial peptide cramp is essential for colon homeostasis by maintaining microbiota balance. J Immunol 2018;200:2174–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darnaud M, Dos Santos A, Gonzalez P, et al. Enteric delivery of regenerating family member 3 alpha alters the intestinal microbiota and controls inflammation in Mice With Colitis. Gastroenterology 2018;154:e14:1009–23. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L-J, Gallo RL. Antimicrobial peptides. Curr Biol 2016;26:R14–19. [DOI] [PubMed] [Google Scholar]
- 9.Okumura R, Kurakawa T, Nakano T, et al. Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature 2016;532:117–21. [DOI] [PubMed] [Google Scholar]
- 10.Fett JW, Strydom DJ, Lobb RR, et al. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry 1985;24:5480–6. [DOI] [PubMed] [Google Scholar]
- 11.Hooper LV, Stappenbeck TS, Hong CV, et al. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol 2003;4:269–73. [DOI] [PubMed] [Google Scholar]
- 12.Cho S, Beintema JJ, Zhang J. The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics 2005;85:208–20. [DOI] [PubMed] [Google Scholar]
- 13.Walker CR, Hautefort I, Dalton JE, et al. Intestinal intraepithelial lymphocyte-enterocyte crosstalk regulates production of bactericidal angiogenin 4 by Paneth cells upon microbial challenge. PLoS One 2013;8:e84553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bond MD, Vallee BL. Isolation and sequencing of mouse angiogenin DNA. Biochem Biophys Res Commun 1990;171:988–95. [DOI] [PubMed] [Google Scholar]
- 15.Dyer KD, Rosenberg HF. The mouse RNase 4 and RNase 5/ang 1 locus utilizes dual promoters for tissue-specific expression. Nucleic Acids Res 2005;33:1077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes JB, Hellmann JJ. The application of rarefaction techniques to molecular inventories of microbial diversity. Methods Enzymol 2005;397:292–308. [DOI] [PubMed] [Google Scholar]
- 17.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coyte KZ, Rakoff-Nahoum S. Understanding competition and cooperation within the mammalian gut microbiome. Curr Biol 2019;29:R538–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva MJB, Carneiro MBH, dos Anjos Pultz B, et al. The multifaceted role of commensal microbiota in homeostasis and gastrointestinal diseases. J Immunol Res 2015;2015:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukhopadhya I, Hansen R, El-Omar EM, et al. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol 2012;9:219–30. [DOI] [PubMed] [Google Scholar]
- 21.Kaakoush NO, Day AS, Huinao KD, et al. Microbial dysbiosis in pediatric patients with Crohn’s disease. J Clin Microbiol 2012;50:3258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagao-Kitamoto H, Kamada N. Host-Microbial cross-talk in inflammatory bowel disease. Immune Netw 2017;17:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin N-R, Whon TW, Bae J-W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 2015;33:496–503. [DOI] [PubMed] [Google Scholar]
- 24.Spees AM, Lopez CA, Kingsbury DD, et al. Colonization resistance: battle of the bugs or ménage à trois with the host? PLoS Pathog 2013;9:e1003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchesi JR, Adams DH, Fava F, et al. The gut microbiota and host health: a new clinical frontier. Gut 2016;65:330–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halfvarson J, Brislawn CJ, Lamendella R, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017;2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013;504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013;341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: networks, competition, and stability. Science 2015;350:663–6. [DOI] [PubMed] [Google Scholar]
- 31.McNally L, Brown SP. Microbiome: ecology of stable gut communities. Nat Microbiol 2016;1:15016. [DOI] [PubMed] [Google Scholar]
- 32.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol 2012;22:1845–50. [DOI] [PubMed] [Google Scholar]
- 33.Mitri S, Foster KR. The genotypic view of social interactions in microbial communities. Annu Rev Genet 2013;47:247–73. [DOI] [PubMed] [Google Scholar]
- 34.Koprivnjak T, Peschel A. Bacterial resistance mechanisms against host defense peptides. Cell Mol Life Sci 2011;68:2243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wehkamp J, Stange EF. An update review on the Paneth cell as key to ileal Crohn’s disease. Front Immunol 2020;11:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khaloian S, Rath E, Hammoudi N, et al. Mitochondrial impairment drives intestinal stem cell transition into dysfunctional Paneth cells predicting Crohn’s disease recurrence. Gut 2020. doi: 10.1136/gutjnl-2019-319514. [Epub ahead of print: 28 Feb 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt M, Schewe M, Sacchetti A, et al. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep 2018;24:2312–28. [DOI] [PubMed] [Google Scholar]
- 38.Berkowitz L, Pardo-Roa C, Ramírez G, et al. The absence of interleukin 10 affects the morphology, differentiation, granule content and the production of cryptidin-4 in Paneth cells in mice. PLoS One 2019;14:e0221618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagpal R, Wang S, Solberg Woods LC, et al. Comparative microbiome signatures and short-chain fatty acids in mouse, rat, non-human primate, and human feces. Front Microbiol 2018;9:2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hugenholtz F, de Vos WM. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci 2018;75:149–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao L, Feng Q, Liang S, et al. A catalog of the mouse gut metagenome. Nat Biotechnol 2015;33:1103–8. [DOI] [PubMed] [Google Scholar]
- 42.Strydom DJ. The angiogenins. Cell Mol Life Sci 1998;54:811–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewies A, Du Plessis LH, Wentzel JF. Antimicrobial peptides: the Achilles’ heel of antibiotic resistance? Probiotics Antimicrob Proteins 2019;11:370–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bacterial 16S rDNA sequences were deposited to the sequence read archive. The accession number for the 16S sequencing data reported in this paper is PRJNA559351.
See online supplementary materials and methods for additional information.
Data are available in a public, open access repository. The bacterial 16S rDNA sequences were deposited to the sequence read archive. The accession number for the 16S sequencing data reported in this paper is PRJNA559351.