Abstract
Hepatoblastoma (HB) is one of the most common liver malignancies in children, while the molecular basis of the disease is largely unknown. Therefore, this study aims to explore the key genes and molecular mechanisms of the pathogenesis of HB using a bioinformatics approach. The gene expression dataset GSE131329 was used to find differentially expressed genes (DEGs). Functional and enrichment analyses of the DEGs were performed by the EnrichR. Then, the protein-protein interaction (PPI) network of the up-regulated genes was constructed and visualized using STRING database and Cytoscape software, respectively. MCODE was used to detect the significant modules of the PPI network, and cytoHubba was utilized to rank the important nodes (genes) of the PPI modules. Overall, six ranking methods were employed and the results were validated by the Oncopression database. Moreover, the upstream regulatory network and the miRNA-target interactions of the up-regulated DEGs were analyzed by the X2K web and the miRTarBase respectively. A total of 594 DEGs, including 221 up- and 373 down-regulated genes, were obtained, which were enriched in different cellular and metabolic processes, human diseases, and cancer. Furthermore, 15 hub genes were screened, out of which, 11 were validated. Top 10 transcription factors, kinases, and miRNAs were also determined. To the best of our knowledge, the association of RACGAP1, MKI67, FOXM1, SIN3A, miR-193b, and miR-760 with HB was reported for the first time. Our findings may be used to shed light on the underlying mechanisms of HB and provide new insights for better prognosis and therapeutic strategies.
Keywords: Hepatoblastoma, Gene regulatory network, Biomarker, Protein kinase, miRNAs, Bioinformatic analysis
Introduction
Hepatoblastoma (HB) is one of the most common liver malignancies in children, which originates from the undifferentiated hepatic progenitor cells during embryogenesis and mainly affects infants under three years of age (Feng et al. 2019; Yang et al. 2020). Although it is a rare neoplasm, accounting for less than 1% of all pediatric tumors, it has increased notably in the last three decades (Carrillo-Reixach et al. 2020; De Ioris et al. 2008). Modern imaging, surgical and chemotherapy techniques have considerably improved the survival rates of HB to the range of 70–80% (Czauderna and Garnier 2018). Despite these advances, major challenges remain undissolved as the 3-year event-free survival rate for advanced tumors is only 34%, and prognosis for patients with advanced or chemotherapy-refractory disease is still relatively poor (Semeraro et al. 2013; Sumazin et al. 2017). Furthermore, the molecular basis of the disease is largely unknown, and the rarity of the HB further complicates the study of the disease (Bell et al. 2017; Magrelli et al. 2009). Therefore, it is vital to further explore the molecular mechanisms of HB to develop more effective treatment methods and potential markers for early diagnosis.
Microarray analysis has been widely used to produce huge quantities of gene expression and other functional genomics data, which can help demonstrate crucial biomarkers, pathways, and gene functions associated with diseases (Mokhlesi and Talkhabi 2020; Talkhabi et al. 2017). Microarray data can be obtained from online repositories such as the NCBI Gene Expression Omnibus (GEO), which provides different forms of high-throughput functional genomic data such as microarray and next-generation sequencing (NGS) (Barrett et al. 2012). Moreover, bioinformatic tools can be implemented for predicting miRNA-target gene interactions, as many studies have indicated that miRNAs are involved in hepatocarcinogenesis (Callegari et al. 2015; Chou et al. 2018).
In the present study, we applied a microarray gene expression profile to identify the potential TFs, protein kinases, and hub genes involved in hepatoblastoma. Thus, differentially expressed genes (DEGs) were analyzed between HB and noncancerous liver tissue samples. Protein-protein interaction (PPI) and upstream regulatory networks were constructed for the up-regulated genes, and the hub genes were screened and validated with/by other resources. Also, we performed functional and pathway enrichment analyses for the DEGs and hub genes. Finally, miRNA-target gene interactions were discovered. These findings/results can be applied to better understand underlying molecular mechanisms of the progression of HB and provide new insights for their prognosis and therapeutic strategies.
Materials and methods
Microarray data
The gene expression profile dataset with the accession number GSE131329 was downloaded from the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) (Edgar et al. 2002). The array data, based on the GPL6244 platform (Affymetrix Human Gene 1.0 ST Array), contains 67 samples, including 53 HB and 14 noncancerous liver tissue samples.
DEGs identification
The GEO online tool GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to screen the DEGs. GEO2R is an online tool that allows users to compare two or more groups of samples in a GEO series (Barrett et al. 2012). We divided the samples into cancerous and noncancerous groups and compared them by GEO2R. Benjamini and Hochberg false discovery rate method was applied for p value adjustment to help correct false-positives. The cut-off criteria for DEGs selection were defined as |log2fold-change (FC)| ≥1.5 and adjusted p value <0.05.
Gene ontology and pathway enrichment analyses
Enrichment analysis is a commonly used approach for analyzing gene sets generated by genome experiments and classifying characteristic biological attributes. To investigate the probable biological functions and signaling pathways correlated with the DEGs, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed using EnrichR (https://amp.pharm.mssm.edu/Enrichr/), a comprehensive web-based resource for analyzing gene sets (Ashburner et al. 2000; Kanehisa and Goto 2000; Kuleshov et al. 2016). An adjusted p value <0.05 was considered statistically significant. Visualization of the top 10 results of the analyses was performed using GraphPad Prism version 8.2.1 for Windows (http://www.graphpad.com).
Gene regulatory network analysis
The eXpression2Kinases (X2K) (https://amp.pharm.mssm.edu/X2K/) is a web-based application that computationally predicts the involvement of upstream cell signaling pathways and identifies the regulatory associations between transcription factors(TFs), protein kinases, and their target genes (Clarke et al. 2018). We utilized X2K to analyze the upstream regulatory network responsible for the regulation of gene expression of the up-regulated genes with default parameters. Top 10 most enriched TFs and kinases were ranked based on hypergeometric p value, and the inferred network was constructed and visualized.
PPI network and module screening
The potential interactions between proteins were demonstrated by submitting the up-regulated genes list into the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://www.string-db.org), which collects and integrates information of functional interactions between expressed proteins (Szklarczyk et al. 2019). The high confidence score (0.7) was set as the minimum required interaction score. The PPI network was visualized using Cytoscape software version 3.7.2 (https://cytoscape.org/) (Shannon et al. 2003). In addition, the Molecular Complex Detection (MCODE), a Cytoscape plug-in, was used to identify more significant modules of the PPI network. The criteria were set as: Degree Cutoff = 2, Node Score Cutoff = 0.2, K-Core = 2, and Max. Depth = 100 (Bader and Hogue 2003).
Hub genes analysis and validation
To predict the hub genes, we used Cytoscape plug-in CytoHubba for ranking and exploring important nodes in the PPI network modules (Chin et al. 2014). Top 40 genes were selected according to centrality measures including Degree, Closeness, Betweenness, Stress, Radiality, and EcCentricity. Any overlap from the six ranking methods was regarded as a hub gene. An online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to extract the overlapping genes. To validate the results, we compared the expression of the hub genes in HB and normal liver tissue samples using Oncopression (http://oncopression.com/), a web-based integrated gene expression profile that uses single sample normalization method UPC (Lee and Choi 2017). GraphPad Prism was utilized for visualizing the results. GO and KEGG enrichment analyses were performed for the hub genes using EnrichR with a criterion of an adjusted p value <0.05.
miRNA-target gene identification
miRTarBase is a database containing miRNA-target interactions (MTIs) that are validated experimentally by reporter assay, western blot, microarray, and next-generation sequencing experiments (Chou et al. 2018). To identify the MTIs, we submitted the up-regulated genes into EnrichR and exported the results from the miRTarBase section. An adjusted p value <0.05 was considered as the cut-off value.
Results
Identification of HB-related genes
A total of 594 DEGs, including 221 up-regulated (HB-related genes) and 373 down-regulated genes (normal tissue-related genes), were obtained comparing HB and normal tissue samples. Top 10 down-regulated genes were CYP2C8, CYP2B7P, CYP2B6, C9, HSD17B13, F9, SLC22A1, CRP, CYP2A13, and CYP2A7. In addition, top 10 up-regulated genes were DKK1, REG3A, GPC3, DLK1, SNORD113–4, LGR5, AFP, EPCAM, PEG10, and SNORD114–26.
GO function and KEGG pathway enrichment analysis of HB-related genes.
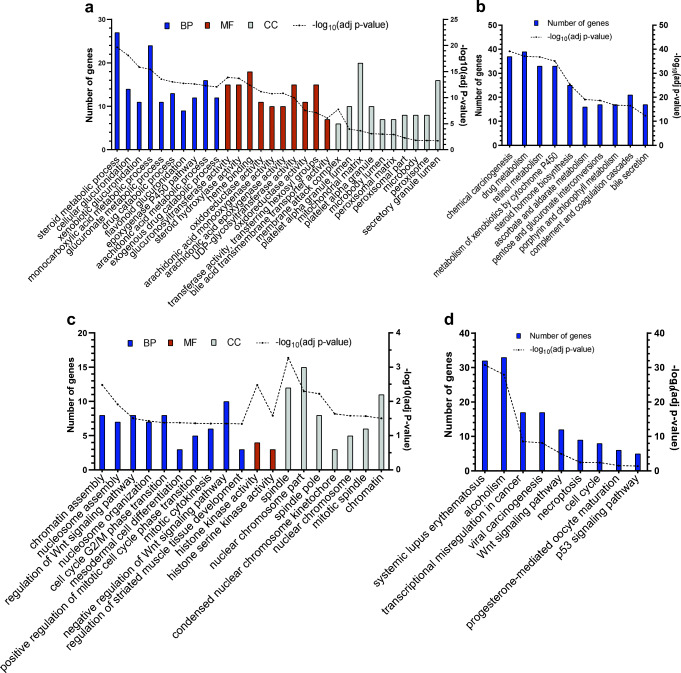
GO enrichment analysis is classified into three functional categories: biological process (BP), molecular function (MF), and cellular component (CC). Here, top 10 enriched GO and KEGG were determined (Fig. 1). These results were mainly associated with the following list for up-regulated and down-regulated genes respectively: cellular processes and biological regulation, cellular and metabolic processes for BP; kinase activity, catalytic activity for MF; spindle and chromosome, organelles especially microbody for CC; human diseases and cellular processes, metabolism and cancer for KEGG.
Fig. 1.
Gene Set Enrichment and Pathway Analyses of the down- (a, b) and up-regulated (c, d) genes. (a) and (c) indicate the top ten GO analysis results (if any) of the DEGs. (b) and (d) indicate the top ten KEGG pathway analysis results of the DEGs. BP: Biological Process, MF: Molecular Function, CC: Cellular Component
Construction of PPI and upstream regulatory network
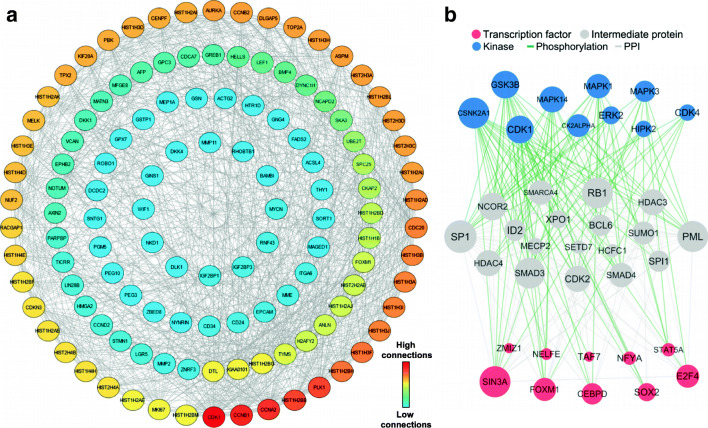
The PPI network of the up-regulated genes was screened via STRING and Cytoscape, and included 123 nodes (genes) and 919 edges (interactions) (Fig. 2a). CDK1, CCNB1, CCNA2, HIST1H2BB and PLK1 had the highest number of connectivities with other upregulated genes. We also applied X2K to construct and visualize the upstream regulatory network of the up-regulated genes produced between top 10 transcription factors and kinases, and their intermediate proteins (Fig. 2b). These TFs and kinases were connected to 18 intermediate proteins with an overall 323 edges. SIN3A, FOXM1, E2F4, CEBPD and SOX2 were most important TFs that regulate upregulated gene expression. In addition, CSNK2A1, GSK3B, CDK1, MAPK13 and MAPK1 were identified as most important kinases that control the expression of upregulated genes (Fig. 2b).
Fig. 2.
A graphic representation of the networks based on Upregulated genes. (a) Protein- protein interaction (PPI) and (b) Upstream regulatory network of the up-regulated genes. The color (in A) and size of nodes (in B) is proportional to their degree
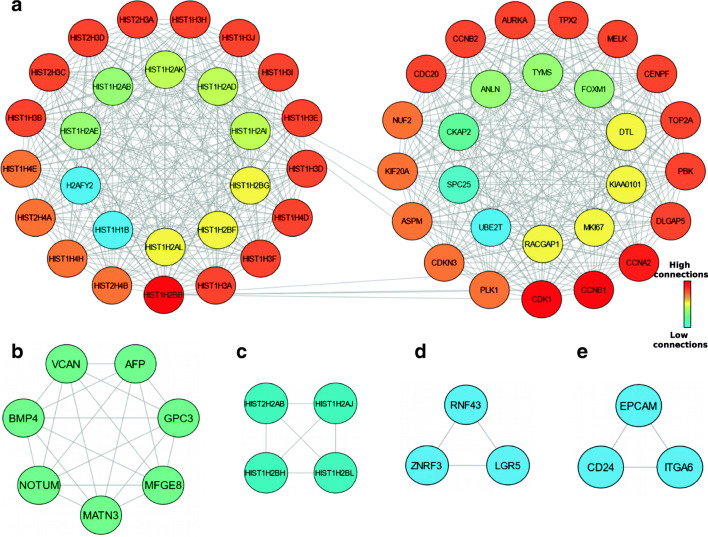
Using MCODE, five modules were retrieved from the PPI network constructed using upregulated genes (Fig. 3). Module 1 includes 54 nodes and 651 edges with a cluster score (density multiplied by the number of members) of 24.566 (Fig. 3a); Module 2, 7 nodes, 21 edges with score 7 (Fig. 3b); Module 3, 4 nodes, 6 edges with score 4 (Fig. 3c); Module 4 and 5, 3 nodes, 3 edges with score 3 (Fig. 3d and e).
Fig. 3.
Graphic representation of Top five significant modules of the PPI network. (a) Module 1, (b) Module 2, (c) Module 3, (d) Module 4, (e) Module 5
Hub genes selection
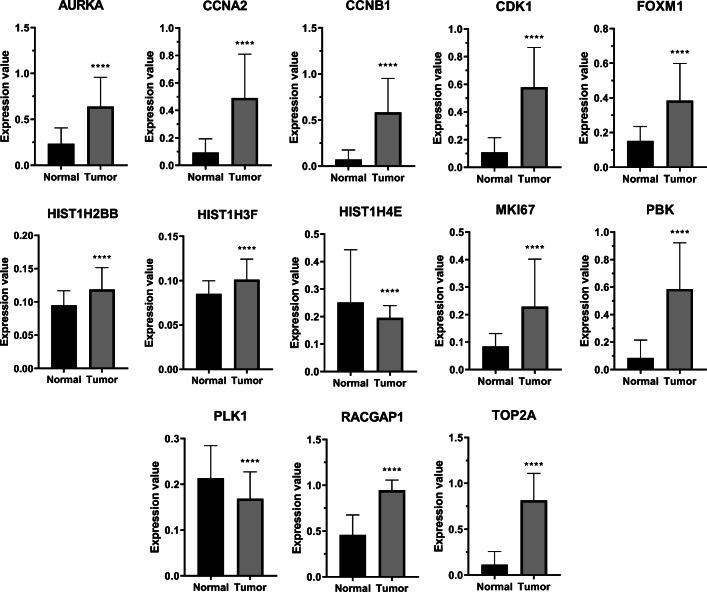
Using Cytohubba plug-in, 15 hub genes were identified from the PPI network modules (Table 1). The functional annotations of hub genes were performed by EnrichR. Top 10 GO analysis results revealed that these genes were mostly enriched in items regarding cell cycle for BP, kinase binding and activity for MF, and spindle and cytoskeleton for CC. Also, KEGG analysis demonstrated that the genes were significantly enriched in terms associated with human diseases and cell growth and death (Table 2). Moreover, to verify the expression of candidate hub genes, Oncopression database was utilized. Results confirmed that the expression of 11 hub genes in tumor samples is significantly higher than that of normal samples (Fig. 4).
Table 1.
The selected Hub genes overlapping in all six centrality ranking methods
| Gene | Degree | Closeness | Betweenness | Stress | Radiality | EcCentricity |
|---|---|---|---|---|---|---|
| CDK1 | 40 | 54 | 425.28385 | 10,828 | 4.57143 | 0.33333 |
| AURKA | 39 | 53.5 | 712.13238 | 10,850 | 4.55714 | 0.33333 |
| CCNB1 | 37 | 52.5 | 292.22128 | 8338 | 4.52857 | 0.33333 |
| CCNA2 | 35 | 51.5 | 235.17959 | 7338 | 4.5 | 0.33333 |
| HIST1H4E | 35 | 49.83333 | 72.79362 | 3918 | 4.31429 | 0.25 |
| HIST1H2BB | 34 | 49.33333 | 62.37444 | 3146 | 4.3 | 0.25 |
| TOP2A | 32 | 50 | 166.35268 | 5026 | 4.45714 | 0.33333 |
| HIST1H3F | 31 | 47.83333 | 13.79695 | 798 | 4.25714 | 0.25 |
| HIST1H3H | 31 | 47.83333 | 13.79695 | 798 | 4.25714 | 0.25 |
| HIST1H3J | 31 | 47.83333 | 13.79695 | 798 | 4.25714 | 0.25 |
| PLK1 | 31 | 49.5 | 138.94383 | 3390 | 4.44286 | 0.33333 |
| MKI67 | 30 | 47.83333 | 681.70006 | 7428 | 4.32857 | 0.33333 |
| PBK | 29 | 48.5 | 134.92199 | 2566 | 4.41429 | 0.33333 |
| FOXM1 | 27 | 46.66667 | 787.57813 | 19,446 | 4.31429 | 0.33333 |
| RACGAP1 | 26 | 45.5 | 1.85942 | 76 | 4.24286 | 0.33333 |
Table 2.
Gene Set Enrichment and Pathway Analyses of the Hub Genes
| Biological Process | Count | Adj P value | Cellular Component | Count | Adj P value |
|---|---|---|---|---|---|
| cell cycle G2/M phase transition | 6 | 7.09E-07 | spindle | 5 | 8.19E-05 |
| G2/M transition of mitotic cell cycle | 6 | 1.35E-06 | spindle midzone | 3 | 3.05E-04 |
| regulation of cell cycle process | 5 | 8.13E-06 | nuclear chromosome part | 5 | 8.03E-04 |
| mitotic cell cycle phase transition | 6 | 9.99E-06 | spindle microtubule | 3 | 9.16E-04 |
| DNA damage response, signal transduction by p53 class mediator | 4 | 2.94E-04 | mitotic spindle | 3 | 2.33E-03 |
| anaphase-promoting complex-dependent catabolic process | 4 | 3.04E-04 | condensed nuclear chromosome kinetochore | 2 | 2.57E-03 |
| regulation of ubiquitin protein ligase activity | 3 | 1.75E-03 | spindle pole | 3 | 4.12E-03 |
| regulation of G2/M transition of mitotic cell cycle | 4 | 2.42E-03 | centrosome | 4 | 1.73E-02 |
| mitotic nuclear envelope disassembly | 3 | 2.52E-03 | microtubule | 3 | 2.11E-02 |
| nuclear envelope disassembly | 3 | 2.59E-03 | microtubule organizing center | 4 | 2.21E-02 |
| Molecular Function | Count | Adj P value | KEGG pathway | Count | Adj P value |
| protein kinase binding | 7 | 3.40E-05 | progesterone-mediated oocyte maturation | 5 | 2.39E-06 |
| kinase binding | 6 | 1.97E-04 | systemic lupus erythematosus | 5 | 5.29E-06 |
| cyclin-dependent protein kinase activity | 3 | 7.73E-04 | alcoholism | 5 | 1.60E-05 |
| protein serine/threonine kinase activity | 5 | 1.52E-03 | oocyte meiosis | 4 | 1.16E-04 |
| histone kinase activity | 2 | 4.34E-03 | cell cycle | 4 | 1.40E-04 |
| protein kinase activity | 5 | 5.07E-03 | cellular senescence | 4 | 2.58E-04 |
| cyclin-dependent protein serine/threonine kinase activity | 2 | 4.50E-02 | viral carcinogenesis | 4 | 5.45E-04 |
Fig. 4.
Hub genes expression analysis in normal and HB tissues. The expression values are UPC-normalized, ranging from 0 to 1.0 where 0.0 indicates no expression and 1.0 the highest expression among other genes. The expression levels of all hub genes in tumor samples were significantly higher than normal samples, except for PLK1 and HIST1H4E. The plot represents Mean with SEM. ****: p value <0.0001
miRNA-target gene interactions
To find the MTIs, we used mirTarBase database from EnrichR. Here, we listed the top 10 important miRNAs that target the highest number of up-regulated genes (Table 3). hsa-miR-34a-5p, hsa-miR-193b-3p, hsa-miR-760, and hsa-miR-98-5p target 42, 27, 26 and 22 genes of the up-regulated genes, respectively.
Table 3.
The experimentally validated microRNA-gene interactions
| miRNA | adjPval | Target genes |
|---|---|---|
| hsa-miR-6822-3p | 6.76E-19 | H2AC13, H3C12, H2BC13, H2AC14, H2AC21, H3C15, H3C1, H2AC8, H2BC7, H3C7, H2AC7, H2BC9, H3C10, H2BC8, H2BC3, H3C13, H3C2, H3C14, H1–5, H3C4, H3C6 |
| hsa-miR-760 | 1.27E-17 | H3C12, MAGED1, H2BC13, H2AC21, AURKA, H3C1, NT5DC2, H3C7, H2AC8, H2AC7, H3C10, H3C2, H3C4, H1–5, H3C6, H2AC13, H2AC14, HMGA2, H3C15, BAMBI, H2BC7, H2BC9, H2BC8, H3C13, H2BC3, H3C14 |
| hsa-miR-34a-5p | 1.24E-15 | H3C12, LEF1, H2AC21, FIGN, GXYLT2, MKI67, TYMS, CDC20, H4C14, H3C1, H4C15, PEG10, STMN1, H3C7, H2AC8, H2AC7, H3C10, IGF2BP3, H3C11, H3C2, H3C4, H1–5, H3C6, H2AC4, H2AC16, H2AC13, SORT1, H2AC15, H2AC14, HMGA2, ACSL4, AXIN2, H3C15, MYCN, H4C8, ITGA6, H3C13, CD24, H2BC3, H3C14, H4C4, H4C5 |
| hsa-miR-1276 | 5.03E-14 | H2AC13, H3C12, H2BC13, H2AC14, H2AC21, SKA3, H3C15, H3C1, H2AC8, H2BC7, H3C7, H2AC7, H2BC9, H3C10, H2BC8, H2BC3, H3C13, H3C2, H3C14, H3C4, H1–5 |
| hsa-miR-8057 | 6.29E-14 | H2AC13, H3C12, H2BC13, H2AC14, H2AC21, H3C15, H3C1, H2AC8, H2BC7, H3C7, H2AC7, H2BC9, H3C10, H2BC8, H2BC3, H3C13, H3C2, H3C14, H3C4, H1–5, H3C6 |
| hsa-miR-4766-5p | 1.46E-13 | H2AC13, H3C12, H2BC13, H2AC14, H3C15, H3C1, H2AC8, H3C7, FLVCR1, H2AC7, H2BC9, H3C10, H2BC8, H2BC3, H3C13, H3C2, H3C14, H3C4, H1–5 |
| hsa-miR-146a-3p | 3.01E-13 | H2AC13, H3C12, H2BC13, H2AC14, H3C15, H3C1, H2AC8, H3C7, H2AC7, H2BC9, H3C10, H2BC8, H2BC3, H3C13, H3C2, H3C14, H3C4, H1–5 |
| hsa-miR-5693 | 1.35E-06 | H3C12, H2AC13, H2BC13, GREB1, H2AC14, H3C15, H3C1, RACGAP1, BAMBI, H3C7, H2AC8, H2AC7, H3C10, H2BC9, H2BC8, H3C2, H2BC3, H3C14, H3C4, H1–5 |
| hsa-miR-193b-3p | 3.17E-04 | TOP2A, LEF1, CDCA7, APCDD1, SKA3, TYMS, CDC20, RACGAP1, PEG10, STMN1, H3C10, H3C2, H3C4, H1–5, HELLS, H2AC14, TICRR, CCNA2, TPX2, ASPM, SLC7A6, H3C15, MELK, PTK7, CDK1, NCAPD2, SPC25 |
| hsa-miR-98-5p | 2.02E-02 | PRKAA2, H2BC13, HMGA2, FIGN, NREP, DUSP9, H4C14, CCNA2, SLC7A6, H4C15, CCND2, PEG10, H2BC7, IGF2BP1, H4C8, OLR1, H2BC9, IGF2BP3, H2BC8, IGF2BP2, H3C4, H2BC5 |
Discussion
Although the survival rates of HB are relatively high, the main treatment methods being chemotherapy and surgical resection, present a great burden on the patient’s quality of life (Czauderna and Garnier 2018). Additionally, the treatment of high-risk patients is not satisfactory (Hiyama 2014). Therefore, exploring the molecular mechanisms of HB is pivotal for early diagnosis and improved treatment.
To better understand the pathogenesis and development of HB, we performed a bioinformatic analysis of microarray data GSE131329, which included 53 HB and 14 noncancerous liver tissue samples. Using GEO2R, we identified 221 up- and 373 down-regulated genes, comparing HB and normal tissue samples (Barrett et al. 2012). Biological process analysis of up-regulated DEGs showed that chromatin and nucleosome assembly, Wnt signaling pathway, cell cycle, and other enriched terms, as shown in Fig. 1/(Fig. 1) might play key roles in the progression of HB. It is well known that cell cycle processes serve an essential role in the development of many cancers (Evan and Vousden 2001). These genes are also involved in the cellular composition of spindle and kinetochore, which have critical functions in mitotic events (Sharp et al. 2000). Also, KEGG pathway analysis was enriched in transcriptional misregulation in cancer, Wnt signaling pathway, cell cycle, and p53 signaling pathway. Recently, researchers have found the effects of Wnt signaling on cancer stem cells and metastasis (Zhan et al. 2017). Mavila et al. revealed that Wnt/beta-catenin signaling abnormality has the highest rate in HB among human cancers (Mavila and Thundimadathil 2019). Moreover, many studies have been focused on cell cycle regulators such as CDKs and PLKs, as attractive targets in cancer therapy (Otto and Sicinski 2017). In summary, some of our results, especially the ones named above, are established terms related to carcinogenesis and tumor proliferation, thereby suggesting that our results may play key roles in the occurrence and development of HB, and shed light on the path of further research.
The PPI network was constructed and included 123 nodes (genes) and 919 edges (interactions). The significance of the PPI enrichment p value demonstrates that the proteins are at least partially biologically connected as a group. From the network, 5 modules were detected, and 15 genes were selected as hub genes. Furthermore, we validated 11 hub genes using Oncopression database: AURKA, CCNA2, CCNB1, CDK1, FOXM1, HIST1H2BB, HIST1H3F, MKI67, PBK, RACGAP1, TOP2A. MKI67 (Marker Of Proliferation Ki-67) is usually used to detect and quantify cell proliferation. Our BP analysis also revealed MKI67 to be involved in the regulation of the cell cycle process. Earlier research has shown that MKI67 is significantly increased in thyroid cancer, gastric carcinogenesis, breast cancer, and HCC (Cheah et al. 2008; Griffiths et al. 2008; Smallridge et al. 2009; Von Minckwitz et al. 2008). Cyclin family proteins bind and activate CDK kinases and function as regulators of the cell cycle (Pagano et al. 1992). Cyclin-A2 protein encoded by CCNA2, is a member of the cyclin family that is used as a marker of cell proliferation, similar to MKI67. It has also been shown that dysregulation of CCNA2 is associated with the epithelial-mesenchymal transition (Loukil et al. 2015). Previous studies have indicated that the overexpression of CCNA2 is associated with the development of different cancers, including gastric and ER+ breast cancer (T. Gao et al. 2014; Mrena et al. 2006). Shin et al. have also shown that CCNA2 is significantly up-regulated in HB, which is consistent with our findings (Shin et al. 2011). Our BP analysis further indicate that CCNA2 is enriched in G2/M transition of the mitotic cell cycle, binding to CDK1 while cells transition from G2 to M phase (Pagano et al. 1992). Cyclin-B1 (CCNB1) is also another member of the cyclin family and regulates the cell cycle. Prior to the present study, few studies have also addressed the association of the up-regulation of CCNB1 with HB (Bandopadhyay et al. 2016; Zhang et al. 2018b). GO and KEGG enrichment analysis showed that CCNB1 is associated with the cell cycle (G2/M transition), and early mitotic events including microtubule organization, spindle pole assembly, and nuclear envelope disassembly. Cyclin A2-Cdk1 triggers cyclin B1-Cdk1 activation and these complexes together bring about early mitotic events (13). CDK1 (Cyclin Dependent Kinase 1), along with some other CDKs, are involved in the progression of the cell cycle, and it is noteworthy that there has been an extensive research in the pursuit of drugs for CDK inhibitors for cancer therapy in recent years (Malumbres et al. 2008). Goga et al. demonstrated that CDK1 inhibitor treatment of MYC-driven hepatoblastoma transgenic mouse models, decreased tumor growth and prolonged their survival (Goga et al. 2007).
MF and CC analyses revealed that PLK1 (Polo Like Kinase 1) is involved in protein serine/threonine kinase activity, spindle pole, and microtubule organizing center. Due to its role in the cell cycle, PLK1 is involved in several cancers, such as melanomas, lymphomas, and carcinomas in different tissues (van de Weerdt and Medema 2006). Yamada et al. have reported that PLK1 oncogene is overexpressed in HB patients (Yamada et al. 2004). Plk1 promotes mitotic entry events by activating Cyclin B-Cdk1 and centrosome maturation in G2/M phase transition (14). BP analysis illustrated that PLK1 is also enriched in nuclear envelope disassembly, regulation of ubiquitin protein ligase activity, and anaphase-promoting complex-dependent catabolic process.
Aurora Kinase A, a protein serine/threonine kinase that is encoded by the AURKA gene, is involved in spindle formation and chromosome segregation (Goos et al. 2013). It begins to accumulate at centrosomes in the S phase, contributes to activation of the Cyclin B-Cdk1 complex, and is activated at the boundary between the G2 and M phases (Nikonova et al. 2013). Katsha et al. showed that AURKA controls STAT3 activation by regulating the expression and phosphorylation of JAK2 in vitro and has essential roles in gastric and esophageal cancers (El-Rifai 2014). In another study, it is reported that the overexpression of miR-26a-5p repressed HB cell proliferation and colony formation through its inhibition of the oncogenic LIN28B–RAN–AURKA pathway (Zhang et al. 2018c). RACGAP1 and TOP2A were two other hub genes, enriched in protein kinase binding according to MF analysis. RACGAP1 encodes a GTPase-activating protein (GAP), a component of the centralspindlin complex that is required for the contractile ring formation during cytokinesis (21). Overexpression of RacGAP1 was observed in the gastric cancer cells, with a significant correlation with age, tumor size, and lymph node metastasis (Saigusa et al. 2015). Another study reported that RACGAP1 was up-regulated in the recurrent HCC liver samples (Wang et al. 2007). DNA topoisomerase 2-alpha encoded by the TOP2A controls DNA topological structure, chromosome segregation, and cell cycle progression (Panvichian et al. 2015). Overexpression of TOP2A has been shown in many cancers such as liver, breast, colon, ovarian, prostate, and small cell lung cancers and HB itself to be a valuable prognostic marker for tumor advancements and recurrences (Panvichian et al. 2015; Shin et al. 2011; Wong et al. 2009).
SIN3A, FOXM1, and E2F4 were the TFs with the highest interactions with other genes. The protein encoded by SIN3A (Paired amphipathic helix protein) is a transcription regulator, which contains paired amphipathic helix (PAH) domains important for protein-protein interactions (26). It regulates gene expression by histone deacetylase (HDAC) activity and acts as a negative regulator of several cancer-related factors such as p53, Rb, and E2F (Das et al. 2013). Sin3a represses the MYC genes and saves the protein from oncogenic activities (26). It also interacts with STAT3 and promotes its deacetylation. FOXM1 (Forkhead Box M1), which was also among hub genes, is a proliferation-associated transcription factor that regulates critical biological processes, including cell proliferation, cell cycle progression, cell differentiation, angiogenesis, and apoptosis (Koo et al. 2012). Due to its overexpression in many cancers such as liver, prostate, breast, lung, colon, pancreas, and cervix, it is plausible that FOXM1 has a crucial role in tumorigenesis (Koo et al. 2012; Mei et al. 2017). FOXM1 transcription levels increase at the entry to the S-phase of the cell cycle and peaks during the G2/M transition which is necessary for the expression of Cdc25B, Ccnb1, Aurora B kinase, Plk-1, CENP-A, B, F, and survivin to allow mitotic progression, assembly of the mitotic spindles, accurate chromosome segregation, and cytokinesis (Halasi and Gartel 2013).
Among the top predicted protein kinases of the inferred upstream regulatory network, Cyclin-dependent kinases (CDKs) and mitogen-activated protein kinases (MAPKs) were enriched more than others. Consistent with our results, other studies have shown CDKs and MAPKs to be frequently overexpressed in HB tumors and suggested CDK-inhibitors for HB treatment (Adesina et al. 2009; Eichenmüller et al. 2012). CDKs are serine/threonine protein kinases and play a central role in cell cycle progression (Z. Wang et al. 2014). The Cdk4/cyclin D complex is required for progression through G1 by the response to the growth factors (Morgan 1997). Cdk2/cyclin E complex functions at the beginning of S phase to induce the initiation of DNA synthesis and then binds to the cyclin A throughout S phase (Morgan 1997). Cdk1/cyclin B complex, which is also known as M-phase promoting factor (MPF), regulates the entry into mitosis, nuclear envelope disassembly, and centrosome separation (Morgan 1997). MAPKs are serine/threonine protein kinases similar to CDKs and share common features with them, and are important components of pathways controlling cell differentiation, proliferation, and death (Pearson et al. 2001). ERK1 (MAPK3) and ERK2 (MAPK1) are activated by a broad spectrum of factors, including growth factors, insulin, GPCRs, and cytokines (Cargnello and Roux 2011). The ERK1/2 module which is activated principally by cell surface receptors such as RTKs plays a key role in the G1 to S phase progression (Cargnello and Roux 2011). MAPK14 (p38) negatively regulates cell cycle progression at both the G1/S and G2/M transitions by downregulation of cyclins and upregulation of CDK inhibitors (Cargnello and Roux 2011). Also, it is reported that p38 activity is associated with the induction of apoptosis by cellular stresses (Cargnello and Roux 2011).
Our miRNA-gene target analysis revealed hsa-miR-34a-5p, hsa-miR-193b-3p, and hsa-miR-760 to be the top miRNAs targeting the highest number of genes. miR-34a-5p has been reported to be a tumor suppressor in several cancers, including pancreatic, prostate, breast, colon cancer, and glioblastoma (Zhang et al. 2018a). In pancreatic cancer and glioblastoma, miR-34a inhibits cell proliferation by regulating Notch and TGF-β signaling networks, respectively (Zhang et al. 2018a). In colorectal cancer, miR-34a-5p induced cell cycle arrest at G1 phase and induced apoptosis in vitro by activation of p53/p21 and caspase-dependent pathways, respectively (Gao et al. 2015). Dong et al. have also reported that TUG1/miR-34a-5p/VEGFA network participates in regulating hypervascularity via VEGFA induction, HB cell function, and tumor progression and angiogenesis (Dong et al. 2016). miR-193b is down-regulated in many cancers such as lung, liver, prostate, breast cancer, neuroblastoma, and ovarian carcinoma. An in vitro study on the non-small cell lung cancer revealed that overexpression of miR-193b repressed the expressions of cyclin D1 and urokinase-type plasminogen activator and led to decreased proliferation, migration, and invasion of cells Hu et al. 2012). Another study showed that miR-193b functions as a tumor suppressor in HCC in vitro, as it induced cell cycle arrest and inhibited the invasion and migration of hepatoma cells by regulating CCND1 and ETS1 (Xu et al. 2010). It is reported that miR-760 is involved in the pathogenesis of various diseases and can be used as a biomarker to predict the progression of various cancers, including gastric, breast, colorectal, and colon cancer (Sun et al. 2018). Recently, a study demonstrated that miR-760 regulates breast cancer stem cell metastasis and gene expression by targeting Nanog dependent pathways (Han et al. 2016). Sun et al. showed that low expression of miR-760 was associated with higher overall survival for HCC patients (Sun et al. 2018). Conversely, miR-760 expression is down-regulated in HCC cell lines, while overexpression of miR-760 leads to an increase in cytotoxicity and apoptosis, probably by down-regulating Notch1 and increasing PTEN expression (Manvati et al. 2020).
While our study provides potential new insights for a better understanding of HB and improving current therapies, it is sensible to highlight some of the limitations of the study. Firstly, although we validated the expression levels of our hub genes through the Oncopression database, our study lacked any experimental analyses; thus, experimental verification of our results is necessary. In addition, due to the accessibility of data by bioinformatic arrays, our sample size was limited; therefore, large-scale research regarding the topic is needed. Finally, we cannot eliminate the possibility that the methods used to determine the hub genes or other results have missed the possible roles of some DEGs; consequently, other methods must be also considered.
Conclusion
In conclusion, we have revealed potential DEGs, PPI modules, hub genes, pathways, upstream regulators, and MTIs which may provide valuable insights into the underlying molecular events that lead to the occurrence and progression of HB. Of these, to the best of our knowledge, the association of RACGAP1, MKI67, FOXM1, SIN3A, miR-193b, and miR-760 with HB has been reported for the first time. These findings may emerge as useful therapeutic targets or prognostic indicators for future investigations on HB.
Acknowledgments
This study was supported by a grant from Shahid Beheshti University.
Compliance with ethical standards
Conflict of interests
The authors declare that there are no conflict of interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Taha Aghajanzadeh and Kiarash Tebbi contributed equally to this work.
References
- Adesina AM, Lopez-Terrada D, Wong KK, Gunaratne P, Nguyen Y, Pulliam J, Margolin J, Finegold MJ. Gene expression profiling reveals signatures characterizing histologic subtypes of hepatoblastoma and global deregulation in cell growth and survival pathways. Hum Pathol. 2009;40(6):843–853. doi: 10.1016/j.humpath.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4(1):2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandopadhyay M, Sarkar N, Datta S, Das D, Pal A, Panigrahi R, et al. Hepatitis B virus X protein mediated suppression of miRNA-122 expression enhances hepatoblastoma cell proliferation through cyclin G1-p53 axis. Infect Agents Cancer. 2016;11(1):40. doi: 10.1186/s13027-016-0085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2012;41(D1):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell D, Ranganathan S, Tao J, Monga SP. Novel advances in understanding of molecular pathogenesis of hepatoblastoma: a Wnt/β-catenin perspective. Gene Expression J Liver Res. 2017;17(2):141–154. doi: 10.3727/105221616X693639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegari E, Gramantieri L, Domenicali M, D'Abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differentiation. 2015;22(1):46–57. doi: 10.1038/cdd.2014.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75(1):50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Reixach J, Torrens L, Simon-Coma M, Royo L, Domingo-Sàbat M, Abril-Fornaguera J, et al. Epigenetic footprint enables molecular risk stratification of hepatoblastoma with clinical implications. J Hepatol. 2020;73:328–341. doi: 10.1016/j.jhep.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah P-L, Looi L-M, Nazarina AR, Mun K-S, Goh K-L. Association of Ki67 with raised transaminases in hepatocellular carcinoma. Malay. 2008;4:12.19. [PubMed] [Google Scholar]
- Chin C-H, Chen S-H, Wu H-H, Ho C-W, Ko M-T, Lin C-Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC systems biology. 2014;8(S4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C-H, Shrestha S, Yang C-D, Chang N-W, Lin Y-L, Liao K-W, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DJB, Kuleshov MV, Schilder BM, Torre D, Duffy ME, Keenan AB, et al. eXpression2Kinases (X2K) web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Res. 2018;46(W1):W171–W179. doi: 10.1093/nar/gky458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna P, Garnier H (2018) Hepatoblastoma: current understanding, recent advances, and controversies. F1000Research 7 [DOI] [PMC free article] [PubMed]
- Das T, Sangodkar J, Negre N, Narla G, Cagan R. Sin3a acts through a multi-gene module to regulate invasion in Drosophila and human tumors. Oncogene. 2013;32(26):3184–3197. doi: 10.1038/onc.2012.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ioris M, Brugieres L, Zimmermann A, Keeling J, Brock P, Maibach R, et al. Hepatoblastoma with a low serum alpha-fetoprotein level at diagnosis: the SIOPEL group experience. Eur J Cancer. 2008;44(4):545–550. doi: 10.1016/j.ejca.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Dong R, Liu G, Liu B, Chen G, Li K, Zheng S, Dong K. Targeting long non-coding RNA-TUG1 inhibits tumor growth and angiogenesis in hepatoblastoma. Cell Death Dis. 2016;7(6):e2278–e2278. doi: 10.1038/cddis.2016.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenmüller M, von Schweinitz D, Kappler R. Therapeutic potential of CDK-inhibitors in hepatoblastoma. Klin Padiatr. 2012;224(03):A34. [Google Scholar]
- El-Rifai W (2014) AURKA regulates JAK2eSTAT3 activity in human gastric and esophageal cancers [DOI] [PMC free article] [PubMed]
- Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- Feng J, Polychronidis G, Heger U, Frongia G, Mehrabi A, Hoffmann K. Incidence trends and survival prediction of hepatoblastoma in children: a population-based study. Cancer Communications. 2019;39(1):62. doi: 10.1186/s40880-019-0411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Han Y, Yu L, Ao S, Li Z, Ji J (2014) CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen resistance. PLoS One, 9(3) [DOI] [PMC free article] [PubMed]
- Gao J, Li N, Dong Y, Li S, Xu L, Li X, et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. 2015;34(31):4142–4152. doi: 10.1038/onc.2014.348. [DOI] [PubMed] [Google Scholar]
- Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med. 2007;13(7):820–827. doi: 10.1038/nm1606. [DOI] [PubMed] [Google Scholar]
- Goos JA, Coupé VM, Diosdado B, Delis-van Diemen PM, Karga C, Beliën JA, et al. Aurora kinase a (AURKA) expression in colorectal cancer liver metastasis is associated with poor prognosis. Br J Cancer. 2013;109(9):2445–2452. doi: 10.1038/bjc.2013.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths EA, Pritchard S, McGrath S, Valentine HR, Price PM, Welch I, West CM. Hypoxia-associated markers in gastric carcinogenesis and HIF-2α in gastric and gastro-oesophageal cancer prognosis. Br J Cancer. 2008;98(5):965–973. doi: 10.1038/sj.bjc.6604210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halasi M, Gartel AL. Targeting FOXM1 in cancer. Biochem Pharmacol. 2013;85(5):644–652. doi: 10.1016/j.bcp.2012.10.013. [DOI] [PubMed] [Google Scholar]
- Han M-l, Wang F, Gu Y-t, Pei X-H, Ge X, Guo G-c, et al. MicroR-760 suppresses cancer stem cell subpopulation and breast cancer cell proliferation and metastasis: by down-regulating NANOG. Biomed Pharmacother. 2016;80:304–310. doi: 10.1016/j.biopha.2016.03.024. [DOI] [PubMed] [Google Scholar]
- Hiyama E. Pediatric hepatoblastoma: diagnosis and treatment. Transl Pediatr. 2014;3(4):293–299. doi: 10.3978/j.issn.2224-4336.2014.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Li S, Liu J, Ni B. MicroRNA-193b modulates proliferation, migration, and invasion of non-small cell lung cancer cells. Acta Biochim Biophys Sin. 2012;44(5):424–430. doi: 10.1093/abbs/gms018. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo C-Y, Muir KW, Lam EW-F. FOXM1: from cancer initiation to progression and treatment. Biochimica et Biophysica Acta (BBA)-Gene Regul Mech. 2012;1819(1):28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Choi C. Oncopression: gene expression compendium for cancer with matched normal tissues. Bioinformatics. 2017;33(13):2068–2070. doi: 10.1093/bioinformatics/btx121. [DOI] [PubMed] [Google Scholar]
- Loukil A, Cheung CT, Bendris N, Lemmers B, Peter M, Blanchard JM. Cyclin A2: at the crossroads of cell cycle and cell invasion. World J Biol Chem. 2015;6(4):346–350. doi: 10.4331/wjbc.v6.i4.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrelli A, Azzalin G, Salvatore M, Viganotti M, Tosto F, Colombo T, et al. Altered microRNA expression patterns in hepatoblastoma patients. Transl Oncol. 2009;2(3):157–163. doi: 10.1593/tlo.09124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Pevarello P, Barbacid M, Bischoff JR. CDK inhibitors in cancer therapy: what is next? Trends Pharmacol Sci. 2008;29(1):16–21. doi: 10.1016/j.tips.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Manvati MKS, Khan J, Verma N, Dhar PK (2020) Association of miR-760 with cancer: an overview. Gene 144648 [DOI] [PubMed]
- Mavila N, Thundimadathil J. The emerging roles of Cancer stem cells and Wnt/Beta-catenin signaling in Hepatoblastoma. Cancers. 2019;11(10):1406. doi: 10.3390/cancers11101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Yang J-P, Qian C-N. For robust big data analyses: a collection of 150 important pro-metastatic genes. Chin J Cancer. 2017;36(1):16. doi: 10.1186/s40880-016-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokhlesi A, Talkhabi M (2020) Comprehensive transcriptomic analysis identifies novel regulators of lung adenocarcinoma. J Cell Commun Signal [DOI] [PMC free article] [PubMed]
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13(1):261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Mrena J, Wiksten JP, Kokkola A, Nordling S, Haglund C, Ristimäki A. Prognostic significance of cyclin a in gastric cancer. Int J Cancer. 2006;119(8):1897–1901. doi: 10.1002/ijc.21944. [DOI] [PubMed] [Google Scholar]
- Nikonova AS, Astsaturov I, Serebriiskii IG, Dunbrack RL, Golemis EA. Aurora a kinase (AURKA) in normal and pathological cell division. Cell Mol Life Sci. 2013;70(4):661–687. doi: 10.1007/s00018-012-1073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17(2):93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin a is required at two points in the human cell cycle. EMBO J. 1992;11(3):961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panvichian R, Tantiwetrueangdet A, Angkathunyakul N, Leelaudomlipi S (2015) TOP2A amplification and overexpression in hepatocellular carcinoma tissues. BioMed Res Int [DOI] [PMC free article] [PubMed]
- Pearson G, Robinson F, Beers Gibson T, Xu B-e, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22(2):153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- Saigusa S, Tanaka K, Mohri Y, Ohi M, Shimura T, Kitajima T, et al. Clinical significance of RacGAP1 expression at the invasive front of gastric cancer. Gastric Cancer. 2015;18(1):84–92. doi: 10.1007/s10120-014-0355-1. [DOI] [PubMed] [Google Scholar]
- Semeraro M, Branchereau S, Maibach R, Zsiros J, Casanova M, Brock P, et al. Relapses in hepatoblastoma patients: clinical characteristics and outcome–experience of the international childhood liver tumour strategy group (SIOPEL) Eur J Cancer. 2013;49(4):915–922. doi: 10.1016/j.ejca.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407(6800):41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- Shin E, Lee K-B, Park S-Y, Kim S-H, Ryu H-S, Park Y-N, Yu E, Jang J-J. Gene expression profiling of human hepatoblastoma using archived formalin-fixed and paraffin-embedded tissues. Virchows Arch. 2011;458(4):453–465. doi: 10.1007/s00428-011-1043-8. [DOI] [PubMed] [Google Scholar]
- Smallridge RC, Marlow LA, Copland JA. Anaplastic thyroid cancer: molecular pathogenesis and emerging therapies. Endocr Relat Cancer. 2009;16(1):17–44. doi: 10.1677/ERC-08-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazin P, Chen Y, Treviño LR, Sarabia SF, Hampton OA, Patel K, et al. Genomic analysis of hepatoblastoma identifies distinct molecular and prognostic subgroups. Hepatology. 2017;65(1):104–121. doi: 10.1002/hep.28888. [DOI] [PubMed] [Google Scholar]
- Sun D, Lu J, Hu C, Zhang Q, Wang X, Zhang Z, Hu S. Prognostic role of miR-760 in hepatocellular carcinoma. Oncol Lett. 2018;16(6):7239–7244. doi: 10.3892/ol.2018.9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkhabi M, Razavi SM, Salari A. Global transcriptomic analysis of induced cardiomyocytes predicts novel regulators for direct cardiac reprogramming. J Cell Commun Signal. 2017;11(2):193–204. doi: 10.1007/s12079-017-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Weerdt BC, Medema RH. Polo-like kinases: a team in control of the division. Cell Cycle. 2006;5(8):853–864. doi: 10.4161/cc.5.8.2692. [DOI] [PubMed] [Google Scholar]
- Von Minckwitz G, Sinn H-P, Raab G, Loibl S, Blohmer J-U, Eidtmann H, et al. Clinical response after two cycles compared to HER2, Ki-67, p53, and bcl-2 in independently predicting a pathological complete response after preoperative chemotherapy in patients with operable carcinoma of the breast. Breast Cancer Res. 2008;10(2):R30. doi: 10.1186/bcr1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SM, Ooi LLP, Hui KM. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin Cancer Res. 2007;13(21):6275–6283. doi: 10.1158/1078-0432.CCR-06-2236. [DOI] [PubMed] [Google Scholar]
- Wang Z, Fan M, Candas D, Zhang T-Q, Qin L, Eldridge A, et al. Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev Cell. 2014;29(2):217–232. doi: 10.1016/j.devcel.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N, Yeo W, Wong WL, Wong NLY, Chan KYY, Mo FKF, et al. TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer. 2009;124(3):644–652. doi: 10.1002/ijc.23968. [DOI] [PubMed] [Google Scholar]
- Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J, Xing R, Jin Y, Sun Z, Zheng X. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer. 2010;46(15):2828–2836. doi: 10.1016/j.ejca.2010.06.127. [DOI] [PubMed] [Google Scholar]
- Yamada S-i, Ohira M, Horie H, Ando K, Takayasu H, Suzuki Y, et al. Expression profiling and differential screening between hepatoblastomas and the corresponding normal livers: identification of high expression of the PLK1 oncogene as a poor-prognostic indicator of hepatoblastomas. Oncogene. 2004;23(35):5901–5911. doi: 10.1038/sj.onc.1207782. [DOI] [PubMed] [Google Scholar]
- Yang Z, Deng Y, Zhang K, Bai Y, Zhu J, Zhang J, Xin Y, Li L, He J, Wang W. LIN28B gene polymorphisms modify hepatoblastoma susceptibility in Chinese children. J Cancer. 2020;11(12):3512–3518. doi: 10.7150/jca.42798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36(11):1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HF, Wang YC, Han YD. MicroRNA-34a inhibits liver cancer cell growth by reprogramming glucose metabolism. Mol Med Rep. 2018;17(3):4483–4489. doi: 10.3892/mmr.2018.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Jin Y, Zheng K, Wang H, Yang S, Lv C, et al. Whole-genome sequencing identifies a novel variation of was gene coordinating with heterozygous Germline mutation of Apc to enhance Hepatoblastoma Oncogenesis. Front Genet. 2018;9:668. doi: 10.3389/fgene.2018.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao Y, Wu J, Liangpunsakul S, Niu J, Wang L. MicroRNA-26-5p functions as a new inhibitor of hepatoblastoma by repressing lin-28 homolog B and aurora kinase a expression. Hepatol Commun. 2018;2(7):861–871. doi: 10.1002/hep4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]