Abstract
(+)-Borneol is a desirable monoterpenoid with effective anti-inflammatory and analgesic effects that is known as soft gold. (+)-bornyl diphosphate synthase is the key enzyme in the (+)-borneol biosynthesis pathway. Despite several reported (+)-bornyl diphosphate synthase genes, relatively low (+)-borneol production hinders the attempts to synthesize it using microbial fermentation. Here, we identified the highly specific (+)-bornyl diphosphate synthase CbTPS1 from Cinnamomum burmanni. An in vitro assay showed that (+)-borneol was the main product of CbTPS1 (88.70% of the total products), and the Km value was 5.11 ± 1.70 μM with a kcat value of 0.01 s–1. Further, we reconstituted the (+)-borneol biosynthetic pathway in Saccharomyces cerevisiae. After tailored truncation and adding Kozak sequences, the (+)-borneol yield was improved by 96.33-fold to 2.89 mg⋅L–1 compared with the initial strain in shake flasks. This work is the first reported attempt to produce (+)-borneol by microbial fermentation. It lays a foundation for further pathway reconstruction and metabolic engineering production of this valuable natural monoterpenoid.
Keywords: (+)-borneol, (+)-bornyl diphosphate synthase, Cinnamomum burmanni, metabolic engineering, Saccharomyces cerevisiae
Introduction
The monoterpene borneol is a highly desirable natural product widely used in medicine, spice, and chemical fields since ancient times (Wojtunik-Kulesza et al., 2019). It has a broad spectrum of bidirectional regulation on the central nervous system (Zhang et al., 2017; Zheng et al., 2018); anti-inflammatory (Zou et al., 2017; Ji et al., 2020) and antimicrobial activities (Xin et al., 2020); and increases biofilm barrier permeability (Song et al., 2018; Chen et al., 2019). Borneol is divided into (+)-borneol and (−)-borneol according to optical rotations. Natural (+)-borneol has primarily been extracted from Cinnamomum camphora (L.) Presl and C. burmanni (Nees et T.Nees) Blume (borneol-type) since the 1980s in China (Chen et al., 2010). However, the slow growth rate, low (+)-borneol levels, and restricted cultivation area mean that the yield of natural (+)-borneol is far from meeting the market demand. Borneol synthesized by chemical methods thus occupies most of the market share; however, a certain number of toxic compounds, such as isoborneol may exist in synthetic borneol. Thus, it is necessary to explore other methods to produce natural (+)-borneol.
Due to the clear genetic background and lack of susceptibility to phage infections, Saccharomyces cerevisiae is the preferred host for metabolic engineering (Kirby and Keasling, 2009; Liu et al., 2019; Nielsen, 2019). Many monoterpenoids, such as geraniol, limonene, linalool, and α-terpineol (Figure 1) have been produced in S. cerevisiae (Amiri et al., 2016; Cao et al., 2016, 2017; Zhang et al., 2019, 2020). Isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP) derived from the mevalonate pathway (MVA) are the precursors of all terpenoids in S. cerevisiae, and geranyl diphosphate (GPP) is the direct precursor of monoterpenes catalyzed by farnesyl diphosphate synthase (ERG20) (Jiang et al., 2017). Thus, in order to produce monoterpenes in yeast, ERG20 is usually mutated or rationally designed into GPP synthase (Ignea et al., 2014; Zhao et al., 2016; Jiang et al., 2017). The upstream MVA pathway genes, tHMG1 and IDI1, are frequently overexpressed in yeast (Zhao et al., 2016; Zhang et al., 2019). Guo et al. (2018) even overexpressed all MVA pathway genes (ERG10, ERG13, tHMG1, ERG12, ERG8, ERG19, IDI1, ERG20) to increase the GPP pool. Modification of the monoterpene synthase, including translational fusion and truncation of transit peptides at the N-terminus of the enzymes, is also an effective strategy for increasing the production of terpenes (Jongedijk et al., 2015; Ignea et al., 2019; Hu et al., 2020).
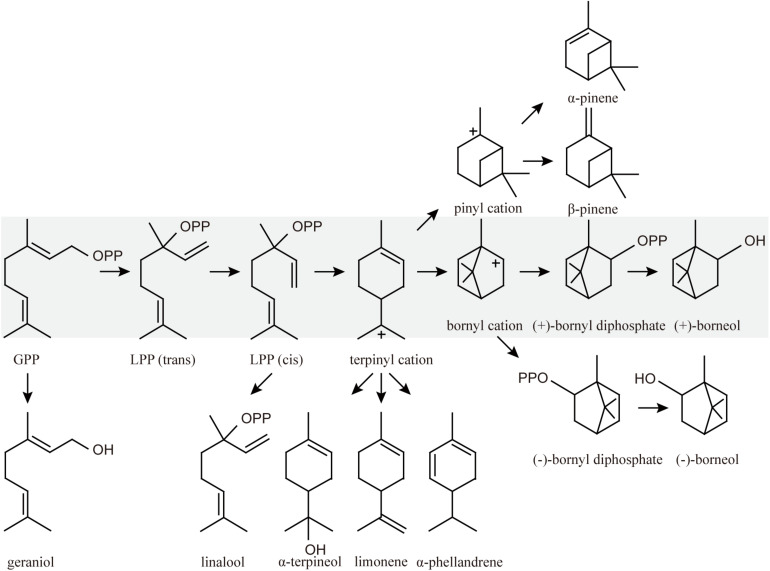
FIGURE 1.
Proposed mechanism for BPPS. The primary pathway leads to the formation of borneol (gray) and other monoterpenoid products.
Like other monoterpenes, bornyl diphosphate synthase (BPPS) is the key enzyme involved in (+)-borneol biosynthesis. It catalyzes the universal precursor GPP to form (+)-bornyl diphosphate, and is then dephosphorylated to produce the target product (+)-borneol (Figure 1). BPPSs have been identified from several plants, including Salvia officinalis (SBS), Lavandula angustifolia (LaBPPS), Lippia dulcis (LdBPPS), and Amomum villosum (AvBPPS) (Wise et al., 1998; Despinasse et al., 2017; Hurd et al., 2017; Wang et al., 2018). However, all these enzymes produced multiple products, such as α-pinene, β-pinene, camphene, and limonene, with the largest amount of (+)-borneol produced by SBS, accounting for 57.8% of the total products. However, there have been no attempts to produce this valuable product by microbial cell factories.
Here we report a high-specificity (+)-borneol BBPS gene (CbTPS1) from C. burmanni. Among the products with GPP as substrate in an in vitro assay, (+)-borneol accounted for 88.70% of the total. We thus aimed to construct a (+)-borneol biosynthesis pathway in S. cerevisiae. To reach the target, eight genes involved in the MVA pathway were overexpressed. Truncated transit peptides and adding the Kozak sequence of CbTPS1 further improved the (+)-borneol production. Our work provides a good example for (+)-borneol production in microbial fermentation.
Materials and Methods
Plant Materials and Chemicals
Leaves of Cinnamomum burmanni (Nees et T.Nees) Blume were obtained from Guangdong Huaqingyuan Technology Co., Ltd. Plant leaf material grown in natural conditions was picked in May 2019. C. burmannii was identified by Prof. Cui Guanghong of China Academy of Chinese Medical Sciences and stored at −80°C for further usage (Storage Number: YXS201905). GPP, geraniol, α-pinene, β-pinene, α-phellandrene, limonene, α-terpineol, (+)-borneol and (−)-borneol standards were purchased from Sigma-Aldrich Chemical Co., United States.
RNA Extraction, cDNA Synthesis
The total RNA from C. burmannii leaves was extracted using a quick RNA isolation kit (HuaYueYang Biotechnology, China) based on the manufacturer’s protocol, and then digested and purified by RNase-free DNase I (TaKaRa, Japan). An aliquot containing 1 μg total RNA was used to synthesize the first-strand cDNA with TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotechnology, China) according to the manufacturer’s guidelines.
BPPS Candidate Selection and Analysis
Transcriptomic libraries of the C. burmannii leaves were shipped to the Novogene Company1 for library construction and RNA-seq. The Illumina-derived nucleotide sequences reported in this paper have been submitted to China National Center for Bioinformation2 under accession number CRA003558. To mine the BPPS candidate genes, TBLASTN analysis of BPPSs in the C. burmannii transcriptome was carried out using BioEdit software (Su et al., 2018). SBS (GenBank Accession Number: AAC26017), LaBPPS (GenBank Accession Number: AJW68082), LdBPPS (GenBank Accession Number: ATY48638), and AvBPPS (GenBank Accession Number: AWW87313) were used as the query sequences. The CbTPS1 (GenBank Accession Number: MW196671) sequence was analyzed using NCBI3. The open reading frames (ORFs) were identified using the ORF Finder4, and deduced amino acid sequences were identified using ExPASy5. Multiple sequence alignments were conducted using CLC Bio Sequence Viewer 66. The chloroplast transit peptide of CbTPS1 was predicted by ChloroP7.
All statistical analyses were conducted using SPSS version 23.0 (SPSS Inc., Chicago, IL, United States) for windows. One-way analysis of variance was used to compare the mean difference in (+)-borneol of strains. The P-value of less than 0.05 considered statistically significant.
Gene Cloning, Protein Expression and Purification
The ORF was cloned using specially designed primers (Supplementary Table 1). Phusion High-Fidelity PCR Master Mix (New England BioLabs, United States) was used for amplification reaction according to the included protocol. PCR products were purified, and then ligated into the pEASY®-Blunt Simple Cloning Vector (TransGen Biotech, China) and transformed into E. coli DH5α cells. Positive colonies were verified by sequencing (Beijing RuiBo Biotechnology Co., Ltd., China) and then subcloned into the pET-32a (+) expression vector (Novagen, United States) according to the protocol of the pEASY®-Uni Seamless Cloning and Assembly Kit (TransGen Biotech, China) (Supplementary Table 1).
Recombinant proteins were expressed and purified following the methods described previously (Ma et al., 2020), with some modifications as follows: the 200 mL bacterial solution was centrifuged (5,000 × g, 5 min, 4°C) to collect the cell pellets, and resuspended in 5 ml assay buffer (50 mM HEPES, pH 7.2, 10 mM MgCl2, 5 mM dithiothreitol), and then a sonicator was used to lyse cells. The lysates were centrifuged (12,000 × g, 30 min, 4°C) to produce crude protein. And then the His-tagged purified proteins were eluted using a buffer equivalent to the binding buffer but supplemented with different concentrations of imidazole (50, 100, 250, 350, and 500 mM). Fractions containing the target protein were pooled together and concentrated to a volume of 1 mL using an Amicon Ultra-15 centrifugal filter unit with an Ultracel-30 membrane (Merck Millipore, Germany). Protein concentrations were determined using the Bradford Assay (Cowin Biotech, China). The protein samples were assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
In vitro Enzyme Assays and Kinetic Assays
In vitro enzyme assays followed the method described below: enzyme assays were performed in 300 μL, containing 50 mM HEPES (Ph 7.2), 10 mM MgCl2, 5 mM DTT, 1 mM PMSF, 380 nM of the enzyme and 50 μM GPP, incubated for 1 h at 30°C. Then 1.5 μL calf intestinal alkaline phosphatase (CIAP) (TaKaRa, Japan) was added, followed by incubation for 2 h at 37°C to allow enzymatic dephosphorylation. Time-course experiments were carried out to obtain the initial speed of the enzymatic reaction from 1 to 180 min (Supplementary Figure 1). Then, 3 min was used in the kinetic assays. The enzyme assays were performed in a 300 μL reaction volume at 30°C. A concentration that ranged from 0.125 to 150 μM GPP substrate was used. After 3 min incubation, the reaction was terminated at 80°C for 3 min, followed by quenching in ice, and then added 1.5 μL CIAP, followed by incubation for 30 min at 37°C. Assay products were extracted twice with 300 μL of hexane and samples were concentrated under a gentle nitrogen flow. The samples were then redissolved with 100 μL of hexane before analysis with gas chromatography coupled with mass spectrometry (GC-MS) (described below).
The GraphPad Prism version 5 for Windows (GraphPad Software, La Jolla California United States)8 was used to obtain kinetic parameters by fitting the obtained data to the Michaelis-Menten equation. All assays were performed in triplicate.
Construction of (+)-Borneol Producing Strains
The initial strain used in this study was CEN.PK2-1D derived from S. cerevisiae (Table 1). All the endogenous genes (ERG10, ERG13, tHMG1, ERG12, ERG8, ERG19, IDI1, ERG20) involved in the MVA pathway were amplified from CEN.PK2-1D genomic DNA. The mutant of ERG20, ERG20F96W–N127W, used in this work was reported to possess higher efficiency for monoterpene production (Jiang et al., 2017). The M2S integration method was applied to integrate gene expression cassettes into the yeast chromosome (Li et al., 2016). Briefly, ERG10 and ERG13 were amplified with the addition of a BsaI digestion site and ligated with head-to-head promoters (pGAL1-pGAL10) into the terminator vector T1-(TPI1-PGI1), resulting in the plasmid T1-(ERG10-ERG13). Two terminators were inserted into the scaffold plasmid, with dedicated homologous arms L1 and L2 lying on both sides. Similarly, plasmids T2-(tHMG1-tHMG1), T3-(tHMG1-ERG12), T4-(ERG8-ERG19), and T5-(IDI1-ERG20F96W–N127W) were generated with dedicated homologous arms L2 and L3, L3 and L4, L4 and L5, L5 and L6, respectively. Each expression cassette with designed homologous arms was amplified individually. The integration site YPRCΔ15 was chosen as the target locus, and URA3 was chosen as the selection marker. The upstream homologous arm YPRCΔ15-UP was amplified from CEN.PK2-1D genomic DNA; URA3 cassette including the promoter was amplified from pESC-URA vector; and L1 arm was amplified from terminator vector T1. These three parts were assembled to form the selection marker module YPRCΔ15UP-URA3-L1 through overlap extension PCR. The downstream homologous arm YPRCΔ15DOWN was amplified from CEN.PK2-1D genomic DNA and the L6 arm was amplified from terminator vector T5, and they were then combined to generate the downstream homologous arm module L6-YPRCΔ15DOWN. All the amplified fragments were used to co-transform CEN.PK2-1D for assembly and integration, and transformants were selected on synthetic drop in medium-Ura (SD-Ura) containing 20 g⋅L–1 glucose and 18 g⋅L–1 agar. Positive transformants were verified by sequencing, yielding the strain MD.
TABLE 1.
Information of strains and vectors used in this study.
| Strains or vectors | Description | Source |
| CEN.PK2-1D | MATα, URA3-52, TRP1-289, LEU2-3112, HIS3Δ1, MAL2-8C, SUC2 | EUROSCARF |
| MD | CEN.PK2-1D, YPRC△15 URA3-PGAL1-ERG10-TTPI1-PGAL10-ERG13-TPGI-PGAL1-tHMG1-TADH1-PGAL10-tHMG1-TCYC1-PGAL1-tHMG1-TFBA1-PGAL10-ERG12-TPDC1-PGAL1-ERG8-TRPS2-PGAL10-ERG19-TTDH1-PGAL1-IDI1-TCCW12-PGAL10-ERG20F96W–N127W-TRPL9A | This study |
| MD-1 | MD, pESC-LEU::CbTPS1 | This study |
| MD-2 | MD, pESC-LEU::CbTPS1K | This study |
| MD-3 | MD, pESC-LEU::t10-CbTPS1 | This study |
| MD-4 | MD, pESC-LEU::t10-CbTPS1K | This study |
| MD-5 | MD, pESC-LEU::t32-CbTPS1 | This study |
| MD-6 | MD, pESC-LEU::t32-CbTPS1K | This study |
| MD-7 | MD, pESC-LEU::t37-CbTPS1 | This study |
| MD-8 | MD, pESC-LEU::t37-CbTPS1K | This study |
| T1-(TPI1-PGI) | Terminator vector with terminators TPI1 and PGI | This study |
| T2-(ADH1-CYC1) | Terminator vector with terminators ADH1 and CYC1 | This study |
| T3-(FBA1-PDC1) | Terminator vector with terminators FBA1 and PDC1 | This study |
| T4-(RPS2-TDH1) | Terminator vector with terminators RPS2 and TDH1 | This study |
| T5-(CCW12-RPL9A) | Terminator vector with terminators CCW12 and RPL9A | This study |
For (+)-borneol production, the yeast codon-optimized CbTPS1 as well as three truncated variants of CbTPS1 (at positions S10, S32 and C37) were cloned into the BamHI site of the pESC-Leu vector (Agilent Technologies, United States) according to the pEASY-Uni Seamless Cloning and Assembly Kit (TransGen Biotech, Beijing, China), yielding the plasmids pESC-LEU:CbTPS1, pESC-LEU:t10-CbTPS1, pESC-LEU:t32-CbTPS1, and pESC-LEU:t37-CbTPS1. Further, yeast-specific Kozak sequence was added in front of the START codon ATG of CbTPS1 and the three truncated variants, generating pESC-LEU:CbTPS1K, pESC-LEU:t10-CbTPS1K, pESC-LEU:t32-CbTPS1K, and pESC-LEU:t37-CbTPS1K. Plasmids with the correct sequence were transferred to the host strain MD using Frozen-EZ Yeast Transformation IITM (Zymo Research, United States) to obtain the (+)-borneol producing strains (Table 1). All the primers used are listed in Supplementary Table 1.
Shake Flask Fermentation
For shake flask fermentation, the positive strains were cultured in flasks (50 ml) containing 10 ml of synthetic drop-out medium without leucine and uracil (SD-Leu-Ura) (FunGenome, China) at 30°C and 200 rpm for 48 h. Next, the cells were collected and induced by GAL promoters in 10 ml of YPL (1% yeast extract, 2% peptone, and 2% galactose) medium at 30°C and 200 rpm for 48 h. The fermentation products were extracted with an equal volume of ethyl acetate for 1 h, and centrifuged at 13,000 × g for 10 min to separate the upper organic phase for analyzing by GC-MS (described below). The calibration curves for content determination are shown in Supplementary Figure 2. All assays were performed in triplicate.
Analysis Using GC-MS
The assay was carried out using a Trace 1310 series GC with a TSQ8000 MS detector (Thermo Fisher Scientific, United States). A TR-5 ms capillary column (30 m × 0.25 mm i.d., 0.25 μm film thickness; Thermo Fisher Scientific, United States) was used. The carrier gas for GC was helium at a flow rate of 1.0 mL⋅min–1. The oven program was as follows: 50°C for 2 min, linear ramp up at a rate of 5°C⋅min–1 to 230°C, held at 230°C for 5 min, followed with a linear ramp up at a rate of 10°C⋅min–1 to 300°C, held at 300°C for 2 min. The injector temperature and transfer line temperature were 280°C.
A chiral column, Agilent CycloSil-B (30 m × 0.25 mm i.d., 0.25 μm film thickness), was used to identify the chirality of the assay product and the content of borneol and camphor in C. camphora leaves. The carrier gas for GC was helium at a flow rate of 1.0 mL⋅min–1. The oven program was as follows: 50°C for 2 min, followed by a gradient from 50°C to 180°C at 5°C⋅min–1, then 10°C⋅min–1 to 230°C, held at 230°C for 2 min. The injector temperature was 200°C, and the transfer line temperature was 230°C.
Results
Transcriptome-Based Discovery of (+)-Bornyl Diphosphate Synthase in 2
Based on the high abundance of (+)-borneol in the leaves of C. burmanni (Shi et al., 2013), we used RNA isolated from young leaves to produce the transcriptome sequences. The reported BPPS genes were further queried against the de novo assembly of these sequences, showing that trinity_1267_c0_g1_i1 had the highest identity with all the reported genes. Trinity_1267_c0_g1_i1 was present as full-length sequence. It was further cloned using specific primers and annotated as CbTPS1.
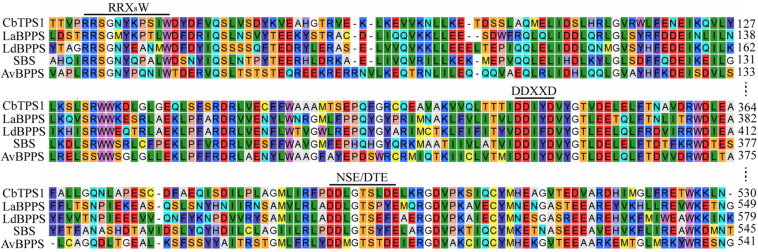
CbTPS1 has an open reading frame of 1,812 bp that encodes a 603-residue enzyme with a calculated molecular mass of 69.1 kDa (Figure 2A). It was classified into the TPS-b subfamily, which contains three motifs of typical terpene synthases, namely the RRX8W motif responsible for monoterpenoid cyclization (Chen et al., 2011); and the DDXXD and NSE/DTE motifs in the C-terminal domain, which are responsible for metal-dependent ionization and substrate binding (Chen et al., 2011). Homologous alignment analysis showed that CbTPS1 shared highest sequence identities with SBS (41.75%) from Salvia officinalis (Wise et al., 1998; Figure 3), followed by AvBPPS (40.10%) from Amomum villosum (Wang et al., 2018), LaBPPS (38.68%) from Lavandula angustifolia (Despinasse et al., 2017), and LdBPPS (36.20%) from Lippia dulcis (Hurd et al., 2017).
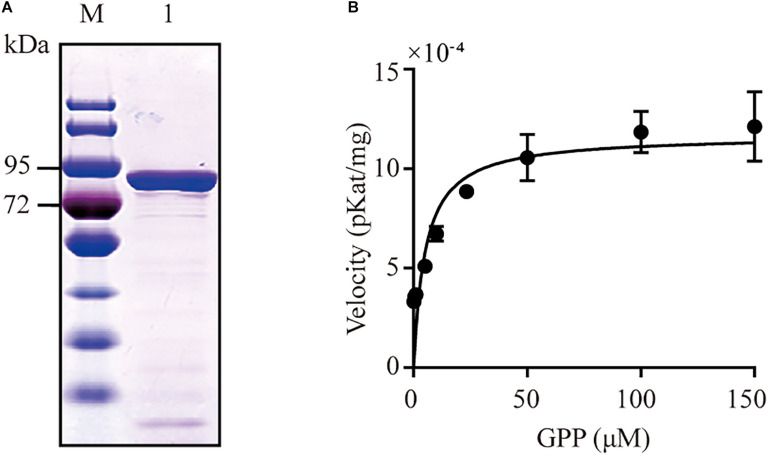
FIGURE 2.
SDS-PAGE and kinetic assays analysis of CbTPS1. (A) SDS-PAGE of CbTPS1 in vitro assays. (B) Velocity of CbTPS1 at increasing GPP concentrations.
FIGURE 3.
Partial alignment of CbTPS1 protein sequence with those of other known BPPSs. LaBPPS (accession No. AJW68082); LdBPPS (accession No. ATY48638); SBS (accession No. AAC26017); AvBPPS (accession No. AWW87313). The conserved RRX8W, DDXXD and NSE/DTE motifs are underlined.
Functional Analysis of CbTPS1
The recombinant protein of CbTPS1 was expressed in E. coli Transetta (DE3) cells using the pET-32a (+) expression vector, and then its function was identified with GPP as a substrate. CIAP was then added to remove the diphosphate group from the intermediate product. The purified CbTPS1 produced several monoterpenes (Figure 4A). Borneol was predominant (88.70%) with small amounts of α-pinene (2.70%), β-pinene (0.76%), α-phellandrene (1.20%), limonene (2.37%), and other minor monoterpenoids (4.27%). CbTPS1 was further examined for its catalytic properties and the Km value was 5.11 ± 1.70 μM with a kcat value of 0.01 s–1 (Figure 2B). In parallel, no product formation was found when the empty vector was transformed into E. coli Transetta (DE3) cells, and no product was produced in the absence of CIAP.
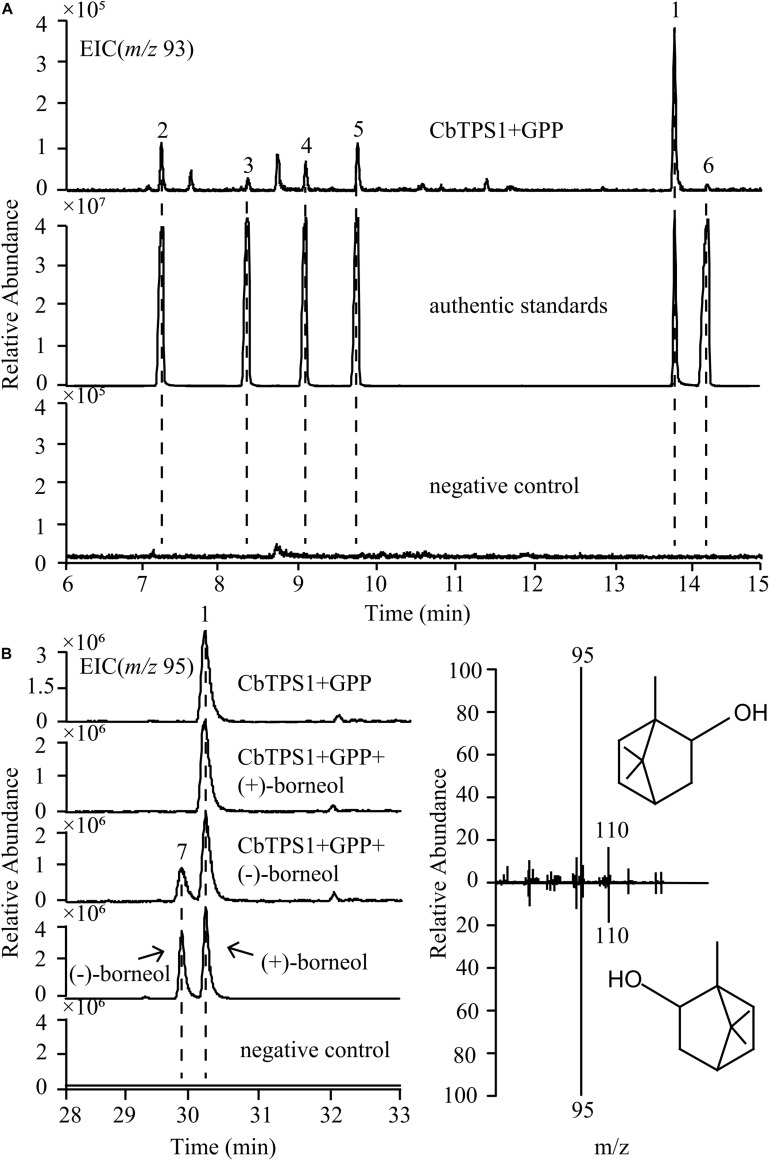
FIGURE 4.
GC-MS analysis of in vitro assays with CbTPS1. (A) Extracted ion chromatograms of m/z 93 in vitro assays with purified CbTPS1 and GPP as a substrate. Peak 1, (+)-borneol, Peak 2, α-pinene, Peak 3, β-pinene, Peak 4, α-phellandrene, Peak 5, limonene, Peak 6, α-terpineol. (B) Chromatogram of borneol product compared with authentic standards (+)- and (−)-borneol. Peak 1, (+)-borneol, Peak 7, (−)-borneol. Corresponding mass spectrum of (+)-borneol (upper halves) and (−)-borneol (lower halves). EIC, Extracted ion chromatograms.
A chiral column was used to identify the chirality of borneol. Based on the results of GC-MS analysis (Figure 4B), a single product (peak 1) corresponding to the authentic standard (+)-borneol was detected. When the authentic standard (+)-borneol was added to the reaction product, only peak 1 was detected. However, a new product (peak 7) was detected when the authentic standard (−)-borneol was added. This result further proved that (+)-borneol was produced with GPP as a substrate.
Reconstituting the MVA Pathway in Yeast for (+)-Borneol Production
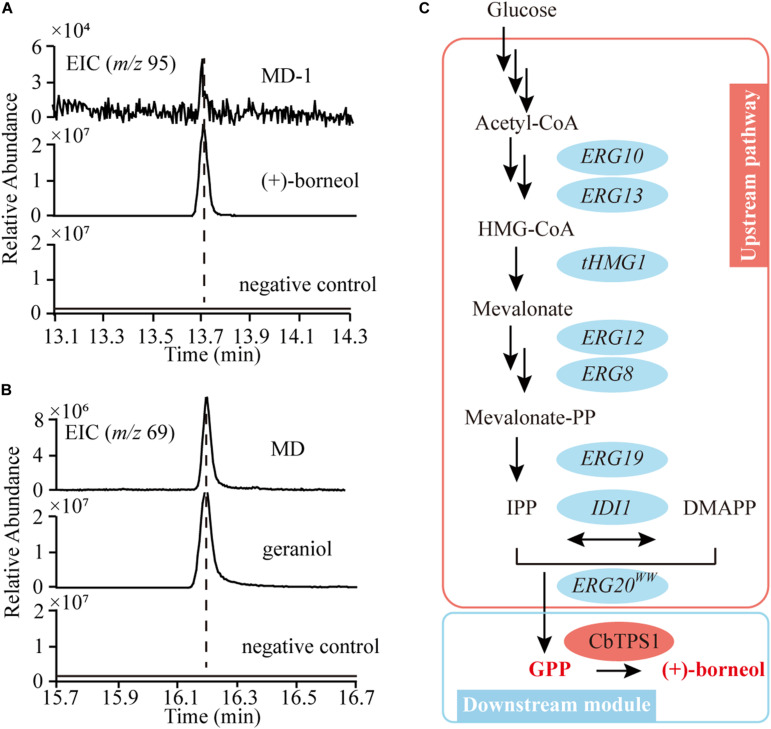
When the codon-optimized CbTPS1 was overexpressed in yeast CEN.PK2-1D, (+)-borneol could not be detected (Figure 5A). In addition, geraniol (the dephosphorylated GPP, precursor of (+)-borneol) was not detected in CEN.PK2-1D (Figure 5B). Hence, we reconstituted the MVA pathway in CEN.PK2-1D by overexpressing all the MVA pathway genes (ERG10, ERG13, tHMG1, ERG12, ERG8, ERG19, IDI1, ERG20F96W–N127W) to increase the precursor pool (Figure 5C). The obtained chassis strain MD can accumulate 12.52 mg⋅L–1 geraniol (Figure 5B). Then CbTPS1 was overexpressed in strain MD, and (+)-borneol was generated with a yield of 0.03 mg⋅L–1 (strain MD-1) (Figure 5A).
FIGURE 5.
The biosynthetic pathway of GPP and (+)-borneol in S. cerevisiae. (A) Extracted ion chromatograms of m/z 95 of (+)-borneol production in CEN.PK2-1D (negative control) and the MD-1 strain. (B) Extracted ion chromatograms of m/z 69 of geraniol production in CEN.PK2-1D (negative control) and the MD strain. (C) Reconstitution of the MVA pathway in yeast for GPP production (pink ellipses), and the biosynthetic pathway of (+)-borneol (blue ellipses).
Improving the (+)-Borneol Yield by Tailored Truncations
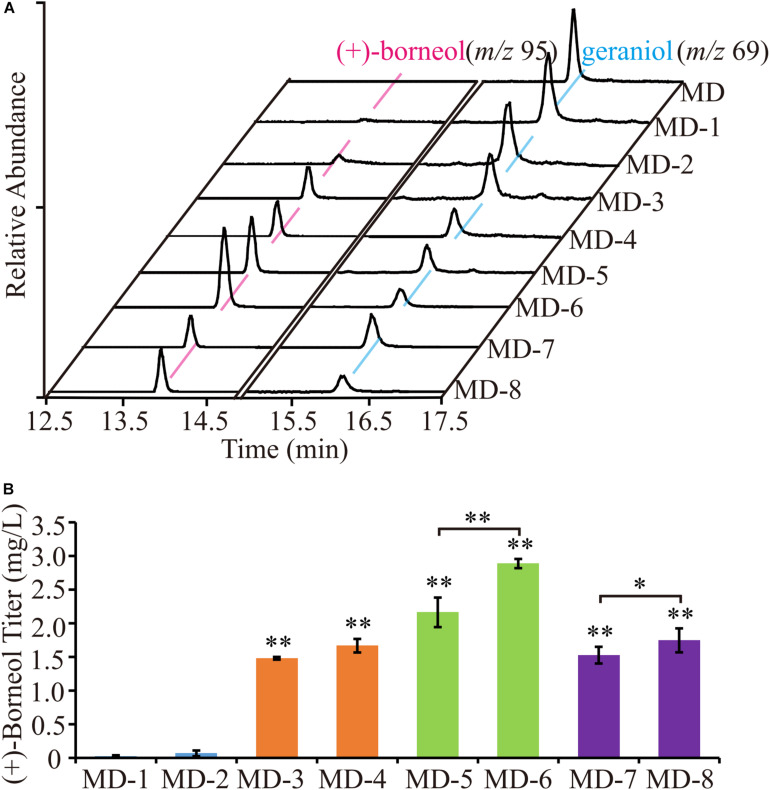
To obtain a higher (+) borneol titer, we engineered the CbTPS1 by further structure optimization. Most terpene synthases in plants have N-terminal plastidic transit peptidases, and will be hydrolyzed after the protein is targeted to the plastid (Bohlmann et al., 1998; Zybailov et al., 2008; Rowland et al., 2015). However, this affects the catalytic activity because yeast cannot digest the transit peptide. Thus, we truncated the chloroplast transit peptide according to the prediction of ChloroP7; CbTPS1 was truncated at the C37 position in the N-terminus, and named t37-CbTPS1. (+)-borneol was detected by GC-MS (Figure 6A). The truncated t37-CbTPS1 showed a significant increase of (+)-borneol production to 1.53 mg⋅L–1 (strain MD-7) (Figure 6B).
FIGURE 6.
The (+)-borneol production of strains expressing truncations of the CbTPS1. (A) GC-MS analysis of the fermentation products of strains expressing the truncated proteins. (B) The titer of (+)-borneol product in strains expressing the truncated proteins. (∗) represent means which are significantly different at p < 0.05; (∗∗) represent means which are significantly different at p < 0.01.
Hamilton compared 96 Saccharomyces cerevisiae sequences, and analyzed the window of 100 bases around the START codon (Hamilton et al., 1987). They found that 50% of highly expressed genes use the UCU serine codon as the second triplet, which indicated that UCU following the START codon ATG could increase gene expression. Therefore, we designed two truncated proteins with ATG followed by the UCU codon. Both amino acids at positions 10 (TCC) and 32 (TCA) of CbTPS1 are serine, which is the same as the amino acid encoded by UCU, so the codon corresponding to the truncated site was mutated to TCT to increase the (+) borneol titer, resulting in t10-CbTPS1 and t32-CbTPS1, respectively. The (+) borneol titer of the two truncated proteins increased significantly. The titer of truncated t10-CbTPS1 was 1.48 mg⋅L–1 (strain MD-3), which was 49.33-fold higher than untruncated CbTPS1, and the titer of truncated t32-CbTPS1 was 72-fold higher than CbTPS1, up to 2.16 mg⋅L–1 (strain MD-5) (Figure 6B).
Improving the (+)-Borneol Yield by Adding Kozak Sequence
The Kozak sequence is roughly the first six important nucleotides upstream of the START codon in S. cerevisiae, which are used for gene translation and expression. In yeast, the Kozak sequence is mostly “AAAAAA” (Hamilton et al., 1987; Li et al., 2017; Hernández et al., 2019). On the basis of truncation, yeast-specific Kozak sequence was added in front of START codon ATG of the codon-optimized CbTPS1 and three truncated proteins to increase (+)-borneol yield. The modified proteins were named CbTPS1K, t10-CbTPS1K, t32-CbTPS1K, and t37-CbTPS1K. The results showed that the yield increased at different levels after adding the Kozak sequence. The highest (+)-borneol titer was achieved in strain MD-6 containing t32-CbTPS1K, which is 96.33-fold higher than that in the strain harboring wild-type CbTPS1, producing 2.89 mg⋅L–1 (+)-borneol (Figure 6B).
Discussion
Due to the insufficient supply of natural products, the role of microbial production of valuable compounds has emerged as an attractive alternative source. Microbial production is a promising choice to substitute for chemical synthesis or phytoextraction (Kirby and Keasling, 2009; Nielsen, 2019). High-efficiency gene elements are vital for metabolic engineering. In this study, we identified a (+)-bornyl diphosphate synthase (CbTPS1) from C. burmannii that catalyzed GPP to form (+)-borneol under the hydrolysis of CIAP. This is the first time an enzyme related to (+)-borneol synthesis was mined from C. burmannii, and it has the highest specificity for (+)-borneol production (Wise et al., 1998; Despinasse et al., 2017; Hurd et al., 2017; Wang et al., 2018). The Km value of CbTPS1 (5.11 μM) for GPP is consistent with SBS (3.0 μM) (Wise et al., 1998) and slightly lower than other reported monoterpene synthases (13.10–26.12 μM), which indicated CbTPS1 had a higher affinity for GPP. Its kcat/Km (1.99 × 10–3 s–1/μM) is similar to that of other efficient and highly specific monoterpene synthases (3.55 × 10–3–1.23 × 10–2 s–1/μM) (Morehouse et al., 2017; Ignea et al., 2019; Dusséaux et al., 2020). Thus, it gives us an opportunity to reconstruct the (+)-borneol biosynthetic pathway in S. cerevisiae.
In S. cerevisiae, GPP is mainly produced by FPP synthase (ERG20) to serve as the intermediate product of FPP synthesis, thus, it should be consumed rapidly. As a result, when there is no engineering of ERG20, no (+)-borneol or geraniol was detected in CEN.PK2-1D. In order to increase the GPP pool, we further overexpressed all MVA pathway genes and mutated the 96F and 127N of ERG20 to obtain the strain MD, which generated the target product (+)-borneol. However, compared with the accumulation of geraniol (12.52 mg⋅L–1), the yield of (+)-borneol product was relatively low (0.03 mg⋅L–1). Thus, modified proteins were used to improve the expression and activity of CbTPS1. After steady modification, strain MD6 was obtained with the highest yield of (+)-borneol (2.89 mg⋅L–1). Thus, the combination of truncation and using Kozak sequence is an effective strategy for improving (+)-borneol productivity.
Though more than 20 mg⋅L–1 of linalool, α-terpineol, and limonene were produced in yeast (Cao et al., 2016; Zhang et al., 2019, 2020), the yields of most monoterpenes are still lower than the sesquiterpenes and diterpenes (Zebec et al., 2016; Zhao et al., 2016; Jiang et al., 2017), such as artemisinic acid (25 g⋅L–1) (Paddon et al., 2013) and miltiradiene (3.5 g⋅L–1) (Hu et al., 2020). The efficiency of forming the final product is influenced by many factors. Reduced efficiency is partially due to the high toxicity of many monoterpenes, such as pinene and limonene, to S. cerevisiae because they alter membrane properties or damage the cell wall (Brennan et al., 2013; Demissie et al., 2019). Two-phase extractive fermentation is usually used to alleviate the toxicity of monoterpenes (Brennan et al., 2012). We next will attempt more protein modification of CbTPS1, such as translational fusion (Ignea et al., 2019) and directed evolution of enzymes (Qu et al., 2019). In addition, optimizing the fermentation strategy, by selecting suitable solvent, and optimizing the carbon sources and fermentation parameters will further enhance production (Zhou et al., 2019). Thus, we have good reason to believe that S. cerevisiae could be a promising platform for a feasible, scalable, and economic route to the overproduction of (+)-borneol derivatives in the future.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2017) in BIG Data Center (Nucleic Acids Res 2018), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers CRA003558 (http://bigd.big.ac.cn/gsa).
Author Contributions
LH and GC conceived and designed the experiment. JG and BJ operated GC-MS. QM, HZ, and LC analyzed the data. LM provided the materials. MT purified protein. RM and PS were involved in all experimental steps and wrote this manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- BPPS
bornyl diphosphate synthase
- CIAP
calf intestinal alkaline phosphatase
- DMAPP
dimethylallyl diphosphate
- GC-MS
gas chromatography coupled with mass spectrometry
- GPP
geranyl diphosphate
- IPP
Isopentenyl diphosphate
- MVA
mevalonate pathway
- ORF
open reading frame
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Funding. This work was supported by the National Key R&D Program of China (2020YFA0908000 and 2018YFA0900600), Major Program of National Natural Science Foundation of China (81891010 and 81891013), the National Natural Science Foundation of China (81822046), and Key project at central government level: The ability to establish sustainable use of valuable Chinese medicine resources (2060302).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.631863/full#supplementary-material
References
- Amiri P., Shahpiri A., Asadollahi M. A., Momenbeik F., Partow S. (2016). Metabolic engineering of Saccharomyces cerevisiae for linalool production. Biotechnol. Lett. 38 503–508. 10.1007/s10529-015-2000-4 [DOI] [PubMed] [Google Scholar]
- Bohlmann J., Meyer-Gauen G., Croteau R. (1998). Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. U.S.A. 95 4126–4133. 10.1073/pnas.95.8.4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan T. C. R., Krömer J. O., Nielsen L. K. (2013). Physiological and transcriptional responses of Saccharomyces cerevisiae to d-limonene show changes to the cell wall but not to the plasma membrane. Appl. Environ. Microbiol. 79 3590–3600. 10.1128/AEM.00463-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan T. C. R., Turner C. D., Krömer J. O., Nielsen L. K. (2012). Alleviating monoterpene toxicity using a two-phase extractive fermentation for the bioproduction of jet fuel mixtures in Saccharomyces cerevisiae. Biotechnol. Bioeng. 109 2513–2522. 10.1002/bit.24536 [DOI] [PubMed] [Google Scholar]
- Cao X., Lv Y. B., Chen J., Imanaka T., Wei L. J., Hua Q. (2016). Metabolic engineering of oleaginous yeast Yarrowia lipolytica for limonene overproduction. Biotechnol. Biofuels 9 1–11. 10.1186/s13068-016-0626-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Wei L. J., Lin J. Y., Hua Q. (2017). Enhancing linalool production by engineering oleaginous yeast Yarrowia lipolytica. Bioresour. Technol. 245 1641–1644. 10.1016/j.biortech.2017.06.105 [DOI] [PubMed] [Google Scholar]
- Chen F., Tholl D., Bohlmann J., Pichersky E. (2011). The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66 212–229. 10.1111/j.1365-313X.2011.04520.x [DOI] [PubMed] [Google Scholar]
- Chen M., Hua Y., Huang L., Lin S., Yang G. (2010). Analysis on chemical components of leaf oil from reproductive progenies of Cinnamomum camphora chvar. Borneo. Chin. J. Tradit. Chin. Med. Inf. 17 37–40. 10.3969/j.issn.1005-5304.2010.08.017 [DOI] [Google Scholar]
- Chen Z.-X., Xu Q.-Q., Shan C.-S., Shi Y.-H., Wang Y., Chang R. C.-C., et al. (2019). Borneol for regulating the permeability of the blood-brain barrier in experimental ischemic stroke: preclinical evidence and possible mechanism. Oxid. Med. Cell. Longev. 2019:2936737. 10.1155/2019/2936737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demissie Z. A., Tarnowycz M., Adal A. M., Sarker L. S., Mahmoud S. S. (2019). A lavender ABC transporter confers resistance to monoterpene toxicity in yeast. Planta 249 139–144. 10.1007/s00425-018-3064-x [DOI] [PubMed] [Google Scholar]
- Despinasse Y., Fiorucci S., Antonczak S., Moja S., Bony A., Nicolè F., et al. (2017). Bornyl-diphosphate synthase from Lavandula angustifolia: a major monoterpene synthase involved in essential oil quality. Phytochemistry 137 24–33. 10.1016/j.phytochem.2017.01.015 [DOI] [PubMed] [Google Scholar]
- Dusséaux S., Wajn W. T., Liu Y., Ignea C., Kampranis S. C. (2020). Transforming yeast peroxisomes into microfactories for the efficient production of high-value isoprenoids. Proc. Natl. Acad. Sci. U.S.A. 117 31789–31799. 10.1073/pnas.2013968117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. J., Xiao W. H., Wang Y., Yao M. D., Zeng B. X., Liu H. (2018). Metabolic engineering of Saccharomyces cerevisiae for 7-dehydrocholesterol overproduction. Biotechnol. Biofuels 11:192. 10.1186/s13068-018-1194-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton R., Watanabe C. K., De Boer H. A. (1987). Compilation and comparison of the s ce context around the AUG startcodons in Saccharomyces cemisiae mRNAs. Nucleic Acids Res. 15 3581–3593. 10.1093/nar/15.8.3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G., Osnaya V. G., Pérez-Martínez X. (2019). Conservation and variability of the AUG initiation codon context in Eukaryotes. Trends Biochem. Sci. 44 1009–1021. 10.1016/j.tibs.2019.07.001 [DOI] [PubMed] [Google Scholar]
- Hu T., Zhou J., Tong Y., Su P., Li X., Liu Y., et al. (2020). Engineering chimeric diterpene synthases and isoprenoid biosynthetic pathways enables high-level production of miltiradiene in yeast. Metab. Eng. 60 87–96. 10.1016/j.ymben.2020.03.011 [DOI] [PubMed] [Google Scholar]
- Hurd M. C., Kwon M., Ro D. K. (2017). Functional identification of a Lippia dulcis bornyl diphosphate synthase that contains a duplicated, inhibitory arginine-rich motif. Biochem. Biophys. Res. Commun. 490 963–968. 10.1016/j.bbrc.2017.06.147 [DOI] [PubMed] [Google Scholar]
- Ignea C., Pontini M., Maffei M. E., Makris A. M., Kampranis S. C. (2014). Engineering monoterpene production in yeast using a synthetic dominant negative geranyl diphosphate synthase. ACS Synth. Biol. 3 298–306. 10.1021/sb400115e [DOI] [PubMed] [Google Scholar]
- Ignea C., Raadam M. H., Motawia M. S., Makris A. M., Vickers C. E., Kampranis S. C. (2019). Orthogonal monoterpenoid biosynthesis in yeast constructed on an isomeric substrate. Nat. Commun. 10 1–15. 10.1038/s41467-019-11290-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Zhang R., Li H., Zhu J., Pan Y., Guo Q. (2020). Analgesic and anti-inflammatory effects and mechanism of action of borneol on photodynamic therapy of acne. Environ. Toxicol. Pharmacol. 75:103329. 10.1016/j.etap.2020.103329 [DOI] [PubMed] [Google Scholar]
- Jiang G. Z., Yao M. D., Wang Y., Zhou L., Song T. Q., Liu H., et al. (2017). Manipulation of GES and ERG20 for geraniol overproduction in Saccharomyces cerevisiae. Metab. Eng. 41 57–66. 10.1016/j.ymben.2017.03.005 [DOI] [PubMed] [Google Scholar]
- Jongedijk E., Cankar K., Ranzijn J., van der Krol S., Bouwmeester H., Beekwilder J. (2015). Capturing of the monoterpene olefin limonene produced in Saccharomyces cerevisiae. Yeast 32 159–171. 10.1002/yea.3038 [DOI] [PubMed] [Google Scholar]
- Kirby J., Keasling J. D. (2009). Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu. Rev. Plant Biol. 60 335–355. 10.1146/annurev.arplant.043008.091955 [DOI] [PubMed] [Google Scholar]
- Li J., Liang Q., Song W., Marchisio M. A. (2017). Nucleotides upstream of the Kozak sequence strongly influence gene expression in the yeast S. cerevisiae. J. Biol. Eng. 11 1–14. 10.1186/s13036-017-0068-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ding W., Zhang X., Jiang H., Bi C. (2016). Development of a modularized two-step (M2S) chromosome integration technique for integration of multiple transcription units in Saccharomyces cerevisiae. Biotechnol. Biofuels 9 1–11. 10.1186/s13068-016-0645-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhang Y., Nielsen J. (2019). Synthetic biology of yeast. Biochemistry 58 1511–1520. 10.1021/acs.biochem.8b01236 [DOI] [PubMed] [Google Scholar]
- Ma R., Su P., Jin B., Guo J., Tian M., Mao L., et al. (2020). Molecular cloning and functional identification of a high-efficiency (+)-borneol dehydrogenase from Cinnamomum camphora (L.) Presl. Plant Physiol. Biochem. 158 363–371. 10.1016/j.plaphy.2020.11.023 [DOI] [PubMed] [Google Scholar]
- Morehouse B. R., Kumar R. P., Matos J. O., Olsen S. N., Oprian D. D. (2017). Functional and structural characterization of a (+)-Limonene Synthase from Citrus sinensis. Biochemistry 56 1706–1715. 10.1021/acs.biochem.7b00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. (2019). Yeast systems biology: model organism and cell factory. Biotechnol. J. 14:e1800421. 10.1002/biot.201800421 [DOI] [PubMed] [Google Scholar]
- Paddon C. J., Westfall P. J., Pitera D. J., Benjamin K., Fisher K., McPhee D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496 528–532. 10.1038/nature12051 [DOI] [PubMed] [Google Scholar]
- Qu G., Li A., Acevedo-Rocha C. G., Sun Z., Reetz M. T. (2019). The crucial role of methodology development in directed evolution of selective enzymes. Angew. Chem. 59 13204–13231. 10.1002/anie.201901491 [DOI] [PubMed] [Google Scholar]
- Rowland E., Kim J., Bhuiyan N. H., Van Wijk K. J. (2015). The Arabidopsis chloroplast stromal n-terminome: complexities of amino-terminal protein maturation and stability. Plant Physiol. 169 1881–1896. 10.1104/pp.15.01214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Wu Q., Su J., Li C., Zhao X., Xie J., et al. (2013). Composition analysis of volatile oils from flowers, leaves and branches of Cinnamomum camphora chvar, Borneol in china. J. Essent. Oil Res. 25 395–401. 10.1080/10412905.2013.809323 [DOI] [Google Scholar]
- Song H., Wei M., Zhang N., Li H., Tan X., Zhang Y., et al. (2018). Enhanced permeability of blood-brain barrier and targeting function of brain via borneol-modified chemically solid lipid nanoparticle. Int. J. Nanomed. 13 1869–1879. 10.2147/IJN.S161237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P., Guan H., Zhao Y., Tong Y., Xu M., Zhang Y., et al. (2018). Identification and functional characterization of diterpene synthases for triptolide biosynthesis from Tripterygium wilfordii. Plant J. 93 50–65. 10.1111/tpj.13756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ma D., Yang J., Deng K., Li M., Ji X., et al. (2018). An integrative volatile terpenoid profiling and transcriptomics analysis for gene mining and functional characterization of avbpps and avps involved in the monoterpenoid biosynthesis in Amomum villosum. Front. Plant Sci. 9:846. 10.3389/fpls.2018.00846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise M. L., Savage T. J., Katahira E., Croteau R. (1998). Monoterpene synthases from common sage (Salvia officinalis). J. Biol. Chem. 273 14891–14899. 10.1074/jbc.273.24.14891 [DOI] [PubMed] [Google Scholar]
- Wojtunik-Kulesza K. A., Kasprzak K., Oniszczuk T., Oniszczuk A. (2019). Natural monoterpenes: much more than only a scent. Chem. Biodivers. 16:e1900434. 10.1002/cbdv.201900434 [DOI] [PubMed] [Google Scholar]
- Xin Y., Zhao H., Xu J., Xie Z., Li G., Gan Z., et al. (2020). Borneol-modified chitosan: antimicrobial adhesion properties and application in skin flora protection. Carbohydr. Polym. 228:115378. 10.1016/j.carbpol.2019.115378 [DOI] [PubMed] [Google Scholar]
- Zebec Z., Wilkes J., Jervis A. J., Scrutton N. S., Takano E., Breitling R. (2016). Towards synthesis of monoterpenes and derivatives using synthetic biology. Curr. Opin. Chem. Biol. 34 37–43. 10.1016/j.cbpa.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Zhang C., Li M., Zhao G. R., Lu W. (2019). Alpha-terpineol production from an engineered Saccharomyces cerevisiae cell factory. Microb. Cell Fact. 18 1–9. 10.1186/s12934-019-1211-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.-L., Fu B. M., Zhang Z.-J. (2017). Borneol, a novel agent that improves central nervous system drug delivery by enhancing blood-brain barrier permeability. Drug Deliv. 24 1037–1044. 10.1080/10717544.2017.1346002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang J., Cao X., Liu W., Yu H., Ye L. (2020). High-level production of linalool by engineered Saccharomyces cerevisiae harboring dual mevalonate pathways in mitochondria and cytoplasm. Enzyme Microb. Technol. 134:109462. 10.1016/j.enzmictec.2019.109462 [DOI] [PubMed] [Google Scholar]
- Zhao J., Bao X., Li C., Shen Y., Hou J. (2016). Improving monoterpene geraniol production through geranyl diphosphate synthesis regulation in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 100 4561–4571. 10.1007/s00253-016-7375-1 [DOI] [PubMed] [Google Scholar]
- Zheng Q., Chen Z.-X., Xu M.-B., Zhou X.-L., Huang Y.-Y., Zheng G.-Q., et al. (2018). Borneol, a messenger agent, improves central nervous system drug delivery through enhancing blood-brain barrier permeability: a preclinical systematic review and meta-analysis. Drug Deliv. 25 1617–1633. 10.1080/10717544.2018.1486471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Hu T., Gao L., Su P., Zhang Y., Zhao Y., et al. (2019). Friedelane-type triterpene cyclase in celastrol biosynthesis from Tripterygium wilfordii and its application for triterpenes biosynthesis in yeast. New Phytol. 223 722–735. 10.1111/nph.15809 [DOI] [PubMed] [Google Scholar]
- Zou L., Zhang Y., Li W., Zhang J., Wang D., Fu J., et al. (2017). Comparison of chemical profiles, anti-inflammatory activity, and UPLC-Q-TOF/MS-based metabolomics in endotoxic fever rats between synthetic Borneol and natural Borneol. Molecules 22:1446. 10.3390/molecules22091446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., et al. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3:1994. 10.1371/journal.pone.0001994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2017) in BIG Data Center (Nucleic Acids Res 2018), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers CRA003558 (http://bigd.big.ac.cn/gsa).