Highlights
-
•
The cytotoxic protein PRF1 is essentially involved in anti-tumour immunity.
-
•
PRF1 is overexpressed in advanced HPV+ HNSCC and correlates with better OS.
-
•
PRF1 expression correlates with tumour infiltrating CD8+ t cells and dendritic cells in HPV+ HNSCC.
-
•
PRF1 expression correlates with gene markers of t cell exhaustion in HNSCC.
Key words: PRF1, prognosis, head and neck squamous cell carcinoma, tumor immune infiltration, HPV
Abstract
Purpose
Head and neck squamous cell carcinoma (HNSCC) is a highly invasive malignancy with poor survival. Perforin (PRF1) plays essential roles in host immunity. Our research intended to identify the correlations of PRF1 with clinical prognosis and tumor immune infiltration in HNSCC.
Methods
We explored PRF1 expression and its associations with the clinical features of HNSCC via the Tumor Immune Estimation Resource (TIMER), Oncomine and The Cancer Genome Atlas (TCGA) databases. The prognostic value of PRF1 for HNSCC was further explored by Kaplan–Meier plotter and TIMER. Finally, the relation between PRF1 and immune infiltration in HNSCC was estimated via CIBERSORT and TIMER.
Results
PRF1 expression was remarkably elevated in HNSCC and associated with clinical stage and HPV infection. High PRF1 expression predicted favorable outcomes in HNSCC, especially in HPV+ HNSCC. Moreover, higher infiltration of CD8+ T cells and CD4+ T cells were found in the PRF1high group of HNSCC. PRF1 expression in HNSCC was strongly correlated with infiltrating CD8+ T cells and dendritic cells (DCs), with higher relevance in HPV+ HNSCC.
Conclusion
Our findings suggested that PRF1 could be a novel prognostic biomarker in HNSCC and that its expression was related to immune cell infiltration, which was impacted by HPV status.
Introduction
Head and neck squamous cell carcinoma (HNSCC), representing approximately 90% of head and neck cancers, ranks ninth in the global incidence of malignant tumors.[1,2] Tobacco exposure, alcohol consumption and human papilloma virus (HPV) infection are the most common risk factors for HNSCC. The incidence of smoking-related HNSCCs has declined over the past several decades, while HPV-related HNSCCs have significantly increased in frequency.[3] HNSCC has the characteristics of high recurrence and metastasis rates, which lead to poor survival. The traditional treatments of HNSCC include surgery, radiation therapy and chemotherapy.[4]
The key role of immune dysfunction in HNSCC has been widely recognized in recent decades and the remarkable advancement of immunotherapy has altered the therapeutic patterns of HNSCC. Some scholars have pointed that HNSCC is a type of immunosuppressive disease with disordered immune effector cells and abnormal secretion of pro-inflammatory factors in the immune microenvironment.[5] Immunotherapy based on immune checkpoint blockade for HNSCC has shown promising clinical results. Two PD-1 antibodies, pembrolizumab and nivolumab, have been approved for the clinical therapy of recurrent/metastatic HNSCC by the US Food and Drug Administration (FDA).[6] Tumor infiltrating lymphocytes (TILs), a prominent feature of HNSCC, has been reported to be an independent prognostic marker. Based on the differences in infiltrating immune cells and the expression of inflammatory markers in tumor and stromal compartments, Chen YP et al. defined two different subtypes of HNSCC, namely, the active immune class and exhausted immune class. The former was correlated with enhanced cytolytic activity, enriched M1 macrophages, abundant TILs, HPV infection and better prognosis, while the latter was characterized by enriched M2 macrophages, activated stroma and poor prognosis.[7] Abundant CD8+ T cell, CD4+ T cell, CD3+ T cell and Foxp3+ T cell infiltration in HNSCC tumor tissues was correlated with favorable prognosis.[8,9] As the main component of the host immune microenvironment, accumulating evidence highlights the value of TILs in predicting tumor progression and response to immunotherapy.[10,11] Therefore, a better understanding of immune infiltration in HNSCC will help us clarify the mechanisms of tumor immunity, define predictive biomarkers and identify new therapeutic targets.
The pore-forming protein perforin (PRF1), belonging to the membrane-attack-complex/PRF (MACPF) protein family, is essentially involved in the granule-dependent killing activities of cytotoxic T lymphocytes (CTLs) and NK cells.[12] Serving as a definite marker of the killing ability of immune cells, PRF1 participates in the establishment of immune homeostasis, elimination of pathogens and tumor surveillance.[13] However, there are only a few studies concerning the correlation of PRF1 with cancer, and its role in the tumor immune microenvironment and related mechanisms remain unclear. Alcaraz-Sanabria A et al. reported that in advanced ovarian cancer, the combination of PRF1, IFNG, CXCL13 and CD30 expression was related to favorable outcome.[14] Additionally, a meta-analysis showed that increasing levels of cytotoxic proteins, including PRF1 and GZMA, indicated better survival in multiple cancers, such as melanoma and bladder cancer.[15] Here, we explored PRF1 expression in HNSCC and its relations with clinical parameters and overall survival (OS) via databases with a good reputation, such as Oncomine, Tumor Immune Estimation Resource (TIMER) and Kaplan–Meier plotter. Moreover, we analyzed the associations between PRF1 expression and tumor immune infiltration and immune marker sets in HNSCC by TIMER and CIBERSORT. Our findings showed the prognostic value of PRF1 in HNSCC and provided novel insight into its role in cancer immunity.
Materials and methods
Data and patients
HNSCC patients datasets (n = 527), including transcriptome sequencing data and clinical materials, were obtained from TCGA utilizing the R/Bioconductor/TCGA package. We excluded cases with insufficient information on gene expression data, TNM stage and OS. Finally, 505 cases were included in the study. 25 pairs of tumor and normal samples came from 25 HNSCC patients diagnosed and hospitalized in Shanghai Jiaotong University School of Medicine, Ruijin Hospital from Oct 2018 to Dec 2019, who experienced surgery and adjuvant chemoradiotherapy. Our research was approved by the institutional review board of Ruijin Hospital. Informed content was obtained from all recruited patients.
Oncomine database analysis
We investigated PRF1 expression in a variety of cancers via Oncomine database.[16] 1.5 fold change, P value = 0.001, and all gene ranking were set as the threshold.
Kaplan–Meier plotter database analysis
Kaplan–Meier plotter database could estimate the impact of gene expression in prognosis in 21 types of cancers including HNSCC. The association between PRF1 expression and patient survival in HNSCC was explored via Kaplan–Meier plotter.[17] In addition, log-rank P value as well as its hazard ratio (HR) with 95% confidence intervals were analyzed.
CIBERSORT algorithm analysis
CIBERSORT is a deconvolution algorithm that calculates immune cell infiltration proportions on the basis of gene expression profiles in the sample.[18] We used a white blood cell gene signature matrix LM22 that includes 547 genes to identify 22 kinds of infiltrating immune cells. In our current analysis, the CIBERSORT tool was utilized to analyze the infiltrating fractions of immune cells of 505 cases of HNSCC from TCGA. We divided patients into the PRF1high and PRF1low group according to the cutoff point (111.0) for PRF1 expression, which was calculated through the automatic Cutoff Finder platform.
TIMER database analysis
TIMER database could analyze the abundance of infiltrating immune cells in 32 types of cancers from TCGA.[19] We studied PRF1 expression in different tumor tissues relative to normal controls and the correlation between PRF1 expression and OS in HNSCC overall, HPV+ HNSCC and HPV- HNSCC by Kaplan–Meier curves. Furthermore, we explored associations of PRF1 expression with tumor immune infiltration, and with gene markers of immune cells in HNSCC overall, HPV+ HNSCC and HPV- HNSCC.
Gene correlation assessment via GEPIA
Gene correlation could be assessed in Gene Expression Profiling Interactive Analysis (GEPIA) using RNA sequencing datasets from TCGA.[20] We explored the correlation of PRF1 with markers of T cell exhaustion signature in HNSCC tumor tissues via GEPIA.
Immunohistochemical analysis
We performed immunohistochemistry (IHC) of PRF1 on the formalin-fixed, paraffin-embedded tumor and matched normal specimens of HNSCC. For antigen retrieval and PRF1 detection, specimen sections were deparaffinized and autoclaved in 10 mM citrate buffer at 121 °C for 20 min. The primary monoclonal mouse anti-PRF1 antibody (Zisbio, ZM-0151,Beijing, China) packaged in working solution was used for staining. We scanned the images via Image Pro Plus 6.0 (Media Cybernetics, Bethesda, USA). The results were presented with the average optical density (AOD) fo PRF1 in intratumoral and normal tissues.
Statistical analysis
SPSS 22.0 and GraphPad Prism 8.0 were utilized for statistical analysis and graph plotting. Mann-Whitney U test was applied to detect the comparisons between two groups. Differences among multiple groups were examined by Kruskal-Wallis one-way ANOVA. The survival curves were created via Kaplan–Meier plotter and TIMER, which included corresponding P values and HR. Cox regression was used to perform univariate and multivariate analysis. Spearman's correlation analysis was applied to estimate correlation coefficients. The degree of correlation was determined according to the following absolute r values: 0.00–0.19 “very weak”, 0.20–0.39 “weak”, 0.40–0.59 “moderate”,0.60–0.79 “strong” and 0.80–1.0 “very strong”. p value less than 0.05 was set as a statistical significance.
Results
PRF1 expression in different cancers
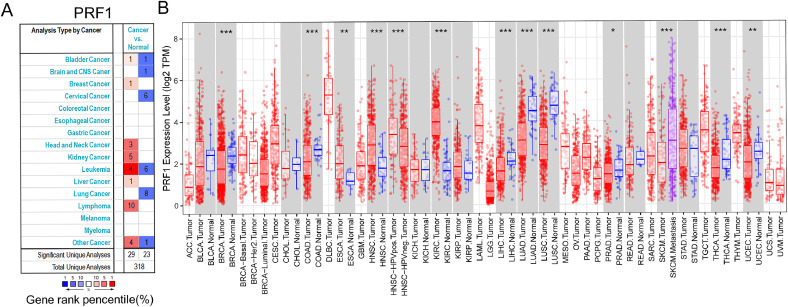
We compared the levels of PRF1 mRNA in various cancers by Oncomine. The results revealed that compared to normal tissues, PRF1 was increased in kidney cancer, liver cancer and breast cancer but was decreased in lung cancer, colorectal cancer and bladder cancer (Fig. 1A). We further investigated PRF1 expression in particular tumors through TIMER. The expression level of PRF1 in HNSCC, ESCA (oesophageal carcinoma) and KIRC (kidney renal clear cell carcinoma) was significantly higher than that in normal tissues. In LUAD (lung adenocarcinoma), LIHC (liver hepatocellular carcinoma), LUSC (lung squamous cell carcinoma), BRCA (breast invasive carcinoma), COAD (colon adenocarcinoma), UCEC (uterine corpus endometrial carcinoma) and THCA (thyroid carcinoma), PRF1 expression was significantly lower (Fig. 1B). Notably, PRF1 was significantly more highly expressed in HPV+ HNSCC in comparison to HPV- HNSCC.
Fig. 1.
The levels of PRF1 expression in multiple cancers. (A) PRF1 expression of various cancers relative to normal controls in Oncomine. (B) PRF1 expression of particular types of cancers in TIMER (*p < 0.05, **p < 0.01, ***p < 0.001).
PRF1 expression profiles of HNSCC patients
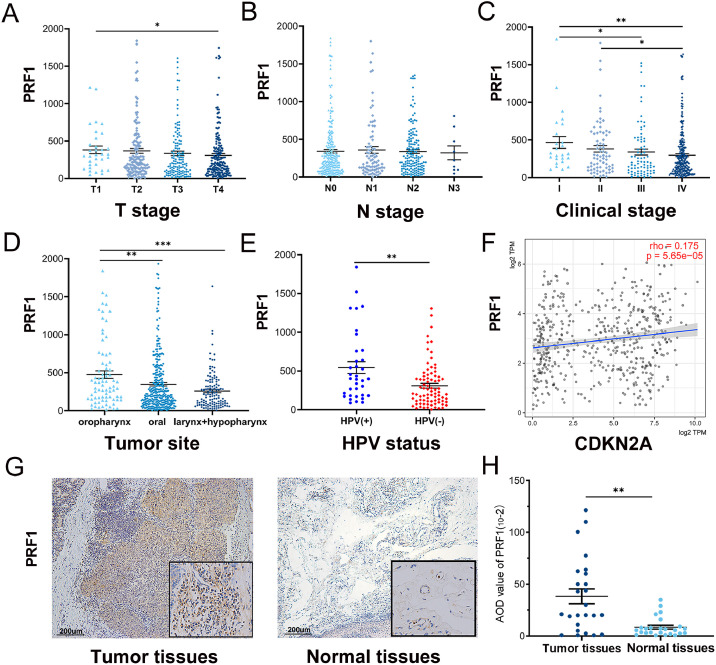
From oncomine and TIMER database, we found that PRF1 expression was significantly increased in tumor tissues compared to normal controls in HNSCC. Then we studied the relationship of PRF1 expression with the clinical parameters of HNSCC in the TCGA cohort (n = 505) (Fig. 2A–E). The results displayed that PRF1 expression was significantly different in terms of tumor T stage, clinical stage, tumor site and HPV status. With the increase in T stage and clinical stage, PRF1 expression showed a downward trend. Specifically, the results indicated significant differences in PRF1 expression between T1 and T4, between stages I and III, between stages II and IV, and between stages I and IV (p < 0.05). Regarding different N stages, no significant difference in PRF1 expression was observed. According to different tumor sites, we divided the patients with HNSCC into three groups: oropharyngeal squamous cell carcinoma (OPSCC), oral squamous cell carcinoma (OSCC), and laryngeal and hypopharyngeal squamous cell carcinoma (LSCC + HPSCC). The results revealed that PRF1 expression was significantly elevated in OPSCC compared to OSCC (p < 0.01) and LSCC + HPSCC (p < 0.001). PRF1 was more highly expressed in OSCC than in LSCC + HPSCC although the difference was not significant. Interestingly, higher PRF1 expression was observed in HPV+ HNSCC than in HPV- HNSCC (p < 0.01). Thus, we explored the correlation between PRF1 and the common HPV molecular marker P16 (CDKN2A) through TIMER and found that PRF1 and P16 expression were significantly related in HNSCC (p < 0.05) (Fig. 2F). Additionally, we performed IHC for PRF1 detection in 25 pairs of tumor and normal samples of HNSCC and found that the protein expression of PRF1 was elevated in tumor tissues. Representative images of IHC are shown in Fig. 2G and Figure S1. Quantitative analysis showed there was an increase of PRF1 expression in tumor tissues compared with normal tissues (p < 0.05) (Fig. 2H).
Fig. 2.
PRF1 expression and association with clinical parameters in HNSCC. PRF1 expression in the different T stage (A), N stage (B), clinical stage (C), tumor site (D) and HPV status (E). (F) Correlation of PRF1 and CDKN2A (P16) expression from TIMER. (G) Representative IHC images of PRF1 protein expression in tumor tissues and normal tissues in HNSCC. (H) Quantitative analysis of PRF1 in tumor and normal tissues. AOD, average optical density. (*p < 0.05, **p < 0.01, ***p < 0.001).
Prognostic value of PRF1 in HNSCC
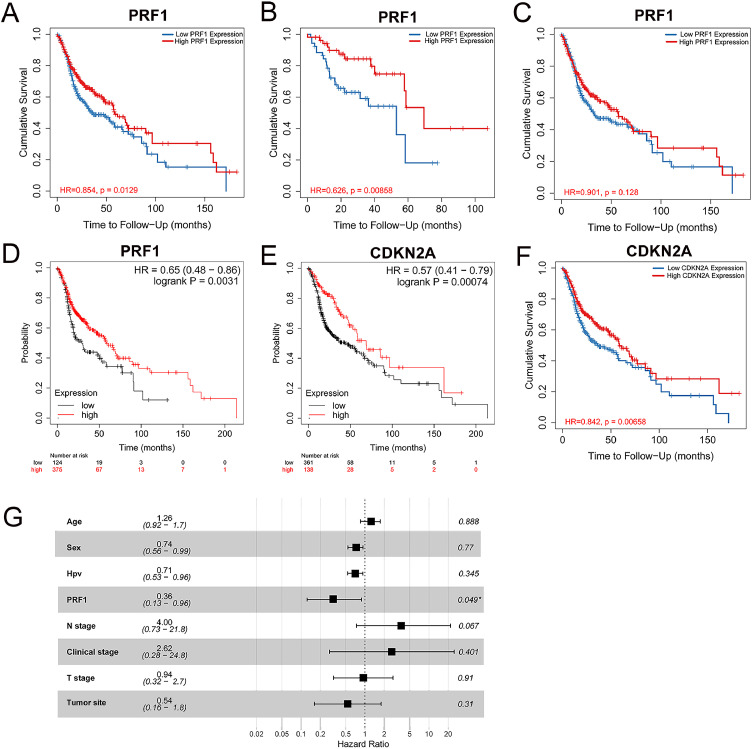
The expression profiles of PRF1 displayed that low expression of PRF1 correlated with advanced T and clinical stages and HPV negative infection status, which were unfavorable prognostic factors of HNSCC. Next, we utilized Kaplan–Meier plotter and TIMER to investigate the influence of PRF1 expression on OS in HNSCC. High PRF1 expression was related to better OS in HNSCC significantly, as analyzed by TIMER (HR = 0.854, p = 0.0129) (Fig. 3A). Consistent results were obtained in Kaplan–Meier plotter, wherein better OS in HNSCC was significantly associated with high PRF1 expression (HR = 0.65, p = 0.0031) (Fig. 3D). Of note, the prognostic value of PRF1 in HNSCC was impacted by HPV status. Survival curves from TIMER demonstrated that compared with HNSCC overall, PRF1 exhibted a stronger prognostic value in HPV+ HNSCC. There was a significant association between high PRF1 expression and better prognosis in HPV+ HNSCC (HR = 0.676, p = 0.00858), while in HPV- HNSCC, no obvious correlation was observed (Fig. 3B,C). Additionally, we also explored the impact of P16 expression on the outcomes of HNSCC. High P16 expression was significantly related to better OS in HNSCC according to TIMER (HR = 0.842, p = 0.00658) (Fig. 3E) and Kaplan–Meier plotter (HR = 0.57, p = 0.00074) (Fig. 3F), which was consistent with the better prognosis of HPV+ HNSCC than HPV- HNSCC. Moreover, univariate analysis displayed that age, sex, PRF1, N stage and T stage were significantly related to OS (p < 0.05) (Table S1), and further multivariate analysis showed that high PRF1 expression was a favorable independent prognostic factor (HR = 0.36, p < 0.05) (Fig. 3G).
Fig. 3.
Prognostic value of PRF1 and CDKN2A in HNSCC. Kaplan–Meier survival curves based on PRF1 expression in HNSCC overall (A), HPV+ HNSCC (B) and HPV- HNSCC (C) in TIMER. Correlation of PRF1 expression (D) and CDKN2A expression (E) with OS of HNSCC in Kaplan–Meier plotter. (F) Correlation of CDKN2A expression with OS of HNSCC in TIMER. (G) Multivariate Cox analysis for PRF1 expression.
Relationship between PRF1 expression and tumor immune infiltration in HNSCC
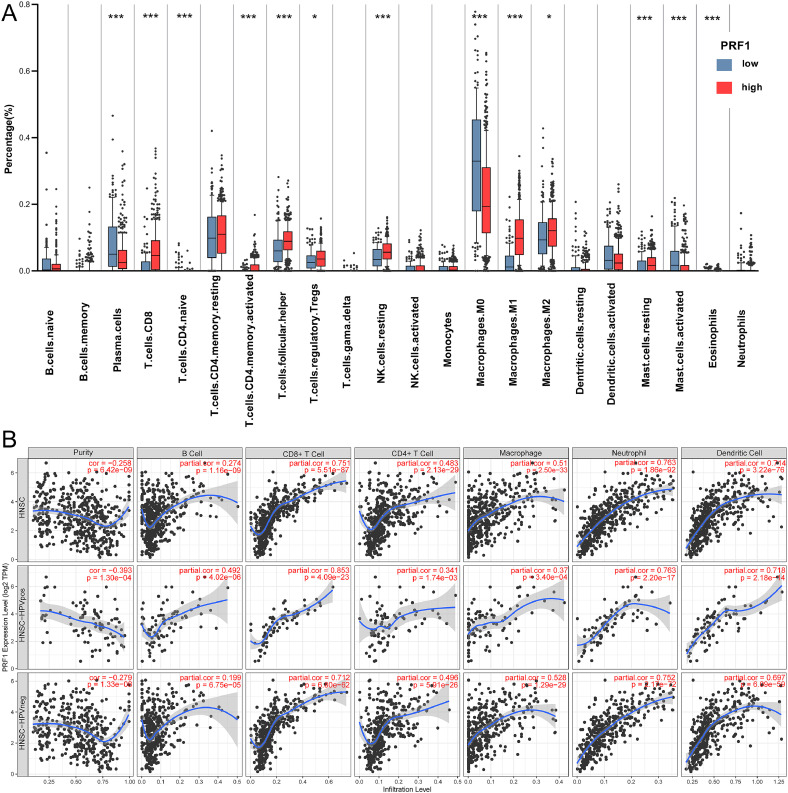
The Kaplan–Meier survival curves showed high PRF1 expression correlated with better prognosis. Since PRF1 is an essential cytotoxic protein involved in anti-tumor immunity, we next explore the relationship of PRF1 expression with immune cell infiltration in HNSCC. The infiltrating proportions and differences of immune cells between the PRF1high and PRF1low groups were estimated via the CIBERSORT tool (Fig. 4A). The results displayed that CD8+ T cells (p < 0.0001), activated CD4+ memory T cells (p < 0.0001), naive CD4+ T cells (p < 0.0001), follicular T helper cells (Tfhs, p < 0.0001), resting NK cells (p < 0.0001), M1 macrophages (p < 0.0001), M2 macrophages (p < 0.01) and regulatory T cells (Tregs, p < 0.01) were significantly elevated in the PRF1high group in comparison to the PRF1low group, while M0 macrophages, eosinophils and plasma cells were significantly decreased (p < 0.0001).
Fig. 4.
Relationship of PRF1 expression to tumor infiltrating immune cells in HNSCC. (A) Comparison of the infiltrating levels of immune cells between the PRF1high and PRF1low groups. (B) Associations of PRF1 expression with tumor immune infiltration in HNSCC overall, HPV+ HNSCC and HPV- HNSCC. (*p < 0.01, **p < 0.001, ***p < 0.0001).
PRF1 was differentially expressed according to HPV status. Therefore, we confirmed the relationship of PRF1 expression to infiltrating immune cells in HNSCC overall, HPV+ HNSCC and HPV- HNSCC through TIMER (Fig. 4B). In whole HNSCC, positive correlations were observed between PRF1 expression and 6 kinds of immune cells, especially neutrophils (r = 0.763, P = 1.86e-92), CD8+ T cells (r = 0.751, P = 5.51e-87) and DCs (r = 0.714, P = 3.22e-76). The associations of PRF1 expression with immune cell infiltration differed by HPV infection status. In HPV+ HNSCC, PRF1 expression showed significant associations with CD8+ T cell (r = 0.853, P = 4.09e-23), neutrophil (r = 0.763, P = 2.20e-27), DC (r = 0.718, P = 2.18e-14) and B cell (r = 0.492, P = 4.02e-6) infiltration. Compared with HNSCC overall, PRF1 expression in HPV+ HNSCC showed stronger associations with infiltrating CD8+ T cells and DCs. Moreover, the associations of PRF1 expression with infiltrating neutrophils (r = 0.752, P = 2.17e-72), CD8+ T cells (r = 0.712, P = 6.80e-62) and DCs (r = 0.697, P = 6.09e-59) were weaker in HPV- HNSCC than in HPV+ HNSCC. Our findings suggested that PRF1 was related to tumor immune infiltration in HNSCC and that the associations were closely influenced by HPV status.
Association assessment between PRF1 and gene markers of immune cells
Since PRF1 expression exhibited significant correlations with infiltrating immune cells in HNSCC, which was influenced by HPV status. We finally estimated the associations between PRF1 and gene markers of various immune cells in HNSCC overall, HPV+ HNSCC and HPV- HNSCC (Table 1). The findings presented that PRF1 expression was significantly related to a majority of immune markers in HNSCC overall and HPV- HNSCC after adjusting for tumor purity. In HPV+ HNSCC, PRF1 expression exhibited significant associations with CD8+ T cell, monocyte, T cell (general), B cell and NK cell markers but poor correlations with DC, TAM, and M1 and M2 macrophage markers. Moreover, regarding different functional T cells, PRF1 expression presented significant correlations with Th1 markers (TBX21, STAT4, STAT1, IFNG, TNF) but poor correlations with Th2 markers (GATA3, STAT6, IL-13), Tfh markers (BCL6, IL-21), Th17 markers (STAT3, IL17A) and Treg markers (CCR8, STAT5B, TGF-B1).
Table 1.
Associations assessment between PRF1 and the gene markers of immune cells in TIMER.
| Description | Gene markers | HNSCC |
HPV (+) HNSCC |
HPV (-) HNSCC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| None |
Purity |
None |
Purity |
None |
Purity |
||||||||
| Cor | P | Cor | P | Cor | P | Cor | P | Cor | P | Cor | P | ||
| B cell | CD19 | 0.456 | *** | 0.421 | *** | 0.599 | *** | 0.550 | ** | 0.392 | *** | 0.344 | *** |
| CD79A | 0.448 | *** | 0.415 | *** | 0.541 | ** | 0.487 | * | 0.411 | *** | 0.366 | *** | |
| CD8+ T cell | CD8A | 0.861 | *** | 0.862 | *** | 0.917 | *** | 0.909 | *** | 0.845 | *** | 0.846 | *** |
| CD8B | 0.799 | *** | 0.792 | *** | 0.775 | *** | 0.789 | *** | 0.793 | *** | 0.784 | *** | |
| T cell (general) | CD3D | 0.822 | *** | 0.811 | *** | 0.867 | *** | 0.862 | *** | 0.809 | *** | 0.795 | *** |
| CD3E | 0.837 | *** | 0.832 | *** | 0.877 | *** | 0.864 | *** | 0.828 | *** | 0.818 | *** | |
| CD2 | 0.828 | *** | 0.818 | *** | 0.855 | *** | 0.840 | *** | 0.822 | *** | 0.811 | *** | |
| Monocyte | CD86 | 0.649 | *** | 0.624 | *** | 0.632 | *** | 0.537 | * | 0.644 | *** | 0.623 | *** |
| CD115(CSF1R) | 0.650 | *** | 0.623 | *** | 0.596 | *** | 0.513 | * | 0.656 | *** | 0.627 | *** | |
| Neutrophil | CD66b (CEACAM8) | 0.014 | 2.730 | 0.003 | 3.469 | 0.107 | 1.089 | 0.128 | 0.853 | 0.068 | 0.611 | 0.089 | 1.007 |
| CD11b (ITGAM) | 0.410 | *** | 0.373 | *** | 0.515 | * | 0.485 | * | 0.362 | *** | 0.317 | *** | |
| CCR7 | 0.575 | *** | 0.534 | *** | 0.636 | *** | 0.568 | ** | 0.546 | *** | 0.496 | *** | |
| Dendritic cell | HLA-DPB1 | 0.727 | *** | 0.706 | *** | 0.753 | *** | 0.717 | *** | 0.705 | *** | 0.680 | *** |
| HLA-DQB1 | 0.618 | *** | 0.590 | *** | 0.641 | *** | 0.595 | *** | 0.565 | *** | 0.595 | *** | |
| HLA-DRA | 0.752 | *** | 0.732 | *** | 0.760 | *** | 0.737 | *** | 0.734 | *** | 0.709 | *** | |
| HLA-DPA1 | 0.750 | *** | 0.730 | *** | 0.761 | *** | 0.725 | *** | 0.732 | *** | 0.707 | *** | |
| BDCA-1(CD1C) | 0.277 | *** | 0.201 | * | 0.263 | 0.458 | 0.164 | 0.456 | 0.255 | ** | 0.174 | 0.084 | |
| BDCA-4(NRP1) | 0.323 | *** | 0.297 | *** | 0.134 | 0.695 | 0.211 | 0.636 | 0.444 | *** | 0.416 | *** | |
| CD11c(ITGAX) | 0.526 | *** | 0.488 | *** | 0.559 | ** | 0.463 | 0.012 | 0.518 | *** | 0.479 | *** | |
| TAM | CCL2 | 0.457 | *** | 0.425 | *** | 0.293 | 0.177 | 0.208 | 0.689 | 0.492 | *** | 0.461 | *** |
| CD68 | 0.323 | *** | 0.302 | *** | 0.264 | 0.436 | 0.141 | 0.684 | 0.333 | *** | 0.316 | *** | |
| IL10 | 0.497 | *** | 0.462 | *** | 0.544 | ** | 0.452 | 0.021 | 0.483 | *** | 0.446 | *** | |
| M1 Macrophage | INOS(NOS2) | 0.219 | * | 0.239 | * | 0.359 | 0.058 | 0.380 | 0.044 | 0.152 | 0.087 | 0.165 | 0.165 |
| IRF5 | 0.218 | * | 0.232 | * | 0.121 | 0.868 | 0.133 | 0.784 | 0.203 | 0.018 | 0.208 | 0.019 | |
| COX2(PTGS2) | 0.056 | 0.754 | 0.032 | 1.773 | 0.038 | 2.601 | 0.079 | 1.641 | 0.077 | 0.412 | 0.048 | 1.127 | |
| M2 Macrophage | CD163 | 0.574 | *** | 0.553 | *** | 0.413 | 0.019 | 0.308 | 0.166 | 0.609 | *** | 0.584 | *** |
| VSIG4 | 0.517 | *** | 0.499 | *** | 0.347 | 0.093 | 0.261 | 0.181 | 0.559 | *** | 0.532 | *** | |
| MS4A4A | 0.588 | *** | 0.563 | *** | 0.434 | 0.025 | 0.316 | 0.128 | 0.615 | *** | 0.591 | *** | |
| NK cell | KIR2DL1 | 0.462 | *** | 0.472 | *** | 0.528 | ** | 0.497 | * | 0.420 | *** | 0.431 | *** |
| KIR2DL3 | 0.618 | *** | 0.614 | *** | 0.644 | *** | 0.638 | *** | 0.594 | *** | 0.583 | *** | |
| KIR2DL4 | 0.788 | *** | 0.797 | *** | 0.769 | *** | 0.783 | *** | 0.775 | *** | 0.781 | *** | |
| KIR3DL1 | 0.517 | *** | 0.513 | *** | 0.539 | ** | 0.509 | * | 0.488 | *** | 0.475 | *** | |
| KIR3DL2 | 0.655 | *** | 0.641 | *** | 0.745 | *** | 0.711 | *** | 0.620 | *** | 0.601 | *** | |
| KIR3DL3 | 0.329 | *** | 0.322 | *** | 0.500 | * | 0.469 | * | 0.260 | * | 0.256 | *** | |
| KIR2DS4 | 0.426 | *** | 0.404 | *** | 0.479 | * | 0.499 | * | 0.395 | *** | 0.366 | *** | |
| Th1 | T-bet(TBX21) | 0.818 | *** | 0.812 | *** | 0.884 | *** | 0.879 | *** | 0.798 | *** | 0.788 | *** |
| STAT4 | 0.632 | *** | 0.602 | *** | 0.760 | *** | 0.705 | *** | 0.595 | *** | 0.564 | *** | |
| STAT1 | 0.672 | *** | 0.661 | *** | 0.617 | *** | 0.591 | ** | 0.686 | *** | 0.674 | *** | |
| IFN-γ (IFNG) | 0.799 | *** | 0.796 | *** | 0.852 | *** | 0.844 | ** | 0.779 | *** | 0.775 | *** | |
| TNF-α (TNF) | 0.234 | * | 0.213 | * | 0.399 | 0.036 | 0.400 | 0.019 | 0.212 | * | 0.195 | 0.058 | |
| Th2 | GATA3 | 0.348 | *** | 0.303 | *** | 0.454 | * | 0.350 | 0.144 | 0.337 | *** | 0.297 | ** |
| STAT6 | 0.270 | ** | 0.300 | *** | 0.204 | 0.604 | 0.257 | 0.206 | 0.257 | ** | 0.280 | ** | |
| STAT5A | 0.568 | *** | 0.556 | *** | 0.626 | *** | 0.608 | ** | 0.543 | *** | 0.520 | *** | |
| IL-13 | 0.403 | *** | 0.386 | *** | 0.365 | 0.040 | 0.298 | 0.228 | 0.398 | *** | 0.380 | *** | |
| Tfh | BCL6 | 0.008 | 3.178 | 0.051 | 0.942 | 0.074 | 1.718 | 0.098 | 1.328 | −0.032 | 1.906 | 0.017 | 2.730 |
| IL-21 | 0.494 | *** | 0.47 | *** | 0.509 | * | 0.451 | 0.023 | 0.482 | *** | 0.456 | *** | |
| Th17 | STAT3 | 0.299 | *** | 0.297 | *** | 0.276 | 0.303 | 0.312 | 0.146 | 0.276 | * | 0.267 | * |
| IL17A | 0.526 | *** | 0.488 | *** | 0.372 | 0.029 | 0.366 | 0.076 | 0.229 | * | 0.191 | 0.021 | |
| Treg | FOXP3 | 0.664 | *** | 0.639 | *** | 0664 | *** | 0.606 | ** | 0.650 | *** | 0.621 | *** |
| CCR8 | 0.560 | *** | 0.529 | *** | 0.463 | * | 0.384 | 0.037 | 0.566 | *** | 0.534 | *** | |
| STAT5B | 0.280 | ** | 0.274 | ** | 0.176 | 1.135 | 0.061 | 2.101 | 0.293 | ** | 0.296 | ** | |
| TGF-β(TGF-B1) | 0.008 | 3.156 | 0.024 | 2.204 | 0.003 | 3.587 | −0.110 | 1.129 | 0.054 | 0.986 | 0.050 | 1.155 | |
| T cell exhaustion | PD-1(PDCD1) | 0.860 | *** | 0.856 | *** | 0.896 | *** | 0.893 | *** | 0.853 | *** | 0.846 | *** |
| CTLA4 | 0.761 | *** | 0.745 | *** | 0.821 | *** | 0.800 | *** | 0.737 | *** | 0.719 | *** | |
| LAG3 | 0.863 | *** | 0.858 | *** | 0.919 | *** | 0.912 | *** | 0.847 | *** | 0.841 | *** | |
| TIM-3(HAVCR2) | 0.752 | *** | 0.738 | *** | 0.772 | *** | 0.735 | *** | 0.741 | *** | 0.724 | *** | |
| GZMB | 0.879 | *** | 0.874 | *** | 0.917 | *** | 0.899 | *** | 0.862 | *** | 0.857 | *** | |
HNSCC, head and neck squamous cell carcinoma; Cor, R value of Spearman's correlation; None, correlation without adjustment; Purity, correlation adjusted by purity; TAM, tumor-associated macrophage; Th, T helper cell; Tfh, Follicular helper T cell; Treg, regulatory T cell. (*p < 0.01, **p < 0.001, ***p < 0.0001).
In addition, there were significant strong correlations between PRF1 and exhausted T cell markers (LAG-3, GZMB, PD-1, CTLA4, TIM-3). We further confirmed this result via the GEPIA database and also found that PRF1 expression exhibited very strong associations with the gene markers of the T cell exhaustion series (r = 0.83, p < 0.0001). Among the series, PRF1 had very strong correlations with PD-1 (r = 0.86, p < 0.0001), LAG3 (r = 0.84, p < 0.0001) and TIGIT (r = 0.82, p < 0.0001), and strong correlations with TIM-3 (r = 0.75, p < 0.0001) and CXCL13 (r = 0.62, p < 0.0001), while the correlation with LAYN was weak (r = 0.29, p < 0.05) (Table 2).
Table 2.
Relationship analysis between PRF1 and the gene signatures of T cell exhaustion in GEPIA.
| Description | Gene markers | HNSCC |
|
|---|---|---|---|
| Cor | P | ||
| T cell exhaustion | PD-1(PDCD1) | 0.86 | *** |
| LAG3 | 0.84 | *** | |
| TIM-3(HAVCR2) | 0.75 | *** | |
| LAYN | 0.29 | 0.047 | |
| TIGIT | 0.82 | *** | |
| CXCL13 | 0.62 | *** | |
| T cell exhaustion signatures | 0.83 | *** | |
Cor, R value of Spearman's correlation. (*p < 0.01, **p < 0.001, ***p < 0.0001).
Discussion
PRF1, a cytotoxic protein expressed in CTLs and NK cells, could serve as a definitive marker of immune cells with killing ability. PRF1 is involved in the granule-dependent killing pathway, which is the primary effector mechanism to clear infected or malignant host cells by CTLs and NK cells in innate and adaptive immune responses.[21] In terms of tumor immunity, PRF1 is closely correlated with the maintenance of immune homeostasis, tumor surveillance and tumor regression etc. In our study, we reported that PRF1 was overexpressed in HNSCC tumor tissues and related to clinical parameters, including T stage, clinical stage, tumor site and HPV infection status. High PRF1 expression indicated a better survival rate in HNSCC, the prognostic value of which was stronger in HPV+ HNSCC. Further analysis suggested that PRF1 expression was related to tumor immune infiltration and associated gene markers in HNSCC, of which the correlation degree was affected by HPV infection status. Our studies highlight the potential role of PRF1 in the immune network of HNSCC.
In this study, we first assessed PRF1 expression in various cancers. Our findings demonstrated that PRF1 expression was markedly increased in tumor tissues of multiple cancers in comparison to corresponding normal tissues, especially in HNSCC and KIRC. Further analysis of 505 cases of HNSCC from TCGA showed that high PRF1 expression was remarkably related to advanced HNSCC. Survival analysis via TIMER and Kaplan–Meier plotter displayed that high PRF1 expression predicted better prognosis of HNSCC. These findings suggested that PRF1 expression was related to the clinical features and survivial rates of HNSCC and could serve as a prognostic biomarker.
According to the analysis of the differential expression of PRF1, PRF1 was significantly increased in HPV+ HNSCC relative to HPV- HNSCC and was also significantly increased in OPSCC compared with OSCC and LSCC+ HPSCC, which demonstrated the association between PRF1 expression and HPV infection. HPV infection represents a critical risk factor for HNSCC, especially OPSCC. Over the past half century, HPV-related HNSCC has increased in incidence. In contrast to HPV- HNSCC patients, HPV+ HNSCC patients present with different clinical behaviours, histologies, molecular landscapes, immune profiles and prognosis.[3] P16 is a common molecular marker of HPV. Previous studies reported that P16 expression was related to the clinical features and survival rates of OSCC.[22,23] In our study, high P16 expression was also found to be associated with better OS in HNSCC. Correlation analysis indicated a positive correlation between PRF1 and P16 expression in HNSCC, which was consistent with high PRF1 expression in HPV+ HNSCC. More particularly, our results demonstrated that the prognostic value of PRF1 in HPV+ HNSCC was higher than that in both HNSCC overall and HPV- HNSCC. In view of the fact that PRF1 is a cytotoxic protein involved in tumor immune activity, we speculate that the association between PRF1 expression and HPV infection may be one of the critical factors facilitating the better survival of HPV+ HNSCC.
The TIL levels and immune status of the TME are key factors affecting tumor progression, therapeutic effects and recurrence. With a highly invasive and heterogeneous nature, HNSCC is believed to be an immunogenic tumor.[24] It has been reported that increased TILs are favorable prognostic factors for HNSCC.[25] Our findings displayed that PRF1 expression was related to the tumor immune infiltration of HNSCC, especially in HPV+ HNSCC. By comparing the differences of TILs between the PRF1high group and PRF1low group, we showed that tumors with high PRF1 expression had elevated levels of CD8+ T cell, CD4+ T cell, NK cell and M1 macrophage infiltration. The increased CD8+ T cell and CD4+ T cell infiltration indicated high levels of TILs in the PRF1high group, which further illustrated the prognostic value of PRF1. NK cells, playing an essential part in the immune effector responses and tumor surveillance, have been reported to give rise to a better prognosis in HNSCC.[26]
Previous studies have pointed out that HPV infection profoundly affects the host immunity in HNSCC and that HPV+ HNSCC presents specific immune profiles, of which CD8+ T cell and CD4+ T cell infiltration plays critical roles and correlates with a better prognosis.[27,28] Our findings displayed that PRF1 expression was strongly related to CD8+ T cell, neutrophil and DC infiltration and moderately related to CD4+ T cell and macrophage infiltration in HNSCC. The associations of PRF1 expression with CD8+ T cell, neutrophil and DC infiltration were stronger in HPV+ HNSCC than in HPV- HNSCC. The effector phenotype of CD8+ T cells could directly kill target cells by expressing perforin, which might partly explain its correlations with PRF1.[13] Moreover, the strong correlation between PRF1 expression and CD8+ T cell infiltration suggested that CD8+ T cells had higher cytotoxicity in HPV+ HNSCC than in HPV- HNSCC, which was related to favorable prognosis. Neutrophils and DCs are important components of innate immunity and can initiate the adaptive immune system in response to antigen stimuli. Our findings indicated the potential function of PRF1 in the recruitment and activation of CTLs by neutrophils and DCs.[29,30] In addition, the significant associations between PRF1 expression and most immune marker sets suggested that PRF1 was closely involved in regulating the tumor immune network in HNSCC. Compared with HPV+ HNSCC, PRF1 expression exhibited higher significant correlations with TAM, M2 Macrophage and Treg markers in HPV- HNSCC. TAMs, M2 Macrophages and Tregs play promotive roles in the establishment of the immunosuppressive TME, which facilitates tumor immune escape.[31] Of note, PRF1 expression was significantly associated with exhausted T cell markers in HNSCC, especially in HPV+ HNSCC. Traditionally, as a marker of immune cytotoxic activity (CYT), PRF1 was expected to play an opposite role against T cell exhaustion. However, Cai M et al. reported that the immune markers of exhausted T-cells were significantly positively associated with CYT and inflamed T-cell markers, which suggested the complicated crosstalk between immune attack and immune protection in the TME.[32] Further works are necessary to elucidate the related mechanisms of the interrelations between T-cell exhaustion and CYT.
To summarize, high PRF1 expression had significant associations with improved survival and with CD8+ T cell, CD4+ T cell, DC and neutrophil infiltration in HNSCC, especially in HPV+ HNSCC. The prominent associations between PRF1 and multiple immune marker sets highlight the important role of PRF1 in tumor immunity in HNSCC. Therefore, PRF1, which is related to immune cell infiltration and HPV infection, may be served as a novel prognostic biomarker in HNSCC. There are some limitations of our work that we are lack of validation model and multiple examinations of PRF1 expression, which need to be studied in future work. Further understanding of PRF1-related immune infiltration mechanisms will favor new progress for immunotherapy in HNSCC.
Author contributions
All authors contributed to the reported work significantly in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas, or in drafting, revising, or critically reviewing the article. All authors gave final approval of the version to be published, have agreed on the journal to which the article has been submitted, and agree to be accountable for all aspects of the work.
Declaration of Competing Interest
The author reports no conflicts of interest in this work.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant nos. 91949119 and 81670926) and Shanghai Sailing Program (Grant nos. 20YF1426400 and 19YF1430300).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101042.
Contributor Information
Bin Ye, Email: aydyebin@126.com.
Mingliang Xiang, Email: mingliangxiang@163.com.
Appendix. Supplementary materials
References
- 1.Siegel R., Miller K., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69(1):7–34. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Gupta B., Johnson N., Kumar N. Global Epidemiology of head and neck cancers: a continuing challenge. Oncology. 2016;91(1):13–23. doi: 10.1159/000446117. [DOI] [PubMed] [Google Scholar]
- 3.McDermott J., Bowles D. Epidemiology of head and neck squamous cell carcinomas: impact on staging and prevention strategies. Curr. Treat. Options Oncol. 2019;20(5):43. doi: 10.1007/s11864-019-0650-5. [DOI] [PubMed] [Google Scholar]
- 4.Marur S., Forastiere A. Head and neck cancer: changing epidemiology, diagnosis, and treatment. Mayo Clin. Proc. 2008;83(4):489–501. doi: 10.4065/83.4.489. [DOI] [PubMed] [Google Scholar]
- 5.Gavrielatou N., Doumas S., Economopoulou P., Foukas P.G., Psyrri A. Biomarkers for immunotherapy response in head and neck cancer. Cancer Treat. Rev. 2020;84 doi: 10.1016/j.ctrv.2020.101977. [DOI] [PubMed] [Google Scholar]
- 6.Cohen E.E.W., Bell R.B., Bifulco C.B. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC) J. Immunother. Cancer. 2019;7(1):184. doi: 10.1186/s40425-019-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y.P., Wang Y.Q., Lv J.W. Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy. Ann. Oncol. 2019;30(1):68–75. doi: 10.1093/annonc/mdy470. [DOI] [PubMed] [Google Scholar]
- 8.Seminerio I., Descamps G., Dupont S. Infiltration of FoxP3+ Regulatory T Cells is a strong and independent prognostic factor in head and neck squamous cell carcinoma. Cancers. 2019;11(2):227. doi: 10.3390/cancers11020227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Ruiter E.J., Ooft M.L., Devriese L.A., Willems S.M. The prognostic role of tumor infiltrating T-lymphocytes in squamous cell carcinoma of the head and neck: a systematic review and meta-analysis. Oncoimmunology. 2017;6(11) doi: 10.1080/2162402x.2017.1356148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanna G.J., Lizotte P., Cavanaugh M. Frameshift events predict anti-PD-1/L1 response in head and neck cancer. JCI Insight. 2018;3(4):e98811. doi: 10.1172/jci.insight.98811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lei Y., Xie Y., Tan Y.S. Telltale tumor infiltrating lymphocytes (TIL) in oral, head & neck cancer. Oral Oncol. 2016;61:159–165. doi: 10.1016/j.oraloncology.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolitho P., Voskoboinik I., Trapani J.A., Smyth M.J. Apoptosis induced by the lymphocyte effector molecule perforin. Curr. Opin. Immunol. 2007;19(3):339–347. doi: 10.1016/j.coi.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Pipkin M.E., Rao A., Lichtenheld M.G. The transcriptional control of the perforin locus. Immunol. Rev. 2010;235:55–72. doi: 10.1111/j.0105-2896.2010.00905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alcaraz-Sanabria A., Baliu-Piqué M., Saiz-Ladera C. Genomic signatures of immune activation predict outcome in advanced stages of ovarian cancer and basal-like breast tumors. Front. Oncol. 2019;9:1486. doi: 10.3389/fonc.2019.01486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roufas C., Chasiotis D., Makris A. The expression and prognostic impact of immune cytolytic activity-related markers in human malignancies: a comprehensive meta-analysis. Front. Oncol. 2018;8:27. doi: 10.3389/fonc.2018.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lánczky A., Nagy Á., Bottai G. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 2016;160(3):439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 18.Gentles A.J., Newman A.M., Liu C.L. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21(8):938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li T., Fan J., Wang B. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.can-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucl. Acids. Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voskoboinik I., Dunstone M.A., Baran K., Whisstock J.C., Trapani J.A. Perforin: structure, function, and role in human immunopathology. Immunol. Rev. 2010;235(1):35–54. doi: 10.1111/j.0105-2896.2010.00896.x. [DOI] [PubMed] [Google Scholar]
- 22.Abdelhakam D.A., Huenerberg K.A., Nassar A. Utility of p16 and HPV testing in oropharyngeal squamous cell carcinoma: an institutional review. Diagn. Cytopathol. 2020 Aug:1–6. doi: 10.1002/dc.24593. [DOI] [PubMed] [Google Scholar]
- 23.Liu S.Z., Zandberg D.P., Schumaker L.M., Papadimitriou J.C., Cullen K.J. Correlation of p16 expression and HPV type with survival in oropharyngeal squamous cell cancer. Oral Oncol. 2015;51(9):862–869. doi: 10.1016/j.oraloncology.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Chen S.M.Y., Krinsky A.L., Woolaver R.A., Wang X., Chen Z., Wang J.H. Tumor immune microenvironment in head and neck cancers. Mol. Carcinog. 2020;59(7):766–774. doi: 10.1002/mc.23162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Q., Wang C., Yuan X., Feng Z., Han Z. Prognostic value of tumor-infiltrating lymphocytes for patients with head and neck squamous cell carcinoma. Transl Oncol. 2017;10(1):10–16. doi: 10.1016/j.tranon.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bisheshar S.K., De Ruiter E.J., Devriese L.A., Willems S.M. The prognostic role of NK cells and their ligands in squamous cell carcinoma of the head and neck: a systematic review and meta-analysis. Oncoimmunology. 2020;9(1) doi: 10.1080/2162402x.2020.1747345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen A.S., Koldjaer Sølling A.S., Ovesen T., Rusan M. The interplay between HPV and host immunity in head and neck squamous cell carcinoma. Int. J. Cancer. 2014;134(12):2755–2763. doi: 10.1002/ijc.28411. [DOI] [PubMed] [Google Scholar]
- 28.Lechien J.R., Seminerio I., Descamps G. Impact of HPV infection on the immune system in oropharyngeal and non-oropharyngeal squamous cell carcinoma: a systematic review. Cells. 2019;8(9):1061. doi: 10.3390/cells8091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaul M.E., Fridlender Z.G. Neutrophils as active regulators of the immune system in the tumor microenvironment. J. Leukoc. Biol. 2017;102(2):343–349. doi: 10.1189/jlb.5MR1216-508R. [DOI] [PubMed] [Google Scholar]
- 30.Gardner A., Ruffell B. Dendritic cells and cancer immunity. Trends Immunol. 2016;37(12):855–865. doi: 10.1016/j.it.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mantovani A., Macrophages Sica A. innate immunity and cancer: balance, tolerance, and diversity. Curr. Opin. Immunol. 2010;22(2):231–237. doi: 10.1016/j.it.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Cai M.C., Zhao X., Cao M. T-cell exhaustion interrelates with immune cytolytic activity to shape the inflamed tumor microenvironment. J. Pathol. 2020;251(2):147–159. doi: 10.1002/path.5435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.