Abstract
In a post-pandemic scenario, indoor air monitoring may be required seeking to safeguard public health, and therefore well-defined methods, protocols, and equipment play an important role. Considering the COVID-19 pandemic, this manuscript presents a literature review on indoor air sampling methods to detect viruses, especially SARS-CoV-2. The review was conducted using the following online databases: Web of Science, Science Direct, and PubMed, and the Boolean operators “AND” and “OR” to combine the following keywords: air sampler, coronavirus, COVID-19, indoor, and SARS-CoV-2. This review included 25 published papers reporting sampling and detection methods for SARS-CoV-2 in indoor environments. Most of the papers focused on sampling and analysis of viruses in aerosols present in contaminated areas and potential transmission to adjacent areas. Negative results were found in 10 studies, while 15 papers showed positive results in at least one sample. Overall, papers report several sampling devices and methods for SARS-CoV-2 detection, using different approaches for distance, height from the floor, flow rates, and sampled air volumes. Regarding the efficacy of each mechanism as measured by the percentage of investigations with positive samples, the literature review indicates that solid impactors are more effective than liquid impactors, or filters, and the combination of various methods may be recommended. As a final remark, determining the sampling method is not a trivial task, as the samplers and the environment influence the presence and viability of viruses in the samples, and thus a case-by-case assessment is required for the selection of sampling systems.
Keywords: Biological air sampler, Cyclone, COVID-19, Impactor, Impinger
Introduction
According to the Centers for Disease Control and Prevention (CDC-Centers for Disease Control and Prevention 2019), coronaviruses are a great family of viruses commonly present in human beings and many other species, including camels, cattle, cats, and bats. Coronaviruses native to non-human species rarely infect and spread through human beings; however, such has occurred with MERS-CoV, the virus responsible for Middle East respiratory syndrome, which arose in Saudi Arabia in 2012 and is a relative of the coronavirus family.
In China, in 2002, a virus was identified as the cause of severe acute respiratory syndrome (SARS), and at present in the same country, it has been attributed to be the cause of COVID-19. In January 2020, the etiological agent responsible for severe pneumonia cases in Wuhan, China, has been identified as a new betacoronavirus which differs from SARS-CoV and MERS-CoV. According to the Centers for Disease Control and Prevention (CDC-Centers for Disease Control and Prevention 2019), some cause minor diseases that affect the upper respiratory tract. However, SARS-CoV-2 is a new coronavirus strain that had not been previously observed on humans and presents potential infection by the inhalation of small droplets exhaled by an infected person, which can transport its viral charge and travel the distance of up to dozens of meters in the air.
The new coronavirus responsible for the beginning of the pandemic in 2019 (COVID-19) is stable for up to 4 h in copper surfaces, up to 24 h in cardboard, and between 2 and 3 days in plastic and stainless steel, according to the paper by the National Institutes of Health (NIH), Centers for Disease Control and Prevention (CDC), University of California, Los Angeles (UCLA), and Princeton University (PU), published in The New England Journal of Medicine (NIH National Institutes of Health 2020). Scientists discovered that SARS-CoV-2 was detectable in aerosols. The results offer important data on SARS-CoV-2 stability and suggest that people may acquire the virus through the air, as well as by touching contaminated objects. Papers report on the mechanisms of its transportation, and there is evidence that this is a significant route of infection in indoor environments (Ijaz et al. 2016; Bae et al. 2020; Bourouiba 2020; Qureshi et al. 2020). The virus present in the biggest aerial droplets penetrates the human organism adhering to the eye, nose, and mouth mucosae or even through contact with previously contaminated surfaces that are then concluded via touching the eyes, nose, and mouth (WHO 2020a).
Leclerc et al. (2020) reported the possibility of SARS-CoV-2 transmission via aerosols, combined with transportation via droplets, primarily in restaurants (Lu et al. 2020) or in gym classrooms (Jang et al. 2020). Transmission via short-range aerosols cannot be disregarded, particularly in confined environments where there is high human density and inadequate ventilation, in case there are infected people for a prolonged time.
The World Health Organization (WHO) declared, on January 30, 2020, that the outbreak caused by the new coronavirus (COVID-19) constitutes a Public Health Emergency of International Concern, the highest WHO alert level, as foreseen in the International Sanitary Regulations (OPAS – Organização Pan-Americana da Saúde 2020).
Infectious diseases represent a permanent threat to public health and safety. The confrontation of the current global COVID-19 pandemic (caused by the new SARS-CoV-2 coronavirus) has shown itself to be a great challenge to this century’s societies, as it can be observed by the amount of information concerning this event ever since March 2020. Scientists from all over the world are making efforts to comprehend the characteristics and behavior of the virus’ transmission. Several research topics have been addressed ranging from genetics and viral detection methods to disinfection processes to avoid dissemination and contamination of people both in indoor and outdoor environments. There is a need for comprehension of the behavior of SARS-CoV-2 in the air using field data involving monitoring and detection of this virus in ambient air.
The authors of the paper Airborne transmission of SARS-CoV-2: The world should face the reality, Morawska and Cao (2020), recommend that Australian authorities acknowledge the fact that the virus spreads through the air and that adequate control methods are implemented to hinder its dissemination, primarily on the removal of droplets that carry the virus in indoor air through ventilation.
The WHO (2020b) performed trials with the generation of aerosols from infectious samples using high potency jet nebulizers in controlled lab conditions. These papers revealed that the SARS-CoV-2 viral RNA remains in the air for up to 3 h (van Doremalen et al. 2020) and detected viruses with the capacity to replicate in 16 h (Fears et al. 2020). However, such papers do not accurately reflect the typical conditions of aerosols caused by human cough.
There is still little data on the characteristics of aerosols that contain SARS-CoV-2 and on both their concentration and behavioral patterns during airborne transmission. Besides the inhalation of liquid droplets and close contact with infected people or surfaces, the transmission of the virus via aerosols is an important factor to be considered (Liu et al. 2020). Nevertheless, there are hindrances to the sampling of aerosols bearing the virus and to the challenges of it quantifying in low concentration levels. Research points to the complications in the replication stage of viable organisms collected in the air (Lindsley et al. 2017). This lack of scientific knowledge limits effective COVID-19 risk evaluations, prophylactic measures, and outbreak control.
Previous papers regarding other viruses have been described by Otter et al. (2016). The SARS-CoV, MERS-CoV, and transmissible gastroenteritis virus (TGEV) survival times are usually measured in days, weeks, or months. The differences in the survival capability of the influenza virus, SARS-CoV, and MERS-CoV were rightly illustrated by van Doremalen et al. (2013), which tested the H1N1 flu virus and MERS-CoV. The viable MERS-CoV virus was recovered after 48 h, with a half-life varying between ± 0.5 and 1 h. On the other hand, no viable H1N1 virus was recovered after 1 h in any of the conditions tested. In these regards, it is observed that there is a certain difficulty in establishing the various survival times of the different types of viruses that were studied, both airborne and on material surfaces.
It has been recently shown that the SARS-CoV-2 virus lasts in aerosols for periods ranging over 3 h, with a half-life estimated to be 1.1 h. The persistent virus genome can be detected 24 h after its exposure to this environment, according to van Doremalen et al. (2020). The monitoring of bioaerosols in occupational environments is one of the many tools that the industrial hygienist utilizes for the evaluation of internal air quality, infectious disease outbreaks, agricultural exposures, and occupational health. Bioaerosol sampling may be a useful tool in studying occupational exposures, possible dangers, and the transmission of infectious diseases, but it has significant limitations. This information must be accounted for when collecting samples, when preparing a sampling routine, and when interpreting results, understanding that there are no standards to be compared to.
Therefore, a literature review has been produced aiming for discussing the methods for air sampling in indoor environments to detect the presence of SARS-CoV-2.
Air sampling methods for viruses detection
According to Pan et al. (2019), viruses that affect human beings, other animals, and plants are frequently dispersed and transmitted via airways. The precise identification of airborne viral presence is challenging. The hindrances on sampling procedures limit the effective collection capacity of viruses in the upkeep of viability and in identifying their distribution in aerosol particles. Therefore, the wide understanding of viral transmission through the air is of fundamental importance to the establishment of better infection control strategies.
Sampler types
The U.S. Environmental Protection Agency (Cao et al. 2010) researched the capacity that the NIOSH bioaerosol sampler had at collecting viable airborne viruses as well as the development of techniques to preserve virus feasibility during and after collecting. The virus of the A flu was used as a surrogate for testing purposes, due to its newly emerging strains having the potential to create a pandemic. It was determined that viable infectious viruses were present in all three of the particle fractions collected. The highest viable influenza virus percentages were found in the fraction of 1–4 μm (48–55%) and the fraction smaller than 1 μm (26–41%), while lower percentages were found in the fraction of particles bigger than 4 μm (11–19%).
Pan et al. (2019) presented conceptual schematic diagrams of various air samplers dedicated to collecting viruses transmitted through the air and their various collection methods, such as an impactor, a cyclone, a filter, an electrostatic precipitator, and an impinger-type collector. Verreault et al. (2008) presented an extensive compilation of papers up to 2008 regarding viral particles. Among the samplers are the cyclonic sampler, which separates particles by their dimensions, the single-stage Andersen sampler, the slit sampler, the liquid AGI-30 impactors, and filtering systems such as polycarbonate, polytetrafluorethylene (PTFE), and gelatin filters.
On the impactor, the air is sucked in by a vacuum pump, and the air current flows through small nozzles (shaped as orifices or slits) towards the impact surface, where the particles are then separated from the air current by their inertia. On the cascade impactor, by successively diminishing the nozzle sizes, particles are categorized by their inertia and are collected in different stages by their size (Pan et al. 2019). Larger particles gather on the impaction surface, while smaller particles that do not impact remain in the air current and are not quantified. The impaction surface usually consists of a lubricated tape or plate, filter material, or growth medium (agar) contained in Petri dishes (Verreault et al. 2008).
The Sioutas impactor, utilized by Liu et al. (2020), consists of four impaction stages and a post-filter that allows for the separation and collection of airborne particles into five size ranges. This equipment is adequate for personal and area sampling in closed environments, and it can be used for open-air sampling in special arrangements. The samples separated by size can be analyzed gravimetrically, chemically, and microscopically.
The slit samplers are used primarily to determine the concentrations of bacteria present in aerosol as a function of time. The accelerated particles impact a rotating Petri dish containing a specific culture medium, as described by Colbeck and Whitby (2019).
On the impinger system, the bioaerosol particles are sampled by a vacuum pump as they shoot through the equipment’s nozzle at high speed, impacting a collector liquid, which is then later withdrawn for polymerase chain reaction (PCR) analysis (Pan et al. 2019; Lindsley et al. 2017).
The aerosol filters are commonly used to collect particles with smaller dimensions (bioaerosols) due to their simplicity and low cost. During filtration, the particles are collected at the filter through interception, inertial impaction, and diffusion. Filter-based sampling is particularly useful for personal bioaerosol sampling, as the filter-based collectors are small, light, and work well with personal sampling pumps. Filters can be preceded by selective particle size openings, such as a cyclone or impactor, to remove larger particles and provide bioaerosol particle size classifications (Lindsley et al. 2017).
Electrets filters, used by Cai et al. (2020), are made from a combination of positively and negatively charged fibers, which substantially increase the effectiveness of collection, allowing for higher sampling rates during longer periods, using batteries.
On the electrostatic precipitator, particles are charged via corona effect to create electrostatic attraction, as the charged particles are directed towards the collection plates (opposite charge) (Pan et al. 2019).
Regarding the cyclone, when the aerosol current enters it, the air flows by the interior curved wall and into a spiral system. If the aerosol particles are bigger than the set diameter, they collide into the cyclone wall and deposit at its bottom. The airflow with lighter particles climbs through the cyclone’s center and exits through the top of the equipment (Pan et al. 2019). Coriolis μ (BERTIN INSTRUMENTS 2020) is based on a cyclonic technology, combined with a high airflow rate (Zhou et al. 2020). The airflow rate from this equipment can degrade viral RNA during its collection when superior to 200 L/min; thus, the recommended airflow rate is 200 L/min and 1 m3 of air during each experiment. The ideal collective liquid medium recommended for the sampling of aerosols that contain viruses is the saline phosphate buffer.
Liquid mediums, such as impinger-type sterilized liquid mediums (Faridi et al. 2020), the SASS 2300 Wetted Wall Cyclone Sampler (Guo et al. 2020), and bioaerosol samplers such as the NIOSH BC 251 and SKC® Biosampler are also utilized in some of the papers presented, according to Ahn et al. (2020) and Kenarkoohi et al. (2020).
According to Chen et al. (2017), each device possesses notable advantages and limits itself to the sampling of a specific environment and analysis method. Despite the wide range of bioaerosol sampling devices, most samplers that collect airborne microorganisms still have many limitations.
Alonso et al. (2017) compared the performance of two air samplers with different sizes in disease surges that occurred in porcine and poultry installations considered to be at risk regarding viral presences. The analyzed viruses were porcine respiratory syndrome virus (PRRSV), porcine epidemic diarrhea virus (PEDV), and highly pathogenic avian influenza virus (HPAIV). Air samples were collected using two sampler types: the Andersen impactor (ACI; Thermo Electron, Waltham, MA) and the Tisch cascade impactor (TCI; Model 230, Tisch Environmental, Village de Cleves, OH). The TCI is a high-volume air sampler and both it and the ACI are air samplers with size differentiation that distributes the particles that are captured into different aerodynamic diameters. The ACI sampled 28.3 L/min for 1 h and separated the particles into eight diameter intervals according to their aerodynamics: 0.4 < D ≤ 0.7, 0.7 < D ≤ 1.1, 1.1 < D ≤ 2.1, 2.1< D ≤ 3.3, 3.3 < D ≤ 4.7, 4.7 < D ≤ 5.8, 5.8 < D ≤ 9.0, and D > 9.0 μm. The TCI sampler collected samples at 1130 L/min for 30 min, separating the particles into four stages relative to their aerodynamic diameter of 0.95, 1.5, 3.0, and 7.2 μm. Particles were captured using P/N TE-230-GF (New Star Environmental, Roswell, GA) equipment, and those that were smaller than 0.95 μm were captured by a backup filter. All three of the viruses were detectable and associated with the aerosolized particles. The proportion of positive sampling events was 69% for PEDV, 61% for HPAIV, and 8% for PRRSV. Higher viral concentrations were detected on PEDV, followed by HPAIV and PRRSV. Both air collectors performed equally at detecting the total concentration of the viruses.
In the paper by Tan et al. (2020), two types of aerosol samplers were utilized for the sampling of respiratory viruses in patients present in waiting areas from three clinics in Singapore in the year of 2016. A BioSampler SKC with a BioLite air sampling pump was utilized for 60 min at 8 L/min, and SKC AirChek TOUCH personal air samplers with Teflon® filter cassettes were utilized for 180 min at 5 L/min. The research was conducted by using molecular assays with the influenza A virus, the influenza B virus, the adenovirus, and the coronavirus. In 33.3% of the 48 samples, there were signs of at least one respiratory pathogen, with a positive for the influenza A virus, three (6%) positives for the influenza B virus, and twelve (25%) positives for the adenovirus. The authors suggested the combination of aerosol sampling with molecular assays for the management of contaminations in clinical environments.
Lindsley et al. (2017) conclude that filters, regardless of causing more damage to viruses compared to other methods, are more effective at determining viral contents in aerosols. Whichever the techniques or recommendations proposed for the sampling of aerosols that contain the virus, it is important to consider the collecting of a representative sample, which must contain both nano and larger airborne particles.
Kim et al. (2018) utilized a modified Coriolis μ (BERTIN INSTRUMENTS 2020) equipment—using a cyclone-type collector for the collection of air samples in an assay prepared with the generation of an aerosol environment containing viruses. The authors detected influenza antigen with great effectiveness and high sensitivity. During a SARS outbreak, Booth et al. (2005) detected airborne coronavirus in several rooms of a hospital in Toronto, Canada, using a sampler in a liquid medium, as well as Teflon® membrane filters. The sampling of humid air was done through a system of a high-resolution slit sampler developed by Defense Research and Development Canada (DRDC). The air was sucked in through a 0.15 × 48 mm slit at a flow rate of 30 L/min and was impacted to a 150 mm Petri dish with a gelatin base (12%) and coated with a viral collection medium (sterile phosphate buffer with 7.5% bovine serum albumin, 10,000 U/mL of penicillin G, 10,000 mg/mL of streptomycin sulfate, and 25 mg/mL of amphotericin B). Other samples were collected in a polytetrafluoroethylene (Teflon®) membrane with 0.3 μm pores in a disposable three-piece closed face plastic cassette and with a personal sampling pump operating at approximately 2 L/min. The swab and humid air samples were evaluated by reverse transcriptase polymerase chain reaction (RT-PCR), being that the ribonucleic acid (RNA) was extracted from 100 μL of a viral transportation fluid using the RNeasy Mini Kit (QIAGEN) with an added RNA transporter.
Sampling challenges
The detection of viruses in air samples presents many challenges. In comparison to other microorganisms, viruses are present in the air only at extremely low concentrations, which translates to the necessity of sampling relatively larger air volumes, in cubic meters, to obtain trustworthy analysis results, unlike the samplings of chemical agents. There is another issue since the viral integrity must also be maintained to obtain trustworthy estimates on viral feasibility. Therefore, to evaluate the presence of viruses in air samples, adequate sampling is required, despite the various issues associated with traditional sampling methods. The first issue is punctual sampling, that is, it represents a single instant, being that it cannot be used to evaluate long-term bioaerosol—particles contain fungi, bacteria, and viruses (Reponen 2017).
According to Verreault et al. (2008), studies on the aerobiology of infectious diseases, including viral diseases, have been greatly limited. This is mainly due to the difficulty in collecting and analyzing airborne biological contaminants, especially viruses. Several sampling devices can be used to collect the airborne virus, the most common being liquid and solid impactors, as well as filters and electrostatic precipitators.
Pan et al. (2019) presented several challenges when evaluating the environmental concentration of viruses in the air: ineffectiveness at collecting fine particles; viral dehydration during the collection process; viral damage occurring due to impact forces, resulting in the loss of some or all viral viability; reaerosolization of viral during collection; and losses due to the viral retention at the samplers’ inputs or walls.
Air sampling for SARS-CoV-2 detection
Some studies conducted in health institutions on symptomatic patients contaminated by COVID-19 have reported the presence of the SARS-CoV-2 RNA in air samples (Chia et al. 2020; Guo et al. 2020; Santarpia et al. 2020; Zhou et al. 2020; Liu et al. 2020; Ma et al. 2020; Lednicky et al. 2020a; Kenarkoohi et al. 2020). Other similar investigations did not find viable SARS-CoV-2 virus in air samples (Faridi et al. 2020; Cheng et al. 2020a, b; Ong et al. 2020; Yamagishi 2020; Döhla et al. 2020; Wu et al. 2020; Ding et al. 2020; Lei et al. 2020; Li et al. 2020; Ahn et al. 2020; Lane et al. 2020; Cai et al. 2020).
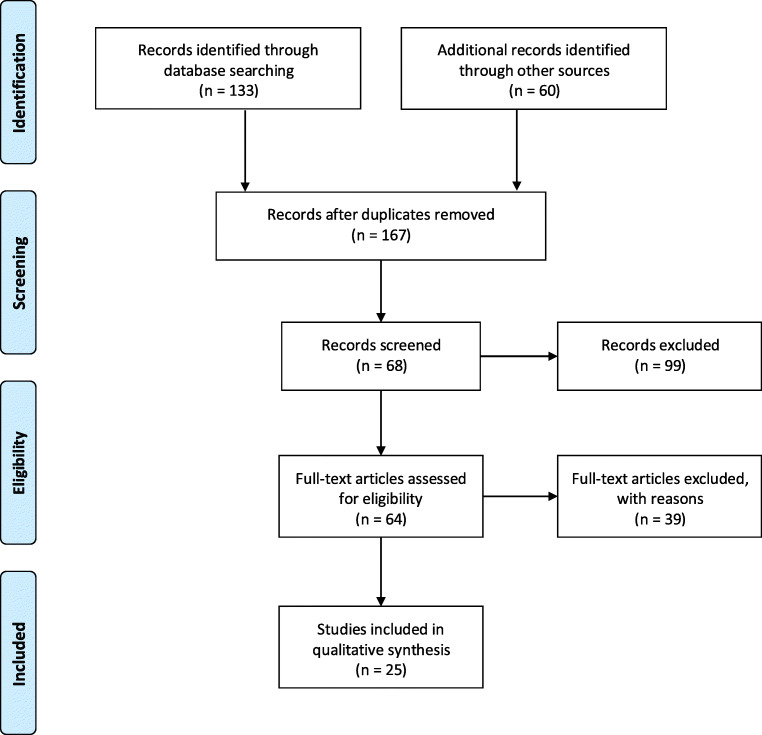
Seeking to conduct a literature review of methods for air sampling in indoor environments to detect SARS-CoV-2, search engines of academic databases were used to select papers describing sampling methods for SARS-CoV-2 detection. On PubMed, Science Direct, and Web of Science database, the Boolean operators “AND” and “OR” were used to combine the following keywords: air sampler, coronavirus, COVID-19, indoor, and SARS-CoV-2. A total of 193 articles published until October 2020 were found with potential interest from the initial search, and their titles were screened based on their research context. Following duplicate removal, 167 papers were selected, and after initial screening, 68 articles were selected, and their abstracts were appropriately reviewed. The final exclusion was performed based on the following criteria: (i) air sampler was not assessed; (ii) study was not conducted in indoor environments. Finally, 25 papers showed qualitative results for COVID-19 in air samples and were summarized in this manuscript. The PRISMA flow diagram (Moher et al. 2009) was applied using only qualitative synthesis to the final inclusion, as shown in Fig. 1.
Fig. 1.
Flow diagram based on PRISMA method
Solid impactors
Lane et al. (2020) utilized 2-stage cyclone samplers (NIOSH BC 251) and a cassette filter containing a Teflon® filter measuring 37 mm in diameter and having 2 μm in porosity. The sampling took place in an isolated room of the ICU in the Serious Communicable Disease Unit at Emory University Hospital in Atlanta (20 air exchanges/h with HEPA filters). Each sampler was connected to an air sampling pump (PCXR-4, SKC, Eighty-Four, PA) at a flow rate of 3.5 L/min, between 315 and 360 min. None of the 28 samples tested positive for SARS-CoV-2.
In Xiangya Hospital (H-X), Chenzhou Second People’s Hospital (H-C), and Shaoyang Central Hospital (H-S), the exposure risk to SARS-CoV-2 was evaluated by Ge et al. (2020). All air samples were collected for 30 min using the NIOSH BC251 bioaerosol sampler (National Institute for Occupational Safety and Health) and air pumps (XR5000, SKC) at 3.5 L/min. The NIOSH sampler segregates air sample into large particles (> 4 μm), medium particles (1–4 μm), and small particles (< 1 μm). Environmental surfaces were sampled using swabs by scrubbing the object surfaces. Positive rate of SARS-CoV-2 nucleic acid was 7.7% in COVID-19 respiratory investigation wards and 82.6% in ICUs with confirmed COVID-19 patients.
In the study by Chia et al. (2020), six bioaerosol NIOSH BC 251 samplers were utilized in samplings distributed into three particle size fractions. All of the NIOSH samplers were connected to SKC AirCheck Touch pumps or SKC Universal air sampling pumps adjusted at a flow rate of 3.5 L/min and working for 4 h, collecting a total of 5040 L of samples from each room. Results revealed that 66.7% of the air samples were positive for SARS-CoV-2. According to the authors, there is great difficulty in cultivating viruses due to the low viral concentrations in the air samples, as well as their compromised integrity due to the stress caused by sampling impacts, even in ICU environments and hospital areas.
Liquid impactors
A total of 14 air samples were taken from different places in the infirmaries of the Shahid Mustafa Khomeini hospital in Iran. The impinger SKC type biosampler was used with a flow rate of 12 L/min at a height of 1.5 m from the floor. Two air samples presented positive results for SARS-CoV-2, indicating that the virus has several air transmission paths besides transmission from person to person (Kenarkoohi et al. 2020).
In the research by Li et al. (2020), 135 air samples were collected with an impinger-type sampler (BIO-Capturer-6, Bioenrichment Co., Hangzhou, China). All samples from the 45 places presented negative results for SARS-CoV-2 RNA.
Air and surface samples were collected in the isolated rooms of three COVID-19 patients that presented severe pneumonia. The air samplers were operated for 20 min with a flow rate of 12.5 L/min and 10 L/min for the SKC BioSampler. The air samples tested negative for SARS-CoV-2 (Ahn et al. 2020).
High-volume air samples were collected in Jiangjunshan Hospital at Guizhou Province, Southwest China, using a WA 400 Portable viral aerosol sampler (Dingblue Tech, Inc.), at 400 L/min for 15 min. Air samples were taken 0.5 m away from the patient’s bedside in the isolation room and in the middle of the staff PPE dressing room. The sampling height for both locations was 1.5 m. Their findings revealed the presence of SARS-CoV-2 in the indoor air of the ICU and indicate that the virus may be shed via aerosol for days, even after a patient has tested negative (Jin et al. 2020).
Filters
Cheng et al. (2020a) performed air sampling with samplers that had sterile gelatin filters close to six COVID-19 patients, either asymptomatic or symptomatic, with or without surgical masks. To increase the proportion of sampled air and to reduce the proportion of environmental air from the air conditioning system with 12 air exchanges per hour, an umbrella equipped with a transparent plastic curtain was used as an air shelter to cover patients during sample collection. Inside this aerial shelter, samples were collected using the airscan Sartorius MD8 sampling device (Sartorius AG, Germany), with sterile gelatin filters (80 mm in diameter and 3 μm pore size (type 17528-80-ACD)) (Sartorius AG). The environmental surfaces that were always touched by people and near 21 patients had swab samplings before daily environmental disinfection. The correlation between the viral charges present in the patients’ clinical samples and environmental samples was analyzed. All air samples tested negative for SARS-CoV-2 RNA in the six patients that were in isolation rooms.
Ong et al. (2020) evaluated rooms with good ventilation—12 air exchanges per hour—of three patients infected with SARS-CoV-2 in Singapore, using SKC Universal pumps (37 mm Teflon® filter, 0.3 μm porosity, 4 h at 5 L/min) in the room and waiting room. They utilized a Sartorius MD8 air sampler with a gelatin membrane filter for 15 min at 6 m3/h. All of the air samples presented negative for viruses.
Air was sampled for 4 days in the East–West Lake Fangcang Shelter Hospital using an air virus collection equipment (NingBo iGene TecTM) with a 0.1μm gelatin membrane filter for 10 min at 6 m3/h. A total of 48 air and environmental surface samples were collected. All PCR results were negative for SARS-CoV-2 RNA (Zhang et al. 2020).
In the study by Cheng et al. (2020b), it was not possible to detect the presence of SARS-CoV-2 in the air sampled at 10 cm from the chin of a patient, with or without a mask and in a single environment, even among symptomatic patients with high viral charges and in a patient receiving oxygen at the ICU. By contrast, Santarpia et al. (2020) performed COVID-19 samplings at the Nebraska hospital and detected viral contamination in all samples, indicating that SARS-CoV-2 can spread itself through direct means (droplets from person to person) and also by indirect means (contaminated objects and airborne transmission).
Yamagishi (2020) performed a study on a cruise ship that was cruising with around 3700 passengers and crew members that were under quarantine in the Yokohama harbor, Japan, and submitted to health exams. Among the results of the 601 samples that were tested, SARS-CoV-2 RNA was detected in 58 samples (10%). The RNA of SARS-CoV-2 was not detected in all of the 14 air samples.
Air samples were collected using a sampler (FSC-1V; Hongrui, Suzhou, China) with filter membranes that had pore sizes of 0.22 μm for 15 min at a flow rate of 100 L/min. The air sampler was put at around 0.6 m away from every patient and at 1 m above the ground, in each room. The filter membranes were cleaned with pre-humidified and sterile swabs (Copan). The air was sampled from all of the six patient rooms, but all of the samples presented negative results for SARS-CoV-2. However, three of the six samples from the exhaustion system outputs presented positive for SARS-CoV-2 (Wei et al. 2020).
In the study by Cai et al. (2020), 15 air samples and 128 environmental surface swabs were collected from 14 patients in four departments with temporary infirmaries at the ICU destined to COVID-19 in a hospital in Wuhan. The real-time polymerase chain reaction (RT-PCR)/quantitative polymerase chain reaction (qPCR) confirmed the existence of COVID-19 pathogens. All air samples presented negative results for SARS-CoV-2 through RT-PCR. The 15 air samples were collected using an air sampler with a dry filter (52 mm electret filter, InnovaPrep ACD-200 Bobcat, America), operating at a flow rate of 200 L/min for 60 min in the 14 temporary infirmaries at the ICU.
Other/various sampling methods
Air and surface samples were collected in areas close to critically or gravely ill COVID-19 patients as well as in areas used by the intensive care unit (ICU) team, for 16 days (Lei et al. 2020). The authors used two cyclonic sampling devices: a two-stage cyclonic bioaerosol sampler developed by NIOSH and a liquid aerosol particle concentrator (model W-15, Beijing Dingblue Technology Co., Ltd.). The air was continuously collected for 4 h at a flow rate of 3.5 L/min in three size fractions: > 4 μm (collected in a 15 mL tube), 1–4 μm (collected in a 1.5 mL tube), and < 1 μm (collected in a Teflon® membrane filter with 3.0 μm pores). In this study, all air samples collected in the semi-contaminated and clean areas were negative.
Lednicky et al. (2020a) presented the results in a pilot study of air sampling for the detection of SARS-CoV-2, which took place in the Student Health Care Center (SHCC) of the University of Florida (Gainesville, FL, EUA). The virus was detected in an air sample, and a phylogenetic inference on the complete genome sequence of SARS-CoV-2 was observed. The analysis through real-time reverse transcription polymerase chain reaction (rtRT-PCR) detected viral RNA (vRNA) in an air sample, and the amount of virus present in 390 L of the sampled air was low (approximately 340 equivalents in the virus’s genome). Consequently, the estimated SARS-CoV-2 concentration was that of 0.87 viral genomes by liter of air. The result of this environmental sampling provided evidence of an aerosol containing SARS-CoV-2 at the SHCC because the sample was collected 3 m from the closest patient traffic rout, in other words, farther than 2 m, defined by the WHO for viral transmissions via aerosols (WHO 2020b).
Air samples were collected in the room of two COVID-19 patients at the University of Florida Health Shands Hospital, and one patient was positive for SARS-CoV-2. Three serial 3-h samplings were performed using a prototype VIVAS air sampler and a BioSpot-VIVAS BSS300P (commercial version of the VIVAS-Aerosol Devices Inc., Ft. Collins, CO). The airborne particles were collected using a water-vapor condensation method. Viable virus was isolated from air samples collected from 2 to 4.8 m away from the patients (Lednicky et al. 2020b).
Ding et al. (2020) realized environmental sampling in four COVID-19 isolation rooms at the Hospital of Nanjing-China and analyzed the association between the sampling results and the environment, as well as the SARS-CoV-2 transmission risk, and they concluded that the concentrations were too low at the hospital that was studied. Forty-six samples were evaluated, and four sampler types were used, yielding only one positive sample.
Jiang et al. (2020) collected 19 air samples in high-risk areas using natural sedimentation and a MAS-100 ECO air sampler, finding one positive sample.
In Table 1 and Fig. 2, the sample collection methods and the results obtained on the evaluations regarding the presence of SARS-CoV-2 in air samples analyzed through reverse transcription polymerase chain reaction (RT-PCR) are summarized. Reverse transcriptase polymerase chain reaction (RT-PCR) is considered the gold standard for the detection of SARS-CoV-2 because of its high sensitivity, being the most used method for this purpose. The RT-PCR can detect 4–8 copies of the virus, considering a 95% confidence interval (Rahmani et al. 2020), and has been used to detect SARS-CoV-2 in a variety of samples, including air and surfaces, besides saliva, blood, stool, and urine (Rahmani et al. 2020; Park et al. 2020; Kenarkoohi et al. 2020). Optimization of primer sets and detection protocols for SARS-CoV-2 using PCR and real-time PCR was evaluated by Park et al. (2020).
Table 1.
Sample collection methods and results in investigations for the presence of viruses focusing on SARS-CoV-2 in air samples analyzed by reverse transcriptase polymerase chain reaction (RT-PCR).
| Sample collection method | Result summary | Reference | |
|---|---|---|---|
| Solid impactors | Air samples were collected in 3 COVID-19 patient rooms, using 6 NIOSH BC 251 bioaerosol samplers in each room, placed at different heights from the floor (1.2, 0.9, and 0.7 m), and between 1 and 2.1 m from patients. 245 surface samples were collected from 30 COVID-19 patient rooms | 66.7% of the air samples were positive for SARS-CoV-2 (1.84 × 103 to 3.38 × 103 RNA copies/m3). Rooms with viral particles in the air also presented surface contamination | Chia et al. (2020) |
| 33 air samples were collected for 30 min using the NIOSH BC 251 bioaerosol sampler (National Institute for Occupational Safety and Health) with air pumps (XR5000, SKC) at 3.5 L/min. The NIOSH sampler segregates air sample into large (≥ 4 μm), medium (1–4 μm), and small (< 1 μm) particles | 7.7% and 82.6% of samples were positive for SARS-CoV-2 in COVID-19 respiratory investigation wards and ICUs with confirmed COVID-19 patients, respectively | Ge et al. (2020) | |
| 28 samples were collected using 2-stage cyclone samplers (NIOSH BC 251), filter cassette containing a Teflon® filter (D = 37 mm and 2 μm porosity), and a sampling pump (PCXR-4, SKC, Eighty-Four, PA) at a flow rate of 3.5 L/min for 315–360 min | All samples were negative for SARS-CoV-2 | Lane et al. (2020) | |
| Liquid impactors | Air samples were collected in COVID-19 isolation rooms used for three patients with severe pneumonia, using a SKC BioSampler at 12.5 L/min and at 10 L/min, both for 20 min, placed 1.2 m from the floor, and at a 1-m distance from patients | All samples were negative for SARS-CoV-2 | Ahn et al. (2020) |
| 10 air samples were collected during 1 h, using a vacuum pump at 1.5 L/min, in SKC sterile standard midget impingers at a height of 1.5–1.8 m from the floor and at a distance of 2–5 m from beds of patients with severe and critical symptoms | All samples were negative for SARS-CoV-2 | Faridi et al. (2020) | |
| Air samples were collected using a SASS 2300 wetted wall cyclone sampler (Research International, Inc., https://www.resrchintl.com) at 300 L/min for 30 min | SARS-CoV-2 was detected in air at a 4-m distance from patients | Guo et al. (2020) | |
| Air samples were collected using a high-volume WA 400 Portable viral aerosol sampler (Dingblue Tech, Inc.), at 400 L/min for 15 min | SARS-CoV-2 was detected in 01 out of 02 air samples | Jin et al. (2020) | |
| Air samples were collected using the SKC impinger-type biosampler at a flow rate of 12 L/min, placed 1.5 m from the floor | 02 out of 14 air samples were positive for SARS-CoV-2 | Kenarkoohi et al. (2020) | |
| 135 air samples were collected using an impinger sampler (BIO-Capturer-6, Bioenrichment Co., Hangzhou, China), for 30 min at 80 L/min, and placed 1.0–1.5 m from the floor | All samples were negative for SARS-CoV-2 | Li et al. (2020) | |
| 26 air samples were collected into 3-mL virus culture liquid (MT0301, Yocon Biology Inc., Beijing, China) using the WA-15 and WA-400 impactors (Beijing dBlueTech, Inc.) at flow rates of 15 L/min and 400 L/min, respectively | One air sample was positive for SARS-CoV-2 | Ma et al. (2020) | |
| Filters | 15 air samples were collected using a dry filter air sampler (52-mm electret filters, InnovaPrep ACD-200 Bobcat, America) for 60 min at 49 L/min | All samples were negative for SARS-CoV-2 | Cai et al. (2020) |
| 1000-L air samples from air shelters with patients were collected using the Sartorius MD8 sampler with sterile gelatin filters (D = 80 mm and pore size = 3 μm) (Sartorius AG, Germany) for 20 min at a flow rate of 50 L/min, being the air sampler perpendicularly positioned 10 cm from the patient’s chin | All samples were negative for SARS-CoV-2 | Cheng et al. (2020a) | |
| 1000-L air samples were collected using a SAS Super ISO 180 model 86834 (VWR International PBI Srl, Milan, Italy), being the air sampler perpendicularly positioned 10 cm from the patient’s chin | All samples were negative for SARS-CoV-2 | Cheng et al. (2020b) | |
| Air samples were collected using SKC Universal pumps (with 37 mm filter cassettes and 0.3-μm Teflon® filters) for 4 h at 5 L/min, and a Sartorius MD8 microbiological sampler (with gelatin membrane filter) for 15 min at 6 m3/h | All samples were negative for SARS-CoV-2 | Ong et al. (2020) | |
| 05 air samples were collected using an AirPort MD8 with gelatin membrane filters (Sartorius, Varedo, MB, Italy) for 40 min at 50 L/min, placed 1.5 m from the floor | All samples collected from contaminated area were positive for SARS-CoV-2 | Razzini et al. (2020) | |
| 24 high-volume air samples were collected using a Sartorius Airport MD8 air sampler at 50 L/min for 15 min, and gelatin filters (D = 80 mm); and 08 low-volume personal air samples were collected at 4 L/min using Personal Button Samplers (SKC, Inc.), AirChek pumps (SKC, Inc.), and gelatin filters (D = 25 mm) | 14 out of 24 high-volume air samples were positive for SARS-CoV-2. All personal air samples were positive for SARS-CoV-2 | Santarpia et al. (2020) | |
| 06 air samples were collected using a FSC-1V air sampler (Hongrui, Suzhou, China) with filter membranes (0.22-μm pore size) for 15 min at 100 L/min, placed 1 m from the floor and 0.6-m away from patients | All samples were negative for SARS-CoV-2 | Wei et al. (2020) | |
| 04 air samples from ship cabins were collected using a Sartorius Airport MD8 air sampler at 50 L/min for 20 min and a Sartorius gelatin filter type 175 (area = 38.5 cm2) | One air sample was positive for SARS-CoV-2 | Yamagishi (2020) | |
| Air samples were collected in the East–West Lake Fangcang Shelter Hospital, using an air virus collection equipment (NingBo iGene TecTM) with a 0.1 μm gelatin membrane filter for 10 min at 6 m3/h. A total of 48 air samples were collected | All samples were negative for SARS-CoV-2 | Zhang et al. (2020) | |
| Other/various sampling methods | Four bioaerosol samplers were used: an Andersen one-stage viable impactor (QuickTake-30, SKC, USA), an AirPort MD8 with gel film (Sartorius, Germany), an ASE-100 (Langsi Medical Technology, Shenzhen, China) in liquid medium, and a WA-15 (Dinglan Technology, Beijing, China) | 45 out of 46 samples were negative for SARS-CoV-2 | Ding et al. (2020) |
| 28 air samples were collected by natural sedimentation and using a microbial air sampler (MAS-100 ECO) at 100 L/min | One air sample was positive for SARS-CoV-2 | Jiang et al. (2020) | |
| Three serial 3-h samplings using a prototype VIVAS air sampler and a BioSpot-VIVAS BSS300P. The airborne particles were collected using a water-vapor condensation method | Viable virus was isolated from air samples collected from 2 to 4.8 m away from the patients | Lednicky et al. (2020b) | |
| Air samples were collected using a two-stage cyclonic bioaerosol sampler (NIOSH) for 4 h at a flow rate of 3.5 L/min, into three size fractions: > 4 μm (15-mL tube), 1–4 μm (1.5–mL tube) and < 1 μm (Teflon® membrane filter with a pore size of 3.0 μm), and a cyclonic aerosol particle liquid concentrator (model W-15, Beijing Dingblue Technology Co., Ltd.) for 30 min at a flow rate of 14 L/min | 02 air samples were positive for SARS-CoV-2 | Lei et al. (2020) | |
| Aerosol samples were collected consisting of 03 different types: total suspended particles, segregated aerosol, and deposition. Total suspended particles were collected on 25 mm styrene filter cassettes (SKC Inc., USA) at 5 L/min using a portable pump (APEX2, Casella, USA). Segregated aerosol samples were collected using a cascade impactor (Sioutas impactor, SKC Inc., USA) at 9 L/min. Aerosol deposition samples were collected using filters (D = 80 mm) placed on the floor and intact for 7 days | 19 out of 35 samples were positive for SARS-CoV-2, being 113 copies/m3 the highest RNA concentration | Liu et al. (2020) | |
| Air samples were collected into conical tubes containing 5 mL of Dulbecco’s minimum essential medium (DMEM) using a Coriolis μ air sampler (BERTIN INSTRUMENTS 2020) | 14 out of 31 air samples were positive for SARS-CoV-2 RNA | Zhou et al. (2020) |
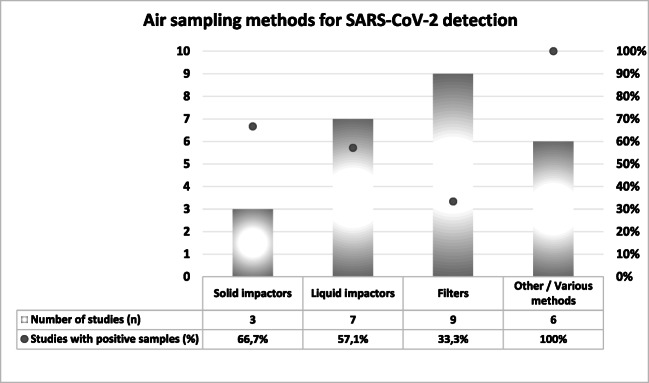
Fig. 2.
Summary of air sampling mechanisms for SARS-CoV-2 detection and percentage of studies with positive samples
As shown in Table 1, different air samplers based on various mechanisms have been used to collect air samples for SARS-CoV-2 detection, and positive results were obtained using the following samplers: NIOSH BC 251 bioaerosol sampler; ASE-100 biological aerosol special-collection liquid; SASS 2300 wetted wall cyclone sampler; microbial air sampler MAS-100 ECO; high-volume WA 400 Portable viral aerosol sampler; SKC impinger-type bio sampler; prototype VIVAS air sampler and BioSpot-VIVAS BSS300P; filter cassettes; WA-15 and WA-400 impactors; AirPort MD8 with gelatin membrane filters; and Coriolis μ air sampler. Importantly, negative results are not exclusively related to the type of sampler, given that a concentration below the detection limit or even the absence of the virus may occur.
As shown in Fig. 2, most of the studies included in this review collected air samples for SARS-CoV-2 detection using variations of filtration systems (n = 9), while liquid impactors represented the second most used mechanism (n = 7), followed by other/various methods (n = 6), being solid impactors the least used mechanism. Regarding the efficacy of each mechanism as measured by the percentage of studies with positive results, the literature review indicates that solid impactors are more effective than liquid impactors, or filters, and the combination of various methods may be recommended.
Overview and final remarks
To evaluate possible microbiological air, surface, and hospital equipment contamination, reliable sampling methods are required. However, as reported by Kowalski (2012), this type of sampling is yet unregulated by the diverse legislations from around the world. Secondly, according to the Centers for Disease Control and Prevention (CDC), airborne particles containing fungi, bacteria, and viruses (bioaerosols) require new sampling techniques, regarding both reduced handling and increased risk detection processes, through more precise ribonucleic acid (RNA), deoxyribonucleic acid (DNA), and immunologic assay identifications.
According to Napoli et al. (2012), through correct air sampling and adequate detection methods, it is possible to evaluate microbial contamination in environments that are at high risk of infection, which can then be used to identify ventilation systems and hygiene procedures efficacies. According to the authors, there are several international standards combined with sampling techniques (active or passive) and many different types of samplers, meaning that the users are free to choose systems. The great challenge is standardizing the viral collection methods, especially the ones used for COVID-2, for, as it is presented in Table 1, several different methods have been utilized.
In active monitoring, a microbiological air sampler sucks in a known air volume through or over a particle collection device that can be either a liquid or solid medium or a filtering membrane, and the quantity of present microorganisms is measured in colony-forming units (CFU)/m3 of air. This system is applicable when the microorganism concentration is not too high, as is in a surgical intervention room and other controlled hospital environments. In passive monitoring, Petri dishes containing specific culture mediums are exposed to air for a period to collect biological particles that “deposit” themselves. In both cases, samples are incubated for posterior microorganism detection and quantifying, expressing their results in CFU/dish/time or CFU/m2/hour (Napoli et al. 2012). Despite the costs for both methods being moderate, there is not a dominant sample collection method, especially regarding the aerial medium. A fact that stands out is that liquid mediums in the particle collection system helps to maintain viral integrity and viability and can be analyzed directly, without the need to prepare and extract the viral target from the filtering medium (Pan et al. 2019).
In active sampling, dry filters—such as polyvinyl chloride (PVC) and polytetrafluoroethylene (PTFE) used by Ong et al. (2020) and Booth et al. (2005), glass fiber, cellulose ester (MCE), and polycarbonate (PC)—and gelatin filters that can be used in the MD-8 Air Scanner (Santarpia et al. 2020; Cheng et al. 2020a; Ong et al. 2020; Razzini et al. 2020; Yamagishi 2020), as well as impinger-type liquids and other mediums, can be utilized. As presented in Table 1, active sampling was used in all works, with various equipment. Sartorius MD 8 equipment with gelatin filters was utilized in five papers, NIOSH BC 251® in four studies, and SKC impinger-type BioSampler® in three works.
Filters have been pointed out as the most effective means to capture particles with sizes that are smaller than 5 μm. Viruses can be identified through reaction assay methods in a polymerase chain reaction (PCR) to detect the presence of viral DNA. However, the challenge is at collecting, upkeeping, and performing the PCR that yields representative results. For the COVID-19 virus to be detected precociously in the body using real-time RT-PCR, scientists need to convert RNA into DNA. This is a process called “reverse transcription.” They must do so because only DNA can be copied—or amplified—which is a fundamental part of the RT-PCR process in real-time for viral detection (IAEA 2020).
To collect sufficient airborne viruses for detection via current molecular techniques, longer active sampling times may be required (Lednicky et al. 2020a, b). The methods that utilize real-time polymerase chain reaction (RT-PCR) can be used to identify viruses much faster than traditional PCR methods (Carter et al. 2020; Chan et al. 2020).
It is important to mention that the elution and separation of the sampled viruses is a critical step in the detection process. The use of filtering mediums can present less efficiency at the time of viral elution. Besides that, environmental factors and the desiccation of viral particles caused by the airflow are variables that justify deeper inquiries before considering using a high-volume air sampler with filters under field conditions (Alonso et al. 2017). The collection mediums that dissect the organisms (such as gelatin or impinger filters in which the water evaporates during sampling) may not be adequate, as is reported by Lednicky et al. (2020a, b). According to what was pointed out by Reponen (2017), as each technique has its unique advantages and disadvantages, usually, it is beneficial to employ several techniques at a study. This way, a technique’s limitations can be compensated by the advantages of others. Professionals should understand the limitations of viruses sampling methods to properly interpret their results. Due to the variations in the performances of several samplers, the results between different studies can only be compared if the same sampling methods are utilized. The sampling device is a critical step in all bioaerosol studies (Mbareche et al. 2018).
It is important to point out that no single sampling process is appropriate for all bioaerosol types. There are many bioaerosols with different behaviors associated with particle size, and, therefore, the choosing of an appropriate sampling procedure is fundamental. To sample a specific type of airborne virus, different types of sampling devices, suction pumps, sampling volumes, sampling times, culture mediums, and incubation conditions must be taken into account. Each one of the sampling methods can provide different performances for sampling and detection of several types of viruses (Rahmani et al. 2020). Some of the important characteristics of aerosol samplers are size, mass, airflow rate, sampling effectiveness, energy requirements, sampling mechanisms, volume, and others (Mbareche et al. 2018).
Environmental conditions play a fundamental role in the sampling and viral detection processes in a confined hospital environment, such as ventilation, air movement, distance from the patient to the sampler, room occupation, patient activities during sampling, relative humidity, temperature, number of patients, sampling flow rate, sampling time, and sampling medium, which may affect results. All of the research listed for airborne COVID-19 studies presented limitations.
The investigations of viruses in aerosols report the environmental contamination of indoor areas and potential transmission to adjacent areas, commonly in closed environments, such as hospitals and clinics, while sampling the air for viruses in open environments is not a common practice due to the low levels of viruses normally present in such environments.
Conclusions
In a post-pandemic scenario, regardless of chemical or physical decontamination methods, air and surface monitoring may be required to safeguard public health, and therefore well-defined protocols and equipment, as well as qualified personnel, play an important role.
A literature review on sampling methods to detect SARS-CoV-2 in indoor air was conducted. Overall, papers report several sampling methods for SARS-CoV-2 detection, using different approaches for distance, height from the floor, flow rates, and sampled air volumes. Most of the studies included in this review collected air samples for SARS-CoV-2 detection using variations of filtration systems, while liquid impactors represented the second most used mechanism, followed by other/various methods, being solid impactors the least used mechanism. Only four studies performed segregation of aerosol particle sizes.
Out of 25 studies, 10 found negative results in all collected air samples, and 15 found at least one positive sample. The literature review indicates that solid impactors are more effective than liquid impactors, or filters, and the combination of various methods may be recommended. Therefore, the challenge of selecting the best sampling method is illustrated in this systematic review.
As a final remark, determining the sampling method is not a trivial task, as the samplers and the environment influence the presence and viability of viruses in the samples, and thus a case-by-case assessment is required for the selection of sampling equipment.
Availability of data and material
Not applicable.
Author contribution
JTB: conceptualization, investigation, and writing-original draft. LYKN: investigation and writing-review and editing. MGM: writing-review and editing. JRG: conceptualization and supervision. All authors read and approved the final manuscript.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahn JY, An S, Sohn Y, Cho Y, Hyun JH, Baek YJ, Kim MH, Jeong SJ, Kim JH, Ku NS, Yeom J-S, Smith DM, Lee H, Yong D, Lee Y-J, Kim JW, Kim HR, Hwang JH, Choi JY. Environmental contamination in the isolation rooms of COVID-19 patients with severe pneumonia requiring mechanical ventilation or high-flow oxygen therapy. J Hosp Infect. 2020;106:570–576. doi: 10.1016/j.jhin.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C, Raynor PC, Goyal S, Olson BA, Alba A, Davies PR, Torremorell M. Assessment of air sampling methods and size distribution of virus-laden aerosols in outbreaks in swine and poultry farms. J Vet Diagn Investig. 2017;29(3):298–304. doi: 10.1177/1040638717700221. [DOI] [PubMed] [Google Scholar]
- Bae S, Kim MC, Kim JY, Cha HH, Lim JS, Jung J, Kim MJ, Oh DK, Lee MK, Choi SH, Sung M, Hong SB, Chung JW, Kim SH. Effectiveness of surgical and cotton masks in blocking SARS-CoV-2: a controlled comparison in 4 patients. Ann Intern Med. 2020;173:W22–W23. doi: 10.7326/M20-1342. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- BERTIN INSTRUMENTS (2020) Air monitoring - Coriolis air samplers collect biological particles in the air which offer new perspectives for the control of airborne contamination thanks to its liquid sample. https://www.bertin-instruments.com/products-range/air-samplers/. Accessed 21 August 2020
- Booth TF, Kournikakis B, Bastien N, Ho J, Kobasa D, Stadnyk L, Li Y, Spence M, Paton S, Henry B, Mederski B, White D, Low DE, McGeer A, Simor A, Vearncombe M, Downey J, Jamieson FB, Tang P, Plummer F. Detection of airborne severe acute respiratory syndrome (SARS) coronavirus and environmental contamination in SARS outbreak units. J Infect Dis. 2005;191(9):1472–1477. doi: 10.1086/429634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourouiba L. Turbulent gas clouds and respiratory pathogen emissions: potential implications for reducing transmission of COVID-19. JAMA. 2020;323(18):1837–1838. doi: 10.1001/jama.2020.4756. [DOI] [PubMed] [Google Scholar]
- Cai Y, Wu X, Zhang Y, Xia J, Li M, Feng Y, Yu X, Duan J, Weng X, Chen Y, Cheng Z, Zhan Q (2020) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) contamination in air and environment in temporary COVID-19 ICU wards. Res Square. 10.21203/rs.3.rs-21384/v1
- Cao G, Blachere FM, Lindsley WG, Noti JD, Beezhold DH. Development of a methodology to detect viable airborne virus using personal aerosol samplers. Washington, DC: U.S. Environmental Protection Agency; 2010. [Google Scholar]
- Carter LJ, Garner LV, Smoot JW, Li Y, Zhou Q, Saveson CJ, Sasso JM, Gregg AC, Soares DJ, Beskid TR, Jervey SR, Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent Sci. 2020;6(5):591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC-Centers for Disease Control and Prevention (2019) COVID-19 and Animals. https://www.cdc.gov/coronavirus/2019-ncov/daily-life-coping/animals.html. Accessed 30 August 2020
- Chan JFW, Yip CCY, To KKW, Tang THC, Wong SCY, Leung KH, Fung AYF, Ng ACK, Zou Z, Tsoi HW, Choi GKY, Tam AR, Cheng VCC, Chan KH, Tsang OTY, Yuen KY (2020) Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol 58(5). 10.1128/JCM.00310-20 [DOI] [PMC free article] [PubMed]
- Chen P, Du Y, Xu YZ, Liu Z, Yan K. Review: bioaerosol collection. IJPEST. 2017;11(1):52–55. doi: 10.34343/ijpest.2017.11.01.052. [DOI] [Google Scholar]
- Cheng VCC, Wong SC, Chan VWM, So SYC, Chen JHK, Yip CCY, Chan KH, Chu H, Chung TWH, Sridhar S, To KKW, Chan JFW, Hung IFN, Ho PL, Yuen KY (2020a) Air and environmental sampling for SARS-CoV-2 around hospitalized patients with coronavirus disease 2019 (COVID-19). Infect Control Hosp Epidemiol. 10.1017/ice.2020.282 [DOI] [PMC free article] [PubMed]
- Cheng VCC, Wong SC, Chen JHK, Yip CCY, Chuang VWM, Tsang OTY, Sridhar S, Chan JFW, Ho PL, Yuen KY. Escalating infection control response to the rapidly evolving epidemiology of the coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 in Hong Kong. Infect Control Hosp Epidemiol. 2020;41(5):493–498. doi: 10.1017/ice.2020.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia PY, Coleman KK, Tan YK, Ong SWX, for the Singapore Novel Coronavirus Outbreak Research T Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbeck I, Whitby C. Biological particles in the indoor environment. In: Indoor air pollution. Issues in environmental science and technology, R.M. Harrison and R.E. Hester (ed) R Soc Chem. 2019;48:127–157. doi: 10.1039/9781788016179. [DOI] [Google Scholar]
- Ding Z, Qian H, Xu B, Huang Y, Miao T, Yen HL, Xiao S, Cui L, Wu X, Shao W, Song Y, Sha L, Zhou L, Xu Y, Zhu B, Li Y. Toilets dominate environmental detection of SARS-CoV-2 virus in a hospital. Sci Total Environ. 2020;141710:141710. doi: 10.1016/j.scitotenv.2020.141710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhla M, Wilbring G, Schulte B, Kümmerer BM, Diegmann C, Sib E, Richter E, Haag A, Engelhart S, Eis-Hübinger AM, Exner M, Streeck H, Schmithausen RM (2020) SARS-CoV-2 in environmental samples of quarantined households. MedRxiv. 10.1101/2020.05.28.20114041
- Faridi S, Niazi S, Sadeghi K, Naddafi K, Yavarian J, Shamsipour M, Jandaghi NZS, Sadeghniiat K, Nabizadeh R, Yunesian M, Momeniha F, Mokamel A, Hassanvand MS, MokhtariAzad T. A field indoor air measurement of SARS-CoV-2 in the patient rooms of the largest hospital in Iran. Sci Total Environ. 2020;725:138401. doi: 10.1016/j.scitotenv.2020.138401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fears AC, Klimstra WB, Duprex P, Hartman A, Weaver SC, Plante KS, Mirchandani D, Plante JA, Aguilar PV, Fernández D, Nalca A, Totura A, Dyer D, Kearney B, Lackemeyer M, Bohannon JK, Johnson R, Garry RF, Reed DS, Roy CJ. Persistence of severe acute respiratory syndrome coronavirus 2 in aerosol suspensions. Emerg Infect Dis. 2020;26(9):2168–2171. doi: 10.3201/eid2609.201806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XY, Pu Y, Liao CH, Huang WF, Zeng Q, Zhou H, Yi B, Wang AM, Dou QY, Zhou PC, Chen HL, Liu HX, Xu DM, Chen X, Huang X. Evaluation of the exposure risk of SARS-CoV-2 in different hospital environment. Sustain Cities Soc. 2020;61:102413. doi: 10.1016/j.scs.2020.102413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZD, Wang ZY, Zhang SF, Li X, Li L, Li C, Cui Y, Fu RB, Dong YZ, Chi XY, Zhang MY, Liu K, Cao C, Liu B, Zhang K, Gao YW, Lu B, Chen W. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg Infect Dis. 2020;26(7):1583–1591. doi: 10.3201/eid2607.200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IAEA (2020) How is the COVID-19 virus detected using real time RT-PCR? IAEA Office of Public Information and Communication https://www.iaea.org/newscenter/news/how-is-the-covid-19-virus-detected-using-real-time-rt-pcr. Accessed 10 August 2020
- Ijaz MK, Zargar B, Wright KE, Rubino JR, Sattar SA. Generic aspects of the airborne spread of human pathogens indoors and emerging air decontamination technologies. Am J Infect Control. 2016;44(9):S109–S120. doi: 10.1016/j.ajic.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Han SH, Rhee JY. Cluster of coronavirus disease associated with fitness dance classes, South Korea. Emerg Infect Dis. 2020;26(8):1917–1920. doi: 10.3201/eid2608.200633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wang H, Chen Y, He J, Chen L, Liu Y, Hu X, Li A, Liu S, Zhang P, Zou H, Hua S (2020) Clinical data on hospital environmental hygiene monitoring and medical staff protection during the coronavirus disease 2019 outbreak. MedRxiv. 10.1101/2020.02.25.20028043
- Jin T, Li J, Yang J, Li J, Hong F, Long H, Deng Q, Qin Y, Jiang J, Zhou X, Song Q, Pan C, Luo P (2020) SARS-CoV-2 presented in the air of an intensive care unit (ICU). Sustain Cities Soc. 10.1016/j.scs.2020.102 [DOI] [PMC free article] [PubMed]
- Kenarkoohi A, Noorimotlagh Z, Falahi S, Amarloei A, Mirzaee SA, Pakzad I, Bastani E. Hospital indoor air quality monitoring for the detection of SARS-CoV-2 (COVID-19) virus. Sci Total Environ. 2020;748:141324. doi: 10.1016/j.scitotenv.2020.141324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Kabir E, Jahan SA. Airborne bioaerosols and their impact on human health. J Environ Sci. 2018;67:23–35. doi: 10.1016/j.jes.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski W. Hospital Airborne Infection Control. Boca Raton: CRC Press; 2012. [Google Scholar]
- Lane MA, Brownsword EA, Morgan JS, Babiker A, Vanairsdale AS, Lyon GM, Mehta AK, Ingersoll JM, Lindsley WG, Kraft CS. Bioaerosol sampling of a ventilated patient with COVID-19. Am J Infect Control. 2020;000:1–3. doi: 10.1016/j.ajic.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc QJ, Fuller NM, Knight LE, Funk S, Knight LE, CMMID COVID-19 Working Group. Funk S, Knight GM. What settings have been linked to SARS-CoV-2 transmission clusters? Wellcome Open Res. 2020;5:83. doi: 10.12688/wellcomeopenres.15889.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky JA, Shankar SN, Elbadry MA, Gibson JC, Alam MM, Stephenson CJ, Eiguren-Fernandez A, Morris JG, Mavian CN, Salemi M, Clugston JR, Wu CY. Collection of SARS-CoV-2 virus from the air of a clinic within a university student health care center and analyses of the viral genomic sequence. Aerosol Air Qual Res. 2020;20:1167–1171. doi: 10.4209/aaqr.2020.05.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky JA, Lauzardo M, Fan ZH, Jutla AS, Tilly TB, Gangwar M, Usmani M, Shankar SN, Mohamed K, Eiguren-Fernandez A, Stephenson CJ, Alam MM, Elbadry MA, Loeb JC, Subramaniam K, Waltzek TB, Cherabuddi K, Morris-Jr JG, Wu CY (2020b) Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. MedrXiv. 10.1101/2020.08.03.20167395 [DOI] [PMC free article] [PubMed]
- Lei H, Ye F, Liu X, Huang Z, Ling S, Jiang Z, Cheng J, Huang X, Wu Q, Wu S, Xie Y, Xiao C, Ye D, Yang Z, Li Y, Leung NHL, Cowling BJ, He J, Wong SS, Zanin M. SARS-CoV-2 environmental contamination associated with persistently infected COVID-19 patients. Influenza Other Respir Viruses. 2020;00:1–12. doi: 10.1111/irv.12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Fan YZ, Jiang L, Wang HB. Aerosol and environmental surface monitoring for SARS-CoV-2 RNA in a designated hospital for severe COVID-19 patients. Epidemiol Infect. 2020;148:e154. doi: 10.1017/S0950268820001570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley WG, Green BJ, Blachere FM, Martin SB, Law B, Jensen P, Schafer M (2017) Sampling and characterization of bioaerosols. NIOSH manual of analytical methods, 5th edn. National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention, Cincinnati
- Liu Y, Ning Z, Chen Y, Guo M, Liu Y, Gali NK, Sun L, Duan Y, Cai J, Westerdahl D, Liu X, Xu K, K-f H, Kan H, Fu Q, Lan K. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582(7813):557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Lu J, Gu J, Li K, Xu C, Su W, Lai Z, Zhou D, Yu C, Xu B, Yang Z. COVID-19 outbreak associated with air conditioning in restaurant, Guangzhou, China, 2020. Emerg Infect Dis. 2020;26(7):1628–1631. doi: 10.3201/eid2607.200764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Qi X, Chen H, Li X, Zhang Z, Wang H, Sun L, Zhang L, Guo J, Morawska L, Grinshpun SA, Biswas P, Flagan RC, Yao M (2020) Exhaled breath is a significant source of SARS-CoV-2 emission. medRxiv. 10.1101/2020.05.31.20115154
- Mbareche H, Veillette M, Bilodeau GJ, Duchaine C. Bioaerosol sampler choice should consider efficiency and ability of samplers to cover microbial diversity. Appl Environ Microbiol. 2018;84:e01589–e01518. doi: 10.1128/AEM.01589-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L, Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ Int. 2020;139:105730. doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C, Marcotrigiano V, Montagna MT. Air sampling procedures to evaluate microbial contamination: a comparison between active and passive methods in operating theatres. BMC Public Health. 2012;12:594. doi: 10.1186/1471-2458-12-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH National Institutes of Health (2020) New coronavirus stable for hours on surfaces. https://www. nih. gov/news-events/news-releases/new-coronavirus-stable-hours-surfaces. Accessed 26 August 2020
- Ong SWX, Tan YK, Chia PY, Lee TH, Ng OT, Wong MSY, Marimuthu K. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323(16):1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OPAS – Organização Pan-Americana da Saúde (2020) Folha informativa COVID-19 - Escritório da OPAS e da OMS no Brasil. https://www.paho.org/pt/covid19. Accessed 24 August 2020
- Otter JA, Donskey C, Yezli S, Douthwaite S, Goldenberg S, Weber DJ. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: the possible role of dry surface contamination. J Hosp Infect. 2016;92(3):235–250. doi: 10.1016/j.jhin.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Lednicky JA, Wu CY. Collection, particle sizing and detection of airborne viruses. J Appl Microbiol. 2019;127:1596–1611. doi: 10.1111/jam.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Won J, Choi BY, Lee CJ. Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR. Exp Mol Med. 2020;52:963–977. doi: 10.1038/s12276-020-0452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi Z, Jones N, Temple R, Larwood JPJ, Greenhalgh T, Bourouiba L (2020) What is the evidence to support the 2-metre social distancing rule to reduce COVID-19 transmission? The Centre for Evidence-Based Medicine develops, promotes and disseminates better evidence for healthcare. https://www.cebm.net/covid-19/what-is-the-evidence-to-support-the-2-metre-social-distancing-rule-to-reduce-covid-19-transmission/. Accessed 17 August 2020
- Rahmani AR, Leili M, Azarian G, Poormohammadi A. Sampling and detection of corona viruses in air: a mini review. Sci Total Environ. 2020;740:140207. doi: 10.1016/j.scitotenv.2020.140207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzini K, Castrica M, Menchetti L, Maggi L, Negroni L, Orfeo NV, Pizzoccheri A, Stocco M, Muttini S, Balzaretti CM. SARS-CoV-2 RNA detection in the air and on surfaces in the COVID-19 ward of a hospital in Milan, Italy. Sci Total Environ. 2020;742:140540. doi: 10.1016/j.scitotenv.2020.140540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reponen T. Sampling for Microbial Determinations. In: Viegas C, Viegas S, Gomes A, Täubel M, Sabino R, editors. Exposure to microbiological agents in indoor and occupational environments. Berlin: Springer; 2017. pp. 85–96. [Google Scholar]
- Santarpia JL, Rivera DN, Herrera VL, Morwitzer MJ, Creager HM, Santarpia GW, Crown KK, Brett-Majo DM, Schnaubelt ER, Broadhurst MJ, Lawler JV, Reid SP, Lowe JJ (2020) Aerosol and surface transmission potential of SARS-CoV-2. medRxiv. 10.1101/2020.03.23.20039446
- Tan L, Ma B, Lai X, Han L, Cao P, Zhang J, Fu J, Zhou Q, Wei S, Wang Z, Peng W, Yang L, Zhang X. Air and surface contamination by SARS-CoV-2 virus in a tertiary hospital in Wuhan, China. Int J Infect Dis. 2020;99:3–7. doi: 10.1016/j.ijid.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N, Bushmaker T, Munster VJ. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Euro Surveill. 2013;18(38):20590. doi: 10.2807/1560-7917.ES2013.18.38.20590. [DOI] [PubMed] [Google Scholar]
- van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, de Wit E, Munster VJ. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.2807/1560-7917.es2013.18.38.20590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault D, Moineau S, Duchain C. Methods for sampling of airborne viruses. Microbiol Mol Biol Rev. 2008;72(3):413–444. doi: 10.1128/MMBR.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Lin J, Duan X, Huang W, Lu X, Zhou J, Zong Z. Asymptomatic COVID-19 Patients Can Contaminate Their Surroundings: an Environment Sampling Study. mSphere. 2020;5:e00442–e00420. doi: 10.1128/mSphere.00442-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020a) Surface sampling of coronavirus disease (COVID-19): a practical “how to” protocol for health care and public health professionals. https://www.who.int/publications/i/item/surface-sampling-of-coronavirus-disease-(-covid-19)-a-practical-how-to-protocol-for-health-care-and-public-health-professionals. Accessed 12 August 2020
- WHO (2020b) Transmission of SARS-CoV-2: implications for infection prevention precautions. https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions. Accessed 12 August 2020
- Wu S, Wang Y, Jin X, Tian J, Liu J, Mao Y. Environmental contamination by SARS-CoV-2 in a designated hospital for coronavirus disease. Am J Infect Control. 2020;48(8):910–914. doi: 10.1016/j.ajic.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi T (2020) Environmental sampling for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) during a coronavirus disease (COVID-19) outbreak aboard a commercial cruise ship. MedRxiv. 10.1101/2020.05.02.20088567
- Zhang M, Wang L, Yu S, Sun G, Lei H, Wu W. Status of occupational protection in the COVID-19 Fangcang Shelter Hospital in Wuhan, China. Emerg Microbes Infect. 2020;9(1):1835–1842. doi: 10.1080/22221751.2020.1803145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Otter J, Price JR, Cimpeanu C, Garcia DM, Kinross J, Boshier P, Mason S, Bolt F, Holmes AH, Barklay W (2020) Investigating SARS-CoV-2 surface and air contamination in an acute healthcare setting during the peak of the COVID-19 pandemic in London. Clin Infect Dis. 10.1093/cid/ciaa905 [DOI] [PMC free article] [PubMed]