Abstract
Epidemiologic studies recognize that trauma and posttraumatic stress are associated with heightened suicidal behavior severity, yet examination of these associations from a genetic perspective is limited. We performed a multivariate gene-by-environment genome-wide interaction study (GEWIS) of suicidality in 123,633 individuals using a covariance matrix based on 26 environments related to traumatic experiences, posttraumatic stress, social support, and socioeconomic status. We discovered five suicidality risk loci, including the male-associated rs2367967 (CWC22), which replicated in an independent cohort. All GEWIS-significant loci exhibited interaction effects where at least 5% of the sample had environmental profiles conferring opposite SNP effects from the majority. We identified PTSD as a primary driving environment for GxE at suicidality risk loci. The male suicidality GEWIS was enriched for three middle-temporal-gyrus inhibitory neuron transcriptomic profiles: SCUBE- and PVALB-expressing cells (β = 0.028, p = 3.74 × 10−4), OPRM1-expressing cells (β = 0.030, p = 0.001), and SPAG17-expressing cells (β = 0.029, p = 9.80 × 10−4). Combined with gene-based analyses (CNTN5 passociation = 2.38 × 10−9, pinteraction = 1.51 × 10−3; PSMD14 passociation = 2.04 × 10−7, pinteraction = 7.76 × 10−6; HEPACAM passociation = 2.43 × 10−6, pinteraction = 3.82 × 10−7) including information about brain chromatin interaction profiles (UBE2E3 in male neuron p = 1.07 × 10−5), our GEWIS points to extracellular matrix biology and synaptic plasticity as biological interactors with the effects of potentially modifiable lifetime traumatic experiences on genetic risk for suicidality. Characterization of molecular basis for the effects of traumatic experience and posttraumatic stress on risk of suicidal behaviors may help to identify novel targets for which more effective treatments can be developed for use in high-risk populations.
Keywords: Suicide, Trauma, Posttraumatic stress, Social support, Socioeconomic status, Gene-by-environment
Abbreviations: GEWIS, gene-by-environment genome-wide interaction study; MICE, Multivariate Imputation via Chained Equations; StructLMM, Structured Linear Mixed Model; MAGMA, Multi-marker Analysis of GenoMic Annotation; FUMA, Functional Mapping and Annotation of Genome-Wide Association Studies; BF, Bayes factor; EE, Environmental enrichment; ECM, Extracellular matrix
Highlights
-
•
Investigate interaction of traumatic events, posttraumatic stress, and suicidality.
-
•
Male suicidality risk loci interact with physically violent trauma.
-
•
Female suicidality risk locus interacts with posttraumatic stress.
-
•
Functional annotation highlights brain cell types and extracellular matrix biology.
-
•
Modifiable suicidality risk environments may mitigate suicidality genetic risk.
1. Introduction
Suicide is a critical public health concern with nearly 45,000 people dying by suicide per year in the United States alone (Hedegaard et al., 2020). Over 70% of these individuals had a prior diagnosed mental disorder (Krysinska and Lester, 2010). Genome-wide studies have estimated the single nucleotide polymorphism (SNP)-based heritability of death by suicide at 25–48% (Docherty et al., 2019; Otsuka et al., 2019). Recent large-scale studies have focused on understanding the genetic risk for thoughts and behaviors leading up to the commission of acts to end ones’ life: ideation (Kimbrel et al., 2018), planning, and attempt (Levey et al., 2019). In this study we use the term suicidality to collectively refer to these thoughts and behaviors (Strawbridge et al., 2019).
Suicidality is interpersonally and situationally complex; and 33–98% of individuals exhibiting non-fatal suicidal behaviors have a prior psychiatric diagnosis (Krysinska and Lester, 2010). With ~7.6% h2, a recent large-scale GWAS detected several genome-wide significant (GWS) loci for suicidality (Strawbridge et al., 2019). Under the stress-diathesis model, suicide and its preceding thoughts and behaviors are the result of interaction between a stressor, environment, or state-of-being and a susceptibility to suicidal behaviors. It remains unclear which stressors are most important in influencing suicidality risk for this stress-diathesis model (van Heeringen and Mann, 2014). Epidemiologic studies associate trauma and posttraumatic stress (PTS) with heightened suicidal behavior severity (LeBouthillier et al., 2015), yet examination of these associations from a genetic perspective is limited.
Gene-by-environment (GxE) interaction studies historically rely on a priori hypotheses regarding SNP or gene effects on a phenotype given specific environmental conditions (Van der Auwera et al., 2018). Large-scale genetic studies fail to support historical candidate genes and GxE interactions (Border et al., 2019). Genome-wide gene-by-environment interaction studies (GEWIS) are an advantageous alternative approach capable of detecting GxE at risk loci across the genome without relying on a priori hypotheses and while assessing interactions with multiple, potentially phenotypically and/or genetically correlated, environmental factors (Moore et al., 2019).
The goal of this study was to identify GxE between traumatic experiences, PTS, social support, and socioeconomic status, and suicidality risk loci. We performed a multivariate GEWIS of a derived ordinal suicidality phenotype in 123,633 participants (Strawbridge et al., 2019) (55.9% female) from the UK Biobank (UKB) using a linear mixed model to investigate a covariance matrix based on 26 environments. Motivated by robust evidence that males and females experience traumatic events differently, we aimed to uncover GxE in sex-stratified analyses. Using the computational tool StructLMM (Moore et al., 2019), we performed a series of follow-up analyses to describe GxE at these detected loci and previously described suicidality risk loci in relation to environmental profiles in the UKB.
2. Materials and methods
2.1. Discovery cohort
The UK Biobank (UKB) is a large population cohort comprised of nearly 502,000 participants. UKB participants range in age from 37 to 73 years at time of recruitment and represent a general sampling of the UK population with no enrichment for specific disorders. Assessments included physical health, anthropometric measurements, and sociodemographic characteristics. By August 2017, 157,366 participants in the UK Biobank completed an ancillary online mental health questionnaire (Davis et al., 2020) covering topics of self-reported mental health and well-being.
From the UKB mental health questionnaire, we defined a suicidality phenotype for 123,633 individuals described below and following the sample inclusion pipeline of Strawbridge et al. (Strawbridge et al., 2019) plus selection of traumatic event and trauma response endorsements. The total sample included 123,633 UKB participants. Of these, 84,196 (52.7% female) were coded as “no suicidality” controls (rank = 0), 21,176 (60.7% female) as “thought life not worth living” (rank = 1), 13,078 (63.6% female) as “contemplated self-harm or suicide” (rank = 2), 2487 (70.7% female) as “deliberate self-harm” (rank = 3), and 2696 (68.1% female) as “attempted suicide” (rank = 4). Complete cohort description is provided in Supplementary Materials and Methods.
UKB genetic data were filtered to include SNPs with imputation score >0.8, minor allele frequency >0.01, Hardy-Weinberg Equilibrium p-values >1 × 10−6, and missingness >0.1.
2.2. Environments
To study the effects of traumatic events, responses to trauma, and diagnosis of posttraumatic stress disorder (PTSD) we included 26 environments from the UKB mental health questionnaire (Tables S1 and S2). (Nievergelt et al., 2018) These relate to traumatic events, PTS, social support, and socioeconomic status. Detailed descriptions of each environment are described in Supplementary Materials and Methods. Environments were imputed using the MICE (Multivariate Imputation via Chained Equations) (van Buuren and Groothuis-Oudshoorn, 2011) package in R using ten iterations of 5 multiple imputations (see Supplementary Materials and Methods). Imputation quality was evaluated by comparing the relative change in standard error (dSE) between original unimputed and imputed data (He et al., 2010).
2.3. Neuroimaging data
UKB neuroimaging data (Field IDs 25005 through 25920) represent magnetic resonance imaging (MRI) data for 32,915–39,755 suicidality study participants (mean N per neuroimaging trait = 37,799). Neuroimaging data include T1-weighted structural MRI, T2-weighted MRI, diffusion MRI, resting functional MRI, task functional MRI, and susceptibility weighted MRI.
Neuroimaging phenotypes were associated with suicidality in the full cohort and in sex-stratified analyses. Linear models were covaried for age, sex, genotype batch, and ten principal components of ancestry. For each significantly associated neuroimaging phenotype, we tested for evidence of GxE with respect to suicidality risk loci including the same set of environments and covariates as the main analysis (Materials and Methods Section 2.4).
2.4. Gene-by-environment genome-wide interaction analyses (GEWIS) using StructLMM
GEWIS was performed using StructLMM (Moore et al., 2019), a linear mixed-model approach to efficiently detect interactions between loci and one or more potentially correlated environments. Here we evaluated whether traumatic events, response to trauma, PTSD, or socioeconomic status individually and collectively interact with respect to risk for suicidality. GEWIS was performed using age, genotyping batch, and 10 principal components of ancestry as covariates. For the full sample combining males and females, sex was included as a covariate.
Marginal log likelihoods (log (Bayes factor (BF)) between the full model and the reduced model with environments removed were used to identify which environments are most relevant for the detected locus interaction effects. To prioritize sets of environments that drive GxE and to account for observed correlation between environments, we performed a greedy backwards elimination procedure prioritizing sequential removal of the strongest individual environments until the change in log marginal likelihood (Δlog (BF)) < 1 (Moore et al., 2019).
Allele effects were predicted in 50% of the UKB participants using a model trained with the remaining 50% of the sample based on the best linear unbiased predictor (BLUP) (Moore et al., 2019) method accounting for potentially different relative importance of each environment.
2.5. Functional annotation
Association results from GEWIS were mapped to genes using MAGMA (Multi-marker Analysis of GenoMic Annotation) (de Leeuw et al., 2015) implemented in FUMA v1.3.6a (Functional Mapping and Annotation of Genome-Wide Association Studies) (Watanabe et al., 2017, 2019) based on positional mapping of each variant to genes in a 2 kb window upstream and downstream of the variant (i.e., 4 kb window size) (Watanabe et al., 2017, 2019). We evaluated enrichment of gene-ontology, tissue-transcriptomic (see Supplementary Materials and Methods), and cell-type transcriptomic profiles. Enrichments, gene-based tests, and expression quantitative locus (eQTL) tests were adjusted for multiple testing using a false discovery rate (FDR) of 5%. By FUMA default, chromatin interaction tests were adjusted for multiple testing using an FDR q < 1 × 10−6. Functional insights of brain-tissue enrichments were further tested using Hi-C coupled MAGMA (H-MAGMA) (Sey et al., 2020). Further annotation of sex-stratified gene sets was performed using the ShinyGO platform (v0.61). (Ge and Jung, 2018).
2.6. Replication
We replicated the GxE effects of genomic risk regions discovered by GEWIS using the Yale-Penn cohort. Mapping of each UKB suicidality phenotype into Yale-Penn and selection of environments (Table S3) is described in Supplementary Materials and Methods).
Replication was performed for all five GEWIS-discovered genomic risk regions in the UKB. Where Yale-Penn did not contain suitable information for the genomic risk region, we performed GEWIS for the first 75 SNPs up and downstream of the region. For example, the chromosome 9 genomic risk region tests 150 SNPs from chr9:137112902-chr9:13749377.
3. Results
3.1. Cohort and phenotype summary
After sample and variant quality control we analyzed 7,284,651 SNPs for suicidality (Strawbridge et al., 2019) GxE effects in 123,633 participants (55.9% female) of European ancestry from the UKB (Fig. 1). (Strawbridge et al., 2019)
Fig. 1.
Sample sizes used in this GEWIS of suicidality by sex and in the full cohort and those used in the GWAS by Strawbridge et al. (Strawbridge et al., 2019) Suicidality is coded as follows: 0 = no suicidality controls, 1 = “thought life not worth living,” 2 = “contemplated self-harm or suicide,” 3 = “deliberate self-harm,” and 4 = “attempted suicide.”
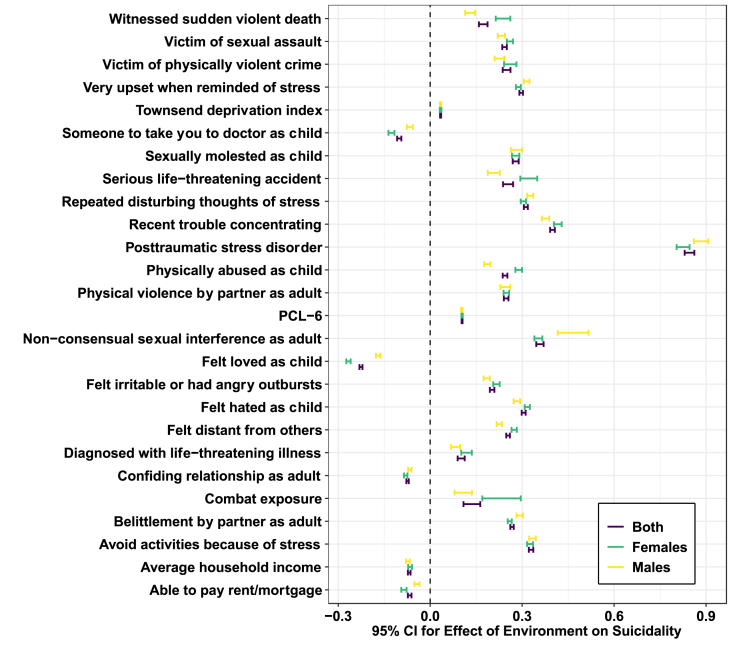
For 24 out of 26 environments (described in Tables S1 and S2), less than 10% of participants required imputation. Two environments, “felt irritable or had angry outbursts in past month” and “felt distant from other people in past month,” had missing information for 68,972 and 68,699 individuals, respectively. After imputation of all 26 environments with MICE, there was a relative decrease in standard error estimates indicating that the data were reliably imputed (Figs. S1 and S2, Tables S1 and S2) (van Buuren and Groothuis-Oudshoorn, 2011; He et al., 2010). After multiple testing correction (FDR<5%), all environments were associated with suicidality in males, females, and the full cohort in generalized linear models (Fig. 2 and S3). Genetic correlations recapitulated phenotype correlations (Supplementary Results Cohort and Phenotype Summary).
Fig. 2.
Association between suicidality and 26 environments related to traumatic experiences, responses to trauma, posttraumatic stress, social support, and socioeconomic status. Shown are 95% confidence intervals surrounding point estimates from a generalized linear model of suicidality adjusting for age, sex, genotyping batch, and ten principal components of ancestry.
3.2. Risk locus discovery using multivariable GEWIS
The discovery GEWIS of 126,633 individuals revealed one risk locus in association and interaction tests whose lead SNP was rs12589041 (passociation (pa) = 2.43 × 10−8, pinteraction (pint) = 2.04 × 10−9; Table 1 & Fig. S4). In sex-stratified GEWIS we discovered 1 significant risk locus for suicidality in females (rs118118557 pa = 2.37 × 10−8, pint = 1.14 × 10−7; Fig. S5) and three LD independent risk loci for suicidality in males (rs2367967 pa = 2.47 × 10−9, pint = 5.91 × 10−10, rs72619337 pa = 1.08 × 10−8, pint = 1.63 × 10−9; and rs6854286 pa = 1.98 × 10−8, pint = 9.41 × 10−9; Fig. S6). The rs118118557 risk locus detected in females positionally mapped to the transcription start site of CHST14 and the rs2367967 locus discovered in males positionally mapped to the CWC22 locus. The male-specific genomic risk locus mapping to CWC22 replicated (N = 1936 males; rs2367967, Yale-Penn pa = 0.019, pint = 0.099; Tables S4–8; Supplementary Results: Locus Replication in Yale-Penn using Multivariable GEWIS).
Table 1.
Lead SNP information for five risk loci, including tested alleles (A1) and their frequencies (AF) in UKB, detected in a GEWIS of suicidality.
| RSID | Sex | Chr | Position | Passociation | Pinteraction | AF (A1) | Nearest Gene |
|---|---|---|---|---|---|---|---|
| rs12589041 | Both | 14 | 71172013 | 2.43 × 10−8 | 2.04 × 10−9 | 0.25 (G) | RP6-65G23.1 |
| rs118118557 | Female | 15 | 40762879 | 2.37 × 10−8 | 1.14 × 10−7 | 0.01 (G) | CHST14 |
| rs2367967 | Male | 2 | 180948951 | 2.47 × 10−9 | 5.91 × 10−10 | 0.32 (G) | CWC22 |
| rs6854286 | Male | 4 | 12652325 | 1.98 × 10−8 | 9.41 × 10−8 | 0.37 (C) | RP11-145E17.2 |
| rs72619337 | Male | 9 | 137175269 | 1.08 × 10−8 | 1.63 × 10−9 | 0.03 (G) | RP11-352E6.1 |
3.3. Comparing GWAS vs. multivariable GEWIS
In multivariable GEWIS interaction tests, Strawbridge, et al. (Strawbridge et al., 2019) GWAS loci showed nominally significant GxE interactions with the covariance matrix tested (rs62535711 pint = 6.76 × 10−5; rs598046 pint = 2.00 × 10−6; rs7989250 pint = 0.023). In the multivariable GEWIS association tests, rs598046 also was associated with suicidality (pa = 0.020) (Table S9). Because of the GxE effects at these loci (Strawbridge et al., 2019), we included them in the subsequent GxE characterization analyses. The effects of loci identified in our multivariate GEWIS were not detected in Strawbridge et al. (Strawbridge et al., 2019) (Table S10).
3.4. Environmental risk factors
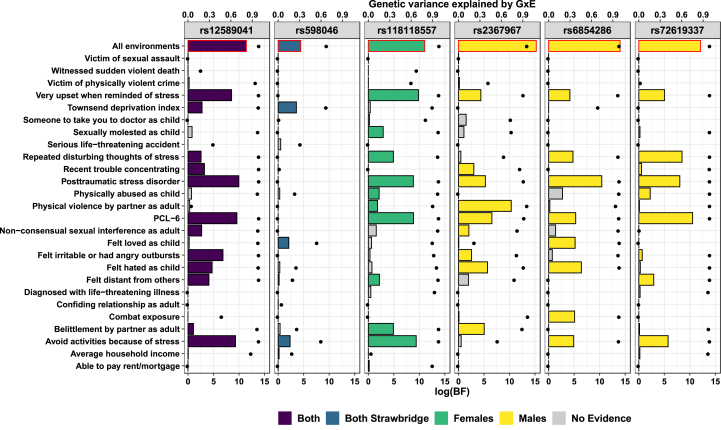
All GEWIS-discovered loci showed compelling evidence for interaction with PTSD and/or a quantitative PTSD risk score (the PTSD Checklist 6-item subset, PCL-6) (Figs. 3 and S7 and Supplementary Results: Genetic Variance Explained by GxE). GxE for rs12589041 in both sexes had log (Bayes factor (BF)) = 9.97 for PTSD and log (BF) = 9.59 for PCL-6; GxE for rs118118557 in females had log (BF) = 8.82 for PTSD and log (BF) = 8.86 for PCL-6; in males, GxE at rs2367967 had log (BF) = 5.27 for PTSD and log (BF) = 6.56 for PCL-6, GxE at rs6854286 had log (BF) = 10.41 for PTSD and log (BF) = 5.31 for PCL-6, and GxE at rs72619337 had log (BF) = 8.04 for PTSD and log (BF) = 10.57 for PCL-6.
Consistent with Moore et al. (Moore et al., 2019), we identified many environments driving the detected GxE interactions (Fig. 3) but also observed distinct GxE architectures per SNP after greedy backwards elimination procedures. All driving environment sets contained PCL-6 and/or PTSD. In females, two additional environments drove GxE at rs118118557: UKB Field ID 20498 felt very upset when reminded of stressful experience in past month log (BF) = 9.82 and UKB Field ID 20495: avoided activities or situations because of previous stressful experience in past month log (BF) = 9.39. Implicated driving environments in males include UKB Field ID 20523: physical violence by partner or ex-partner as an adult (log (BF) = 10.34 at rs2367967), UKB Field ID 20487: felt hated by family member as a child (log (BF) = 6.42 at rs6854286), and UKB Field ID 20497: repeated disturbing thoughts of stressful experience in past month (i.e., re-experiencing; log (BF) = 8.47 at rs72619337).
Fig. 3.
Individual relevance of the environment included in the covariance matrix used to detect gene-by-environment (GxE) interactions with suicidality. Each bar represents the log marginal likelihood (log (Bayes factor (BF))) of models for the indicated environment relative to models lacking that environment. Black data points indicate the genetic variance explained by GxE at each locus for the indicated environment. Bars outlined in red highlight the full GxE model at each locus; colored bars indicate environments with positive evidence for their contribution as part of a driving set of environments underlying the detected GxE (Fig. S7). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
One suicidality risk locus from Strawbridge et al. (Strawbridge et al., 2019) showed evidence of GxE interaction with the environments evaluated in this study (rs598046 full model log (BF) = 4.43; rs62535711 full model log (BF) = 0; and rs7989250 full model log (BF) = 0). The GxE at rs598046 (CNTN5 intronic variant) was driven primarily by UKB Field ID 189: Townsend deprivation index at recruitment (log (BF) = 3.58, Fig. S7).
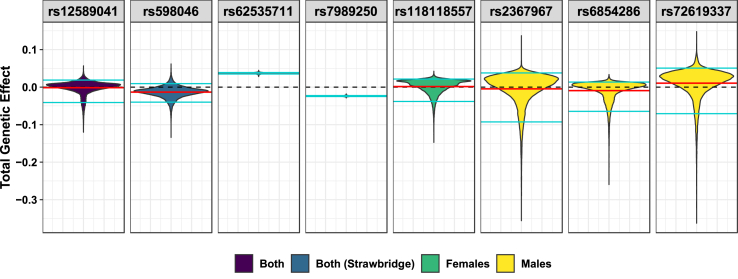
3.5. Effect size predictions
All five suicidality risk loci discovered by GEWIS exhibited qualitative interactive effects in which the effect of each SNP on suicidality switches direction depending on an individual's environmental profile (Fig. 4). Two loci exhibited positive persistent genetic effects on suicidality: rs118118557 in females (β = 1.39 × 10−3) and rs72619337 in males (β = 0.011) and three loci exhibited negative persistent genetic effects: rs12589041 in the full cohort (β = −1.49 × 10−3), rs2367967 in males (β = −4.62 × 10−3), and rs6854286 in males (β = −9.35 × 10−3).
Fig. 4.
Predicted genetic effects of GEWIS loci for suicidality. Violin plots of the distribution of estimated allelic effect sizes at each locus in 61,816 unrelated individuals (“Both”), 27,256 unrelated males, and 34,560 unrelated females of European ancestry given full models of traumatic events and posttraumatic stress environments. Persistent genetic effect estimates are shown in solid red lines; zero effect is shown in black dashed lines; cyan lines indicate the top and bottom 5% of each distribution. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Effect sizes for the loci that Strawbridge et al. (Strawbridge et al., 2019) detected by GWAS should show effects on suicidality unbiased by GxE effects. GEWIS persistent genetic effects for rs62535711 (βGEWIS = 0.037, βStrawbridge = 0.105, pdiff = 1.70 × 10−4) and rs7989250 (βGEWIS = −0.024, βStrawbridge = −0.052, pdiff = 0.002) were consistent, yet significantly smaller, estimates compared to GWAS. The locus rs598046 (CNTN5) demonstrated qualitative interactive effects with a persistent genetic effect significantly different than, and in the opposite direction from, that reported by Strawbridge et al. (Strawbridge et al., 2019) (βGEWIS = −0.013, βStrawbridge = 0.053, pdiff = 1.84 × 10−13; Fig. 4).
3.6. Neuroimaging
We investigated potential neurobiological underpinnings of the detected GxE relationships by testing each GEWIS locus for GxE effects in brain phenotypes from UKB (872 brain-imaging phenotypes; N ≤ 39,755). After multiple testing correction per sex (FDR<5%), we detected 285 neuroimaging correlates of suicidality in the full cohort, 189 in females, and 33 in males (Tables S11–13). We detected nominally significant GxE-adjusted neuroimaging associations at rs12589041 in the full sample putatively implicating various regions of the brain involved in memory (e.g., T1-weighted MRI volume of hippocampus (left) pa = 0.037), emotions (e.g., T1-weighted MRI volume of grey matter in amygdala (left) pa = 0.040), and goal-oriented behaviors (e.g., diffusion MRI weighted-mean fractional anisotropy in tract inferior fronto-occipital fasciculus (right) pa = 0.049) (Tables S14–16).
3.7. Gene-based association tests
HEPACAM (za = 4.36, pa = 6.44 × 10−6, zint = 4.62, pint = 1.93 × 10−6) was the only gene associated with suicidality in association and interaction tests (FDR q < 0.05; Table S17). Three additional genes were associated with suicidality after multiple testing correction (FDR q < 0.05) in either association or interaction tests (CNTN5 (za = 5.30, pa = 5.85 × 10−8, zint = 2.99, pint = 1.66 × 10−4), PSMD14 (za = 4.68, pa = 1.44 × 10−6, zint = 3.96, pint = 3.74 × 10−5), and HEPN1 (za = 4.20, pa = 1.35 × 10−5, zint = 4.52, pint = 3.05 × 10−6)). After multiple testing correction, there were no genes surviving multiple testing correction in the sex-stratified analyses.
We detected 9 Hi-C coupled gene-neuron associations in male suicidality GEWIS (Table S18). Two of these have been previously associated with psychopathology: UBE2E3-neuron (za = 4.25, pa = 1.07 × 10−5, zint = 4.42, pint = 4.89 × 10−6) previously associated with bipolar disorder, smoking behavior, and hurt (McCarthy et al., 2014) feelings; MORC3-neuron (za = 4.16, pa = 1.58 × 10−5, zint = 4.42, pint = 4.99 × 10−6) previously associated with frequency of tiredness and depressive symptoms; and DOP1B-neuron (za = 4.16, pa = 1.58 × 10−5, zint = 4.42, pint = 4.99 × 10−6) previously implicated in neurological disorders and dystonias (Al-Mubarak et al., 2020).
3.8. Functional annotation and regulatory effects
The GEWIS association test in males was enriched for the gene set mature B-cell differentiation (FDR q = 0.027) (Table S19). Three middle temporal gyrus (MTG) cell-type transcriptomic profiles were enriched in the GEWIS of suicidality in males (FDR q < 0.05; Table S20): (1) SCUBE3-and PVALB-expressing inhibitory neurons from MTG layers 2–5 (association tests: β = 0.028, FDR q = 0.028; interaction tests: β = 0.018, FDR q = 0.049), (2) OPRM1-expressing neurons from MTG layers 1–4 (β = 0.030, FDR q = 0.049), and (3) SPAG17-expressing inhibitory neurons from MTG layers 2–4 (β = 0.029, FDR q = 0.049).
In the full UKB sample, the genomic risk locus containing rs12589041 was a PsychENCODE eQTL for TTC9 (FDR q = 0.026) but did not exhibit brain-associated chromatin interactions (Fig. S8 and Table S21). In females, the genomic risk locus containing rs118118557 exhibited significant chromatin interactions in fetal and adult cortex, the most significant of which involved the genomic region containing CCDC32 (fetal cortex FDR q = 2.61 × 10−35; adult cortex FDR q = 1.25 × 10−46; Fig. S9 and Table S22). In males, two SNPs in the genomic risk locus containing rs2367967 were PsychENCODE eQTLs for UBE2E3 (FDR q = 0.044 and 0.046; Fig. S10 and Table S23). This same genomic risk locus exhibited significant chromatin interactions with the region of chromosome 2 containing NEUROD1 and CERKL (dorsolateral prefrontal cortex cells FDR q = 6.75 × 10−10), ITGA4 (neural progenitor cells FDR q = 6.31 × 10−7), and ZFN385B (neural progenitor cells FDR q = 4.07 × 10−19; Fig. S10 and Table S24). In males, the genomic risk locus containing rs6854286 exhibited chromatin interaction with the chromosome 2 region encoding HS3ST1 (neural progenitor cells FDR q = 1.18 × 10−7; Fig. S11 and Table S24).
4. Discussion
We conducted a comprehensive analysis of suicidality describing (i) five GEWIS risk loci with interaction and association effects with specific environments including sex-stratified GxE effects, (ii) environments with strong evidence as drivers of GxE interactions at these loci, (iii) replication of locus-suicidality associations in an independent dataset, (iv) description of GxE effects that may have confounded prior large-scale studies of suicidality, and (v) annotation of genome-wide suicidality genetic architecture uncovering relevant neuronal cell types and brain-circuitry. In aggregate, these data identify the extracellular matrix (ECM) as a molecular target for suicidality that interacts with traumatic experiences and PTS.
Of the five suicidality risk loci discovered by GEWIS, two mapped to genic regions: rs118118557 mapped to CHST14 and rs2367967 mapped to CWC22. CHST14 encodes carbohydrate sulfotransferase 14 and is responsible for sulfation of N-acetylgalactosine residues of dermatan sulfate in the ECM (Kosho et al., 2014). ECM in the brain plays a major role in the storage of information through learning and thus, ECM maintenance, regulation, and composition are reportedly highly relevant for disorders related to, or exacerbated by, prior experiences including traumatic events (Bach et al., 2019). Describing the GxE at the CHST14 SNP rs118118557 demonstrated interaction with physically and sexually violent experiences in childhood and adulthood (Moore et al., 2019). This relationship suggests that synaptic plasticity mechanisms are possible targets for treating suicidality and perhaps psychiatric disorders with a strong influence from memories of prior experiences (Whitlock et al., 2006). Although they do not survive multiple testing correction, our neuroimaging-based analyses seem to point to brain regions involved in emotion and memory processing such as hippocampus, amygdala, and the inferior fronto-occipital fasciculus (Chaaya et al., 2018). Based on this preliminary evidence, we hypothesize that one possible suicidality therapeutic target is decentering, the capacity to observe items arising in the mind (e.g., thoughts, feelings, memories, or bodily sensations) with physiological distance, heightened self-awareness, and perspective thinking (Fresco et al., 2017). One successful example of decentering is the application of mindfulness in controlling symptoms of anxiety, depression, and PTSD (Boyd et al., 2018; Parmentier et al., 2019). More statistically powerful follow-up studies are required to confirm our suggestive results and test further our hypothesis.
CWC22 encodes the “CWC22 spliceosome associated protein homolog” protein, which is the only protein-coding component of the large (127 kb) Rd2 segmental duplication. CWC22 is one of the protein-coding regions of the genome that is most often duplicated (Pezer et al., 2015). CWC22 and the Rd2 segmental duplication have previously been implicated in brain microstructural changes in response to environmental factors such as social engagement (Dause and Kirby, 2019), which has memory-inducing effects on the ECM (Koskinen et al., 2019). Environmental enrichment (EE) describes exposure to situations that facilitate enhanced sensory, cognitive, and motor stimulation and is known to affect synaptic plasticity in humans (Dause and Kirby, 2019). EE approaches have shown favorable clinical outcomes with respect to developmental disorders (Woo et al., 2015) and stroke (McDonald et al., 2018); we hypothesize that this association between CWC22 and suicidality suggests that such a noninvasive, non-pharmacological, and personalizable treatment strategy such as EE may effectively mitigate the effects of genetic liability to suicidality.
Using gene-based association testing, we discovered four genes associated with suicidality: HEPACAM (hepatocyte cell adhesion molecule), HEPN1 (HEPACAM Opposite Strand 1), PSMD14 (26S proteasome non-ATPase regulatory subunit 14), and CNTN5 (contactin-5). HEPACAM encodes GlialCAM, which is responsible for promoting interactions between ECM components and glia (Moh et al., 2009). Though previously associated with psychiatric disorders, detection of HEPN1 is likely attributed to colocalization of HEPN1 and HEPACAM (Chung Moh et al., 2005). The ubiquitin-proteasome system, of which the PSMD14 gene product is a component, plays a critical role in the expression of ECM-associated genes (Ramos de Carvalho et al., 2018). CNTN5 was previously detected by GWAS of suicidality also performed in the UKB – a finding that is consistent, but not wholly independent from the information included herein. Its product, contactin-5 is part of the glycosylphosphatidylinositol-bound protein family that has often been implicated in autism spectrum disorder (Sharom and Lehto, 2002). Contactin-5 is secreted into the ECM, where it facilitates the formation and growth of neuronal projections (Mercati et al., 2013). Strawbridge et al. (Strawbridge et al., 2019), described the rs598046 risk locus, mapped to CNTN5, as conferring increased risk for suicidality (β = 0.053) (Strawbridge et al., 2019). However, using GEWIS, we observed significant interaction effects at rs598046. With respect to Townsend deprivation index, rs598046 conferred increased and decreased suicidality risk. The confounding nature of socioeconomic status has been observed extensively in GWAS of psychiatric disorders including PTS and PTSD (Wendt et al., 2020) and it is therefore unsurprising to observe similar effects by GEWIS. This finding highlights the importance GxE characterization of GWAS risk loci to adequately evaluate the effects of environmental heterogeneity, especially via socioeconomic status.
Detected via functional annotation of the suicidality GEWIS in males only, we recapitulate two cell types previously implicated in suicidal behaviors: SPAG17-expressing and OPRM1-expressing inhibitory neurons from the middle temporal gyrus (Hodge et al., 2019). SPAG17 (sperm-associated antigen 17) was previously identified by whole exome sequencing of suicide deaths as a locus enriched for rare variants (Tombacz et al., 2017). OPRM1 encodes the mu-opioid receptor 1 and has garnered considerable attention in candidate gene and large-scale genetic studies of several psychiatric disorders and suicidal ideation (Nobile et al., 2019) and suicidal behavior (Arias et al., 2012). We also detected inhibitory neurons from layers 2–5 of the middle temporal gyrus expressing both SCUBE3 and PVALB. PVALB encodes parvalbumin, a calcium-binding protein that supports neuroplasticity and other functional properties of fast-spiking interneurons in cortical and subcortical regions; in humans, pathology in parvalbumin neurons was implicated in schizophrenia (Dienel and Lewis, 2019) and depression (Fogaca and Duman, 2019). The OPRM1, SCUBE3, and SPAG17 cell-type transcriptomic enrichments converged with nominally significant Hi-C chromatin-informed gene-based association thereby reinforcing their importance with respect to suicidality and pinpointing possible cellular and developmental trajectories of particular relevance. These findings highlight genetic and biological overlap between suicidality and key genetic targets for other psychiatric disorders, especially those exacerbated by adverse prior experiences, and suggests that there are pervasive environmental interaction effects underlying genetic risk for the trait.
Several regulatory interactions were detected that further implicate the ECM and synaptic plasticity in the GxE effect of PTS and genetic risk for suicidality. Reduced TTC9 (tetratricopeptide repeat domain 9) expression was associated with higher anxiety-like behaviors (Nepon et al., 2010) and regulatory pathways implicated in adult neuroplasticity and serotonin signaling. NEUROD1 (neuronal differentiation 1) is a potent transcription factor capable of controlling neuronal fate under some circumstances. For example, NEUROD1 alone was capable of converting reactive glia into functional neurons in vivo (Guo et al., 2014). NEUROD1 activates neuronal development genes though acetylation and its transient activity during development was sufficient for long-term epigenetic memory (Pataskar et al., 2016). HS3ST1 encodes heparan sulfate 3-O-sulfotransferase 1, which is responsible for sulfation of heparan in the ECM. Heparan sulfate has been previously implicated in ECM contributions to schizophrenia and bipolar disorder (Woo et al., 2017). Finally, UBE2E3 was detected via eQTL and chromatin interaction mapping in FUMA and Hi-C coupled gene-based association in the neuron. This ubiquitin-conjugating enzyme participates in efficient degradation of DNA binding-proteins in the frontal lobe whose accumulation contributes to neurodegeneration and associated psychopathologies (Rabinovici and Miller, 2010). These findings reinforce suicidality as a trans-diagnostic psychiatric trait (Strawbridge et al., 2019), demonstrating substantial overlap between regulatory effects across disorders including major depressive disorder (Mullins et al., 2019), schizophrenia, and bipolar disorder.
Convergent evidence that ECM biology underlies suicidality is intriguing given recent evidence that ECM is involved in memory retention and synaptic plasticity following traumatic experiences (Bach et al., 2019). Rather than identifying a single environment or state-of-being that interact with the loci discovered herein, we uncover patterns of highly correlated environments that contributed to a locus' effect on suicidality. Many of the environments that we studied are modifiable, lending additional evidence that an EE approach may mitigate suicidality risk. For example, attachment style (e.g., an individual's perception and expectations of the availability and/or responsiveness of meaningful others during times of stress (Mikulincer and Shaver, 2012)) is a potentially modifiable risk factor for PTS, PTSD, and suicidality. A secure attachment style is clinically a protective factor for PTSD (Woodhouse et al., 2015) but also protects against PTSD polygenic risk (Tamman et al., 2020). Possible future directions for this work include the investigation of relationships between attachment style and social support, PTS, and longitudinal reports of suicidal behaviors. Future work may also consider how these factors influence the putatively informative brain regions identified herein with well-characterized roles in memory and emotion. Based on the present findings, we hypothesize that future studies can be most impactful if focused on the longitudinal effects of traumatic events identified by our sex stratified analyses (e.g., male suicidality genetic risk and specific incidence traumas such sexual violence by partner in adulthood).
Our study detected novel biological underpinnings of suicidality but has limitations. First, replication of GEWIS was limited to independent samples with comparable environments. Indeed, our replication with the Yale-Penn cohort lacked 1-to-1 matched environments; however, we successfully included variables from Yale-Penn that capture the same domains of environment or state-of-being: traumatic experiences, PTS, social support, and socioeconomic status. The highly correlated nature of these environments suggests that GxE tests in Yale-Penn should be well suited to detect comparable association and interaction effects. Second, our findings rely on self-reported thoughts and behaviors of suicide and may suffer from reporting biases due to stigma associated with these responses. Third, these data reflect European-ancestry risks for suicidality that require dedicated investigation in other ancestries. Finally, although Struct-LMM is a powerful tool for GEWIS locus discovery, additional work is required to understand best practices for validating GEWIS functional annotations using dynamic SNP effect estimates in place of static GWAS effect sizes.
5. Conclusions
We identified interactions between suicidality risk variants and potentially modifiable environments that point to the ECM as a possible source of suicidality risk in relation to learned behavior following traumatic experiences across the lifespan. There were shared genetic effects between the identified loci and brain circuitry associated with planning behaviors and cell types that contribute to synaptic plasticity and axonal innervation. Characterization of the molecular basis for the effects of traumatic experience and PTS on the risk of suicidal behaviors may help to identify novel targets for which more effective treatments can be developed for use in high-risk populations.
CRediT authorship contribution statement
Frank R. Wendt: Conceptualization, Data curation, Funding acquisition, Formal analysis, Writing - original draft, Writing - review & editing. Gita A. Pathak: Data curation, Writing - review & editing. Daniel F. Levey: Data curation, Writing - review & editing. Yaira Z. Nuñez: Data curation, Writing - review & editing. Cassie Overstreet: Data curation, Writing - review & editing. Chelsea Tyrrell: Data curation, Writing - review & editing. Keyrun Adhikari: Software, Writing - review & editing. Flavio De Angelis: Writing - review & editing. Daniel S. Tylee: Writing - review & editing. Aranyak Goswami: Writing - review & editing. John H. Krystal: Writing - review & editing. Chadi G. Abdallah: Writing - review & editing. Murray B. Stein: Writing - review & editing. Henry R. Kranzler: Writing - review & editing. Joel Gelernter: Writing - review & editing, Resources, Funding acquisition. Renato Polimanti: Conceptualization, Resources, Funding acquisition, Supervision, Writing - original draft, Writing - review & editing.
Declaration of competing interest
Dr. Krystal reports compensation as the Editor of Biological Psychiatry. He also serves on the Scientific Advisory Boards for Bioasis Technologies, Inc., Biohaven Pharmaceuticals, BioXcel Therapeutics, Inc. (Clinical Advisory Board), Cadent Therapeutics (Clinical Advisory Board), PsychoGenics, Inc, Stanley Center for Psychiatric research at the Broad Institute of MIT and Harvard and the Lohocla Research Corporation. He owns stock in ArRETT Neuroscience, Inc., Biohaven Pharmaceuticals, Sage Pharmaceuticals, and Spring Care, Inc. and stock options in Biohaven Pharmaceuticals Medical Sciences, BlackThorn Therapeutics, Inc. and Storm Biosciences, Inc. He is a co-inventor on multiple patents as listed below: (1) Seibyl JP, Krystal JH, Charney DS. Dopamine and noradrenergic reuptake inhibitors in treatment of schizophrenia. US Patent #:5,447,948. September 5, 1995, (2) Vladimir, Coric, Krystal, John H, Sanacora, Gerard—Glutamate Modulating Agents in the Treatment of Mental Disorders US Patent No. 8,778,979 B2 Patent Issue Date: July 15, 2014. US Patent Application No. 15/695,164: Filing Date: May 09, 2017, (3) Charney D, Krystal JH, Manji H, Matthew S, Zarate C.—Intranasal Administration of Ketamine to Treat Depression United States Application No. 14/197,767 filed on March 5, 2014; United States application or Patent Cooperation Treaty (PCT) International application No. 14/306,382 filed on June 17, 2014, (4): Zarate, C, Charney, DS, Manji, HK, Mathew, Sanjay J, Krystal, JH, Department of Veterans Affairs “Methods for Treating Suicidal Ideation”, Patent Application No. 14/197.767 filed on March 5, 2014 by Yale University Office of Cooperative Research, (5) Arias A, Petrakis I, Krystal JH.—Composition and methods to treat addiction. Provisional Use Patent Application no.61/973/961. April 2, 2014. Filed by Yale University Office of Cooperative Research, (6) Chekroud, A., Gueorguieva, R., & Krystal, JH. “Treatment Selection for Major Depressive Disorder” [filing date June 3, 2016, USPTO docket number Y0087.70116US00]. Provisional patent submission by Yale University, (7) Gihyun, Yoon, Petrakis I, Krystal JH—Compounds, Compositions and Methods for Treating or Preventing Depression and Other Diseases. U. S. Provisional Patent Application No. 62/444,552, filed on January 10, 2017 by Yale University Office of Cooperative Research OCR 7088 US01, (8) Abdallah, C, Krystal, JH, Duman, R, Sanacora, G. Combination Therapy for Treating or Preventing Depression or Other Mood Diseases. U.S. Provisional Patent Application No. 047162–7177P1 (00754) filed on August 20, 2018 by Yale University Office of Cooperative Research OCR 7451 US01. Dr. Kranzler is a member of the American Society of Clinical Psychopharmacology's Alcohol Clinical Trials Initiative, which for the past three years was supported by AbbVie, Alkermes, Amygdala Neurosciences, Arbor, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, and Pfizer. Dr. Kranzler is paid for his editorial work on the journal Alcoholism: Clinical and Experimental Research. Drs. Kranzler and Gelernter are named as inventors on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018. Dr. Murray Stein is paid for his editorial work on the journals Biological Psychiatry and Depression and Anxiety, and the health professional reference Up-To-Date. Drs. Polimanti and Gelernter are paid for their editorial work on the journal Complex Psychiatry. The other authors declare no competing interests.
Acknowledgements
This study was supported by the National Center for PTSD of the U.S. Department of Veterans Affairs, the American Foundation for Suicide Prevention (YIG-1-109-16), the National Institutes of Health (F32 MH122058, R21 DC018098, R21 DA047527, R01 DA12690), and the U.S. Department of Veterans Affairs Office of Academic Affiliations Advanced Fellowship Program in Mental Illness Research and Treatment and the VISN 4 Mental Illness Research, Education and Clinical Center. The content is solely the responsibility of the authors and do not necessarily represent the views of the funding agencies. This research has been conducted using the UK Biobank Resource (application reference no. 58146).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2021.100309.
Role of the funder/sponsor
The funders had no role in the designing, collecting, managing, analyzing, interpreting, writing, reviewing, or approving the materials presented in this study. Funders had no role in the decision to submit the manuscript for publication.
Data sharing
All data discussed in this study are provided in the article and in the Supplementary Material.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Al-Mubarak B.R., Omar A., Baz B. Whole exome sequencing in ADHD trios from single and multi-incident families implicates new candidate genes and highlights polygenic transmission. Eur. J. Hum. Genet. 2020;28(8):1098–1110. doi: 10.1038/s41431-020-0619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias A.J., Chan G., Gelernter J., Farrer L., Kranzler H.R. Variation in OPRM1 and risk of suicidal behavior in drug-dependent individuals. Am. J. Addict. 2012;21(1):5–10. doi: 10.1111/j.1521-0391.2011.00195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera S., Peyrot W.J., Milaneschi Y. Genome-wide gene-environment interaction in depression: a systematic evaluation of candidate genes: the childhood trauma working-group of PGC-MDD. Am J Med Genet B Neuropsychiatr Genet. 2018;177(1):40–49. doi: 10.1002/ajmg.b.32593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach D., Brown S.A., Kleim B., Tyagarajan S. Extracellular matrix: a new player in memory maintenance and psychiatric disorders. Swiss Med. Wkly. 2019;149:w20060. doi: 10.4414/smw.2019.20060. [DOI] [PubMed] [Google Scholar]
- Border R., Johnson E.C., Evans L.M. No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. Am. J. Psychiatr. 2019;176(5):376–387. doi: 10.1176/appi.ajp.2018.18070881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J.E., Lanius R.A., McKinnon M.C. Mindfulness-based treatments for posttraumatic stress disorder: a review of the treatment literature and neurobiological evidence. J. Psychiatry Neurosci. 2018;43(1):7–25. doi: 10.1503/jpn.170021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Buuren S., Groothuis-Oudshoorn K. Mice: multivariate imputation by chained Equations in R. 2011;45(3):67. 2011. [Google Scholar]
- Chaaya N., Battle A.R., Johnson L.R. An update on contextual fear memory mechanisms: transition between Amygdala and Hippocampus. Neurosci. Biobehav. Rev. 2018;92:43–54. doi: 10.1016/j.neubiorev.2018.05.013. [DOI] [PubMed] [Google Scholar]
- Chung Moh M., Hoon Lee L., Shen S. Cloning and characterization of hepaCAM, a novel Ig-like cell adhesion molecule suppressed in human hepatocellular carcinoma. J. Hepatol. 2005;42(6):833–841. doi: 10.1016/j.jhep.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Dause T., Kirby E. Aging gracefully: social engagement joins exercise and enrichment as a key lifestyle factor in resistance to age-related cognitive decline. Neural Regeneration Research. 2019;14(1):39–42. doi: 10.4103/1673-5374.243698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K.A.S., Coleman J.R.I., Adams M. Mental health in UK Biobank – development, implementation and results from an online questionnaire completed by 157 366 participants: a reanalysis. BJPsych Open. 2020;6(2):e18. doi: 10.1192/bjo.2019.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienel S.J., Lewis D.A. Alterations in cortical interneurons and cognitive function in schizophrenia. Neurobiol. Dis. 2019;131:104208. doi: 10.1016/j.nbd.2018.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty A.R., Shabalin A.A., DiBlasi E. Genome-wide association study of suicide death and polygenic prediction of clinical antecedents. bioRxiv. 2019:234674. doi: 10.1176/appi.ajp.2020.19101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogaca M.V., Duman R.S. Cortical GABAergic dysfunction in stress and depression: new insights for therapeutic interventions. Front. Cell. Neurosci. 2019;13:87. doi: 10.3389/fncel.2019.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresco D.M., Roy A.K., Adelsberg S. Distinct functional connectivities predict clinical response with emotion regulation therapy. Front. Hum. Neurosci. 2017;11:86. doi: 10.3389/fnhum.2017.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S.X., Jung D. ShinyGO: a graphical enrichment tool for ani-mals and plants. bioRxiv. 2018:315150. [Google Scholar]
- Guo Z., Zhang L., Wu Z., Chen Y., Wang F., Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer's disease model. Cell Stem Cell. 2014;14(2):188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zaslavsky A.M., Landrum M.B., Harrington D.P., Catalano P. Multiple imputation in a large-scale complex survey: a practical guide. Stat. Methods Med. Res. 2010;19(6):653–670. doi: 10.1177/0962280208101273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedegaard H., Curtin S., Warner M. National Center for Health Statistics; Hyattsville, MD: 2020. Increase in Suicide Mortality in the United States, 1999-2018. NCHS Data Brief, No 362. [PubMed] [Google Scholar]
- van Heeringen K., Mann J.J. The neurobiology of suicide. Lancet Psychiatry. 2014;1(1):63–72. doi: 10.1016/S2215-0366(14)70220-2. [DOI] [PubMed] [Google Scholar]
- Hodge R.D., Bakken T.E., Miller J.A. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573(7772):61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbrel N.A., Garrett M.E., Dennis M.F., Hauser M.A., Ashley-Koch A.E., Beckham J.C. A genome-wide association study of suicide attempts and suicidal ideation in U.S. military veterans. Psychiatr. Res. 2018;269:64–69. doi: 10.1016/j.psychres.2018.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosho T., Mizumoto S., Sugahara K. Carbohydrate (N-acetylgalactosamine 4-O) sulfotransferase 14 (CHST14) In: Taniguchi N., Honke K., Fukuda M., Narimatsu H., Yamaguchi Y., Angata T., editors. Handbook of Glycosyltransferases and Related Genes. Springer Japan; Tokyo: 2014. pp. 1135–1148. [Google Scholar]
- Koskinen M.-K., van Mourik Y., Smit A.B., Riga D., Spijker S. From stress to depression: development of extracellular matrix-dependent cognitive impairment following social stress. bioRxiv. 2019:806935. doi: 10.1038/s41598-020-73173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysinska K., Lester D. Post-traumatic stress disorder and suicide risk: a systematic review. Arch. Suicide Res. 2010;14(1):1–23. doi: 10.1080/13811110903478997. [DOI] [PubMed] [Google Scholar]
- LeBouthillier D.M., McMillan K.A., Thibodeau M.A., Asmundson G.J. Types and number of traumas associated with suicidal ideation and suicide attempts in PTSD: findings from a U.S. Nationally representative sample. J. Trauma Stress. 2015;28(3):183–190. doi: 10.1002/jts.22010. [DOI] [PubMed] [Google Scholar]
- de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11(4) doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey D.F., Polimanti R., Cheng Z. Genetic associations with suicide attempt severity and genetic overlap with major depression. Transl. Psychiatry. 2019;9(1):22. doi: 10.1038/s41398-018-0340-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M.J., Liang S., Spadoni A.D., Kelsoe J.R., Simmons A.N. Whole brain expression of bipolar disorder associated genes: structural and genetic analyses. PloS One. 2014;9(6) doi: 10.1371/journal.pone.0100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M.W., Hayward K.S., Rosbergen I.C.M., Jeffers M.S., Corbett D. Is environmental enrichment ready for clinical application in human post-stroke rehabilitation? Front. Behav. Neurosci. 2018;12:135. doi: 10.3389/fnbeh.2018.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercati O., Danckaert A., Andre-Leroux G. Contactin 4, -5 and -6 differentially regulate neuritogenesis while they display identical PTPRG binding sites. Biol Open. 2013;2(3):324–334. doi: 10.1242/bio.20133343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulincer M., Shaver P.R. An attachment perspective on psychopathology. World Psychiatr. 2012;11(1):11–15. doi: 10.1016/j.wpsyc.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moh M.C., Tian Q., Zhang T., Lee L.H., Shen S. The immunoglobulin-like cell adhesion molecule hepaCAM modulates cell adhesion and motility through direct interaction with the actin cytoskeleton. J. Cell. Physiol. 2009;219(2):382–391. doi: 10.1002/jcp.21685. [DOI] [PubMed] [Google Scholar]
- Moore R., Casale F.P., Jan Bonder M. A linear mixed-model approach to study multivariate gene-environment interactions. Nat. Genet. 2019;51(1):180–186. doi: 10.1038/s41588-018-0271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins N., Bigdeli T.B., Børglum A.D. GWAS of suicide attempt in psychiatric disorders and association with major depression polygenic risk scores. Am. J. Psychiatr. 2019;176(8):651–660. doi: 10.1176/appi.ajp.2019.18080957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepon J., Belik S.L., Bolton J., Sareen J. The relationship between anxiety disorders and suicide attempts: findings from the National Epidemiologic Survey on Alcohol and Related Conditions. Depress. Anxiety. 2010;27(9):791–798. doi: 10.1002/da.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievergelt C.M., Maihofer A.X., Klengel T. Largest genome-wide association study for PTSD identifies genetic risk loci in European and African ancestries and implicates novel biological pathways. bioRxiv. 2018:458562. [Google Scholar]
- Nobile B., Ramoz N., Jaussent I. Polymorphism A118G of opioid receptor mu 1 (OPRM1) is associated with emergence of suicidal ideation at antidepressant onset in a large naturalistic cohort of depressed outpatients. Sci. Rep. 2019;9(1):2569. doi: 10.1038/s41598-019-39622-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka I., Akiyama M., Shirakawa O. Genome-wide association studies identify polygenic effects for completed suicide in the Japanese population. Neuropsychopharmacology. 2019;44(12):2119–2124. doi: 10.1038/s41386-019-0506-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier F.B.R., García-Toro M., García-Campayo J., Yañez A.M., Andrés P., Gili M. Mindfulness and symptoms of depression and anxiety in the general population: the mediating roles of worry, rumination, reappraisal and suppression. Front. Psychol. 2019;10:506. doi: 10.3389/fpsyg.2019.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataskar A., Jung J., Smialowski P. NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J. 2016;35(1):24–45. doi: 10.15252/embj.201591206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezer Z., Harr B., Teschke M., Babiker H., Tautz D. Divergence patterns of genic copy number variation in natural populations of the house mouse (Mus musculus domesticus) reveal three conserved genes with major population-specific expansions. Genome Res. 2015;25(8):1114–1124. doi: 10.1101/gr.187187.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovici G.D., Miller B.L. Frontotemporal lobar degeneration: epidemiology, pathophysiology, diagnosis and management. CNS Drugs. 2010;24(5):375–398. doi: 10.2165/11533100-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos de Carvalho J.E., Verwoert M.T., Vogels I.M.C. Involvement of the ubiquitin-proteasome system in the expression of extracellular matrix genes in retinal pigment epithelial cells. Biochem Biophys Rep. 2018;13:83–92. doi: 10.1016/j.bbrep.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sey N.Y.A., Hu B., Mah W. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat. Neurosci. 2020;23(4):583–593. doi: 10.1038/s41593-020-0603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharom F.J., Lehto M.T. Glycosylphosphatidylinositol-anchored proteins: structure, function, and cleavage by phosphatidylinositol-specific phospholipase C. Biochem. Cell. Biol. 2002;80(5):535–549. doi: 10.1139/o02-146. [DOI] [PubMed] [Google Scholar]
- Strawbridge R.J., Ward J., Ferguson A. Identification of novel genome-wide associations for suicidality in UK Biobank, genetic correlation with psychiatric disorders and polygenic association with completed suicide. EBioMedicine. 2019;41:517–525. doi: 10.1016/j.ebiom.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamman A.J.F., Wendt F.R., Pathak G.A. Attachment style moderates polygenic risk for posttraumatic stress in United States military veterans: results from the national health and resilience in veterans study. Biol. Psychiatr. 2020;S0006-3223(20):31947–31948. doi: 10.1016/j.biopsych.2020.09.018. [DOI] [PubMed] [Google Scholar]
- Tombacz D., Maroti Z., Kalmar T. High-coverage whole-exome sequencing identifies candidate genes for suicide in victims with major depressive disorder. Sci. Rep. 2017;7(1):7106. doi: 10.1038/s41598-017-06522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Umicevic Mirkov M., de Leeuw C.A., van den Heuvel M.P., Posthuma D. Genetic mapping of cell type specificity for complex traits. Nat. Commun. 2019;10(1):3222. doi: 10.1038/s41467-019-11181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt F.R., Pathak G.A., Lencz T., Krystal J.H., Gelernter J., Polimanti R. Multivariate genome-wide analysis of education, socioeconomic status, and brain phenome. Nature Human Behavior. 2020 doi: 10.1038/s41562-020-00980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J.R., Heynen A.J., Shuler M.G., Bear M.F. Learning induces long-term potentiation in the hippocampus. Science. 2006;313(5790):1093–1097. doi: 10.1126/science.1128134. [DOI] [PubMed] [Google Scholar]
- Woo C.C., Donnelly J.H., Steinberg-Epstein R., Leon M. Environmental enrichment as a therapy for autism: a clinical trial replication and extension. Behav. Neurosci. 2015;129(4):412–422. doi: 10.1037/bne0000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.J., Yu C., Kumar K., Reifman J. Large-scale interaction effects reveal missing heritability in schizophrenia, bipolar disorder and posttraumatic stress disorder. Transl. Psychiatry. 2017;7(4) doi: 10.1038/tp.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse S., Ayers S., Field A.P. The relationship between adult attachment style and post-traumatic stress symptoms: a meta-analysis. J. Anxiety Disord. 2015;35:103–117. doi: 10.1016/j.janxdis.2015.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.