Abstract
Acute myeloid leukemia (AML) affects tens of thousands of patients a year, yet survival rates are as low as 25% in certain populations. This poor survival rate is partially due to the vast genetic diversity of the disease. Rarely do 2 patients with AML have the same mutational profile, which makes the development of targeted therapies particularly challenging. However, a set of recurrent mutations in chromatin modifiers have been identified in many patients, including mutations in the cohesin complex, which have been identified in up to 20% of cases. Interestingly, the canonical function of the cohesin complex in establishing sister chromatid cohesin during mitosis is unlikely to be the affected role in leukemogenesis. Instead, the cohesin complex's role in DNA looping and gene regulation likely facilitates disease. The epigenetic mechanisms by which cohesin complex mutations promote leukemia are not completely elucidated, but alterations of enhancer-promoter interactions and differential histone modifications have been shown to drive oncogenic gene expression changes. Such changes commonly include HoxA upregulation, which may represent a common pathway that could be therapeutically targeted. As cohesin mutations rarely occur alone, examining the impact of common co-occurring mutations, including those in NPM1, the core-binding factor complex, FLT3, and ASXL1, will yield additional insight. While further study of these mutational interactions is required, current research suggests that the use of combinatorial genetics could be the key to uncovering new targets, allowing for the treatment of AML patients based on their individual genetic profiles.
Keywords: Acute Myeloid Leukemia, Cohesin, gene expression, Epigenetics, Mutations
Introduction
Acute myeloid leukemia (AML) is a bone marrow malignancy resulting from a failure of normal hematopoiesis in which excessive proliferation of abnormal myeloid cells occurs. The accumulation of myeloid cells at the expense of other cell types results in pancytopenia, the clinical manifestations of which are wide and include fever, shortness of breath, fatigue, increased risk of infection, excessive bleeding/bruising, and enlargement of the liver and/or spleen. AML represents 32% of leukemias in adults (>19 y), and the American Cancer Society estimates 19,940 new cases being diagnosed in 2020, with 11,180 (56%) resulting in death.1 AML occurs in people of all ages, but is more common in adults >65 y, with a median age of diagnosis of 68. The average 5-y survival rate is 28.7%, with wide variation depending on the age group. This older population (>60 y) fares worse, with a 5 y survival rate of 23% compared to the younger population (< 60 y) that has a 5 y survival rate of 53%.2 It should be noted that newer therapies for the elderly may be improving the long-term survival for this subgroup of patients.3
Since the 1960s there has been modest improvement in survival for AML patients, however, current therapy with curative intent remains intensive cytotoxic chemotherapy with or without an allogeneic stem cell transplant. Such aggressive measures have been shown not to be tolerated in older adults irrespective of performance status.4 Survivors also often incur side effects such as sterility, cardiotoxicity, endocrinopathies, and secondary malignancies, all of which highlight the need for more effective therapies with fewer long-term sequelae.
A major challenge for developing AML therapeutics is the genetic diversity of the disease. A landmark study published in 2013 by The Cancer Genome Atlas (TCGA) Research Network sequenced the genomes or exons of 200 adult patients with de novo AML. The group discovered that while AML genomes have fewer mutations in genes than most common adult cancers (ranging from nearly 50 in breast, 100 in colon and up to 300 and lung squamous cell carcinomas), they have more genetic variability from patient to patient. On average, 13 mutations in genes were identified per patient. Additionally, they identified 23 mutations that were recurrently mutated in the 200 patients they sequenced, with an additional 237 mutations present in at least 2 samples. The common mutations that they identified fall into various functional categories: signaling genes, chromatin modifying genes, the nucleophosmin gene (NPM1), myeloid transcription factor genes and transcription factor fusions, tumor suppressor genes, spliceosome complex genes, and cohesin complex genes.5 These data demonstrate that rarely do 2 patients with AML have the same mutational spectrum, making therapeutic development challenging. This was the first study to identify the presence of cohesin mutations in up to 20% of AML patients,6 and subsequently lead to an explosion of research on the importance of cohesin mutations in leukemogenesis.
The primary focus of our review is the recent data surrounding the role of cohesin mutations in AML. Expansive recent work since the seminal studies of the TCGA has provided important insights into how cohesin mutations promote key aspects of the AML phenotype by altering gene expression. Importantly, while key aspects have been elucidated, a range of critical questions remain within the field.
The cohesin complex
Normal structure and canonical function
The cohesin complex is composed of 4 subunits: SMC1A, SMC3, RAD21, and either STAG1 or STAG2 (STAG1/2). SMC1A, SMC3 and RAD21 form a core ring-like structure with STAG1/2 serving as accessory protein. SMC1A and SMC3 (Structural Maintenance of Chromosomes) are composed of anti-parallel coiled-coils and are attached at their hinge domains. RAD21 connects the nucleotide binding domains of the SMC proteins, completing the ring structure (Fig. 1).
Fig. 1.
Structure of the cohesin complex. The core subunits making up the ring-like structure include SMC1A, SMC3, and RAD21. SMC1A and SMC3 both are composed of antiparallel coiled-coil domains joining each other at their hinge domains. RAD21 connects the nucleotide binding domains to close the ring. STAG1/2 joins the complex by associating with RAD21. Mutations in myeloid malignancies are commonly found in SMC1A, SMC3, RAD21, and STAG2.
Components of the complex were initially identified in yeast for their canonical role in mitosis and meiosis7 in the late 1980s and early 1990s.8., 9., 10. The cohesin complex is loaded onto chromosomes during G1 via an ATP-dependent process11 by the loading factor NIPBL. During S phase, the complex encircles the replicated sister chromatids and holds the 2 sister chromatids together from G2 through metaphase. Once sister chromatids are aligned at the metaphase plate and checkpoints are met to proceed, cohesin is removed by separase, allowing the sister chromatids to separate during anaphase and telophase.12
In mammals, cohesin has been localized to various regions across the chromosome during interphase. Cohesin has been identified in focal points at the centromere (containing STAG2) and telomere (containing STAG1)13 during mitosis, however whether this is true during interphase remains unclear. Additionally, cohesin has been identified at other genomic locations including active enhancers and core promoters of transcribed genes (along with Mediator and NIPBL) and sites containing CCCTC-factor (CTCF).14 These observations and subsequent studies have demonstrated that cohesin plays a critical role in nuclear architecture, chromatin looping, and gene expression. This aspect of cohesin biology is generally considered cohesin's altered role during AML development, as patients with cohesin-mutated AML are rarely aneuploidy,5 and will be described in more detail below.
Cohesin's role in DNA looping, accessibility and gene expression
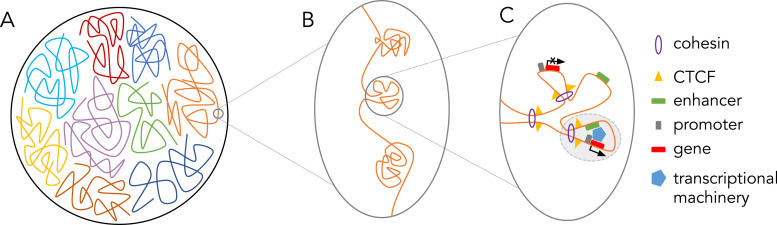
The advent of chromosome capture technologies has allowed the identification of more interactions between chromosomal regions than ever before (reviewed by Agrawal et al.15). A new field emerged through the rise of these techniques: nuclear architecture, the 3-dimensional organization of chromatin within nuclear space (Fig. 2A). These studies have identified regions termed topologically associated domains (TADs, Fig. 2B) and insulated neighborhoods (Fig. 2C). Sequences inside a TAD are more likely to interact with one another than with locations outside of the TAD, often resulting in the coregulation of genes within a TAD. In fact, alteration of TAD borders leads to changes in gene expression, likely by way of altered enhancer-promoter interactions,16,17 and have been identified in multiple diseases.16,18 Insulated neighborhoods are defined more specifically as chromatin loops formed by CTCF-CTCF homodimers surrounding a gene and at least one regulatory element (Fig. 2C). However, as insulated neighborhoods and TADs were identified using techniques with a different resolution, some in the field question if they are truly distinct entities.
Fig. 2.
(A) Depiction of a nucleus with compacted chromosomes. (B) Depiction of 3 topologically associated domains (TADs) within one chromosome. (C) Within one TAD, CTCF and cohesin interaction promote the formation of DNA loops. One active insulated neighborhood (shaded in light grey) results from and enhancer interacting with a promoter and gene body as well as the transcriptional machinery necessary to transcribe the activated gene.
In 2014, Baranello et al. discovered that cohesin colocalizes with CTCF throughout the genome19 where they function together as insulators.20 Cohesin is loaded onto DNA by NIPBL and then in an ATP-dependent process chromatin is extruded through the cohesin ring.11 The result is a looped chromatin conformation that is often bounded by CTCF to insulate genomic neighborhoods and promote enhancer-promoter interactions at euchromatin. Pugacheva et al. recently showed that the N-terminus of CTCF, as well as the 3D structure CTCF makes with DNA, form a “roadblock” that determines the size and position of chromatin loops being extruded through the cohesin ring.21 This correlates with the identification of CTCF binding site enrichment at the borders of TADs.22 While not always located at TAD borders, CTCF sites have been identified across multiple cell types and are evolutionarily conserved, indicating an importance of these regions in chromatin organization and gene expression across organisms. Interestingly, the directionality of the CTCF site is important, as site inversion leads to changes in topology.23 While cohesin and CTCF function together to promote chromatin looping, they associate with DNA independently24 and depletion of CTCF and cohesin lead to differential effects on chromatin loops of different sizes. Depletion of CTCF leads to loss of interactions within 100kb, while depletion of cohesin leads to loss of larger, 100-220kb interactions.25 It should also be noted that cohesin is often enriched at enhancers and promoters, independent of the presence of CTCF.24
Further investigation of cohesin's effects on DNA looping led to the discovery that depletion of cohesin results in the elimination of chromatin loops, with rapid recovery upon cohesin reintroduction. More specifically, upon cohesin loss, enhancer-promoter interactions are weakened.26 However, in erythroid cells, key enhancer-promoter looping occurs independently of cohesin levels, arguing that cohesin's role in enhancer-promoter looping is variable, and perhaps cell-type specific.27 One debate that remains in the field is whether cohesin depletion eliminates TAD architecture,26,28 or whether this action is limited to intra-TAD loops.25,29,30 Furthermore, how cohesin and CTCF play a role in determining chromosomal organization is still an active area of investigation. Specifically, how they manage to promote both global versus intra-TAD organization in such a wide variety of cell types remains to be elucidated.
As cohesin can be found occupying promoters and enhancers and facilitates their interaction, a logical next question is if cohesin plays a role in gene regulation. Multiples studies indicate this is indeed the case. Many groups have investigated the DNA accessibility and transcriptomic changes that result from cohesin loss in hematopoietic stem and progenitor cells (HSPCs).31., 32., 33., 34., 35. Overall, both gene downregulation and upregulation has been observed. Perhaps the most interesting transcriptomic change identified is sustained expression of genes within the HOXA cluster, specifically HOXA7 and HOXA9. The HOXA cluster has a known role in driving hematopoietic stem cell transcriptional profiles and is normally silenced during differentiation. This silencing occurs via polycomb repressive complex (PRC2) mediated trimethylation of lysine 27 of histone 3 (H3K27me3).35., 36., 37., 38. Many studies have identified that cohesin depletion leads to decreased PRC2 recruitment.31,33,35 The epigenetic-mediated gene silencing caused by cohesin loss likely occurs in a variety of contexts and can be assessed in further detail by looking at changes in DNA accessibility.
Both increased and decreased DNA accessibility upon cohesin loss have been identified by multiple groups.31., 32., 33. Viny et al. correlated the transcriptomic and accessibility changes they identified, showing that the downregulation of certain genes was associated decreased DNA accessibility.31 However, DNA accessibility increased for key myeloid and erythroid genes,32,39 as well as for consensus binding sites of myeloid related transcription factors (RUNX1, ERG, and GATA2).33 The exact effects of the accessibility changes that result from cohesin loss are still being elucidated. Additionally, whether cohesin loss directly leads to changes in gene accessibility, or if such changes are influenced by cell-type specific differentiation factors remains to be determined. These data suggest that functional loss of cohesin leads to changes in accessibility that skew the ability of a cell to proceed properly through hematopoietic development. On the other hand, perhaps differentiation factors act first, leading to changes in DNA accessibility and cohesin localization that reinforce a cell's differentiation status.
Cohesin in disease
As mentioned above, cohesin mutations have been identified in up to 20% of patients with AML.5,6,40,41 In a unique subtype of AML, acute Down Syndrome-associated Acute Megakaryocytic Leukemia (DS-AMKL), cohesin mutations have been identified in 50% of cases.42 Furthermore, cohesin mutations are also commonly identified in 15% of myelodysplastic syndromes (MDS).43 Uniquely, STAG2 has been identified in a variety of solid tumors, described further in the following section.
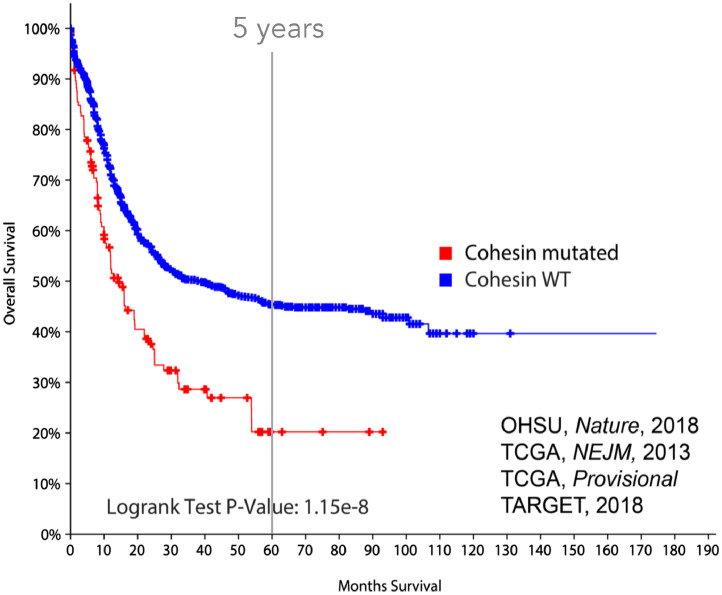
While it was underappreciated in 2013, there is now a vast amount of knowledge regarding the role of cohesin loss in myeloid malignancies (reviewed by many6,40,41). Cohesin mutations are mutually exclusive, heterozygous, distributed throughout the gene body, and typically result in loss of function (17% frameshift, 70% missense),5 and therefore the working model is that mutations in autosomal subunits (SMC3, RAD21) operate through haploinsufficiency, whereas STAG2 mutations may be complete loss-of-function. This is challenging with STAG2 or SMC1A loss, since both are X-linked and currently no evidence of a male predominance for these mutations has been observed. Cohesin-mutant AML is rarely aneuploid, with more than half of AML patients presenting with a normal karyotype, indicating mitotic defects are likely not involved in leukemogenesis. Cohesin-mutant patients do not have a higher than normal presence of chromosomal translocations, which are common oncogenic processes in myeloid malignancies.40 Most interestingly, cohesin-mutant patients have a lower 5-y survival rate (20%) compared to cohesin-WT AML patients (48%, Fig. 3).
Fig. 3.
Survival curve for AML patients with versus without cohesin complex mutations. Cohesin-mutated patients have a significantly lower 5 y survival rate (20% versus 48%, log rank test P-value: 1.15e-18).
Many groups have investigated the effects of cohesin loss in hematopoietic stem and progenitor cells (HSPCs).31., 32., 33., 34., 35. Different experimental systems have been employed, including shRNA-mediated depletion of cohesin complex members,32,34 lentiviral introduction of cohesin mutants,33 and Smc3 and Stag2 conditional knockouts.31,44 These studies have also investigated both human (CD34+ cells) and murine (HSPCs and bone marrow cells) systems. The individual conclusions of these studies have been reviewed nicely,40 but they all agree that cohesin loss confers increased self-renewal. This increased self-renewal is likely due to the increase in HOXA expression and increased accessibility for key myeloid transcription factors (RUNX1, ERG, GATA2) that is described above. Enhanced self-renewal is a common finding in HSPCs with other leukemic driver mutations (NPM1c, TET2, FLT3mut) and serves as an indicator of an immortalized, cancer-like phenotype. While these studies show that cohesin loss confers enhanced self-renewal, cohesin disruption alone does not lead to leukemogenesis, suggesting that cohesin loss cooperates with other mutations to promote AML.
Differential actions of cohesin subunits?
Historically, the effect of alteration in one subunit of the cohesin complex has been considered widely interchangeable with the effects of alteration in any other subunit, but recent studies have revealed this may not be entirely accurate. While mutations in the core cohesin components (RAD21, SMC1A, and SMC3) have been mainly identified in myeloid malignancies, STAG2 mutations have been identified in up to 2% of all human cancer types, recently reviewed by Romero-Pérez et al.45 In fact, STAG2 is considered one of the 12 genes that is significantly mutated in 4 or more types of cancer.46 Mutations in STAG2 have been identified in bladder cancers,47 Ewing sarcoma,48 glioblastomas,49,50 and melanomas, as well as carcinomas of the breast, gallbladder, and lung.45 It is specifically notable that STAG1 mutations have not been observed at a significant frequency in cancer.
Other biological differences exist between STAG1/2 and the core cohesin complex members. There are 2 different STAG subunits: STAG1 and STAG2. STAG2 is encoded on the X chromosome and maintains sister centromeres during metaphase, while STAG1 is autosomal and more commonly found at telomeres, supported by the morphologic changes observed in cytogenetic observation with genetic manipulation in vitro.13 There appears to be some compensatory effects between STAG1 and STAG2, as deletion of one subunit results in increased expression of the other.51,53 Several studies have discovered synthetic lethality between the 2 subunits in various cancer contexts.52,53
There are functional similarities between STAG1/2 and the core cohesin complex members, such as the requirement for hematopoiesis. Concurrent knockout of both Stag1 and Stag2 is lethal, with mice rapidly developing bone marrow aplasia and pancytopenia,44 a situation which phenocopies Smc3 loss.31 However, many recent studies have investigated subunit-based differences in double-stranded DNA repair51 as well as gene looping and transcriptional control in various cell types, including murine HSPCs,44 ESCs54 and AML cell lines.55 In 2018, the first study to describe differential roles for STAG1 versus STAG2 in chromosome organization (in a variety of human cell types) identified differential interactions with CTCF sites, where both STAG1 and STAG2 are known to bind. Kojic et al. concluded that STAG1 mainly works with CTCF to promote and maintain TAD boundaries.56 More recently the direct interface of STAG2 and CTCF-specific polarity has been described, with CTCF orientation functioning to permit cohesin to anchor to the CTCF N-terminus, yet slide past the inverted C-terminus conformation.21
In contrast, a subset of enhancers is occupied by STAG2-containing cohesin and lack CTCF. These enhancers do not become occupied by STAG1-containing cohesin during STAG2-deficiency, and are responsible for tissue-specific transcription.56 Such STAG2-specific sites are likely important in leukemogenesis, as STAG2 loss alone in murine HSPCs leads to decreased chromatin accessibility and decreased transcription of hematopoietic lineage-specification genes.44 The downstream effect is increased self-renewal, indicating a role in transformation for STAG2. Interestingly, while deletion of either STAG1/2 alone leads to differential gene expression, deletion of both subunits enhances gene dysregulation.54
In summary, it is clear that there are both redundant and specific roles for STAG1 and STAG2 in chromosomal organization. Although there are sites within the genome that will bind both STAG1 and STAG2 to drive DNA looping, STAG1 seems to be involved in larger interactions between chromatin, cooperating with CTCF to promote and/or maintain TADs, while STAG2 seems to have a role in short-range, intra-TAD interactions, especially enhancer-promoter interactions. Thus, STAG2 likely plays a unique role in gene regulation, with STAG2 loss resulting in transcriptional changes that promote oncogenic transformation. While some of the studies described above begin to investigate the effects of STAG2 loss on gene expression, direct comparison of these changes to transcriptional profiles resulting from core cohesin subunit loss has not been performed. As there are unique effects of STAG2 loss on chromosomal organization, it is likely that there are also genes uniquely affected by STAG2 loss versus loss of a core cohesin subunit.
Mutations that cooperate with cohesin
As many studies have described,31., 32., 33., 34., 35. cohesin loss alone is insufficient to promote leukemic transformation. This is likely because AML requires multiple mutations to drive leukemogenesis, agreeing with the classical 2-hit hypothesis.5 To b1egin identifying some of the other candidate mutations that may be responsible for promoting leukemia along with cohesin mutations, we can look at mutations that are enriched in cohesin-mutant AML. The most common co-occurring mutation is with NPM1. Other common co-occurring mutations have been identified in RUNX1, CBFB, FLT3, and ASXL1. Below, and in Table 1, we discuss these mutations and how each may cooperate with cohesin mutations to drive AML.
Table 1.
List of mutations that co-occur or are enriched with cohesin mutations in AML
| Comutation rate | Sufficient to generate AML? | Mutation class | Potential mechanism(s) of co-op. with cohesin mutations | Key Refs | |
|---|---|---|---|---|---|
| NPM1 | 21–57% | Yes | NPM1 | Elevation of HOXA/MEIS1 Enhanced HSPC self-renewal | 31–35, 60, 62–65 |
| CBF complex | 18–27%-t(8;21) 0–4%-inv(16) | No-t(8;21) Yes-inv(16) | TF fusions | Effects on myeloid gene expression Altered epigenetic regulation | 75–76, 80–81 |
| RUNX1 | 27–52% | No | Myeloid TF | Altered chromatin structure/gene expression Altered enhancer-promoter looping | 87–90 |
| FLT3 | 21–26% | No | Signaling | Enhanced HSPC self-renewal Altered chromatin structure/gene expression | 31, 92–93 |
| ASXL1 | 25–52% | No | Chromatin modifying | Altered PRC2 binding/epigenetic marks Changes in gene expression | 35, 97–98 |
The comutation rates indicate the co-occurrence rate of each mutation with cohesin mutations, with the ranges representing data from referenced large-scale patient studies. Sufficiency to generate AML is based on if a single mutation of the indicated gene results in AML development in a mouse model. The functional category (class) of each mutation and the potential mechanisms by which each may cooperate with cohesin mutations in AML are provided. TF = transcription factor. HSPC = hematopoietic stem and progenitor cell.
NPM1
Approximately 30% of AML patients harbor a mutation in NPM1 that results in the mislocalization of the NPM1 protein from the nucleolus to the cytoplasm (termed NPM1c).57,58 NPM1 has roles in centrosome duplication and the DNA damage response, and also shuttles proteins from the nucleus to the cytoplasm.59 NPM1c can drive AML development in a mouse model, however disease develops with a prolonged latency and incomplete penetrance,60 suggesting that additional mutations may facilitate leukemic transformation of NPM1-mutated hematopoietic cells. Several studies have observed a correlation between NPM1 and cohesin mutations in adult patients with AML.5,61 Thol et al. reported that 57% of AML patients with a cohesin mutation also harbored an NPM1 mutation. The allelic ratios of mutated NPM1 or cohesin to wildtype are similar, suggesting both mutations exist within the same clone.61
While the exact mechanism by which NPM1 mislocalization facilitates AML development is unclear, several studies report that NPM1 mutation results in an elevated expression of the HOXA and HOXB gene clusters as well as MEIS1 in AML patients and in model systems.60,62., 63., 64., 65., 66. As discussed above, overexpression of these genes has been linked to aberrant HSPC self-renewal and myeloid leukemogenesis and also occurs with cohesin mutation.31., 32., 33., 34.,37,38,60,63,64,66,67 For instance, HOXA9 and MEIS1 overexpression is sufficient for the leukemic transformation of primary bone marrow cells.37 As both cohesin and NPM1 mutations drive HOXA gene expression and NPM1 mutation additionally drives MEIS1 expression, cells harboring both mutations may have enhanced or reinforced self-renewal, which may facilitate AML development. Mechanistically, both NPM1 mutation and cohesin deficiency have been linked to epigenetic changes at HOXA loci. Interestingly, forced nuclear relocalization of NPM1 results in a loss of active chromatin marks, such as H4K4me3 and H3K27ac, at the HOXA and MEIS1 loci, although no changes in the repressive mark H3K27me3 were observed.62 In contrast, loss of cohesin has been shown to reduce H3K27me3 at HOXA7/9 through a failure to recruit the silencing complex PRC2, as described above.35 These results provide additional support to the hypothesis that NPM1 and cohesin mutations may cooperate to drive aberrant self-renewal in AML, synergistically reinforcing HOX7/9 and MEIS1 expression by affecting different epigenetic pathways. Adding further support to this hypothesis, HOXA genes are activated by H3K79 methylation through DOT1L, and NPM1 mutated cell lines have increased H3K79 methylation at HOXA genes. Because the PRC2 complex inhibits DOT1L's activity, NPM1c;cohesinmut cells may also cooperatively drive HOXA gene expression through effects on H3K79me2/3 levels.35,63
Interestingly, NPM1c has been shown to interact with CTCF, resulting in a partial mislocalization of CTCF to the cytoplasm.68 Mislocalization disrupts CTCF's insulator function, driving aberrant gene expression.68 Therefore, in addition to effects on HOXA and MEIS1 gene expression, NPM1 and cohesin mutations may also cooperate to deregulate gene expression in AML through disruptions of CTCF function. While the discussed hypotheses may explain why cohesin and NPM1 mutations commonly co-occur, definitive mechanistic confirmation awaits the development of a combined cohesin/NPM1 mutant model system.
The core-binding factor complex
The core-binding factor complex is a heterodimeric complex composed of an alpha and a beta subunit. The alpha subunit can be one of 3 RUNX proteins while there is only one beta subunit (referred to simply as CBFB). RUNX1, also known as AML1, is the imperative alpha subunit for hematopoiesis and is responsible for the DNA binding activity of the complex.69 The beta subunit enhances the DNA binding activity of the alpha subunit but does not directly bind DNA itself.70 The CBF complex plays a role in both fetal and adult hematopoiesis, acting through a variety of epigenetic pathways. RUNX1-null or CBFB-null mouse embryos die from lack of definitive hematopoiesis at day E12.5-13.5.71., 72., 73., 74.
CBF AMLs account for 25-30% of pediatric and 15% of adult AML.75,76 In general, patients have a good prognosis in comparison with other AML subtypes, however relapse still occurs in 40% of patients,77., 78., 79. indicating a need for better therapeutic options and the likely existence of genetic heterogeneity. More than a dozen chromosomal rearrangements in both RUNX1 and CBFB have been identified in patients with AML, but the most common are t(8;21) and inv(16) which lead to the fusion oncoproteins AML1-ETO (RUNX1-RUNX1T1) and CBFB-MYH11, respectively.69 The AML1-ETO oncoprotein retains the N-terminal ability of RUNX1 to bind DNA and is fused with full-length ETO at the C-terminus, which is known to recruit repressive factors such as the nuclear receptor corepressor (N-CoR) and histone deacetylases.80,81 This fusion protein is thought to predominantly function by repressing gene expression thereby preventing normal myeloid differentiation. The most widely discussed role of CBFB-MYH11 in leukemogenesis is a dominant negative phenotype, that mimics RUNX1 loss of function models,74,82 while some newer studies delve into RUNX1-independent functions.83
The genetics of CBF leukemias have recently been profiled by Faber et al. and Duployez et al.75,76 Interestingly, studies of CBF leukemias indicate a varying interaction with mutations in the cohesin complex depending on the driver oncogene. Both Duployez and Faber identified cohesin mutations cooccurring with t(8;21) in 18% to 27% of cases, but failed to identify a single patient with a mutation in cohesin and inv(16). This observation is likely related to the enrichment of STAG2 mutations in AML arising from antecedent MDS,84 while inv(16) typically occurs in younger patients with de novo AML.75,76 While the importance of the difference in interaction frequency of cohesin mutations in CBF leukemogenesis is unknown, the overlapping roles that the fusion proteins have with cohesin could be meaningful. Currently, CBF leukemias are referred to in the clinic as one group, both prognostically and perhaps more importantly treatment-wise. If the presence of cohesin mutations is deemed important for leukemogenesis, and the interaction is different between AML1-ETO and inv(16) driven leukemias, this would indicate the need for treatment stratification based on an individual's mutational status.
RUNX1
As discussed above, RUNX1 is involved in the t(8;21) chromosomal translocation in some cases of AML, however the gene can also be affected by loss-of-function point mutations and deletions. In a study encompassing a range of hematopoietic disorders including MDS, MPN, and primary and secondary AML, Thota et al. noted that mutations in RUNX1 were enriched in patients with cohesin mutations.85 A similar finding was described in Tsai et al. in a large cohort of de novo AML patients, with STAG2 and RUNX1 mutations frequently co-occurring.86 Studies have indicated that cohesin regulates RUNX1 expression in both zebrafish and a human promyelocytic leukemia cell line.87,88 Both enhancer and promoter regions of the RUNX1 gene contain multiple binding sites for cohesin, which have been experimentally verified in zebrafish, mouse, and human cells.88,89 Mazumdar et al. also reported that hematopoietic cells with RAD21 mutations have increased chromatin accessibility at promoter regions containing RUNX1 binding sites, as well as enhanced RUNX1 binding to the genome.33 These data suggest that cohesin mutation results in deregulation of not only RUNX1 itself, but also of RUNX1 target genes.
While cohesin mutation alone may result in exacerbated RUNX1 expression, it is unclear why mutations in both genes commonly co-occur in AML. Recently, Ochi et al. provided insight to this question discovering a functional relationship between STAG2 and RUNX1 in regulating chromatin structure and gene expression in hematopoietic cells.90 The authors examined the effects of Runx1 and Stag2 double knockout in a mouse model. Interestingly, all mice that were transplanted with double knockout-derived bone marrow cells developed MDS within 6 mo, while none of the single knockout animals did, experimentally verifying that Runx1 and cohesin mutations cooperate to drive disease development. Mechanistically, the authors showed that wild type Runx1 and the cohesin complex can interact, and ChIP sequencing studies showed that Stag2 and Runx1 co-occupy active enhancer sites. While some alterations in enhancer-enhancer or enhancer-promoter loops are observed upon Stag2 knockout, the authors show that the addition of Runx1 deficiency causes a further disruption in certain enhancer-promoter loops. Such loops were predominantly associated with genes that exhibit a high rate of transcriptional pausing, such as p53 pathway and interferon response genes as well as some involved in ribosomal translation and DNA repair.90 Thus, STAG2 and RUNX1 co-mutation may result in the deregulation of a specific set of genes that are critical for maintaining DNA stability and mounting an appropriate immune response to transformed cells.
While this study lends insight into how RUNX1 and STAG2 mutations may cooperate in AML, it does not accurately depict the disease setting. STAG2 mutations are not homozygous in female AML patients, and RUNX1 mutations in AML are also typically not homozygous nulls. While many RUNX1 mutations disrupt gene function, some result in dominant negative effects, some result in RUNX1 overexpression, and some impart new functions, such as those that occur upon RUNX1 translocation, as discussed above (AML1-ETO).91 Because the effects of the inv(16) fusion mimic RUNX1 loss of function, and inv(16) has not been observed to co-occur with cohesin, comparing the effects of gain of function RUNX1 point mutations with those of AML1-ETO in the presence of cohesin may be particularly helpful, as altered RUNX1 DNA binding may uniquely impact cohesin function. It is additionally of interest that STAG2 mutations are less common in t(8;21) AMLs but readily co-occur with RUNX1 mutations, highlighting a complicated relationship between RUNX1 and cohesin. Additional models that more accurately represent the disease-associated mutations will thus be critical in delineating the relationship between cohesin and RUNX1 mutations in AML.
FLT3
Several studies have uncovered an enrichment of FLT3 mutations in cohesin-mutated AML.5,86 FLT3 is a receptor tyrosine kinase that is expressed on HSPCs and regulates myeloid and lymphoid lineage development, proliferation, and survival.92,93 The kinase activity of FLT3 activates downstream signaling molecules including PI3K, RAS, and STAT5. Mutations occur at a rate of approximately 30% in AML.92 The most common type of FLT3 mutation is an internal tandem duplication (FLT3-ITD), however point mutations in the tyrosine kinase domain are also observed.92,93 Both types of mutation result in chronic activation of FLT3 which drives aberrant proliferation and a block in HSPC differentiation. However, FLT3 mutations are not sufficient to drive AML development.31,93
Mouse models of Smc3 haploinsufficiency combined with Flt3-ITD result in an acute leukemia phenotype and have been used to decipher the cooperative pathogenic effects.31 Mice transplanted with bone marrow carrying single Smc3−/+ or Flt3-ITD mutations did not develop AML, whereas all of the Smc3−/+;Flt3-ITD mice died of AML with a median survival of 167 d. Smc3−/+;Flt3-ITD cells exhibited enhanced serial replating compared to cells with either single mutation. Consistent with this result, double mutant animals had increased numbers of hematopoietic stem cells (HSCs). ATAC sequencing of HSCs showed an increase in accessible sites in the Smc3−/+;Flt3-ITD cells, which were enriched for Stat family transcription factor binding sites. FLT3-ITD has been previously shown to alter gene expression through STAT5 family transcription.31,92 As an enrichment of the Stat5 gene expression signature was observed in Smc3−/+;Flt3-ITD cells, these data collectively suggest that cohesin mutation results in an opening of chromatin at sites that STAT5 is able to bind, and permits amplification of the FLT3-ITD enforced STAT-driven transcriptional program.31 The specific downstream STAT5 targets that are critical for AML development are currently unknown.
Interestingly, the presence of a STAG2 mutation is predictive of a poor response to the pan-FLT3-inhibitor Crenolanib in FLT3-ITD AML patients.94 The variant allele frequency of STAG2 increased in patients treated with Crenolanib, suggesting that STAG2 mutation may contribute to drug resistance or may drive clonal outgrowth in response to treatment.94 Therefore, further understanding of the FLT3-cohesin interaction may aid in the development of more effective therapies. Additionally, it is important to consider that FLT3-ITD mutations often co-occur with NPM1 mutations.5 As both mutations are enriched in cohesin-mutated patients, it may be useful to study the effect of all 3 mutations (FLT3-ITD, NPM1, and cohesin) in combination in an effort to more carefully tease apart the impact each has on disease development and progression.
ASXL1
Mutations in ASXL1 and cohesin have also been linked by several studies. ASXL1 is involved in epigenetic regulation through its interactions with polycomb complex proteins.95 Mutations in ASXL1 commonly co-occur with cohesin mutations in MDS, MPN, and primary and secondary AML patients.86,96 In AML, a trend toward co-occurrence was observed between ASXL1 and STAG2 mutations.86 Interestingly, Li et al. recently published a study describing an interaction between Asxl1 and the cohesin complex, with ChIP sequencing studies indicating significant overlap in binding sites between Asxl1, Rad21, and Smc1a.97 Loss of Asxl1 resulted in reduced genomic occupancy of Rad21 and Smc1a as well as alterations in the expression of genes involved in apoptosis, proliferation, and myeloid differentiation.97 Interestingly, ASXL1 can recruit PRC2, which deposits the repressive histone mark H3K27me3.98 As mentioned above, Fisher et al. have previously shown that the cohesin complex also recruits PRC2 to regulate HoxA gene expression.35 These data collectively suggest that ASXL1, cohesin, and PRC2 may, at times, all be present within the same complex and may cooperatively affect gene transcription. Mutations in PRC2 complex members SUZ12 and EZH2 are also found in AML, suggesting that the PRC2 complex is a common target in AML.
Although ASXL1 and cohesin may share some functions, Li et al. showed that some genomic regions are uniquely bound by Asxl1 or cohesin, suggesting that their roles are not entirely redundant.97 One can thus speculate that the loss of ASXL1 in an AML setting would result in the deregulation of a certain set of genes, and that a further loss of cohesin would compound gene dysregulation. It will be interesting to learn if ASXL1 cooperates with cohesin in genomic looping or organization events, or if both ASXL1 and cohesin can independently recruit PRC2 to different genomic locations.
Conclusions
It has become increasingly clear that a one-size-fits-all approach to treatment for AML patients may not be applicable to this disease, greatly underscoring the need for targeted therapies. The advent of next generation sequencing and its application to patient cohorts has uncovered the vast genetic complexity of AML, including the prevalence of cohesin mutations in AML as well as the role of cohesin mutations in driving leukemogenesis. Some have begun to investigate whether targeting cohesin may be a successful therapeutic strategy. For example, inhibition of HDAC8 (which is necessary for the removal of cohesin from sister chromatids during mitosis) may be useful in treating cohesin-mutated AML by preventing normal cell cycle progression.85,99
While moderate success has been achieved with some targeted therapies, such as with FLT3 or IDH1/2 inhibitors, different mutational combinations may have unique effects on the underlying molecular environment. The development of model systems that allow researchers to study how different genetic lesions influence one another has yielded some promising results, particularly in the case of cohesin mutations. Themes are beginning to emerge, suggesting several mutations target the same molecular pathway. For instance, while EZH2 and SUZ12 mutations in AML directly disrupt the PRC2 complex, new research points to cohesin and ASXL1 mutations also affecting PRC2. Further, NPM1 and cohesin mutations both affect HSPC self-renewal and HOXA gene expression as do MLL-rearrangements, another common driver mutation in AML. Several studies indicate that inhibition of the DOT1L complex (responsible for H3K79 methylation) may normalize HOXA upregulation, which is commonly found in leukemic gene expression profiles, including loss of cohesion.63,100,101 Clinical trials have shown moderate success with use of pinometostat (a DOT1L inhibitor),102 but perhaps efficacy could be improved by selecting patients with known HoxA dysregulation). The use of combinatorial genetic models thus offers insight into underlying commonalities that may be exploited for therapeutic development. Teasing apart the complex genetic interactions in AML seems daunting, but if several different mutations affect a common pathway, it may be possible to target the pathway rather than each individual mutation, which would make a therapy more widely applicable. Cases of clear mutual exclusivity, such as occurs with cohesin and inv(16), are also interesting to consider. If the 2 mutations result in synthetic lethality, this may be exploitable for therapeutic use by identifying potential precision medicine targets. It is thus important to determine why the cohesin and t(8;21) mutational combination is permissive yet the cohesin and inv(16) combination is not.
Cohesin is unique in that it can alter genomic organization and ultimately affect gene transcription. Several studies have indicated differences in gene transcription and looping between cohesin mutations alone and in combination with other AML lesions, making it important to examine the effects of cohesin mutation in different genetic backgrounds. The differences between STAG1 and STAG2 mutation must also be considered. Although research on cohesin mutations in AML development and progression is still young, the development of new combinatorial genetic models will not only provide insight into how cohesin functions in normal versus disease states, but also holds promise in the generation of tailored therapies.
Author contribution
Initial writing was done by KEH, AEM, and PA. ADV and SR participated in the development of the early drafts. All authors participated in editing the manuscript and approved the final submission.
Conflicts of interest
None of the authors had any relevant financial conflicts of interests to report.
Acknowledgments
This work was supported by the National Institute of Health (NIH) grants R01 CA204231 (to SR), K08 CA215317 (to ADV), F30 CA236322 (to KEH), and F30 DK120152 (to PA). Additional funding was received from the NIH T32 GM080202 (to KEH and PA), the Midwest Athletes Against Childhood Cancer Fund (to SR), and the EvansMDS foundation (to ADV).
Footnotes
Abbreviations: AML, acute myeloid leukemia, HSPC, hematopoietic stem and progenitor cell, MDS, myelodysplastic syndromes, TF, transcription factor.
References
- 1.Cancer Facts & Figures: 2020. Atlanta, Georgia; 2019:1–76.
- 2.Burnett A, Wetzler M, Löwenberg B. Therapeutic Advances in Acute Myeloid Leukemia. JCO. 2011;29(5):487–494. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- 3.DiNardo CD, Pratz K, Pullarkat V, Jonas BA, Arellano M, Becker PS, Frankfurt O, Konopleva M, Wei AH, Kantarjian HM. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood. 2019;133(1):7–17. doi: 10.1182/blood-2018-08-868752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Estey EH. How I treat older patients with AML. Blood. 2000;96(5):1670–1673. doi: 10.1182/blood.V96.5.1670. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson AG, Hoadley K, Triche TJ, Laird PW. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leeke B, Marsman J, O'Sullivan JM, Horsfield JA. Cohesin mutations in myeloid malignancies: underlying mechanisms. Exp Hematol Oncol. 2014;3:13. doi: 10.1186/2162-3619-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooker AS, Berkowitz KM. The roles of cohesins in mitosis, meiosis, and human health and disease. Methods Mol Biol. 2014;1170:229–266. doi: 10.1007/978-1-4939-0888-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohkura H, Adachi Y, Kinoshita N, Niwa O, Toda T, Yanagida M. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. The EMBO Journal. 1988;7(5):1465–1473. doi: 10.1002/j.1460-2075.1988.tb02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes & Development. 1999;13(3):307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rockmill B, Roeder GS. The yeast med1 mutant undergoes both meiotic homolog nondisjunction and precocious separation of sister chromatids. Genetics. 1994;136(1):65–74. doi: 10.1093/genetics/136.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters J-M, Nishiyama T. Sister chromatid cohesion. Cold Spring Harb Perspect Biol. 2012;4(11) doi: 10.1101/cshperspect.a011130. a011130-a011130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters J-M, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes & Development. 2008;22(22):3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 13.Canudas S, Smith S. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol. 2009;187(2):165–173. doi: 10.1083/jcb.200903096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agrawal P, Heimbruch KE, Rao S. Genome-Wide Maps of Transcription Regulatory Elements and Transcription Enhancers in Development and Disease. Compr Physiol. 2018;9(1):439–455. doi: 10.1002/cphy.c180028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell. 2015;161(5):1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan W-L. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538(7624):265–269. doi: 10.1038/nature19800. [DOI] [PubMed] [Google Scholar]
- 18.Matharu N, Ahituv N. Minor Loops in Major Folds: Enhancer-Promoter Looping, Chromatin Restructuring, and Their Association with Transcriptional Regulation and Disease. PLoS Genet. 2015;11(12) doi: 10.1371/journal.pgen.1005640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baranello L, Kouzine F, Levens D. CTCF and cohesin cooperate to organize the 3D structure of the mammalian genome. Proc Natl Acad Sci USA. 2014;111(3):889–890. doi: 10.1073/pnas.1321957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Z, Wang X. Roles of cohesin in chromosome architecture and gene expression. Semin Cell Dev Biol. 2018;90:187–193. doi: 10.1016/j.semcdb.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Pugacheva EM, Kubo N, Loukinov D, Tajmul M, Kang S, Kovalchuk AL, Strunnikov AV, Zentner GE, Ren B, Lobanenkov VV. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proceedings of the National Academy of Sciences. 2020;117(4):2020–2031. doi: 10.1073/pnas.1911708117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162(4):900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20(5):578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuin J, Dixon JR, van der Reijden MIJA, Ye Z, Kolovos P, Brouwer RWW, van de Corput MPC, van de Werken HJG, Knoch TA, van IJcken WFJ. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA. 2014;111(3):996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao SSP, Huang S-C, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon K-R, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID. Cohesin Loss Eliminates All Loop Domains. Cell. 2017;171(2) doi: 10.1016/j.cell.2017.09.026. 305-320.e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krivega I, Dean A. LDB1-mediated enhancer looping can be established independent of mediator and cohesin. Nucleic Acids Research. 2017;45(14):8255–8268. doi: 10.1093/nar/gkx433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551(7678):51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23(12):2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sofueva S, Yaffe E, Chan W-C, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. The EMBO Journal. 2013;32(24):3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viny AD, Ott CJ, Spitzer B, Rivas M, Meydan C, Papalexi E, Yelin D, Shank K, Reyes J, Chiu A. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J Exp Med. 2015;212(11):1819–1832. doi: 10.1084/jem.20151317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, Wang K, Kayembe C, Rocha PP, Raviram R, Gong Y. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med. 2015;212(11):1833–1850. doi: 10.1084/jem.20151323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazumdar C, Shen Y, Xavy S, Zhao F, Reinisch A, Li R, Corces MR, Flynn RA, Buenrostro JD, Chan SM. Leukemia-Associated Cohesin Mutants Dominantly Enforce Stem Cell Programs and Impair Human Hematopoietic Progenitor Differentiation. Cell Stem Cell. 2015;17(6):675–688. doi: 10.1016/j.stem.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galeev R, Baudet A, Kumar P, Rundberg Nilsson A, Nilsson B, Soneji S, Törngren T, Å Borg, Kvist A, Larsson J. Genome-wide RNAi Screen Identifies Cohesin Genes as Modifiers of Renewal and Differentiation in Human HSCs. Cell Reports. 2016;14(12):2988–3000. doi: 10.1016/j.celrep.2016.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher JB, Peterson J, Reimer M, Stelloh C, Pulakanti K, Gerbec ZJ, Abel AM, Strouse JM, Strouse C, McNulty M. The cohesin subunit Rad21 is a negative regulator of hematopoietic self-renewal through epigenetic repression of Hoxa7 and Hoxa9. Leukemia. 2017;31(3):712–719. doi: 10.1038/leu.2016.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS, Largman C, Lawrence HJ, Humphries RK. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci USA. 1994;91(25):12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. The EMBO Journal. 1998;17(13):3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, Humphries K, Sauvageau G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99(1):121–129. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 39.Sasca D, Yun H, Giotopoulos G, Szybinski J, Evan T, Wilson NK, Gerstung M, Gallipoli P, Green AR, Hills R. Cohesin-dependent regulation of gene expression during differentiation is lost in cohesin-mutated myeloid malignancies. Blood. 2019;134(24):2195–2208. doi: 10.1182/blood.2019001553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fisher JB, McNulty M, Burke MJ, Crispino JD, Rao S. Cohesin Mutations in Myeloid Malignancies. TRENDS in CANCER. 2017;3(4):282–293. doi: 10.1016/j.trecan.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Losada A. Cohesin in cancer: chromosome segregation and beyond. Nat Rev Cancer. 2014;14(6):389–393. doi: 10.1038/nrc3743. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A, Sanada M, Park M-J, Terui K, Suzuki H. The landscape of somatic mutations in Down syndrome–related myeloid disorders. Nat Genet. 2013;45(11):1293–1299. doi: 10.1038/ng.2759. [DOI] [PubMed] [Google Scholar]
- 43.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viny AD, Bowman RL, Liu Y, Lavallée V-P, Eisman SE, Xiao W, Durham BH, Navitski A, Park J, Braunstein S. Cohesin Members Stag1 and Stag2 Display Distinct Roles in Chromatin Accessibility and Topological Control of HSC Self-Renewal and Differentiation. Cell Stem Cell. 2019;25(5) doi: 10.1016/j.stem.2019.08.003. 682-696.e688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romero-Pérez L, Surdez D, Brunet E, Delattre O, Grünewald TGP. STAG Mutations in Cancer. TRENDS in CANCER. 2019;5(8):506–520. doi: 10.1016/j.trecan.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Hill VK, Kim J-S, Waldman T. Cohesin mutations in human cancer. Biochim Biophys Acta. 2016;1866(1):1–11. doi: 10.1016/j.bbcan.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balbás-Martínez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Márquez M, Vazquez M, Lapi E, Castro-Giner F, Beltran S, Bayés M. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45(12):1464–1469. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beaino El M, Liu J, Wasylishen AR, Pourebrahim R, Migut A, Bessellieu BJ, Huang K, Lin PP. Loss of Stag2 cooperates with EWS-FLI1 to transform murine Mesenchymal stem cells. BMC Cancer. 2020;20(1):3. doi: 10.1186/s12885-019-6465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333(6045):1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey ML, O'Neil NJ, van Pel DM, Solomon DA, Waldman T, Hieter P. Glioblastoma cells containing mutations in the cohesin component STAG2 are sensitive to PARP inhibition. Mol Cancer Ther. 2014;13(3):724–732. doi: 10.1158/1535-7163.MCT-13-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kong X, Ball AR, Pham HX, Zeng W, Chen H-Y, Schmiesing JA, Kim J-S, Berns M, Yokomori K. Distinct functions of human cohesin-SA1 and cohesin-SA2 in double-strand break repair. Mol Cell Biol. 2014;34(4):685–698. doi: 10.1128/MCB.01503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Lelij P, Lieb S, Jude J, Wutz G, Santos CP, Falkenberg K, Schlattl A, Ban J, Schwentner R, Hoffmann T, Kovar H. Synthetic lethality between the cohesin subunits STAG1 and STAG2 in diverse cancer contexts. eLife. Sciences. 2017;6:1464. doi: 10.7554/eLife.26980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benedetti L, Cereda M, Monteverde L, Desai N, Ciccarelli FD. Synthetic lethal interaction between the tumour suppressor STAG2 and its paralog STAG1. Oncotarget. 2017;8(23):37619–37632. doi: 10.18632/oncotarget.16838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arruda NL, Carico ZM, Justice M, Liu YF, Zhou J, Stefan HC, Dowen JM. Distinct and overlapping roles of STAG1 and STAG2 in cohesin localization and gene expression in embryonic stem cells. Epigenetics Chromatin. 2020;13(1):32. doi: 10.1186/s13072-020-00353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith JS, Lappin KM, Craig SG, Liberante FG, Crean CM, McDade SS, Thompson A, Mills KI, Savage KI. Chronic loss of STAG2 leads to altered chromatin structure contributing to de-regulated transcription in AML. J Transl Med. 2020;18(1):339. doi: 10.1186/s12967-020-02500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kojic A, Cuadrado A, De Koninck M, Giménez-Llorente D, Rodríguez-Corsino M, Gómez-López G, Le Dily F, Marti-Renom MA, Losada A. Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat Struct Mol Biol. 2018;25(6):496–504. doi: 10.1038/s41594-018-0070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolli N, Nicoletti I, De Marco MF, Bigerna B, Pucciarini A, Mannucci R, Martelli MP, Liso A, Mecucci C, Fabbiano F. Born to be exported: COOH-terminal nuclear export signals of different strength ensure cytoplasmic accumulation of nucleophosmin leukemic mutants. Cancer Res. 2007;67(13):6230–6237. doi: 10.1158/0008-5472.CAN-07-0273. [DOI] [PubMed] [Google Scholar]
- 58.Falini B, Nicoletti I, Bolli N, Martelli MP, Liso A, Gorello P, Mandelli F, Mecucci C, Martelli MF. Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias. Haematologica. 2007;92(4):519–532. doi: 10.3324/haematol.11007. [DOI] [PubMed] [Google Scholar]
- 59.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, Bigerna B. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 60.Vassiliou GS, Cooper JL, Rad R, Li J, Rice S, Uren A, Rad L, Ellis P, Andrews R, Banerjee R. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet. 2011;43(5):470–475. doi: 10.1038/ng.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thol F, Bollin R, Gehlhaar M, Walter C, Dugas M, Suchanek KJ, Kirchner A, Huang L, Chaturvedi A, Wichmann M. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood. 2014;123(6):914–920. doi: 10.1182/blood-2013-07-518746. [DOI] [PubMed] [Google Scholar]
- 62.Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang Y-H, Ramabadran R, Gionfriddo I, Mezzasoma F, Milano F, Nabet B. Mutant NPM1 Maintains the Leukemic State through HOX Expression. Cancer Cell. 2018;34(3) doi: 10.1016/j.ccell.2018.08.005. 499-512.e499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kühn MWM, Song E, Feng Z, Sinha A, Chen C-W, Deshpande AJ, Cusan M, Farnoud N, Mupo A, Grove C. Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia. Cancer Discov. 2016;6(10):1166–1181. doi: 10.1158/2159-8290.CD-16-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woolthuis CM, Han L, Verkaik-Schakel RN, van Gosliga D, Kluin PM, Vellenga E, Schuringa JJ, Huls G. Downregulation of MEIS1 impairs long-term expansion of CD34+ NPM1-mutated acute myeloid leukemia cells. Leukemia. 2012;26(4):848–853. doi: 10.1038/leu.2011.277. [DOI] [PubMed] [Google Scholar]
- 65.Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP, Meani N, Diverio D, Bernard L, Tizzoni L. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106(3):899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 66.Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O'Laughlin M, Fronick C, Magrini V, Demeter RT, Miller CA. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia. 2015;29(6):1279–1289. doi: 10.1038/leu.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meyer AE, Rao S, Fisher JB. Cohesin mutations: contributors to myeloid malignancies. Oncotarget. 2017;8(46):80107–80108. doi: 10.18632/oncotarget.21050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang AJ, Han Y, Jia N, Chen P, Minden MD. NPM1c impedes CTCF functions through cytoplasmic mislocalization in acute myeloid leukemia. Leukemia. 2020;34(5):1278–1290. doi: 10.1038/s41375-019-0681-8. [DOI] [PubMed] [Google Scholar]
- 69.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2(7):502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 70.Tang Y-Y, Shi J, Zhang L, Davis A, Bravo J, Warren AJ, Speck NA, Bushweller JH. Energetic and Functional Contribution of Residues in the Core Binding Factor β (CBFβ) Subunit to Heterodimerization with CBFα. J Biol Chem. 2000;275(50):39579–39588. doi: 10.1074/jbc.M007350200. [DOI] [PubMed] [Google Scholar]
- 71.Wang Q, Stacy T, Binder M, Marín-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93(8):3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87(4):697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 73.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 74.Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci USA. 1996;93(22):12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faber ZJ, Chen X, Gedman AL, Boggs K, Cheng J, Ma J, Radtke I, Chao J-R, Walsh MP, Song G, Andersson AK. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet. 2016;48(12):1551–1556. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duployez N, Marceau-Renaut A, Boissel N, Petit A, Bucci M, Geffroy S, Lapillonne H, Renneville A, Ragu C, Figeac M. Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood. 2016;127(20):2451–2459. doi: 10.1182/blood-2015-12-688705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Appelbaum FR, Kopecky KJ, Tallman MS, Slovak ML, Gundacker HM, Kim HT, Dewald GW, Kantarjian HM, Pierce SR, Estey EH. The clinical spectrum of adult acute myeloid leukaemia associated with core binding factor translocations. Br J Haematol. 2006;135(2):165–173. doi: 10.1111/j.1365-2141.2006.06276.x. [DOI] [PubMed] [Google Scholar]
- 78.Schlenk RF, Benner A, Krauter J, Büchner T, Sauerland C, Ehninger G, Schaich M, Mohr B, Niederwieser D, Krahl R. Individual patient data-based meta-analysis of patients aged 16 to 60 years with core binding factor acute myeloid leukemia: a survey of the German Acute Myeloid Leukemia Intergroup. JCO. 2004;22(18):3741–3750. doi: 10.1200/JCO.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 79.Jourdan E, Boissel N, Chevret S, Delabesse E, Renneville A, Cornillet P, Blanchet O, Cayuela J-M, Recher C, Raffoux E. Prospective evaluation of gene mutations and minimal residual disease in patients with core binding factor acute myeloid leukemia. Blood. 2013;121(12):2213–2223. doi: 10.1182/blood-2012-10-462879. [DOI] [PubMed] [Google Scholar]
- 80.Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, Huynh KD, Bardwell VJ, Lavinsky RM, Rosenfeld MG. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18(12):7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18(12):7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Castilla LH, Wijmenga C, Wang Q, Stacy T, Speck NA, Eckhaus M, Marín-Padilla M, Collins FS, Wynshaw-Boris A, Liu PP. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87(4):687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 83.Hyde RK, Kamikubo Y, Anderson S, Kirby M, Alemu L, Zhao L, Liu PP. Cbfb/Runx1 repression-independent blockage of differentiation and accumulation of Csf2rb-expressing cells by Cbfb-MYH11. Blood. 2010;115(7):1433–1443. doi: 10.1182/blood-2009-06-227413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lindsley RC. Mutational complexity in myelodysplasia. Best Pract Res Clin Haematol. 2017;30(4):290–294. doi: 10.1016/j.beha.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 85.Thota S, Viny AD, Makishima H, Spitzer B, Radivoyevitch T, Przychodzen B, Sekeres MA, Levine RL, Maciejewski JP. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood. 2014;124(11):1790–1798. doi: 10.1182/blood-2014-04-567057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsai C-H, Hou H-A, Tang J-L, Kuo Y-Y, Chiu Y-C, Lin C-C, Liu C-Y, Tseng M-H, Lin T-Y, Liu M-C. Prognostic impacts and dynamic changes of cohesin complex gene mutations in de novo acute myeloid leukemia. Blood Cancer J. 2017;7(12):663. doi: 10.1038/s41408-017-0022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Horsfield JA, Anagnostou SH, Hu JK-H, Cho KHY, Geisler R, Lieschke G, Crosier KE, Crosier PS. Cohesin-dependent regulation of Runx genes. Development. 2007;134(14):2639–2649. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- 88.Marsman J, O'Neill AC, Kao BR-Y, Rhodes JM, Meier M, Antony J, Mönnich M, Horsfield JA. Cohesin and CTCF differentially regulate spatiotemporal runx1 expression during zebrafish development. Biochim Biophys Acta. 2014;1839(1):50–61. doi: 10.1016/j.bbagrm.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 89.Antony J, Gimenez G, Taylor T, Khatoon U, Day R, Morison IM, Horsfield JA. BET inhibition prevents aberrant RUNX1 and ERG transcription in STAG2 mutant leukaemia cells. J Mol Cell Biol. 2020;12(5):397–399. doi: 10.1093/jmcb/mjz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ochi Y, Kon A, Sakata T, Nakagawa MM, Nakazawa N, Kakuta M, Kataoka K, Koseki H, Nakayama M, Morishita D. Combined Cohesin-RUNX1 Deficiency Synergistically Perturbs Chromatin Looping and Causes Myelodysplastic Syndromes. Cancer Discov. 2020;10(6):836–853. doi: 10.1158/2159-8290.CD-19-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bellissimo DC, Speck NA. RUNX1 Mutations in Inherited and Sporadic Leukemia. Front Cell Dev Biol. 2017;5:111. doi: 10.3389/fcell.2017.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33(2):299–312. doi: 10.1038/s41375-018-0357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lagunas-Rangel FA, Chávez-Valencia V. FLT3-ITD and its current role in acute myeloid leukaemia. Med Oncol. 2017;34(6):114. doi: 10.1007/s12032-017-0970-x. [DOI] [PubMed] [Google Scholar]
- 94.Zhang H, Savage S, Schultz AR, Bottomly D, White L, Segerdell E, Wilmot B, McWeeney SK, Eide CA, Nechiporuk T. Clinical resistance to crenolanib in acute myeloid leukemia due to diverse molecular mechanisms. Nature Communications. 2019;10(1):244. doi: 10.1038/s41467-018-08263-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gelsi-Boyer V, Brecqueville M, Devillier R, Murati A, Mozziconacci M-J, Birnbaum D. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol. 2012;5(1):12. doi: 10.1186/1756-8722-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kon A, Shih L-Y, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45(10):1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 97.Li Z, Zhang P, Yan A, Guo Z, Ban Y, Li J, Chen S, Yang H, He Y, Li J. ASXL1 interacts with the cohesin complex to maintain chromatid separation and gene expression for normal hematopoiesis. Sci Adv. 2017;3(1) doi: 10.1126/sciadv.1601602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, Pandey S, Patel JP, Chung YR, Koche R. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22(2):180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dasgupta T, Antony J, Braithwaite AW, Horsfield JA. HDAC8 Inhibition Blocks SMC3 Deacetylation and Delays Cell Cycle Progression without Affecting Cohesin-dependent Transcription in MCF7 Cancer Cells. J Biol Chem. 2016;291(24):12761–12770. doi: 10.1074/jbc.M115.704627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, Allain CJ, Klaus CR, Raimondi A, Scott MP. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122(6):1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stein EM, Garcia-Manero G, Rizzieri DA, Tibes R, Berdeja JG, Savona MR, Jongen-Lavrenic M, Altman JK, Thomson B, Blakemore SJ. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood. 2018;131(24):2661–2669. doi: 10.1182/blood-2017-12-818948. [DOI] [PMC free article] [PubMed] [Google Scholar]