Abstract
The regulation of myeloid-derived suppressor cells (MDSCs) function is key for effective tumor immunotherapy. Recent lipidomics data revealed that MDSCs accumulate lipid species thereby promote their immunosuppressive activity on T cells. However, genetic manipulation of fatty acid transport protein 2 in mice reduced lipid accumulation in polymorphonuclear MDSCs. Herein we present for the first time lipidome of splenic MDSCs from B16F10 melanoma-bearing mice treated with FATP2 inhibitor – lipofermata compared to the control group. B16F10 were subcutaneously injected into the left flank of wild-type C57BL/6 mice, either lipofermata or vehicle was administered to the mice every day starting from day 7 post-tumor injection for 2 weeks. CD11b+Gr1+ cells from the spleen referred to as MDSCs were sorted on a flow cytometer machine for lipid extraction. Lipid was extracted using methyl‑tert‑butyl ether as previously described with slight modification, followed by liquid chromatography-mass spectrophotometry lipid profiling using a Q-Exactive instrument coupled with HPLC. The raw scans were identified and quantified with LipidSearch while raw data for various lipid species available on the Mendeley Data repository [1]. The lipid profiles reveal change in lipid species following blockade of FATP2 expression in MDSCs compared to the control. These data were collected in connection to a co-submitted paper [2].
Keywords: Lipids, MDSCs, Tumor immunotherapy, Fatty acid transport protein 2 (FATP2), LC-MS
Specifications Table
| Subject | Immunology |
| Specific subject area | Lipid in splenic myeloid-derived suppressor cells from melanoma-bearing mice |
| Type of data | Image Graph Chart |
| How data were acquired | Liquid Chromatography Q-Exactive Mass Spectrophotometry system |
| Data format | Raw Analyzed Filtered |
| Parameters for data collection | Flow cytometry CD11b+Gr1+ MDSCs isolated from the spleen of B16F10 tumor-bearing mice treated with or without fatty acid transport protein 2 (FATP2) inhibitor - lipofermata were subjected to lipid extraction. |
| Description of data collection | Wild-type C57BL/6 mice were injected with 1 × 106 B16F10 cells and allowed to grow until the tumor was palpable. Starting from day 7, mice were treated with vehicle (PBS) or lipofermata for 2 weeks before sacrificing on day 20 after tumor injection. Spleens were isolated from both groups and purified MDSCs sorted for lipid extraction according to a previously described method with slight modifications before LC-MS analysis. The data obtained were in accordance with earlier information of lipid profiling described in splenic MDSCs from FATP2 deletion in Lewis lung carcinoma (LLC) bearing mice [3]. |
| Data source location | Southern University of Science and Technology, China. Nanshan/Shenzhen/Guangdong China |
| Data accessibility | Repository Name: Mendeley data Data identification number: 10.17632/3sncj38wds.1 Direct URL to data: https://data.mendeley.com/datasets/3sncj38wds/draft?a=41a7ccea-0194-4c95-8561-3bbe62920295 |
| Related research article | Adeleye Oluwatosin Adeshakin, Wan Liu, Funmilayo O. Adeshakin, Lukman O. Afolabi, Mengqi Zhang, Guizhong Zhang, Lulu Wang, Zhihuan Li, Lilong Lin, Qin Cao, Dehong Yan*, Xiaochun Wan*, Regulation of ROS in myeloid-derived suppressor cells through targeting fatty-acid transport protein 2 enhanced anti-PD-L1 tumor immunotherapy, Cellular Immunology 362 (2021) 104,286. https://doi.org/10.1016/j.cellimm.2021.104286 |
Value of the Data
-
•
The data gives information on changes in lipid species present in splenic MDSCs from melanoma-bearing mice that were treated with either fatty acid transport protein 2 inhibitor (lipofermata) or PBS.
-
•
The data is useful for future works of MDSCs and other related immune cells using pharmacological inhibitors of FATPs.
-
•
The data can be applied for the development of databases for future lipidomics experiments of MDSCs in tumor or other pathological conditions.
1. Data Description
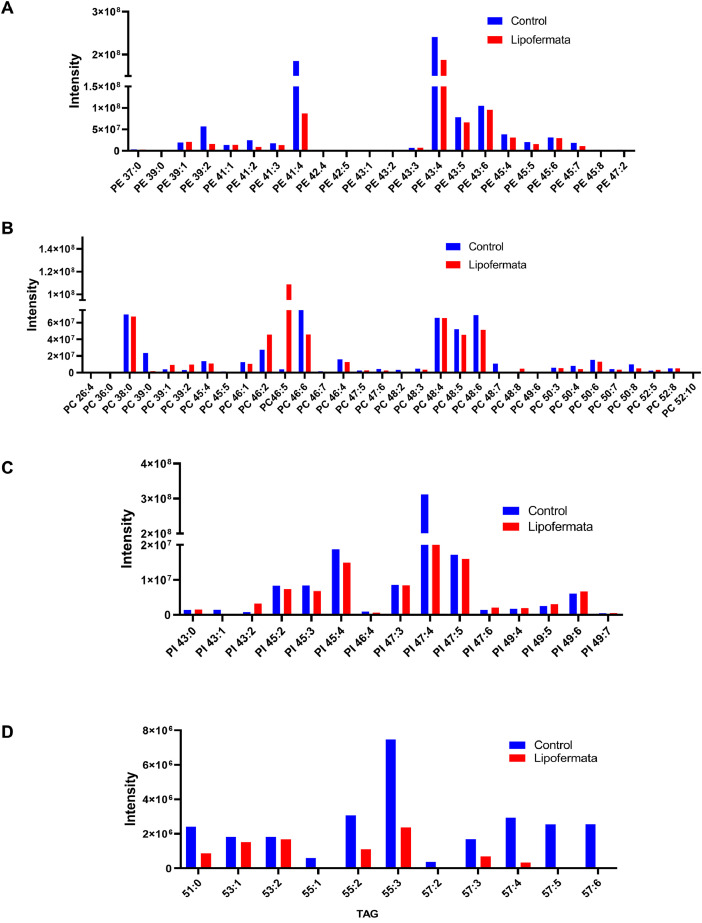
Lipid profiling was performed for MDSCs isolated from the spleens of B16F10 melanoma-bearing mice 20 days post-tumor injection which were subjected to treatment with an intraperitoneal injection of either lipofermata or vehicle (control) daily for 2 weeks starting on day 7 when the tumor was palpable on the skin of mice (Fig. 1). Single-cell suspension from the spleen was stained with CD11b and Gr1 antibodies to purify MDSCs by flow cytometry cell sorter for lipidomics. Next, lipid extraction was done using methyl‑tert‑butyl ether (MBTE) as previously described with slight modification [4]. Splash lipidomix mass spectrophotometry standard from Avanti polar served as control and samples analyzed using reversed-phase high-performance liquid chromatography (HPLC) coupled to a Q-Exactive mass spectrometer. Lipid identification and relative intensity were performed in LipidSearch software to obtain the different lipid species from negative and positive mode full scan operation of the mass spectrometer. The transformed raw dataset is shown in supplementary data (Figs. S1-S4). Fig. S1 and Fig. S2 show the lipid species for splenic MDSCs from B16F10 melanoma-bearing mice receiving PBS while Fig. S3 and Fig. S4 represent the lipid species for splenic MDSCs from the mice treated with lipofermata. From the raw data, the lipid species that were most differentially expressed between MDSCs from tumor-bearing mice treated with lipofermata and PBS were phospholipids (Phosphatidylethanolamine, phosphatidylcholine, and phosphatidylinositol) and triglycerides (Fig. 2A-D).
Fig. 1.
Schematic showing experimental procedure.
Chart illustrating steps from tumor model development to lipidomics analysis. B16F10 melanoma cell line was injected subcutaneously into the left flank of wild type C57BL/6 mice on day 0, tumors were allowed to grow on the skin until it was palpable. Starting from day 7, mice were administered either PBS or lipofermata daily until day 20 when mice were sacrificed. After sacrificing the mice, spleens were excised to prepare single-cell suspension and obtain purified MDSCs via a flow cytometer cell sorting machine. Lipid was then isolated from the MDSCs and subjected to LC-MS analysis to detect the different lipid species present in splenic MDSCs from PBS or lipofermata treated mice. Abbreviations: s.c – subcutaneous, i.p – intraperitoneal, LC-MS – liquid chromatography-mass spectrophotometry.
Fig. 2.
Profile for LC-MS analysis of lipid molecular species in splenic MDSCs from B16F10-bearing mice treated with FATP2 inhibitor.
LC-MS lipid analysis for flow cytometry sorted CD11b+ Gr1+ MDSCs isolated from the spleen of B16F10 tumor-bearing mice treated with or without 2.5 mg/kg lipofermata or PBS daily. Lipids were extracted from purified MDSCs with methyl-tert-butyl ether (MBTE) followed by LC-MS analysis. (A-D) Lipid Molecular species were detected on both the positive and negative mode – Phosphatidylethanolamine – PE (A) Phosphatidylcholine – PC (B) Phosphatidylcholine – PC (C) Triacylglycerol - TAG (D). Data represents Means ± SEM of relative intensity. MDSCs were CD11b+Gr-1+.
2. Experimental Design, Materials and Methods
2.1. Cell culturing
B16F10 melanoma cell line kept in the cell bank of Shenzhen Institutes of Advanced Technology (SIAT), Chinese Academy of Sciences was thawed and cultured in Dulbecco's modified eagle medium (DMEM) with 10% fetal bovine serum album (FBS) and 1% penicillin-streptomycin. Cells were grown in a 37 °C, 5% incubator, and the complete medium was replenished every 2–3 days or cells sub-cultured if growth had reached about 90% confluence.
2.2. Animal experiments
All mice used in this research were approved by the SIAT, Chinese Academy of Sciences ‘Animal Care and Use Committee’ according to the protocol - SIAT-IACUC-190,403-YYS-YXL-A0732. C57BL/6 mice (6–8-weeks-old females) obtained from Guangdong Province Animal Care Facilities were kept under pathogen-free conditions at the animal house facilities of SIAT throughout the experiment. 1 × 106 B16F10 cells were injected into the left flank of each mice to establish subcutaneous tumors. Mice were divided into two groups (n = 6 mice per group) and 2.5 mg/kg lipofermata per day was administered intraperitoneally starting from day 7 post-tumor inoculation while PBS was used as a vehicle in the control group. The last doses of lipofermata or PBS were administered on day 20 about eight (8) hours before sacrificing the mice. Spleens were isolated from the mice after sacrificing for further experiment.
2.3. Flow cytometry cell sorting
To obtain purified MDSCs, spleen from PBS or lipofermata treated mice were re-suspended in ammonium-chloride-potassium lysing buffer to remove red blood cells. The cell suspension was incubated on ice for 5 min. PBS was added to stop the lysis reaction and centrifuge at 1500 rpm, 4 °C for 5 min. The cell pellet was re-suspended in PBS to obtain a single-cell suspension. The single-cell suspension was subjected to mojosort magnetic cell separation according to the manufacturer's procedure and followed by cell surface staining with CD11b and Gr1 monoclonal antibodies for cell sorting on BD FACS Aria III cell sorter (BD Biosciences) as previously described [5].
2.4. Lipid extraction
CD11b+ Gr1+ flow cytometry sorted MDSCs from the spleens of B16F10 tumor-bearing mice treated with or without FATP2 inhibitor (lipofermata) were resuspended in 200μl of PBS and lipid was extracted according to a previously described method [4]. 1.5 ml of methanol was added to the samples and vortexed vigorously for 30 s. 5 ml of MTBE was added to the vortexed mixture and incubated for 1 hour on a shaker at room temperature. Phase separation was carried out by adding 1.25 ml of MS-grade water to the above mixture. The samples were incubated for 10 min at room temperature and then centrifuged at 1000 g for another 10 min. The upper phase was collected and placed in a new tube while the lower phase was re-extracted with 2 ml of the solvent mixture - MTBE/methanol/water (10:3:2.5, v/v/v). The upper phase was collected and added to the previously collected upper phase. The combined organic phase was dried in a vacuum centrifuge overnight. The vacuum dried samples were reconstituted in ACN/IPA/H2O (65:30:5, v/v/v) containing 5 mM ammonium acetate, and 5μL was injected into the LC-ESI-MS system to identify and quantify different lipid species present in MDSCs.
2.5. Liquid chromatography-mass spectrophotometry
A reversed-phase BEH C8 column (2.1 mm 100 mm, 1.7 μm, Waters, Milford, MA, U.S.A.) was used for the chromatographic separation of lipids. Mobile phases A and B were ACN/H2O (60:40, v/v) and IPA/ACN (90:10, v/v) respectively, both containing 10 mM ammonium acetate. The flow rate was 0.26 mL/min while the column temperature was at 55 °C. The elution started with 68% mobile phase A (ACN: H2O = 6:4, 10 mM ammonium acetate) and 32% mobile phase B (IPA: ACN = 9:1, 10 mM ammonium acetate) and maintained for 1.5 min. Mobile phase B was then linearly increased to 85% for 10 min and further to 97% in the next 0.1 min followed by maintenance for 1.5 min. Afterward, it was decreased to 32% B in 0.1 min and kept for 2 min till the next injection. The temperature of the sample manager was set at 10 °C.
The mass spectrometer was operated with a capillary voltage of 3.5 kV in positive mode and 3.0 kV in negative mode. The capillary temperature was set at 300 °C. Sheath gas flow rate and aux gas flow rate were set at 30 and 10 (in arbitrary units) respectively. Aux gas heater temperature was 310 °C while the S-lens RF level was 50.0. The resolutions of 70,000 and 17,500 were set for full scan MS and data-dependent MS/MS (ddMS2) in both modes. AGC target and maximum IT were 3 × 106 ions capacity and 200 ms in full-scan MS settings while their values were 1 × 105 ions capacity and 50 ms in ddMS2 settings. The Top N (N, the number of topmost abundant ions for fragmentation) was set to 10. The normalized collision energy (NCE) was set at 20, 35, 70 eV, respectively while the scan range was set at m/z 133.4 − 2000. The raw dataset for a full scan of negative and positive modes in the different treatment groups are shown in supplementary data (Figs. S1-S4).
Ethics Statement
Study involved the use of animals (female mice only). All animal experiments were approved by the ‘Animal Care and Use Committee’ of the Shenzhen Institutes of Advanced Technology which complied with the National Institutes of Health guide for the care and use of laboratory animals
CRediT Author Statement
Adeleye Oluwatosin Adeshakin: Investigation, Data curation, Visualization, Methodology, Software, Writing - Original draft preparation; Funmilayo O. Adeshakin: Animal experiment, Visualization, Writing - original draft preparation; Wan Liu: Visualization, Validation, Data analysis; Hua Li: Data curation, Software, Writing - Original draft preparation, Supervision; Dehong Yan: Conceptualization, Supervision, Writing - Reviewing and Editing; Xiaochun Wan: Supervision, Resources, Writing - Reviewing, and Editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have or could be perceived to have influenced the work reported in this article.
Acknowledgments
Adeleye Oluwatosin Adeshakin is sponsored by the University of Chinese Academy of Sciences (UCAS) and Shenzhen Institute of Advanced Technology scholarship for international students. We thank members of the Public Technology Service Cores at the Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, for their technical assistance. We also appreciate the support from Zhan Wugen of the Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, for flow cytometry cell sorting.
This work was supported by the National Key R&D Program of China (2019YFA0906100), National Natural Science Foundation of China (Grants 82071772, 81501356, and 81373112), Key-Area Research and Development Program of Guangdong Province (2019B020201014), the Shenzhen Basic Science Research Project (Grants JCYJ20190807161419228, JCYJ20170818155135838, JCYJ20170818164619194, and JCYJ20170413153158716), China Postdoctoral Science Foundation (2019M660220), Basic and Applied Basic Research Foundation of Guangdong Province (2019A1515110359), Nanshan pilot team project (LHTD20160004), Start-up funding (CYZZ20180307154657923), and the SIAT-GHMSCB Biomedical Laboratory for Major Diseases and Dongguan Introduction Program of Leading Innovative and Entrepreneurial Talents (to Z. L.).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.17632/3sncj38wds.1 and doi:10.1016/j.dib.2021.106882.
Contributor Information
Dehong Yan, Email: dh.yan@siat.ac.cn.
Xiaochun Wan, Email: xc.wan@siat.ac.cn.
Appendix. Supplementary materials
References
- 1.Adeshakin, Adeleye; Adeshakin, Funmilayo; Liu, Wan; Li, Hua; Yan, Dehong; Wan, Xiaochun (2021), “LC-MS dataset showing changes in MDSCs lipid species following FATP2 inhibition”, Mendeley Data, v1, doi: 10.17632/3sncj38wds.1. [DOI]
- 2.Adeshakin A.O., Liu W., Adeshakin F.O., Afolabi L.O., Zhang M., Zhang G., Wang L., Li Z., Lin L., Cao Q., Yan D., Wan X. Regulation of ROS in myeloid-derived suppressor cells through targeting fatty-acid transport protein 2 enhanced anti-PD-L1 tumor immunotherapy. Cell. Immunol. 2021;362 doi: 10.1016/j.cellimm.2021.104286. [DOI] [PubMed] [Google Scholar]
- 3.Veglia F., Tyurin V.A., Blasi M., De Leo A., Kossenkov A.V., Donthireddy L., To T.K.J., Schug Z., Basu S., Wang F., Ricciotti E., DiRusso C., Murphy M.E., Vonderheide R.H., Lieberman P.M., Mulligan C., Nam B., Hockstein N., Masters G., Guarino M., Lin C., Nefedova Y., Black P., Kagan V.E., Gabrilovich D.I. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature. 2019;569:73–78. doi: 10.1038/s41586-019-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matyash V., Liebisch G., Kurzchalia T.V., Shevchenko A., Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adeshakin A.O., Yan D., Zhang M., Wang L., Adeshakin F.O., Liu W., Wan X. Blockade of myeloid-derived suppressor cell function by valproic acid enhanced anti-PD-L1 tumor immunotherapy. Biochem. Biophys. Res. Commun. 2020;522:604–611. doi: 10.1016/j.bbrc.2019.11.155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.