Abstract
Rapid, sensitive, and precise multiplexed assays for serological analysis during candidate COVID-19 vaccine development would streamline clinical trials. The VaxArray Coronavirus (CoV) SeroAssay quantifies IgG antibody binding to 9 pandemic, potentially pandemic, and endemic human CoV spike antigens in 2 h with automated results analysis. IgG antibodies in serum bind to the CoV spike protein capture antigens printed in a microarray format and are labeled with a fluorescent anti-species IgG secondary label. The assay demonstrated excellent lower limits of quantification ranging from 0.3 to 2.0 ng/mL and linear dynamic ranges of 76 to 911-fold. Average precision of 11 % CV and accuracy (% recovery) of 92.5 % over all capture antigens were achieved over 216 replicates representing 3 days and 3 microarray lots. Clinical performance on 263 human serum samples (132 SARS-CoV-2 negatives and 131 positives based on donor-matched RT-PCR and/or date of collection) produced 98.5 % PPA and 100 % NPA.
Keywords: COVID-19, SARS-CoV-2, Human coronavirus, Serology, Immunoassay, VaxArray
1. Introduction
A novel coronavirus (nCoV) was first identified in December 2019 in Wuhan, China, and was declared a worldwide pandemic by the World Health Organization on March 11, 2020. Coronaviruses infecting humans belong to either the alpha or beta coronaviruses. 229E and NL63 are alphacoronaviruses, while coronaviruses HKU1, OC43, SARS-CoV-1 (the virus causing Severe Acute Respiratory Syndrome that first circulated in 2003), MERS-CoV (the virus causing Middle East Respiratory Syndrome), and the current SARS-CoV-2 virus causing COVID-19, formerly referred to as nCoV, belong to the betacoronaviruses (Chen et al., 2020; Ye et al., 2020). While 229E, HKU1, OC43, and NL63, known to continually circulate in humans, generally produce mild symptoms, MERS-CoV, SARS-CoV-1, and SARS-CoV-2 more recently have crossed over from animal reservoirs into humans (Chen et al., 2020; Ye et al., 2020) and can produce symptoms that are quite severe. Moreover, SARS-CoV-2 has been shown to effectively transmit person-to-person and cause significant morbidity and mortality, as evidenced by the over 28.4 million US and 113.1 million worldwide confirmed cases of the virus as of February 26, 2021 and the associated 2.2 % global mortality rate (Johns Hopkins University Center for Systems Science and Engineering (CSSE), 2020).
The role of serological testing to measure antibodies to SARS-CoV-2 has been the subject of recent debate (Theel et al., 2020; Bryant et al., 2020), given that we do not yet fully understand the antibody levels required for seroprotection or how long protective antibodies may persist. In addition, the Food and Drug Administration’s waiver of the requirement for Emergency Use Authorization (EUA) for SARS-CoV-2 serological assays as long as internal validation has been conducted and appropriate limitations language are included in product labeling (Food and Drug Administration, 2020) resulted in a large number of serological tests introduced into the market for diagnostic use. As of August 18, 2020, 397 commercialized antibody detection immunoassays were listed on FIND’s website (Foundation for Innovative New Diagnostics, 2020), and numerous reports of serological assays with variable performance have been recently highlighted (Cohen, 2020; Bastos et al., 2020; Weissleder et al., 2020; Cassaniti et al., 2020).
Regardless of the utility and role of SARS-CoV-2 serology assays for diagnostic and seroprevalence applications, a critical application of serological testing for SARS-CoV-2 is monitoring antibody response to COVID-19 vaccine candidates during pre-clinical and clinical trials to enable a full understanding of the immune response post-vaccination (Theel et al., 2020; Lipsitch et al., 2020). There are currently 202 SARS-CoV-2 candidate vaccines in pre-clinical or clinical development (Milken Institute, 2020; Amanat and Krammer, 2020). Binding assays including ELISAs are an alternative to the gold standard virus neutralization assays, as they do not require cell culture or significant biosafety containment measures, are straightforward to conduct, and have been shown to correlate with virus neutralization assays for other coronaviruses (Chan et al., 2009). In addition, antigen binding assays currently commercially available or in clinical use typically establish specificity using pre-pandemic serum samples which cannot conclusively show that pre-existing antibodies to the endemic coronaviruses do not cross react with SARS-CoV-2.
Serological testing that assesses binding to a variety of coronaviruses is important for: (1) screening enrollees before trial admission to establish baseline antibody titers, (2) monitoring longitudinal profiles of antibody response post-vaccination, and (3) comparing pre- and post-vaccination immune responses in pediatric vs. adult cohorts. Recent reports and workshops have highlighted the unknowns about pre-existing immunity to SARS-CoV-2 (Sette and Crotty, 2020; Lerner et al., 2020). In light of these unknowns and recent discussion around the potential for antibody dependent enhancement (Francesco, 2020; Iwasaki and Yang, 2020; Peeples, 2020), monitoring the serological responses before and after immunization to other human coronaviruses, such as SARS, MERS, and the endemic coronaviruses including HKU1, OC43, NL63, and 229E, and comparing these responses to those from natural infection as a function of disease severity, will be critical for understanding the immune response and ultimately delivering a safe and effective vaccine.
For effective application in vaccine clinical trials, a highly specific and highly quantitative assay is required to enable accurate quantitative assessment of antibody responses. Given the rapid timelines for vaccine development already underway, a multiplexed assay that can measure vaccine-induced antibody response to a variety of related antigens simultaneously is highly desirable for both time and cost savings. The VaxArray platform (InDevR, Inc., Boulder, CO) is a microscale, multiplexed, microarray-based immunoassay platform that has been well-validated for use in influenza vaccine antigen characterization (Kuck et al., 2018; Byrne-Nash et al., 2019), and has been adapted for serological analysis of coronaviruses with the recent availability of the Coronavirus (CoV) SeroAssay. Specifically, nine unique coronavirus spike protein antigens are printed in replicate in a microarray format, providing the ability to perform simultaneous analysis of antibody responses to all 9 antigens in a single, 2 h assay. In comparison, one would have to run 9 parallel ELISA plates to obtain the same information content. The 9 proteins represented on the microarray are full-length spike, receptor binding domain (RBD), and the S2 extracellular domain of SARS-CoV-2, and the spike proteins from SARS, MERS, HKU1, OC43, NL63, and 229E. In addition, the platform is antigen-sparing, requiring ∼200x less antigen to manufacture than a traditional plate-based ELISA, which is particularly important during this time of strained supply chains. Lastly, the CoV SeroAssay is provided as a validated, off-the-shelf kit to minimize user-to-user and laboratory-to-laboratory variability associated with in-house immunoassays, and the associated software provides automated analysis of the results to further increase ease of use.
This study reports on the VaxArray CoV SeroAssay linear dynamic range, limit of detection, specificity, reproducibility, accuracy, and investigates assay performance on a retrospective set of 263 blinded, de-identified human serum and plasma specimens to demonstrate positive and negative percent agreement to a mixed reference method of RT-PCR on a patient-matched specimen and collection date prior to the COVID-19 outbreak. An easy-to-use, high information content assay with the capability to evaluate antibody response to a variety of coronavirus spike proteins will aid in monitoring the immune response during COVID-19 candidate vaccine clinical trials and ultimately facilitate the delivery of a safe and effective vaccine.
2. Materials and methods
2.1. VaxArray coronavirus SeroAssay standard procedure
The VaxArray Coronavirus SeroAssay Kit (#VXCV-5100, InDevR, Inc.) contains four microarray slides, printed with 16 replicate arrays per slide, an optimized Protein Blocking Buffer (VX-6305), Wash Buffer 1 concentrate (VX-6303), and Wash Buffer 2 concentrate (VX-6304). Prior to use, microarray slides were equilibrated to room temperature for 30 min in the provided foil pouch. Prepared standards and specimens were diluted at least 1:100 in Protein Blocking Buffer and applied to the microarray and allowed to incubate in a humidity chamber (VX-6200) on an orbital shaker at 80 rpm for 60 min. After incubation, samples were removed using an 8-channel pipette, and the microarray was subsequently washed by applying 50 μL of prepared Wash Buffer 1. Slides were washed for 5 min on an orbital shaker at 80 RPM after which the wash solution was removed via 8-channel pipette. During sample incubation, Anti-human IgG Label (VXCV-7623) and/or anti-mouse IgG Label (VXCV-7620) were prepared by first diluting the label 1:10 in PBB, and aliquoting into 8-tube PCR strips after which 50 μL of label mixture was added to each array using an 8-channel pipette. Detection label was incubated on the slides in the humidity chamber for 30 min before subsequent, sequential washing in Wash Buffer 1, Wash Buffer 2, 70 % Ethanol, and finally ultrapure water. Slides were dried using a compressed air pump system and imaged using the VaxArray Imaging System (VX-6000).
2.2. Linear dynamic range and lower limit of quantification analysis
A study to determine the lower limit of quantification and linear dynamic range of the different capture antigens represented was executed using monoclonal antibodies that target the spike proteins of SARS-CoV-1 (MRO-1214LC, CR3022, Creative Biolabs), SARS-CoV-2 (GTX632604, Genetex), MERS (40069-MM23, Sino Biological), and HKU1 (40021-MM07, Sino Biological). The CR3022 antibody targeting SARS-CoV-1 is known to bind the the nCoV(ii) RBD antigen, the SARS antigen (and the nCoV(i) full-length spike antigen to a much weaker extent), and the SARS-CoV-2 Genetex antibody is known to bind the nCoV(i) full-length spike antigen (and the nCoV(iii) S2 antigen to a much weaker extent). The four antibodies were mixed, and a 13-point serial dilution in Protein Blocking Buffer and three blank wells containing Protein Blocking Buffer without antibody were prepared, with each sample subsequently analyzed on the VaxArray CoV SeroAssay according to the operation manual with one exception: because the anti-SARS-CoV1 antibodies are human antibodies and the other three antibodies are mouse antibodies, antibodies were detected with a mixture of anti-mouse and anti-human IgG secondary antibody labels (VXCV-7620 and VXCV-7326, InDevR, Inc., respectively). After analysis, the median signals extracted from the VaxArray Imaging System software for each relevant capture antigen for each dilution as well as for the blanks were analyzed, with the serial dilutions plotted as a function of the known concentration of the antibody and a series of moving 4-point linear fits applied to the data. The upper limits of quantification (ULOQ) were calculated for each of the relevant capture agents as the back-calculated concentration at the highest RFU signal that was within the highest 4-point fit with an R2 value exceeding 0.95. The lower limits of quantification (LLOQ) for each of the relevant capture antigens was calculated for each blank sample as the back-calculated concentration at the background-subtracted median signal of the blank plus 5 standard deviations of the blank. This value was then averaged over the 3 blanks. In addition, the linear dynamic range (LDR) was calculated as ULOQ/LLOQ for each relevant capture antigen.
Because monoclonal antibodies binding to the OC43, NL63, or 229E capture antigens were not available at the time of testing, linearity and limit of detection for these 3 capture antigens was explored using a limiting endpoint dilution series of a pooled human serum sample previously shown to produce positive responses on the CoV SeroAssay for all four human CoVs: OC43, NL63, HKU1, and 229E. The pooled human serum sample was used to create a 13-point serial dilution in Protein Blocking Buffer with all samples subsequently analyzed in duplicate on the VaxArray CoV SeroAssay according to the operation manual. Data was extracted in the same manner as for the mixed monoclonal antibody analysis to determine the signals at the ULOQ and LLOQ (without back-calculating to concentration based on the linear fits, as the concentration in each of the dilutions is unknown). The LDR was expressed as the signal at the ULOQ divided by the signal at the LLOQ. The limiting endpoint dilution titer was also presented as the highest dilution factor at which the signal exceeded the signal at the LLOQ.
2.3. Specificity
Specificity of the capture antigens was investigated using the same 4 monoclonal antibodies described for use in the LLOQ and LDR analysis. No monoclonal antibodies were available at the time of testing that target OC43, NL63, or 229E, and so specificity for these capture antigens was not assessed. However, we did assess specimens positive for all 4 endemic coronaviruses that were collected prior to the SARS-CoV-2 outbreak to examine potential cross-reactivity with any of the 3 SARS-CoV-2 antigens. A total of 132 serum samples known to be negative for the presence of antibodies to SARS-CoV-2 based on date of collection prior to December 2019, including 33 specimens from pediatric donors age 2–16, were analyzed via the standard VaxArray CoV SeroAssay procedure at a 1:100 dilution in PBB.
2.4. Reproducibility and accuracy
To assess reproducibility and accuracy, a pooled human serum sample known to be positive for antibodies to SARS-CoV-2 (and known to bind all 3 SARS-CoV-2 antigens on the microarray) and all 4 of the endemic coronaviruses (HKU1, OC43, NL63, and 229E) was prepared in adequate volume to run a large number of replicates. This sample did not contain any antibody reactive to the MERS capture antigen, and therefore, reproducibility and accuracy of the assay’s ability to detect antibodies that bind to the MERS spike protein was not assessed. This study examined a single operator over three days of testing, as previous studies (data not shown) indicated little user-to-user or instrument-to-instrument variability. On days 1 and 2 of testing, a single slide containing an 8-point calibration curve (7 standards and a blank, analyzed at arbitrary relative concentrations of 1.0, 0.8, 0.6, 0.4, 0.2, 0.1, 0.05, and 0) was run alongside 8 replicates of an intermediate dilution of the same sample (expected to be at 0.4). On the 3rd testing day, three additional slides of 16 replicates each were analyzed alongside the first slide, for a total of 72 replicate microarrays (the standard curve on slide 1 was used to analyze data on all 4 slides in the experiment). The above testing was performed on 3 unique slide manufacturing lots for a total of 216 replicate analyses over the 3 days and 3 lots. The quantitative mode of the VaxArray Imaging System software was used to generate back-calculated concentrations for each of the replicates analyzed for data analysis, using the relevant standard curve for each 1-slide or 4-slide analysis on each day. Averages over each set of 8 replicates (each slide) as well as over all 216 replicates were calculated and are presented as % coefficient of variation (%CV). To assess accuracy or % recovery (% of expected value), the same data were analyzed as the calculated concentration divided by the expected concentration of 0.4, expressed as a percentage. The % recovery over all 216 replicates analyzed are presented. Precision and accuracy data can be found in Table 3.
Table 3.
Precision, Accuracy of all 9 CoV SeroAssay Capture Antigens.a
| Capture Antigen | Average Accuracy (measured/expected concentration)b | Average Precision, expressed as % CV |
|---|---|---|
| nCoV(i) | 94 % (0.377/.0.400) | 10 % |
| nCoV(ii) | 97 % (0.387/0.400) | 11 % |
| nCoV(iii) | 88 % (0.354/0.400) | 19 % |
| SARS | 92 % (0.367/0.400) | 11 % |
| HKU1 | 90 % (0.358/0.400) | 10 % |
| OC43 | 96 % (0.383/0.400) | 10 % |
| 229E | 95 % (0.378/0.400) | 7 % |
| NL63 | 91 % (0.366/0.400) | 11 % |
Data represents n = 216 pooled serum samples analyzed (single user, 3 slide lots over 3 days). MERS capture antigen was not evaluated in this study.
Expected concentration of the replicates analyzed was 0.4 relative to the highest standard which was assigned an arbitrary concentration of 1.0.
2.5. Positive and negative percent agreement
To determine the positive (PPA) and negative (NPA) percent agreement of the VaxArray CoV SeroAssay with a known result, 263 retrospective, deidentified human specimens (260 serum, 3 plasma) were obtained from the authors’ institutions, collaborators, and commercial sources. Commercial specimen sources included Boca Biolistics (Pompano Beach, FL), US Biolab (Rockville, MD), and Lee BioSolutions (Maryland Heights, MO). Specimens from Colorado Children’s Hospital (Denver, CO) were collected under a Colorado Multiple IRB (COMIRB) approved protocol. Specimens obtained from Lankenau Institute for Medical Research (LIMR, Wynnewood, PA) were collected under a Main Line Hospitals IRB approved protocol. Deidentified specimens from Veritas, PA (Belton, TX) were obtained under an AspireIRB-approved protocol. Deidentified specimens from Mount Sinai Icanh School of Medicine (New York, NY) were obtained under a license agreement which indicates appropriate IRB approval and informed consent for the specimens provided. Additional details regarding clinical specimens included can be obtained from the authors upon request.
The reference method used for specimens positive for SARS-CoV-2 antibodies was an RT-PCR result for a donor-matched specimen. The reference method used for specimens negative for antibodies to SARS-CoV-2 was either a negative RT-PCR result for a donor-matched specimen or a known collection date prior to the COVID-19 outbreak in late 2019. All specimens obtained from Colorado Children’s Hospital were also analyzed independently by ELISA in a CLIA-certified laboratory using either an NP-based ELISA (Epitope Diagnostics, San Diego, CA) or by an S1-based ELISA (Euroimmun Ag, Germany), with a significant number of these specimens analyzed by both additional ELISAs. These ELISA results were used as orthogonal information to further investigate discrepant results. Testing personnel were blinded to these orthogonal results prior to completing VaxArray CoV SeroAssay analysis.
The sample set included 132 negatives and 131 positives, with all positives collected at least 14 days after onset of symptoms. Ninety-two (92) of the specimens were received with a known donor age. Of note, 33 of the negative specimens were from pediatric donors between the ages of 2 and 16. All specimens were internally blinded by someone not involved with the testing prior to analysis. Specimens were diluted 1:100 in PBB and carried through the VaxArray CoV SeroAssay procedure as described in the Methods section. After imaging was complete, median signal data from the VaxArray Imaging System for the sample set were exported into MS Excel and analyzed. From the median signal data, the signal to background value (S/B) was calculated for each of the 9 antigens on the chip by taking the median of 9 replicate spots for each antigen and dividing by the background of the microarray.
The diagnostic algorithm used a multi-antigen approach. Specimens were considered positive for SARS-CoV-2 antibodies by the VaxArray CoV Assay if both of the following two conditions were met: 1) the S/B on the nCoV(i) antigen exceeded the average S/B of the bank of negative specimens + 3 standard deviations of the average, AND 2) the sum of the S/B values for all 3 nCoV antigens exceeded the sum of the average S/B of the bank of negative specimens + 6 standard deviations. Once VaxArray CoV SeroAssy results were determined, PPA and NPA were calculated compared to the reference method and reported with associated 95 % confidence intervals. Confidence intervals reported are two-sided Wilson Scores with no continuity correction.
3. Results
3.1. Assay principles
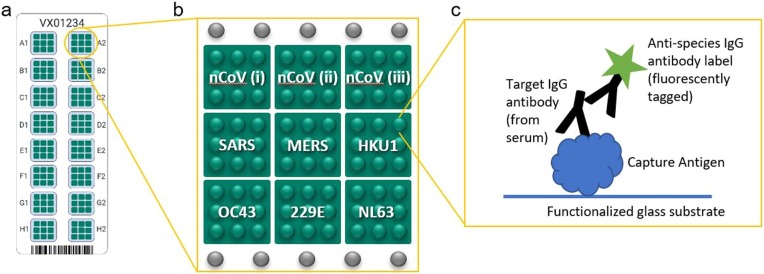
The VaxArray CoV SeroAssay is a multiplexed immunoassay that consists of 9 coronavirus spike proteins printed in a microarray format as schematically illustrated in Fig. 1 . Each of 9 antigens are printed in 9 replicate spots in a single microarray, with 16 identical microarrays printed on each slide. Details regarding the capture antigens are found in Table 1 . The nCoV(i) full-length spike protein and nCoV(ii) receptor binding domain (RBD) protein were licensed from Icahn School of Medicine at Mount Sinai, and are described in Amanat et al. (2020). All other antigens were obtained from commercial sources. For quantitative analysis, serum samples diluted in a blocking buffer are analyzed alongside a serial dilution of an appropriate standard material, which is utilized to quantify the antibody binding to each of the 9 antigens on the microarray. Antibodies in serum are captured by the printed antigens and are subsequently labeled with a fluorescent species-specific IgG label for detection. Spatial separation of the 9 antigens enables multiplexed analysis of antibody binding to a variety of coronavirus antigens. For qualitative analysis, serum specimens are diluted in a blocking buffer and analyzed at a single dilution factor and compared to an established cutoff value based on responses from a bank of known negative samples.
Fig. 1.
(a) Schematic illustration of VaxArray CoV SeroAssay slide with 16 identical microarrays labeled A1 through H2, (b) Layout of an individual microarray including 9 unique capture antigens labeled nCoV(i) through NL63, each printed as 9 replicate spots. Fiducial markers are shown as grey spots in rows above and below capture antigens. (c) Schematic of the immunoassay principle in which capture antigen binds target antibodies from serum, and target antibodies are labeled using a species-specific IgG secondary antibody label that contains a fluorescent tag. S refers to full-length spike, and S1, S2, and RBD refer to corresponding portions of spike as called out in Table 1.
Table 1.
Identifying Information for Nine Human Coronavirus Spike Antigens Represented on the CoV SeroAssay.
| Name on chip | Expression System | Antigen | Source |
|---|---|---|---|
| nCoV(i) | Mammalian | Full length spike (S1 + S2) | Licensed from Mount Sinai (Amanat et al., 2020) |
| nCoV(ii) | Mammalian | Receptor Binding Domain (RBD) | Licensed from Mount Sinai (Amanat et al., 2020) |
| nCoV(iii) | Insect | S2 extracellular domain | commercially available |
| SARS | Mammalian | S1 | commercially available |
| MERS | Mammalian | S1 | commercially available |
| HKU1 | Mammalian | S1 | commercially available |
| OC43 | Mammalian | S1 | commercially available |
| 229E | Insect | Full length spike (S1 + S2) | commercially available |
| NL63 | Mammalian | S1 | commercially available |
3.2. VaxArray CoV SeroAssay is quantitative with low ng/mL sensitivity
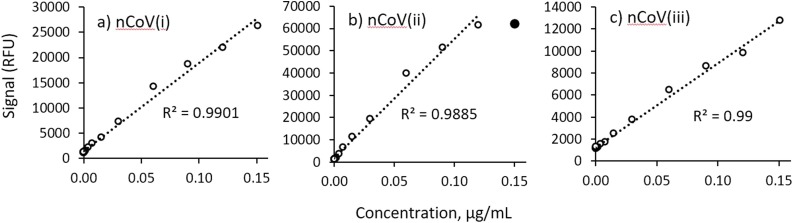
To demonstrate the quantitative ability of the assay, a study was executed using a mixture of mouse and human monoclonal antibodies (mAbs) that target SARS-CoV-1, SARS-CoV-2, MERS, and HKU1 (see the Methods section for details). The 4 antibodies were mixed, and a 13-point dilution was analyzed using a mixture of anti-mouse and anti-human IgG labels. Fig. 2 shows representative 13-point dilutions demonstrating linearity with dilution for the nCoV(i), (ii), and (iii) antigens, respectively. Lower limits of quantification (LLOQ) for the 6 targeted antigens ranged from 0.32 ng/mL to 1.99 ng/mL, with associated linear dynamic ranges (LDR) from 76 to 911 × . Because there were no available monoclonal antibodies for OC43, NL63, or 229E at the time of testing, the linearity with dilution of the assay for antibody binding to these antigens was investigated by determining a limiting endpoint dilution titer using a pooled human serum sample known to be positive for all 4 endemic human coronaviruses. Table 2 shows the calculated LLOQ, upper limit of quantification (ULOQ), and LDR for the captures for which mAbs were available, as well as the limiting endpoint dilution titers (which range from 4000 to 16,000) and LDRs for OC43, NL63, and 229E (which range from 16x to 32x).
Fig. 2.
13-point dilution series of monoclonal antibodies reactive to the SARS-CoV-2 spike protein capture antigens along with associated R2 values for the linear regressions (n = 1 for each datapoint): a) nCoV(i), b) nCoV(ii), and c) nCoV(iii). In panel b), the filled circle at the highest concentration was not included in the linear fit since it is outside the linear range.
Table 2.
Sensitivity, Linear Dynamic Range of all 9 CoV SeroAssay Capture Antigens.
|
Metrics determined with monoclonal antibodies | |||
|---|---|---|---|
| Capture Antigen | LLOQ (ng/mL) | ULOQ (ng/mL) | LDR (no units) |
| nCoV(i) | 1.40 | 150 | 107 |
| nCoV(ii) | 0.47 | 120 | 255 |
| nCoV(iii) | 1.99 | 151 | 76 |
| SARS | 0.55 | 150 | 273 |
| MERS | 1.00 | 911 | 911 |
| HKU1 | 0.32 | 59 | 184 |
|
Metrics determined with pooled human serum | ||
|---|---|---|
| Capture Antigen | Limiting Endpoint Dilution Titer | LDR (no units) |
| OC43 | 8000 | 32 |
| 229E | 16,000 | 32 |
| NL63 | 4000 | 16 |
3.3. VaxArray CoV SeroAssay demonstrates reasonable specificity between SARS-CoV-2 and endemic coronaviruses
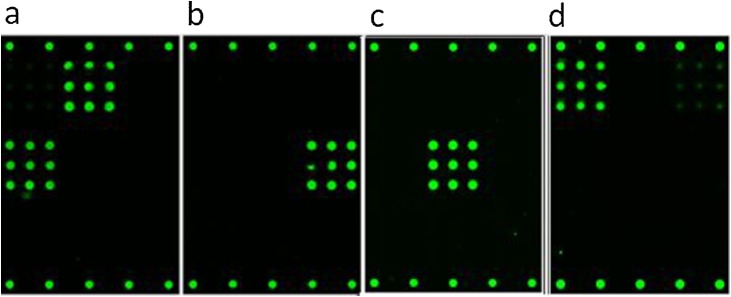
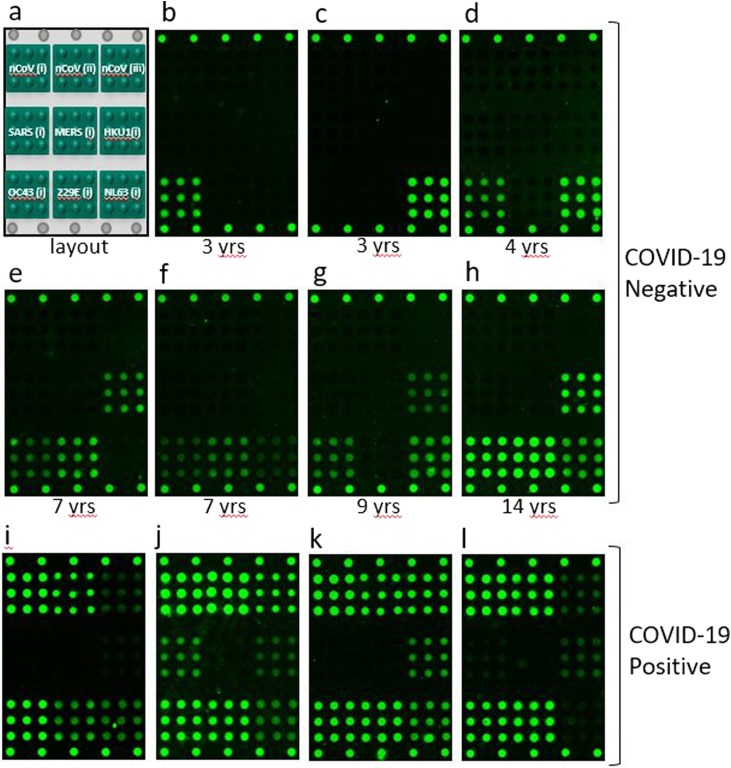
Monoclonal antibodies reactive to SARS-CoV-2, SARS-CoV-1, MERS, and HKU1 were analyzed to investigate specificity, with representative images shown in Fig. 3 . No monoclonal antibodies were available at the time of testing that specifically target OC43, NL63, or 229E. One hundred thirty-two (132) human serum specimens negative for antibodies to SARS-CoV-2 were analyzed, and none showed reactivity to the nCoV(i) full-length spike or nCoV(ii) RBD antigens. Thirteen (13) specimens did show some reactivity to the nCoV(iii) (S2) antigen, indicating some degree of non-specificity. Thirty-three (33) of the 132 negative specimens were pediatric samples from donors aged 2–16, one of which was cross-reactive on the nCoV(iii) S2 antigen. Representative microarray images of human serum samples negative for SARS-CoV-2 with different reactivities to the endemic CoV capture antigens on the microarray are shown in Fig. 4 (panels b through h) along with the associated donor ages; also in Fig. 4 (panels i through l) are representative images of human serum samples from donors of unknown age known to be positive for SARS-CoV-2 antibodies for comparison. A reminder of the microarray layout is included as Fig. 4 panel a for clarity. In addition, for donor-matched serum from SARS-CoV-2 RT-PCR positive patients, we do observe some cross-reactivity of SARS-CoV-2 antibodies with the SARS-CoV-1 antigen.
Fig. 3.
Fluorescence microarray images illustrating binding of monoclonal antibodies to the CoV SeroAssay. (a) CR3022 SARS-CoV-1 antibody from Creative Biolabs binding to the nCoV(ii) and SARS antigens, (b) 40021-MM07 HKU1 antibody from Sino Biological binding to HKU1 antigen, (c) 40069-MM23 MERS antibody from Sino Biological binding to the MERS antigen, and (d) GTX632604 SARS-CoV-2 antibody from Genetex binding to the nCoV(i) and nCoV(iii) antigens.
Fig. 4.
(a) Schematic illustration of microarray layout, and representative fluorescence images of the VaxArray CoV SeroAssay microarray in (b) through (l). Images (b) through (h) show the variety of responses to the endemic human CoV capture antigens for specimens from pediatric donors (ages noted for each image), and images (i) through (l) show responses from 4 unique COVID-19 positive donors of unknown age.
3.4. VaxArray CoV SeroAssay shows 11 % overall %CV for replicate measurements
To assess reproducibility and accuracy, a pooled human serum sample positive for antibodies to SARS-CoV-2, and also known to be reactive to the SARS antigen and the 4 endemic coronaviruses was prepared (no reactivity to MERS antigen), and used to create a serial dilution to be used as a standard curve as well as replicate aliquots of an intermediate dilution to be analyzed in replicate. Seventy-two (72) replicates of the intermediate dilution were tested by a single operator over 3 days on each of 3 manufacturing lots of microarray slides, for a total of 216 replicate analyses. Previous studies indicated little user-to-user or instrument-to-instrument variability (data not shown). The replicates analyzed on each day were quantified in arbitrary concentration units using the day- and lot-specific calibration curve, with the highest concentration of the serum sample arbitrarily assigned a value of 1. Replicates were run at an expected concentration of 0.4.
Table 3 shows the % CV in the back-calculated concentration value obtained on each relevant capture antigen for all 216 replicate measurements over all 3 days and all three lots of slides, with values ranging from 7 to 19 %CV for the 9 antigens. In addition, the %CV of the 8 replicates run on each slide in the study was analyzed to assess the intra-slide precision, resulting in an average intra-slide CV that ranged from 5 % to 8 % for each of the 9 antigens, for an overall intra-slide CV of 6 %.
3.5. VaxArray CoV SeroAssay accuracy (% recovery) ranges from 88 % to 97 %
The data generated for the reproducibility study were also used to assess accuracy expressed as % of expected result (% recovery), where the expected result for all replicates was 0.4 arbitrary concentration units (highest standard assigned a value of 1, and other standards assigned based on dilution factor). The calibration curves generated using the quantitative mode of the VaxArray Imaging System software were utilized to back-calculate the measured concentration present for all the replicates for each of the relevant antigens. The concentrations determined were then averaged for all 216 replicates and compared to the expected concentration. These accuracy data are presented in Table 3 and range from 88 to 97 % for the various capture antigens for an overall average accuracy of 92.5 %.
3.6. VaxArray CoV SeroAssay demonstrated PPA of 98.5 % and NPA of 100 %
Two hundred sixty-three (263) deidentified specimens (260 serum, 3 plasma) were received for testing. The sample set included 132 specimens known to be negative for COVID-19 by RT-PCR of a donor-matched specimen or were collected prior to the COVID-19 outbreak in late 2019, and 131 specimens known to be positive for COVID-19 by RT-PCR of a donor-matched specimen. All serum specimens from COVID-19 positive donors were collected at least 14 days after the initial onset of symptoms, and representative fluorescence microarray images from positive specimens are shown in Fig. 4. This study resulted in positive percent agreement (PPA) of 98.5 % (95 % CI of 94.6–99.6 95 %, Wilson Score, two-sided with no continuity correction) and negative percent agreement (NPA) of 100 % (95 % CI of 97.2–100 %, Wilson Score, two-sided with no continuity correction) with the mixed reference method described. This dataset resulted in zero false positives and 2 false negatives by the VaxArray CoV SeroAssay.
As a comparison, of the 151 samples obtained from Children’s Hospital of Colorado, 119 were also analyzed by the Euroimmun ELISA, with these specimens producing PPA of 90.6 % (58 true positives and 6 false negatives) and NPA of 94.6 % (52 true negatives and 3 false positives) with the mixed reference method described herein. In addition, all 151 samples from Children’s Hospital of Colorado were analyzed by the Epitope Diagnostics ELISA and produced PPA of 84.4 % (81 true positives and 15 false negatives) and NPA of 94.6 % (52 true negatives and 3 false positives).
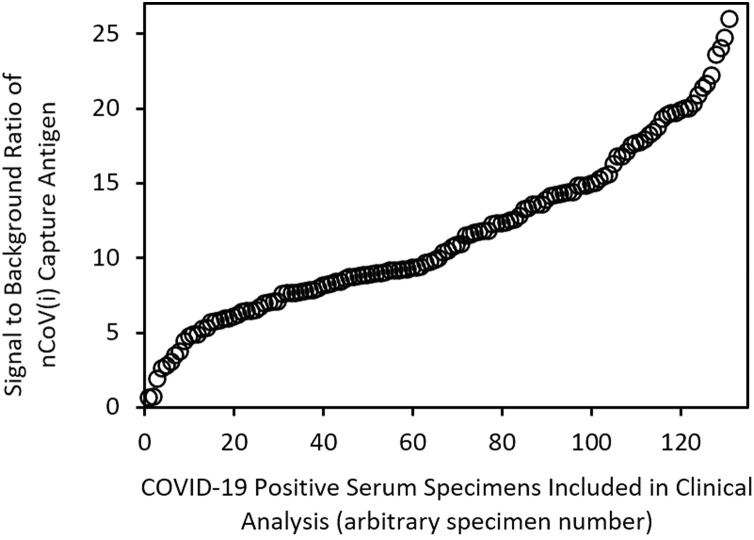
To demonstrate the range of underlying quantitative responses obtained from these clinical specimens, Fig. 5 shows a scatter plot of signal to background ratios obtained for the nCoV(i) capture antigen for the 131 specimens from donors expected to be positive by RT-PCR, with signal to background ratios sorted from low to high. The data show that the antibody responses of these specimens from COVID-19 positive donors are highly variable.
Fig. 5.
Signal to background ratio of the nCoV(i) antigen for the 131 human serum specimens included in the clinical analysis that were from donors positive for SARS-CoV-2 by RT-PCR, sorted from lowest to highest signal to background ratio.
4. Discussion
The availability of an accurate, precise, sensitive and specific multiplexed antibody quantification assay for COVID-19 vaccine development is an important tool for vaccine manufacturers to enhance and speed up pre-clinical and clinical trials and to ultimately aid in the delivery of a safe and effective vaccine. Much of the development of antibody-based tests for COVID-19 has focused on the development of qualitative ELISA and lateral flow-type tests for applications in diagnostics and seroprevalence to assess an individual’s antibody response post-infection. In contrast, this assay was specifically developed to provide information regarding antibody response for the wide variety of almost 200 COVID-19 vaccines currently under development (Milken Institute, 2020; Amanat and Krammer, 2020). The VaxArray CoV SeroAssay capture antigens represent not only different forms of the spike protein from SARS-CoV-2, including full-length spike, receptor binding domain (RBD), and the S2 extracellular domain, but also includes spike proteins from a variety of other endemic and potentially pandemic coronaviruses as outlined in Table 1 to enable comprehensive analysis of the polyclonal antibody responses produced post-vaccination or post-infection.
The VaxArray CoV SeroAssay is quantitative with a linear response over a wide range of concentration. In addition, further dilution of the starting specimen can easily enable samples above the linear dynamic range to be effectively quantified. It is difficult to compare analytical sensitivity of the VaxArray CoV SeroAssay to the wide variety of available IgG ELISA assays for measuring antibodies to SARS-CoV-2 based on published data, as most of these assays are qualitative and are not intended for quantitative analysis. As a result, performance metrics for other IgG immunoassays focus on clinical sensitivity and specificity and typically do not report analytical sensitivity. Limits of quantification of the VaxArray CoV SeroAssay are comparable to other fluorescence- and chemiluminescence-based immunoassays that are typically in the sub- to low-ng/mL range (Zhang et al., 2013). This sensitivity is clearly adequate for measuring antibody responses in human serum specimens after natural infection, as evidenced by the data summarized in Table 4 , in which all 263 serum samples were diluted 100-fold in Protein Blocking Buffer prior to analysis. For the 131 known positive specimens in this dataset, 80 % of the specimens had a signal close to fluorescence saturation on the nCoV(i) capture sequence (a signal exceeding 60,000 RFU with fluorescence saturation occurring at 65535 RFU).
Table 4.
Positive and Negative Percent Agreement between VaxArray CoV SeroAssay for the detection of SARS-CoV-2 compared to a Mixed Reference Method.a
| Positive Percent Agreement (PPA) |
Negative Percent Agreement (NPA) |
||||
|---|---|---|---|---|---|
| TP/(TP + FN) | % | 95 % CI (LCL – UCL) | TN/(TN + FP) | % | 95 % CI (LCL-UCL) |
| 129/(129 + 2) | 98.5 % | 94.6–99.6 | 132/(132 + 0) | 100 % | 97.2−100 |
Reference method was RT-PCR of donor-matched specimen or date of collection. Sample set included 263 deidentified specimens (260 serum, 3 plasma). Results shown with associated 95 % Confidence Intervals (Wilson method).
Using monoclonal antibodies, we demonstrated the VaxArray CoV SeroAssay shows the expected specific response for 6 of the 9 antigens, with the monoclonal antibody to SARS-CoV-1 (CR3022) producing signal on both the nCoV(ii) RBD antigen and the SARS antigen (SARS-CoV-1 spike protein). This is not surprising given that SARS-CoV-1 and SARS-CoV-2 share a cellular receptor and have significant sequence homology (Ye et al., 2020). The SARS-CoV-2 monoclonal antibody (Genetex) bound to both the nCoV(i) full-length spike protein and nCoV(iii) S2 extracellular domain. Importantly, though, neither of these antibodies bound to any of the human endemic coronavirus antigens on the array. As there were no specimens available from donors previously infected with COVID-19 but known to be negative for previous infections with all 4 endemic coronaviruses (likely due to the very high proportion of seropositivity to some or all of the endemic CoVs in the human population) (Gorse et al., 2010), examining specificity of the polyclonal antibody response in humans in this manner is difficult. However, in our analysis of 132 human serum specimens from COVID-19 negative donors, no samples produced a signal to background value exceeding the threshold for the nCoV(i) or nCoV(ii) antigens. Thirteen (13) samples produced a signal to background value exceeding the threshold for nCoV(iii), indicating some cross-reactivity of COVID-19 negative human serum on this antigen. Only one of the 13 specimens from COVID-19 negative donors that produced signal on nCoV(iii) was from a pediatric patient. In further analyzing pediatric specimens from COVID-19 negative donors, we note that a variety of responses to the endemic CoVs are present as shown in Fig. 4. As expected, younger children in general tend to have antibodies that bind to some of the 4 endemic coronaviruses, whereas older children and adolescents often show antibodies to all 4 in most cases. This is consistent with reports that seroconversion increases with age, with most adults showing seroconversion to all 4 human CoVs (Gorse et al., 2010).
Importantly, in a pre-clinical or clinical trial for a candidate COVID-19 vaccine, specimens can be analyzed prior to vaccine administration to determine a baseline response to each of the antigens on the array. Therefore, serum reactivity to the nCoV(iii) antigen pre-vaccination can be accounted for by this baseline response. This response can then be compared to the response as a function of time post-vaccination, and any changes in reactivity to all 9 antigens can be quantitatively assessed to provide a broader profile of the antibodies produced after vaccination. That this can be accomplished in a single test with a 2 -h turnaround time means that serology analysis during a clinical trial can be completed in less time and for less cost.
In a set of experiments involving 216 replicate analyses, the VaxArray CoV SeroAssay demonstrated good precision and accuracy, as shown in Table 3. The data represented a single user testing over 3 days and on 3 unique lots of microarray slides (a total of 6 unique slides per lot). Within a single slide of 8 replicates, the average % CV of the measurements was 6 %, indicating excellent precision on a single slide. This %CV ranged from 7 % to 19 % for the 9 antigens, averaging over all 3 days and all 3 lots. A higher %CV representing day-to-day and lot-to-lot variation is reasonable given the variables represented. As shown in Table 3, nCoV(iii) demonstrated the highest variation. This was due to a single lot of microarray slides (lot 1) that produced a higher than expected 25 % CV variation for this antigen, whereas lots 2 and 3 produced only 10 % and 13 % CV, respectively. Interestingly, however, the other 7 capture antigens investigated in this study did not experience a higher %CV for lot 1. This single lot may have experienced a printing artifact for this antigen, and the absolute signals generated on this nCoV(iii) antigen for this study were only ∼2x LLOQ. Regardless of the root cause, printing optimization efforts are underway to improve the lot-to-lot consistency in performance of this antigen. Accuracy expressed as % recovery (% of expected result) was also quite good, ranging from 88 % to 97 % over the entire dataset of n = 216 replicates. Unsurprisingly, nCoV(iii) also suffered from the lowest accuracy on the same lot of slides showing lower than expected precision. The average accuracy of the nCoV(iii) result was 80 %, 85 %, and 100 % for lots 1, 2, and 3, respectively.
The clinical specimen analysis summarized in Table 4 indicates the VaxArray CoV SeroAssay has excellent positive and negative percent agreement compared to a mixed reference method of RT-PCR status for a matched donor specimen or collection date prior to the COVID-19 outbreak in late 2019. The cutoff established for the VaxArray CoV SeroAssay takes advantage of the unique multiplexed capability of the assay by using a multi-antigen approach to thresholding to increase the confidence in a positive or negative call. Specifically, the signal on the nCoV(i) antigen was used as a first ‘gate’ to a positive call, and the sum of the three nCoV antigens was used as a secondary ‘gate’ to a positive call. Both cutoffs had to be exceeded to make a positive call, providing additional confidence against false positive results. This thresholding methodology resulted in 98.5 % positive agreement (129/131) and 100 % negative agreement (132/132) with the mixed reference method. The two specimens that produced false negative results by the VaxArray CoV SeroAssay were obtained from Children’s Hospital of Colorado, and both were independently found to be negative by two alternative IgG-based ELISA assays, and both had very low titers by a virus neutralization assay (data not shown). All orthogonal analyses were conducted by Children’s Hospital of Colorado, and InDevR staff analyzing the clinical specimens by the VaxArray CoV SeroAssay were blinded to these results. These additional serological analyses indicate concordance with the VaxArray CoV SeroAssay results, likely indicating that either the donors produced little to no antibody response after infection, or that the associated RT-PCR results were false positives.
As an additional comparison, we also analyzed the PPA and NPA with the mixed reference method for both the Euroimmun and Epitope Diagnostics ELISAs that were run on the Children’s Hospital of Colorado samples. For the Euroimmun ELISA, PPA of 90.6 % and NPA of 94.6 % were obtained for the analysis of 119 samples. For the Epitope Diagnostics ELISA, PPA of 84.4 % and NPA of 94.6 % were obtained for the analysis of 151 samples. Both assays showed reduced performance and a higher incidence of both false negatives and false positives as compared to the VaxArray CoV SeroAssay.
To convey the benefit of using this multi-antigen diagnostic approach for the VaxArray CoV SeroAssay over a single antigen approach such as that used in a standard singleplex ELISA, we also compared the PPA and NPA for that would result if each of the nCoV antigens on the CoV SeroAssay were assessed separately. In this case, nCoV(i) resulted in PPA of 98.5 % and NPA of 100 %, nCoV(ii) resulted in PPA of 96.2 % and NPA of 100 %, and nCoV(iii) produced the biggest difference from the multi-antigen approach with PPA of 83.2 % and NPA of 91.7 %. These data highlight that for this dataset, the performance of nCoV(i) alone produces the same PPA and NPA as the multi-antigen approach. In addition, these data highlight the previously mentioned cross-reactivity of the nCoV(iii) antibody in specimens known to be COVID-19 negative. While these data highlight that a multi-antigen approach can produce more optimal performance than analysis of a single antigen response in a diagnostic algorithm, we note that there is also value in the ability to examine individual responses to each individual capture agent for assessment of comparative binding of vaccine antigens.
To highlight the underlying quantitative response generated for these clinical serum specimens, Fig. 5 shows the signal to background ratio produced on the nCoV(i) capture antigen for all 131 serum specimens from patients known to be COVID-19 positive by RT-PCR, sorted from lowest to highest. Given that all specimens were analyzed at the same 1:100 dilution and the analytical data indicating the quantitative ability of the assay, these data are indicative of relative SARS-CoV-2 antibody concentrations. The two clinical specimens that produced false negative results described above are the first two datapoints in the lower left closest to the origin, with corresponding signal to background ratios of 0.63 and 0.68. The remainder of these data show that a wide range of antibody responses were observed in the positive specimens, highlighting the quantitative capabilities of the assay for assessing antibody response in COVID-19 vaccine pre-clinical and clinical trials. In addition, tools such as the VaxArray CoV SeroAssay could easily be used to correlate severity of disease with antibody titer produced, and for a wide variety of other SARS-CoV-2 applications to add to our current understanding.
5. Conclusion
Developers and manufacturers of candidate COVID-19 vaccines face the daunting challenge of bringing a safe and effective vaccine to market in record time to put a halt to the current global pandemic. As such, tools that empower developers and manufacturers to conduct vaccine clinical trials efficiently and to obtain the maximum amount of information in a rapid turnaround time are critical. The collective studies presented herein demonstrate that the VaxArray CoV SeroAssay is one of those tools, offering excellent analytical performance in terms of limits of quantification, high precision and accuracy and high clinical sensitivity and specificity. While most of the data herein demonstrated applicability to measurement of IgG antibodies in human serum, applicability to other animal models such as mouse or non-human primates is readily enabled using alternative anti-species label antibodies. We hope this tool will be utilized to maximize information content in the critical effort of delivering a safe and effective SARS-CoV-2 vaccine in record time.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Erica D. Dawson: Project administration, Supervision, Methodology, Resources, Formal analysis, Writing - original draft, Visualization. Laura R. Kuck: Methodology, Investigation, Validation. Rebecca H. Blair: Conceptualization, Methodology, Resources, Writing - review & editing. Amber W. Taylor: Supervision, Methodology, Formal analysis, Visualization, Writing - review & editing. Evan Toth: Investigation, Writing - review & editing. Vijaya Knight: Resources, Writing - review & editing. Kathy L. Rowlen: Conceptualization, Project administration, Resources, Writing - review & editing.
Declaration of Competing Interest
E. Dawson, K. Rowlen, and L. Kuck are stockholders of InDevR, Inc. E. Dawson, K. Rowlen, R. Blair, A. Taylor, and E. Toth are employed by InDevR, Inc. L. Kuck was employed by InDevR, Inc. at the time the work described was conducted. V. Knight has no declarations of interest.
Acknowledgements
We acknowledge specimens received under a materials transfer agreement from Dr. Scott Dessain at Lankenau Institute for Medical Research (Wynnewood, PA), specimens received from Icahn School of Medicine at Mount Sinai under a license agreement, and specimens from Dr. Aleta Bonner at Veritas, PA (Belton, TX).
References
- Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmieier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Flerer D.S., Lugo L.A., Kojic E.M., Stoever J., Liu S.T.H., Cunningham-Rundles C., FElgner P.L., Moran T., Garcia-Sastre A., Caplivski D., CHeng A.C., Kedzlerska K., Vapalahti O., Hepojoki J.M., Simon V., Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans nat. Medicine. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos M., Tavaziva G., Abidi S., Campbell J., Haraoui L.-P., Johnston J., Lan Z., Law S., MacLean E., Trajman A., Menzies D., Benedetti A., Khan F. Diagnostic accuracy of serological tests for COVID-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant J., Azman A., Ferrari M., Arnold B., Boni M., Boum Y., Hayford K., Luquero F., Mina M., Rodriguez-Barraquer I., Wu J., Wade D., Vernet G., Leung D. Serology for SARS-CoV-2: apprehensions, opportunities, and the path forward. Science Immun. 2020;5 doi: 10.1126/sciimmunol.abc6347. eabc6347. [DOI] [PubMed] [Google Scholar]
- Byrne-Nash R., Gillis J., Miller D., Bueter K., Kuck L., Rowlen K. A neuraminidase potency assay for quantitative assessment of neuraminidase in influenza vaccines. Npj Vaccines. 2019;3:1–10. doi: 10.1038/s41541-019-0099-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassaniti I., Novazzi F., Giardina F., Salinaro F., Sachs M., Perlini S., Bruno R., Mojoli F., Baldanti F. Performance of VivaDiag COVID-19 IgM/IgG rapid test is inadequate for diagnosis of COVID-19 in acute patients referring to the emergency room department. J. Med. Virol. 2020:1–4. doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C., Tse H., Wong S., Woo P., Lau S., Chen L., Zheng B., Huang J., Yuen K. Examination of seroprevalence of coronavirus HKU1 infection with S protein-based ELISA and neutralization assay against viral spike pseudotyped virus. J. Clin. Virol. 2009;45:54–60. doi: 10.1016/j.jcv.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Li X., Li S., Xiao Y., Ye M., Yan X., Li X. How related is SARS-CoV-2 to other coronaviruses? Vet. Record. 2020;185(15):496. doi: 10.1136/vr.m1452. [DOI] [PubMed] [Google Scholar]
- Cohen, E. Coronavirus antibody tests have’ really terrible’ accuracy, researcher says. CNN online, https://www.cnn.com/2020/04/28/health/coronavirus-antibody-tests-terrible/index.html, Accessed 30 July 2020.
- Food and Drug Administration, Policy for Coronavirus Disease-2019 Tests during the Public Health Emergency (Revised), issued on the web May 11, 2020 and accessed via FDA website 30 July 2020.
- Foundation for Innovative New Diagnostics, SARS-CoV-2 Diagnostic Pipeline, accessed 18 August 2020 at: https://www.finddx.org/covid-19/pipeline.
- Francesco N. Is antibody-dependent enhancement playing a role in COVID-19 pathogenesis? Swiss Med. 2020;150 doi: 10.4414/smw.2020.20249. [DOI] [PubMed] [Google Scholar]
- Gorse G., Patel G., Vitale J., O’Connor T. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin. Vaccine Immunol. 2010;17(12):1875–1880. doi: 10.1128/CVI.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Yang Y. The potential danger of suboptimal antibody responses in COVID-19 nat. Rev. Immun. 2020;20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns Hopkins University Center for Systems Science and Engineering (CSSE) at https://coronavirus.jhu.edu/map.html (2020), accessed 26 February, 2021.
- Kuck L., Byrne-Nash R., Gillis J., Bueter K., Couzens L., Eichelberger M., Rowlen K. VaxArray for hemagglutinin and neuraminidase potency testing of influenza vaccines. Vaccine. 2018;36:2937–2945. doi: 10.1016/j.vaccine.2018.04.048. [DOI] [PubMed] [Google Scholar]
- Lerner A.M., Eisinger R.W., Lowy D.R., Petersen L.R., Humes R., Hepburn M., Cassetti M.C. Meeting report: the COVID-19 serology studies workshop: recommendations and challenges. Immunity. 2020;53:1–5. doi: 10.1016/j.immuni.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Kahn R., Mina M. Antibody testing will enhance the power and accuracy of COVID-19-prevention trials. Nature Med. 2020;26:814–821. doi: 10.1038/s41591-020-0887-3. [DOI] [PubMed] [Google Scholar]
- Milken Institute, COVID-19 Treatment and Vaccine Tracker, Accessed 18 August 2020 at: https://covid-19tracker.milkeninstitute.org/.
- Peeples L. Avoiding pitfalls in the pursuit of a COVID-19 vaccine. Proc. Nat. Acad. Sci. U. S. A. 2020;117(15):8218–8221. doi: 10.1073/pnas.2005456117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Pre-existing Immunity to SARS-CoV-2: The Knowns and Unknowns. Nature Rev. Immun. 2020;20:457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theel E., Slev P., Wheeler S., Couturier M., Wong S., Kadkhoda K. The role of antibody testing for SARS-CoV-2: is there one? J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R., Lee H., Ko J., Pittet M. COVID-19 diagnostics in context. Sci. Trans. Med. 2020;12 doi: 10.1126/scitranslmed.abc1931. eabc1932. [DOI] [PubMed] [Google Scholar]
- Ye Z.-W., Yuan S., Yuen K.-S., Fung S.-Y., Chan C.-P., Jin D.-Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020;16:1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Garcia-D’Angeli A., Brennan J., Huo Q. Predicting detection limits of enyzme-linked immunosorbent assay (ELISA) and bioanalytical techniques in general. Analyst. 2013;139:439–445. doi: 10.1039/c3an01835k. [DOI] [PubMed] [Google Scholar]