Highlights
-
•
NSE expression was positively correlated with distant metastasis of SCLC.
-
•
NSE promotes migration, invasion and EMT process of SCLC cells.
-
•
Wnt/β-catenin signaling pathway is activated in the NSE-overexpressing SCLC cells.
-
•
NSE interacts with β-catenin and inhibits the degradation of β-catenin.
Keywords: Small cell lung cancer, Neuron-specific enolase, Metastasis, Epithelial-mesenchymal transition, Wnt/β-catenin
Abstract
Neuron-specific enolase (NSE) has been used as a specific biomarker for small cell lung cancer (SCLC) patients. Nevertheless, the biological function and mechanism of NSE in SCLC are still unclear. In this study, we clarified the role of NSE in the progression of SCLC and found that NSE expression was positively correlated with distant metastasis. Functional analysis showed that overexpression of NSE promoted migration and invasion of SCLC cells. Mechanism analysis showed that NSE overexpression induced epithelial-mesenchymal transition (EMT) of SCLC cells. Moreover, overexpression of NSE increased the protein expression of β-catenin and its downstream target genes, and silencing β-catenin eliminated NSE-mediated cell migration, invasion and EMT process. Furthermore, NSE interacted with β-catenin and inhibited the degradation of β-catenin. Besides, the animal experiments also indicated that NSE could promote the EMT process and distant metastasis of SCLC cells in vivo. In summary, our results revealed that NSE could promote the EMT process of SCLC cells by activating the Wnt/β-catenin signaling pathway, thereby promoting cell migration, invasion and distant metastasis, which might serve as a potential target for the therapy of SCLC patients.
Background
Lung cancer is the most common cancer worldwide and the leading cause of cancer-related death [1]. SCLC is the most malignant lung cancer type, and accounts for 10–25% of lung cancers [2], [3], [4], [5]. Chemotherapy and radiation therapy are the main treatments for patients with SCLC [4]. In addition, there are targeted therapy and immune checkpoint inhibitor therapy for partial SCLC patients [5,6]). However, the 5-year survival rate of SCLC is still less than 7%, and the main reasons for treatment failure are high aggressiveness and distant metastasis [2]. Therefore, we urgently need to clarify the molecular mechanism of distant metastasis of SCLC and provide novel and effective ideas for the treatment of SCLC patients.
NSE is an important tumor marker of SCLC, and its serum concentration was closely correlated to tumor progression, recurrence and treatment effect of SCLC patients [7], [8], [9], [10]. There has been considerable research concerning NSE as a biomarker in tumors [11,12]. However, the effect and molecular mechanism of NSE in promoting distant metastasis of SCLC are still unclear. In addition, our previous research also found that knocking down NSE can inhibit the proliferation and migration of SCLC cells [13]. Therefore, we would like to further study whether NSE can promote SCLC cell migration, invasion and distant metastasis, and to clarify its molecular mechanisms.
In this study, we proved that NSE could promote migration, invasion and EMT process of SCLC cells in vitro, and promote tumor metastasis in vivo. Mechanism analysis revealed that the Wnt/β-catenin pathway was activated by up-regulating the expression of β-catenin and inhibiting the protein degradation of β-catenin in NSE-overexpressing SCLC cells, and played a key role in promoting EMT, migration and invasion of SCLC cells. All in all, our research revealed that NSE promoted invasion, migration, EMT process and distant metastasis by activating Wnt/β-catenin pathway, which might make contribution to preventing the metastases of SCLC patients.
Materials and methods
Clinical data information and ethical statement
The clinical data of SCLC was gathered from Guangzhou First People's Hospital. We then analyzed the association between NSE concentration and the distant metastasis of SCLC. This study was approved by the Research Ethics Committee of Guangzhou First People's Hospital. And all the patients were informed and agreed to participate in this study.
Cell lines and cell culture
The human SCLC cell lines H209 (ATCCⓇ HTB-172™), H446 (ATCCⓇ HTB-119™) and H69 (ATCCⓇ HTB-171™) were obtained from Genetimes ExCell Technology (Shanghai, China) and maintained in RMPI-1640 supplement with 10% fetal bovine serum (FBS) at 37 °C in 5% CO2. H69 is from lung tissue and H446 is derived from pleural effusion and H209 is derived from bone marrow of SCLC patients.
Plasmids and establishment of stable cells
The CDS sequence of human NSE and β-catenin gene were cloned into pSIN-3 × Flag lentiviral vector. NSE and β-catenin shRNA sequence were inserted into the PLKO vector. The shRNA targeting sequences are listed in Supplementary Table 1. All lentiviruses were produced on HEK293T cells. H446 and H69 cells were infected with lentivirus in the presence of polybrene, and selected with puromycin to establish stably transfected cells. In addition, a lentivirus with a luciferase gene was used to infect SCLC cells for animal experiments.
Western blot analysis
All cells were collected and lysed in 100 μl lysis buffer with protease inhibitor cocktail. BCA Protein Assay Kit (Thermo Fisher Scientific, USA) was used to measure the total protein concentration. Equal proteins were separated into a 10% SDS-PAGE gel and then transferred to a PVDF membrane (Millipore, Germany). The membranes were blocked for 1 h using 5% no-fat milk and then incubated overnight in 4 °C with antibodies. Then the membranes were incubated with secondary antibodies at room temperature for 1 h. Relative expression of proteins was detected using a GE Amersham Imager 600 system. Anti-E-cadherin (AF0131), anti-N-cadherin (AF4039), anti-SNAIL (AF6032), anti-c-MYC (AF0358) were purchased from Affinity Biosciences. Anti-SNAI2 (12,129–1-AP) and anti-β-actin (60,008–1-Ig) were purchased from Proteintech. Anti-NSE (8171), anti-β-catenin (8480) were purchased from CST. Image J software was used for detection and quantitative analysis.
Wound-healing assay
Cells were seeded on a 6-well plate and were grown to get 90% confluence. Wounds were created by using 10 µl pipette tips to scratch a straight line. The wound areas were imaged at 0 h, 24 h under a microscope. Image J software (NIH, USA) was used to calculate the wound areas [14,15].
Transwell assay
The cell migration and invasion assays were applied by using 12 well plates with 8 µm inserts (Corning, NY, USA). For tumor cell invasion assay, the inserts were coated with Matrigel (BD, USA) at 37 °C overnight. 5 × 10^5 cells in 200 µl serum free medium were seeded into the inserts. A volume of 600 µl medium with 10% FBS was added to the lower chamber. After being incubated at 37 °C, the migration or invasion cells were fixed by 4% formaldehyde, stained with 0.1% crystal violet and then being counted under a microscope.
Real-time quantitative PCR analysis (qRT-PCR)
Total RNA of cells or xenograft cancer tissues was extracted by using Trizol (Invitrogen, USA). cDNA was synthesized from isolated RNA by using HiScript II 1st Strand cDNA Synthesis Kit (Vazyme biotech, Nanjing, China). 18 s was used as internal control. ChamQ Universal SYBR qPCR Master Mix (Vazyme biotech, Nanjing, China) was applied for RT-qPCR to measure the levels of mRNAs. The tests were performed on a Roche LightCyclerⓇ 96 system and were repeated in triplicate. The sequences of the primers were shown in Supplementary Table 2, which were synthesized by Sangon Biotech (Shanghai, China). The expression of mRNAs was calculated using the 2−ΔΔCt method.
GSEA
Gene Set Enrichment Analysis (GSEA; http://www.broadinstitute.org/gsea/index.jsp) was a computational method, which was used to assess whether a gene set showed a statistical difference between two biological conditions. [16,17] GSEA was performed to investigate the enriched pathways between NSE low expression group and NSE high expression group based on GSE43580 dataset.
Co-immunoprecipitation (Co-IP)
Cells were lysed using a low-salt IP buffer containing cocktails. The supernatants were obtained by high speed centrifugation. The corresponding antibody was added to the supernatant and incubated overnight. Then the supernatants were added to protein A/G conjugated beads and incubated for 2 h. Finally, beads were collected and washed and analyzed by western blot assay.
Half-life analysis
Cycloheximide (CHX, Sigma) was added to the cell culture medium at a concentration of 20 μg/ml. After blocking protein synthesis, we collected cells for western blotting assay at 0,6,12,18,24 h. [18]
Immunohistochemistry
As mentioned earlier, the slides were incubated with the primary antibody (NSE, 1:100, Abcam, ab231114) overnight after antigen retrieval. Then diaminobenzidine (DAB) was used for immunohistochemical staining. After hematoxylin counterstaining, the slides were dehydrated and sealed. Finally, the images were collected under a 400X microscope.
Animal experiments
All the animal experiments were done according to the protocols approved by the Animal Care and Use Committee of South China University of Technology. Female Balb/c nude mice at the age of 6 weeks were purchased from Hunan SJA Laboratory Animal Co, Ltd. Cells were resuspended in PBS. A total of 10^5 SCLC cells in 100 µl PBS were injected into the left cardiac ventricle of anesthetized mice. The weights and survival information of mice were measured every week. The bioluminescence images analysis was conducted to evaluate SCLC distant metastasis by using an IVIS imaging system (Bruker MI SE) [19].
Statistics
All statistical analyses were performed using the SPSS 24.0 version and Graphpad prism 7. Data were presented as the mean ± SD of at least three independent experiments. The independent Student's t test was used to compare the differences between two groups. Univariate and multivariate survival analysis were performed by likelihood ratio test of stratified Cox proportional risk regression analysis. Kaplan–Meier survival plot was used to draw survival curves, and the log-rank test was used for comparison. Two-tailed p-values less than 0.05 are considered statistically significant.
Results
NSE expression was positively correlated with distant metastasis of SCLC
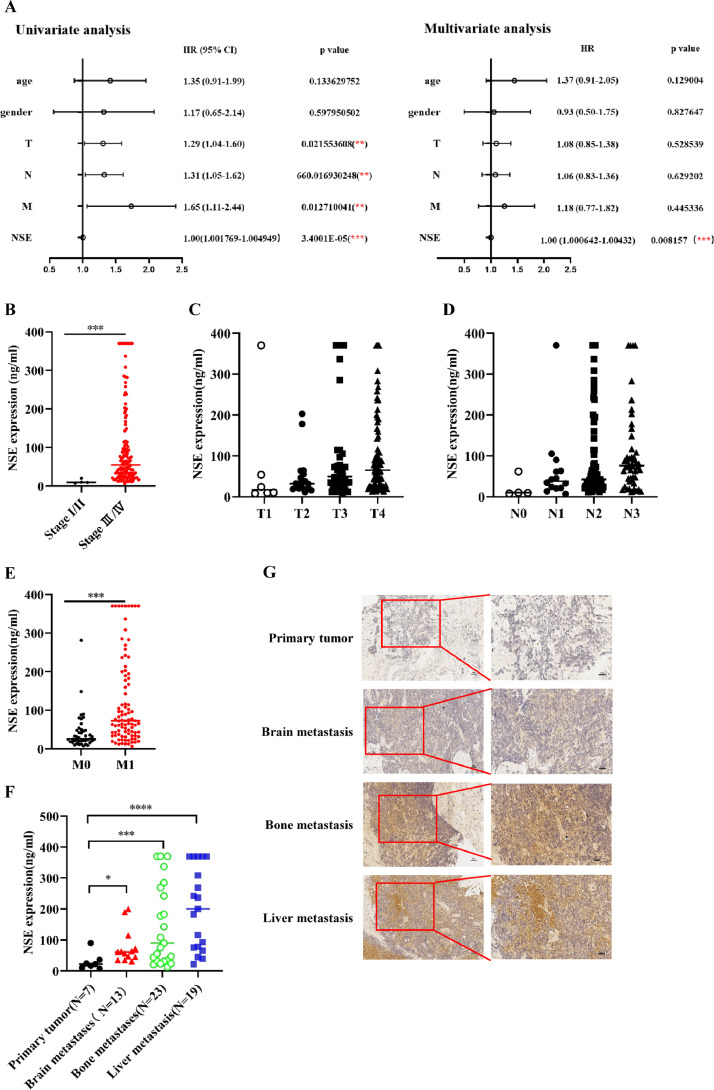
In order to clarify the biological function of NSE in SCLC, we firstly collected clinical data of 138 cases of SCLC patients, and defined serum NSE concentrations below 25 ng/ml as "low-NSE", while serum NSE concentrations above 25 ng/ml as "high-NSE". Univariate and multivariate analysis results showed that the serum concentration of NSE was significantly correlated with the prognosis of SCLC patients. Moreover, NSE can be used as an independent indicator of the prognosis for SCLC patients (p < 0.05, Fig. 1A). In addition, the serum concentration of NSE was significantly increased in SCLC with advanced stage and distant metastasis (p < 0.05, Fig. 1B–F). The results of immunohistochemistry also showed that the expression level of NSE was higher in SCLC patients with distant metastasis than patients without distant metastasis (Fig. 1G).
Fig. 1.
Upregulated expression of NSE was positively correlated with distant metastasis of SCLC patients. (A) The relationship between serum NSE concentration and the prognosis of SCLC patient was evaluated by univariate and multivariate COX regression analysis. The correlations between NSE serum concentration and TNM stage (B), T stage (C), N stage (D), and M stage (E) were evaluated. (F) The correlations between NSE serum concentration and different sites of metastasis. (G) The expression difference of NSE in SCLC with or without distant metastasis was detected by immunohistochemistry (Left scale bars: 50 um, Right scale bars: 25 um). Data is represented as mean ± SD. The significance level is represented by ns, p < 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
NSE promoted sclc cell migration and invasion
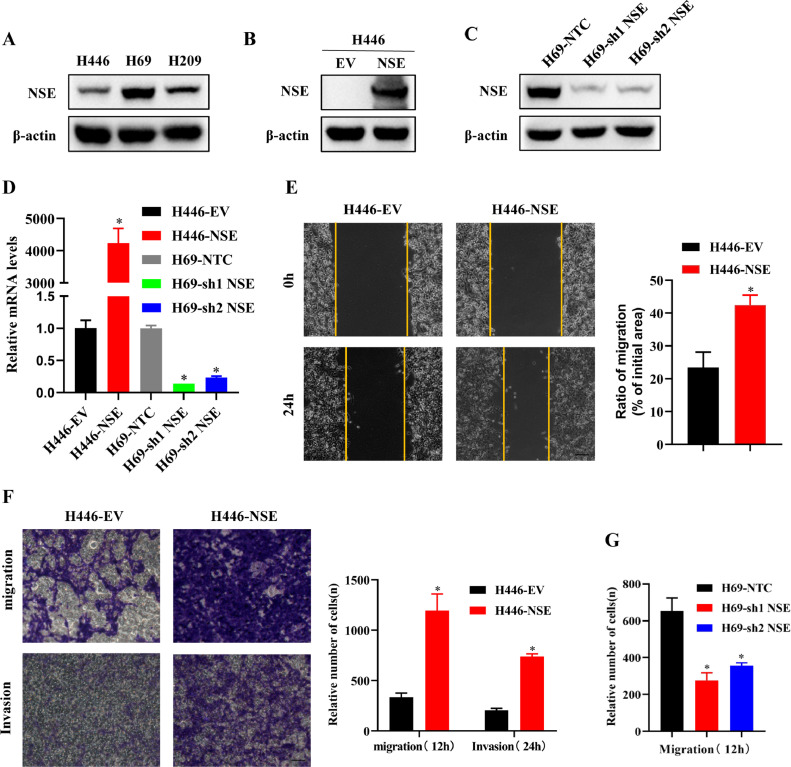
To further clarify the phenomenon that NSE promoting the distant metastasis of SCLC, we conducted a series of cell functional experiments to explore the influence of NSE on the migration and invasion of SCLC cells. The differences in the expression of NSE were detected in 3 SCLC cell lines (H446, H69 and H209). The results showed that the expression level of NSE was the lowest in H446 cells and the highest in H69 cells (Fig. 2A). Therefore, H446 and H69 cell lines were chosen for further experiments. The overexpression or knockdown of NSE in two cell lines was confirmed by western blot (p < 0.05, Fig. 2B, C) and qRT-PCR (p < 0.05, Fig. 2D).
Fig. 2.
NSE promoted migration and invasion in SCLC cells. (A) The protein expressions of NSE in different SCLC cell lines (H446, H69, H209) were detected by western blotting. (B–D) The efficiency of NSE overexpression or knockdown was verified by western blotting (B and C) and qRT-PCR (D). (E) migration ability of the H446 cells overexpressing NSE or control cells was detected by wound-healing assay (Scale bar: 100 um). (F) Transwell assay was used to analyze the migration and invasion capabilities of NSE-overexpressed H446 cells (Scale bar: 50 um). (G) The effect of NSE knockdown on the migration capability of H69 cells was analyzed by Transwell assay. The results are expressed as the mean ± SD of three independent experiments. *p < 0.05 using the two-sided Student's t-test.
Next, a wound healing assay was conducted to test the migration capability. The result showed that overexpression of NSE promoted cell migration in H446 cells (p < 0.05, Fig. 2E). Moreover, Transwell assay also showed that overexpression of NSE promoted cell migration and invasion in H446 cells (p < 0.05, Fig. 2F), while knockdown of NSE suppressed cell migration in H69 cells (p < 0.05, Fig. 2G).
These results indicated that NSE could enhance the migration and invasion capabilities of SCLC cells.
NSE induced EMT process of SCLC cells
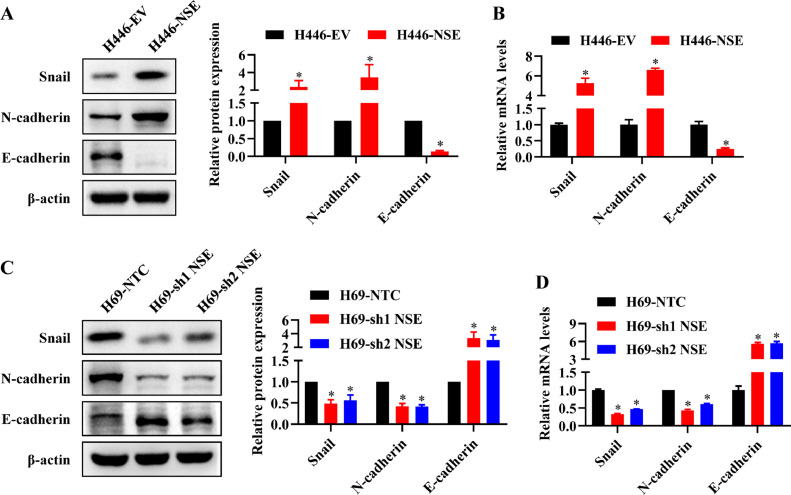
EMT phenotype has been shown to play a vital role in cell migration and invasion of cancer cells. Thus, we detected the expression levels of EMT-related markers to clarify whether NSE enhanced the migration and invasion ability of SCLC cells by promoting EMT process. The results from western blot and qRT-PCR demonstrated that higher expression of mesenchymal markers (N-cadherin, Snail) and lower expression of epithelial marker (E-cadherin) in NSE-overexpressing H446 cells compared control cells (p < 0.05, Fig. 3A, B). Meanwhile, knocking down NSE in H69 cells gave the opposite result (p < 0.05, Fig. 3C, D). These findings indicated that NSE could promote EMT process of SCLC cells.
Fig. 3.
NSE induced EMT in SCLC cells. (A, B) NSE overexpression promoted the EMT process of SCLC. (A) Western blot assay was performed to measure the protein expression levels of the EMT-related markers (Snail, N-cadherin and E-cadherin). (B) qRT-PCR was performed to measure the mRNA expression levels of the EMT markers. (C, D) NSE knockdown represses the EMT process of SCLC. Protein levels (C) and mRNA levels (D) of EMT-related markers were measured using western blot or qRT-PCR, respectively. These results were repeated of three independent experiments.
Wnt/β-catenin signaling pathway was activated in NSE-overexpressing sclc cells
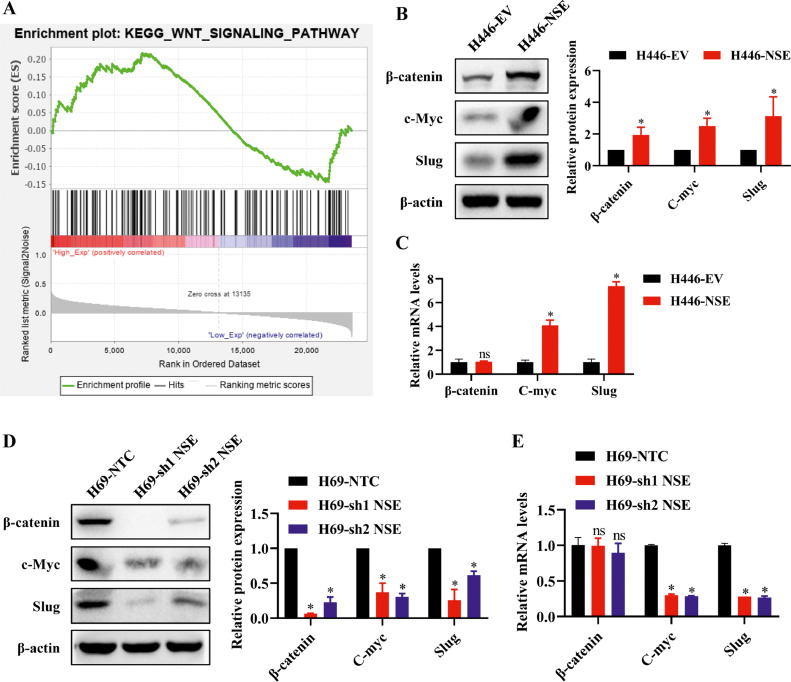
To explore the mechanism of NSE in promoting EMT process, we performed GSEA to identify the potential signaling pathway. There were several pathways enriched in high NSE groups, among them the Wnt/β-catenin pathway was mostly related to EMT phenotype (Fig. 4A). Based on this, we further detected the expression level of β-catenin and its downstream target genes in NSE-mediated EMT process of SCLC cells. The western blot results showed that NSE overexpression upregulated the level of β-catenin and its downstream target genes (c-myc, slug) in H446 cells (p < 0.05, Fig. 4B), while NSE knockdown restrained this effect in H69 cells (p < 0.05, Fig. 4D). As expected, the mRNA expression levels of C-myc and Slug were consistent with the western blot results, but modulation of NSE failed to change the mRNA expression level of β-catenin (Fig. 4C, E).
Fig. 4.
Wnt/β-catenin signaling pathway was activated in NSE-overexpressing SCLC cells. (A) Signal pathway enrichment investigation between high NSE expression group and low NSE expression group was enriched by GSEA. (B, C) NSE overexpression activated Wnt/β-catenin pathway and upregulated the expression of downstream target genes (c-Myc and Slug). The protein (B) and mRNA (C) expression of β-catenin, c-Myc and Slug in NSE-overexpressing H446 cells were tested by western blotting and qRT-PCR, respectively. (D, E) Silencing NSE repressed Wnt/β-catenin signaling pathway. The protein (D) and mRNA (E) expression of β-catenin, c-Myc and Slug in NSE-silencing H69 cells. Results was representative of three independent experiments. Data are presented as mean values ± SD. *p < 0.05 using the two-sided Student's t-test. ns: no significance.
These findings indicated that NSE could activate the Wnt/β-catenin signaling pathway. Moreover, NSE may only change the protein level of β-catenin but not its mRNA expression level.
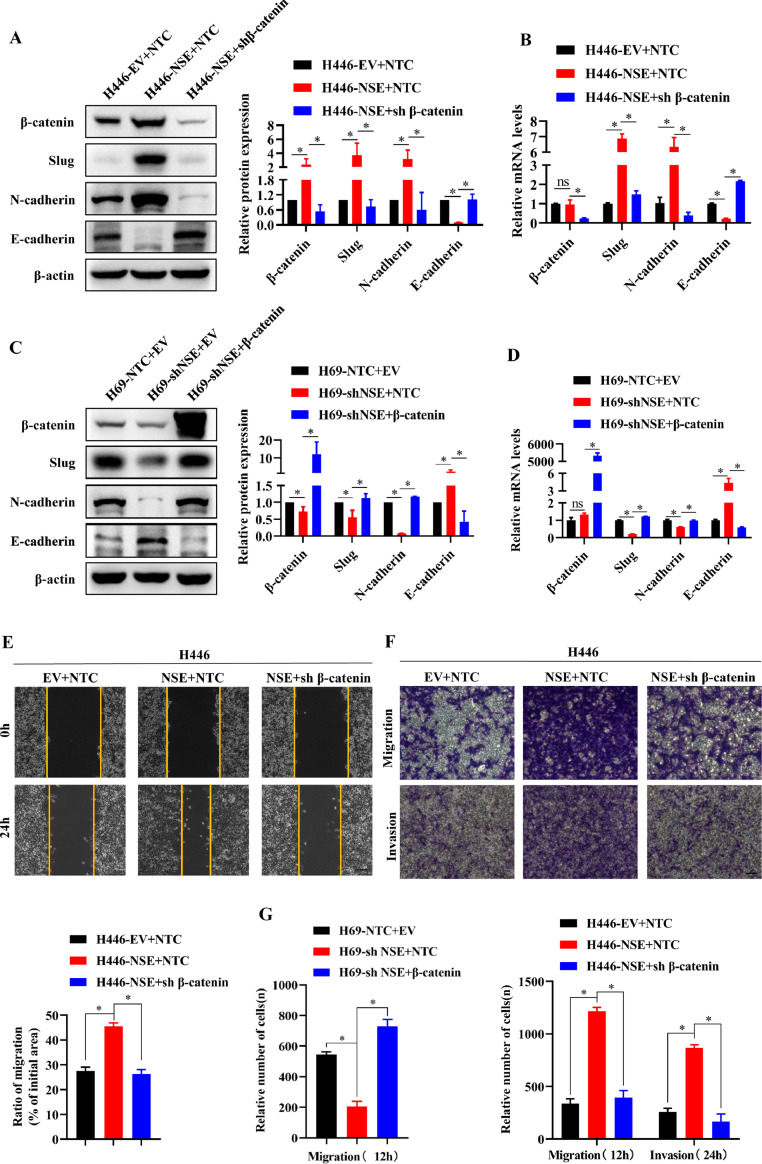
β-catenin was required in nse promoting EMT, migration and invasion
To further verify that NSE promotes the EMT, migration and invasion of SCLC cells by activating the Wnt/β-catenin signaling pathway, the β-catenin shRNA or control shRNA (NTC) was transfected into the NSE-overexpressing H446 cells and the Flag-β-catenin or control Flag-EV was transfected into the NSE-silencing H69 cells.
As shown in Fig. 5A and B, when β-catenin was suppressed by shRNA, the effect that overexpression of NSE activated the Wnt/β-catenin pathway and promoted the EMT process in H446 cells was eliminated (p < 0.05). As expected, when β-catenin was overexpressed, the effect that knockdown NSE inhibited the Wnt/β-catenin pathway and EMT process in H69 cells was abolished (p < 0.05, Fig. 5C, D). Furthermore, knocking down β-catenin abolished the enhancing migration and invasion of overexpression NSE in H446 cell (p < 0.05, Fig. 5E, F). Consistently, overexpression β-catenin eliminated the attenuating migration of knockdown NSE in H69 cell (p < 0.05, Fig. 5G).
Fig. 5.
NSE required β-catenin to promote migration, invasion, and EMT of SCLC cells. (A-D) Western blotting and qRT-PCR assay for the expression of β-catenin, slug, E-cadherin, and N-cadherin in NSE-overexpressing H446 cells in the presence of shβ-catenin (A, B) and in NSE-silencing H69 cells in the presence of β-catenin (C, D). (E) Wound healing assay for the migration of NSE-overexpressing H446 cells in the presence of shβ-catenin (Scale bar: 100 um). (F, G) Transwell assay for the invasion and migration of NSE-overexpressing H446 cells which were further infected with virus expressing shβ-catenin (F) and NSE-silencing H69 cells which were further infected with virus expressing β-catenin (G) (Scale bar: 50 um). Data are presented as mean values ± SD of three independent experiments. *p < 0.05 using the two-sided Student's t-test. ns: no significance.
In summary, these findings indicated that β-catenin played a vital role in NSE's promotion of EMT, migration and invasion of SCLC cells.
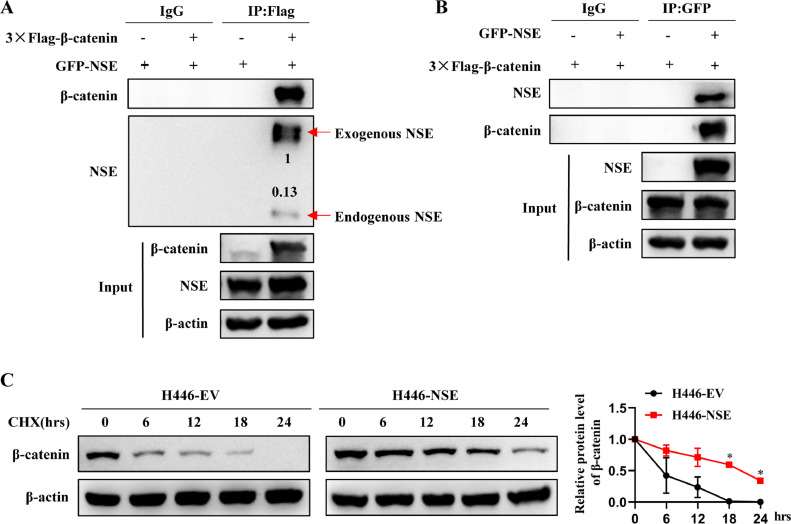
NSE interacted with β-catenin and inhibited its protein degradation
As mentioned earlier, NSE mainly regulates the protein level of β-catenin. We verified the interaction between NSE and β-catenin through Co-IP experiments (Fig. 6A, B). Moreover, overexpression of NSE inhibited the protein degradation of β-catenin in SCLC cells (p < 0.05, Fig. 6C). This indicated that NSE is likely to inhibit β-catenin protein degradation by interacting with β-catenin, thereby activating the Wnt/β-catenin pathway.
Fig. 6.
NSE interacted with β-catenin and inhibited its protein degradation. (A, B) H446 cells were transfected with Flag-β-catenin or GFP-NSE or both vectors. IP was performed with anti-Flag (A) or anti-GFP (B), followed by Western blot analysis. (C) H446-EV and H446-NSE cells were cultured in cell culture medium and treated with 20 μg/ml cycloheximide (CHX) for 0, 6, 12, 18, and 24 h, and then collected total cell for western blot analysis. Data are presented as mean values ± SD. *p < 0.05 using the two-sided Student's t-test.
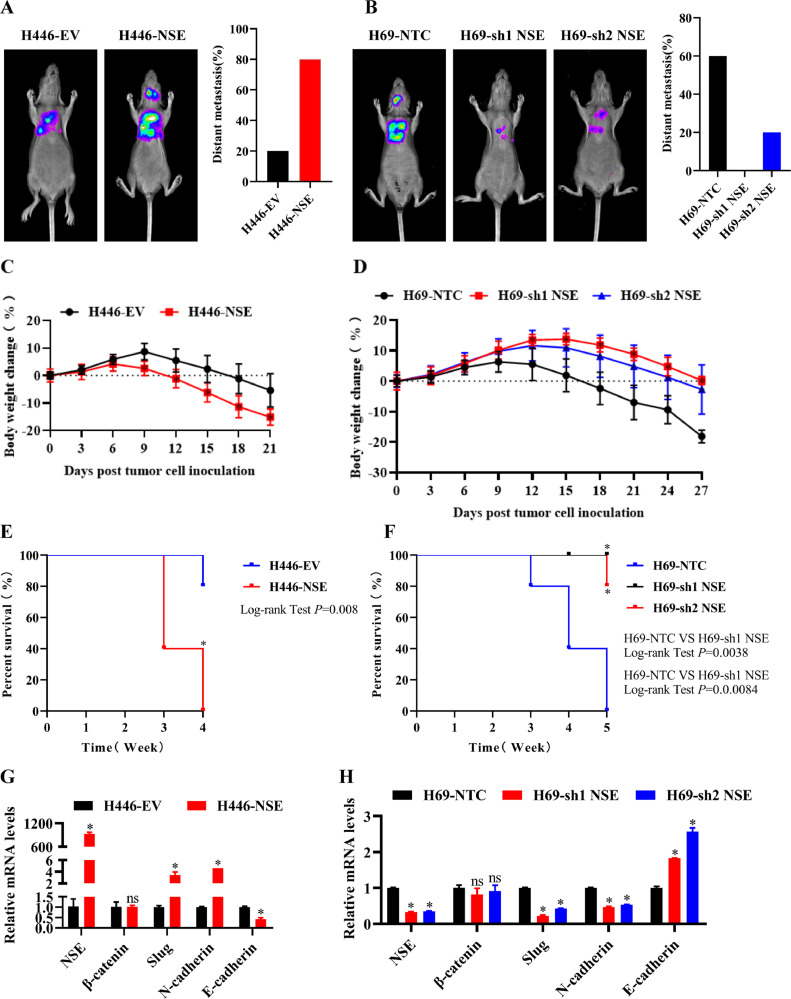
NSE promoted EMT process and tumor metastasis by activating the WNT/β-catenin pathway in vivo
Our above studies have confirmed that NSE can promote the migration, invasion and EMT process of SCLC cells by activating Wnt/β-catenin pathway in vitro. Therefore, we constructed a metastasis mouse model through cardiac injection to verify whether NSE promoted EMT process and distant metastasis of SCLC cells in vivo.
As shown in Fig. 7A, the distant metastasis rate was higher in the NSE-overexpressing group, compared with the control group. However, knockdown of NSE considerably reduced the distant metastasis rate in H69 cells (Fig. 7B). Meanwhile, overexpression NSE accelerated body weight loss of mice (p < 0.05, Fig. 7C) and showed disbenefit in animal survival (p < 0.05, Fig. 7E), while knockdown NSE decelerated body weight loss of mice (p < 0.05, Fig. 7D) and showed benefit in animal survival (p < 0.05, Fig. 7F). In addition, extracting RNA from tumor of mice, the qRT-PCR analyses shown that overexpression of NSE promoted EMT process in H446 cells (p < 0.05, Fig. 7G), while knockdown of NSE suppressed these effects in H69 cells (p < 0.05, Fig. 7H).
Fig. 7.
NSE promoted EMT and tumor metastasis by activating the Wnt/β-catenin pathway in vivo. H446 cells and H69 cells were transfected with Luciferase and GFP-NSE or sh1NSE or sh2NSE or vectors and then injected through the left cardiac ventricle of anesthetized nude mice. (A, B) The bioluminescence images analysis was conducted by using an IVIS imaging system in nude mice models of NSE-overexpressing H446 cells (A) and NSE-silencing H69 cells (B). (C, D) Body weight loss was observed in nude mice models of NSE-overexpressing H446 cells (C) and NSE-silencing H69 cells (D). (E, F) Kaplan–Meier plot of survival in the experiment of NSE-overexpressing group (E) and NSE-silencing group (F) (*p < 0.05, log-rank test). (G, H) qRT-PCR assay for the expression of NSE, β-catenin and EMT markers in tumor tissues of nude mice models of NSE-overexpressing H446 cells (G) and NSE-silencing H69 cells (H). Data are presented as mean values ± SD. *p < 0.05 using the two-sided Student's t-test. ns: no significance.
Taken together, our findings indicated that NSE could promote EMT process and tumor distant metastasis of SCLC cells in vivo.
Discussion
In this study, we found that high concentration of serum NSE was significantly correlated with metastasis in SCLC patients. Overexpression NSE enhanced migration and invasion of SCLC cells in vitro, and promoted tumor distant metastasis in vivo, while knockdown NSE inhibited these effects. Molecular mechanism studies revealed that NSE promoted migration, invasion, EMT and distant metastasis of SCLC cells by activating the Wnt/β-catenin pathway through its interaction with β-catenin.
NSE has always been used as a specific marker for the diagnosis and monitoring of treatment effects and recurrence in SCLC patients [[7], [8], [9],20]. However, the function and mechanism of NSE in SCLC were still unclear. Our previous research found that silencing NSE could inhibit the proliferation and migration of SCLC cells [13]. Our clinical data also indicated that high levels of serum NSE were positively correlated with tumor metastasis in SCLC patients. This prompted us to further explore the role and molecular mechanism of NSE in promoting SCLC cell migration, invasion and distant metastasis.
NSE has good sensitivity and specificity for SCLC diagnosis [21]. Nevertheless, the function of NSE to promote SCLC metastasis remained unclear. The main way for small cell lung cancer to develop distant metastasis is through the blood system. Therefore, we used the method of injecting tumor cells into the left ventricle of mice to construct a tumor metastasis model, which is also a common experimental method for studying tumor metastasis in many reports [22], [23], [24]. The disadvantage of this method is that it cannot evaluate the ability of tumor cells to enter the blood system from the tumor in situ, but it can evaluate the ability of tumor cells to colonize specific regions or organs and form metastases. The ability of tumor cells to colonize specific regions or organs is a prerequisite for tumors to form metastases. In addition, we evaluate the migration and invasion capabilities of tumor cells through the Transwell and Wound-healing Assay. Our research showed that overexpression NSE promoted migration, invasion and tumor metastasis of SCLC cells in vitro and vivo. Moreover, overexpression NSE promoted tumor progression, resulting in accelerated weight loss and shortened survival of mice, while knockout NSE inhibited these effects.
Overexpression NSE promoted EMT process of SCLC cells. EMT was an important part of cancer metastasis [25]. It was characterized by the transformation from epithelial phenotype to mesenchymal phenotype, accompanied by the down-regulation of epithelial markers and the up-regulation of mesenchymal markers [26], such as down-regulation of E-cadherin and up-regulation of N-cadherin, Vimentin, Snail and Slug ([27,28]). Recent studies shown that in many types of cancer, including lung cancer, distant metastasis of tumors was associated with EMT [29], [30], [31]. Consistent with these previous reports, we found that NSE-overexpressing SCLC cells showed the typical characteristics of EMT, with reduced expression of E-cadherin and increased expression of N-cadherin, Slug and Snail. However, silencing NSE significantly increased the expression of E-cadherin, and decreased the expression of N-cadherin, and Snail.
The Wnt /β-catenin pathway was activated in NSE-overexpressing SCLC cells and played a decisive role in distant metastasis of tumor. The Wnt/β-catenin pathway has been studied extensively [32], including a large number of studies on promoting EMT ([27], [33,34]). Specifically, β-catenin-mediated transcription could induce the expression of c-Myc and Slug [35], [36], [37]. c-Myc could promote EMT by up-regulating the expression of Snail. Snail could directly bind to the E-boxes of the E-cadherin promoter and inhibit the transcription of E-cadherin [38]. Slug was a close relative of Snail and could inhibit the transcription of E-cadherin, which led to decrease in cell adhesion during EMT [39]. Our study results showed that overexpression NSE could up-regulate the protein expression of β-catenin and its downstream genes c-myc, Snail and Slug, thereby promoting EMT in SCLC cells. On the contrary, knockdown NSE inhibits these effects. However, the mRNA level of β-catenin did not change significantly. The interaction between NSE and β-catenin inhibited the degradation of β-catenin protein, thereby increasing the protein level of β-catenin. Moreover, we found that silencing β-catenin eliminated the promotion effect of overexpression NSE in migration, invasion and EMT process of SCLC cells. These indicated that Wnt/β-catenin pathway was activated in NSE-overexpressing SCLC cells and played an indispensable role in NSE's promotion of migration, invasion and EMT process of SCLC cells.
In summary, our study clarified that NSE promoted cell invasion, migration, EMT process and distant metastasis by inhibiting the protein degradation of β-catenin and activating Wnt/β-catenin pathway in SCLC cells, and was a potential target for the therapy of SCLC patients.
Conclusion
This research uncovered the role of NSE in promoting distant metastasis of SCLC and clarified its molecular mechanism. Above all, our research shows that NSE is associated with tumor metastasis in clinical SCLC patients. In addition, NSE enhances the migration, invasiveness and metastasis of SCLC cells in terms of cell function. Finally, in the molecular mechanism, NSE interacts with β-catenin and inhibits the protein degradation of β-catenin. Furthermore, NSE can induce EMT of SCLC cells by up-regulating the protein level of β-catenin through inhibiting the protein degradation of β-catenin and activating the Wnt/β-catenin pathway, thereby promoting tumor metastasis, which provided a novel approach for preventing and treating distant metastasis of SCLC.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgments
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of Guangzhou First People's Hospital and the Institutional Animal Care and Use Committee of South China University of Technology.
Funding
This research was supported by the National Natural Science Foundation of China (Grant numbers 81672900), Guangzhou Planned Project of Science and Technology (Grant numbers 201904010427, 201707010262), Guangzhou General Science and technology project of Health and Family Planning (Grant number 20201A011007).
CRediT authorship contribution statement
Zhiqiang Zha: Methodology, Software, Validation, Formal analysis, Visualization, Writing - Original Draft. Dailing Li: Methodology, Software, Validation, Formal analysis, Writing - Original Draft. Peiling Zhang:Methodology, Software, Validation, Formal analysis. Peipei Wang: Investigation. Xisheng Fang: Investigation. Xia Liu: Investigation. Chengyin Weng: Investigation. Baoxiu Li: Investigation. Yong Wu: Investigation. Haibo Mao: Investigation. Lina Wang: Investigation. Lin Xu: Investigation. Mingmei Guan: Investigation. Jiaming Dong: Investigation. Lin Lu: Conceptualization, Supervision, Resources, Data Curation, Writing - Review & Editing, Project administration, Funding acquisition. Guolong Liu: Conceptualization, Supervision, Resources, Data Curation, Writing - Review & Editing, Project administration, Funding acquisition.
Acknowledgments
We gratefully acknowledge the Gene Expression Omnibus.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101039.
Contributor Information
Lin Lu, Email: eylinlv@scut.edu.cn.
Guolong Liu, Email: eyglliu@scut.edu.cn.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Karachaliou N., Pilotto S., Lazzari C., Bria E., de Marinis F., Rosell R. Cellular and molecular biology of small cell lung cancer: an overview. Transl. Lung Cancer Res. 2016;5(1):2–15. doi: 10.3978/j.issn.2218-6751.2016.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsoukalas N., Aravantinou-Fatorou E., Baxevanos P., Tolia M., Tsapakidis K., Galanopoulos M. Advanced small cell lung cancer (SCLC): new challenges and new expectations. Ann. Transl. Med. 2018;6(8):145. doi: 10.21037/atm.2018.03.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunn P.A., Minna J.D., Augustyn A., Gazdar A.F., Ouadah Y., Krasnow M.A. Small Cell Lung Cancer: can Recent Advances in Biology and Molecular Biology Be Translated into Improved Outcomes? J. Thorac. Oncol. 2016;11(4):453–474. doi: 10.1016/j.jtho.2016.01.012. Jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Meerbeeck J.P., Fennell D.A., De Ruysscher D.K. Small-cell lung cancer. Lancet. 2011;378(9804):1741–1755. doi: 10.1016/S0140-6736(11)60165-7. [DOI] [PubMed] [Google Scholar]
- 6.Waqar S.N., Morgensztern D. Treatment advances in small cell lung cancer (SCLC) Pharmacol. Ther. 2017;180:16–23. doi: 10.1016/j.pharmthera.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Pujol J.L., Quantin X., Jacot W., Boher J.M., Grenier J., Lamy P.J. Neuroendocrine and cytokeratin serum markers as prognostic determinants of small cell lung cancer. Lung Cancer. 2003;39(2):131–138. doi: 10.1016/s0169-5002(02)00513-5. [DOI] [PubMed] [Google Scholar]
- 8.Pujol J.L., Boher J.M., Grenier J., Quantin X. Cyfra 21-1, neuron specific enolase and prognosis of non-small cell lung cancer: prospective study in 621 patients. Lung Cancer. 2001;31(2–3):221–231. doi: 10.1016/s0169-5002(00)00186-0. [DOI] [PubMed] [Google Scholar]
- 9.Scagliotti G.V., Piani M., Gatti E., Gozzelino F., Albera C., Pozzi E. Combined measurements of neuron specific enolase and bombesin/gastrin releasing peptide in lung cancer. Eur. Respir. J. 1989;2(8):746–750. [PubMed] [Google Scholar]
- 10.Akoun G.M., Scarna H.M., Milleron B.J., Bénichou M.P., Herman D.P. Serum neuron-specific enolase. A marker for disease extent and response to therapy for small-cell lung cancer. Chest. 1985;87(1):39–43. doi: 10.1378/chest.87.1.39. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Z.F., Wang M., Xu J.L. Thymidine kinase 1 combined with CEA, CYFRA21-1 and NSE improved its diagnostic value for lung cancer. Life Sci. 2018;194:1–6. doi: 10.1016/j.lfs.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 12.Isgrò M.A., Bottoni P., Scatena R. Neuron-specific enolase as a biomarker: biochemical and clinical aspects. Adv. Exp. Med. Biol. 2015;867:125–143. doi: 10.1007/978-94-017-7215-0_9. [DOI] [PubMed] [Google Scholar]
- 13.Liu X., Liu S., Fu J., Huang J., Weng C., Fang X. Knockdown of neuron-specific enolase suppresses the proliferation and migration of NCI-H209 cells. Oncol. Lett. 2019;18(5):4809–4815. doi: 10.3892/ol.2019.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang W.C., Wong C.W., Liang P.P., Shi M., Cao Y., Rao S.T. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20(1):84. doi: 10.1186/s13059-019-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song P., Li Y., Dong Y., Liang Y., Qu H., Qi D. Estrogen receptor β inhibits breast cancer cells migration and invasion through CLDN6-mediated autophagy. J. Exp. Clin. Cancer Res. 2019;38(1):354. doi: 10.1186/s13046-019-1359-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J.P. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23(23):3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 18.Yang F., Xu J., Li H., Tan M., Xiong X., Sun Y. FBXW2 suppresses migration and invasion of lung cancer cells via promoting β-catenin ubiquitylation and degradation. Nat. Commun. 2019;10(1):1382. doi: 10.1038/s41467-019-09289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mustafa D.A.M., Pedrosa R., Smid M., van der Weiden M., de Weerd V., Nigg A.L. T lymphocytes facilitate brain metastasis of breast cancer by inducing Guanylate-Binding Protein 1 expression. Acta Neuropathol. 2018;135(4):581–599. doi: 10.1007/s00401-018-1806-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carney D.N., Marangos P.J., Ihde D.C., Bunn P.A., Cohen M.H., Minna J.D. Serum neuron-specific enolase: a marker for disease extent and response to therapy of small-cell lung cancer. Lancet. 1982;1(8272):583–585. doi: 10.1016/s0140-6736(82)91748-2. Jr. [DOI] [PubMed] [Google Scholar]
- 21.Molina R., Marrades R.M., Augé J.M., Escudero J.M., Viñolas N., Reguart N. Assessment of a combined panel of six serum tumor markers for lung cancer. Am. J. Respir. Crit. Care Med. 2016;193(4):427–437. doi: 10.1164/rccm.201404-0603OC. [DOI] [PubMed] [Google Scholar]
- 22.Zhuo W., Liu Y., Li S., Guo D., Sun Q., Jin J. Long noncoding RNA GMAN, up-regulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of Ephrin A1 by competitively binding GMAN-AS. Gastroenterology. 2019;156(3):676–691. doi: 10.1053/j.gastro.2018.10.054. [DOI] [PubMed] [Google Scholar]
- 23.Ren D., Yang Q., Dai Y., Guo W., Du H., Song L. Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-κB signaling pathway. Mol. Cancer. 2017;16(1):117. doi: 10.1186/s12943-017-0688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian H., Lian R., Li Y., Liu C., Liang S., Li W. AKT-induced lncRNA VAL promotes EMT-independent metastasis through diminishing Trim16-dependent Vimentin degradation. Nat. Commun. 2020;11(1):5127. doi: 10.1038/s41467-020-18929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol. 2018;13:395–412. doi: 10.1146/annurev-pathol-020117-043854. [DOI] [PubMed] [Google Scholar]
- 26.Liu L., Zhu H., Liao Y., Wu W., Liu L., Liu L. Inhibition of Wnt/β-catenin pathway reverses multi-drug resistance and EMT in Oct4 (+)/Nanog (+) NSCLC cells. Biomed. Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110225. [DOI] [PubMed] [Google Scholar]
- 27.Qu C., He D., Lu X., Dong L., Zhu Y., Zhao Q. Salt-inducible Kinase (SIK1) regulates HCC progression and WNT/β-catenin activation. J. Hepatol. 2016;64(5):1076–1089. doi: 10.1016/j.jhep.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Bae W.J., Lee S.H., Rho Y.S., Koo B.S., Lim Y.C. Transforming growth factor β1 enhances stemness of head and neck squamous cell carcinoma cells through activation of Wnt signaling. Oncol. Lett. 2016;12(6):5315–5320. doi: 10.3892/ol.2016.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pastushenko I., Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Yeh H.W., Hsu E.C., Lee S.S., Lang Y.D., Lin Y.C., Chang C.Y. PSPC1 mediates TGF-β1 autocrine signalling and Smad2/3 target switching to promote EMT, stemness and metastasis. Nat. Cell Biol. 2018;20(4):479–491. doi: 10.1038/s41556-018-0062-y. [DOI] [PubMed] [Google Scholar]
- 31.Chockley P.J., Chen J., Chen G., Beer D.G., Standiford T.J., Keshamouni V.G. Epithelial-mesenchymal transition leads to NK cell-mediated metastasis-specific immunosurveillance in lung cancer. J. Clin. Invest. 2018;128(4):1384–1396. doi: 10.1172/JCI97611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang T., Ma Z., Liu L., Sun J., Tang H., Zhang B. DDX39 promotes hepatocellular carcinoma growth and metastasis through activating Wnt/β-catenin pathway. Cell Death. Dis. 2018;9(6):675. doi: 10.1038/s41419-018-0591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H., Lu X.X., Wang J.R., Yang T.Y., Li X.M., He X.S. TRAF6 inhibits colorectal cancer metastasis through regulating selective autophagic CTNNB1/β-catenin degradation and is targeted for GSK3B/GSK3β-mediated phosphorylation and degradation. Autophagy. 2019;15(9):1506–1522. doi: 10.1080/15548627.2019.1586250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y.R., Wang J.L., Xu C., Li Y.M., Sun B., LY Yang. HEG1 indicates poor prognosis and promotes hepatocellular carcinoma invasion, metastasis, and EMT by activating Wnt/β-catenin signaling. Clin. Sci. 2019;133(14):1645–1662. doi: 10.1042/CS20190225. [DOI] [PubMed] [Google Scholar]
- 35.Dai Y., Liu L., Zeng T., Liang J.Z., Song Y., Chen K. Overexpression of MUC13, a poor prognostic predictor, promotes cell growth by activating wnt signaling in hepatocellular carcinoma. Am. J. Pathol. 2018;188(2):378–391. doi: 10.1016/j.ajpath.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 36.Conacci-Sorrell M., Simcha I., Ben-Yedidia T., Blechman J., Savagner P., Ben-Ze'ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J. Cell Biol. 2003;163(4):847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J., Weinberg R.A. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 2008;14(6):818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000;2(2):84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 39.Hajra K.M., Chen D.Y., Fearon E.R. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62(6):1613–1618. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.