Abstract
Lung adenocarcinoma (LUAD) is the most common subtype of lung cancer. Nevertheless, the detailed molecular mechanisms of the progression of LUAD remain largely unknown. The present bioinformatics analysis reported that FAM83A and FAM83A-AS1 were upregulated in LUAD tissues and associated with prognosis in patients with LUAD. The purpose of the current study was to investigate the role of FAM83A and its antisense long non-coding (lnc)RNA FAM83A-AS1 in LUAD. Gene Expression Profiling Interactive Analysis was used to screen for potential oncogenes in LUAD and to analyze the clinical significance of FAM83A and FAM83A-AS1. Small interfering RNAs were constructed and transfected into LUAD cells to knock down the expression of FAM83A and FAM83A-AS1. EdU, Cell Counting Kit-8, Transwell and Matrigel assays were performed to detect the proliferation, migration and invasion of LUAD cells. The interaction between FAM83A-AS1, microRNA (miR)-495-3p and FAM83A was explored using a luciferase reporter assay. FAM83A and FAM83A-AS1 were both overexpressed in LUAD tissues compared with adjacent normal tissues. High expression of FAM83A and FAM83A-AS1 predicted worse survival and more advanced clinical stage. Knockdown of FAM83A or FAM83A-AS1 could inhibit the proliferation, migration and invasion of LUAD cells. Moreover, lncRNA FAM83A-AS1 regulated the expression of FAM83A by functioning as competing endogenous RNA for miR-495-3p. These results implicated that FAM83A and FAM83A-AS1 both played oncogenic roles in LUAD and FAM83A-AS1 could regulate the expression of FAM83A by sponging miR-495-3p. The study revealed a novel regulatory mechanism of tumor development in LUAD and FAM83A and FAM83A-AS1 may be novel biomarkers and therapeutic targets for LUAD.
Keywords: FAM83A, FAM83A-AS1, miR-495-3p, lung adenocarcinoma
Introduction
Lung cancer is the most common malignancy worldwide and one-quarter of all cancer-associated deaths are due to lung cancer globally (1). As the predominant subtype of lung cancer, non-small cell lung cancer (NSCLC) is furtherly divided into adenocarcinoma and squamous cell carcinoma, of which lung adenocarcinoma (LUAD) is the most common type and comprises ~40% of all lung cancer globally in 2020 (2). Although treatments for LUAD are plentiful, including surgery, radiotherapy, chemotherapy, targeted therapy and immunotherapy, the prognosis of patients with LUAD remains poor, with a 5-year overall survival rate <20% globally in 2020 (3). Thus, exploring novel genes functioning in the development of LUAD is important and may provide potential targets for the diagnosis and treatment of LUAD.
The Gene Expression Profiling Interactive Analysis (GEPIA) website (http://gepia.cancer-pku.cn/) is an online tool for visually analyzing gene expression in all types of cancer. The database was explored to seek for potential oncogenes in LUAD. FAM83A belongs to the FAM83 family of proteins, which are characterized by a highly conserved domain of unknown function (4). It has been demonstrated that FAM83 proteins exhibit oncogenic properties and have significantly elevated levels of expression in multiple human tumor types, including breast, lung, bladder, testis and ovarian cancer (5). Further research showed that ablation of numerous FAM83 proteins resulted in a marked suppression of cancer-associated signaling and loss of tumorigenic potential (6). Therefore, it is important to study the function and mechanism of FAM83A in LUAD.
Long non-coding (lnc)RNAs are a type of transcript that are >200 nucleotides in length and with limited or no protein-coding capacity (7,8). It has been reported that lncRNAs exert vital roles in cancer, including cell cycle procession, apoptosis, drug resistance and stemness maintenance (9,10). Gene-editing tools to target lncRNA have already shown the feasibility of gene editing in treating cancer (11). FAM83A-AS1 is the anti-sense lncRNA of FAM83A, and the association of FAM83A-AS1 with the development of LUAD was analyzed by GEPIA.
The aim of the present study was to explore the roles of FAM83A and FAM83A-AS1 in LUAD. Public databases were analyzed and experiments were performed to uncover the expression and function of FAM83A and FAM83A-AS1 in LUAD. The oncogenic role of FAM83A and FAM83A-AS1 might provide diagnostic markers and therapeutic targets for patients with LUAD.
Materials and methods
Bioinformatics
The GEPIA website (http://gepia.cancer-pku.cn/) was used to screen for potential oncogenes in LUAD and to analyze the correlation between gene expression and clinical characteristics. The differentially expressed genes in LUAD were obtained by searching the ‘Differential Expression Analysis’ section of GEPIA website (http://gepia2.cancer-pku.cn/#degenes) with the following terms: Cancer name, LUAD; |log2 fold-change (FC)|≥1; q-value ≤0.01 and differential methods, ANOVA. The top 500 differential survival genes in LUAD were obtained by searching the ‘Most Differential Survival Genes’ section of GEPIA website (http://gepia2.cancer-pku.cn/#survival) with the following terms: Cancer name, LUAD and group cut-off, median. The Kaplan-Meier curves were downloaded from the ‘Survival Analysis’ section of GEPIA website (http://gepia2.cancer-pku.cn/#survival) with the following terms: Gene, FAM83A or FAM83A-AS1; group cut-off, median and cancer name, LUAD. The staging boxplots were downloaded from the ‘Expression DIY-Stage Plot’ section of GEPIA website (http://gepia2.cancer-pku.cn/#analysis) with the following terms: Gene, FAM83A or FAM83A-AS1 and cancer name, LUAD. The median expression (transcripts per million) of genes in normal and LUAD tissue was obtained from ‘Differential Expression Analysis’ section of GEPIA website (http://gepia2.cancer-pku.cn/#degenes). The hierarchical clustering map in Fig. 1B was drafted with these data using GraphPad Prism version 6 (GraphPad Software). The Kaplan-Meier curves were downloaded from the ‘Survival Analysis’ section of GEPIA website (http://gepia2.cancer-pku.cn/#survival) with the following terms: Gene, FAM83A or FAM83A-AS1; group cut-off, median and cancer name, LUAD. The violin plots in Fig. 2B and E were downloaded from the ‘Expression DIY-Stage Plot’ section of GEPIA website (http://gepia2.cancer-pku.cn/#analysis) with the following terms: Gene, FAM83A or FAM83A-AS1 and cancer name, LUAD. The boxplots in Figs. 3A and 4A were downloaded from the CCLE dataset (https://portals.broadinstitute.org/ccle) by searching for FAM83A or FAM83A-AS1. The Ensembl genome browser (http://asia.ensembl.org/index.html) was scanned to observe the genomic location of FAM83A and FAM83A-AS1. Gene clusters highly correlated with FAM83A and FAM83A-AS1 (Pearson score >0.3) were submitted to the database for Annotation, Visualization and Integrated Discovery Bioinformatics resources version 6.8 (http://david.abcc.ncifcrf.gov/) (12) for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, and GO terms and KEGG pathways with a gene count ≥3 were included in subsequent analysis. The mRNA expression data of FAM83A and FAM83A-AS1 in different types of cancer cells was downloaded from the CCLE dataset (https://portals.broadinstitute.org/ccle). The histograms in Figs. 3B and 4B were constructed using these data and GraphPad. The secondary structure of FAM83A-AS1 was downloaded from the HUGO Gene Nomenclature Committee (http://www.genenames.org/) (13). The cBioPortal website (http://www.cbioportal.org/) (14) was utilized to analyze the correlation between FAM83A and FAM83A-AS1. LncBase version 2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php) (15) and StarBase version 2.0 (http://starbase.sysu.edu.cn/starbase2/browseNcRNA.php) (16) were browsed to identify the miRNAs that bind FAM83A and FAM83A-AS1.
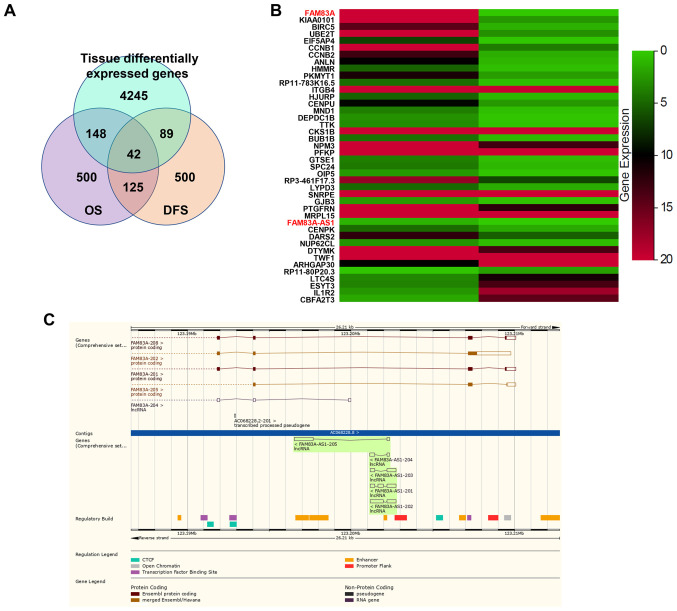
Figure 1.
Gene Expression Profiling Interactive Analysis website predicts that FAM83A and FAM83A-AS1 are potential oncogenes in LUAD. (A) Venn Diagram shows that 42 genes are differentially expressed and associated with OS and DFS in LUAD. (B) FAM83A was the most differentially expressed gene in these 42 genes and he antisense lncRNA of FAM83A, FAM83A-AS1. (C) Genomic location of FAM83A and FAM83A-AS1 in Ensembl genome browser. LUAD, lung adenocarcinoma; OS, overall survival; DFS, disease-free survival.
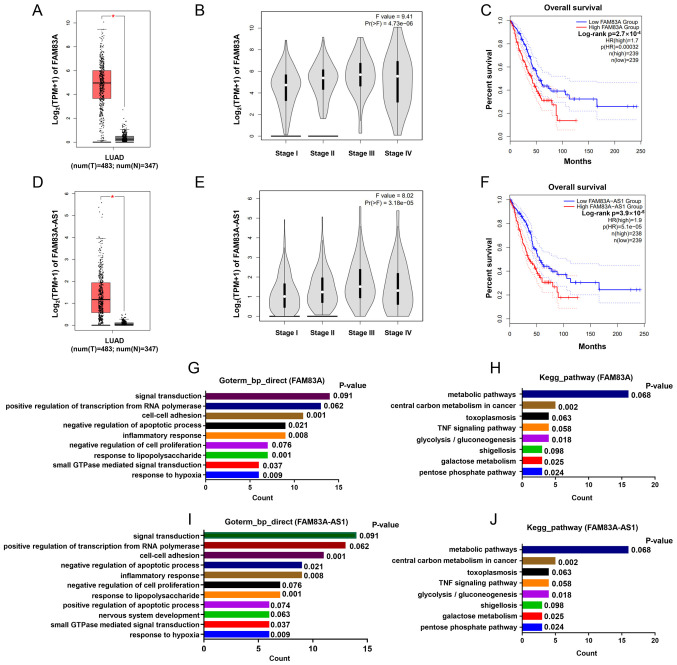
Figure 2.
FAM83A and FAM83A-AS1 are both overexpressed in LUAD and relevant to tumor stage and patients' survival. (A) FAM83A overexpressed in LUAD. (B) Tissues with more advanced stages exhibit higher expression of FAM83A. (C) Patients in the high FAM83A group showed worse OS compared with those of low FAM83A group. (D) FAM83A-AS1 is overexpressed in LUAD. (E) Tissues with more advanced stages exhibit higher expression of FAM83A-AS1. (F) Patients of high FAM83A-AS1 group show worse OS compared with those of low FAM83A group. Enrichments of (G) GO and (H) KEGG for FAM83A. Enrichments of (I) GO and (J) KEGG for FAM83A-AS1 bear strong similarities in biological function. *P<0.05. LUAD, lung adenocarcinoma; OS, overall survival; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
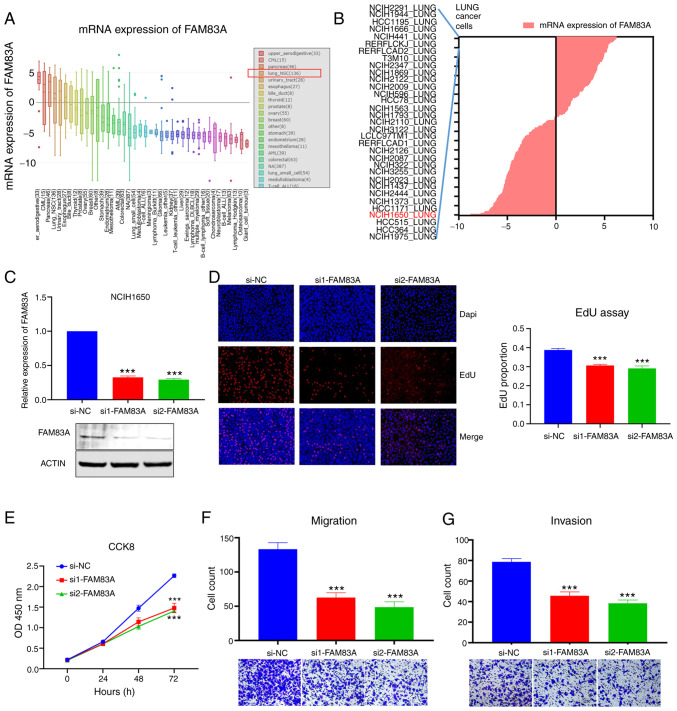
Figure 3.
Knockdown of FAM83A inhibits the malignant phenotype of LUAD in NCIH1650 cells. (A) Expression plot FAM83A in multiple cancer cell lines. (B) Expression of FAM83A in some lung_NSC cells and the common LUAD cell line NCIH1650 was chosen for further research. (C) Transfection efficiency of siRNAs were confirmed using reverse transcirption-quantitative PCR and western blotting. (D) EdU (magnification, ×400) and (E) CCK-8 assays indicated that silencing FAM83A suppressed proliferation of NCIH1650 cells. (F) Transwell (×400) and (G) Matrigel (magnification, ×400) assays suggested that knockdown of FAM83A inhibited migration and invasion of NCIH1650 cells. ***P<0.001 vs. si-NC. lung_NSC, non-small cell lung cancer; CCK-8, Cell Counting Kit-8; si, small interfering; NC, negative control.
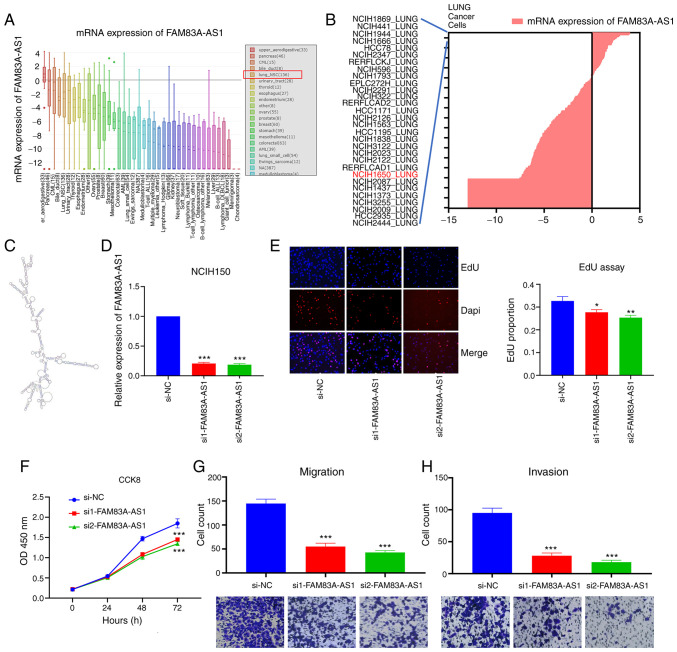
Figure 4.
Knockdown of FAM83A-AS1 inhibits the malignant phenotype of LUAD in NCIH1650 cells. (A and B) Expression of FAM83A-AS1 in multiple cancer cell lines and NCIH1650 cells. (C) Predicted secondary structure of FAM83A-AS1. (D) Two siRNAs effectively downregulated the expression of FAM83A-AS1 in NCIH1650 cells. (E and F) Silencing FAM83A-AS1 suppressed proliferation of NCIH1650 cells (EdU magnification ×400). (G and H) Knockdown of FAM83A-AS1 inhibited migration and invasion of NCIH1650 cells (magnification ×400). *P<0.05; **P<0.01; ***P<0.001. si, small interfering; NC, negative control.
Cell culture and transfection
NCIH1650 and A549 cells was purchased from the Shanghai Institutes for Biological Science and were cultured in RPMI1640 (Nanjing KeyGen Biotech Co., Ltd.) medium containing 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.) in a humidified incubator containing 5% CO2 at 37°C. LUAD cells were seeded in 6-well plates with a density of 5×105 cells per well. Two small interfering (si)RNAs targeting FAM83A/FAM83A-AS1, miR-495-3p mimics and miR-495-3p inhibitors were constructed and purchased from Guangzhou RiboBio Co., Ltd, and si-NC or miR-NC (non-targeting) was used as a control. Transfection of siRNA and miRNA mimics was performed in room temperature for 6 h according to the Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) protocol. For 6-well plates, 5 µl siRNAs (20 nM) or miRNAs mimics (20 nM) were transfected in each well. Subsequent experiments were performed after 48 h of transfection. The rescue experiments were performed by transfecting siRNA of FAM83A/FAM83A-AS1 and miR-495-3p inhibitor. The sequence of siRNAs and miRNA mimics were as follows: si-NC, forward: 5′-UUCUCCGAACGUGUCACGUTT-3′and reverse: 5′-ACGUGACACGUUCGGAGAATT-3′; si1-FAM83A, forward: 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse: 5′-ACGUGACACGUUCGGAGAATT-3′; si2-FAM83A, forward: 5′-UGAACUUCUCCCGGAUUUGTT-3′ and reverse: 5′-CAAAUCCGGGAGAAGUUCATT-3′; si1-FAM83A-AS1, forward: 5′-GCUGCCACCUACAAGAUAATT-3′ and reverse: 5′-UUAUCUUGUAGGUGGCAGCTT-3′; si2-FAM83A-AS1, forward: 5′-GGCCCUGGGCUGAAUAAUUTT-3′ and reverse: 5′-AAUUAUUCAGCCCAGGGCCTT-3′; miR-NC, 5′-UUCUCCGAACGUUCACGUTT-3′; miR-495-3p mimics, 5′-AGGAUGUCUAAAUGUUUGUUA-3′ and miR-495-3p inhibitor, 5′-AAGAAGUGCACCAUGUUUGUUU-3′.
RNA extraction and reverse transcription-quantitative (RT-q)PCR
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract the total RNAs from cells transfected as aforementioned. cDNAs were obtained by reverse transcription using a Reverse Transcription kit (cat. no. RR036A; Takara Bio, Inc.) at 37°C for 15 min and 85°C for 5 min. RT-qPCR was conducted with SYBR Select Master mix (cat. no. 4472908; Applied Biosystems; Thermo Fisher Scientific, Inc.) and the relative expression was calculated using the 2−ΔΔCq method (17), with ACTB as a reference gene. The thermocycling conditions for RT-qPCR were as follows: Initial denaturation, 95°C for 30 sec; denaturation, 95°C for 5 sec, 60°C for 35 sec, for a total of 40 cycles; annealing and extension, 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. The primers used are shown in Table SI.
Western blotting
For protein extraction, cells transfected as aforementioned were lysed in RIPA lysis buffer with freshly added protease inhibitor PMSF (1:100; Nanjing KeyGen Biotech Co., Ltd.). Protein concentration was determined using a BCA kit (Nanjing KeyGen Biotech Co., Ltd.) and 30 µg of lysate protein was loaded per lane for western blotting. Briefly, protein extracts were loaded on 10% SDS-PAGE gels for electrophoresis and then were electrotransferred to a PVDF membrane. The membrane was blocked at room temperature with non-fat milk for 2 h and incubated overnight with respective primary antibodies at 4°C. The following primary antibodies were used: Anti-β-actin (1:1,000; cat. no. 3700; Cell Signaling Technology, Inc.) and anti-FAM83A (1:1,000; cat. no. ab214014; Abcam). Then secondary antibodies (goat anti-rabbit antibody, 1:10,000; cat. no. ab6721; Abcam, or goat anti-mouse antibody, 1:10,000; cat. no. ab6789; Abcam) were used to incubate the membrane for 2 h. Blots were visualized using ECL detection with an ECL Substrate kit (cat. no. ab133406; Abcam) and ImageJ (version 1.48; National Institutes of Health) software was used to quantify protein expression.
EdU assay
An EdU Apollo® 488 In Vitro Imaging kit (Guangzhou RiboBio Co., Ltd.) was used to detect proliferation rate of NCIH1650 and A549 cells. After transfection of 24 h, 8,000 cells/100 µl were plated in 96-well plates overnight and then incubated with 100 µl 50 µM EdU solution for 2 h. The cells were then fixed with 50 µl 4% paraformaldehyde at room temperature for 10 min and neutralized using 50 µl 2 mg/ml glycine. After permeabilization with 0.5% Triton X-100 for 20 min, the cells were stained for DAPI with 100 µl 1× Apollo solution and stained for Edu with 100 µl 1× Hoechst33342 solution, both at room temperature for 30 min. Images were captured using fluorescence microscopy with 40 times magnification and proliferation cells ratios were counted from three random fields of view.
Cell Counting Kit (CCK)-8 assay
The CCK-8 assay was performed to evaluate the relative number of NCIH1650 and A549 cells. After transfection of 24 h, 2,000 cells/100 µl were plated in 96-well plates and total of 20 µl CCK-8 reagent (Guangzhou RiboBio Co., Ltd.) was added to each test well and cells were then incubated for 2 h at 37°C. The absorbance at 450 nm was measured every 24 h.
Migration and invasion assay
Transwell assay inserts (8 µM PET, 24-well Millicell) and Matrigel-coated membranes (BD Biosciences) were used to detect the migration and invasion of cells, respectively. In total, 200 µl serum-free medium was added to the upper chamber and 800 µl 10% DMEM serum-containing medium to the lower chamber. Matrigel pre-coating was performed at 37°C for 30 min. The NCIH1650 and A549 cells were harvested 24 h after transfection and added to the upper chamber (50,000 cells). After incubation at 37 C for 24 h (migration assay) or 48 h (invasion assay), the migrated or invaded cells were fixed with 4% polyformaldehyde and stained with crystal violet, both at room temperature for 30 min. Images were captured with light microscopy and cells were counted by ImageJ software.
Luciferase reporter assay
A Dual Luciferase Reporter Assay system (Promega Corporation) was used for luciferase assays. The binding sites of FAM83A-AS1 with FAM83A 3′UTR were identified using StarBase as aforementioned. A fragment of the FAM83A-AS1 or FAM83A 3′UTR sequence containing a putative target site or mutated target site for miR-495-3p was amplified and cloned into the pGL3 reporter vector (Guangzhou RiboBio Co., Ltd.). The luciferase reporter plasmids were co-transfected miR-495-3p mimics or miR control (Guangzhou RiboBio Co., Ltd.) as aforementioned. After transfection for 48 h, Renilla and Firefly luciferase activities were detected in the dual luciferase reporter gene kit (Promega Corporation). Firefly/Renilla value was used to measure relative luciferase activity and results were normalized to the activity of the control. The sequences of miRNA mimic/inhibitor were as follows: miR-NC, 5′-UUCUCCGAACGUUCACGUTT-3′; miR-495-3p mimics, 5′-AGGAUGUCUAAAUGUUUGUUA-3′ and miR-495-3p inhibitor, 5′-AAGAAGUGCACCAUGUUUGUUU-3′
Statistical analysis
Generally experiments were repeated three times and data are presented as the mean ± standard deviation (unless otherwise shown). Unpaired student's t-tests were used to determine statistical significance between two groups. ANOVA test and Dunnett's post hoc were used for the comparison of multiple groups. Spearman's test was used for the correlation analysis of two genes. Graphs were made using the GraphPad Prism 6.0 software package. Statistical analysis was performed using SPSS version 20.0 (IBM Corp) and P<0.05 was considered to indicate a statistically significant difference.
Results
FAM83A and FAM83A-AS1 are potential oncogenes according to GEPIA analysis
To screen for genes abnormally expressed and associated with survival of patients with LUAD, the following filter criterion was used: Cancer name, LUAD; gene expression, |log2 FC|≥1, q-value ≤0.01 (tumor vs. normal); differential survival genes with overall survival (OS) rates (top 500) and disease-free survival (top 500) rates. Using this method, 42 genes were obtained (Fig. 1A). To sort these genes by differences in expression, it was demonstrated that FAM83A was the most differentially expressed gene and its antisense lncRNA, FAM83A-AS1, was also among the list (Fig. 1B). As shown in Ensembl genome browser, FAM83A-AS1 is the antisense of FAM83A and both have several different transcripts (Fig. 1C).
FAM83A and FAM83A-AS1 are correlated with clinical characteristics of LUAD
Further analysis of GEPIA indicated that FAM83A was significantly upregulated in LUAD (tumor, n=483 vs. normal, n=347) (Fig. 2A) and its expression was relatively higher in patients with LUAD with more advanced stages (Fig. 2B). Survival analysis showed that patients of high FAM83A (n=239) had a worse OS (P<0.001) compared paired with patients with low FAM83A expression (n=239) (Fig. 2C). Analogously, FAM83A-AS1 was also overexpressed in LUAD and associated with tumor stage and survival of patients (Fig. 2D-F). Moreover, to predict the potential biological function of FAM83A and FAM83A-AS1, genes highly co-expressed with these were submitted for GO and KEGG pathway analysis, respectively. Results suggested that FAM83A and FAM83A-AS1 bear strong similarities in biological processes, mainly including ‘cell-cell adhesion’ and ‘central carbon metabolism in cancer’ (Fig. 2G-J). FAM83A and FAM83A-AS1 both associated with clinical characteristics of LUAD and it is important to deeply study their roles in LUAD.
Knockdown of FAM83A inhibits proliferation, migration and invasion of LUAD cells
The CCLE dataset was searched to evaluate the expression of FAM83A in different cancer cells and the results indicated that the expression of FAM83A in NSCLC (lung_NSC) and multiple other types of cancer cell was elevated (Fig. 3A). In the list of lung_NSC cell lines with high expression of FAM83A, the common LUAD cell line, NCIH1650, has been used for research in a previous study (18) and was selected for subsequent experiments (Fig. 3B). To explore the biological role of FAM83A, two siRNAs targeting FAM83A were transfected into NCIH1650 and both siRNAs effectively silenced the expression of FAM83A, confirmed by RTq-PCR and western blotting (Fig. 3C). CCK-8 and EdU assays validated that knockdown of FAM83A inhibited proliferation of NCIH1650 cells (Fig. 3D and E). Transwell and Matrigel assays revealed that silencing FAM83A suppressed the ability of NCIH1650 cells to migrate and invade (Fig. 3F and G). Additionally, knockdown of FAM83A inhibited proliferation, migration and invasion of A549 cells (Fig. S1). These results suggested that FAM83A played a vital role in the carcinogenesis of LUAD.
Knockdown of FAM83A-AS1 inhibits the malignant phenotype of LUAD cells
The average expression of FAM83A-AS1 in lung_NSC cells was high, ranking 5th in all types of cancer (Fig. 4A). The expression of FAM83A-AS1 in NCIH1650 was also elevated compared with other common LUAD cell lines (Fig. 4B). FAM83A-AS1 is an antisense transcript and with limited ability to encode proteins (19,20). Two siRNAs were transfected to knock down the expression of FAM83A-AS1 in NCIH1650 cell and to further study its function (Fig. 4D). Consistently, silencing FAM83A-AS1 inhibited the proliferation, migration and invasion of NCIH1650 cells (Fig. 4E-H). Moreover, knockdown of FAM83A-AS1 also suppressed the proliferation, migration and invasion of A549 cells (Fig. S2). Based on these results, it was concluded that both FAM83A and FAM83A-AS1 played oncogenic roles in LUAD.
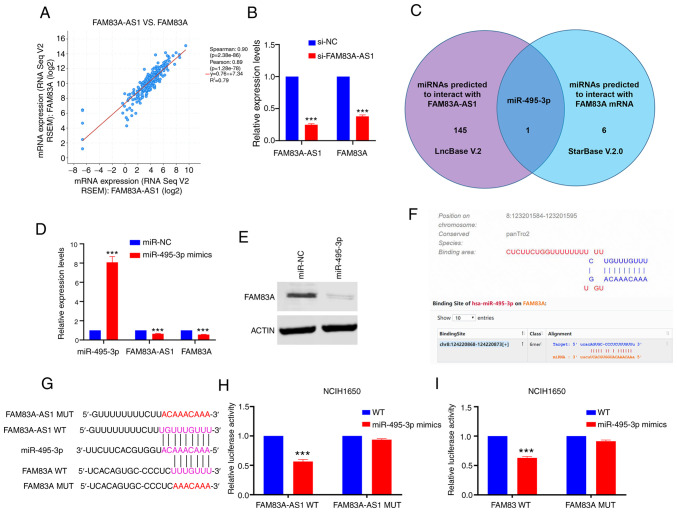
FAM83A-AS1 functions as a competing endogenous (ce)RNA to regulate FAM83A by sponging miR-495-3p
Previous studies have shown that numerous antisense lncRNAs exert biological roles via regulating its natural antisense transcript (21–23). Thus, it was hypothesized that FAM83A-AS1 might promote the progression of LUAD by influencing the expression of FAM83A. Using the cBioPortal website, it was reported that the expression of FAM83A-AS1 positively correlated with FAM83A (Spearman score=0.90; Fig. 5A). After knockdown of FAM83A-AS1, the expression of FAM83A was decreased (Fig. 5B). Accumulating evidence has highlighted the important roles of lncRNAs acting as ceRNAs in cancer development (24–26), and FAM83A-AS1 might regulate FAM83A via sponging specific miRNA. Using LncBase and StarBase, miR-495-3p was predicted to interact with both FAM83A-AS1 and FAM83A (Fig. 5C). Consistent with the assumption that FAM83A-AS1 might regulate FAM83A via sponging specific miRNA, treatment of miR-495-3p mimics significantly decreased mRNA levels of FAM83A-AS1 and FAM83A in NCIH1650 cells (P<0.001; Fig. 5D and E). To clarify their potential interactions, luciferase vectors (wild-type and mutant type) were constructed according to the putative binding sites (Fig. 5F and G). The luciferase reporter assay showed that ectopic expression of miR-495-3p decreased the luciferase activity of FAM83A/FAM83A-AS1 wild-type reporters, but not mutant type (Fig. 5H and I). Furthermore, the rescue experiment showed that miR-495-3p counteracted the effect of FAM83A and FAM83A-AS1 in NCIH1650 cells. The influence of FAM83A/FAM83A-AS1-knockdown on proliferation, migration and invasion in LUAD cells could be restored by miR-495-3p inhibitor (Fig. S3). These results indicated that FAM83A-AS1 functioned as a ceRNA through binding miR-495-3p, thereby decreasing the levels of FAM83A mRNA transcripts.
Figure 5.
FAM83A-AS1 functions as competing endogenous RNA to regulate FAM83A by sponging miR-495-3p. (A) Expression of FAM83A-AS1 positively correlated with the expression of FAM83A in LUAD. (B) Knockdown of FAM83A-AS1 downregulated the expression of FAM83A. (C) Predicted using LncBase and StarBase, miR-495-3p could interact with both FAM83A-AS1 and FAM83A. (D and E) miR-495-3p mimics significantly reduced the expression of FAM83A-AS1 and FAM83A. (F) Potential binding sites of miR-495-3p with FAM83A-AS1 and FAM83A. (G) WT and MUT FAM83A-AS1/FAM83A sequences were constructed according to the putative binding sites. (H and I) miR-495-3p decreased the luciferase activity of wild-type FAM83A-AS1 and FAM83A reporter plasmids. ***P<0.001 miRNA/miR, microRNA; NC, negative control; WT, wild-type; MUT, mutant.
Discussion
The poor prognosis of LUAD is largely due to that most cases are diagnosed in advanced stages and lose the opportunity of surgical treatment (27). Therefore, it is critical to identify novel molecular drivers of LUAD and thus to provide diagnostic markers and therapeutic targets for patients. The present study identified that coding gene FAM83A and lncRNA FAM83A-AS1 were both upregulated in LUAD tissues, and the overexpression of FAM83A and FAM83A-AS1 was associated with more advanced clinical stages and poorer prognosis of patients with LUAD. Further experiments indicated that knockdown of FAM83A or FAM83A-AS1 could inhibit proliferation, migration and invasion of LUAD. Moreover, it was confirmed that FAM83A-AS1 regulated the expression of its natural antisense transcript FAM83A by acting as a ceRNA through binding miR-495-3p.
FAM83A belongs to FAM83 family of proteins, which contain a conserved domain of unknown function, namely the DUF1669 domain (28). This family consists of eight members (A-H), and a study demonstrated that some of the FAM83 family proteins have oncogenic properties and have significantly elevated expression levels in multiple human tumor types (29), including cervical, prostate, liver and ovarian cancer. For example, FAM83B promotes endometrial cancer cell proliferation and invasion by inhibiting autophagy via activating the PI3K/AKT/mTOR pathway (30). FAM83D is associated with sex, American Joint Committee on Cancer stage, OS and DFS in hepatocellular carcinoma (31). FAM83H is involved in progression of osteosarcoma via stabilization of β-catenin (32). As for FAM83A, previous studies have indicated that FAM83A conferred EGFR-tyrosine kinase inhibitor resistance in breast cancer cells by interacting with and inducing phosphorylation of c-RAF and PI3K p85 (33). FAM83A expression is significantly elevated in human and murine pancreatic cancer and is essential for the proliferation and tumorigenesis of pancreatic cancer cells (34). In conformity with these studies, the present study identified the oncogenic role of FAM83A in LUAD and deepened the cognizance of FAM83 family proteins in cancer development.
lncRNAs are RNA molecules >200 nucleotides in length with limited ability to encode proteins and participate in the regulation of various biological processes, such as embryonic development, tissue differentiation and cell proliferation (35). As a type of lncRNA, natural antisense transcripts (NATs) are non-coding RNA sequences that are complementary to and overlap with either protein-coding or non-coding transcripts (36). NATs have diverse transcriptional and post-transcriptional regulatory mechanisms with a wide variety of biological roles, such as alternative splicing, chromatin remodeling and transcriptional regulation (37). In particular, NATs show potential for use as therapeutic targets because of their highly locus-specific effects (38). In the present study, FAM83A-AS1, the antisense transcript of FAM83A, was overexpressed in LUAD and involved in the malignant properties of LUAD cells. A previous review of studies identified the tumor-promoting function of FAM83A-AS1 in lung tumorigenesis, but the mechanism is still unclear (39,40). It has been demonstrated that NATs are abundant non-coding RNAs that regulate their corresponding sense mRNA through a variety of molecular mechanisms (41). For instance, receptor-type tyrosine-protein phosphatase ε (PTPRE)-AS1 binds to WD repeat-containing protein 5 and mediates H3 lysine 4 trimethylation of the PTPRE promoter region, thereby epigenetically activating PTPRE gene expression (42). NCK1-AS1 competed with NCK1 mRNA for miR-137 binding to boost NCK1 mRNA expression and thus to facilitate tumorigenesis and chemo-resistance in ovarian cancer (43). Metastasis-associated in colon cancer protein 1 (MACC1)-AS1 promoted MACC1 mRNA stability and expression through AMPK activation and subsequent Lin28 translocation from the nucleus to the cytoplasm (44). The most common mode of lncRNA function is as ceRNAs to sponge certain miRNAs, hence relieving repression of target mRNAs at a post-transcriptional level (45), and the present study validated that FAM83A-AS1 regulated the expression of FAM83A via a ceRNA network by sponging miR-495-3p. miR-495-3p act as a tumor suppressor in multiple cancer types (46–48). In lung cancer, miR-495-3p is associated with prolonged OS time of patients treated with nivolumab (49) and is a novel biomarker to predict radiotherapy response clinically (50). The FAM83A-AS1/miR-495-3p/FAM83A axis played important roles in the regulation of LUAD progression and the current results provided new evidence for the pathogenesis of LUAD. The limitation of the present study is the lack of an interaction network involving FAM83A-AS1 and miR-495-3p. Mapping the proposed network of ceRNAs will determine whether miR-495-3p is part of a wider ceRNA network. Additional studies should be conducted to further our understanding of the role of this ceRNA network in lung cancer.
In conclusion, the current study demonstrated that FAM83A and FAM83A-AS1 were both upregulated in LUAD and their expression is associated with advanced stages and poor prognosis of patients with LUAD. Moreover, knockdown of FAM83A or FAM83A-AS1 could inhibit proliferation, migration and invasion of LUAD and FAM83A-AS1 regulated the expression of FAM83A via sponging miR-495-3p. These results highlighted that FAM83A and FAM83A-AS1 are promising novel targets for the diagnosis and therapy of patients with LUAD.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The study was funded by The National Natural Science Foundation for Youth of China (grant no. 81902354), The Science and Technology Program of Xu Zhou (grant no. KC18037), The Xuzhou Clinical Technology Key Research Project (grant no. 2019GG021) and The Excellent Talents Fund Project of Xuzhou Medical University (grant no. XYFY2020017).
Funding
The study was funded by The National Natural Science Foundation for Youth of China (grant no. 81902354), The Science and Technology Program of Xu Zhou (grant no. KC18037), The Xuzhou Clinical Technology Key Research Project (grant no. 2019GG021) and The Excellent Talents Fund Project of Xuzhou Medical University (grant no. XYFY2020017).
Availability of data and materials
The datasets used/and or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YS and JL designed and supervised the study. GW, XL and YY performed most experiments and wrote the manuscript. ZJ and HZ helped to perform parts of experiments. KX analyzed the data and revised the article. YS and JL confirm the authenticity of all raw data. All authors contributed to the article and approved the submitted version. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Jr, Wu YL, Paz-Ares L. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 4.Cipriano R, Bryson BL, Miskimen KL, Bartel CA, Hernandez-Sanchez W, Bruntz RC, Scott SA, Lindsley CW, Brown HA, Jackson MW. Hyperactivation of EGFR and downstream effector phospholipase D1 by oncogenic FAM83B. Oncogene. 2014;33:3298–3306. doi: 10.1038/onc.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol Cancer Res. 2014;12:1156–1165. doi: 10.1158/1541-7786.MCR-13-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel CA, Parameswaran N, Cipriano R, Jackson MW. FAM83 proteins: Fostering new interactions to drive oncogenic signaling and therapeutic resistance. Oncotarget. 2016;7:52597–52612. doi: 10.18632/oncotarget.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 9.Blokhin I, Khorkova O, Hsiao J, Wahlestedt C. Developments in lncRNA drug discovery: Where are we heading? Expert Opin Drug Discov. 2018;13:837–849. doi: 10.1080/17460441.2018.1501024. [DOI] [PubMed] [Google Scholar]
- 10.Lin C, Yang L. Long noncoding RNA in cancer: Wiring signaling circuitry. Trends Cell Biol. 2018;28:287–301. doi: 10.1016/j.tcb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9:1354–1366. [PMC free article] [PubMed] [Google Scholar]
- 12.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braschi B, Denny P, Gray K, Jones T, Seal R, Tweedie S, Yates B, Bruford E. Genenames.org: The HGNC and VGNC resources in 2019. Nucleic Acids Res. 2019;47:D786–D792. doi: 10.1093/nar/gky930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P, Floros E, Dalamagas T, Hatzigeorgiou AG. DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44:D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Sun H, Liu K, Huang J, Sun Q, Shao C, Luo J, Xu L, Shen Y, Ren B. FAM111B, a direct target of p53, promotes the malignant process of lung adenocarcinoma. Onco Targets Ther. 2019;12:2829–2842. doi: 10.2147/OTT.S190934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He J, Yu J. Long noncoding RNA FAM83A-AS1 facilitates hepatocellular carcinoma progression by binding with NOP58 to enhance the mRNA stability of FAM83A. Biosci Rep. 2019;39:BSR20192550. doi: 10.1042/BSR20192550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang GM, Zang HL, Geng YX, Li YH. LncRNA FAM83A-AS1 aggravates the malignant development of esophageal cancer by binding to miR-495-3p. Eur Rev Med Pharmacol Sci. 2020;24:9408–9415. doi: 10.26355/eurrev_202009_23024. [DOI] [PubMed] [Google Scholar]
- 21.Wu F, Zhong Y, Lang XB, Tu YL, Sun SF. MNX1-AS1 accelerates the epithelial-mesenchymal transition in osteosarcoma cells by activating MNX1 as a functional oncogene. Eur Rev Med Pharmacol Sci. 2019;23:8194–8202. doi: 10.26355/eurrev_201910_19126. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, Wang X, Fu C, Wang X, Zou J, Hua H, Bi Z. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol Biol Rep. 2016;43:427–436. doi: 10.1007/s11033-016-3975-1. [DOI] [PubMed] [Google Scholar]
- 23.Wu LM, Wu SG, Chen F, Wu Q, Wu CM, Kang CM, He X, Zhang RY, Lu ZF, Li XH, et al. Atorvastatin inhibits pyroptosis through the lncRNA NEXN-AS1/NEXN pathway in human vascular endothelial cells. Atherosclerosis. 2019;293:26–34. doi: 10.1016/j.atherosclerosis.2019.11.033. [DOI] [PubMed] [Google Scholar]
- 24.Bi X, Guo XH, Mo BY, Wang ML, Luo XQ, Chen YX, Liu F, Olsen N, Pan YF, Zheng SG. LncRNA PICSAR promotes cell proliferation, migration and invasion of fibroblast-like synoviocytes by sponging miRNA-4701-5p in rheumatoid arthritis. EBioMedicine. 2019;50:408–420. doi: 10.1016/j.ebiom.2019.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong X, Duan Y, Sang Y, Li Y, Zhang H, Liang Y, Liu Y, Zhang N, Yang Q. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234:9105–9117. doi: 10.1002/jcp.27587. [DOI] [PubMed] [Google Scholar]
- 26.Xu F, Zha G, Wu Y, Cai W, Ao J. Overexpressing lncRNA SNHG16 inhibited HCC proliferation and chemoresistance by functionally sponging hsa-miR-93. Onco Targets Ther. 2018;11:8855–8863. doi: 10.2147/OTT.S182005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355:479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 28.Bozatzi P, Sapkota GP. The FAM83 family of proteins: From pseudo-PLDs to anchors for CK1 isoforms. Biochem Soc Trans. 2018;46:761–771. doi: 10.1042/BST20160277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snijders AM, Lee SY, Hang B, Hao W, Bissel MJ. FAM83 family oncogenes are broadly involved in human cancers: An integrative multi-omics approach. Mol Oncol. 2017;11:167–179. doi: 10.1002/1878-0261.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Q, Chen H, Zhang M, Xiong H, Jiang Q. Knocking down FAM83B inhibits endometrial cancer cell proliferation and metastasis by silencing the PI3K/AKT/mTOR pathway. Biomed Pharmacother. 2019;115:108939. doi: 10.1016/j.biopha.2019.108939. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Gao H, Zhang J, Xue D. FAM83D is associated with gender, AJCC stage, overall survival and disease-free survival in hepatocellular carcinoma. Biosci Rep. 2019;39:BSR20181640. doi: 10.1042/BSR20181640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim KM, Hussein UK, Park SH, Park SH, Kang MA, Moon YJ, Zhang Z, Song Y, Park HS, Bae JS, et al. FAM83H is involved in stabilization of β-catenin and progression of osteosarcomas. J Exp Clin Cancer Res. 2019;38:267. doi: 10.1186/s13046-019-1274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SY, Meier R, Furuta S, Lenburg ME, Kenny PA, Xu R, Bissell MJ. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest. 2012;122:3211–3220. doi: 10.1172/JCI60498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parameswaran N, Bartel CA, Hernandez-Sanchez W, Miskimen KL, Smigiel JM, Khalil AM, Jackson MW. A FAM83A positive feed-back loop drives survival and tumorigenicity of pancreatic ductal adenocarcinomas. Sci Rep. 2019;9:13396. doi: 10.1038/s41598-019-49475-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng WX, Koirala P, Mo YY. LncRNA-Mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balbin OA, Malik R, Dhanasekaran SM, Prensner JR, Cao X, Wu YM, Robinson D, Wang R, Chen G, Beer DG, et al. The landscape of antisense gene expression in human cancers. Genome Res. 2015;25:1068–1079. doi: 10.1101/gr.180596.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wanowska E, Kubiak MR, Rosikiewicz W, Makałowska I, Szcześniak MW. Natural antisense transcripts in diseases: From modes of action to targeted therapies. Wiley Interdiscip Rev RNA. 2018;9:e1461. doi: 10.1002/wrna.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 39.Shi R, Jiao Z, Yu A, Wang T. Long noncoding antisense RNA FAM83A-AS1 promotes lung cancer cell progression by increasing FAM83A. J Cell Biochem. 2019;120:10505–10512. doi: 10.1002/jcb.28336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao G, Wang P, Zheng X, Liu D, Sun X. FAM83A-AS1 promotes lung adenocarcinoma cell migration and invasion by targeting miR-150-5p and modifying MMP14. Cell Cycle. 2019;18:2972–2985. doi: 10.1080/15384101.2019.1664225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 42.Han X, Huang S, Xue P, Fu J, Liu L, Zhang C, Yang L, Xia L, Sun L, Huang SK, Zhou Y. LncRNA PTPRE-AS1 modulates M2 macrophage activation and inflammatory diseases by epigenetic promotion of PTPRE. Sci Adv. 2019;5:eaax9230. doi: 10.1126/sciadv.aax9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang H, Li B, Zhang X, Meng X. NCK1-AS1 promotes NCK1 expression to facilitate tumorigenesis and chemo-resistance in ovarian cancer. Biochem Biophys Res Commun. 2020;5:292–299. doi: 10.1016/j.bbrc.2019.11.014. [DOI] [PubMed] [Google Scholar]
- 44.Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang S, Dong S, Wen Z, Rao J, Liao W, Shi M. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. 2018;17:69. doi: 10.1186/s12943-018-0820-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19:1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao G, Zhang L, Qian D, Sun Y, Liu W. MiR-495-3p inhibits the cell proliferation, invasion and migration of osteosarcoma by targeting C1q/TNF-related protein 3. Onco Targets Ther. 2019;12:6133–6143. doi: 10.2147/OTT.S193937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S, Wu J, Jiao K, Wu Q, Ma J, Chen D, Kang J, Zhao G, Shi Y, Fan D, Zhao G. MicroRNA-495-3p inhibits multidrug resistance by modulating autophagy through GRP78/mTOR axis in gastric cancer. Cell Death Dis. 2018;9:1070. doi: 10.1038/s41419-018-0950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eun JW, Kim HS, Shen Q, Yang HD, Kim SY, Yoon JH, Park WS, Lee JY, Nam SW. MicroRNA-495-3p functions as a tumor suppressor by regulating multiple epigenetic modifiers in gastric carcinogenesis. J Pathol. 2018;244:107–119. doi: 10.1002/path.4994. [DOI] [PubMed] [Google Scholar]
- 49.Halvorsen AR, Sandhu V, Sprauten M, Flote VG, Kure EH, Brustugun OT, Helland Å. Circulating microRNAs associated with prolonged overall survival in lung cancer patients treated with nivolumab. Acta Oncol. 2018;57:1225–1231. doi: 10.1080/0284186X.2018.1465585. [DOI] [PubMed] [Google Scholar]
- 50.Chen X, Xu Y, Liao X, Liao R, Zhang L, Niu K, Li T, Li D, Chen Z, Duan Y, Sun J. Plasma miRNAs in predicting radiosensitivity in non-small cell lung cancer. Tumour Biol. 2016;37:11927–11936. doi: 10.1007/s13277-016-5052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used/and or analyzed during the current study are available from the corresponding author on reasonable request.