Abstract
Background
This study aims to characterize, the gut and oral microbiome in Asian subjects with Crohn’s disease (CD) using whole genome shotgun sequencing, thereby allowing for strain-level comparison.
Methods
A case–control study with age, sex and ethnicity matched healthy controls was conducted. CD subjects were limited to well-controlled patients without oral manifestations. Fecal and saliva samples were collected for characterization of gut and oral microbiome respectively. Microbial DNA were extracted, libraries prepared and sequenced reads profiled. Taxonomic diversity, taxonomic association, strain typing and microbial gene pathway analyses were conducted.
Results
The study recruited 25 subjects with CD and 25 healthy controls. The oral microbe Streptococcus salivarius was found to be enriched and of concordant strains in the gut and oral microbiome of Crohn’s disease subjects. This was more likely in CD subjects with higher Crohn’s Disease Activity Index (184.3 ± 2.9 vs 67.1 ± 82.5, p = 0.012) and active disease status (Diarrhoea/abdominal pain/blood-in-stool/fever and fatigue) (p = 0.016). Gut species found to be significantly depleted in CD compared to control (Relative abundance: Median[Range]) include: Faecalibacterium prausnitzii (0.03[0.00–4.56] vs 13.69[5.32–18.71], p = 0.010), Roseburia inulinivorans (0.00[0.00–0.03] vs 0.21[0.01–0.53], p = 0.010) and Alistipes senegalensis (0.00[0.00–0.00] vs 0.00[0.00–0.02], p = 0.029). While Clostridium nexile (0.00[0.00–0.12] vs 0.00[0.00–0.00], p = 0.038) and Ruminococcus gnavus (0.43[0.02–0.33] vs 0.00[0.00–0.13], p = 0.043) were found to be enriched. C. nexile enrichment was not found in CD subjects of European descent. Microbial arginine (Linear-discriminant-analysis: 3.162, p = 0.001) and isoprene (Linear-discriminant-analysis: 3.058, p < 0.001) pathways were found at a higher relative abundance level in gut microbiome of Crohn’s disease.
Conclusions
There was evidence of ectopic gut colonization by oral bacteria, especially during the active phase of CD. Previously studied gut microbial differences were detected, in addition to novel associations which could have resulted from geographical/ethnic differences to subjects of European descent. Differences in microbial pathways provide possible targets for microbiome modification.
Keywords: Crohn’s disease, Gastrointestinal microbiome, Oral microbiome, Metagenomics
Background
Inflammatory bowel diseases (IBD) are a group of digestive tract disease that affect millions of people worldwide. There are signs that incidence rates of IBD are increasing and presenting earlier in life [1]. IBD is divided into 2 main disease processes: Crohn’s Disease (CD), which may affect any segment of the gastro-intestinal tract, and Ulcerative Colitis (UC), which is limited to the large intestine [2]. Patients with CD frequently present with severe abdominal pain, fever, and clinical signs of bowel obstruction or diarrhea [3]. Inflammation modulation is the mainstay of medical management and ranges from the use of anti-inflammatories, to corticosteroids and immunomodulators [4]. In severe cases, surgery may be required. Unfortunately, the need for surgery has not decreased despite advancements in diagnostic and treatment protocols [5].
Despite theorizing that CD arises from an impaired interaction between commensal microbiome and the human host, the distinction between primary driver events and secondary occurrences remains murky. However, the recent focus on gut microbiome dysbiosis in CD has led to the discovery of new diagnostic and therapeutic directions [6]. Murine models have shown that the disease only manifests in susceptible genotypes and is driven by microbial dysbiosis [7], while human studies have found a general decrease in alpha diversity with clade-specific changes in CD patients such as increased Enterobacteriaceae and decreased Firmicutes [8, 9].
One of the limitations of previous microbiome studies has been the sequencing strategy. Most of the studies employed 16S rRNA sequencing, which limits their findings to bacteria at the genus level. This gap can be addressed with whole genome shotgun (WGS) sequencing, which is able to detect the presence of microbes with better accuracy and provide data at species level [10]. Additionally, most of these studies were in western populations and with subjects of European descent. This limited the scope of previous results as ethnicity, eating habits and living environment are variables that affect the gut microbiome [11].
Previous work on the oral-gut axis has demonstrated a connection between oral inflammation and its contribution to gut inflammation in animal models. Oral pathobiont-reactive inflammatory cells arising from oral inflammation were found to migrate to the gut, promoting and contributing to colitis [12]. The effect of the oral microbiome on CD is by comparison relatively understudied; however, its impact cannot be ignored [13]. A recent study found that oral Klebsiella can colonize the gut and result in severe gut inflammation in susceptible individuals, thereby exacerbating inflammatory disease [14]. Another study found increased presence of species found abundantly in oral communities in the gut microbiomes of CD subjects having diarrhea, suggesting that the oral cavity may serve as a reservoir for opportunistic gut pathogens [15].
Previous studies examining the oral microbiome were also obfuscated by having different sampling sites, such as sampling from the tongue [16], plaque [17] and saliva [18, 19]. Of these sampling sites, the salivary microbiome appears to offer the most diagnostic value without the excessive influence of local factors [20]. These studies found enrichment of Veillonellaceae and depletion of Haemophilus in CD subjects, highlighting the diagnostic value of the oral microbiome [18, 19]. Moreover, as a diagnostic tool, saliva samples are non-invasive unlike colonoscopy, and easy to collect at any time unlike fecal samples.
This study aims to characterize, for the first time, the oral and gut microbiome in Asian subjects with CD using whole genome shotgun technique. A case–control analysis was conducted with matched healthy controls to investigate the presence of altered community structure and different community ecotypes in the oral and gut microbiomes of a mixed Asian population consisting of Han Chinese, Malay and Indian.
Results
The study recruited 25 subjects with CD and 25 healthy controls who were age, sex and ethnicity matched. The subjects’ demographics, clinical and oral conditions are summarized in Table 1. Most of the subjects were well controlled with Crohn’s Disease Activity Index (CDAI) scores lower than 150. There were no differences found between the caries and periodontal status between the control and CD groups. To examine the oral and gut microbiome, 50 saliva and 50 fecal samples were processed, DNA extracted and sequenced using whole genome shotgun sequencing.
Table 1.
Demographics, clinical and oral conditions of subjects (n = 50)
| Crohn's disease (CD) | Healthy controls (HC) | |
|---|---|---|
| Mean age* (Range) | 40 (23–67) | 40 (21–66) |
| Race | ||
| Han Chinese | 13 | 13 |
| Malay | 3 | 3 |
| Indian | 9 | 9 |
| Sex | ||
| Male | 10 | 10 |
| Female | 15 | 15 |
| Oral condition | ||
| Mean DMFT^ | 6.5 ± 7.0 | 4.6 ± 5.0 |
| Periodontitis present | 5 | 5 |
| Smoking | ||
| Non-smoker | 20 | 21 |
| Previous/current smoker | 5 | 4 |
| CD condition | ||
| Length of diagnosis in years (range) | 7.8 (1–20) | |
| Crohn’s Disease Activity Index (CDAI) | 86.2 ± 87.1 | |
| Current disease activity: Active (Diarrhoea, Abdominal pain, Blood-in-stool, fever and fatigue) | 10 | |
| Drug regimen# | ||
| Steroids/Anti-inflammatory (Mesalazine) | 8 | |
| Azathioprine | 13 | |
| Biologics | 11 | |
| Others (Tacrolimus, sulphasalazine) | 2 | |
*Matched controls are ± 3 years
^p-value = 0.29
#some subjects are on multiple drug regimen
Gut microbiome profile and differentially abundant species
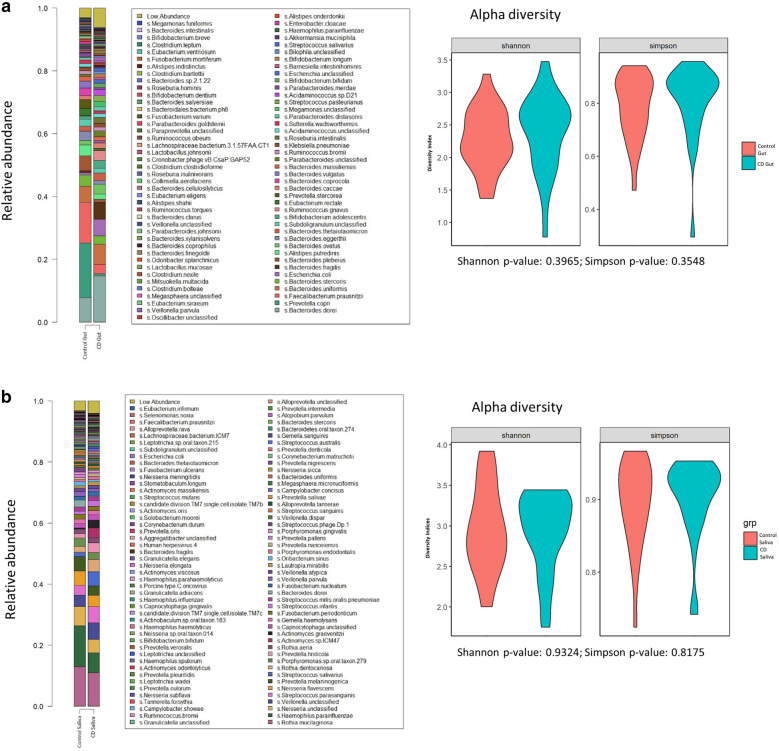
Many commensal gut microbial species were found present at high abundances (> 5%) in both CD and healthy controls. These include Bacteroides dorei (11.2%), Prevotella copri (9.1%), Faecalibacterium prausnitzii (8.0%), and Bacteroides uniformis (5.8%) (Fig. 1a). There was no difference noted in alpha diversity at species level.
Fig. 1.
Microbiome diversity at species level in Healthy (Control) vs Crohn’s Disease (CD). a Gut microbiome. b Salivary Microbiome
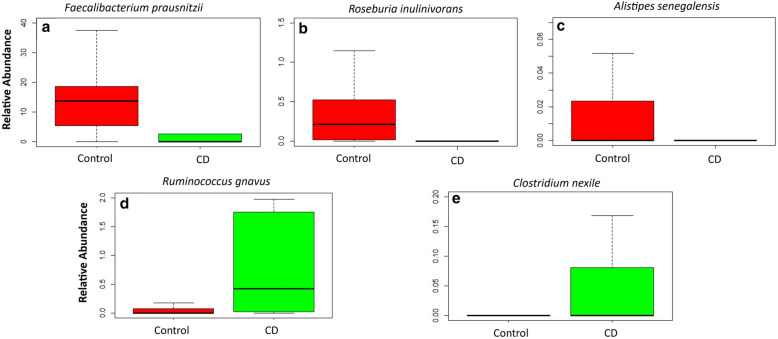
The species that were found to be significantly depleted (relative abundance) in CD compared to control include: F. prausnitzii (Median [Range]: 0.03 [0.00–4.56] vs 13.69 [5.32–18.71], p = 0.010), Roseburia inulinivorans (Median [Range]: 0.00 [0.00–0.03] vs 0.21 [0.01–0.53], p = 0.010) and Alistipes senegalensis (Median [Range]: 0.00 [0.00–0.00] vs 0.00 [0.00–0.02], p = 0.029), while Clostridium nexile (Median [Range]: 0.00 [0.00–0.12] vs 0.00 [0.00–0.00], p = 0.038) and Ruminococcus gnavus (Median [Range]: 0.43 [0.02–0.33] vs 0.00 [0.00–0.13], p = 0.043) were found to be significantly enriched in subjects with CD compared to control (Fig. 2) (Table 2).
Fig. 2.
Gut microbiome species relative abundance in Healthy (Control) vs Crohn’s Disease (CD). a Faecalibacterium prausnitzii. b Roseburia inulinivorans. c Alistipes senegalensis. d Ruminococcus gnavus. e Clostridium nexile
Table 2.
Species level differences between Crohn's disease and Control
| Depleted in crohn's disease | Enriched in crohn's disease | ||||
|---|---|---|---|---|---|
| Species | p-value | Adjusted p-value | Species | p-value | Adjusted p-value |
| Gut | |||||
| Faecalibacterium prausnitzii* | < 0.001 | 0.010 | Clostridium nexile* | < 0.001 | 0.038 |
| Roseburia inulinivorans* | < 0.001 | 0.010 | Ruminococcus gnavus* | 0.001 | 0.043 |
| Alistipes senegalensis* | < 0.001 | 0.029 | Bifidobacterium breve | 0.001 | 0.060 |
| Bacteroides faecis | 0.001 | 0.060 | Escherichia unclassified | 0.003 | 0.105 |
| Coprococcus catus | 0.002 | 0.083 | Escherichia coli | 0.004 | 0.115 |
| Ruminococcus callidus | 0.002 | 0.099 | Anaerostipes unclassified | 0.005 | 0.115 |
| Eubacterium rectale | 0.003 | 0.100 | Streptococcus anginosus | 0.005 | 0.115 |
| Parabacteroides merdae | 0.005 | 0.115 | Streptococcus pasteurianus | 0.005 | 0.115 |
| Bacteroides vulgatus | 0.006 | 0.125 | Coprobacillus unclassified | 0.009 | 0.149 |
| Eubacterium eligens | 0.007 | 0.140 | Atopobium parvulum | 0.010 | 0.149 |
| Dorea longicatena | 0.007 | 0.140 | Fusobacterium nucleatum | 0.010 | 0.149 |
| Megamonas hypermegale | 0.010 | 0.149 | Blautia producta | 0.021 | 0.208 |
| Alistipes putredinis | 0.010 | 0.149 | Erysipelotrichaceae bacterium | 0.021 | 0.208 |
| Catenibacterium mitsuokai | 0.010 | 0.149 | Morganella morganii | 0.021 | 0.208 |
| Bacteroides coprocola | 0.012 | 0.163 | Streptococcus macedonicus | 0.021 | 0.208 |
| Paraprevotella unclassified | 0.013 | 0.163 | Proteus mirabilis | 0.021 | 0.208 |
| Coprococcus comes | 0.013 | 0.163 | Veillonella atypica | 0.026 | 0.232 |
| Peptostreptococcaceae unclassified | 0.014 | 0.170 | Acidaminococcus fermentans | 0.041 | 0.268 |
| Bacteroides plebeius | 0.018 | 0.208 | Acidaminococcus sp D21 | 0.041 | 0.268 |
| Eubacterium ventriosum | 0.021 | 0.208 | Clostridium innocuum | 0.041 | 0.268 |
| Bacteroides stercoris | 0.023 | 0.219 | Lachnospiraceae bacterium | 0.041 | 0.268 |
| Alistipes finegoldii | 0.024 | 0.222 | Lactobacillus gasseri | 0.041 | 0.268 |
| Alistipes shahii | 0.025 | 0.228 | Lactobacillus mucosae | 0.041 | 0.268 |
| Bacteroides nordii | 0.027 | 0.236 | Peptostreptococcus anaerobius | 0.041 | 0.268 |
| Odoribacter splanchnicus | 0.028 | 0.236 | Prevotella histicola | 0.041 | 0.268 |
| Eubacterium siraeum | 0.031 | 0.260 | Scardovia wiggsiae | 0.041 | 0.268 |
| Holdemania unclassified | 0.037 | 0.268 | Streptococcus mutans | 0.041 | 0.268 |
| Lachnospiraceae bacterium 5_1_63FAA | 0.038 | 0.268 | Rothia dentocariosa | 0.043 | 0.273 |
| Barnesiella intestinihominis | 0.049 | 0.297 | Clostridium ramosum | 0.048 | 0.297 |
| Roseburia hominis | 0.049 | 0.297 | |||
| Saliva | |||||
| Bacteroides uniformis | < 0.001 | 0.117 | Human herpesvirus 4 | 0.002 | 0.117 |
| Bacteroides dorei | 0.001 | 0.117 | Streptococcus anginosus | 0.002 | 0.117 |
| Haemophilus parainfluenzae | 0.003 | 0.128 | Streptococcus oligofermentans | 0.002 | 0.117 |
| Subdoligranulum unclassified | 0.023 | 0.526 | Streptococcus vestibularis | 0.002 | 0.117 |
| Prevotella salivae | 0.033 | 0.526 | Streptococcus cristatus | 0.004 | 0.134 |
| Prevotella intermedia | 0.039 | 0.526 | Peptostreptococcus stomatis | 0.010 | 0.326 |
| Streptococcus salivarius | 0.020 | 0.526 | |||
| Actinomyces sp. ICM47 | 0.021 | 0.526 | |||
| Actinomyces naeslundii | 0.032 | 0.526 | |||
| Stomatobaculum longum | 0.033 | 0.526 | |||
| Actinomyces georgiae | 0.041 | 0.526 | |||
| Lactobacillus fermentum | 0.041 | 0.526 | |||
| Olsenella_uli | 0.041 | 0.526 | |||
| Oribacterium sp. Oral taxon 078 | 0.041 | 0.526 | |||
| Parvimonas micra | 0.041 | 0.526 | |||
| Slackia unclassified | 0.041 | 0.526 | |||
| Streptococcus peroris | 0.046 | 0.564 | |||
*Adjusted p-value < 0.05
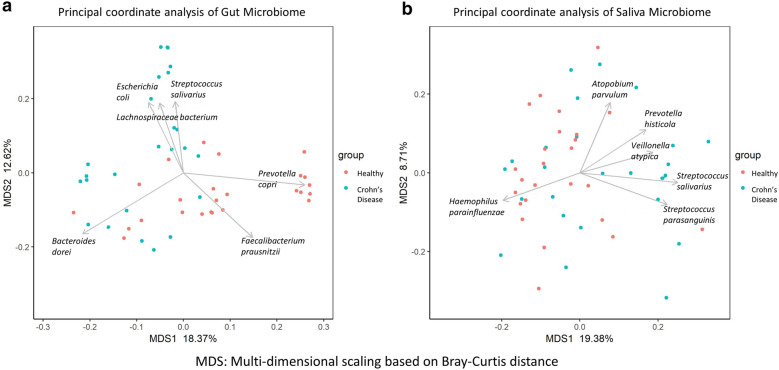
Principal coordinate analysis (Fig. 3a) revealed visually-derived groups of healthy controls driven by Prevotella copri and F. prausnitzii, while a group of CD subjects was characterized by Escherichia coli, Streptococcus salivarius and Lachnospiraceae (Wilcoxon test, p-value ≤ 0.0025).
Fig. 3.
Principal coordinate analysis of microbiome in Healthy (Control) vs Crohn’s Disease (CD). a Gut microbiome. b Salivary Microbiome. The arrows drawn are for the top 6 most significant species at p-value ≤ 0.0025
Oral microbiome profile and differentially abundant species
The oral microbiome was less dominated by specific species as compared to the gut microbiome. Microbes present at high abundance include Rothia mucilaginosa (12.0%), Haemophilus parainfluenzae (10.3%), and Neisseria sp. (5.7%). There was also no difference noted in alpha diversity at species level between CD and control (Fig. 1b).
Interestingly, several Streptoccocus sp. and Human Herpesvirus 4 were found to be enriched and Bacteroides sp found to be depleted in subjects with CD; however, they were not significantly different after multiple testing correction (Table 2).
Principal coordinate analysis conducted on the species found in the oral microbiome did not reveal distinct visual clusters based on CD status. The microbiome also did not cluster based on oral conditions such as caries or periodontal disease (Fig. 3b).
Relationship between gut and oral microbiome
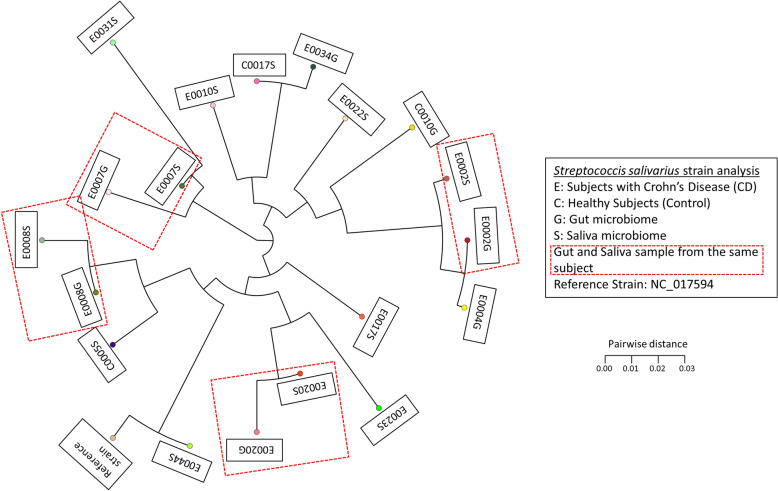
Out of the 50 pairs of gut and oral samples, Streptoccous salivarius were detected in 19 libraries; 7 gut and 12 oral samples. Among these, S. salivarius was detected in both the gut and oral samples in 4 subjects, all with CD. A strain typing analysis based on multiple sequence alignment of marker genes of S. salivarius in the subjects showed that the gut and saliva strains from the same subject clustered in close distance despite the samples being sampled separately (Fig. 4 and Additional file 1: Table S1). This suggests the S. salivarius found in the saliva and gut were colonised by similar strains. This finding was not seen in the healthy controls.
Fig. 4.
Phylogenetic tree plot of Streptococcus salivarius strain analysis for all Gut and Saliva samples
Clinical characteristics of the subjects with concordant gut and oral S. salivarius strains were compared (Table 3). Subjects with higher CDAI scores (< 150) indicating active disease were significantly more likely to have concordant strains of S. salivarius (p = 0.012). This was also reflected in subjects with signs and symptoms of active disease (Diarrhoea/abdominal pain/blood-in-stool/fever and fatigue) at the time of sampling (p = 0.016). Other clinical characteristics such as age, sex, race and oral conditions did not result in having more likely concordant gut and oral strains. Additionally, the use of proton pump inhibitors did not result in having more likely concordant gut and oral strains.
Table 3.
Clinical characteristics of subjects with concordant gut and oral S. salivarius strains
| CD subjects with concordant strains (n = 4) | CD subjects with no concordant strain (n = 21) | p-value | |
|---|---|---|---|
| Crohn’s Disease Activity Index (CDAI)# | 184.3 ± 2.9 | 67.1 ± 82.5 | *0.012 |
| Disease status | |||
| Active disease at time of sampling (Diarrhoea/abdominal pain/blood-in-stool/fever and fatigue) | 4 | 6 | *0.016 |
| Inactive disease at time of sampling | 0 | 15 | |
| Proton Pump Inhibitor use | 1.000 | ||
| Yes | 1 | 7 | |
| No | 3 | 14 | |
| Mean age# | 38 ± 14 | 40 ± 14 | 0.774 |
| Race | 0.056 | ||
| Han Chinese | 4 | 9 | |
| Malay | 0 | 3 | |
| Indian | 0 | 9 | |
| Sex | 0.142 | ||
| Male | 3 | 7 | |
| Female | 1 | 14 | |
| Oral condition | 0.661 | ||
| DMFT# | 5.0 ± 4.2 | 6.8 ± 7.4 | |
| Periodontitis | 0.451 | ||
| Present | 1 | 4 | |
| Absent | 3 | 17 | |
All Fisher-Exact test except #T-test
*p-value < 0.05
Differentially abundant microbial pathways
The HUMAnN2 analysis detected 395 pathways in the gut microbiome, of which 33 had Linear-discriminant-analysis (LDA) scores ≥ 3 (Table 4). The top 4 pathways with higher relative abundance levels found in CD compared to control were related to the biosynthesis of arginine: L-arginine biosynthesis II (acetylcycle) (LDA = 3.162, p = 0.001), L-ornithine biosynthesis (LDA = 3.084, p = 0.001), L-arginine biosynthesis I (via L-ornithine) (LDA = 3.077, p = 0.006) and L-arginine biosynthesis IV (archaebacterial) (LDA = 3.059, p = 0.008). The other higher relative abundance level pathways involve isoprene biosynthesis (2 pathways) (LDA = 3.058 and 3.042, p = < 0.001 and < 0.001) and glycolysis (2 pathways) (LDA = 3.038 and 3.023, p = < 0.001 and 0.001). While the lower relative abundance level pathways found in CD compared to control include the S-adenosyl-L-methionine cycle I (LDA = 3.550, p < 0.001), UMP biosynthesis (LDA = 3.434, p < 0.001) and 2 L-lysine biosynthesis pathways (LDA = 3.471 and 3.421, p = < 0.001 and 0.001).
Table 4.
Microbial pathways with LDA effect > 3
| Most common in control group | Most common in crohn's disease group | ||||
|---|---|---|---|---|---|
| Pathway | LDA effect | P-value | Pathway | LDA effect | P-value |
| Gut | |||||
| PWY 6151 S-adenosyl-L-methioninecycle I | 3.550 | < 0.001 | ARGSYNBSUB PWY L-arginine biosynthesis II (acetylcycle) | 3.162 | 0.001 |
| PWY 5097 L-lysine biosynthesis VI | 3.471 | < 0.001 | GLUTORN PWY L-ornithine biosynthesis | 3.084 | 0.001 |
| PWY 5686 UMP biosynthesis | 3.434 | < 0.001 | ARGSYN PWY L-arginine biosynthesis I (via L-ornithine) | 3.077 | 0.006 |
| PWY 2942 L-lysine biosynthesis III | 3.421 | 0.001 | PWY-7400 L-arginine biosynthesis IV (archaebacterial) | 3.059 | 0.008 |
| PWY 6700 queuosine biosynthesis | 3.414 | 0.002 | PWY 6270 isoprene biosynthesis I | 3.058 | < 0.001 |
| NONMEVIPP PWY methylerythritol phosphate pathway I | 3.414 | < 0.001 | PWY 7560 methylerythritol phosphate pathway II | 3.042 | < 0.001 |
| PWY 6737 starch degradation V | 3.335 | 0.003 | GLYCOLYSIS glycolysis I from glucose-6-phosphate | 3.038 | < 0.001 |
| PWY 7219 adenosine ribonucleotides de novo biosynthesis | 3.296 | 0.001 | PWY66 400 glycolysis VI (metazoan) | 3.023 | 0.001 |
| PWY 6386 UDP N-acetylmuramoyl pentapeptide biosynthesis II (lysine containing) | 3.234 | 0.003 | PWY 5484 glycolysis II from fructose-6-phosphate | 3.017 | 0.000 |
| PWY 7221 guanosine ribonucleotides de novo biosynthesis | 3.219 | 0.004 | |||
| PEPTIDOGLYCANSYN PWY peptidoglycan biosynthesis I (meso-diaminopimelate containing) | 3.212 | 0.003 | |||
| ILEUSYN PWY L-isoleucine biosynthesis I (from threonine) | 3.212 | 0.006 | |||
| VALSYN PWY L-valine biosynthesis | 3.212 | 0.006 | |||
| PWY 6387 UDP N-acetylmuramoyl pentapeptide biosynthesis I (meso-diaminopimelate containing) | 3.170 | 0.003 | |||
| BRANCHED CHAIN AA SYN PWY superpathway of branched aminoacid biosynthesis | 3.132 | 0.008 | |||
| GALACTUROCAT PWY D-galacturonate degradation I | 3.113 | < 0.001 | |||
| PWY 6897 thiamin salvage II | 3.104 | 0.001 | |||
| PWY 7111 pyruvate fermentation to isobutanol engineered | 3.098 | 0.015 | |||
| PWY 5973 cis-vaccenate biosynthesis | 3.096 | < 0.001 | |||
| GALACT GLUCUROCAT PWY superpathway of hexuronide and hexuronate degradation | 3.087 | 0.001 | |||
| GLUCUROCAT PWY superpathway of beta D-glucuronide and D-glucuronate degradation | 3.078 | 0.001 | |||
| PWY 5103 L-isoleucine biosynthesis III | 3.062 | 0.012 | |||
| PWY 6507 4-deoxy-L-threo-hex-4-enopyranuronate degradation | 3.040 | 0.001 | |||
| PWY 7242 D-fructuronate degradation | 3.019 | 0.003 | |||
| Saliva | |||||
| FASYN INITIAL PWY superpathway of fatty acid biosynthesis initiation (E-coli) | 3.036 | 0.002 | PWY66 409 superpathway of purine nucleotide salvage | 3.035 | 0.018 |
In the oral microbiomes, 365 pathways were detected, of which 2 had LDA scores ≥ 3 (Table 3). In CD, the superpathway of purine nucleotide salvage was found to have a higher relative abundance level while the superpathway of fatty acid biosynthesis initiation (E. coli) had a lower relative abundance level than the control group.
Discussion
This study represents a metagenomic insight into the oral and gut microbiome in CD patients, conducted in an Asian population, consisting subjects of Chinese, Indian and Malay descent (Table 1), which may have innate differences in gut and oral microbiome. Although some studies have examined the gut microbiome in CD [10, 21], none of the previous studies on the oral microbiome employed the shotgun metagenomic technique used in this study. Furthermore, this is the first metagenomic study of matched oral and gut samples in CD subjects, allowing matching of the gut and oral microbiome at the strain level.
Crucially, this study found a cluster of CD subjects whose gut microbiome were characterized by S. salivarius, a prominent oral microbe. In health, Streptococcus genus typically constitute less than 4% of gut microbiome [13], thus it was note-worthy to find S. salivarius enriched in the gut microbiome of CD subjects. Although the ectopic colonization of oral bacteria such as Klebsiella pneumoniae has been found to induce intestinal inflammation and can result in the progression of CD [14], it was only demonstrated in mouse models. Other studies suggesting roles in CD by oral bacteria were largely based on finding typically oral bacteria in the gut microbiome of CD subjects. In the current study, matched oral and gut samples were taken and shown for the first time in CD subjects that S. salivarius were of similar strains. This increases the likelihood that the oral microbiome was the source for ectopic gut colonization in CD. This study also found that subjects in the active phase of disease were more likely to have ectopic gut colonization from the oral microbiome. A previous study found that oral microbes can colonize the gut after diarrheal episodes [15]. The authors hypothesized that the loss of gut microbes during diarrheal episodes reduces bacterial competition, this coupled with a transient increase in oxygen, allows for the ectopic colonization of oral microbes. Diarrhea is a major symptom during the active phase CD, suggesting that the oral microbiome can serve as a reservoir for pathogenic recolonization of the gut. A factor in ectopic gut colonization is the use of proton pump inhibitors which decreases the gastric acidity, allowing for ectopic gut colonization of oral microbes [22]. However, this study did not find that the use of proton pump inhibitors and low gastric acid state increases the likelihood of ectopic gut colonization. A limitation of the study is the small number of subjects in which concordant gut and oral strains were found. Furthermore, although not statistically significant, all CD subjects with concordant gut and oral S. salivarius were Han Chinese. Therefore, it cannot be ruled out that racial differences [11] contributed to the finding of ectopic gut colonization. Larger follow up studies collecting both oral and gut samples will be required to expand upon this finding.
The gut microbial species found at high abundances (> 5%) in both CD and healthy controls in this study were similar to previous studies employing WGS [10, 23]. Contrary to previous studies, this study did not find significant reduction of alpha-diversity in the gut microbiome of CD subjects [24, 25]. However, most of these studies used 16S rRNA sequencing, which may be the reason for the difference. Additionally, another aspect of the study was that most of the CD subjects were in remission (CDAI < 150) and well-controlled with medication, which may be a reason for their microbiome being less divergent from control. However, this would mean that any changes detected are more likely to be key changes driving microbiome dysbiosis.
The differentially abundant species found in gut microbiomes here were similar to previous studies. The depletion of butyrate-producing species such as F. prausnitzii and R. inulinivorans has been described in CD subjects [26]. F. prausnitzii in particular has been proposed to have protective effects in CD [27, 28], as the butyrate produced by these microbes aid in mucosal barrier function and maintenance of gut health [23]. Although Prevotella sp. has been studied extensively and some species have been shown to induce colitis in mice, it has yet been found to be associated with CD in humans [29]. In this study, it was found that P. copri was detected in a discreet cluster of healthy controls, similar to the previous WGS study [10]. This suggests that P. copri may be protective in some individuals against CD instead. A. senegalensis is a newly delineated species [30], which has yet to be implicated in CD. A related species, Alistipes putredinis has been described to contribute to transcriptional pathways in IBD [23]. Interestingly, it was also found to be significantly depleted in CD subjects in a previous WGS study [10] warranting further investigation. Additionally, a commonly implicated microbe Ruminococcus gnavus [23, 31] was found to be significantly enriched in CD subjects in this study. A recent mechanistic study into R. gnavus showed that it produces a complex polysaccharide that can result in the pattern of inflammation seen in CD [32]. Another species implicated in the pathogenesis of CD is E. coli, which was found to work in conjunction with other microbes as well as by exhibiting virulence features in CD subjects [31]. Although Clostridium nexile was found to be enriched in CD subjects in this study, it was found to be decreased in previous studies [33]. Comparing this to another WGS study, C. nexile was also found to be enriched in CD subjects, although not significantly [10]. C. nexile is able to produce short-chain fatty acids which decreases inflammation and alleviate colitis in experimental models [34]. However, the exact mechanism of that action is still unknown and does not explain why C. nexile was enriched instead in CD. This suggests that C. nexile has a geographically or ethnically specific role and its function warrants further investigation.
Experts in the field have suggested of use the salivary microbiome as a non-invasive tool in the diagnosis of CD [18, 19]. This study did not find any significant differences between the oral microbiome in CD and controls after adjustment, which could be due to the CD subjects being under treatment, with microbiome alterations due to medications rather than a signature of disease. However, when examining the genus differences, the study found similar enrichment of Streptococcus, and depletion of Haemophilus and Prevotella in CD subjects from previous 16S studies [18, 19]. Additionally, the ability to detect changes at species level provided better resolution than previous studies showing that the enrichment of Streptococcus genus involved several species such as S. anginosus, S. oligofermentans, S. vestibularis, S. cristatus and S. salivarius, while the depletion of Prevotella involved P. salivae and P. intermedia.
Another interesting finding from the salivary analysis is the enrichment of Human Herpesvirus 4, also known as Epstein-Barr virus (EBV), found in CD subjects. Recent studies have found a significant correlation between EBV presence in the gut with clinical disease severity [35]. This could be due to increased susceptibility of the CD patients on immunosuppressive treatment and the ability of EBV to induce inflammation. Being able to detect this in the oral microbiome may allow it to serve as a marker for disease severity.
The number of microbial pathways detected in this study was similar to a previous WGS study which detected 255 pathways in CD subjects [10]. The microbial pathway analysis found that 4 pathways in the gut microbiome with higher relative abundance levels found in CD compared to control are related to arginine biosynthesis. The arginine pathway is a major contributor to host inflammatory processes via inducible nitric oxide (NO) production, with NO providing protective cytostatic/cytotoxic antimicrobial action [36]. Administration of L-arginine has even been shown to reduce intestinal inflammation and pathology [37]. However, the specific role of microbial arginine metabolism has yet to be explored in the context of CD. It could be that these bacteria proliferate in the arginine poor environment in CD subjects as they are able to produce their own arginine. Additionally, isoprene biosynthesis was also found to be at higher relative abundance levels in CD subjects. The increase of isoprene in expired air from IBD subjects has been linked to the activity of disease, and is at a higher level than healthy individuals [38]. The microbial dysbiosis in CD favors a shift towards arginine and isoprene forming microbes providing a possible target to modify the microbiome.
Conclusions
The use of metagenomics has highlighted the relationship of the gut and oral microbiome in well-controlled CD subjects. The study found the evidence of ectopic gut colonization by oral bacteria in CD subject during active disease, suggesting that the oral microbiome could be a reservoir for pathogens in CD patients. In addition to corroborating previously implicated gut microbial differences such as R. gnavus and F. prausnitzii, this study also detected the depletion of the newly described A. senegalensis not previously implicated in CD. Moreover, microbial arginine and isoprene pathways were found to be commonly present in CD gut microbiome, indicating that further study in this area is warranted.
Methods
Subject inclusion and recruitment
The study was conducted at the National University Hospital, Singapore from May 2017 to January 2019. Subjects with CD (n = 25) were recruited from the IBD clinic, Division of Gastroenterology & Hepatology. Healthy controls (n = 25) without any signs and symptoms of CD were age, sex and ethnicity matched and recruited from the Dental Centre, University Dental Cluster. The control subjects presented at the Dental Centre for routine dental care such as check-up, cleaning and minor restoration. Demographic information, history of tobacco use, other co-morbidities, current drug regimen and current disease activity were recorded. Current disease activity was calculated using the Crohn’s Disease activity index (CDAI) as well as the presence of common signs and symptoms (Diarrhoea/abdominal pain/blood-in-stool/fever and fatigue) [39]. Subjects who had antibiotic/probiotic/prebiotic use within a month, actively treated for a malignancy with chemotherapy, or diagnosed with an indeterminate colitis were excluded from the study. Non-CD controls were further required to have no known gastrointestinal signs and symptoms such as diarrhoea, abdominal pain and blood-in-stool.
The study received ethics approval from the institutional review boards of the National Healthcare Group, Singapore (DSRB reference E/2016/01,285). Informed consent was obtained from all participants. Additionally, all experiments were performed in accordance with relevant guidelines and regulations.
Oral condition of subjects
Oral involvement of CD is well documented; with studies reporting between 20 and 50% of cases exhibiting oral manifestations including aphthous ulcers, linear deep ulcers, mucosal tags, mucosal “cobblestoning”, mucogingivitis and lip swelling. The presence of these conditions can affect the oral microbiome [40]. In order to detect differences in the oral microbiome that are key drivers of CD progression, subjects exhibiting oral manifestations were excluded for this study. Other conditions that can affect the oral microbiome such as the presence of caries and periodontal disease were also recorded at sample collection. This was verified by an oral examination conducted by a dentist under artificial light.
Sample collection
Saliva was collected for the examination of the oral microbiome. Subjects were told to refrain from eating, smoking and dental procedures one hour prior to the collection. At least 5 ml of resting whole saliva was collected using the OMNIgene Discover Kit 505 (DNA Genotek, Ottawa, ON, Canada) according to manufacturer’s instructions.
Fecal sample was collected for the examination of the gut microbiome. Subjects were instructed and issued the OMNIgene•GUT (OMR-200) (DNA Genotek, Ottawa, ON, Canada) kit for sample collection. This was completed within two weeks of the saliva collection.
The oral and fecal samples were stored at room temperature as per manufacturer’s recommendation and sent for DNA extraction within 4 weeks from collection.
DNA extraction and purification
Community DNA was extracted from saliva and fecal samples after mechanical lysis via bead-beating using Exgene™ Clinic SV Mini kits (GeneAll Biotechnology, Dongnam-ro, Seoul, Korea), and QIAmp® FAST DNA Stool Mini kits (Qiagen, Hilden, Germany) respectively according to manufacturer’s instructions. Extracted DNA was purified using AMPure XP beads (Beckman Coulter, Indianapolis, IN, USA). The quantity and quality of DNA was examined using a NanoDrop 8000 Spectrophotometer (Thermo Fisher Scientific, Carlsbad, CA, United States). Extracted DNA samples were stored at -80 °C for up to 6 weeks prior to library preparation.
Library preparation and Illumina sequencing
Indexed sequencing libraries were prepared using QIAGEN® QIAseq FX DNA Library Kit (Qiagen, Hilden, Germany) following manufacturer’s instructions and sequenced as paired-end reads 2 × 151 bp on the Illumina HiSeq 4000 sequencer (Illumina, San Diego, CA, USA). On average, 15.5 million raw read pairs were obtained for each sample.
Sequence data processing
Sequenced reads for each library were de-multiplexed into individual fastq file, and analysed using a pipeline (https://github.com/gis-rpd/pipelines/tree/master/metagenomics/shotgun-metagenomics) for processing paired-end shotgun metagenomic sequencing data. Firstly, raw reads was analyzed using Skewer to trim-off adapter sequences and low-quality bases from each read [41]. After trimming, reads were decontaminated to remove genomic sequences from the human host by using BWA-MEM to map reads against the hg19 reference [42]. The remaining reads were regarded as likely of microbial origin (average = 7 million) and were used as input for subsequent taxonomic and functional profiling.
Taxonomic profiling
The MetaPhlAn2 software was used to profile the taxonomic composition of the microbial communities from the post quality-filtered reads [43]. Reads with sequences that matched microbial clades were used to normalize and calculate relative abundance for taxa from kingdom to species ranks. For reducing noise from false positive identifications, taxa with total relative abundance < 0.1% were excluded from further statistical analysis.
Strain typing
To examine the relationship of the oral microbiome to the gut microbiome, strain analysis on Streptoccous salivarius, a predominantly oral microbe, was conducted on all gut and oral samples using StrainPhlAn. Using the reads mapped against the marker database of MetaPhlAn2, we extracted the clade specific markers of S. salivarius. Subsequently multiple sequence alignment of the marker sequences of S. salivarius strains was performed among the gut and saliva libraries of samples as well as the reference genome (NC_017594), before building the phylogenetic tree [44].
Statistical analysis
Principal coordinates analysis (PCoA)
Bray–Curtis distance was calculated for PCoA analysis based on the species profiled in the sample set, while the multi-dimensional scaling (MDS) dimensions 1 and 2 were used to visualize the level of similarity between samples based on their microbiome composition.
Diversity analysis
The Vegan package in R was used to calculate Shannon and Simpson’s diversity values at the species level. Subsequently, ggplot2 was used to generate diversity plots. Wilcoxon test in R was used to test for significant difference in the medians of Shannon diversity values between case and control groups.
Association analysis for taxonomic abundances with Crohn’s disease
The Wilcoxon test in R was used to compare the median relative abundance in the cases with Crohn’s disease (CD) versus the non-diseased control group for a significant difference. To analyze the direction of the association, the “alternative” function was used in the model.
Multiple testing correction
We adopted Benjamin Hochberg’s false discovery rate method to correct for multiple testing at a significance threshold of 5%.
Pathway analysis
The HMP Unified Metabolic Analysis Network (HUMAnN2) program was used to determine relative abundance of microbial pathways in gut and saliva microbiomes of CD and control groups. The default Kyoto encyclopedia of genes and genomes (KEGG) orthology catalogue was used as the pathway reference. Subsequently, we extracted the total pathway abundance contributed by the genes present in every taxa of the community for analysis. For association, the Linear discriminant analysis Effect Size (LEfSe) software recognized the relative abundance of each pathway as the feature to test for significant difference in the medians between the two groups. We used the default LDA scores ≥ 2 as the threshold for significance, and to highlight the significance of the association, a more stringent threshold at ≥ 3 was also used for reporting [45].
Supplementary Information
Additional file 1: Table S1. Pairwise distance between S. salivarius strains gene markers analysis.
Acknowledgements
The authors would like to thank Dr Juanda Leo Hartono, Dr Chen Weihao, Ms Coka Yip, Mr Collins Wenhan Chu and Ms Ong Mei Teng for their valuable contributions in subject recruitment.
Authors’ contributions
SH and EP are considered first authors. SH conceived the idea; SH, MG and EO participated in collecting the samples; EP conducted the data analysis; SH, EP and NN led the writing; MG, EO, PFdS and JS revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Funding
This study is supported by a National University of Singapore Start-up grant (Exploring the Oral Microbiome in Crohn’s Disease).
Availability of data and materials
Whole-genome sequencing data that support the findings of this study are available in the European Nucleotide Archive with the primary accession code PRJEB39813.
Ethics approval and consent to participate
The study received ethics approval from the institutional review boards of the National Healthcare Group, Singapore (DSRB reference E/2016/01285). Informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shijia Hu, Email: Shijia_hu@nuhs.edu.sg.
Eileen Png, Email: eileenpng@u.nus.edu.
Michelle Gowans, Email: michelle_angela_gowans@nuhs.edu.sg.
David E. H. Ong, Email: david_eh_ong@nuhs.edu.sg
Paola Florez de Sessions, Email: florezdesessions@gmail.com.
Jie Song, Email: songjie66@hotmail.com.
Niranjan Nagarajan, Email: nagarajann@gis.a-star.edu.sg.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13099-021-00409-5.
References
- 1.Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12(12):720–727. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 2.Baumgart DC, Sandborn WJ. Crohn's disease. Lancet. 2012;380(9853):1590–1605. doi: 10.1016/S0140-6736(12)60026-9. [DOI] [PubMed] [Google Scholar]
- 3.Kappelman MD, Rifas-Shiman SL, Porter CQ, Ollendorf DA, Sandler RS, Galanko JA, et al. Direct health care costs of Crohn's disease and ulcerative colitis in US children and adults. Gastroenterology. 2008;135(6):1907–1913. doi: 10.1053/j.gastro.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheifetz AS. Management of active Crohn disease. JAMA. 2013;309(20):2150–2158. doi: 10.1001/jama.2013.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosnes J, Nion-Larmurier I, Beaugerie L, Afchain P, Tiret E, Gendre JP. Impact of the increasing use of immunosuppressants in Crohn's disease on the need for intestinal surgery. Gut. 2005;54(2):237–241. doi: 10.1136/gut.2004.045294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hold GL, Smith M, Grange C, Watt ER, El-Omar EM, Mukhopadhya I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years? World J Gastroenterol. 2014;20(5):1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9(5):390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2(2):119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17(1):179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Zhang YH, Huang T, Cai YD. Gene expression profiling gut microbiota in different races of humans. Sci Rep. 2016;6:23075. doi: 10.1038/srep23075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitamoto S, Nagao-Kitamoto H, Jiao Y, Gillilland III MG, Hayashi A, Imai J, et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell. 2020;182(2):447–62. e14. [DOI] [PMC free article] [PubMed]
- 13.Kitamoto S, Nagao-Kitamoto H, Hein R, Schmidt TM, Kamada N. The bacterial connection between the oral cavity and the gut diseases. J Dent Res. 2020;99(9):1021–1029. doi: 10.1177/0022034520924633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 2017;358(6361):359–365. doi: 10.1126/science.aan4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The HC, Florez de Sessions P, Jie S, Pham Thanh D, Thompson CN, Nguyen Ngoc Minh C, et al. Assessing gut microbiota perturbations during the early phase of infectious diarrhea in Vietnamese children. Gut Microbes. 2018;9(1):38–54. [DOI] [PMC free article] [PubMed]
- 16.Docktor MJ, Paster BJ, Abramowicz S, Ingram J, Wang YE, Correll M, et al. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2012;18(5):935–942. doi: 10.1002/ibd.21874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelsen J, Bittinger K, Pauly-Hubbard H, Posivak L, Grunberg S, Baldassano R, et al. Alterations of the Subgingival Microbiota in Pediatric Crohn's Disease Studied Longitudinally in Discovery and Validation Cohorts. Inflamm Bowel Dis. 2015;21(12):2797–2805. doi: 10.1097/MIB.0000000000000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Said HS, Suda W, Nakagome S, Chinen H, Oshima K, Kim S, et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res. 2014;21(1):15–25. doi: 10.1093/dnares/dst037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xun Z, Zhang Q, Xu T, Chen N, Chen F. Dysbiosis and ecotypes of the salivary microbiome associated with inflammatory bowel diseases and the assistance in diagnosis of diseases using oral bacterial profiles. Front Microbiol. 2018;9:1136. doi: 10.3389/fmicb.2018.01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, He J, Xue J, Wang Y, Li K, Zhang K, et al. Oral cavity contains distinct niches with dynamic microbial communities. Environ Microbiol. 2015;17(3):699–710. doi: 10.1111/1462-2920.12502. [DOI] [PubMed] [Google Scholar]
- 21.He Q, Gao Y, Jie Z, Yu X, Laursen JM, Xiao L, et al. Two distinct metacommunities characterize the gut microbiota in Crohn's disease patients. Gigascience. 2017;6(7):1–11. doi: 10.1093/gigascience/gix050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, et al. Extensive transmission of microbes along the gastrointestinal tract. Elife. 2019;8. [DOI] [PMC free article] [PubMed]
- 23.Schirmer M, Franzosa EA, Lloyd-Price J, McIver LJ, Schwager R, Poon TW, et al. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat Microbiol. 2018;3(3):337–346. doi: 10.1038/s41564-017-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wills ES, Jonkers DM, Savelkoul PH, Masclee AA, Pierik MJ, Penders J. Fecal microbial composition of ulcerative colitis and Crohn's disease patients in remission and subsequent exacerbation. PLoS ONE. 2014;9(3):e90981. doi: 10.1371/journal.pone.0090981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K, Nishida A, Fujimoto T, Fujii M, Shioya M, Imaeda H, et al. Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in crohn's disease. Digestion. 2016;93(1):59–65. doi: 10.1159/000441768. [DOI] [PubMed] [Google Scholar]
- 27.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15(8):1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 29.Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151(4):363–374. doi: 10.1111/imm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishra AK, Gimenez G, Lagier JC, Robert C, Raoult D, Fournier PE. Genome sequence and description of Alistipes senegalensis sp. nov. Stand Genomic Sci. 2012;6(3):1–16. doi: 10.4056/sigs.2956294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn's Disease. MBio. 2016;7(5). [DOI] [PMC free article] [PubMed]
- 32.Henke MT, Kenny DJ, Cassilly CD, Vlamakis H, Xavier RJ, Clardy J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn's disease, produces an inflammatory polysaccharide. Proc Natl Acad Sci U S A. 2019;116(26):12672–12677. doi: 10.1073/pnas.1904099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuang X, Li T, Li M, Huang S, Qiu Y, Feng R, et al. Systematic review and meta-analysis: short-chain fatty acid characterization in patients with inflammatory bowel disease. Inflamm Bowel Dis. 2019;25(11):1751–1763. doi: 10.1093/ibd/izz188. [DOI] [PubMed] [Google Scholar]
- 35.Nissen LH, Nagtegaal ID, de Jong DJ, Kievit W, Derikx LA, Groenen PJ, et al. Epstein-Barr virus in inflammatory bowel disease: the spectrum of intestinal lymphoproliferative disorders. J Crohns Colitis. 2015;9(5):398–403. doi: 10.1093/ecco-jcc/jjv040. [DOI] [PubMed] [Google Scholar]
- 36.Satriano J. Arginine pathways and the inflammatory response: interregulation of nitric oxide and polyamines: review article. Amino Acids. 2004;26(4):321–329. doi: 10.1007/s00726-004-0078-4. [DOI] [PubMed] [Google Scholar]
- 37.Fritz JH. Arginine cools the inflamed gut. Infect Immun. 2013;81(10):3500–3502. doi: 10.1128/IAI.00789-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurada S, Alkhouri N, Fiocchi C, Dweik R, Rieder F. Review article: breath analysis in inflammatory bowel diseases. Aliment Pharmacol Ther. 2015;41(4):329–341. doi: 10.1111/apt.13050. [DOI] [PubMed] [Google Scholar]
- 39.Best WR, Becktel JM, Singleton JW, Kern F., Jr Development of a Crohn's disease activity index. National Cooperative Crohn's Disease Study Gastroenterology. 1976;70(3):439–444. [PubMed] [Google Scholar]
- 40.O'Brien CL, Kiely CJ, Pavli P. The microbiome of Crohn's disease aphthous ulcers. Gut Pathog. 2018;10:44. doi: 10.1186/s13099-018-0265-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint. arXiv:1303.3997. 2013
- 43.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12(10):902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 44.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27(4):626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8(6):e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Pairwise distance between S. salivarius strains gene markers analysis.
Data Availability Statement
Whole-genome sequencing data that support the findings of this study are available in the European Nucleotide Archive with the primary accession code PRJEB39813.