Abstract
DNA and RNA can fold into a variety of alternative conformations. In recent years, a particular nucleic acid structure was discussed to play a role in malignant transformation and cancer development. This structure is called a G-quadruplex (G4). G4 structure formation can drive genome instability by creating mutations, deletions and stimulating recombination events. The importance of G4 structures in the characterization of malignant cells was currently demonstrated in breast cancer samples. In this analysis a correlation between G4 structure formation and an increased intratumor heterogeneity was identified. This suggests that G4 structures might allow breast cancer stratification and supports the identification of new personalized treatment options. Because of the stability of G4 structures and their presence within most human oncogenic promoters and at telomeres, G4 structures are currently tested as a therapeutic target to downregulate transcription or to block telomere elongation in cancer cells. To date, different chemical molecules (G4 ligands) have been developed that aim to target G4 structures. In this review we discuss and compare G4 function and relevance for therapeutic approaches and their impact on cancer development for three cancer entities, which differ significantly in their amount and type of mutations: pancreatic cancer, leukemia and malignant melanoma. G4 structures might present a promising new strategy to individually target tumor cells and could support personalized treatment approaches in the future.
Keywords: G-quadruplex, genome instability, cancer progression, cancer therapy, malignant melanoma, pancreatic cancer, acute myeloid leukemia
Introduction
Cancer is the world’s second most leading cause of death [1]. Although therapeutic strategies for many cancers have greatly advanced during the last years, still about 9.6 million people died of cancer in 2018 [2]. This highlights the need to strengthen the research on causes of cancer development and to improve diagnostic and anti-tumor treatment options. Different promising therapeutic strategies have been identified in the last decades. Among those, immunotherapy and targeted therapies have revolutionized anti-tumor therapy [3]. The identification of genetic and epigenetic abnormalities as well as tumor-growth promoting oncogenes in tumor cells provided the rationale for molecular targeted therapies [4, 5]. Immune-oncology (IO) aims at boosting the patient´s own immune system to eliminate tumor cells. One example is the immune checkpoint blockade [6]. Both approaches, targeted therapies as well as IO, significantly improved survival outcome in cancer patients, such as in malignant melanoma and other cancer entities [7, 8]. A diverging new strategy aims to modulate the 3-dimensional structure of the DNA with the goal to influence biological processes and genome stability.
Genomic DNA canonically adopts a standard B-DNA conformation. DNA can also fold into alternative structures such as DNA hairpins, holiday junctions, cruciforms, triplexes or G-quadruplexes (G4) [9, 10]. Although the relevance of G4 structures in living cells was controversially discussed in the past, accumulating experimental data now supports the existence and importance of these structures in living cells [10–12].
G4 structures can form within DNA and RNA [13]. In a G4 structure, four guanines are held together by Hoogsteen hydrogen bonds wherein each guanine can act as a donor and acceptor for two hydrogen bonds. Based on in vitro experiments it was predicted that G4 structures form in regions harboring a specific G4 motif: G≥3N1-7G≥3N1-7G≥3N1-7G≥3 [13]. However, current experimental data shows that G4 structures can also form within regions that have longer loops or less than 3 guanines per repeat as well as in regions that do not follow this stringent G4 motif [14, 15]. Among different factors the stability of the G4 structure depends on the numbers of guanines per repeat and the length of the loops [13, 16].
Computational as well as deep-sequencing approaches have demonstrated that in the human genome over 700.000 regions exist that could potentially fold into G4 structures [14, 15, 17, 18]. Also in other organisms including viruses, yeasts and different bacterial genomes regions with a strong potential to fold into G4 were mapped genome-wide [15, 19–21]. To this date, the identification of a G4 motif within the genome does not proof the formation of G4 structures at these regions in vivo, but simply gives a potential to form a G4. G4 formation needs to be experimentally evaluated and depends on different factors (e.g. protein binding, loss of protein binding, cell cycle phase, stress), of which not all are known, yet.
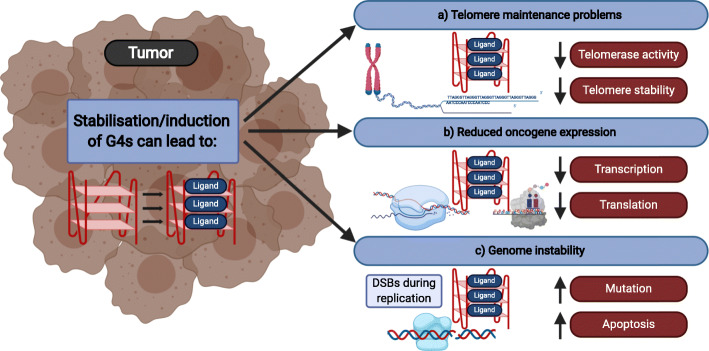
G4 motifs are not randomly distributed throughout the genome, but are enriched in certain regions (e.g. promoters, telomeres, transcription factor binding sites) [14, 22, 23]. More than 40% of human promotor regions harbor at least one G4 motif [24]. The evolutionary conservation, the specific location within the genome [15, 19, 25] as well as different biochemical and molecular experiments underline the current model that G4 structures form in living cells, where they support/affect different biological pathways (e.g. protein expression, telomerase activity and genome stability) (Fig. 1) [10–12].
Fig. 1.
Schematic overview of the effects of G4 ligands on cancer cells. Most G4 ligands cause slow growth. These growth changes are the consequence of alteration within biological processes. Depending on the ligand and cell type G4 stabilization can lead to changes in a telomere maintenance b gene expression of oncogenes c increased genome instability. Created with BioRender.
The physiological relevance of G4 structures is further supported by the existence of proteins that are able to bind and unfold G4 structures [11, 12, 26]. There are three classes of G4-interacting proteins described in the literature: G4 binding, G4 stabilizing and G4 unwinding proteins (e.g. helicases: BLM, WRN, BRIP1/FANCJ and PIF1) [27]. It has been reported that mutations and/or deletions of these proteins (e.g. PIF1) lead to changes in the formation of G4 structures. This in turn can result in changes of biological pathways (transcriptional changes) and can also increase genome instability [28–32]. This agrees with the finding that changes within some G4-interacting helicases are linked to cancer progression and tumorigenesis [27]. However, the link of tumorigenesis and mutations of helicases is not proofed to be related to G4 formation. Antibody stainings as well as G4 ChIP-seq in stomach and liver tissues of immortalized HaCaT cells showed increased levels of G4 structures compared to a normal/healthy state [18, 33, 34]. Similar observations were made for other cancer states. Different lines of evidence demonstrated that changes in G4 formation/stability can alter telomerase activity [35, 36], transcription efficiency (inhibit or promote [11, 37, 38]), stall DNA replication and induce genome instability [39, 40]. Changes can be triggered chemically (G4-ligands) or by proteins that modulate G4 formation. G4 ligands that modulate G4 structure formation or stabilize G4 structures were developed with the idea that G4 formation can be used as an anti-tumor treatment strategy by blocking cellular replication or expression of oncogenes [41, 42]. To date, about 1000 different G4 ligands have been identified [43]. These ligands differ in their specificity, binding surface and cell permeability [44]. Most of them, including MM41 [45], Telomestatin [46], BRACO19 [47], TMPyP4 [48], RHPS4 [49] and PDS [50] preferentially bind G4 structures over duplex DNA. However, most of these binding preferences were tested in vitro and it is difficult to elucidate if and how selective these G4 ligands perform in living cells. Additionally, different lines of evidence indicate that at least some G4 ligands (e.g. BRACO19) also bind to other non-canonical DNA structures such as the i-motif [51, 52], indicating the possibility that some of the G4 ligand-mediated effects in living cells might not only be caused by G4 stabilization. Regardless, some ligands show very promising results as novel therapeutic drugs [41, 42], but their use in clinical applications is not approved, yet. This is mainly due to selectivity problems and the fact that most G4 ligands target multiple G4s and by this effect many different sites in the genome. In the last years, tremendous effort has taken place to investigate G4 ligands that selectively target specific G4 structures with the aim that these ligands have high anti-tumor activity but reduced side effects [53, 54].
G-quadruplex structures and cancer
In the last years, formation and/or stabilization of G4 structures has been discussed as a potential therapeutic tool against tumor cells [41, 42, 55]. In most of these reports, G4 formation/stabilization was supported by G4 ligands. Three main therapeutic strategies have been investigated so far (Fig. 1). First, G4 formation/stabilization at telomeres was used as a tool to block telomerase activity (Fig. 1a). In about 85-90% of all cancer cells telomerase activity is upregulated, which allows the cell to replicate without telomere shortening [56]. G4 structures at telomeres alter telomerase binding and block telomerase activity in vitro [57] and in vivo [58–60]. The working hypothesis is that G4 formation at telomeres can be used to block telomerase in tumors and by this prevent uncontrolled DNA replication, while somatic cells do not express telomerase and are thus not affected. Until now G4 stabilization at telomeres has been studied by using different ligands [35]. Most of them cause reduced growth of targeted tumor cells by influencing different telomere maintenance factors [35, 61]. For example, treatment of cancer cells with the G4 ligands Telomestatin [62] or 2,6-diamidoanthraquinone derivatives [63] leads to telomerase inhibition, whereas the G4 ligand RHPS4 resulted in telomere dysfunction by disrupting the telomere protecting shelterin complex [64]. These uncapped telomeres are repaired in dependency of PARP1. Co-treatment of cells with RHPS4 and the PARP1 inhibitor GPI-15427 increased the growth reduction observed upon RHPS4 treatment, indicating the biomedical significance of this finding [65]. A novel approach is to use specific G4 ligands that act as photosensitizer to facilitate photodynamic therapy (PDT) [66]. Photosensitizer aim to specifically target tumors and cause an increased ROS production after photo-irradiation, which results in a cytotoxic effect for the tumor cells. Porphyrins are well known photosensitizer used in PDT. Specific porphyrin derivates have been developed to target telomeric G4 (TMPipEOPP [67], ZnP1 [68]). TMPipEOPP binds to telomeric G4. After photoinduction, TMPipEOPP-bound sites are cleaved, which leads to increased ROS levels and cell death [67]. ZnP1 acts via a different mechanism and creates a singlet oxygen after photoinduction that drives cleavage, ROS production and cell death [68].
Second, G4 formation was also discussed as a supporting element that impacts gene expression of oncogenes [38, 53, 69, 70] (Fig. 1b). This hypothesis was underlined by the observation that most promoters of oncogenes harbor more G4 motifs than promoters of regulatory or tumor suppressor genes [70, 71]. In vitro and in cellulo experiments revealed that changes within G4 structure formation in promoters correlates with a reduction in gene expression (e.g. MYC [72], VEGF [73], BCL2 [74], KRAS [75] and KIT [72, 76, 77]). Especially the G4-mediated changes in MYC expression [72, 76] have been studied extensively. The MYC gene encodes for the transcription factor MYC, which is upregulated in 70% of all cancers [78] and drives oncogenesis by altering cell proliferation, metabolism, and immune evasion [78–80]. Due to the lack of direct inhibitors, current strategies aim to regulate the gene expression of MYC itself [81]. One attempt is to block its expression by inducing G4 structures that are located within the MYC gene promoter [72, 76, 82]. Different G4 ligands have been tested to regulate MYC expression. Global studies using these G4 ligands resulted in reduced tumor growth, which was correlated to decreased expression of MYC and other oncogenes [54, 83]. But many of these ligands are not selective and it is not clear if the G4 ligand-dependent mechanism is via a MYC-dependent or another, yet unknown mechanism [84].
Third, under specific conditions G4 structures can cause genome instability [10, 11] (Fig. 1c). In detail, mis-regulated G4 structures, which form at the “wrong” time and place in the cell, cause alterations within DNA replication and can induce DNA damage and recombination events [39, 40, 85]. These findings agree with observations that treatment with G4 ligands (e.g. PDS) leads to enhanced DNA double strand breaks, replication pauses, micronuclei formation and telomere maintenance problems [30, 50, 65, 86, 87]. The enhanced mutagenic rate, which is stimulated by G4 structures, is favored in cells that lack functional G4-unwinding helicases (e.g. FANCJ, BLM) [28, 88–91]. This evidence raised the idea that genetic alterations (e.g. point mutations, insertion, deletion, recombination events, telomere addition or even epigenetic changes), which are observed in many cancers, might be stimulated by G4 formation. If this assumption is correct, therapeutic strategies using G4 ligands will cause increased genome instability.
Increased genome instability has a dual role in cancer research. On the one hand it stimulates tumorigenesis, but on the other hand genome instability is used as a therapeutic approach to induce apoptosis and autophagy in tumor cells (e.g. radiation therapy). A number of publications demonstrated that treatment with G4 ligands is correlated with enhanced DNA damage, telomere dysfunction and DNA damage checkpoint activation by ATM [30, 50, 92–94]. Treatment of cells with the G4 ligand 20A led to a G4-mediated upregulation of genes associated with apoptosis and autophagy, which underlines the anti-tumorigenic effect of 20A [93]. Stabilization of G4 structures by PDS in cells that were deficient in homologous recombination (HR) (e.g. BRCA1, BRCA2, Rad51 deficient cells) exhibited slow growth, fragile telomeres, DNA double strand breaks and checkpoint activation [94]. The authors concluded that in the absence of a functional DNA repair machinery G4 stabilization by PDS exacerbates genome instability, which further drives checkpoint activation, G2/M cell-cycle arrest and cell death [86]. This data underlines the hypothesis that G4 stabilization might be a promising tool to target HR-deficient tumors, even for those that are resistant to PARP inhibition (olaparib) [94]. At this point the G4 ligand CX-5461 [95, 96] is in phase I clinical trials for patients with BRCA1/2 deficient tumors, which is deficient for HR. CX-5461 acts as a topoisomerase II inhibitor that impacts DNA break production [97]. Also, other G4 ligands have been tested in combination with DNA damaging therapies. Co-treatment of PDS with the PRKDC inhibitor NU7441 resulted in enhanced growth defects. PRKDC (also known as DNA-PK) is crucial for non-homologous end joining (NHEJ) [98].
ATRX-deficient glioma cells accumulate replication problems and DNA damage due to the lack of the helicase ATRX [90, 99–101]. Most likely the DNA damage is favored by G4 structures [102, 103]. Treatment with the G4 ligands PDS, CX-5461 or CX-3543 leads to even more elevated DNA damage. In combination with DNA damaging agents (IR or hydroxyurea) the cytotoxic effect becomes further accelerated [90]. The question is how and if G4 structure-mediated DNA damage might be a potential tool to modulate DNA damage as a treatment option.
G-quadruplexes - a tool for anti-cancer therapy prospects?
The current knowledge of G4 DNA suggests that the anti-tumor effects of different G4 ligands relies on changes within telomere maintenance (Fig. 1a) (telomere damage or telomerase regulation), changes of oncogene expression (Fig. 1b) and increased genome instability (Fig. 1c). While functional telomeres and active oncogene expression is known to be important for the development of malignancies, DNA-damaging agents are usually considered cancerogenic. Genome instability and mutations are considered one of the ten hallmarks of cancer [104]. Different cancer types vary in the amount of acquired somatic mutations [105]. In consequence, some cancers rely on alterations in a small and defined subset of cellular pathways, while other cancers, such as melanoma, are very versatile. This is considered an important explanation for the development of resistance to targeted therapies and immunotherapies and highlights three important points. First, G4 stabilization drives genome instability and the cytotoxic effect of G4 stabilization is enhanced in cells that accumulate DNA damage (e.g. due to IR treatment or the loss of a functional repair pathway – see above). This raises the question whether the mutational burden of a tumor has an impact on the outcome of G4 ligand treatment. Second, what long-term effects does the treatment with G4 ligands have on tumor development? Third, does the G4 landscape change during therapy and does this contribute to genome instability that supports tumor relapse?
To this date little is known regarding the risks of long-term treatment and how G4 formation is changed over time during treatment and how/if this contributes to tumor relapse. We believe that these questions are of high relevance and that they will be addressed in the near future. Here, we aim to investigate if the current literature allows us to address these questions. We chose three different cancer entities that vary in their mutational burden and have been used to study G4 structure-mediated changes of cancer growth.
G-quadruplexes and melanoma
Melanoma is the most aggressive skin cancer. While the 5-year overall survival (OS) rate of localized melanoma is about 99%, it decreases for distant metastatic melanomas to only about 25% [106]. The development of targeted therapies [107, 108] and new immunotherapeutic approaches [109] have significantly improved survival rates, but most patients still die [110].
Melanomas are generally characterized by a high mutational burden [111, 112] and have frequently been used as an example for a highly mutated cancer that blocks DNA-damage induced apoptosis by mutations in different proteins, including TP53 [113, 114], POU3F2/BRN2 [115], RHOJ [116] and RSK [117]. Despite this, some prevalent mutated genes such as BRAF (50% of non-chronic sun damage melanomas) [118, 119] and NRAS (20-30%) [110, 120] have been identified. NRAS is also known to harbor a potential G4 motif [121].
Many studies have used different melanoma cell lines to test the relevance and effect of G4 structure stabilization on melanoma cell growth. As shown in Table 1, a variety of studies were conducted using different G4 ligands in mouse and human melanoma cell lines.
Table 1.
Overview of studies investigating the effect of G4 ligands on different melanoma cell lines
| Ligand/G4 targeting | Cell line (Xh=human, Xm=mouse) | Treatment duration (in vitro) | Growth Effect | Cellular Effects | Literature | |||
|---|---|---|---|---|---|---|---|---|
| Telomere maintenance | Oncogene regulation | Genome instability | Not tested | |||||
| 5ME | B16F10m | 48 h | IC50: ~100 μM | x | [122] | |||
| Ant1,5 | SKMel-5h, ROSSh, A375h, M14h | 48 h | IC50: 4 μM (ROSS) to 10 μM (SCMel-5) | x | [123] | |||
| BRACO19 | B16m | 24 h | IC50: 28 μM | x | [124] | |||
| C8 | B16m | 72 h | IC50: ~5 μM | x | [125] | |||
|
C14 Derivate from TMPyP4; induces singlet oxygen (1O2) production |
B78-H1m | n.a. |
In vitro: IC50 (metabolic activity): 10 nM In vivo: About 50% tumor growth reduction. |
x | [121] | |||
| c-exNDIs | A375h, SK-MEL-2h | 48 h | IC50: ~8 nM | x | [126] | |||
| CORON | M14h, LOX IMVIh, MALME-3Mh, SK-MEL-2h, SK-MEL-28h, SK-MEL-5h, UACC-62h | n.a. | LC50: mean for all cell lines ~2.5 μM | x | x | x | [127, 128] | |
| CX-3543 (Quarfloxin) | M14h, MALME-3Mh, UACC-257h, UACC-62h | n.a. | IC50: > 1 μM for all cell lines | x | x | [129] | ||
| EMICORON | M14h, LOX IMVIh, MDA-MB-435h, SK-MEL-2h, SK-MEL-28h, SK-MEL-5h, UACC-62h, UACC-257h | n.a. | LC50: mean for all cell lines ~3 μM | x | x | x | [127, 128] | |
| IZCZ-0 | A375h | 24 h | IC50: 2.3 μM | x | [130] | |||
| IZCZ-3 | A375h | 24 h | IC50: 4.2 μM | x | [130] | |||
| IZTC-1 | B16m | 24 h | IC50: ~2.2 μM, ~50-65% reduced melanoma growth in vivo. Binds preferentially to MYC G4 | x | [124] | |||
| N,N'-bis(3,4-dihydroxbenzy lidene)-1,2-diaminobenzene (crosslinker) | B16F1m | n.a. | x | [131] | ||||
|
Naphthalene diimide derivatives (compound 2) |
SKMEL-5h | 48 h | IC50: 1.7 μM | x | x | [132] | ||
| PhenDC3 | A375h | 96 h | GI20: 10 μM | x | [69] | |||
|
Phenyl 1,2,3-triazole-thymidine ligands (L1, L2, L3) |
B16F10m | 24 and 48 h | IC50: 200 μM (L1), 125 μM (L2), 50 μM (L3) | x | [133] | |||
| PPL3C | M14h | 96 h | IC50: 0.8 μM | x | [128, 134] | |||
| Pyridostatin | A375h | 96 h | GI20: 1.5 μM | x | [69] | |||
| RHPS4 | M14h, PLF2h, JR1h, JR8h, SBCL1h, SANh, LPh, LMh, JR5h, M14h | 5 and 7 days |
IC50: 3.1 μM (5 days M14), ~1 μM (5 days PLF2h, JR1, JR8, SBCL1, SAN) ~1 μM (7 days M14, PLF2h, JR1, JR8, SBCL1, SAN) About 50% reduced tumor growth in melanoma xenografts |
x | [135–137] | |||
| TMPyP4 | B78-H1m | 48 h |
IC50 (metabolic activity): 200 nM IC50: 85 μM ~65% reduced tumor growth in vivo (+ light therapy) |
x | [121, 122] | |||
| trans-resveratrol (tRES) | M14h, SKMEL-28h | 48 and 72 h | IC50: 5 μM (M14, 48 h), 2.5 μM (SKMEL-28, 48 h) IC50: ~25 μM (SKMEL-28, 72 h) | x | x | [138, 139] | ||
| Trisubstituted naphthalimides (compounds VII, VIII and IX) | M14h | 5 days | IC50: ~1.5 μM (VII), ~34.7 μM (IX) | x | [140] | |||
Treatment with the G4 ligand RHPS4 leads to the inhibition of cancer cell growth in several different melanoma cell lines [135]. Also, treatment with the imidazole-benzothiazole conjugate IZTZ-1 reduces the growth of melanoma cells [124]. Both ligands target MYC and lead to a reduction of MYC protein expression (up to 80%) after G4 stabilization [124, 131]. A systematic screen of different naphthalene diimides revealed that “G4 ligand 1” targets the G4s within KIT and BCL2 and drives melanoma tumors into a pro-apoptotic environment. In this paper the authors propose that G4 stabilization might be relevant in the context of resistance to targeted therapies of BRAF mutant melanomas [126]. However, if the ligands target other promoters and stimulate changes in other pathways (e.g. telomeres) has not been investigated in this context, yet. A similar growth effect was observed after treatment of B78-H1-tumor-bearing mice with the G4 ligands C14 and TMPyP4 [121]. Both ligands are photosensitizers that caused retarded tumor growth and increased survival time of irradiated mice. Based on their data, the authors propose a model in which photocleavage of G4 RNA at the mitogenic Ras gene was stimulated due to TMPyP4 and C14 binding, which led to reduced levels of RAS protein [141, 142]. Overall these studies measured enhanced apoptosis and necrosis [121]. It is not clear whether the observed effects were only because of changes within the mRNA of the Ras genes or if additional targets such as Myc and other genes also drove this effect.
Note, treatment with either RHPS4 or BRACO19 for 3-21 days led to telomere uncapping and end-to-end fusion of chromosomal ends in both, prostate and melanoma cell lines [65]. In contrast, in a UXF1138L uterus carcinoma cell line telomere uncapping could already be detected after 24 h of treatment with a G4 ligand [64]. The authors speculate that this difference in treatment timing might come from the difference in telomere length between prostate/melanoma cells (4-10 kb) and uterus carcinoma cells (2.7 kb). This raises the hypothesis that telomere length as well as telomerase activity might be useful markers for the success of G4 stabilization by ligands [64].
In an alternative approach guanine-rich oligos (t-oligos) homolog to the telomeric overhang that forms the G4 structures were designed [143]. Within melanoma cells the t-oligos led to a decreased proliferation rate, enhanced apoptosis and reduced expression of the catalytic subunit of the telomerase reverse transcriptase (TERT). Two major telomere-binding proteins, POT1 and TERF2, bind to these t-oligos and might cause the observed effects (107). Another study investigated the effect of TERF2 on cancer proliferation and genome stability [136]. They observed that TERF2 inhibition reduces tumor growth in dependency to its telomere capping potential. This implies that cells with shorter telomeres are targeted more efficiently by TERF2 inhibitor. In subsequent experiments they revealed that G4 stabilization by RHPS4 makes cells with longer telomeres (M14 cells) sensitive to TERF2 inhibition [136]. This indicates that G4 stabilization might induce telomere uncapping, which supports co-treatments that target short or unprotected telomeres (e.g. after TERF2 inhibitors) [136].
Pancreatic cancer
Pancreatic cancer is associated with a poor prognosis. The five-year OS rate of diagnosed patients is only 9% [144]. Once surgery is incapable of removing the tumor, treatment options are very limited. In contrast to melanoma, pancreatic cancer is associated with a lower mutational burden [111] although 97% of pancreatic cancers have gene alterations [145].
RAS genes (KRAS, NRAS and HRAS) represent the most frequently mutated oncogenes in human cancer [146] including pancreatic cancer [147]. Generally, tumor cells harbor multiple genetic and epigenetic abnormalities. Nevertheless, in some cancers, tumor growth depends on one single oncogene and its continued activation. This is named oncogene addiction [148, 149]. For the initiation of pancreatic ductal adenocarcinoma (PDCA), the oncogenic KRAS mutation is indispensable [150]. In pancreatic cancer mutations in the oncogene KRAS are essential for tumorigenesis and impact directly multiple metabolic pathways of PDAC [150].
As KRAS is a driver of oncogene addiction, it is tempting to identify and characterize a therapeutic target to specifically down-regulate KRAS. The expression of all three RAS genes can be reduced by G4 formation [75, 151–153]. G4 stabilization by G4 ligands is a promising strategy to target the activity of the KRAS gene promotor [70]. There are three G4 motifs in the KRAS promoter which are located in the nuclease hypersensitive element upstream of the transcription start site [75, 154–157]. It has been reported that the transcription factors MAZ and HNRNPA1 as well as HMGB1 can upregulate KRAS expression by binding to a G4 structure within KRAS [158, 159]. G4 structure formation itself acts as a gene silencer for KRAS expression [75, 160]. These biochemical studies strengthen the hypothesis that G4 structures within KRAS represent a promising new drug target.
G4 ligands were also used in targeted approaches in pancreatic tumors analog to other cancer entities [72, 161]. These studies revealed that application of different G4 ligands resulted in reduced viability and growth inhibition of pancreatic cancer cells. In detail, the G4 ligand TMPyP4 induced tumor cell death when incubated with MIA Pa-Ca-2 pancreatic cancer cells while causing no effect in non-malignant cells [162]. Additional studies demonstrated that the G4 ligand MM41 led to reduced tumor cell growth in MIA Pa-Ca-2 xenografts [163]. Also, the G4 binding porphyrins Tetrakis and Octaacetyl inhibited proliferation of Ehrlich Ascites Carcinoma (EAC) solid tumors. The authors explained these growth changes of MIA Pa-Ca-2 and EAC tumors by the reduced expression of the KRAS and BCL2 genes in cancer cells. Note, in these experiments the G4 ligands enhanced the expression of pro-apoptotic molecules such as BAX and TP53 and by this promoted apoptosis [164]. Similarly, the G4 ligand nitidine, or TINA-modified oligonucleotides (insertion of (R)-1-O-(4-(1-pyrenylethynyl)phenylmethyl]glycerol to increase stability) that mimic G4 structures of KRAS down-regulate KRAS protein expression and inhibit pancreatic cancer cell growth [155, 160]. One possible explanation for these observations is that KRAS expression is reduced due to the competition of G4 structure formation and MAZ binding [155, 165]. An alternative, but not exclusive hypothesis is that the G4 structures within the promoter are crucial for KRAS expression itself by inducing a positive feedback loop. KRAS stimulates the expression of ILK, which regulates HNRNPA1. HNRNPA1 binds and breaks down the G4 motif in the KRAS promotor and induces KRAS transcription [166, 167].
While most of the G4-related studies in the context of pancreatic cancer focus on changes of KRAS expression, one study demonstrated that G4 stabilization by BMSG-SH-3 in MIA Pa-Ca-2 xenograft tumors led to telomere shortening caused by reduced expression of TERT. In this study neither KRAS nor BCL2 expression changed [168].
Taken together, these studies revealed promising experimental evidence that G4 structure formation alters the expression of RAS genes and by this inhibits tumor growth. This can be explained by the blocking of DNA polymerases and/or altering the binding of proteins in these regions. It is not clear which additional G4 motifs are targeted by G4 ligands and if this induces genome instability, alters transcription or even translation of multiple other sites. It would be interesting to identify proteins that bind to G-rich non-G4 regions whose binding behaviour is altered upon G4 structure formation at oncogenes These proteins would be useful future drug targets to gain specificity of treatments with G4 ligands (Table 2).
Table 2.
Overview of studies investigating the effect of G4 ligands on different pancreatic cancer cell lines
| Ligand/G4 targeting | Cell line (all human) |
Treatment duration (in vitro) | Growth Effect | Cellular Effects | Literature | |||
|---|---|---|---|---|---|---|---|---|
| Telomere maintenance | Oncogene regulation | Genome instability | Not tested | |||||
| 4,11-bis(2-aminoethylamino) anthra[2,3-b]furan-5,10-dione (2a),11-bis(2-aminoethylamino) anthra[2,3-b]thiophene-5,10-dione (2b) | PANC-1 | 72 h | IC50(metabolic activity): 0.26 μM (2a) and 0.9 μM (2b) | x | [169] | |||
| 5ME | PANC-1 | 48 h | IC50: 80 μM | x | [122] | |||
| Alkyl-modified porphyrins | PANC-1 | 72 h | IC50(metabolic activity): ~15 nM | x | [142] | |||
| Azidothymidine (AZT) | MIA PaCa-2 | 4 and 7 days | IC50: >200 μM | x | [162] | |||
| BMSG-SH3 | MIA PaCa-2 | n.a. |
50% decreased tumor growth of MIA-Pa-Ca2 xenografts No in vitro cyto-toxicity assay done |
x | [168] | |||
| C14 | PANC-1 | n.a. | IC50: ~10 nM when irradiated with halogen light | x | [141] | |||
| C-2028 | PANC-1, MIA PaCa-2, BXpC-3, AsPC-1, Capan-2 | 72 h |
IC50 for all cell lines < 100 nm About 80% reduced Panc-1 xenograft growth in vivo |
x | [170] | |||
| CM03 |
MIA PaCa-2, PANC-1 |
96 h | IC50: 7 nM (MIA), 18 nM (PANC-1), reduced tumor growth by ~ 73% | x | [171–173] | |||
|
Copper(ii) l/d-valine-(1,10-phen) complexes (complex 1a, 1b) |
BxPC3, AsPC1 | 72 h | IC50: ~2 μM for both complexes in both cell lines | x | [174] | |||
| CX-3543 (Quarfloxin) | MIA PaCa-2 | n.a. | >50% reduced tumor growth of MIA PaCa-2 xenografts | x | x | [129] | ||
| CX-5461 | Gemcitabine-resistant MIA PaCa-2 (GemMIA-R3) and normal MIA-PA-Ca2 | 96 h | GI50: 90.3 nM (GemMIA-R3), 88.7 nM (MIA-Pa-Ca2) | x | [175] | |||
| MM41 |
MIA PaCa-2, PANC-1 |
96 h | IC50: 11 nM (MIA), 3 nM (PANC-1), ~80% reduced growth of MIA PaCa-2 xenografts in vivo | x | [163, 171] | |||
|
Naphthalene diimide ligands (compounds 3d, 3h) |
MIA PaCa-2 | 96 h | IC50: 10 nM for both compounds | x | - | [45] | ||
| Naphthalene diimide isomer ligands (compounds 2-5) |
MIA PaCa-2 PANC-1 |
96 h | IC50: 5-130 nM (MIA PaCa-2), 2 nM-1.5 μM (PANC-1) | - | [176] | |||
| Nitidine | AsPC-1, BxPC-3, MIA PaCa-2, PANC-1 | 72 h | IC50: 6.1 μM (AsPC-1), 5.2 μM (BxPC-3), 13.4 μM (MIA PaCa-2), 35.3 μM (PANC-1) | x | [160] | |||
| Octaacetyl | Panc-1h, MIA PaCa-2h | 24-72 h |
IC50: 65, 40, 36 μM for PANC-1 (24, 48, 72 h) and 62, 38, 33 μM for MIA PaCa-2 Reduced tumor growth in vivo |
x | [164] | |||
| RHPS4 | PAXF 736 | 15 days colony forming assay | IC50: 0.44 μM | x | [177] | |||
| SOP1812 | MIA PaCa-2, PANC-1, Capan-1, BXPC-3 | 96 h |
GI50: 1.3 nM (MIA PaCa-2), 5.9 nM (Capan-1) Significantly reduced MIA PaCa-2 xenograft growth in vivo |
x | [173] | |||
| Telomestatin | MIA PaCa-2 | 48 h | IC50: 0.5 μM | x | [178] | |||
| Tetrakis | PANC-1, MIA PaCa-2 | 24-72 h |
IC50: 60, 31, 25 μM (PANC-1) and 65, 36, 30 μM (MIA PaCa-2) for 24, 48, 72 h Reduced tumor growth in vivo |
x | [164] | |||
| Tetrasubstituted naphthalene diimide ligands | PANC-1, MIA PaCa-2, HPAC, BxPc-3 | 96 h | IC50: 0.1-0.2 μM (PANC-1, MIA PaCa-2, HPAC), 1.5 μM (BxPc-3) | x | x | [179] | ||
| TMPyP4 | MIA PaCa-2, PANC-1 | 48 h, 4 and 7 days |
IC50: 21.9 μM (MIA PaCa-2 , 4 days), 50 μM (PANC1, 7 days), 50 μM (MIA PaCa-2, 48 h), ~20 μM (PANC-1, 48 h). ~30% KRAS inhibition after 12 and 24 h |
x | [122, 142, 154, 162, 178] | |||
G-quadruplexes and leukemia
Leukemia is caused by the abberant proliferation of blood cells. Different subtypes of leukemia exist according to the WHO classification depending on the affected cell type, the cell phenotype, cytogenetic and molecular genetic alterations. Acute myeloid leukemia (AML) is the most common acute leukemia in adults and is characterized by a relatively low number of mutations [112]. It is a clinically as well as genetically heterogeneous disease and is also popular as a leukemia model in G4 research. Over the past two decades a number of different G4 ligands (e.g. SYUIQ-5 [180], APTO-253 [181], TMPyP4 [182], telomestatin [62] and Tel03 [183]) were used to test their effect on leukemia cell growth. Most of these studies demonstrated that leukemia cell growth was inhibited after G4 structure stabilization by G4 ligands in vitro and in vivo [62, 181, 182]. Similar to previously discussed cancer entities, studies in leukemia revealed that after G4 stabilization the expression of oncogenes changed as well as the function of telomerase. G4 stabilization by telomestatin led to apoptosis and telomere shortening in leukemia cells from four AML patients [184]. Similarily, telomere shortening and senesence was observed with the G4 ligand SYUIQ-5 in K-562 and HL-69 leukemia cells, because of changes in the expression of telomerase (TERT) and TERF2 [180]. In addition to the relevance of G4 structure regulation at telomeres, several important leukemic oncogenes were inhibited after using G4 ligands (including BCL2 [183], MYC [180, 181, 185], MLLT1 and AFDN [186], WT1 [182], KIT [181] and KRAS [185]) in human AML-derived cell lines.
As stated above, G4 structure formation might also challenge genome stability and exhibit tumor/leukemia-promoting properties. Indeed, Tauchi and collegues demonstrated that telomestatin activates the ATM-dependent DNA-damage checkpoint response [62]. In silico analysis revealed that 70% of the rearrenged genes in leukeamia contain a G4 motif [187]. This might indicate that G4 structures stimulate genome instability in these tumors. This was further supported by the finding that sites of rearrangement in TCF3, which promotes leukemia development, colocalize with regions that can form G4 structures [188]. In the future, it would be interesting to determine, if the G4 landscape within these mutagenic tumors changes and to address, which G4 structure newly forms and which are lost in these tumors. One hypothesis is that G4 structure formation contributes to the mutagenic nature of tumors. The determination of the location of G4 structures within these cancers together with deeper knowledge of the function and relevance of these structures will be beneficial to adjust G4 structure-driven treatments.
One of the most common recurring genetic events in AML are mutations in the C-terminal domain of NPM1, which occur in about 30-35% of all AML patients [189]. NPM1 is an abundant non-ribosomal nucleolar protein and essential for ribosome biogenesis. It was reported that NPM1 can bind to G4 structures located within the ribosomal DNA in vitro [190] and in vivo [191]. G4 binding by NPM1 is associated with its cellular localization. Experiments using the G4 ligand TMPyP4 in OCI-AML2 cells demonstrated that after G4 stabilization NPM1 translocates from the nucleolus to the nucleoplasm. The authors explain this effect by the competitive effect of TMPyP4 on G4 structure binding by NPM1 [191]. These findings together with the observation that the most common AML variant of NPM1 no longer binds to G4 structures in ribosomal DNA [191] led to the idea that the disruption of the G4 structure binding capability of NPM1 is linked to nucleolar localization in AML-associated protein variants [191].
As a side note we would like to point out that also G4 structure formation within RNAs could impact tumorigenesis and might be a useful target for cancer therapy. The translational control of oncoprotein expression is considered to be relevant in many solid cancers [192] and leukemias. EIF4A is an RNA helicase that promotes and sustains T-acute lymphoblastic leukemia and preferably targets mRNAs with G4 motifs in their 5’UTR [192]. Silvestrol, hippuristanol and pateamine A are natural compounds that target EIF4A. Inhibition of EIF4A with silvestrol showed anti-tumor efficacy in vitro and in vivo [192]. It was proposed that after EIF4A inhibition by silvestrol more G4 structures are formed within these transcripts leading to altered translation, which caused the slow growth of tumor cells (Table 3).
Table 3.
Overview of studies investigating the effect of G4 ligands on different leukemia cell lines
| Ligand/G4 targeting | Cell line (all human) | Treatment duration (in vitro) | Growth Effect | Cellular Effects | Literature | |||
|---|---|---|---|---|---|---|---|---|
| Telomere maintenance | Oncogene regulation | Genome instability | Not tested | |||||
| 1,8-dipyrazolcarbazole (DPC) derivatives, compound 7b | HL-60 | 48 h | IC50: 1.2 μM | x | [193] | |||
| 7-substituted-5,6-dihydrobenzo[c] acridine derivatives, compound 2b | K-562 | 48 h | IC50: 9.2 μM | x | [194] | |||
| 9-N-substituted berberine derivatives, compound 2j | HL-60 | 8 days | IC50: ~3 μM | x | [195, 196] | |||
| 12-N-Methylated 5,6-dihydrobenzo[c] acridine derivatives, compound 21c | Ramos, CA46 | 96 h | IC50: ~5-10 μM | x | [197] | |||
| Actinomycin D | Ramos, CA46 | 24 h | IC50: ~25 nM (Ramos), ~10 nM (CA46) | x | [198] | |||
| Alkynylplatinum(II) terpyridine complexes (1-3) | K-562 | 48 h | IC50: 4-7.3 μM | x | [199] | |||
| APTO-253 |
AML: EOL-1, HEL92.1.7, MV4-11, KG-1, SKM-1, THP-1, NOMO-1, HL-60, MOLM-13 Leukemia/ Lymphoma: GRANTA-519, Jeko-1, Jurkat, SUDHL-6, Mino, Raji, Ramos |
5 days | LC50: 0.14-1.75 μM (AML), 0.057-0.52 μM (non-AML) | x | x | [181] | ||
| AQ1 | HMC1.2, α155 | n.a. | n.a. | x | x | [200, 201] | ||
| B5 | Ramos, CA45 | 48 h | IC50: 11.3 μM (Ramos), 21.8 μM (CA45) | x | [198, 202] | |||
| Ber8 | HL-60 | 48 h | IC50: 1.7 μM | x | x | x | [196] | |
| BRACO-19 | CLL, AML | n.a. | IC50: 80 μM | [203] | ||||
| C2 | Ramos, CA45 | 48 h | IC50: 1.4 μM (Ramos), >100 μM (CA45) | x | [198, 202] | |||
| CORON | CCRF-CEM, HL-60(TB), K-562, MOLT-4, RPMI-8226, SR | n.a. | LC50: 0.85 μM (K-562), 1.4 μM (MOLT-4), 1.9 μM (HL-60(TB)), > 50 μM (all other) | x | x | x | [127] | |
|
CX-3543 (Quarfloxin) |
A3, CCRF-CEM, D1-1, GDM-1, HL-60, I 9.2, J45-01, Jgamma-1, Jurkat, K-562, Kasumi-1, KG-1, Ku-812, MEG-01, MOLT-3, MOLT-4, MV-4-11, P116, Reh, RPMI-8226, RS4-11, SR, TF-1, THP-1 | n.a. | IC50: ~1 μM | - | - | [129] | ||
| D2 | Ramos, CA45 | 48 h | IC50: 11.7 μM (Ramos), 17.8 μM (CA45) | x | [198, 202] | |||
|
Diquinolinyl- Pyridine Ligands (1a-c) |
K-562, HL60 | 72 h |
IC50: >50 μM (1a), 3 μM (1b), >50 μM (1c, all K-562) IC50: >50 μM (1a), 18 μM (1b), >50 μM (1c, all HL60) |
x | [204] | |||
| Disubstituted quindoline derivatives, compound 74a |
Raji CCRF-CEM U266B2 |
48 h |
IC50: 4.7 μM (Raji), 18.1 μM (CCRF-CEM), 23.0 μM (U266B2) Xenograft of Raji cells >50% reduced tumor growth |
x | [205] | |||
| DNR | K-562 |
48 h 96 h |
IC50: 0.33 μM, 0.067 μM |
x | [206] | |||
| EMICORON | CCRF-CEM, HL-60(TB), K-562, MOLT-4, RPMI-8226, SR | n.a. | LC50: 3.7 μM (HL-60(TB)), > 50 μM (all other) | x | x | x | [127] | |
| GQC-05 | KG-1a, CMK, TF-1 | 24 h | IC50: >1 μM | x | x | [207] | ||
| LZ-11 | HL-60 | 48 h | IC50: 9.6 μM | x | [208] | |||
| MXR | K-562 |
48 h 96 h |
IC50: 0.411 μM IC50: 0.105 μM |
x | [206] | |||
| Pegaharmine D | HL-60 | 72 h | IC50: 3.81 μM | x | [209] | |||
| Pyrazine-based cyclometalated (C^Npz^C)Au(III) carbene complexes, compound 2 and 3 | HL-60 | 72 h |
IC50: 0.31 μM (2), 4.05 μM (3) |
- | [210] | |||
| Pyridine(2,4- dihydroxybenz aldehyde dibenzyl semicarbazone) copper(II) | MOLT-4 |
24 h 72 h |
IC50: 5 μM IC50: 1.5 μM |
x | [185] | |||
| QPB compound 15e | HL-60 | 48 h | IC50: 1.7 μM | x | [211] | |||
| Quinolino-benzo- [5, 6]-dihydro isoquindolium compounds 3a, 3f, 3g, 3j | HL-60 | 48 h | IC50: 2 μM (3j) - 12.6 μM (3g) | x | [212] | |||
| TMPyP4 | K-562, OCI-AML2, OCI-AML3 | 48 and 96 h | IC50: 100-170 μM (K-562, 48 h), 60 μM (OCI-AML2, 96 h), 50 μM (OCI-AML3, 96 h) | x | [182, 191, 213, 214] | |||
| Telomestatin |
Primary blast cells from AML patients OM9;22, K-562 |
10-30 days | IC50: 5 μM (10 days), 2 μM showed growth inhibition (15/30 days) | x | x | [62, 184, 215] | ||
| Substituted salicylaldehyde dibenzyl semicarbazones copper(II) complexes (7, 9, 7-py and 9-py) | MOLT-4 | 24 h | IC50: 3.1 μM (9-py), 18.1 μM (7-py), 5 μM (7), 8.03 μM (9) | x | [215] | |||
| Sysu12d | CA46, HL-60 | 48 h | IC50: 11.2 μM (CA46), 5 μM (HL-60) | x | x | [216] | ||
| SYUIQ-5 | K-562, HL-60 | 72 h | IC50: 5 μM (K-562, HL-60) or 2.65 μM (K-562) | x | x | x | [180, 217] | |
| SYUIQ-FM05 | K-562 | 24 h | IC50: 10.83 nM | x | [218] | |||
Discussion
The research of the past decades links G4 structure formation and unfolding to defects and changes observed in different cancer entities. Current studies revealed that although most G4 ligands result in changes of cancer cell growth, they act via different mechanims and most likely at different targets. It is not clear, if the reduced growth effect on cancer cells after G4 ligand treatment is caused by multiple G4 structure-mediated changes, by a single G4 structure-driven event or by unspecific binding of the ligand. The problem in the current literature is to pinpoint the exact cause that drives growth changes and to monitor where the G4 ligand binds. Currently, there are several trials to establish G4 ligands that are specific to only one target [83, 219] and/or to develop novel photosensitizing G4 ligands that only cause toxic effects to G4 regions after irradiation [66]. These trials are very promising as it is expected that these approaches will reduce side effects. Additionally, the list of G4-interacting and -regulating proteins is increasing, which will not only give insights into G4 function in normal cells but will also provide ideas how to target/regulate specific G4 structures via the protein itself or protein-specific inhibotors. This would be particularly interesting for cancers like pancreatic cancer that for some cases are caused by a single upregulation of one oncogene (e.g. KRAS). In these cancers a G4 structure-based block of transcription is a very promising treatmen option. The current literature demonstrates that G4 stabilization often correlates with reduced cellular growth of cancers. It is not fully understood if the reduced growth is due to changes in telomere maintenance, transcriptional changes or genome stability. We would like to emphasize that co-treatment of G4 stabilization with drugs that either block DNA repair or induce additional genome instability is currently a very promising approach.
An interesting therapeutic strategy could be to combine immune checkpoint inhibitors with G4 structure-stabilizing ligands. It has been demonstrated that the success of immune checkpoint inhibition correlates with the mutational burden of a tumor [111, 112]. Hence, immunotherapeutic approaches might benefit from a combinational therapy with G4 ligands. It seems plausible that G4 ligands, which induce genome instability, might increase the immunogenecity of a tumor and sensitize it for checkpoint inhibition or other immunotherapeutic approaches.
Conclusion
In this review we presented and discussed the relevance of G4 structure formation and stabilization as a therapeutical approach to treat cancer cells based on the current literature. As pointed out, different ligands have often a negative effect on cancer cell growth via different mechanisms. We discussed the hypothesis, if the mutagenic burden of the tumor positivly or negatively influences the outcome of G4 ligand treatment. We summarized the current research results that are linked to changes in G4 levels in melanoma, pancreatic cancer and leukemia cells. The conclusions are not very clear. We oberve that all three tumor entities show reduced cell growth upon treatment with different G4 ligands and, depending on the ligand, also changes in telomere maintenance, gene expression of oncogenes and increased genome instability. A direct comparision is difficult, because different studies were often done using different conditions. Some ligands were used in at least two entities. TMPyP4, EMICORON and CORON were tested in all three selected entities. Although similar effects were documented, the timing of treatment as well as the used concentration differs in these different studies. These differences indicate that lower concentrations of G4 ligands (e.g. EMICORON) are required to induce G4 structure-driven toxicity in cancer cells that have a high mutagenic burden (see Table 1, 2 and 3). A similar trend was observed for BRACO-19 (28 μM in melanoma and 80 μM for leukemia). Further experimental data demonstrated that a treatment with G4 ligands further enhances the cytotoxic effect in cells that have high levels of genome instability (e.g. after radiation therapy, after PARP inhibitor treatment or in BRCA1-deficient cells). However, in studies that used quarfloxin (CX-3543) we could not find this correlation. Future studies with similar culturing conditions and additional molecular studies are required to proof this hypothesis.
Lastly, we would like to disucss the potential impact that G4 structure formation might have on the mutagenic burden of the tumor and what the consequences are for tumor development and tumor relapse after treatment. In general, G4 structure formation, if not regulated efficiently (this includes formation and unfolding), can stimulate genome instability, which includes muations and deletions and complex gross chromosomal rearrangements [28, 30, 220]. A computational study investigated how the mutational burden correlates with G4 structure formation [221]. They revealed by comparing the location of potential G4 forming sites with cancer-associated breakpoints (using the COSMIC database) a significant overlap, in particular in those cancers that harbor a mutation in TP53. This is underlined by compuational studies in melanoma cells that link G4 regions with mutational hot spots [187]. A recent study identified a direct correlation of G4 structure formation with mutational changes in different breast cancer entities [222]. This supports the notion that G4 formation indeed stimulates and influences mutation rates in different cancers and may also contribute to subtype classifications [222]
Acknowledgements
We thank the Paeschke and the Heine group for helpful discussion.
Abbreviations
- 20A
1-ethyl-N-(phenylmethyl)-4-(tetrahydro-2H-pyran-4-ylamino)-1H-pyrazolo[3,4-b]pyridine-5-carboxamide
- AFDN
Afadin, Adherens Junction Formation Factor
- AML
Acute myeloid leukemia
- APTO-253
2-(5-fluoro-2-methyl-1H-indol-3-yl)-1H-imidazo[4,5-f][1,10]phenanthroline
- ATM
Ataxia Telangiectasia Mutated
- ATRX
ATRX Chromatin Remodeler
- BAX
BCL2 Associated X, Apoptosis Regulator
- BCL2
BCL2 Apoptosis Regulator
- BLM
Bloom Syndrome RecQ Like Helicase
- BMSG-SH-3
2,7-Bis-[5-(4-methyl-piperazin-1-yl)-pentyl]-4,9-bis-[3-(4-methyl-piperazin-1-yl)-propylamino]-benzo[lmn][3,8]phenanthroline-1,3,6,8-tetraone
- BRACO19
N,N′-(9-(4-(Dimethylamino)phenylamino)acridine-3,6-diyl)bis(3-(pyrrolidin-1-yl)propanamide) hydrochloride
- BRAF
B-Raf Proto-Oncogene, Serine/Threonine Kinase
- BRCA1
Breast Cancer Type 1 Susceptibility Protein
- BRCA2
Breast Cancer Type 2 Susceptibility Protein
- BRIP1/FANCJ
BRCA1 Interacting Protein C-Terminal Helicase 1
- C14
C14H28-alkyl derivative of PMPyP4
- ChIP-seq
Chromatin Immunoprecipitation and sequencing
- CX-3543
Quarfloxin, 15-fluoro-N-[2-[(2S)-1-methylpyrrolidin-2-yl]ethyl]-18-oxo-14-(3-pyrazin-2-ylpyrrolidin-1-yl)-12-oxa-1-azapentacyclo[11.7.1.02,11.04,9.017,21]henicosa-2,4,6,8,10,13(21),14,16,19-nonaene-19-carboxamide
- CX-5461
2-(4-methyl-1,4-diazepan-1-yl)-N-[(5-methylpyrazin-2-yl)methyl]-5-oxo-[1,3]benzothiazolo[3,2-a][1,8]naphthyridine-6-carboxamide
- EAC
Ehrlich ascites carcinoma
- EIF4A
Eukaryotic translation initiation factor 4A
- G4
G-quadruplex
- GI20/GI50
20%/50% growth inhibition
- GPI-15427
10-((4-Methylpiperazin-1-yl)methyl)chromeno(4,3,2-de)phthalazin-3(2H)-one
- h
Hours
- HaCaT
Human adult low Calcium high Temperature Keratinocytes
- HMGB1
High mobility group box 1
- HNRNPA1
Heterogeneous nuclear ribonucleoprotein A1
- HR
Homologous recombination
- HRAS
HRas Proto-Oncogene, GTPase
- IC50
Half maximal inhibitory concentration
- ILK
Integrin Linked Kinase
- IO
Immune-oncology
- IR
Ionizing radiation
- IZTZ-1
Benzothiazole-2-carboxaldehyde-derivate
- KIT
KIT Proto-Oncogene, Receptor Tyrosine Kinase
- KRAS
KRAS Proto-Oncogene, GTPase
- LC50
Half lethal concentration
- MAZ
MYC-associated zinc finger protein
- MLLT1
MLLT1 Super Elongation Complex Subunit
- MM41
4,9-Bis((3-(4-methylpiperazin-1-yl)propyl)amino)-2,7-bis(3-morpholinopropyl)benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone
- MYC
MYC Proto-Oncogenes c-myc, l-myc, n-myc
- n.a.
Not available
- NHEJ
Non-homologous end joining
- NPM1
Nucleophosmin 1
- NRAS
NRAS Proto-Oncogene, GTPase
- NU7441
8-(4-Dibenzothienyl)-2-(4-morpholinyl)-4H-1-benzopyran-4-one
- olaparib
4-[[3-[4-(cyclopropanecarbonyl)piperazine-1-carbonyl]-4-fluorophenyl]methyl]-2H-phthalazin-1-one
- OS
Overall survival
- PARP1
Poly [ADP-ribose] polymerase 1
- PDCA
Pancreatic ductal adenocarcinoma
- PDS
Pyridostatin, 4-(2-Aminoethoxy)-N2,N6-bis[4-(2-aminoethoxy)-2-quinolinyl]-2,6-pyridinedicarboxamide
- PDT
Photodynamic therapy
- PIF1
Petite Integration Frequency 1
- POT1
Protection of telomeres 1
- Pou3FS/BRN2
Pou class 3 homeobox 2
- PRKDC
Protein Kinase, DNA-Activated, Catalytic Subunit
- Rad51
RAD51 Recombinase
- RAS
NRAS Proto-Oncogene, GTPase
- RHOJ
Ras homolog family member J
- RHPS4
3,11-Difluoro-6,8,13-trimethylquino[4,3,2-kl]acridinium methylsulfate
- ROS
Reactive oxygen species
- RSK
P90 ribosomal S6 kinase
- SYUIQ-5
N-(10H-indolo[3,2-b]quinolin-11-yl)-N',N'-dimethylpropane-1,3-diamine
- TCF3
Transcription Factor 3
- Tel03
N,N¢-Bis-(2-(dimethylamino)ethyl)-3,4,9,10-perylenetetracarboxylic acid diimide
- Telomestatin
(1R)-4,8-Dimethyl-3,7,11,15,19,23, 27-heptaoxa-31-thia-33,34,35, 36,37,38,39,40-octaazanonacyclo[28.2.1.12,5.16,9.110,13.114, 17.118,21.122,25.126,29]tetraconta-2(40),4,6(39),8,10(38), 12,14 (37),16,18(36),20,22(35), 24,26(34),28,30(33)-pentadecaene
- TERF2
Telomeric repeat binding factor 2
- TERT
Telomerase reverse transcriptase
- TINA
Twisted intercalating nucleic acid
- TMPipEOPP
5,10,15,20-tetra-{4-[2-(1-methyl-1-piperidinyl)ethoxy]phenyl} porphyrin
- TMPyP4
Meso-Tetra (N-methyl-4-pyridyl) porphine tetra tosylate
- TP53
Tumor Protein P53
- VEGF
Vascular Endothelial Growth Factor A
- WHO
World Health Organization
- WRN
Werner Syndrome RecQ Like Helicase
- WT1
WT1 Transcription Factor
- ZnP1
Zn(II) 5,10,15,20-tetrakis(N-carboxymethyl-4-pyridinium)porphyrin
Authors’ contributions
NK, SJ, AH, KP literature search, NK, AH, KP initial draft of manuscript, NK, SJ, PB, AH, KP editing and final draft of manuscript, AH and KP supervision and funding aquvisition. All authors approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. Research in the Paeschke laboratory is fund by an ERC Stg Grant (638988-G4DSB) and by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project-ID 369799452 – TRR237”. Research in the Paeschke and Heine lab are funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC2151 – 390873048.
Availability of data and materials
Not applicable
Ethics Approval and Consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Annkristin Heine and Katrin Paeschke contribute equally to this work.
Contributor Information
Annkristin Heine, Email: annkristin.heine@ukbonn.de.
Katrin Paeschke, Email: katrin.paeschke@ukbonn.de.
References
- 1.Hannah Ritchie and Max Roser. Causes of Death - Our World in Data. https://ourworldindata.org/causes-of-death. Accessed 4 Dec 2020.
- 2.Cancer. https://www.who.int/news-room/fact-sheets/detail/cancer. Accessed 4 Dec 2020.
- 3.Couzin-Frankel J. Breakthrough of the year Cancer immunotherapy. Vol. 342. Science. 2013;2013:1432–1433. doi: 10.1126/science.342.6165.1432. [DOI] [PubMed] [Google Scholar]
- 4.Sun W, Shi Q, Zhang H, Yang K, Ke Y, Wang Y, et al. Advances in the techniques and methodologies of cancer gene therapy. Discov Med. 2019;27(146):45–55. [PubMed] [Google Scholar]
- 5.Klinakis A, Karagiannis D, Rampias T. Targeting DNA repair in cancer: current state and novel approaches. 77, Cellular and Molecular Life Sciences. 2020. p. 677–703. [DOI] [PMC free article] [PubMed]
- 6.A R, JD W. Cancer immunotherapy using checkpoint blockade. Science. 2018;359(6382). [DOI] [PMC free article] [PubMed]
- 7.Hochhaus A, Larson RA, Guilhot F, Radich JP, Branford S, Hughes TP, et al. Long-Term Outcomes of Imatinib Treatment for Chronic Myeloid Leukemia. N Engl J Med. 2017;376(10):917–927. doi: 10.1056/NEJMoa1609324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellmann MD, Paz-Ares L, Bernabe Caro R, Zurawski B, Kim S-W, Carcereny Costa E, et al. Nivolumab plus Ipilimumab in Advanced Non–Small-Cell Lung Cancer. N Engl J Med. 2019;381(21):2020–2031. doi: 10.1056/NEJMoa1910231. [DOI] [PubMed] [Google Scholar]
- 9.Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. Journal of Biological Chemistry. 2004;279:47411–4. [DOI] [PubMed]
- 10.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: Stability and function of G-quadruplex structures. Nature Reviews Genetics. 2012;13:770–80. [DOI] [PMC free article] [PubMed]
- 11.Varshney D, Spiegel J, Zyner K, Tannahill D, Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Vol. 21, Nature Reviews Molecular Cell Biology. 2020. p. 459–74. [DOI] [PMC free article] [PubMed]
- 12.Spiegel J, Adhikari S, Balasubramanian S. The Structure and Function of DNA G-Quadruplexes. Vol. 2, Trends in Chemistry. 2020. p. 123–36. [DOI] [PMC free article] [PubMed]
- 13.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006;34(19):5402–5415. [DOI] [PMC free article] [PubMed]
- 14.Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33(8):877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 15.Marsico G, Chambers VS, Sahakyan AB, McCauley P, Boutell JM, Di Antonio M, et al. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019;47(8):3862–3874. doi: 10.1093/nar/gkz179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Vol. 36, Nucleic Acids Research. 2008. p. 5482–515. [DOI] [PMC free article] [PubMed]
- 17.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33(9):2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hänsel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016;48(10):1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 19.Capra JA, Paeschke K, Singh M, Zakian VA. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput Biol. 2010;6(7):9. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan OI, Berber B, Hekim N, Doluca O. G-quadruplex prediction in E. coli genome reveals a conserved putative G-quadruplex-Hairpin-Duplex switch. Nucleic Acids Res. 2016;44(19):9083–9095. doi: 10.1093/nar/gkw769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavezzo E, Berselli M, Frasson I, Perrone R, Palù G, Brazzale AR, et al. G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLOS Comput Biol. 2018;14(12):e1006675. doi: 10.1371/journal.pcbi.1006675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36(4):1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33(9):2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35(2):406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakken S, Rognes T, Hovig E. The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 2009;37(17):5749–5756. doi: 10.1093/nar/gkp590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brázda V, Hároníková L, Liao JCC, Fojta M. DNA and RNA quadruplex-binding proteins. Int J Mol Sci. 2014;15:17493–517. [DOI] [PMC free article] [PubMed]
- 27.Sauer M, Paeschke K. G-quadruplex unwinding helicases and their function in vivo. Vol. 45, Biochemical Society Transactions. 2017. p. 1173–82. [DOI] [PubMed]
- 28.Paeschke K, Capra JA, Zakian VA. DNA Replication through G-Quadruplex Motifs Is Promoted by the Saccharomyces cerevisiae Pif1 DNA Helicase. Cell. 2011;145(5):678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiavone D, Guilbaud G, Murat P, Papadopoulou C, Sarkies P, Prioleau M, et al. Determinants of G quadruplex-induced epigenetic instability in REV 1-deficient cells. EMBO J. 2014;33(21):2507–2520. doi: 10.15252/embj.201488398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Magis A, Manzo SG, Russo M, Marinello J, Morigi R, Sordet O, et al. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc Natl Acad Sci U S A. 2019;116(3):816–825. doi: 10.1073/pnas.1810409116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopes J, Le Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou MP, et al. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30(19):4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puig Lombardi E, Holmes A, Verga D, Teulade-Fichou MP, Nicolas A, Londoño-Vallejo A. Thermodynamically stable and genetically unstable G-quadruplexes are depleted in genomes across species. Nucleic Acids Res. 2019;47(12):6098–6113. doi: 10.1093/nar/gkz463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biffi G, Tannahill D, Miller J, Howat WJ, Balasubramanian S. Elevated levels of G-quadruplex formation in human stomach and liver cancer tissues. PLoS One. 2014;9(7). [DOI] [PMC free article] [PubMed]
- 34.Hänsel-Hertsch R, Spiegel J, Marsico G, Tannahill D, Balasubramanian S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc. 2018;13(3):551–564. doi: 10.1038/nprot.2017.150. [DOI] [PubMed] [Google Scholar]
- 35.Tan J, Lan L. The DNA secondary structures at telomeres and genome instability. Vol. 10, Cell and Bioscience. 2020. [DOI] [PMC free article] [PubMed]
- 36.Bryan TM. G-quadruplexes at telomeres: Friend or foe?. Vol. 25, Molecules. 2020. [DOI] [PMC free article] [PubMed]
- 37.Kim N. The Interplay between G-quadruplex and Transcription. Curr Med Chem. 2017;26(16):2898–2917. doi: 10.2174/0929867325666171229132619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian T, Chen YQ, Wang SR, Zhou X. G-Quadruplex: A Regulator of Gene Expression and Its Chemical Targeting. Vol. 4, Chem. 2018. p. 1314–44.
- 39.Bryan TM. Mechanisms of DNA replication and repair: Insights from the study of G-quadruplexes. Molecules. 2019;24(19). [DOI] [PMC free article] [PubMed]
- 40.Lerner LK, Sale JE. Replication of G quadruplex DNA. Vol. 10, Genes. 2019. [DOI] [PMC free article] [PubMed]
- 41.Ruggiero E, Richter SN. Survey and summary G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Vol. 46, Nucleic Acids Research. 2018. p. 3270–83. [DOI] [PMC free article] [PubMed]
- 42.Carvalho J, Mergny JL, Salgado GF, Queiroz JA, Cruz C. G-quadruplex, Friend or Foe: The Role of the G-quartet in Anticancer Strategies. Vol. 26, Trends in Molecular Medicine. 2020. p. 848–61. [DOI] [PubMed]
- 43.Li Q, Xiang JF, Yang QF, Sun HX, Guan AJ, Tang YL. G4LDB: A database for discovering and studying G-quadruplex ligands. Nucleic Acids Res. 2013;41(D1). [DOI] [PMC free article] [PubMed]
- 44.Haider SM, Neidle S, Parkinson GN. A structural analysis of G-quadruplex/ligand interactions. Vol. 93, Biochimie. 2011. p. 1239–51. [DOI] [PubMed]
- 45.Micco M, Collie GW, Dale AG, Ohnmacht SA, Pazitna I, Gunaratnam M, et al. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J Med Chem. 2013;56(7):2959–2974. doi: 10.1021/jm301899y. [DOI] [PubMed] [Google Scholar]
- 46.Kim MY, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J Am Chem Soc. 2002;124(10):2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 47.Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, et al. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65(4):1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 48.Izbicka E, Wheelhouse RT, Raymond E, Davidson KK, Lawrence RA, Sun D, et al. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999;59(3):639–644. [PubMed] [Google Scholar]
- 49.Gowan SM, Heald R, Stevens MFG, Kelland LR. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol Pharmacol. 2001;60(5):981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez R, Müller S, Yeoman JA, Trentesaux C, Riou JF, Balasubramanian S. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J Am Chem Soc. 2008;130(47):15758–15759. doi: 10.1021/ja805615w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pagano A, Iaccarino N, Abdelhamid MAS, Brancaccio D, Garzarella EU, Di Porzio A, et al. Common G-quadruplex binding agents found to interact with i-motif-forming DNA: Unexpected multi-target-directed compounds. Front Chem. 2018;6. [DOI] [PMC free article] [PubMed]
- 52.Abdelhamid MAS, Gates AJ, Waller ZAE. Destabilization of i-Motif DNA at Neutral pH by G-Quadruplex Ligands. Biochemistry. 2019;58(4):245–249. doi: 10.1021/acs.biochem.8b00968. [DOI] [PubMed] [Google Scholar]
- 53.Asamitsu S, Bando T, Sugiyama H. Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures. Vol. 25, Chemistry - A European Journal. 2019. p. 417–30. [DOI] [PubMed]
- 54.Felsenstein KM, Saunders LB, Simmons JK, Leon E, Calabrese DR, Zhang S, et al. Small Molecule Microarrays Enable the Identification of a Selective, Quadruplex-Binding Inhibitor of MYC Expression. ACS Chem Biol. 2016;11(1):138–148. doi: 10.1021/acschembio.5b00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cimino-Reale G, Zaffaroni N, Folini M. Emerging Role of G-quadruplex DNA as Target in Anticancer Therapy. Curr Pharm Des. 2016;22(44):6612–6624. doi: 10.2174/1381612822666160831101031. [DOI] [PubMed] [Google Scholar]
- 56.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, et al. Specific association of human telomerase activity with immortal cells and cancer. Science (80- ). 1994;266(5193):2011–5. [DOI] [PubMed]
- 57.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DMA structures. Nature. 1991;350(6320):718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 58.Moye AL, Porter KC, Cohen SB, Phan T, Zyner KG, Sasaki N, et al. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat Commun. 2015;6. [DOI] [PMC free article] [PubMed]
- 59.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12(10):847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 60.Paudel BP, Moye AL, Assi HA, El-Khoury R, Cohen SB, Holien JK, et al. A mechanism for the extension and unfolding of parallel telomeric g-quadruplexes by human telomerase at single-molecule resolution. Elife. 2020;9:1–9. [DOI] [PMC free article] [PubMed]
- 61.Neidle S. Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. Vol. 277, FEBS Journal. 2010. p. 1118–25. [DOI] [PubMed]
- 62.Tauchi T, Shin-Ya K, Sashida G, Sumi M, Nakajima A, Shimamoto T, et al. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: Involvement of ATM-dependent DNA damage response pathways. Oncogene. 2003;22(34):5338–5347. doi: 10.1038/sj.onc.1206833. [DOI] [PubMed] [Google Scholar]
- 63.Wheelhouse RT, Sun D, Han H, Han FX, Hurley LH. Cationic porphyrins as telomerase inhibitors: The interaction of tetra- (N-methyl-4-pyridyl)porphine with quadruplex DNA [1]. Vol. 120, Journal of the American Chemical Society. 1998. p. 3261–2.
- 64.Phatak P, Cookson JC, Dai F, Smith V, Gartenhaus RB, Stevens MFG, et al. Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Br J Cancer. 2007;96(8):1223–1233. doi: 10.1038/sj.bjc.6603691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salvati E, Leonetti C, Rizzo A, Scarsella M, Mottolese M, Galati R, et al. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J Clin Invest. 2007;117(11):3236–3247. doi: 10.1172/JCI32461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawauchi K, Urano R, Kinoshita N, Kuwamoto S, Torii T, Hashimoto Y, et al. Photosensitizers based on g-quadruplex ligand for cancer photodynamic therapy. Vol. 11, Genes. 2020. p. 1–13. [DOI] [PMC free article] [PubMed]
- 67.Zhu LN, Zhao SJ, Wu B, Li XZ, Kong DM. A new cationic porphyrin derivative (TMPipEOPP) with large side arm substituents: A highly selective G-quadruplex optical probe. PLoS One. 2012;7(5). [DOI] [PMC free article] [PubMed]
- 68.Beniaminov AD, Novikov RA, Mamaeva OK, Mitkevich VA, Smirnov IP, Livshits MA, et al. Light-induced oxidation of the telomeric G4 DNA in complex with Zn(II) tetracarboxymethyl porphyrin. Nucleic Acids Res. 2016;44(21):10031–10041. doi: 10.1093/nar/gkw947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zyner KG, Mulhearn DS, Adhikari S, Cuesta SM, Di Antonio M, Erard N, et al. Genetic interactions of G-quadruplexes in humans. Elife. 2019;8. [DOI] [PMC free article] [PubMed]
- 70.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat Rev Drug Discov. 2011;10(4):261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol. 2011;18(8):950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A. 2002;99(18):11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33(18):6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J Am Chem Soc. 2006;128(16):5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34(9):2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang D, Hurley L. Structure of the biologically relevant g-quadruplex in the c-MYC promoter. Nucleosides, Nucleotides and Nucleic Acids. 2006;25(8):951–968. doi: 10.1080/15257770600809913. [DOI] [PubMed] [Google Scholar]
- 77.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, et al. Putative DNA Quadruplex Formation within the Human c-kit Oncogene. J Am Chem Soc. 2005;10:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dang C V. MYC on the path to cancer. Vol. 149, Cell. 2012. p. 22–35. [DOI] [PMC free article] [PubMed]
- 79.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bretones G, Delgado MD, León J. Myc and cell cycle control. Vol. 1849, Biochimica et Biophysica Acta - Gene Regulatory Mechanisms. 2015. p. 506–16. [DOI] [PubMed]
- 81.Whitfield JR, Beaulieu ME, Soucek L. Strategies to inhibit Myc and their clinical applicability. Vol. 5, Frontiers in Cell and Developmental Biology. 2017. [DOI] [PMC free article] [PubMed]
- 82.Nasiri HR, Bell NM, Mc Luckie KIE, Husby J, Abell C, Neidle S, et al. Targeting a c-MYC G-quadruplex DNA with a fragment library. Chem Commun. 2014;50(14):1704–1707. doi: 10.1039/C3CC48390H. [DOI] [PubMed] [Google Scholar]
- 83.Calabrese DR, Chen X, Leon EC, Gaikwad SM, Phyo Z, Hewitt WM, et al. Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex. Nat Commun. 2018;9(1):4229–9. [DOI] [PMC free article] [PubMed]
- 84.Boddupally PVL, Hahn S, Beman C, De B, Brooks TA, Gokhale V, et al. Anticancer activity and cellular repression of c-MYC by the G-quadruplex-stabilizing 11-piperazinylquindoline is not dependent on direct targeting of the G-quadruplex in the c-MYC promoter. J Med Chem. 2012;55(13):6076–6086. doi: 10.1021/jm300282c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valton AL, Prioleau MN. G-Quadruplexes in DNA Replication: A Problem or a Necessity? Vol. 32, Trends in Genetics. 2016. p. 697–706. [DOI] [PubMed]
- 86.Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8(3):301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MIR, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149(4):795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 88.Wu Y, Shin-ya K, Brosh RM. FANCJ Helicase Defective in Fanconia Anemia and Breast Cancer Unwinds G-Quadruplex DNA To Defend Genomic Stability. Mol Cell Biol. 2008;28(12):4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu W, Rokutanda N, Takeuchi J, Lai Y, Maruyama R, Togashi Y, et al. HERC2 facilitates BLM and WRN helicase complex interaction with RPA to suppress G-quadruplex DNA. Cancer Res. 2018;78(22):6371–6385. doi: 10.1158/0008-5472.CAN-18-1877. [DOI] [PubMed] [Google Scholar]
- 90.Wang Y, Yang J, Wild AT, Wu WH, Shah R, Danussi C, et al. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat Commun. 2019;10(1). [DOI] [PMC free article] [PubMed]
- 91.London TBC, Barber LJ, Mosedale G, Kelly GP, Balasubramanian S, Hickson ID, et al. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283(52):36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rizzo A, Salvati E, Porru M, D’Angelo C, Stevens MF, D’Incalci M, et al. Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res. 2009;37(16):5353–5364. doi: 10.1093/nar/gkp582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beauvarlet J, Bensadoun P, Darbo E, Labrunie G, Rousseau B, Richard E, et al. Modulation of the ATM/autophagy pathway by a G-quadruplex ligand tips the balance between senescence and apoptosis in cancer cells. Nucleic Acids Res. 2019;47(6):2739–2756. doi: 10.1093/nar/gkz095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zimmer J, Tacconi EMC, Folio C, Badie S, Porru M, Klare K, et al. Targeting BRCA1 and BRCA2 Deficiencies with G-Quadruplex-Interacting Compounds. Mol Cell. 2016;61(3):449–460. doi: 10.1016/j.molcel.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, et al. Inhibition of RNA Polymerase I as a Therapeutic Strategy to Promote Cancer-Specific Activation of p53. Cancer Cell. 2012;22(1):51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O’Neil NJ, et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun. 2017;8. [DOI] [PMC free article] [PubMed]
- 97.Bruno PM, Lu M, Dennis KA, Inam H, Moore CJ, Sheehe J, et al. The primary mechanism of cytotoxicity of the chemotherapeutic agent CX-5461 is topoisomerase II poisoning. Proc Natl Acad Sci U S A. 2020;117(8):4053–4060. doi: 10.1073/pnas.1921649117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McLuckie KIE, Di Antonio M, Zecchini H, Xian J, Caldas C, Krippendorff BF, et al. G-quadruplex DNA as a molecular target for induced synthetic lethality in cancer cells. J Am Chem Soc. 2013;135(26):9640–9643. doi: 10.1021/ja404868t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Clynes D, Jelinska C, Xella B, Ayyub H, Scott C, Mitson M, et al. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat Commun. 2015;6. [DOI] [PMC free article] [PubMed]
- 100.Heaphy CM, De Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. [DOI] [PMC free article] [PubMed]
- 101.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of Telomeres pathway. PLoS Genet. 2012;8:7. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clynes D, Higgs DR, Gibbons RJ. The chromatin remodeller ATRX: A repeat offender in human disease. Trends in Biochemical Sciences. 2013;38:461–6. [DOI] [PubMed]
- 103.Law MJ, Lower KM, Voon HPJ, Hughes JR, Garrick D, Viprakasit V, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143(3):367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 104.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–74. [DOI] [PubMed]
- 105.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Vol. 349, Science. 2015. p. 1483–9. [DOI] [PubMed]
- 106.Survival Rates for Melanoma Skin Cancer. https://www.cancer.org/cancer/melanoma-skin-cancer/detection-diagnosis-staging/survival-rates-for-melanoma-skin-cancer-by-stage.html. Accessed 4 Dec 2020.
- 107.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved Survival with Vemurafenib in Melanoma with BRAF V600E Mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved Survival with MEK Inhibition in BRAF-Mutated Melanoma. N Engl J Med. 2012;367(2):107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 109.Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob J-JJ, Cowey CL, et al. Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med. 2017;377(14):1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kozar I, Margue C, Rothengatter S, Haan C, Kreis S. Many ways to resistance: How melanoma cells evade targeted therapies. Biochimica et Biophysica Acta - Reviews on Cancer. 2019;1871:313–22. [DOI] [PubMed]
- 111.Arora S, Velichinskii R, Lesh RW, Ali U, Kubiak M, Bansal P, et al. Existing and Emerging Biomarkers for Immune Checkpoint Immunotherapy in Solid Tumors. Vol. 36, Advances in Therapy. 2019. p. 2638–78. [DOI] [PMC free article] [PubMed]
- 112.Castle JC, Uduman M, Pabla S, Stein RB, Buell JS. Mutation-derived neoantigens for cancer immunotherapy. Front Immunol. 2019;10(AUG). [DOI] [PMC free article] [PubMed]
- 113.Monnerat C, Chompret A, Kannengiesser C, Avril MF, Janin N, Spatz A, et al. BRCA1, BRCA2, TP53, and CDKN2A germline mutations in patients with breast cancer and cutaneous melanoma. Fam Cancer. 2007;6(4):453–461. doi: 10.1007/s10689-007-9143-y. [DOI] [PubMed] [Google Scholar]
- 114.Flørenes VA, Øyjord T, Holm R, Skrede M, Børresen AL, Nesland JM, et al. TP53 allele loss, mutations and expression in malignant melanoma. Br J Cancer. 1994;69(2):253–259. doi: 10.1038/bjc.1994.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Herbert K, Binet R, Lambert JP, Louphrasitthiphol P, Kalkavan H, Sesma-Sanz L, et al. BRN2 suppresses apoptosis, reprograms DNA damage repair, and is associated with a high somatic mutation burden in melanoma. Genes Dev. 2019;33(5–6):310–332. doi: 10.1101/gad.314633.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ho H, Aruri J, Kapadia R, Mehr H, White MA, Ganesan AK. RhoJ regulates melanoma chemoresistance by suppressing pathways that sense DNA damage. Cancer Res. 2012;72(21):5516–5528. doi: 10.1158/0008-5472.CAN-12-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ray-David H, Romeo Y, Lavoie G, Déléris P, Tcherkezian J, Galan JA, et al. RSK promotes G2 DNA damage checkpoint silencing and participates in melanoma chemoresistance. Oncogene. 2013;32(38):4480–4489. doi: 10.1038/onc.2012.472. [DOI] [PubMed] [Google Scholar]
- 118.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct Sets of Genetic Alterations in Melanoma. N Engl J Med. 2005;353(20):2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 119.Grimaldi AM, Cassidy PB, Leachmann S, Ascierto PA. Novel approaches in melanoma prevention and therapy. Cancer Treat Res. 2014;159:443–455. doi: 10.1007/978-3-642-38007-5_25. [DOI] [PubMed] [Google Scholar]
- 120.Kunz M, Hölzel M. The impact of melanoma genetics on treatment response and resistance in clinical and experimental studies. Cancer Metastasis Rev. 2017;36(1):53–75. doi: 10.1007/s10555-017-9657-1. [DOI] [PubMed] [Google Scholar]
- 121.Rapozzi V, Zorzet S, Zacchigna M, Della Pietra E, Cogoi S, Xodo LE. Anticancer activity of cationic porphyrins in melanoma tumour-bearing mice and mechanistic in vitro studies. Mol Cancer. 2014;13(1):75. doi: 10.1186/1476-4598-13-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chilakamarthi U, Koteshwar D, Jinka S, Vamsi Krishna N, Sridharan K, Nagesh N, et al. Novel Amphiphilic G-Quadruplex Binding Synthetic Derivative of TMPyP4 and Its Effect on Cancer Cell Proliferation and Apoptosis Induction. Biochemistry. 2018;57(46):6514–6527. doi: 10.1021/acs.biochem.8b00843. [DOI] [PubMed] [Google Scholar]
- 123.Orlotti NI, Cimino-Reale G, Borghini E, Pennati M, Sissi C, Perrone F, et al. Autophagy acts as a safeguard mechanism against G-quadruplex ligand-mediated DNA damage. Autophagy. 2012;8(8):1185–1196. doi: 10.4161/auto.20519. [DOI] [PubMed] [Google Scholar]
- 124.Wu TY, Huang Q, Huang ZS, Hu MH, Tan JH. A drug-like imidazole-benzothiazole conjugate inhibits malignant melanoma by stabilizing the c-MYC G-quadruplex. Bioorg Chem. 2020;99. [DOI] [PubMed]
- 125.Lopes-Nunes J, Lifante J, Shen Y, Ximendes EC, Jaque D. Iglesias-de la Cruz MC, et al. Biological studies of an ICG-tagged aptamer as drug delivery system for malignant melanoma. Eur J Pharm Biopharm. 2020;154:228–235. doi: 10.1016/j.ejpb.2020.07.018. [DOI] [PubMed] [Google Scholar]
- 126.Recagni M, Tassinari M, Doria F, Cimino-Reale G, Zaffaroni N, Freccero M, et al. The Oncogenic Signaling Pathways in BRAF-Mutant Melanoma Cells are Modulated by Naphthalene Diimide-Like G-Quadruplex Ligands. Cells. 2019;8:10. doi: 10.3390/cells8101274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Franceschin M, Rizzo A, Casagrande V, Salvati E, Alvino A, Altieri A, et al. Aromatic Core Extension in the Series of N-Cyclic Bay-Substituted Perylene G-Quadruplex Ligands: Increased Telomere Damage, Antitumor Activity, and Strong Selectivity for Neoplastic over Healthy Cells. ChemMedChem. 2012;7(12):2144–2154. doi: 10.1002/cmdc.201200348. [DOI] [PubMed] [Google Scholar]
- 128.Micheli E, Altieri A, Cianni L, Cingolani C, Iachettini S, Bianco A, et al. Perylene and coronene derivatives binding to G-rich promoter oncogene sequences efficiently reduce their expression in cancer cells. Biochimie. 2016;125:223–231. doi: 10.1016/j.biochi.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 129.Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, et al. Anticancer activity of CX-3543: A direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69(19):7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 130.Hu MH, Wang YQ, Yu ZY, Hu LN, Ou TM, Chen S. Bin, et al. Discovery of a New Four-Leaf Clover-Like Ligand as a Potent c-MYC Transcription Inhibitor Specifically Targeting the Promoter G-Quadruplex. J Med Chem. 2018;61(6):2447–2459. doi: 10.1021/acs.jmedchem.7b01697. [DOI] [PubMed] [Google Scholar]
- 131.Yuan L, Tian T, Chen Y, Yan S, Xing X, Zhang Z, et al. Existence of G-quadruplex structures in promoter region of oncogenes confirmed by G-quadruplex DNA cross-linking strategy. Sci Rep. 2013;3. [DOI] [PMC free article] [PubMed]
- 132.Doria F, Nadai M, Folini M, Di Antonio M, Germani L, Percivalle C, et al. Hybrid ligand-alkylating agents targeting telomeric G-quadruplex structures. Org Biomol Chem. 2012;10(14):2798–2806. doi: 10.1039/c2ob06816h. [DOI] [PubMed] [Google Scholar]
- 133.Mahesh Kumar J, Idris MM, Srinivas G, Vinay Kumar P, Meghah V, Kavitha M, et al. Phenyl 1,2,3-Triazole-Thymidine Ligands Stabilize G-Quadruplex DNA, Inhibit DNA Synthesis and Potentially Reduce Tumor Cell Proliferation over 3′-Azido Deoxythymidine. PLoS One. 2013;8(8):e70798. doi: 10.1371/journal.pone.0070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Casagrande V, Salvati E, Alvino A, Bianco A, Ciammaichella A, D’Angelo C, et al. N-cyclic bay-substituted perylene g-quadruplex ligands have selective antiproliferative effects on cancer cells and induce telomere damage. J Med Chem. 2011;54(5):1140–1156. doi: 10.1021/jm1013665. [DOI] [PubMed] [Google Scholar]
- 135.Leonetti C, Scarsella M, Riggio G, Rizzo A, Salvati E, D’Incalci M, et al. G-quadruplex ligand RHPS4 potentiates the antitumor activity of camptothecins in preclinical models of solid tumors. Clin Cancer Res. 2008;14(22):7284–7291. doi: 10.1158/1078-0432.CCR-08-0941. [DOI] [PubMed] [Google Scholar]
- 136.Biroccio A, Rizzo A, Elli R, Koering CE, Belleville A, Benassi B, et al. TRF2 inhibition triggers apoptosis and reduces tumourigenicity of human melanoma cells. Eur J Cancer. 2006;42(12):1881–1888. doi: 10.1016/j.ejca.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 137.Leonetti C, Amodei S, D’Angelo C, Rizzo A, Benassi B, Antonelli A, et al. Biological activity of the G-quadruplex ligand RHPS4 (3, 11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate) is associated with telomere capping alteration. Mol Pharmacol. 2004;66(5):1138–1146. doi: 10.1124/mol.104.001537. [DOI] [PubMed] [Google Scholar]
- 138.Platella C, Raucci U, Rega N, D’Atri S, Levati L, Roviello GN, et al. Shedding light on the interaction of polydatin and resveratrol with G-quadruplex and duplex DNA: a biophysical, computational and biological approach. Int J Biol Macromol. 2020;151:1163–1172. doi: 10.1016/j.ijbiomac.2019.10.160. [DOI] [PubMed] [Google Scholar]
- 139.Platella C, Guida S, Bonmassar L, Aquino A, Bonmassar E, Ravagnan G, et al. Antitumour activity of resveratrol on human melanoma cells: A possible mechanism related to its interaction with malignant cell telomerase. Biochim Biophys Acta - Gen Subj. 2017;1861(11):2843–2851. doi: 10.1016/j.bbagen.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 140.Peduto A, Pagano B, Petronzi C, Massa A, Esposito V, Virgilio A, et al. Design, synthesis, biophysical and biological studies of trisubstituted naphthalimides as G-quadruplex ligands. Bioorganic Med Chem. 2011;19(21):6419–6429. doi: 10.1016/j.bmc.2011.08.062. [DOI] [PubMed] [Google Scholar]
- 141.Faudale M, Cogoi S, Xodo LE. Photoactivated cationic alkyl-substituted porphyrin binding to g4-RNA in the 5′-UTR of KRAS oncogene represses translation. Chem Commun. 2012;48(6):874–876. doi: 10.1039/C1CC15850C. [DOI] [PubMed] [Google Scholar]
- 142.Ferino A, Nicoletto G, D’Este F, Zorzet S, Lago S, Richter SN, et al. Photodynamic Therapy for ras-Driven Cancers: Targeting G-Quadruplex RNA Structures with Bifunctional Alkyl-Modified Porphyrins. J Med Chem. 2020;63(3):1245–1260. doi: 10.1021/acs.jmedchem.9b01577. [DOI] [PubMed] [Google Scholar]
- 143.Chhabra G, Wojdyla L, Frakes M, Schrank Z, Leviskas B, Ivancich M, et al. Mechanism of Action of G-Quadruplex–Forming Oligonucleotide Homologous to the Telomere Overhang in Melanoma. J Invest Dermatol. 2018;138(4):903–910. doi: 10.1016/j.jid.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 144.Bisht S, Brossart P, Feldmann G. Current Therapeutic Options for Pancreatic Ductal Adenocarcinoma. Oncol Res Treat. 2018;41(10):590–594. doi: 10.1159/000493868. [DOI] [PubMed] [Google Scholar]
- 145.Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer. Cancers. 2017;9. [DOI] [PMC free article] [PubMed]
- 146.Waters AM, Der CJ. KRAS: The critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med. 2018;8:9. doi: 10.1101/cshperspect.a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Prior IA, Lewis PD, Mattos C. A comprehensive survey of ras mutations in cancer. Vol. 72, Cancer Research. 2012. p. 2457–67. [DOI] [PMC free article] [PubMed]
- 148.Weinstein IB. Cancer: Addiction to oncogenes - The Achilles heal of cancer. Vol. 297, Science. 2002. p. 63–4. [DOI] [PubMed]
- 149.Weinstein IB. Disorders in cell circuitry during multistage carcinogenesis: The role of homeostasis. Vol. 21, Carcinogenesis. 2000. p. 857–64. [DOI] [PubMed]
- 150.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3(4):218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Membrino A, Cogoi S, Pedersen EB, Xodo LE. G4-DNA formation in the HRAS promoter and rational design of decoy oligonucleotides for cancer therapy. PLoS One. 2011;6(9):24421. doi: 10.1371/journal.pone.0024421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cogoi S, Shchekotikhin AE, Xodo LE. HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res. 2014;42(13):8379–8388. doi: 10.1093/nar/gku574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cogoi S, Paramasivam M, Spolaore B, Xodo LE. Structural polymorphism within a regulatory element of the human KRAS promoter: Formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008;36(11):3765–3780. doi: 10.1093/nar/gkn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cogoi S, Paramasivam M, Filichev V, Géci I, Pedersen EB, Xodo LE. Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells. J Med Chem. 2009;52(2):564–568. doi: 10.1021/jm800874t. [DOI] [PubMed] [Google Scholar]
- 156.Morgan RK, Batra H, Gaerig VC, Hockings J, Brooks TA. Identification and characterization of a new G-quadruplex forming region within the kRAS promoter as a transcriptional regulator. Biochim Biophys Acta - Gene Regul Mech. 2016;1859(2):235–245. doi: 10.1016/j.bbagrm.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 157.Cogoi S, Xodo LE. G4 DNA in ras genes and its potential in cancer therapy. Vol. 1859, Biochimica et Biophysica Acta - Gene Regulatory Mechanisms. 2016. p. 663–74. [DOI] [PubMed]
- 158.Cogoi S, Paramasivam M, Membrino A, Yokoyama KK, Xodo LE. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J Biol Chem. 2010;285(29):22003–22016. doi: 10.1074/jbc.M110.101923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Amato J, Madanayake TW, Iaccarino N, Novellino E, Randazzo A, Hurley LH, et al. HMGB1 binds to the KRAS promoter G-quadruplex: A new player in oncogene transcriptional regulation? Chem Commun. 2018;54(68):9442–9445. doi: 10.1039/C8CC03614D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kaiser CE, Van Ert NA, Agrawal P, Chawla R, Yang D, Hurley LH. Insight into the Complexity of the i-Motif and G-Quadruplex DNA Structures Formed in the KRAS Promoter and Subsequent Drug-Induced Gene Repression. J Am Chem Soc. 2017;139(25):8522–8536. doi: 10.1021/jacs.7b02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Brown RV, Danford FL, Gokhale V, Hurley LH, Brooks TA. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J Biol Chem. 2011;286(47):41018–41027. doi: 10.1074/jbc.M111.274720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rha SY, Izbicka E, Lawrence R, Davidson K, Sun D, Moyer MP, et al. Effect of Telomere and Telomerase Interactive Agents on Human Tumor and Normal Cell Lines. Clin Cancer Res. 2000;6(3). [PubMed]
- 163.Ohnmacht SA, Marchetti C, Gunaratnam M, Besser RJ, Haider SM, Di Vita G, et al. A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci Rep. 2015;5. [DOI] [PMC free article] [PubMed]
- 164.Pattanayak R, Barua A, Das A, Chatterjee T, Pathak A, Choudhury P, et al. Porphyrins to restrict progression of pancreatic cancer by stabilizing KRAS G-quadruplex: In silico, in vitro and in vivo validation of anticancer strategy. Eur J Pharm Sci. 2018;125:39–53. doi: 10.1016/j.ejps.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 165.Cogoi S, Zorzet S, Rapozzi V, Géci I, Pedersen EB, Xodo LE. MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res. 2013;41(7):4049–4064. doi: 10.1093/nar/gkt127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Paramasivam M, Membrino A, Cogoi S, Fukuda H, Nakagama H, Xodo LE. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: Implications for transcription. Nucleic Acids Res. 2009;37(9):2841–2853. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chu PC, Yang MC, Kulp SK, Salunke SB, Himmel LE, Fang CS, et al. Regulation of oncogenic KRAS signaling via a novel KRAS-integrin-linked kinase-hnRNPA1 regulatory loop in human pancreatic cancer cells. Oncogene. 2016;35(30):3897–3908. doi: 10.1038/onc.2015.458. [DOI] [PubMed] [Google Scholar]
- 168.Gunaratnam M, de la Fuente M, Hampel SM, Todd AK, Reszka AP, Schätzlein A, et al. Targeting pancreatic cancer with a G-quadruplex ligand. Bioorg Med Chem. 2011;19(23):7151–7157. doi: 10.1016/j.bmc.2011.09.055. [DOI] [PubMed] [Google Scholar]
- 169.Miglietta G, Cogoi S, Marinello J, Capranico G, Tikhomirov AS, Shchekotikhin A, et al. RNA G-Quadruplexes in Kirsten Ras (KRAS) Oncogene as Targets for Small Molecules Inhibiting Translation. J Med Chem. 2017;60(23):9448–9461. doi: 10.1021/acs.jmedchem.7b00622. [DOI] [PubMed] [Google Scholar]
- 170.Paluszkiewicz E, Horowska B, Borowa-Mazgaj B, Peszyńska-Sularz G, Paradziej-Łukowicz J, Augustin E, et al. Design, synthesis and high antitumor potential of new unsymmetrical bisacridine derivatives towards human solid tumors, specifically pancreatic cancers and their unique ability to stabilize DNA G-quadruplexes. Eur J Med Chem. 2020;204:112599. doi: 10.1016/j.ejmech.2020.112599. [DOI] [PubMed] [Google Scholar]
- 171.Marchetti C, Zyner KG, Ohnmacht SA, Robson M, Haider SM, Morton JP, et al. Targeting Multiple Effector Pathways in Pancreatic Ductal Adenocarcinoma with a G-Quadruplex-Binding Small Molecule. J Med Chem. 2018;61(6):2500–2517. doi: 10.1021/acs.jmedchem.7b01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ahmed AA, Neidle S. A G-Quadruplex-Binding Small Molecule and the HDAC Inhibitor SAHA (Vorinostat) Act Synergistically in Gemcitabine-Sensitive and Resistant Pancreatic Cancer Cells. Molecules. 2020;25(22):5407. doi: 10.3390/molecules25225407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ahmed AA, Angell R, Oxenford S, Worthington J, Williams N, Barton N, et al. Asymmetrically Substituted Quadruplex-Binding Naphthalene Diimide Showing Potent Activity in Pancreatic Cancer Models. ACS Med Chem Lett. 2020;11(8):1634–1644. doi: 10.1021/acsmedchemlett.0c00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Arjmand F, Sharma S, Parveen S, Toupet L, Yu Z, Cowan JA. Copper(ii) l/d-valine-(1,10-phen) complexes target human telomeric G-quadruplex motifs and promote site-specific DNA cleavage and cellular cytotoxicity. Dalt Trans. 2020;49(28):9888–9899. doi: 10.1039/D0DT01527J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ahmed AA, Marchetti C, Ohnmacht SA, Neidle S. A G-quadruplex-binding compound shows potent activity in human gemcitabine-resistant pancreatic cancer cells. Sci Rep. 2020;10:1. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mpima S, Ohnmacht SA, Barletta M, Husby J, Pett LC, Gunaratnam M, et al. The influence of positional isomerism on G-quadruplex binding and anti-proliferative activity of tetra-substituted naphthalene diimide compounds. Bioorganic Med Chem. 2013;21(20):6162–6170. doi: 10.1016/j.bmc.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 177.Cookson JC, Dai F, Smith V, Heald RA, Laughton CA, Stevens MFG, et al. Pharmacodynamics of the G-quadruplex-stabilizing telomerase inhibitor 3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate (RHPS4) in vitro: Activity in human tumor cells correlates with telomere length and can be enhanced, or antagonized, with cytotoxic agents. Mol Pharmacol. 2005;68(6):1551–1558. doi: 10.1124/mol.105.013300. [DOI] [PubMed] [Google Scholar]
- 178.Liu W, Sun D, Hurley LH. Binding of G-quadruplex-interactive agents to distinct G-quadruplexes induces different biological effects in MiaPaCa cells. Nucleosides, Nucleotides and Nucleic Acids. 2005;24(10–12):1801–1815. doi: 10.1080/15257770500267238. [DOI] [PubMed] [Google Scholar]
- 179.Hampel SM, Sidibe A, Gunaratnam M, Riou J-F, Neidle S. Tetrasubstituted naphthalene diimide ligands with selectivity for telomeric G-quadruplexes and cancer cells. Bioorg Med Chem Lett. 2010;20(22):6459–6463. doi: 10.1016/j.bmcl.2010.09.066. [DOI] [PubMed] [Google Scholar]
- 180.Liu JN, Deng R, Guo JF, Zhou JM, Feng GK, Huang ZS, et al. Inhibition of myc promoter and telomerase activity and induction of delayed apoptosis by SYUIQ-5, a novel G-quadruplex interactive agent in leukemia cells. Leukemia. 2007;21:1300–2. [DOI] [PubMed]
- 181.Local A, Zhang H, Benbatoul KD, Folger P, Sheng X, Tsai CY, et al. APTO-253 stabilizes G-quadruplex DNA, inhibits MYC expression, and induces DNA damage in acute myeloid leukemia cells. Mol Cancer Ther. 2018;17(6):1177–1186. doi: 10.1158/1535-7163.MCT-17-1209. [DOI] [PubMed] [Google Scholar]
- 182.Zidanloo SG, Hosseinzadeh Colagar A, Ayatollahi H, Raoof JB. Downregulation of the WT1 gene expression via TMPyP4 stabilization of promoter G-quadruplexes in leukemia cells. Tumor Biol. 2016;37(7):9967–9977. doi: 10.1007/s13277-016-4881-9. [DOI] [PubMed] [Google Scholar]
- 183.Chu B, Zhang Y, Liu HX, Bai YX, Zuo XG, Lu MQ, et al. Induction of apoptosis of leukemic tumor cells in mouse model with G-quadruplex ligand Tel03. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010;18(1):57–60. [PubMed] [Google Scholar]
- 184.Nakajima A, Tauchi T, Sashida G, Sumi M, Abe K, Yamamoto K, et al. Telomerase inhibition enhances apoptosis in human acute leukemia cells: Possibility of antitelomerase therapy. Leukemia. 2003;17(3):560–567. doi: 10.1038/sj.leu.2402825. [DOI] [PubMed] [Google Scholar]
- 185.Goh YY, Yan YK, Tan NS, Goh SA, Li S, Teoh YC, et al. Downregulation of oncogenic RAS and c-Myc expression in MOLT-4 leukaemia cells by a salicylaldehyde semicarbazone copper(II) complex. Sci Rep. 2016;6(1):1–12. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Thandapani P, Song J, Gandin V, Cai Y, Rouleau SG, Garant JM, et al. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. Elife. 2015;4(AUGUST2015):1–30. doi: 10.7554/eLife.06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Katapadi VK, Nambiar M, Raghavan SC. Potential G-quadruplex formation at breakpoint regions of chromosomal translocations in cancer may explain their fragility. Genomics. 2012;100(2):72–80. doi: 10.1016/j.ygeno.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 188.Williams JD, Fleetwood S, Berroyer A, Kim N, Larson ED. Sites of instability in the human TCF3 (E2A) gene adopt G-quadruplex DNA structures in vitro. Front Genet. 2015;6(MAY). [DOI] [PMC free article] [PubMed]
- 189.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic Nucleophosmin in Acute Myelogenous Leukemia with a Normal Karyotype. N Engl J Med. 2005;352(3):254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 190.Federici L, Arcovito A, Scaglione GL, Scaloni F, Lo Sterzo C, Di Matteo A, et al. Nucleophosmin C-terminal leukemia-associated domain interacts with G-rich quadruplex forming DNA. J Biol Chem. 2010;285(48):37138–37149. doi: 10.1074/jbc.M110.166736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chiarella S, De Cola A, Scaglione GL, Carletti E, Graziano V, Barcaroli D, et al. Nucleophosmin mutations alter its nucleolar localization by impairing G-quadruplex binding at ribosomal DNA. Nucleic Acids Res. 2013;41(5):3228–3239. doi: 10.1093/nar/gkt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513(7516):65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chen WJ, Zhou CX, Yao PF, Wang XX, Tan JH, Li D, et al. Disubstituted 1,8-dipyrazolcarbazole derivatives as a new type of c-myc G-quadruplex binding ligands. Bioorganic Med Chem. 2012;20(9):2829–2836. doi: 10.1016/j.bmc.2012.03.031. [DOI] [PubMed] [Google Scholar]
- 194.Guo QL, Su HF, Wang N, Liao SR, Lu YT, Ou TM, et al. Synthesis and evaluation of 7-substituted-5,6-dihydrobenzo[c]acridine derivatives as new c-KIT promoter G-quadruplex binding ligands. Eur J Med Chem. 2017;130:458–471. doi: 10.1016/j.ejmech.2017.02.051. [DOI] [PubMed] [Google Scholar]
- 195.Ma Y, Ou TM, Hou JQ, Lu YJ, Tan JH, Gu LQ, et al. 9-N-Substituted berberine derivatives: Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc. Bioorganic Med Chem. 2008;16(16):7582–7591. doi: 10.1016/j.bmc.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 196.Xiong YX, Su HF, Lv P, Ma Y, Wang SK, Miao H, et al. A newly identified berberine derivative induces cancer cell senescence by stabilizing endogenous G-quadruplexes and sparking a DNA damage response at the telomere region. Oncotarget. 2015;6(34):35625–35635. doi: 10.18632/oncotarget.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Liao SR, Zhou CX, Wu W, Bin OTM, Tan JH, Li D, et al. 12-N-Methylated 5,6-dihydrobenzo[c]acridine derivatives: A new class of highly selective ligands for c-myc G-quadruplex DNA. Eur J Med Chem. 2012;53:52–63. doi: 10.1016/j.ejmech.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 198.Kang HJ, Park HJ. Novel molecular mechanism for actinomycin D activity as an oncogenic promoter G-quadruplex binder. Biochemistry. 2009;48(31):7392–7398. doi: 10.1021/bi9006836. [DOI] [PubMed] [Google Scholar]
- 199.Ou Z, Wang Y, Gao Y, Wang X, Qian Y, Li Y, et al. Targeting human telomeric and c-myc G-quadruplexes with alkynylplatinum(II) terpyridine complexes under molecular crowding conditions. J Inorg Biochem. 2017;166:126–134. doi: 10.1016/j.jinorgbio.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 200.Zorzan E, Elgendy R, Giantin M, Dacasto M, Sissi C. Whole-Transcriptome Profiling of Canine and Human in Vitro Models Exposed to a G-Quadruplex Binding Small Molecule. Sci Rep. 2018;8:1. doi: 10.1038/s41598-018-35516-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zorzan E, Da Ros S, Musetti C, Shahidian LZ, Coelho NFR, Bonsembiante F, et al. Screening of candidate G-quadruplex ligands for the human c-KIT promotorial region and their effects in multiple in-vitro models. Oncotarget. 2016;7(16):21658–21675. doi: 10.18632/oncotarget.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kang HJ, Park HJ. In silico identification of novel ligands for G-quadruplex in the c-MYC promoter. J Comput Aided Mol Des. 2015;29(4):339–348. doi: 10.1007/s10822-014-9826-z. [DOI] [PubMed] [Google Scholar]
- 203.Machireddy B, Sullivan HJ, Wu C. Binding of BRACO19 to a telomeric G-quadruplex DNA probed by all-atom molecular dynamics simulations with explicit solvent. Molecules. 2019;24:6. doi: 10.3390/molecules24061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Das RN, Chevret E, Desplat V, Rubio S, Mergny JL, Guillon J. Design, synthesis and biological evaluation of new substituted diquinolinyl-pyridine ligands as anticancer agents by targeting G-Quadruplex. Molecules. 2018;23(1). [DOI] [PMC free article] [PubMed]
- 205.Liu HY, Chen AC, Yin QK, Li Z, Huang SM, Du G, et al. New Disubstituted Quindoline Derivatives Inhibiting Burkitt’s Lymphoma Cell Proliferation by Impeding c-MYC Transcription. J Med Chem. 2017;60(13):5438–5454. doi: 10.1021/acs.jmedchem.7b00099. [DOI] [PubMed] [Google Scholar]
- 206.Ghazaey Zidanloo S, Hosseinzadeh Colagar A, Ayatollahi H, Bagheryan Z. G-quadruplex forming region within WT1 promoter is selectively targeted by daunorubicin and mitoxantrone: A possible mechanism for anti-leukemic effect of drugs. J Biosci. 2019;44(1):1–9. doi: 10.1007/s12038-018-9837-3. [DOI] [PubMed] [Google Scholar]
- 207.Montoya JJ, Turnidge MA, Wai DH, Patel AR, Lee DW, Gokhale V, et al. In vitro activity of a G-quadruplex-stabilizing small molecule that synergizes with Navitoclax to induce cytotoxicity in acute myeloid leukemia cells. BMC Cancer. 2019;19(1):1251. doi: 10.1186/s12885-019-6464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Li Z, Tan JH, He JH, Long Y, Ou TM, Li D, et al. Disubstituted quinazoline derivatives as a new type of highly selective ligands for telomeric G-quadruplex DNA. Eur J Med Chem. 2012;47(1):299–311. doi: 10.1016/j.ejmech.2011.10.057. [DOI] [PubMed] [Google Scholar]
- 209.Wang KB, Li DH, Hu P, Wang WJ, Lin C, Wang J, et al. A Series of β-Carboline Alkaloids from the Seeds of Peganum harmala Show G-Quadruplex Interactions. Org Lett. 2016;18(14):3398–3401. doi: 10.1021/acs.orglett.6b01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bertrand B, Fernandez-Cestau J, Angulo J, Cominetti MMD, Waller ZAE, Searcey M, et al. Cytotoxicity of Pyrazine-Based Cyclometalated (C^Npz^C)Au(III) Carbene Complexes: Impact of the Nature of the Ancillary Ligand on the Biological Properties. Inorg Chem. 2017;56(10):5728–5740. doi: 10.1021/acs.inorgchem.7b00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.He JH, Liu HY, Li Z, Tan JH, Ou TM, Huang SL, et al. New quinazoline derivatives for telomeric G-quadruplex DNA: Effects of an added phenyl group on quadruplex binding ability. Eur J Med Chem. 2013;63:1–13. doi: 10.1016/j.ejmech.2013.01.051. [DOI] [PubMed] [Google Scholar]
- 212.Ma Y, Ou TM, Tan JH, Hou JQ, Huang SL, Gu LQ, et al. Quinolino-benzo-[5, 6]-dihydroisoquindolium compounds derived from berberine: A new class of highly selective ligands for G-quadruplex DNA in c-myc oncogene. Eur J Med Chem. 2011;46(5):1906–1913. doi: 10.1016/j.ejmech.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 213.Mikami-Terao Y, Akiyama M, Yuza Y, Yanagisawa T, Yamada O, Yamada H. Antitumor activity of G-quadruplex-interactive agent TMPyP4 in K562 leukemic cells. Cancer Lett. 2008;261(2):226–234. doi: 10.1016/j.canlet.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 214.De Cola A, Pietrangelo L, Forlì F, Barcaroli D, Budani MC, Graziano V, et al. AML cells carrying NPM1 mutation are resistant to nucleophosmin displacement from nucleoli caused by the G-quadruplex ligand TmPyP4. Vol. 5, Cell Death and Disease. 2014. p. e1427. [DOI] [PMC free article] [PubMed]
- 215.Munira Haidad Ali S, Yan YK, Lee PPF, Khong KZX, Alam Sk M, Lim KH, et al. Copper(II) complexes of substituted salicylaldehyde dibenzyl semicarbazones: Synthesis, cytotoxicity and interaction with quadruplex DNA. Dalt Trans. 2014;43(3):1449–1459. doi: 10.1039/C3DT52297K. [DOI] [PubMed] [Google Scholar]
- 216.Su L, Zheng H, Li Z, Qiu J, Chen S, Liu J, et al. Mechanistic studies on the anticancer activity of 2,4-disubstituted quinazoline derivative. Biochim Biophys Acta - Gen Subj. 2014;1840(10):3123–3130. doi: 10.1016/j.bbagen.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 217.Zhou JM, Zhu XF, Lu YJ, Deng R, Huang ZS, Mei YP, et al. Senescence and telomere shortening induced by novel potent G-quadruplex interactive agents, quindoline derivatives, in human cancer cell lines. Oncogene. 2006;25(4):503–511. doi: 10.1038/sj.onc.1209067. [DOI] [PubMed] [Google Scholar]
- 218.Shen FH, Jin J, Li J, Wang Y, Zhu SH, Lu YJ, et al. The G-quadruplex ligand, SYUIQ-FM05, targets proto-oncogene c-kit transcription and induces apoptosis in K562 cells. Pharm Biol. 2013;51(4):447–454. doi: 10.3109/13880209.2012.738424. [DOI] [PubMed] [Google Scholar]
- 219.Bhattacharjee S, Chakraborty S, Chorell E, Sengupta PK, Bhowmik S. Importance of the hydroxyl substituents in the B–ring of plant flavonols on their preferential binding interactions with VEGF G–quadruplex DNA: Multi-spectroscopic and molecular modeling studies. Int J Biol Macromol. 2018;118(Pt A):629–39. [DOI] [PubMed]
- 220.Paeschke K, Bochman ML, Daniela Garcia P, Cejka P, Friedman KL, Kowalczykowski SC, et al. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497(7450):458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bacolla A, Ye Z, Ahmed Z, Tainer JA. Cancer mutational burden is shaped by G4 DNA, replication stress and mitochondrial dysfunction. Prog Biophys Mol Biol. 2019;147:47–61. doi: 10.1016/j.pbiomolbio.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hänsel-Hertsch R, Simeone A, Shea A, Hui WWI, Zyner KG, Marsico G, et al. Landscape of G-quadruplex DNA structural regions in breast cancer. Nat Genet. 2020;52(9):878–83. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable