Abstract
Pancreatic cancer is one of the deadliest diseases, due to the lack of early symptoms and resistance to current therapies, including radiotherapy. However, the mechanisms of radioresistance in pancreatic cancer remain unknown. The present study explored the role of microRNA-153 (miR-153) in radioresistance of pancreatic cancer. It was observed that miR-153 was downregulated in pancreatic cancer and positively correlated with patient survival time. Using stably-infected pancreatic cancer cells that overexpressed miR-153 or miR-153 inhibitor, it was found that miR-153 overexpression sensitized pancreatic cancer cells to radiotherapy by inducing increased cell death and decreased colony formation, while cells transfected with the miR-153 inhibitor promoted radioresistance. Further investigation demonstrated that miR-153 promoted radiosensitivity by directly targeting jagged canonical Notch ligand 1 (JAG1). The addition of recombinant JAG1 protein in the cell cultures reversed the therapeutic effect of miR-153. The present study revealed a novel mechanism of radioresistance in pancreatic cancer and indicated that miR-153 may serve as a potential therapeutic target for radioresistance.
Keywords: microRNA-153, jagged canonical Notch ligand 1, pancreatic cancer, radioresistance, cell apoptosis
Introduction
Pancreatic cancer is one of the most lethal diseases, with a 5-year survival rate of ~5% (1). Due to the absence of obvious symptoms during the early stages, patients diagnosed with pancreatic cancer often have late-stage disease, where the tumour may have metastasized to other organs, and have missed the opportunity to be treated with surgical resection. Unfortunately, pancreatic cancer is relatively resistant to chemotherapy and radiotherapy compared with other types of cancer, such as breast or lung cancer. Thus, it is crucial to understand the mechanisms of radioresistance (2,3).
MicroRNA (miRNA/miR) is a type of small non-coding RNA of ~23 nucleotides in length (4). They exert their regulatory roles by binding to the 3′ untranslated region (UTR) of mRNAs and inhibiting their transcription. Due to the extensiveness of their targets, a single miRNA can regulate multiple molecules and serve important roles in a series of physiological and pathological processes, including cancer (5).
Radioresistance is often thought to be regulated by cell stemness. In the process of searching for a candidate miRNA that can modulate radioresistance in pancreatic cancer, it was found that multiple stem cell-related genes were targeted by miR-153, including jagged canonical Notch ligand 1 (JAG1) and Kruppel-like factors (6,7). Of note, miR-153 has previously been reported to regulate radiosensitivity in human glioma (8,9), and to influence the efficacy of other therapies (10). However, the role of miR-153 in the radioresistance of pancreatic cancer remains unknown. In the present study, it was found that miR-153 could sensitize pancreatic cancer to radiotherapy by directly targeting JAG1.
Materials and methods
Cell culture
The human pancreatic cancer cell line SW1990 [American Type Culture Collection (ATCC); cat. no. CRL-2172] was cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and the MIA PaCa-2 cell line (ATCC; cat. no. CRM-CRL-1420) was cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS.
Animal studies
Experimental procedures involving animals were approved by the Taizhou People's Hospital Institutional Animal Care and Use Committee (Taizhou, China). Briefly, 5×106 miR-153-overexpressing or control SW1990 cells were subcutaneously injected into 10, 6–8 week-old (~20 g weight), male BALB/c nude mice, with 5 mice in each group. The mice were supplied by Shanghai SLAC Laboratory Animal Co., Ltd and housed at ~22°C, ~50% humidity with a 12/12 light/dark cycle and free access to the food and water. When the tumours reached 500 mm3, they were irradiated by a single dose of 4 Gy for ~2 min. The tumour volume was measured every 3 days and, 15 days later, the mice were anaesthetized with sodium pentobarbital (40 mg/kg) by intraperitoneal injection, and then sacrificed by cervical dislocation. The largest tumour diameter was 15 mm, and the largest tumour volume was 1,365 mm3.
Lentivirus infection
The miR-153 overexpression and inhibition lentivirus and their controls were purchased from OBiO Technology (Shanghai) Corp., Ltd. Lentivirus was then added to the cell culture media at a cell:virus ratio of 1:10 with 8 µg/ml polybrene (Merck KGaA). Following incubation for 24 h, cells were changed with fresh culture media. The stably transfected cells were selected using puromycin (final concentration, 1 µg/ml) and fluorescence-activated cell sorting.
JAG1 treatment
After plating 2×105 SW1990 cells/well in 6-well plates, the JAG1 recombinant protein (50 ng/ml; cat. no. 1277-JG; R&D Systems, Inc.) was added. The medium was changed every 2 days with fresh medium containing recombinant protein until the cells were used for analysis.
Dual-luciferase reporter assay
The pmirGLO reporter plasmid containing the wild-type and mutant 3′UTR segment of the miR-153 binding site in JAG1, and the control, were constructed by OBiO Technology (Shanghai) Corp., Ltd. The reporter plasmid was co-transfected with miR-153 mimic (5′-UUGCAUAGUCACAAAAGUGAUC-3′) and mimic-negative control (NC) (5′-UUUGUACUACACAAAAGUACUG-3′) (Guangzhou RiboBio Co., Ltd.) into SW1990 cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Six hours following transfection, the culture medium was replaced with fresh culture media, and 72 h following transfection, SW1990 cells were collected and analysed using Dual-Luciferase® Reporter Assay (Promega Corporation), according to the manufacturer's instructions and the results were normalized to Renilla luciferase activity.
Cell viability assay
Cell viability was analysed using Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.), according to the manufacturer's instructions. Briefly, 1×105 SW1990 cells/well were seeded into 6-well plates. Following culture for 6 h, the cells were treated with 4 Gy radiotherapy. Untreated cells were used as control. After 48 h, the cells were incubated with 10 µl CCK-8 per 100 µl culture media for 2 h at 37°C. The absorbance was measured at 450 nm using a Varioskan Flash microplate reader (Thermo Fisher Scientific, Inc.).
Cell apoptosis analysis
Cell apoptosis was analysed by flow cytometry. SW1990 cells were treated as described in the ‘Cell viability assay’ section. At 48 h after radiotherapy, cells were collected and washed twice with cold PBS. Cell apoptosis was evaluated using the Annexin V-FITC/PI apoptosis detection kit (BD Biosciences), according to the manufacturer's instructions. Data were collected by flow cytometry (BD Accuri C6) BD Bioscience) and analysed by FlowJo v.10.5.2 (BD Bioscience).
Colony formation assay
SW1990 cells (1×104) were seeded into 6-well plates. After 6 h of culture, cells were exposed to 0, 2, 4 and 6 Gy radiotherapy. The cell culture medium was replaced with fresh culture media every 3–4 days. After 12–14 days, the colonies were fixed using 4% paraformaldehyde at room temperature for 30 min and stained with crystal violet solution (Beyotime Institute of Biotechnology). Cell plates were scanned and colonies were counted using ImageJ v.1.8.0 (National Institutes of Health).
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR)
Cells were washed twice with PBS and lysed in TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). Briefly, total RNA was separated by chloroform and precipitated by isopropanol. Following washing with 75% ethanol, RNA was dried and resuspended in RNase-free ddH2O. RNA from SW1990 cells was reverse-transcribed with PrimeScript™ RT reagent kit (Takara Bio, Inc.) according to the manufacturer's protocol. qPCR was performed using SYBR® Premix Ex Taq™ (Takara Bio, Inc.), according to the manufacturer's instructions with the following thermocycling conditions: 95°C for 5 sec, 60°C for 34 sec for 40 cycles. The relative expression levels were determined using the −2∆∆Cq method (11). Primers for GAPDH were purchased from Sangon Biotech Co., Ltd. (cat. no. B661104). The other primers used in this study are as follows: miR153, forward, 5′-CATGCTAGCTCTCTCTCCCTCCCTCTTTCCC-3′ and reverse, 5′-GCGGATCCCCGTTAGCAATACAAACCAACCC-3′; RT primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGATCAC-3′; U6, forward, 5′-CAAGGATGACACGCAAA-3′ and reverse, 5′-TCAACTGGTGTCGTGG-3′; RT primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAAATAT-3′; JAG1, forward, 5′-ATTACCAGGATAACTGTGCGAA-3′ and reverse, 5′-CAAATGTGCTCCGTAGTAAGAC-3′.
Western blotting
Cells were subjected to lysis with RIPA buffer (Thermo Fisher Scientific, Inc.) supplemented with the complete protease inhibitor cocktail (Roche Diagnostics) for 10 min on ice. The lysates were then centrifuged at 15,000 × g for 30 min at 4°C to remove cell debris. Protein concentrations were determined using a BCA Protein Assay kit (Beyotime Institute of Biotechnology). A total of 40 µg cellular proteins were then separated by 10% SDS-PAGE and transferred to a nitrocellulose membrane. Membranes were blocked with 5% skimmed milk in TBS at room temperature for 1 h and incubated with primary antibody, anti-GAPDH (Cell Signalling Technology, Inc.; cat. no. 2118; 1:1,000) or anti-JAG1 (Cell Signalling Technology, Inc.; cat. no. 70109; 1:1,000), according to the manufacturer's instructions. Following primary antibody incubation, the secondary antibody IRDye 800CW Goat anti-Rabbit IgG (1:5,000; cat. no. 925-32211; LI-COR Biosciences) were used to incubate the membrane at room temperature for 1 h, and the membranes were then subjected to imaging using the Odyssey Imaging Systems (LI-COR Biosciences).
Bioinformatics
Survival analysis of pancreatic cancer patients with different miR-153 expression was performed on the website http://www.oncolnc.org/, using the data from The Cancer Genome Atlas (TCGA) Program (https://www.cancer.gov/tcga). In addition, expression data for miR-153 and JAG1 were download from the oncolnc website and analysed using GraphPad Prism 7 (GraphPad Software, Inc.).
Statistical analysis
All experiments were repeated at least three times. Data are presented as the mean ± SD. Statistical analysis was performed using a one-way ANOVA followed by Tukey's honest significant difference test on GraphPad Prism 7 (GraphPad Software, Inc.). When comparing two groups, unpaired Student's t test was performed. Pearson's r correlation was used for correlation analysis of miR-153 and JAG1 in pancreatic ductal adenocarcinoma tissues. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-153 is downregulated in pancreatic cancer and associated with poor prognosis
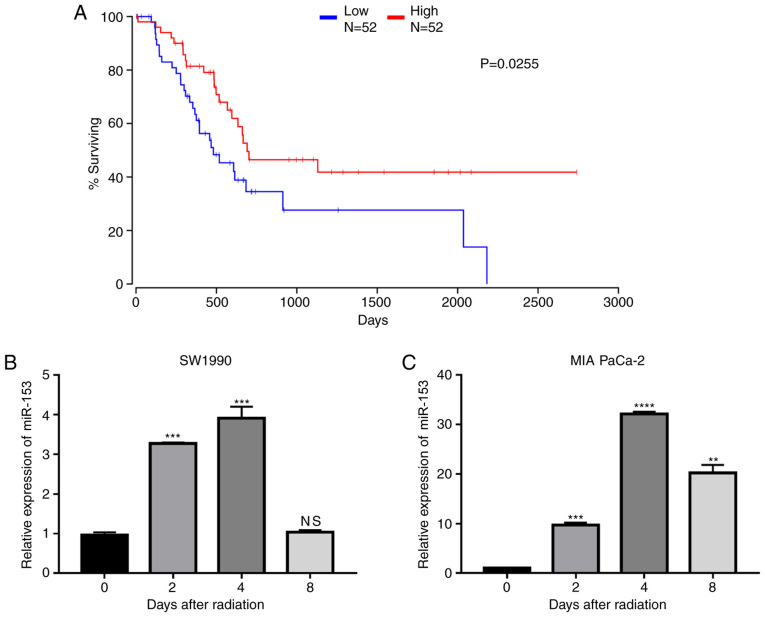
To explore the role of miR-153 in pancreatic cancer, data from TCGA were analysed and it was found that miR-153 expression levels were positively associated with patient survival (Fig. 1A), indicating that miR-153 may serve as a target in the treatment of pancreatic cancer. The expression changes of miR-153 following radiation in two pancreatic cancer cell lines were further investigated. The results demonstrated that miR-153 expression levels were significantly upregulated in both cell lines following radiation (Fig. 1B and C).
Figure 1.
miR-153 is correlated with the prognosis of patients with pancreatic cancer and is upregulated following radiation. (A) Survival rate of patients with pancreatic ductal adenocarcinoma divided by different expression levels of miR-153. (B) miR-153 expression levels in SW1990 and (C) MIA PaCa-2 pancreatic cancer cell lines following radiation. U6 small nuclear RNA was used as an endogenous control. **P<0.01, ***P<0.001 and ****P<0.0001 compared with the 0-day control group. miR-153, microRNA-153; NS, not significant.
miR-153 sensitizes SW1990 cells to radiotherapy
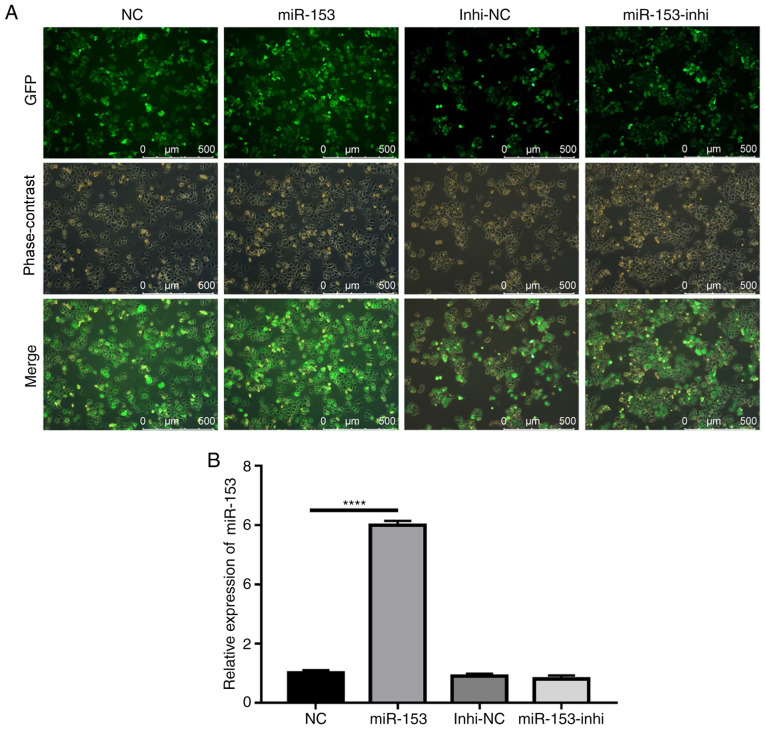
The potential role of miR-153 in the radioresistance of pancreatic cancer was subsequently explored. Using lentiviruses expressing miR-153 mimic or miR-153 inhibitor, SW1990 cell lines that stably overexpressed miR-153 mimic or miR-153 inhibitor were successfully established. The efficiency of stable infection was detected by green fluorescent protein expression (Fig. 2A). The miR-153 expression levels in the stably infected cell lines were confirmed by RT-qPCR (Fig. 2B). The results indicated that miR-153 was significantly overexpressed in the mimic-transduced cells, and miR-153 inhibition did not cause any significant changes to miR-153 expression, as compared with the control.
Figure 2.
Establishment of stably transduced SW1990 cell lines. (A) Representative images showing GFP expression in stably transduced cells. (B) Reverse transcription-quantitative PCR analysis of miR-153 expression levels in the stably transduced cell lines. ****P<0.0001. miR-153, microRNA-153; GFP, green fluorescent protein; NC, negative control; Inhi, inhibitor.
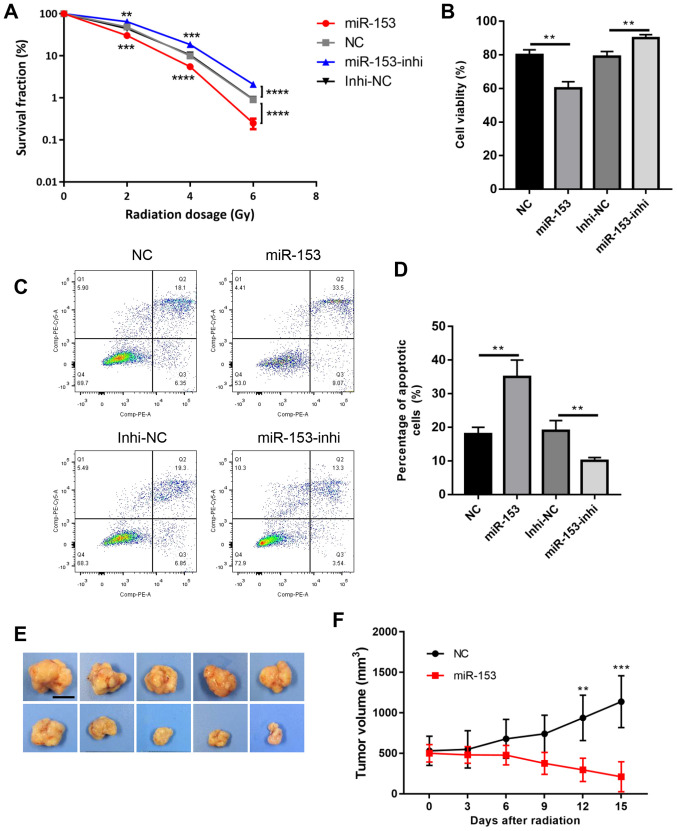
These cells were then subjected to radiotherapy and the colony formation survival assay, which is considered the classic method of evaluating radioresistance. The results revealed that miR-153 overexpression significantly reduced colony formation following radiation, while miR-153 inhibition increased cell survival in a dose-dependent manner (Fig. 3A). Cell viability was then analysed using a CCK-8 assay, and it was found that cell viability was significantly decreased in the miR-153 overexpression group following radiation, while miR-153 inhibition had the opposite effect (Fig. 3B). In addition, cell apoptosis was evaluated following radiation, and the results showed that miR-153 overexpression increased cell apoptosis, while miR-153 inhibition had the opposite effect (Fig. 3C and D). Furthermore, the effect of miR-153 in radioresistance was investigated in vivo. Radiation had little effect in the control group and the tumours continued to grow following radiation treatment. By contrast, miR-153 overexpression significantly inhibited radioresistance and the tumours shrank following radiation treatment (Fig. 3E and F). In combination, the present results indicated that miR-153 sensitized SW1990 cells to radiotherapy.
Figure 3.
miR-153 inhibits radiation resistance. (A) Colony survival fraction at 14 days after 0, 2, 4 and 6 Gy radiation in stably transduced cells. (B) Cell Counting Kit-8 assay showing the percentage of viable cells following 4 Gy radiation in the stably transduced cells. (C) Representative plots and (D) quantification of flow cytometry analysis showing the percentage of apoptotic cells after 4 Gy radiation in stably transduced cells. (E) Representative photographs of the tumours at 15 days following radiation (scale bar, 1 cm). (F) Growth rate of the subcutaneous tumours following irradiation. **P<0.01, ***P<0.001 and ****P<0.0001, with comparisons shown by lines. miR-153, microRNA-153; NC, negative control; Inhi, inhibitor.
miR-153 affects radioresistance by directly targeting JAG1
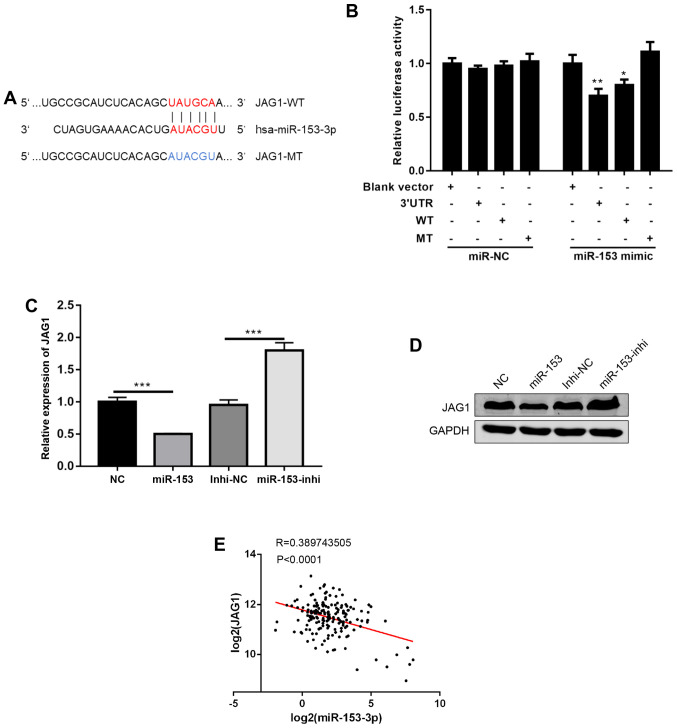
To further investigate the mechanisms of miR-153, a commonly used miRNA target prediction website, TargetScan (http://www.targetscan.org/vert_72/), was used. It was found that miR-153 could bind to the 3′UTR of JAG1, a ligand in NOTCH signalling (Fig. 4A) (12). Using a dual luciferase reporter assay, the direct binding of miR-153 to the 3′UTR of JAG1 was confirmed (Fig. 4B). Next, JAG1 mRNA and protein expression levels were detected following miR-153 overexpression or inhibition, and it was found that miR-153 overexpression decreased JAG1 expression levels, while miR-153 inhibition increased the JAG1 expression levels (Fig. 4C and D). Of note, data from human pancreatic cancer tissues were analysed, and it was found that the expression of miR-153 was negatively correlated with JAG1 expression (Fig. 4E). In combination, the present results suggested that JAG1 is a direct target of miR-153.
Figure 4.
JAG1 is a direct target of miR-153. (A) Predicted binding sites between JAG1 and miR-153-3p (shown in red print). (B) Relative luciferase activity in SW1990 cells co-transfected with miR-153 mimic or miR-NC and JAG1-WT, JAG1-MT or JAG1-3′UTR (the JAG1-3′UTR contains the whole length of 3′UTR). *P<0.05, **P<0.01 compared with the blank vector control. (C) Reverse transcription-quantitative PCR analysis of JAG1 mRNA expression levels in stably transduced cells. ***P<0.001 with comparisons shown by lines. (D) Western blot quantification of JAG1 protein expression levels in stably transduced cells. (E) Pearson's correlation analysis of the expression levels (log2) between miR-153 and JAG1 in pancreatic ductal adenocarcinoma tissues. JAG1, jagged canonical Notch ligand 1; miR-153, microRNA-153; NC, negative control; WT, wild-type; MT, mutant; UTR, untranslated region; Inhi, inhibitor.
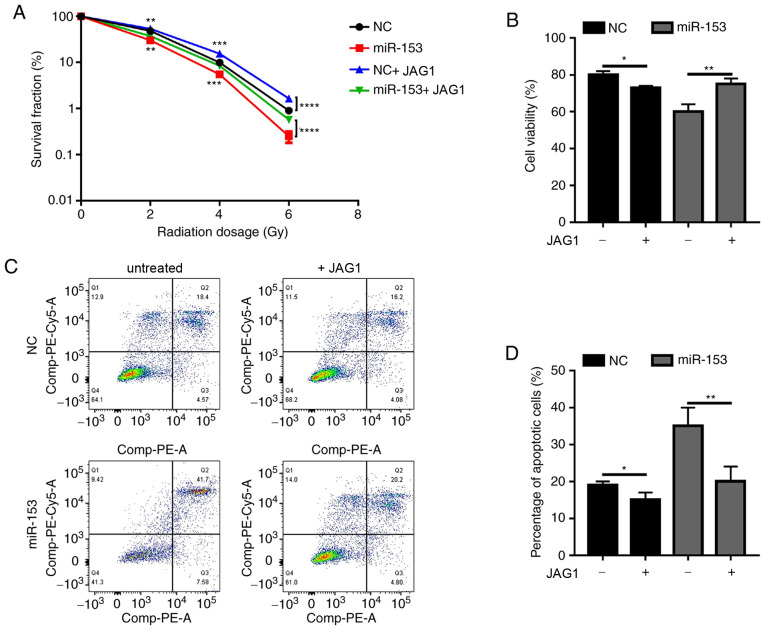
Next, the present study investigated whether miR-153 exerted its function through JAG1. Since JAG1 expression was decreased by miR-153, a recombinant JAG1 protein was used to determine if it could rescue the phenotypic effects of miR-153 overexpression in pancreatic cancer cells. In the miR-153 overexpression group, JAG1 treatment significantly increased the colony formation ability of irradiated cancer cells (Fig. 5A). Of note, when JAG1 was added in the NC group, it was also able to promote radioresistance (Fig. 5A). The results of the cell viability assay (Fig. 5B) and apoptosis analysis (Fig. 5C and D) were similar. Addition of exogenous JAG1 protein in the cultures significantly increased cell viability and decreased the percentage of apoptotic cells. The present results demonstrated that miR-153 sensitized pancreatic cancer cells to radiotherapy by inhibiting JAG1.
Figure 5.
miR-153 sensitizes SW1990 to radiation through JAG1. (A) Colony survival fraction at 14 days after 0, 2, 4 and 6 Gy radiation in miR-153-overexpressing cells with or without JAG1 treatment. (B) Cell Counting Kit-8 assay showing the percentage of viable cells after 4 Gy radiation in miR-153-overexpressing cells with or without JAG1 treatment. (C) Representative plots and (D) quantification of flow cytometry analysis showing the percentage of apoptotic cells after 4 Gy radiation in miR-153-overexpressing cells with or without JAG1 treatment. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 with comparisons shown by lines. miR-153, microRNA-153; JAG1, jagged canonical Notch ligand 1; NC, negative control.
Discussion
Pancreatic cancer is a deadly disease that is resistant to most current treatments for cancer, particularly radiotherapy. Unfortunately, the underlying mechanism of radioresistance in pancreatic cancer remains unclear. In the present study, it was found that miR-153 could sensitize pancreatic cancer cells to radiotherapy by directly targeting and downregulating JAG1.
In the present study, a new target of miR-153 was confirmed. In addition, it was found that miR-153 contributed to radioresistance in pancreatic cancer cells. miR-153 has been previously reported to regulate radiosensitivity (8,9), as well as other processes, in various types of cancer. For example, miR-153 was reported to mediate cell proliferation and apoptosis in renal cancer (13), inhibit migration in breast (14,15) and lung cancer (16), act as a prognostic marker for gastric (17) and cervical cancer (18), mediate immune escape from natural killer cells in pancreatic cancer (19), and enhance chimeric antigen receptor T cell immunotherapy in colon cancer (20). In pancreatic cancer, miR-153 mainly has antitumour properties. miR-153 was reported to act as a prognostic marker in pancreatic cancer, inhibit cell migration and invasion (21), and enhance the therapeutic effect of gemcitabine by targeting snail family transcriptional repressor 1 (22). As multiple cancer stem cell-related genes have been predicted to be targeted by miR-153 (6,7), the effect of miR-153 may be mediated by suppressing cancer cell stemness. Indeed, miR-153 was previously found to inhibit cancer cell stemness and hinder tumour growth (6).
The present study revealed a new role of JAG1 in pancreatic cancer, which was its potential to increase resistance to radiotherapy. JAG1 is one of the ligands for NOTCH signalling, a well-known stem cell signalling pathway. JAG1 also participates in numerous physiological and pathological processes. Consistent with the present results, as the ligand for NOTCH signalling, JAG1 upregulation was previously reported to mediate epithelial-mesenchymal transition and subsequently chemoresistance of pancreatic cancer (23). Furthermore, JAG1 was found to be highly expressed in pancreatic cancer compared with normal pancreatic tissue, and JAG1 silencing by small interfering RNA or miRNA significantly inhibited cell growth, migration, and invasion in pancreatic cancer cells (24,25). A higher JAG1 expression was reported to be associated with a poorer prognosis in patients with pancreatic ductal adenocarcinoma (26).
In conclusion, the present study determined the important role of miR-153 in mediating radioresistance in pancreatic cancer cells and demonstrated that miR-153 exerted its function by directly targeting and inhibiting JAG1. These findings provided two new potential therapeutic targets, miR-153 and JAG1, for pancreatic cancer.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- miR-153
microRNA-153
- WT
wild-type
- MT
mutant
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the TCGA Research Network (https://www.cancer.gov/tcga).
Authors' contributions
ZZ and XS performed experiments, analysed data and wrote the manuscript. DZ, HX, HK, and BY performed experiments and analyzed data. LY designed the experiments, supervised the project and revised the manuscript. ZZ, XS and LY confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Experimental procedures involving animals were approved by the Taizhou People's Hospital Institutional Animal Care and Use Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Halbrook CJ, Lyssiotis CA. Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell. 2017;31:5–19. doi: 10.1016/j.ccell.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, Hruban RH. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 6.Zhao G, Zhang Y, Zhao Z, Cai H, Zhao X, Yang T, Chen W, Yao C, Wang Z, Wang Z, et al. MiR-153 reduces stem cell-like phenotype and tumor growth of lung adenocarcinoma by targeting Jagged1. Stem Cell Res Ther. 2020;11:170. doi: 10.1186/s13287-020-01679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu R, Shi P, Nie Z, Liang H, Zhou Z, Chen W, Chen H, Dong C, Yang R, Liu S, et al. Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics. 2016;6:533–544. doi: 10.7150/thno.14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W, Shen Y, Wei J, Liu F. MicroRNA-153/Nrf-2/GPx1 pathway regulates radiosensitivity and stemness of glioma stem cells via reactive oxygen species. Oncotarget. 2015;6:22006–22027. doi: 10.18632/oncotarget.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun D, Mu Y, Piao H. MicroRNA-153-3p enhances cell radiosensitivity by targeting BCL2 in human glioma. Biol Res. 2018;51:56. doi: 10.1186/s40659-018-0203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Lv X, Fu X, Su L, Yang T, Xu P. MiR-153 inhibits the resistance of lung cancer to gefitinib via modulating expression of ABCE1. Cancer Biomark. 2019;25:361–369. doi: 10.3233/CBM-190094. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Grochowski CM, Loomes KM, Spinner NB. Jagged1 (JAG1): Structure, expression, and disease associations. Gene. 2016;576:381–384. doi: 10.1016/j.gene.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou B, Zheng P, Li Z, Li H, Wang X, Shi Z, Han Q. CircPCNXL2 sponges miR-153 to promote the proliferation and invasion of renal cancer cells through upregulating ZEB2. Cell Cycle. 2018;17:2644–2654. doi: 10.1080/15384101.2018.1553354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang H, Ge F, Xu Y, Xiao J, Zhou Z, Liu R, Chen C. miR-153 inhibits the migration and the tube formation of endothelial cells by blocking the paracrine of angiopoietin 1 in breast cancer cells. Angiogenesis. 2018;21:849–860. doi: 10.1007/s10456-018-9630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Liang S, Duan X. Molecular mechanism of miR-153 inhibiting migration, invasion and epithelial-mesenchymal transition of breast cancer by regulating transforming growth factor beta (TGF-β) signaling pathway. J Cell Biochem. 2019;120:9539–9546. doi: 10.1002/jcb.28230. [DOI] [PubMed] [Google Scholar]
- 16.Shan N, Shen L, Wang J, He D, Duan C. MiR-153 inhibits migration and invasion of human non-small-cell lung cancer by targeting ADAM19. Biochem Biophys Res Commun. 2015;456:385–391. doi: 10.1016/j.bbrc.2014.11.093. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Sun J, Bai Z, Li H, He S, Chen R, Che X. MicroRNA-153 acts as a prognostic marker in gastric cancer and its role in cell migration and invasion. Onco Targets Ther. 2015;8:357–364. doi: 10.2147/OTT.S78236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang D, Zhang Q. miR-152 may function as an early diagnostic and prognostic biomarker in patients with cervical intraepithelial neoplasia and patients with cervical cancer. Oncol Lett. 2019;17:5693–5698. doi: 10.3892/ol.2019.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ou ZL, Luo Z, Wei W, Liang S, Gao TL, Lu YB. Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: Role of circ_0000977/miR-153 axis. RNA Biol. 2019;16:1592–1603. doi: 10.1080/15476286.2019.1649585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Q, Xia J, Wang L, Wang X, Ma X, Deng Q, Lu Y, Kumar M, Zhou Z, Li L, et al. miR-153 suppresses IDO1 expression and enhances CAR T cell immunotherapy. J Hematol Oncol. 2018;11:58. doi: 10.1186/s13045-018-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai Z, Sun J, Wang X, Wang H, Pei H, Zhang Z. MicroRNA-153 is a prognostic marker and inhibits cell migration and invasion by targeting SNAI1 in human pancreatic ductal adenocarcinoma. Oncol Rep. 2015;34:595–602. doi: 10.3892/or.2015.4051. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Liu F, Liu B, Qian J, Wu G, Li J, Ma Z. miR-153 enhances the therapeutic effect of gemcitabine by targeting Snail in pancreatic cancer. Acta Biochim Biophys Sin (Shanghai) 2017;49:520–529. doi: 10.1093/abbs/gmx039. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS, Ali S, Abbruzzese JL, Gallick GE, Sarkar FH. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009;69:2400–2407. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, Lee J, Kim JH. Association of Jagged1 expression with malignancy and prognosis in human pancreatic cancer. Cell Oncol (Dordr) 2020;43:821–834. doi: 10.1007/s13402-020-00527-3. [DOI] [PubMed] [Google Scholar]
- 25.Cao TH, Ling X, Chen C, Tang W, Hu DM, Yin GJ. Role of miR-214-5p in the migration and invasion of pancreatic cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:7214–7221. doi: 10.26355/eurrev_201811_16255. [DOI] [PubMed] [Google Scholar]
- 26.Huang SF, Yang ZL, Li DQ, Liu ZY, Wang CW, Miao XY, Zou Q, Yuan Y. Jagged1 and DLL4 expressions in benign and malignant pancreatic lesions and their clinicopathological significance. Hepatobiliary Pancreat Dis Int. 2016;15:640–646. doi: 10.1016/S1499-3872(16)60110-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the TCGA Research Network (https://www.cancer.gov/tcga).