Abstract
Aim:
In osteoarthritis (OA) there is a need for automated screening systems for early detection of structural progressors. We built a comprehensive machine learning (ML) model that bridges major OA risk factors and serum levels of adipokines/related inflammatory factors at baseline for early prediction of at-risk knee OA patient structural progressors over time.
Methods:
The patient- and gender-based model development used baseline serum levels of six adipokines, three related inflammatory factors and their ratios (36), as well as major OA risk factors [age and bone mass index (BMI)]. Subjects (677) were selected from the Osteoarthritis Initiative (OAI) progression subcohort. The probability values of being structural progressors (PVBSP) were generated using our previously published prediction model, including five baseline structural features of the knee, i.e. two X-rays and three magnetic resonance imaging variables. To identify the most important variables amongst the 47 studied in relation to PVBSP, we employed the ML feature classification methodology. Among five supervised ML algorithms, the support vector machine (SVM) demonstrated the best accuracy and use for gender-based classifiers development. Performance and sensitivity of the models were assessed. A reproducibility analysis was performed with clinical trial OA patients.
Results:
Feature selections revealed that the combination of age, BMI, and the ratios CRP/MCP-1 and leptin/CRP are the most important variables in predicting OA structural progressors in both genders. Classification accuracies for both genders in the testing stage (OAI) were >80%, with the highest sensitivity of CRP/MCP-1. Reproducibility analysis showed an accuracy ⩾92%; the ratio CRP/MCP-1 demonstrated the highest sensitivity in women and leptin/CRP in men.
Conclusion:
This is the first time that such a framework was built for predicting knee OA structural progressors. Using this automated ML patient- and gender-based model, early prediction of knee structural OA progression can be performed with high accuracy using only three baseline serum biomarkers and two risk factors.
Plain language summary
Machine learning model for early knee osteoarthritis structural progression
Knee osteoarthritis is a well-known debilitating disease leading to reduced mobility and quality of life – the main causes of chronic invalidity. Disease evolution can be slow and span many years; however, for some individuals, the progression/evolution can be fast. Current treatments are only symptomatic and conventional diagnosis of osteoarthritis is not very effective in early identification of patients who will progress rapidly. To improve therapeutic approaches, we need a robust prediction model to stratify osteoarthritis patients at an early stage according to risk of joint structure disease progression.
We hypothesize that a prediction model using a machine learning system would enable such an early identification of individuals for whom osteoarthritis knee structure will degrade rapidly. Data were from the Osteoarthritis Initiative, a National Institute of Health (United States) databank, and the robustness and generalizability of the developed model was further evaluated using osteoarthritis patients from an external cohort. Using the supervised machine learning system (support vector machine), we developed an automated patient- and gender-based model enabling an early clinical prognosis for individuals at high risk of structural progressive osteoarthritis. In brief, this model employed at baseline (when the subject sees a physician) easily obtained features consisting of the two main osteoarthritis risk factors, age and bone mass index (BMI), in addition to the serum levels of three molecules. Two of these molecules belong to a family of factors names adipokines and one to a related inflammatory factor. In brief, the model comprising a combination of age, BMI, and the ratios CRP/MCP-1 and leptin/CRP were found very robust for both genders, and the high accuracy persists when tested with an external cohort conferring the gender-based model generalizability. This study offers a new automated system for identifying early knee osteoarthritis structural progressors, which will significantly improve clinical prognosis with real time patient monitoring.
Keywords: adipokines, biomarkers, early prediction, knee osteoarthritis, machine learning, structural progressor
Introduction
Osteoarthritis (OA), a slowly progressive disease with joint structural change, is amongst the most prevalent chronic musculoskeletal diseases. Its clinical burden is considerable and includes pain, functional limitations, and multimorbidity.1 Although early proactive management is essential for OA patients in which the disease will progress rapidly, validated early-stage diagnosis is unavailable at present.
To assess the burden of OA on an individual basis and to accurately monitor and manage the course of the disease, it is important to improve our knowledge and identify early predictors of the structural progression of this disease. This will lead to precision medicine ensuring “the right treatment at the right time”, a key strategy for customized and more efficient therapies. Currently, in clinical practice, the progression of OA relies mainly on clinical judgment, often combined with imaging examinations (usually X-ray images). However, these approaches quite often show a discrepancy between patient symptoms and the extent of joint structural changes,2 limiting prediction of disease progression at an early stage. There is therefore an obvious need for more specific means that not only efficiently predict the structural progression of the disease at an early stage, but also easily allow individualized risk assessment for use in clinical practice.
The understanding of OA processes has led to the search of molecules/factors that could be used as biochemical markers. Biomarkers offer an interesting alternative means to chart the early progression of this disease. During the last few decades, researchers have looked at serum/urinary levels of several biomarkers for an early diagnosis, monitoring and/or prediction of the course of the disease. Yet, none have been shown to be sufficiently specific or sensitive.
Recently, adipokines have prompted much interest as biomarkers for OA.3,4 This family of factors have demonstrated potent modulatory properties on different effector cells in the pathogenesis of OA and have been considered key players in the network of mediators involved in the progression of the disease. The main sources of adipokines are the infrapatellar fat pad (IPFP) and adipocytes, but they are also synthesized by many cells in the joint microenvironment.5 Although there has been work looking at the association of adipokines with knee structural degeneration, there are differing data in the literature when individual adipokine members are studied.3,6–8 An explanation may be the different experimental conditions and patient populations used among studies. However, it also could also be due to the fact that a given adipokine may not reflect a comprehensive and effective evaluation of knee structure progression in OA. Data from different diseases including OA,9–15 suggest that the ratio of adipokine levels, or a combination of adipokines, their ratios, and other biomarkers demonstrated a better association and prediction assessment. In recent years, the concept of a combination of OA biomarkers and patient features has also been considered as being a more successful approach than using individual factors/parameters to identify OA progressors. Hence, it has been shown recently, by using machine learning (ML) methodologies, that a combination of biomarkers and/or ratios of biomarkers and other clinical and demographic variables were associated with OA progressors16 or predicted alteration of an articular tissue,13 the IPFP, reported to be a potential early marker of OA incidence and progression.17–19
As a novel contribution, the present study aimed to develop a model that could predict at an early stage, and in patient- and gender-specific manners, individuals at-risk of being knee OA structural progressors to improve clinical diagnosis with real-time patient monitoring. Our hypothesis suggests that a prediction model using an ML system would enable such an early identification of individuals for whom OA knee structure will degrade rapidly. To this end, the main objective is to find the answer to the following question: can baseline serum biomarker levels of adipokines/related inflammatory factors, in addition to major OA risk factors, be used to classify and predict with high accuracy knee OA progressor individuals?
Materials and methods
Study population
For the ML prediction model, individuals were all from the Osteoarthritis Initiative (OAI). The OAI cohort is an observational study on the natural progression of knee OA. Men and women between the ages of 45 and 79 were enrolled at four centers across the United States (Columbus, OH; Baltimore, MD; Pawtucket, RI; Pittsburgh, PA). The collected information includes clinical evaluations, radiological and magnetic resonance images (MRI), nutritional information, and physical activity monitoring, to name a few. The cohort included 4796 individuals at baseline divided into Control (normal), Incident, and Progression subcohorts (https://nda.nih.gov/oai/study-details). We received, from OAI, the serum at baseline of 700 individuals from the Progression subcohort. The latter include individuals in which there is, at baseline, OA radiographic and/or symptom features (https://oai.nih.gov). Of the 700 individuals, 11 were eliminated as they were missing at least one feature for labeling the probability values of being progressors (PVBSP) and 12 others for which the value for at least one of the studied biomarkers was missing (see below). Then, 677 individuals (364 men and 313 women) were employed for the model development. From these individuals, 22% had a meniscus (18%) and/or ligament (4%) repair. The participant characteristics of the cohort are as described in Table 1.
Table 1.
Participant characteristics. Data are presented as mean ± SD, % and number (n) of patients. Continuous variables were compared using the Student’s t test/Mann–Whitney test.
| OAI cohorta (n = 677) | Reproducibility (Naproxen) (n = 44) | p value* | |
|---|---|---|---|
| Age, years | 60 ± 9 | 59 ± 8 | 0.687 |
| Gender, male, % (n) | 54% (364) | 45% (20) | 0.350** |
| BMI, kg/m2 | 29.2 ± 4.9 | 34.5 ± 3.9 | <0.0001* |
| WOMAC | |||
| Pain (0–20) | 3.7 ± 3.8 | 11.2 ± 2.3 | <0.0001* |
| Function (0–68) | 11.8 ± 12.1 | 37.0 ± 11.2 | <0.0001* |
| Stiffness (0–8) | 2.1 ± 1.8 | 4.6 ± 1.4 | <0.0001* |
| Total (0–96) | 17.6 ± 17.0 | 52.9 ± 13.9 | <0.0001* |
| Kellgren–Lawrence gradeb, % (n) | |||
| 0–1 | 32% (218) | ||
| 2 | 30% (201) | ||
| 3 | 28% (189) | ||
| 4 | 10% (69) | ||
| Joint space width (medial minimum), mm |
3.8 ± 1.8 | 3.2 ± 2.0 | 0.019* |
| Joint space narrowing (score 0–2)c | 0.7 ± 0.8 | 1.1 ± 0.5 | <0.0001* |
| Biomarker levels | |||
| Adiponectin HMW, µg/ml | 4.5 ± 3.2 | ||
| Adiponectin LMW, µg/ml | 2.4 ± 1.1 | ||
| Adipsin, µg/ml | 7.3 ± 2.5 | ||
| Chemerin, ng/ml | 6.4 ± 2.7 | ||
| CRP, µg/ml | 4.1 ± 7.0 | 11.0 ± 15.1 | <0.0001* |
| IL-8, pg/ml | 12.2 ± 7.2 | ||
| Leptin, ng/ml | 23.4 ± 28.6 | 49.2 ± 31.3 | <0.0001* |
| MCP-1, pg/ml | 447.4 ± 305.0 | 578.8 ± 211.3 | <0.0001* |
| Visfatin, pg/ml | 614.0 ± 2139.4 | ||
Individuals were from the OAI cohort (http://oai.nih.gov), and the Reproducibility (Naproxen) cohort from the comparator arm of the Licofelone/Naproxen clinical trial.21
Indicates that one of the inclusion criteria of the trial (reproducibility) was the Kellgren-Lawrence (KL) grade of 2 or 3;21 however, the individual grade (KL 2 or 3) for each patient in the Naproxen group is not available.
The joint space narrowing scoring was as described.22
p values ⩽ 0.050 considered statistically different.; **proportions were compared using the chi-squared test/Fisher’s exact test.
Adiponectin HMW, adiponectin high molecular weight; adiponectin LMW, adiponectin low molecular weight; BMI, body mass index; CRP, C-reactive protein; IL-8, interleukin 8; MCP-1, monocyte chemoattractant protein-1; OAI, osteoarthritis initiative; WOMAC, Western Ontario and McMaster Universities Osteoarthritis Index.
Predictors: serum samples and biomarker determination
The serum was obtained from the baseline time point of both OAI and Naproxen (see Reproducibility section) cohorts.3,13,14 As previously described,13,14 the markers included the two main OA risk factors [age and bone mass index (BMI)], nine serum biomarkers [six adipokines: adiponectin high (H) and low (L) molecular weight (MW), adipsin, chemerin, leptin, visfatin, and three related inflammatory factors: C-reactive protein (CRP), interleukin (IL)-8, monocyte chemoattractant protein-1 (MCP-1)], and their 36 ratios. All biomarkers were determined with specific assays according to the manufacturer specifications.
All methods, including serum measurements, were performed in accordance with the relevant guidelines and regulations.
Probability values of being progressors
In order to assign a label for each participant, the PVBSP were generated using the prediction model developed in our previous study.20 As this model consists of features that could be difficult to obtain for a health care professional, we chose to use this knowledge and build a model that would be more accessible, that is, biomarkers and OA risk factors. In brief, we used to label PVBSP for each participant the baseline medial minimum joint space width, mean cartilage thickness of peripheral, medial and central tibial plateaus as assessed by quantitative MRI, the medial joint space narrowing (JSN) as a score,22 and the outcome JSN ⩾ 1 at 48 months.
The PVBSP for each participant was done as follows. Classification of knees from the OAI cohort was performed, in which the five features at baseline as well as the outcome were available and served to generate the model/code as well as the threshold for discriminating the progressor/no-progressor. The threshold was calculated with the F1 Max from the data model metrics. The F1 score provides a measure for how well a binary classifier can classify positive cases (given a threshold value). The F1 score is calculated from the harmonic mean of the precision as follows:
| (1) |
where: (i) precision represents the positive observations (true positives) of the model correctly identified from all the observations it labeled as positive; and (ii) recall represents the positive observations (true positives) of the model correctly identified from all the actual positive cases. All predicted probabilities greater than or equal to the Max threshold are labeled progressors, and the Max threshold values are labeled no-progressors.
Thus, a prediction value was assigned for each OAI (677) participant and labeled as progressor/no-progressor and named actual. Data showed that 55%/45% of the men were classified as progressors/no-progressors, respectively, and 50% in each group for the women.
ML methodology
To train supervised classifiers, five ML techniques including k-nearest neighbor,23 random forest,24 decision tree,25 extreme learning machine,26 and support vector machine (SVM)27 were investigated. The dataset was divided randomly into 70% of the participants for training and 30% for testing data. Based on prediction accuracy in the training stage, data showed that SVM is the best ML classifier technique to predict the probability of being PVBSP; we then, for further analysis, used only the SVM.
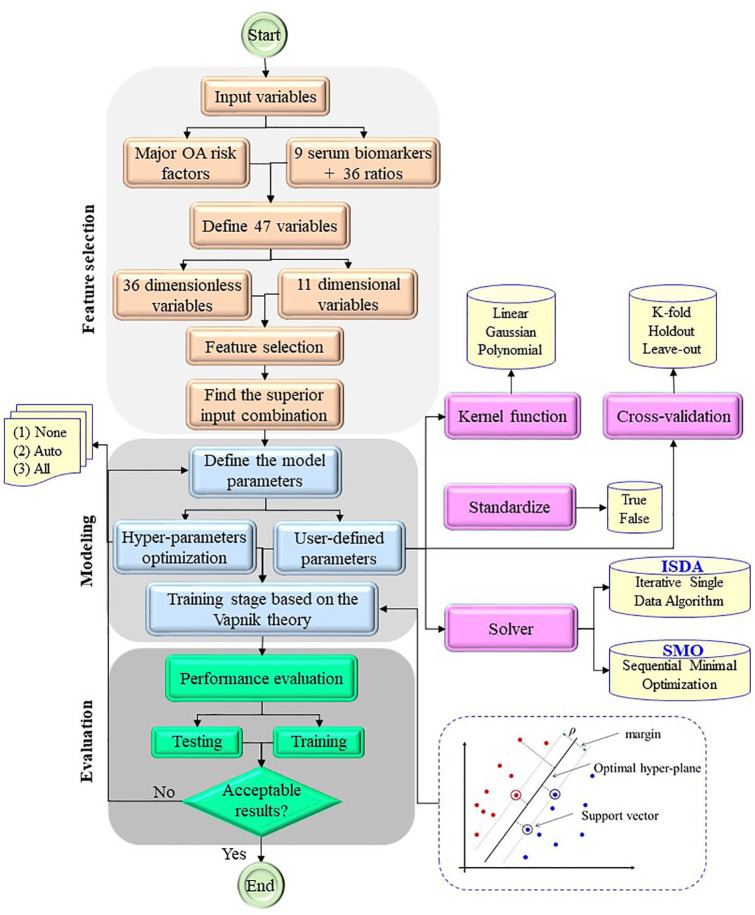
To develop the SVM model for PVBSP prediction (Figure 1), three general steps were performed and included feature selection, modeling, and evaluation.
Figure 1.
Flowchart of developed SVM model for predicting PVBSP based on gender separation.
ISDA, iterative single data algorithm; OA, osteoarthritis; PVBSP, probability values of being structural progressors; SMO, sequential minimal optimization; SVM support, vector machine.
Variable selection
In the feature selection step, out of the 47 input variables, which include 11-dimensional and 36 high dimensionless variables, all possible combinations are checked with 1–47 input variables, and the best combination is selected.
In supervised ML methods, to avoid the overfitting problem, the number of variables should be limited,28 as the application of the fully related variables in ML development not only leads to simplicity of the final model but also can improve prediction results. Therefore, to develop the SVM classification model, one must first apply the feature selection to identify the best input variables for the classifier model. To find the most effective input variables in PVBSP prediction, 47 independent variables were considered. These variables were categorized into three groups; OA risk factors variables (age and BMI), biomarker levels (six adipokines and three related inflammatory factors), and biomarker ratios (36).
Therefore, several input combinations from 1 to 47 parameters exist, and, for developing the ML prediction model, PVBSP, 2.81E + 14 input combinations were checked to find the most important one for the prediction. From an ML approach point of view in a high-dimensional problem, a subset of relevant factors related to all different input combinations should be found to build the final predictive model. By removing extraneous inputs and redundant information, the final model is more interpretable and easier to understand. In addition, the problem of overfitting during training and testing stages will be reduced. In the current study, the particle swarm optimization (PSO)-based feature selection technique was applied to find the most efficient sub-variable producing high prediction accuracy with the least number of input parameters.13 The cost function related to the feature selection problem based on Akaike information criteria (AIC) is defined. The PSO algorithm is then used to find the best input combination. Integrated with the PSO based feature selection model, the k-fold cross-validation approach was considered to prevent overfitting and evaluate the generalizability of the selected sub-variable in PVBSP prediction in all ranges of outcomes. In the k-fold cross-validation technique, all samples are randomly divided into k different categories. In the following, one category is considered as test samples and the other remaining categories (k–1) are considered as training samples. This process is repeated k times, so that, in each iteration, the test samples are different from other iterations. In this study, the value of k is considered to be 10.
Modeling
After selecting the best combination, the modeling step begins. After finding the best sub-variable in PVBSP prediction, the SVM method was applied for PVBSP modeling. In addition, to check the effect of each selected sub-variable on the PVBSP prediction, different SVM models were examined.
The most important part of the ML modeling is to determine the parameters to achieve the optimal one. Two modes were used: user-defined parameters and hyperparameter optimization. In the user-defined mode, the modeling parameters are selected by the user. These parameters include kernel function, cross-validation technique, and standardize mode. For standardize mode, one of the two true and false modes is selected in which the true choice indicates that the parameters are standardized before starting the modeling. For kernel function, there are three choices, linear, Gaussian, and polynomial, then the polynomial degree can also be selected by the user. In addition, there are three different options for cross-validation, including k-fold, holdout, and leave-out. Leave-one is k-fold cross-validation that the k is equal to the number of all samples (i.e., N). The holdout is the simplest type of cross-validation so that k is equal to 2, which are known as training and testing samples. The hyperparameter optimization mode includes three options: none, all, and auto. The “none” option is used when the “user-defined parameters” are considered, while the “auto” and “all” are considered for automatic optimization of the SVM parameters and optimization of all vector machine parameters, respectively.
After defining the model parameters, we further train the model. There are two ways to use the solver to train the model: iterative single data algorithm (ISDA) and sequential minimal optimization (SMO). The SMO method is an algorithm developed to solve the quadratic programming problems generated through SVM model training.29 The ISDA is a good Gauss–Seidel technique to solve linear systems of equations subject to the constraints.30
Evaluation
Finally, the model performance is evaluated by the percent estimation consisting of the ratio of corrected classified participants on the total number for the test and validation dataset. If the results are confirmed, the modeling ends. Otherwise, the modeling is re-run from the model parameter determination stage. The accuracy of the model is checked based on the following index:
| (2) |
where TP and TN are the number of correctly estimated positive and negative samples, respectively, while the FP and FN denote the number of wrongly estimated positive and negative samples, respectively.
The best sub-variables for progressors/no-progressors for both men and women were then selected, and the performance of the models evaluated. Further, the effect of each selected variable as well the sensitivity of the independent variables, that is the impact of a variation of a variable, were also assessed.
Reproducibility of the proposed model
To assess the reproducibility of the developed ML models, we used an external dataset consisting of 44 knee OA patients (20 men and 24 women) from the comparator arm (Naproxen, a cyclooxygenase inhibitor) of patients with primary symptomatic knee OA from a multicenter, randomized, double-blind clinical trial evaluating the effect of Licofelone (a lipoxygenase/cyclooxygenase inhibitor).21 This cohort was further named Naproxen. To label PVBSP for each participant of this cohort, the baseline values of the five X-rays and MRI features as described above were used as well as the outcome JSN ⩾ 1 but at 24 months, since, compared with OAI, Naproxen participants demonstrated a higher level of disease severity (Table 1).
The participant characteristics of the Naproxen cohort is as described in Table 1. Classification of the participants as progressors/no-progressors (named actual) were 60%/40% for the men and 75%/25% for women, respectively. Samples from this cohort were used only for validation and had no role in the modeling development.
Results
Variables selected and performance of the developed model
The feature selection results based on gender separation indicated that the best sub-variable for both men and women PVBSP estimation is as follows:
| (3) |
Using feature selection to identify the most effective ones led to a simple and accurate model based on four input variables as defined in Equation 3. The statistical indices of each gender-based input parameter in the PVBSP model in training and testing stages (OAI cohort) and Naproxen cohort are shown in Table 2. In brief, data showed that, for the men, the mean PVBSP values around 0.60 for all the analyses indicate that the number of progressors is higher than the no-progressors. For the women, the number of progressor and no-progressor subjects are about similar in the training stage (0.48), whereas the number of progressor patients is higher in the test stage (0.58) and more so in reproducibility analysis (0.70). Statistical differences within women showed that when Naproxen (reproducibility) was compared with test or train individuals, there were higher BMI (p < 0.0001) and CRP/MCP-1 (p ⩽ 0.006) (Tables 2). Moreover, a comparison between gender revealed that women demonstrated higher levels of CRP/MCP-1 (p ⩽ 0.035) for each cohort and leptin/CRP (p ⩽ 0.004) for both train and test cohorts.
Table 2.
Statistical indices of the risk factors and biomarker ratios.
| Gender | Stagea | Index | No. | Variable |
Outcome | |||
|---|---|---|---|---|---|---|---|---|
| Age (years) | BMI (kg/m2) | CRP/MCP-1 (×103) | Leptin/CRP (×10–3) | PVBSP | ||||
| Man | Train | Min | 255 | 45.00 | 16.60 | 0.17 | 0.05 | 0.00 |
| Max | 79.00 | 40.70 | 306.47 | 98.76 | 1.00 | |||
| Mean | 61.04 | 29.49 | 10.10 | 9.77 | 0.60 | |||
| SD | 9.76 | 4.09 | 25.50 | 13.37 | 0.38 | |||
| Test | Min | 109 | 45.00 | 20.90 | 0.17 | 0.12 | 0.00 | |
| Max | 78.00 | 43.00 | 57.68 | 216.67 | 1.00 | |||
| Mean | 59.96 | 29.21 | 7.51 | 11.45 | 0.57 | |||
| SD | 8.87 | 4.26 | 11.40 | 23.26 | 0.38 | |||
| Reproducibility | Min | 20 | 42.00 | 28.63 | 0.87 | 1.74 | 0.10 | |
| Max | 74.00 | 42.80 | 48.71 | 26.23 | 1.00 | |||
| Mean | 57.80 | 34.75 | 8.93 | 10.12 | 0.63 | |||
| SD | 8.73 | 3.71 | 11.84 | 6.37 | 0.35 | |||
| Woman | Train | Min | 219 | 45.00 | 16.80 | 0.21 | 0.33 | 0.00 |
| Max | 79.00 | 50.40 | 184.80 | 249.77 | 1.00 | |||
| Mean | 59.18 | 28.71 | 15.03 | 21.61 | 0.48 | |||
| SD | 8.58 | 5.74 | 24.99 | 31.10 | 0.39 | |||
| Test | Min | 94 | 45.00 | 21.00 | 0.28 | 0.35 | 0.00 | |
| Max | 79.00 | 45.60 | 227.72 | 383.61 | 1.00 | |||
| Mean | 60.72 | 29.86 | 14.20 | 25.57 | 0.58 | |||
| SD | 9.59 | 5.51 | 28.64 | 43.60 | 0.38 | |||
| Reproducibility | Min | 24 | 44.00 | 30.18 | 2.03 | 1.12 | 0.06 | |
| Max | 71.00 | 47.18 | 143.91 | 23.39 | 0.99 | |||
| Mean | 60.33 | 34.37 | 34.61 | 11.37 | 0.70 | |||
| SD | 6.69 | 4.05 | 43.32 | 9.05 | 0.31 | |||
Train and test individuals were from the OAI cohort and reproducibility (Naproxen) cohort from the comparator arm of the Licofelone/Naproxen clinical trial.21
Statistical analysis was done using the Student’s t test; p-values ⩽ 0.050 were considered statistically different. Inter-cohort significant differences were obtained when train or test individuals were compared to reproducibility ones for BMI, p < 0.0001 for both man and woman, and CRP/MCP-1, p ⩽ 0.006 for woman. Values between man and woman differ for CRP/MCP-1, p ⩽ 0.035 for each cohort; and leptin/CRP, p ⩽ 0.004 for train and test.
BMI, bone mass index; CRP, C-reactive protein; Max, maximum; MCP-1, monocyte chemoattractant protein-1; Min, minimum; No, number of subjects; OAI, osteoarthritis initiative; PVBSP, probability values of being structural progressor; SD, standard deviation.
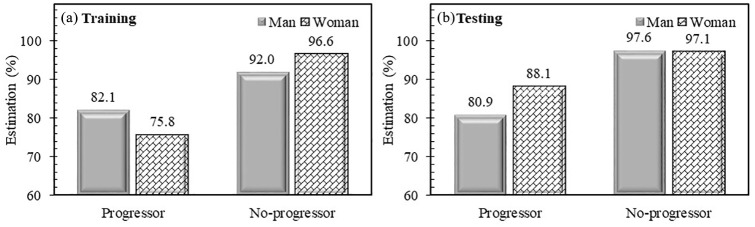
Figure 2 illustrates the performance of the developed models in PVBSP prediction in training and testing stages in regard to actual values. The developed ML classifier training stage for men showed that about 82% of progressors and 92% of no-progressors were correctly estimated, whereas, in the test stage, the developed model estimates 81% and 98%, respectively. For women, the number of estimations in the training and testing stages is about the same as for the men (both at 97%) for the no-progressors, but for the progressors slightly lower in the training stage (76%) and higher in the test stage (88%). Data showed that in the testing stage for the progressor estimation, the performance of the model is almost the same for men, but an increase of 12% for women is observed.
Figure 2.
Performance of developed models in probability values of being structural progressors/no-progressors in training and testing stages [Osteoarthritis Initiative (OAI) cohort]. Percent estimation is ratio of corrected classified participants on total number.
Reproducibility of the developed model
As suggested in the literature,31 we designed two-step supervised ML models, in which we first developed discriminative SVM classifier codes for men and women and then analyzed the developed SVM model for PVBSP prediction on an external cohort. Compared with OAI participants (Table 1), Naproxen patients had higher BMI, WOMAC scores, JSW, JSN, and serum levels of the three biomarkers used for the analysis (CRP, leptin, and MCP-1), all with a p < 0.02, indicating clinically more symptomatic knee OA patients than the OAI cohort.
Estimations for both genders (Figure 3) in the Naproxen cohort indicate that for the men, 92%, and 100% of the patients were estimated correctly for progressors and no-progressors, respectively. For the women, the progressors were estimated correctly at 94% and the no-progressors at 83%. Data then demonstrated that the developed SVM-based model for PVBSP prediction provides high performances for each gender.
Figure 3.
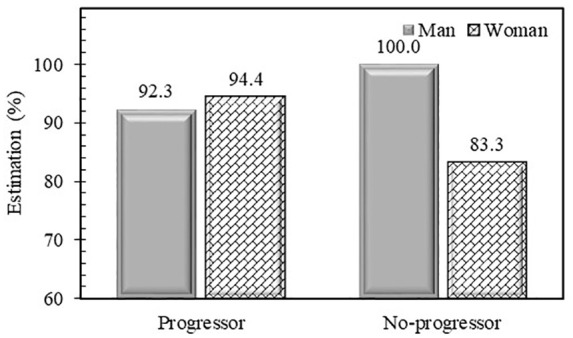
Performance of developed models in probability values of being structural progressors/no-progressors prediction for external cohort Naproxen. Percent estimation is ratio of corrected classified participants on total number.
Study of the effect of each variable in PVBSP prediction
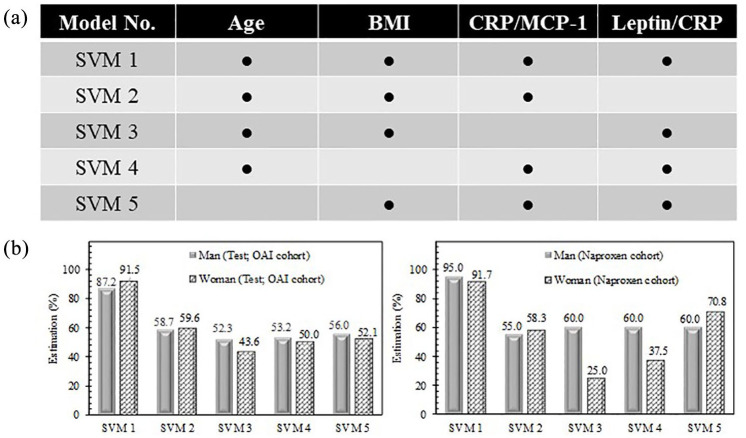
To verify the effect of each of the PVBSP selected variables: age, BMI, CRP1/MCP-1, and leptin/CRP, five different SVM-based models were designed. As illustrated in Figure 4a, with the exception of the first model (SVM 1), which takes into account all the variables as in Equation 3, the other models have only three variables.
Figure 4.
Effect of each variable in probability values. (a) SVM models 1–5 are designed as a function of selected features. (b) Performance of developed models for progressors and no-progressors together for OAI test stage and Naproxen cohorts. Percent estimation is ratio of corrected classified participants on total number.
BMI, bone mass index; CRP, C-reactive protein; MCP-1, monocyte chemoattractant protein-1; OAI, osteoarthritis initiative; SVM, support vector machine.
Performance of developed models was done with progressors and no-progressors together for OAI test stage and Naproxen cohorts (Figure 4b). Data show that, for both cohorts and for either men or women, the removal of a variable results in a significant reduction in the performance of the developed model. For the test stage using the OAI cohort, the order of the significance of the variables is CRP/MCP-1, BMI, age, and leptin/CRP for both genders. Hence, by not considering the CRP/MCP-1 variable, there is a reduction in the accuracy of 35% for men and 48% for women. This indicates the significant importance of the CRP/MCP-1 variable in the developed PVBSP model.
For the reproducibility analysis (Naproxen cohort; Figure 4b), using only three variables also results in reducing the accuracy for both genders. For women, similar to the OAI cohort, the most important variable is the ratio CRP/MCP-1; however, for men, it is the leptin/CRP.
Altogether these results show that all variables from Equation 3 are of importance, but both ratios CRP/MCP-1 and leptin/CRP are of the most significance for at least one gender.
Sensitivity analysis of the developed model to independent variables
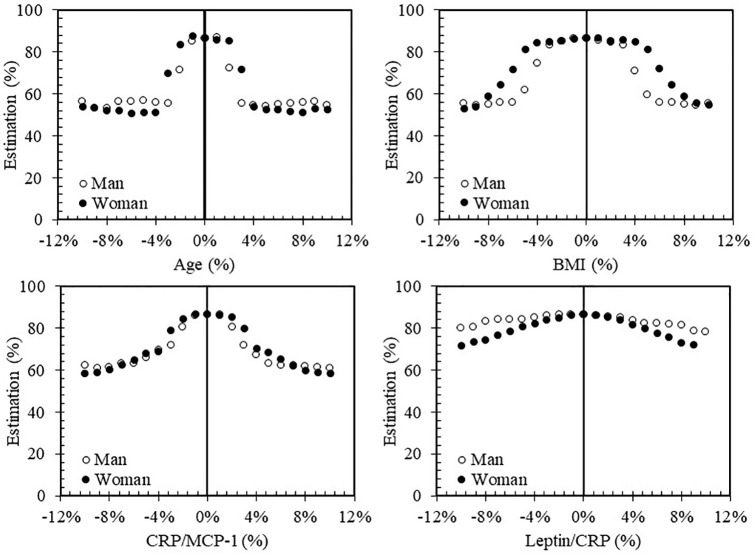
To examine the impact of a variation of a variable on the PVBSP by the developed ML prediction due to, for example, laboratory/clinical measurement errors, the sensitivity of each variable was investigated using all dataset (OAI and Naproxen cohorts) and all participants (progressor and no-progressor) (Figure 5). The variation range was from –10% to 10% with a step of 1%.
Figure 5.
Change effect of each variable on performance of the model. Data employed all datasets: OAI and Naproxen cohorts as well as progressors and no-progressors.
BMI, bone mass index; CRP, C-reactive protein; MCP-1, monocyte chemoattractant protein-1; OAI, osteoarthritis initiative.
For age, a 10% error reduces the accuracy index by about 30%; thus, the value of this index in error-free mode is 86%, but, considering a 10% error, the value of this index for men and women is 54% and 52%, respectively. The highest decrease in accuracy index for age occurs in low percentages, in which a 1–3% error is associated with a steep slope, and, in both men and women, the accuracy index reaches less than 55% and then almost no significant changes occur. Although the effect of error reduction on both genders is almost the same, the performance of this model in different errors is not constant for both genders in the range of 1–3%; the highest error reduction is related to men, but for the 10% error rate, the lowest accuracy index is related to women.
For BMI, the maximum reduction in accuracy index after 10% error, similar results to age with the accuracy index of 55% and 53% for men and women, respectively. The main difference between age and BMI is the downward slope of the accuracy index. In the BMI, in contrast to age, the slope of the accuracy index decreases by 1–3% and gradually increases with the increasing error value. Men are more sensitive to BMI variation, as their decreasing slope of the accuracy index is higher than for women, but the accuracy values for both genders are almost equal at ±10% error.
For CRP/MCP-1, the decreasing trend of the accuracy index is almost the same as for age, in that when the error value increases, the accuracy index decreases, and the highest decreasing of the accuracy index occurs in the 1–3% range. The increase error of this variable has almost the same effect on the performance in men and women.
Leptin/CRP experienced the lowest decrease in accuracy index in comparison with other variables. Increasing the error of this variable demonstrated a greater effect on women in that the accuracy index decreases by about 15%. For men, the 10% error effect leads to only a 7% decrease in accuracy index.
Discussion
In this study, a novel supervised ML model was developed to predict, in knee OA individuals, those at high risk of a rapid progressive disease. An automated assistance ML- and gender-based model to improve clinical diagnosis with real-time patient monitoring was introduced. Data showed that, among the 47 variables used, four of them, including age, BMI, and two ratios of serum biomarkers, CRP/MCP-1 and leptin/MCP-1, could estimate with high accuracy the OA progressors and no-progressors in either men or women. More specifically, the developed methodology could correctly estimate >80% (OAI test stage), in either gender, the individuals at-risk of being progressors. Moreover, the reproducibility experiments done with symptomatic knee OA patients provide face validity that the combination of these factors/biomarker ratios is reflective of OA progression. In addition, this external cohort also demonstrated a high sensitivity (⩾92%). The fact that, in the Naproxen cohort, OA patients had more disease severity than the OAI participants and more men (60%) and women (75%) were classified progressors could provide an explanation for the better performance.
In this study, we chose to investigate the biomarkers adipokines, as this family of factors is increasingly recognized of being of importance for OA progression.3,4,32–35 However, there is only a limited amount of information on the predictive values of such factors on OA structure progression. Moreover, many studies that have examined predictors of OA progressors have focused on an individual factor instead of on a combination and/or ratio as in this study. Hence, in recent years, it has been shown for this disease and others that combinations, as well as ratios, of factors could lead to better accuracy than individual ones.9–16 Data from this study demonstrated that ratios, as well as combinations of some adipokines/related inflammatory factors, contribute to better prediction of OA structural progressors than individual factors.
In regard to the modeling, considering all of the 47 variables in an ML model without sufficient knowledge of the various variable combinations not only does provides the model with proper accuracy but also greatly enhances model complexity. Therefore, a feature selection approach using ML was first employed to assess a pattern recognition for the outcome.
Data revealed that CRP appeared to be an important factor as a biomarker for predicting OA structural progressors in both genders. Moreover, when combined in ratios with either leptin or MCP-1, this study showed that it contributes more accurately to the identification of progressors in both genders. Each of these three biomarkers (CRP, MCP-1, and leptin), all considered inflammatory molecules, have been previously found associated with OA structural alterations and/or symptoms.3,14,36–40 Moreover, MCP-1 and leptin were also found associated with obesity and OA obese individuals.14,41,42
The developed ML model was based on gender separation as in the pathophysiology of knee OA major differences (e.g., knee anatomy, hormones, etc.) exist between genders in that, compared with women, fewer men develop OA.43 Moreover, in this disease, it cannot be assumed that the factors influencing OA outcomes are all similar in both genders; an average effect may not be appropriate or lead to the best accuracy as a prognostic model. The need for gender specific investigation in OA research has been widely highlighted, and our findings confirm evidence of this need. Hence, data revealed that although the same factors (Equation 3) were found for both genders, some demonstrated different weight according to gender. In the Naproxen cohort, more representative of the OA patients routinely seen by physicians, the ratio CRP/MCP-1 showed the highest performance for women, while it was the leptin/CRP for men.
The fact that MCP-1 and leptin, factors involved in both OA and lipid metabolism,3,14,36–38 differ between men and women in discriminating OA progressors is not very surprising as gender differences have been reported in fat metabolism.44,45 In addition, a difference in the circulating levels of leptin and CRP was reported to be increased significantly in OA women compared with men as well as in our study (data not shown).46–48 Moreover, the sensitivity analysis evaluating the impact of each parameter variation on the accuracy of the prediction could also provide an explanation for these differences. Indeed, data revealed that variation in the leptin/CRP ratio has the lowest influence, while the ML model is more sensitive to variation of age and CRP/MCP-1 for both genders compared with other variables, and that, compared with women, men had a higher sensitivity to variation in BMI.
Amongst supervised ML methods, SVM demonstrated the highest accuracy of the other four techniques analyzed. This was not unexpected, as although all five methods used showed some common general properties, SVM in addition minimizes the classification error by not only mapping inputs to a higher dimensional feature space via kernel function, but also by maximizing the margin between different classes.49 This method has shown excellent performance in classification-based problems in recent years compared with other methods.50–53 Moreover, SVM has been used widely to deal with difficult classification problems in digital health technology to provide an accurate, quantitative prediction model to improve clinical diagnosis.53–55
This study has several strengths. The excellent accuracy of the gender-based model persistence, when tested with an external cohort, enhances the robustness and generalizability of our findings. Another strength is that the model used at baseline the serum levels as variables for the discrimination of the progressor/no-progressor, which is relatively easy to obtain in addition to two risk factors that are basics characteristics. This study was based on discriminating knee OA structural progressors instead of pain as this latter outcome is a subjective phenomenon that is highly variable, complex, and multifactorial in nature. Furthermore, having applied the imaging (radiographic and MRI features)-based predictions from our previous study for generating the PVBSP label for each individual,22 provides a comprehensive and effective assessment of knee OA structure.
As in all studies, this one has limitations. One of them could be that our models were developed using the OAI cohort in which participants are at a mild–moderate stage of the disease, and that the reproducibility analysis was performed with OA patients with more disease severity but mimicking clinical routine. However, such a concern about the use of different knee OA patient disease severity could be lessened by the fact that the accuracy of the model persists and is even better in the reproducibility cohort. Moreover, and to add to the generalizability of this study’s findings, a further cohort showing similar characteristics as the OAI (mild–moderate stage) could be used. In addition, cohorts from Europe or Asia, for example, could also be analyzed, as the participants of the two cohorts used in this study were from North America. However, such a cohort should have, in addition to serum and X-rays, MRI quantitative data features at baseline in order to label the subjects. In regard to different cohorts, as an extension of this study, such a model could also be developed on specific OA types, including post-traumatic and congenital musculoskeletal abnormalities. Finally, we tested nine adipokines/related inflammatory factors to develop ML models, others reported serum biomarkers associated with OA could be further tested.
As a next step of this work, we are planning to transform this automated early screening of OA structural progressors into an application that will make it quick and easy for individual usage by health professionals for a given patient. It will also help discriminate progressor individuals for a clinical trial aimed at looking at disease-modifying OA drugs.
In conclusion, this study offers automated OA structural progressor patient screening as a promising step toward OA precision medicine. Hence, based on three baseline serum biomarkers and two major risk factors, we developed sensitive, reliable, and highly accurate gender-based models that can discriminate OA knee structural progression over time. These models offer a significant step forward as a source of decision support, enabling health professionals to be more effective in screening and detecting patients at risk of structural progressive OA. Such a framework will allow the adjustment of the treatment plan and therapeutic approaches that can lead to improved long-term patient outcomes and hopefully delay the time to or stop the need for surgery (e.g., knee replacement). This is important as more serious adverse events are found associated with patients that underwent knee replacement than those having nonsurgical treatment.56 In addition, such a prognosis could also be very useful for the patients; if they are better informed it may promote a more robust shared discussion-making process, particularly if the patient has modifiable factors that place them at risk of a poor outcome.
Acknowledgments
The authors would like to thank the OAI participants and Coordinating Center for their work in generating the clinical and radiological data of the OAI cohort and for making them publicly available. The OAI is a public–private partnership comprised of five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2-2260; N01-AR-2-2261; N01-AR-2-2262) funded by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners include Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer, Inc. Private sector funding for the OAI is managed by the Foundation for the National Institutes of Health. This manuscript was prepared using an OAI public use data set and does not necessarily reflect the opinions or views of the OAI investigators, the NIH, or the private funding partners. None of the authors are part of the OAI investigator team. The authors are grateful to ArthroLab Inc. for having provided the MRI data and the reproducibility cohort data, and to Santa Fiori for preparing the manuscript.
Footnotes
Conflict of interest statement: J-PP and JM-P are shareholders in ArthroLab Inc. and FA is an employee of ArthroLab Inc. The other authors declare that there is no conflict of interest with regard to this work.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by the Osteoarthritis Research Unit of the University of Montreal Hospital Research Centre, the Chair in Osteoarthritis from the University of Montreal, and a bursary (to AJ) from the Canada First Research Excellence Fund through the TransMedTech Institute, Montreal, Canada.
Ethical approval and consent to participate: All OAI participants provided written informed consent for participation in the OAI. Ethics approval was obtained by each OAI clinical site (University of Maryland Baltimore -Institutional Review Board, Ohio State University’s Biomedical Sciences Institutional Review Board, University of Pittsburgh Institutional Review Board, and Memorial Hospital of Rhode Island Institutional Review Board) and the OAI coordinating center (Committee on Human Research at University of California, San Francisco, CA, USA [#10-00532]). The Institutional Ethics Committee Board of the University of Montreal Hospital Research Centre [#20151130] approved the use of the human serum. For the Naproxen cohort, the original study was approved by IRB Services Central Ethics Committee (Toronto, ON, Canada) [#9427-A1706-21C] and all patients gave their oral and written informed consent, including permission for the use of serum to be collected for biomarker assessment.
Data availability statement: Data from the Osteoarthritis Initiative (OAI) cohort used in this study is publicly available (https://data-archive.nimh.nih.gov/oai/). Additional data may be obtained upon reasonable request, as long as the request is evaluated as scientifically relevant and pertinent.
ORCID iD: Johanne Martel-Pelletier  https://orcid.org/0000-0003-2618-383X
https://orcid.org/0000-0003-2618-383X
Contributor Information
Hossein Bonakdari, Osteoarthritis Research Unit, University of Montreal Hospital Research Centre (CRCHUM), Montreal, QC, Canada.
Afshin Jamshidi, Osteoarthritis Research Unit, University of Montreal Hospital Research Centre (CRCHUM), Montreal, QC, Canada; Laval University Hospital Research Centre, Quebec, QC, Canada.
Jean-Pierre Pelletier, Osteoarthritis Research Unit, University of Montreal Hospital Research Centre (CRCHUM), Montreal, QC, Canada.
François Abram, Medical Imaging Research and Development, ArthroLab Inc., Montreal, QC, Canada.
Ginette Tardif, Osteoarthritis Research Unit, University of Montreal Hospital Research Centre (CRCHUM), Montreal, QC, Canada.
Johanne Martel-Pelletier, Osteoarthritis Research Unit, University of Montreal Hospital Research Centre (CRCHUM), 900 Saint-Denis, Suite R11.412, Montreal, QC H2X 0A9, Canada.
References
- 1. Calders P, Van Ginckel A. Presence of comorbidities and prognosis of clinical symptoms in knee and/or hip osteoarthritis: a systematic review and meta-analysis. Semin Arthritis Rheum 2018; 47: 805–813. [DOI] [PubMed] [Google Scholar]
- 2. Raynauld JP, Martel-Pelletier J, Berthiaume MJ, et al. Quantitative magnetic resonance imaging evaluation of knee osteoarthritis progression over two years and correlation with clinical symptoms and radiologic changes. Arthritis Rheum 2004; 50: 476–487. [DOI] [PubMed] [Google Scholar]
- 3. Martel-Pelletier J, Raynauld JP, Dorais M, et al. The levels of the adipokines adipsin and leptin are associated with knee osteoarthritis progression as assessed by MRI and incidence of total knee replacement in symptomatic osteoarthritis patients: a post hoc analysis. Rheumatology (Oxford) 2016; 55: 680–688. [DOI] [PubMed] [Google Scholar]
- 4. Poonpet T, Honsawek S. Adipokines: biomarkers for osteoarthritis? World J Orthop 2014; 5: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Presle N, Pottie P, Dumond H, et al. Differential distribution of adipokines between serum and synovial fluid in patients with osteoarthritis. Contribution of joint tissues to their articular production. Osteoarthritis Cartilage 2006; 14: 690–695. [DOI] [PubMed] [Google Scholar]
- 6. Calvet J, Orellana C, Gratacos J, et al. Synovial fluid adipokines are associated with clinical severity in knee osteoarthritis: a cross-sectional study in female patients with joint effusion. Arthritis Res Ther 2016; 18: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Staikos C, Ververidis A, Drosos G, et al. The association of adipokine levels in plasma and synovial fluid with the severity of knee osteoarthritis. Rheumatology (Oxford) 2013; 52: 1077–1083. [DOI] [PubMed] [Google Scholar]
- 8. de Boer TN, van Spil WE, Huisman AM, et al. Serum adipokines in osteoarthritis; comparison with controls and relationship with local parameters of synovial inflammation and cartilage damage. Osteoarthritis Cartilage 2012; 20: 846–853. [DOI] [PubMed] [Google Scholar]
- 9. Sarray S, Madan S, Saleh LR, et al. Validity of adiponectin-to-leptin and adiponectin-to-resistin ratios as predictors of polycystic ovary syndrome. Fertil Steril 2015; 104: 460–466. [DOI] [PubMed] [Google Scholar]
- 10. Oda N, Imamura S, Fujita T, et al. The ratio of leptin to adiponectin can be used as an index of insulin resistance. Metabolism 2008; 57: 268–273. [DOI] [PubMed] [Google Scholar]
- 11. Norata GD, Raselli S, Grigore L, et al. Leptin:adiponectin ratio is an independent predictor of intima media thickness of the common carotid artery. Stroke 2007; 38: 2844–2846. [DOI] [PubMed] [Google Scholar]
- 12. Satoh N, Naruse M, Usui T, et al. Leptin-to-adiponectin ratio as a potential atherogenic index in obese type 2 diabetic patients. Diabetes Care 2004; 27: 2488–2490. [DOI] [PubMed] [Google Scholar]
- 13. Bonakdari H, Tardif G, Abram F, et al. Serum adipokines/related inflammatory factors and ratios as predictors of infrapatellar fat pad volume in osteoarthritis: applying comprehensive machine learning approaches. Sci Rep 2020; 10: 9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martel-Pelletier J, Tardif G, Rousseau Trépanier J, et al. The ratio adipsin/MCP-1 is strongly associated with structural changes and CRP/MCP-1 with symptoms in obese knee osteoarthritis subjects: data from the osteoarthritis initiative. Osteoarthritis Cartilage 2019; 27: 1163–1173. [DOI] [PubMed] [Google Scholar]
- 15. Gandhi R, Takahashi M, Smith H, et al. The synovial fluid adiponectin-leptin ratio predicts pain with knee osteoarthritis. Clin Rheumatol 2010; 29: 1223–1228. [DOI] [PubMed] [Google Scholar]
- 16. Nelson AE, Fang F, Arbeeva L, et al. A machine learning approach to knee osteoarthritis phenotyping: data from the FNIH biomarkers consortium. Osteoarthritis Cartilage 2019; 27: 994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davis JE, Ward RJ, MacKay JW, et al. Effusion-synovitis and infrapatellar fat pad signal intensity alteration differentiate accelerated knee osteoarthritis. Rheumatology (Oxford) 2019; 58: 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harkey MS, Davis JE, Lu B, et al. Early pre-radiographic structural pathology precedes the onset of accelerated knee osteoarthritis. BMC Musculoskelet Disord 2019; 20: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Masaki T, Takahashi K, Hashimoto S, et al. Volume change in infrapatellar fat pad is associated not with obesity but with cartilage degeneration. J Orthop Res 2019; 37: 593–600. [DOI] [PubMed] [Google Scholar]
- 20. Jamshidi A, Leclercq M, Labbe A, et al. Identification of the most important features of knee osteoarthritis structural progressors using machine learning methods. Ther Adv Musculoskelet Dis 2020; 12: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Raynauld JP, Martel-Pelletier J, Bias P, et al. Protective effects of licofelone, a 5-lipoxygenase and cyclo-oxygenase inhibitor, versus naproxen on cartilage loss in knee osteoarthritis: a first multicentre clinical trial using quantitative MRI. Ann Rheum Dis 2009; 68: 938–947. [DOI] [PubMed] [Google Scholar]
- 22. Altman RD, Gold GE. Atlas of individual radiographic features in osteoarthritis, revised. Osteoarthritis Cartilage 2007; 15(Suppl. A): A1–A56. [DOI] [PubMed] [Google Scholar]
- 23. Davis GA, Nihan NL. Nonparametric regression and short-term freeway traffic forecosting. J Transport Eng 1991; 117: 178–188. [Google Scholar]
- 24. Breiman L. Random forests. Mach Learn 2001; 45: 5–32. [Google Scholar]
- 25. Breiman L, Friedman JH, Olshen RA, et al. Classification and regression trees. Boca Raton, FL: Chapman & Hall, 1993. [Google Scholar]
- 26. Huang G-B, Zhu Q-Y, Mao KZ, et al. Can threshold networks be trained directly? IEEE Trans Circuits Syst II: Express Briefs 2006; 53: 187–191. [Google Scholar]
- 27. Vapnik VN. The nature of statistical learning theory. New York, NY: Springer, 2000, p.314. [Google Scholar]
- 28. Mohr M, von Tscharner V, Emery CA, et al. Classification of gait muscle activation patterns according to knee injury history using a support vector machine approach. Hum Mov Sci 2019; 66: 335–346. [DOI] [PubMed] [Google Scholar]
- 29. Platt J. Sequential minimal optimization: a fast algorithm for training support vector machines. MSR-TR-98-14, Microsoft Research, April 1998. [Google Scholar]
- 30. Kecman V, Vogt M, Huang TM. On the equality of kernel adatron and sequential minimal optimization in classification and regression tasks and alike algorithms for kernel machines. Paper presented at 11th European Symposium on Artificial Neural Networks (ESANN), 23–25 April 2003, Bruges, Belgium, pp.215–222. [Google Scholar]
- 31. Tao W, Radstake T, Pandit A. Using machine learning to molecularly classify systemic sclerosis patients. Arthritis Rheumatol 2019; 71: 1595–1598. [DOI] [PubMed] [Google Scholar]
- 32. Tu C, He J, Wu B, et al. An extensive review regarding the adipokines in the pathogenesis and progression of osteoarthritis. Cytokine 2019; 113: 1–12. [DOI] [PubMed] [Google Scholar]
- 33. Valverde-Franco G, Tardif G, Mineau F, et al. High in vivo levels of adipsin lead to increased knee tissue degradation in osteoarthritis: data from humans and animal models. Rheumatology (Oxford) 2018; 57: 1851–1860. [DOI] [PubMed] [Google Scholar]
- 34. Azamar-Llamas D, Hernández-Molina G, Ramos-Ávalos B, et al. Adipokine contribution to the pathogenesis of osteoarthritis. Mediators Inflamm 2017; 2017: 5468023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Conde J, Scotece M, Lopez V, et al. Differential expression of adipokines in infrapatellar fat pad (IPFP) and synovium of osteoarthritis patients and healthy individuals. Ann Rheum Dis 2014; 73: 631–633. [DOI] [PubMed] [Google Scholar]
- 36. Xu YK, Ke Y, Wang B, et al. The role of MCP-1-CCR2 ligand-receptor axis in chondrocyte degradation and disease progress in knee osteoarthritis. Biol Res 2015; 48: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang P, Zhong ZH, Yu HT, et al. Significance of increased leptin expression in osteoarthritis patients. PLoS One 2015; 10: e0123224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karvonen-Gutierrez CA, Harlow SD, Mancuso P, et al. Association of leptin levels with radiographic knee osteoarthritis among a cohort of midlife women. Arthritis Care Res (Hoboken) 2013; 65: 936–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith JW, Martins TB, Gopez E, et al. Significance of C-reactive protein in osteoarthritis and total knee arthroplasty outcomes. Ther Adv Musculoskelet Dis 2012; 4: 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pearle AD, Scanzello CR, George S, et al. Elevated high-sensitivity C-reactive protein levels are associated with local inflammatory findings in patients with osteoarthritis. Osteoarthritis Cartilage 2007; 15: 516–523. [DOI] [PubMed] [Google Scholar]
- 41. Panee J. Monocyte chemoattractant protein 1 (MCP-1) in obesity and diabetes. Cytokine 2012; 60: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim CS, Park HS, Kawada T, et al. Circulating levels of MCP-1 and IL-8 are elevated in human obese subjects and associated with obesity-related parameters. Int J Obes (Lond) 2006; 30: 1347–1355. [DOI] [PubMed] [Google Scholar]
- 43. Srikanth VK, Fryer JL, Zhai G, et al. A meta-analysis of sex differences prevalence, incidence and severity of osteoarthritis. Osteoarthritis Cartilage 2005; 13: 769–781. [DOI] [PubMed] [Google Scholar]
- 44. Power ML, Schulkin J. Sex differences in fat storage, fat metabolism, and the health risks from obesity: possible evolutionary origins. Br J Nutr 2008; 99: 931–940. [DOI] [PubMed] [Google Scholar]
- 45. Blaak E. Gender differences in fat metabolism. Curr Opin Clin Nutr Metab Care 2001; 4: 499–502. [DOI] [PubMed] [Google Scholar]
- 46. Kraus VB, Stabler TV, Luta G, et al. Interpretation of serum C-reactive protein (CRP) levels for cardiovascular disease risk is complicated by race, pulmonary disease, body mass index, gender, and osteoarthritis. Osteoarthritis Cartilage 2007; 15: 966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hellstrom L, Wahrenberg H, Hruska K, et al. Mechanisms behind gender differences in circulating leptin levels. J Intern Med 2000; 247: 457–462. [DOI] [PubMed] [Google Scholar]
- 48. Couillard C, Mauriège P, Prud’homme D, et al. Plasma leptin concentrations: gender differences and associations with metabolic risk factors for cardiovascular disease. Diabetologia 1997; 40: 1178–1184. [DOI] [PubMed] [Google Scholar]
- 49. Lin G-S, Chai S-K, Li J-Y, et al. Vision-based patient identification recognition based on image content analysis and support vector machine for medical information system. EURASIP J Adv Signal Process 2020; 27: 1–15. [Google Scholar]
- 50. Ganesan P, Sathish BS, Elamaran V, et al. Brain tumour segmentation and measurement based on threshold and support vector machine classifier. Res J Pharm Technol 2020; 13: 2573–2577. [Google Scholar]
- 51. Gao N, Tian S, Li X, et al. Three-dimensional texture feature analysis of pulmonary nodules in CT Images: lung cancer predictive models based on support vector machine classifier. J Digit Imaging 2020; 33: 414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luo J, Gao X, Zhu X, et al. Motor imagery EEG classification based on ensemble support vector learning. Comput Methods Programs Biomed 2020; 193: 105464. [DOI] [PubMed] [Google Scholar]
- 53. Franks JM, Martyanov V, Cai G, et al. A machine learning classifier for assigning individual patients with systemic sclerosis to intrinsic molecular subsets. Arthritis Rheumatol 2019; 71: 1701–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ed-daoudy A, Maalmi K. Breast cancer classification with reduced feature set using association rules and support vector machine. Netw Model Anal Health Inform Bioinform 2020; 9: 34. [Google Scholar]
- 55. Devi RD, Bai A, Nagarajan N. A novel hybrid approach for diagnosing diabetes mellitus using farthest first and support vector machine algorithms. Obes Med 2020; 17: 100152. [Google Scholar]
- 56. Culliford DJ, Maskell J, Kiran A, et al. The lifetime risk of total hip and knee arthroplasty: results from the UK general practice research database. Osteoarthritis Cartilage 2012; 20: 519–524. [DOI] [PubMed] [Google Scholar]